SUMMARY
Clonal hematopoiesis (CH) occurs when blood cells harboring an advantageous mutation propagate faster than others. These mutations confer a risk for hematological cancers and cardiovascular disease. Here, we analyze CH in blood samples from a pair of twin astronauts over 4 years in bulk and fractionated cell populations using a targeted CH panel, linked-read whole-genome sequencing, and deep RNA sequencing. We show CH with distinct mutational profiles and increasing allelic fraction that includes a high-risk, TET2 clone in one subject and two DNMT3A mutations on distinct alleles in the other twin. These astronauts exhibit CH almost two decades prior to the mean age at which it is typically detected and show larger shifts in clone size than age-matched controls or radiotherapy patients, based on a longitudinal cohort of 157 cancer patients. As such, longitudinal monitoring of CH may serve as an important metric for overall cancer and cardiovascular risk in astronauts.
In Brief
Trinchant et al. examined twin astronauts for clonal hematopoiesis (CH). Some high-risk CH clones (TET2 and DNMT3A) were observed two decades before expected, with TET2 decreasing in spaceflight and elevating later post flight. Thus, CH is an important metric for overall cancer and cardiovascular risk in astronauts.
Graphical Abstract
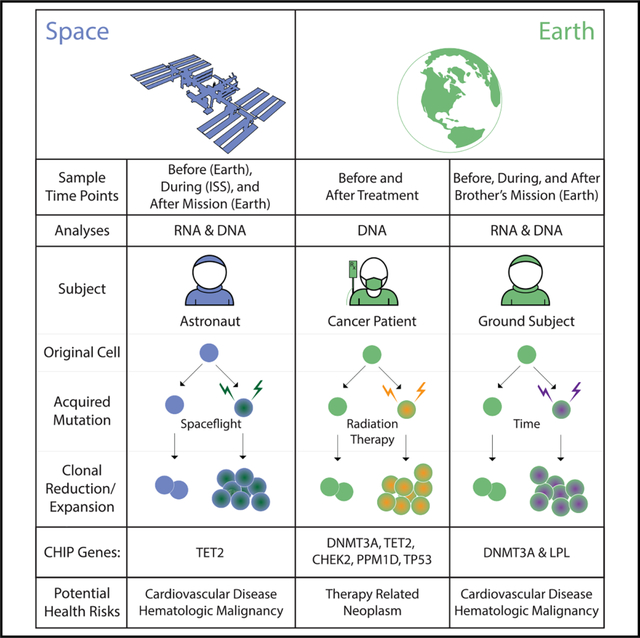
INTRODUCTION
The accumulation of DNA mutations in the cells of healthy tissues occurs during normal aging and begins before birth (Blokzijl et al., 2016; De, 2011; Vattathil and Scheet, 2016; Vilkki et al., 2001). While the majority of these mutations are neutral and do not favor cell expansion (Busque et al., 1996; Genovese et al., 2014; Jaiswal et al., 2014; Jan et al., 2012; Martincorena et al., 2015), certain mutations can cause cells to gain a growth advantage. When such a mutation occurs in the hematopoietic system and provides some growth advantage to blood cells, it causes clonal hematopoiesis (CH). The blood cells harboring this mutation are thought to arise from a single mutated “clone.” While advanced age is associated with higher rates of CH acquisition, certain environmental exposures such as cigarette smoke and radiation have been associated with increased CH as well (Coombs et al., 2017). CH has been shown to be associated with all-cause mortality (Jaiswal et al., 2014; Zink et al., 2017).
A proposed functional definition of CH includes the presence of a dominant somatic mutation at >2% variant allele frequency (VAF), although a scientifically or clinically relevant lower threshold remains to be determined (Steensma, 2018). CH can be pro-inflammatory and, through this mechanism, is thought to impact cardiovascular health based on data from model systems and humans (Calvillo-Argüelles et al., 2019; Fuster et al., 2017; Jaiswal et al., 2017; Sano et al., 2018; Wang et al., 2020). Research has found that the most common mutations driving CH are in epigenetic modifier genes such as DNMT3A, TET2, and ASXL1 (Gertz et al., 2020). The known risks of CH include elevated rates of cardiovascular disease (CVD) and hematologic cancer (Calvillo-Argüelles et al., 2019). Notably, CH is associated with a 1.9-fold increase in coronary heart disease and a 4-fold increase in early-onset myocardial infarction (Jaiswal et al., 2017) as well as an 11-fold increase in hematologic cancer. Case-control studies have shown that CH with increased mutational complexity (i.e., >1 mutation and/or >1 mutation in the same gene) can increase the risk for acute myeloid leukemia (AML) (Desai et al., 2018), and this finding has been independently confirmed in men and women (Abelson et al., 2018).
CH has been studied in elderly twins (>70 years old), revealing that twin pairs were discordant for CH mutations but with no obvious genetic predisposition to CH (Hansen et al., 2020). However, CH has not yet been studied in younger twins or in subjects who have been exposed to the milieu of space radiation, such as astronauts. While the presence of CH, by itself, does not constitute a recognized disease condition, it can inform long-term CVD and cancer risk and thus can serve as a potential useful metric for astronauts preparing for long-term missions as well as a defined metric of acquired somatic mutations. Here, we describe the such data from astronauts (twin subjects TW and HR, spaceflight and ground control, respectively), including metrics during a year-long spaceflight mission for the flight subject (Garrett-Bakelman et al., 2019) and results of a 3-year follow-up study.
RESULTS
Individual TW presented at baseline evaluation with a single non-truncating mutation in TET2 (Cys1273Tyr) at 3.8% VAF, which corresponds to approximately 1 in 12 affected mononuclear cells (Figure 1A). The mutation is predicted to be pathogenic according to FATHMM and M-CAP mutation scoring, and it was present in all bulk mononuclear cell collections (Figure 1B) but absent from T cell populations (CD4, CD8). This lesion was observed twice in the Catalog of Somatic Mutations in Cancer (COSMIC) in association with AML, and a TET2 mutation at the same amino acid (Cys1273Ser) was discovered at 25% VAF in an 86-year-old female who participated in a recent study of CH in solid-tumor patients (Coombs et al., 2017). These data suggest a capacity for progression to high-risk CH, and the mutant clone showed an overall rising VAF trajectory. While in flight, the mutation clone size remained relatively stable but then increased to 7.4% after the end of his last year-long International Space Station (ISS) mission (Garrett-Bakelman et al., 2019). The variation on VAF over time was more pronounced in the 3 months immediately after returning to Earth than during the last 6 months of the space flight (percent deltaVAF [(VAF2−VAF1)/month] was 0.119% versus 0.036%, respectively) (Figure 1A).
Figure 1. Clonal Hematopoiesis Profiling.
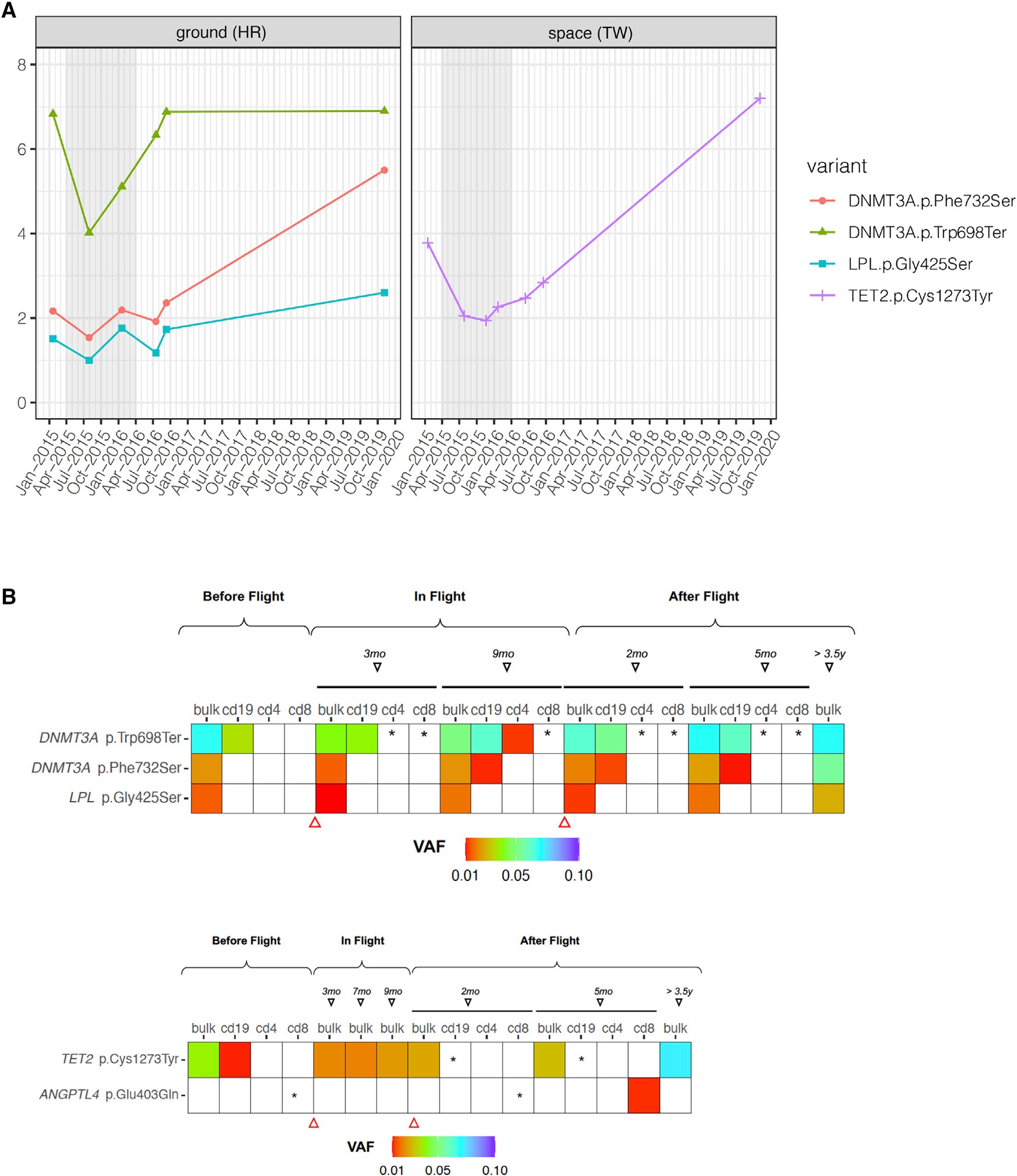
(A) Longitudinal trajectories of CH mutations discovered in the astronaut subjects. Allele frequency (%) is the VAF of each clone (vertical axis) visualized across time (horizontal axis). Subject TW (spaceflight) is compared to subject HR (ground). Flight start and end for the NASA Twins Study are denoted by the gray shaded region (left). Baseline (2015) time point corresponds to lymphocyte-depleted MNC fraction. Inflight time points and after flight 2016 time points were collected and processed by density gradient to obtain peripheral blood mononuclear cells, and the 2019 time point corresponds to buffy coat, spun-down blood cells.
(B) VAF in distinct cell populations. The three significant mutations (rows) in DNMT3A and LPL for HR (top) and the two mutations found in subject TW (bottom). Shown mutations are present in bulk cell populations (bulk), as well as almost all subfractions of T cells (CD4, CD8) and B cells (CD19). VAF range is show in fraction of color (red to blue, up to 0.1), and asterisks (*) indicate detection of the variant but below 1%. Triangles indicate the beginning and end of the year-long mission.
It is noteworthy that subject TW was previously diagnosed with and treated for prostate cancer and that this risk was also present in his germline DNA as a stop-gain mutation in the RNA-SEL gene (p.Glu265Ter). Thus, it is likely relevant to compare his CH data with studies of CH in blood drawn from patients who were also diagnosed with solid tumors (Coombs et al., 2017). From this CH cohort (Coombs et al., 2017), the mean age at which TET2 CH mutations are found in the blood of solid-tumor patients is 68 years of age (n = 8,810 individuals; Coombs et al., 2017). Subject TW is therefore exhibiting this mutation nearly two decades prior to the mean age at which it is typically detected (50 versus 68). By comparison, TET2 mutations in blood were detected in only 1% of persons under 55 years of age in the solid-tumor patient cohort (Coombs et al., 2017), never observed in a separate control, AML-free population in a previous study (n = 212) of pre-AML in persons under 65 (Desai et al., 2018), and also found to be rare (<1%) in another control (n = 678) cohort (Abelson et al., 2018).
Conversely, the HR subject presented at baseline evaluation with two mutations in the DNMT3A gene (Figure 1A), which also were present in B cell (CD19) and T cell (CD4, CD8) populations (Figure 1B). CH mutations in DNMT3A tend to be loss of function, arising either from a truncating event such as a frameshift or from introduction of a stop codon or a dominant-negative mutation (Desai et al., 2018). The major DNMT3A mutation observed in subject HR (Trp698Ter) is a truncating mutation arising from introduction of a stop codon in DNMT3A, causing premature termination of the protein at amino acid position 698. This mutation presented at 6.8% VAF, corresponding to an estimated 1 in 7 affected mononuclear cells. This clone presented a significant increase during and for the first year after the end of the flight. After that point, the clone remained stable. The percent variation in VAF per month for this clone is greater after the end of the mission (0.304%) compared to in-flight time points (0.19%).
The second mutation in DNMT3A (Phe732Ser) converts phenylalanine-732 to serine and initially presented at 2.1% VAF. The mutation occurs in a mutation cluster of the DNA methylase domain, thus likely impacting enzymatic function. This mutation was previously observed three times in COSMIC and was confirmed somatic in both myeloid and lymphoid cancers (Desai et al., 2018). This mutation was also discovered in one study of pre-AML in a healthy woman who later progressed to AML (Desai et al., 2018). Furthermore, this mutation is predicted to be pathogenic according to FATHMM and M-CAP scores. Variation in VAF during flight was also significant for this mutation, and the change in dVAF/month was 0.114% during flight versus 0.243% after flight. Finally, a novel somatic mutation in the LPL gene, which encodes lipoprotein lipase, presented at 1.5% VAF at baseline evaluation and remained stable to 2020. This mutation affects the PLAT domain of the protein but is predicted benign according to variant classification criteria (PolyPhen and SIFT). Loss-of-function mutations in LPL have been shown to increase macrophage foam cell formation and promote atherosclerosis (Babaev et al., 1999). However, the functional impact of this particular LPL mutation is unknown.
Like his brother, HR was diagnosed and treated for prostate cancer (tumors were resected for both twins, with no radiation or chemotherapy) and carried the same risk allele. Thus, it is also relevant to compare his CH mutations in the context of the same solid-tumor study of CH (Coombs et al., 2017). Overall, 5% of people under 55 years of age exhibit a single DNMT3A mutation, and no one in the recent study exhibited two DNMT3A mutations (Coombs et al., 2017). The mean age of people with one DNMT3A mutation was 65.8 years. Thus, the fact that subject HR exhibits two DNMT3A mutations nearly 15 years before the mean age of onset for a single DNMT3A mutation is potentially significant, and the combined VAF is now above 10%. In a mutational analysis of a pre-AML cohort (Desai et al., 2018), two or more DNMT3A mutations were associated with a 12.6-fold increase in the odds of eventual AML (OR: 12.6, 95% CI 3.0–59.2; p < 0.001) compared to a 2.1-fold increase for people with a single DNMT3A mutation (OR: 2.1, 95% CI 1.2–3.8; p = 0.01). Of note, the RNA sequencing data showed that these two variants are not expressed on the same allele (Figure S1), and the linked-read whole-genome sequencing data also showed no evidence to the two mutations on the same linked-molecule across four time points (pre-flight 90×, mid-flight 94× and 88×, and post-flight 77× coverage); thus, these two mutations are likely from two distinct CH events.
To place these variants into a broader context, the astronaut CH VAFs were compared to data from a longitudinal CH study by Bolton and colleagues (Bolton et al., 2019b). This study compared the effect of oncologic treatment (X-ray radiation treatment [XRT]) on CH in 49 patients relative to a longitudinal CH study of 108 controls (n = 157 total). In this cohort, the median age of patients with CH was 68 years old overall and 70 years old for prostate cancer patients with detected CH (including TET2 or DNMT3A mutations). Also, the median age for prostate cancer patients with no detected CH was 65 years old (Bolton et al., 2019b), and CH was associated with cytotoxic drugs, external XRT beam radiation, and radionuclide-based therapy. The most strongly associated genes with oncologic treatment were those involved in DNA-damage repair (PPM1D, TP53, and CHEK2). Conversely, DNMT3A or TET2 mutations were not strongly associated with any tumor type or therapy. Also, prostate cancer was not particularly associated with CH, and no significant enrichment of any particular CH gene was observed in these patients.
However, serial sampling revealed a range of VAF changes in patients over time, which were often smaller than the astronaut CH dynamics (Figure 2). Specifically, a subgroup of patients showed that growth rate of mutations in DTA genes (DNMT3A, TET2, and ASXL1) was lower than that of DNA damage response (DDR)-related genes and that the odds of a mutation expanding after treatment were higher for DDR genes as opposed to DTA genes. The mean percent variation in VAF per month was highest in DDR genes after treatment with external beam radiation (0.16%) and cytotoxic drugs (0.19%). DNMT3A/TET2 mutations after radiation treatment presented a mean percent VAF variation per month higher (0.1%) than any other treatment (range 0.02%–0.08%). While variation in VAF over time for the samples in this study may suffer from unique challenges in collection (e.g., storage on the ISS and return-to-Earth protocols), it was possible to estimate such VAF changes within the bulk cell fractions. Interestingly, the variation observed in the astronauts (1%–2%) was higher than the one observed for DNMT3A/TET2 mutations after oncologic treatment with external beam radiation, while other variants showed a consistent VAF across all time points (Figure S2). Ascertaining the significance of such CH VAF dynamics in astronauts and cancer patients requires larger cohorts.
Figure 2. VAF over Time of CH in Astronauts and Cancer Patients.
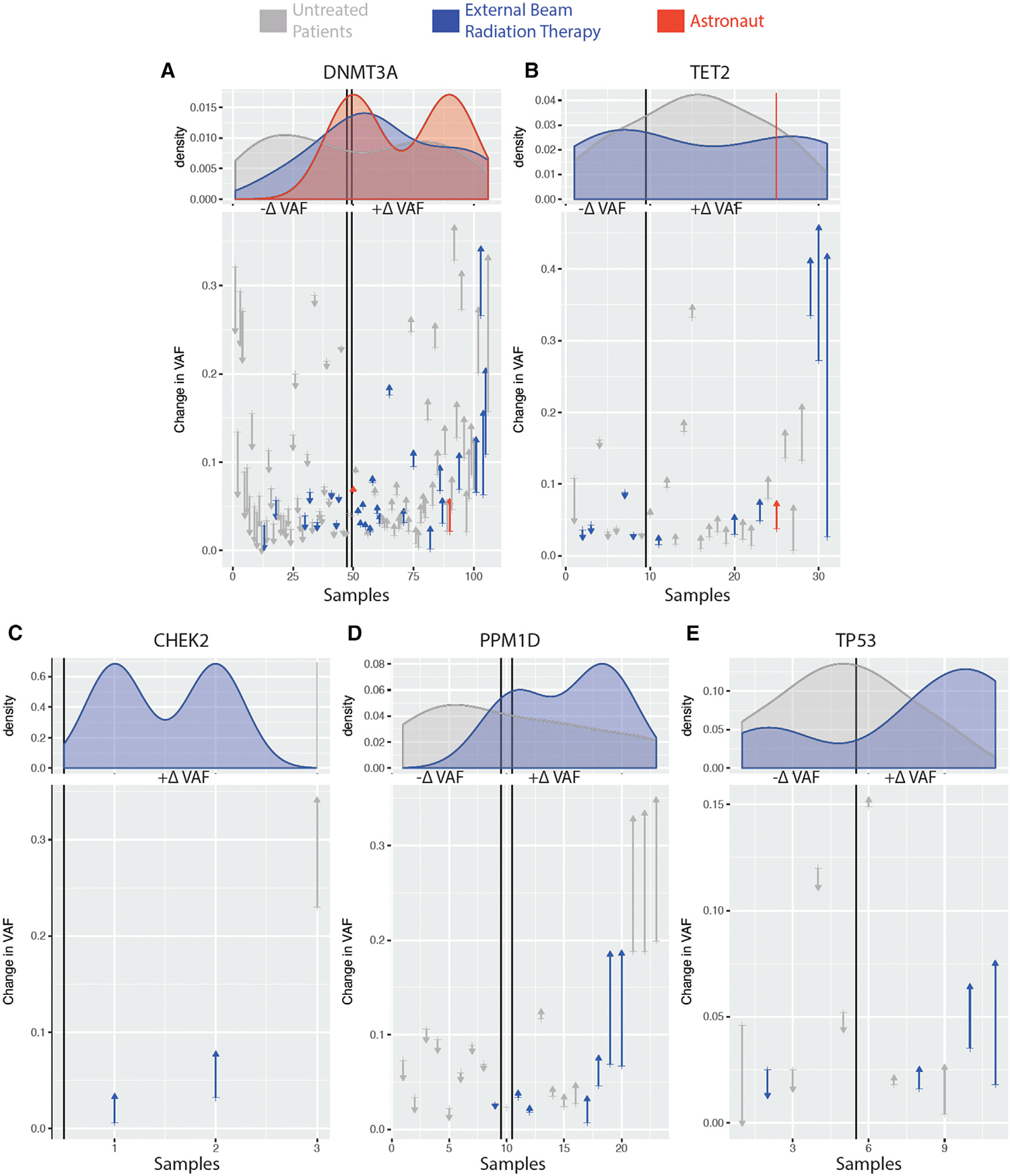
Change in VAF in DNMT3A (A), TET2 (B), CHEK2 (C), PPM1D (D), and TP53 (E) within astronauts (red), cancer patients treated with external beam radiation (blue), and untreated cancer patients (gray). Identified variants in samples are ordered along the x axis based on their VAF change over two time points (astronauts baseline versus 3.5 years after flight and cancer patients before and after treatment) with ones decreasing the most on the left (arrow down) and ones increasing the most on the right (arrow up). Density plots of the variants for each subject group are shown above the line plots
DISCUSSION
Currently, there are no evidence-based guidelines for the management of CH. However, a set of management strategies for incidental findings of CH in solid-tumor patients has recently been suggested (Bolton et al., 2019a). Here, the two astronauts were found to have mutations in DNMT3A and TET2 consistent with the presence of CH. People with CH progress to hematologic malignancies at a rate of 0.5%–1% per year, similar to the rate of progression of frank neoplasia observed in monoclonal gammopathy of uncertain significance (MGUS) or monoclonal B cell lymphocytosis (MBL) (Gertz et al., 2020). However, CH has the potential to give rise to a broader range of blood cancers than either MGUS or MBL because it usually arises in earlier, less committed hematopoietic stem or progenitor cells. While having CH or increased exposure to radiation are both implicated in higher-risk profiles for AML, it should be stressed that the absolute risk of the disease is still low in people in the absence of pre-disposing factors (6 per 100,000 white males per year between ages 50 and 64, SEER statistics, https://seer.cancer.gov/statistics/).
The factors contributing to these CH variations are unknown but may include the known conditions of the ISS and spaceflight, including: reduced gravity, weight loss, baseline genetic risk (e.g., RNASEL), and increased radiation exposure (including the 146 mSv for the year-long mission) (Garrett-Bakelman et al., 2019). While these results are preliminary and require much larger cohorts for proper interpretation, they nonetheless have implications for long-term health management. There is suggestive evidence that people with TET2 mutations exhibit a 1.9-fold risk (95% CI 1.0–3.7; p = 0.06) of coronary heart disease and 8.3-fold increased odds of myocardial infarction (95% CI 1.2–357.5; p = 0.02). Single TET2 mutations also exhibit increased odds of AML compared to matched controls (odds ratio: 3.29, 95% CI 1.38–7.83; p = 0.005) (Desai et al., 2018). The presence of two mutations in DNMT3A in subject HR is suggestive of high-risk CH. The risk of coronary heart disease in patients exhibiting DNMT3A-mutation-driven CH is 1.7 (95% CI 1.1–2.6; p = 0.01), and the pathophysiology of CH in CVD is thought to be inflammation driven (Calvillo-Argüelles et al., 2019). Assuming the two DNMT3A mutations occur in separate cells, then the combined VAF of both DNMT3A mutations is ~10%. A value that approaches high-risk CH for CVD (≥10% VAF), additionally, people with a VAF ≥ 10% exhibit a 12-fold increase in the odds of a coronary artery calcification score ≥ 615 Agatson units (OR: 12.1, 95% CI 2.4–64.0; p < 0.001) (Bolton et al., 2019a; Cimmino et al., 2017). It is unknown how the presence of two DNMT3A mutations affects the risk of other cancers besides AML.
Prior work on these subjects (Garrett-Bakelman et al., 2019) noted a range of molecular and physiological changes using a multimodal and integrated approach, all of which may impact the overall VAF trajectories. Changes during flight or due to the stress of returning to Earth were observed for parameters that could have an impact on CH dynamics, such as spikes in inflammation markers (e.g., IL-6, IL-1ra) (Gertz et al., 2020), T cell function expression changes (Garrett-Bakelman et al., 2019), and cytokine shifts (Gertz et al., 2020). Because CH clones both are pro-inflammatory and expand under inflammatory conditions, it is possible that the fluctuation detected in the levels of specific cytokines has an effect in the expansion/reduction of the mutated clones upon returning to Earth. Previous studies have reported changes in cardiovascular structure and function, as well as exercise and nutritional differences, which can also be a factor (Cimmino et al., 2017). Also, of note, TW presented with signs of vascular remodeling of his carotid artery for the duration of the flight; this subject presented a mutation in TET2, a gene that (when mutated) imposes a significant risk to develop CVD. Therefore, given the changes in the vasculature structure, inflammatory cytokines, and gene expression pathways observed for these individuals, and the association between these parameters and CH, it is reasonable to suggest that monitoring of CH clones, alongside all of these health parameters, should be included as part of the overall health assessment for astronauts.
Finally, while subject HR exhibits overall higher-risk CH, the mutation patterns observed in both astronauts are nonetheless unusual when considering the relatively young age at which the mutations are observed (Steensma et al., 2015). The mutations in both twins had an overall increasing trajectory; however, the dynamics of the clones was different across time points during the mission and differed between the two twins. The genesis of these mutations is unknown, but occupational exposures received during spaceflight from ages 35–52 and 32–45 for TW and HR, respectively, are potential factors. Moreover, the differential rates of these mutations in B cells (CD19) versus T cells (CD4, CD8) appear to be rare from these data, but it is not known how this would appear in other cell types and may relate to distinct risk profiles. Additional data (e.g., red cell distribution width) would enable improved cancer risk stratification, and continued evaluation and a longitudinal monitoring plan for both astronauts could be used to track any change in VAF. These changes can then be examined relative to published CH guidelines (Bolton et al., 2019a; Jaiswal and Ebert, 2019), other CH data from emerging clinical and control cohorts (Busque et al., 2012), and medical guidance from NASA flight surgeons. Indeed, when integrated with other health data such as folic acid levels (Garrett-Bakelman et al., 2019), telomere length (Luxton et al., 2020a, 2020b), heart morphology (Garrett-Bakelman et al., 2019), and long-term stress markers for farther missions (Iosim et al., 2019; Nangle et al., 2020), these measures of CH, including other changes in physiology (Gertz et al., 2020), could serve as an additional, quantitative measure of long-term risk of cardiovascular and hematological diseases for astronauts before, during, and after spaceflight.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact Christopher E. Mason (chm2042@med.cornell.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The NASA Life Sciences Data Archive (LSDA) is the repository for all human and animal research data, including that associated with this study. LSDA has a public facing portal where data requests can be initiated (https://lsda.jsc.nasa.gov/Request/dataRequestFAQ). The LSDA team provides the appropriate processes, tools, and secure infrastructure for archival of experimental data and dissemination while complying with applicable rules, regulations, policies, and procedures governing the management and archival of sensitive data and information. The LSDA team enables data and information dissemination to the public or to authorized personnel either by providing public access to information or via an approved request process for information and data from the LSDA in accordance with NASA Human Research Program and Johnson Space Center (JSC) Institutional Review Board direction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human samples
All samples were collected after obtaining written informed consent. Protocols were approved by all participating institutions. Blood samples from were collected from space and ground subjects into 4 mL CPT vacutainers (BD Biosciences Cat # 362760) as per the manufacturer’s recommendations. Fresh or frozen samples were processed as described in the NASA Twins Study. Cell population separation based on CD4, CD8 or CD19 markers was performed using column-based method as described in Garrett-Bakelmann et al. (2019). Astronaut subjects (both male) began the study at age 50.
All subjects were collected under the Weill Cornell Medicine and NASA Institutional Review Board (IRB) protocols, including those for the NASA Twins Study (IRB#1309014347). For the MSKCC cohort, dosimetric parameters for each course of external beam radiation (i.e., dose, fractionation, 905 technique/modality, target) were extracted from the treatment planning system (ARIA; Varian Medical Systems, Palo Alto, CA), from radiotherapeutic prescriptions and from clinical treatment summaries. For patients treated prior to the implementation of contemporary treatment planning (prior to 2003), receipt and timing of radiation therapy was abstracted from medical billing records, although dose and fractionation were not available for these patients and were set 910 as missing. Other clinical details are online from the Bolton et al. (2019b) paper.
METHOD DETAILS
CH panel, WGS, and RNA-sequencing
Genomic DNA from bulk mononuclear cells (bulk), column-based, purified lymphocytes (CD4, CD8, and CD19) or buffy coat was extracted, purified, and mechanically sheared using the same protocol as the NASA Twins Study (Garrett-Bakelman et al., 2019). Illumina sequencing adapters containing unique dual sample indices (UDIs) were ligated onto fragments according to the Kapa Biosystems HyperPrep protocol. Targeted enrichment using a custom pool of biotinylated baits directed to 93 genes involved in clonal hematopoiesis, cancer, and CVD was performed according to the standard Twist Biosciences capture protocol. Sequencing was performed to > 2000x median unique depth of coverage using NovaSeq6000 using 150×150 S4 chemistry. CH detection follows the strategy employed in our previous study Desai et al. (2018) for quality and filtration of artifacts. Gencore v0.13.0 (Chen et al., 2019) and fastp v0.20.1 (Chen et al., 2018) were used to trim adaptor sequences and remove duplicates. After alignment with BWA MEM v0.7.17 (Li, 2013), VarDict-Java v1.7.0 (Lai et al., 2016) was used to call variants on reads. Bcftools v1.10.2 (Li, 2011), SnpEff v4.3 (6), and Vcflib v1.0.1 (Garrison, 2016) were used to filter out reads for mapping quality, depth, and strand bias. Variant-Effect-Predictor (VEP) v100.3 (McLaren et al., 2016) was used to annotate variants. NA12878 reference DNA was included to detect and remove background. SNP were stringently filtered out when reported as > 0.25% in population allele frequency databases. Variants with VAF ≥ 1% are being reported. The sensitivity of the test is estimated at > 95% for mutations present at VAF > 1% (corresponding to 1/50 mutated cells). Whole genome sequencing (WGS) libraries were prepared with the 10x genomics Chromium Genome kit (2016) and aligned to the GRCv38 human genome with LongRanger (1.0.0) and supporting files (http://support.10xgenomics.com/genome-exome/downloads/latest) RNA-sequencing (RNA-seq) were performed with New England Biolabs (NEB) ribo-depletion RNA-seq and polyA-enrichment kits, as previously described (Garrett-Bakelman et al., 2019), and sequenced Illumina NovaSeq6000 S4 flowcells with 150×150 nt sequencing. Variant calling was completed with the Sentieon instantiation of the Genome Analysis ToolKit (GATK) WGS and RNA-seq variant pipeline (4.1.9.0).
MSK-IMACT Study and Subjects Details
In order to study the growth rate of clonal hematopoiesis mutations over time we collected additional blood samples on patients sequenced using MSK-IMPACT for repeat CH mutation testing (Bolton et al., 2019b). We extracted data on race, smoking, date of birth and cancer history through the MSK cancer registry, chemotherapy and radiation therapy through our electronic health record. We manually reviewed their medical records to capture receipt of oncologic therapy received at outside institutions during the follow-up period. If subjects received therapy outside MSK during the follow-up period, we excluded them from analyses of dose-response relationships since cumulative dose of therapy could not be consistently collected from outside records. This study was approved by the MSKCC Institutional Review Board. Subjects had a tumor and blood sample (as a matched normal) sequenced using MSK-IMPACT, a FDA-authorized hybridization capture-based next-generation sequencing assay encompassing all protein-coding exons of cancer-associated genes. MSK-IMPACT is validated and approved for clinical use by New York State Department of Health Clinical Laboratory Evaluation Program and is used to sequence cancer patients at Memorial Sloan Kettering. Genomic DNA is extracted from de-paraffinized formalin fixed paraffin embedded (FFPE) tumor tissue and patient matched blood sample, sheared and DNA fragments were captured using custom probes. The blood samples in the serial sampling cohort that were obtained for repeat CH testing were sequenced using a comparable capture-based custom panel using 163 genes implicated in myeloid pathogenesis, which included the most commonly mutated genes in our MSK-IMPACT study and this study here, with the exception of ATM. The median sequencing depth was 665X (range = 111–1987X) which was comparable to that obtained in the blood using MSK-IMPACT. We only considered mutations that were present in both the initial and follow-up panel and related to the CH panel for these astronaut samples.
Dosimetric parameters for each course of external beam radiation for each patient (i.e., dose, fractionation, technique/modality, target, n = 157 total, including 49 XRT samples) were extracted from the treatment planning system (ARIA; Varian Medical Systems, Palo Alto, CA), from radiotherapeutic prescriptions and from clinical treatment summaries. For patients treated prior to the implementation of contemporary treatment planning (prior to 2003), receipt and timing of radiation therapy was abstracted from medical billing records, although dose and fractionation were not available for these patients and were set as missing. Given the variety of radiotherapy fractionation schemes and prescribed tumor doses, we calculated the cumulative radiation dose received by each patient prior to blood draw in 2-Gy per fraction equivalents (EQD2) using an α/β of 3 Gy, considering CH to be a late-responding tissue effect. We calculated tertiles of dose based on the distribution of cumulative EQD2 received over the entire cohort and assigned each individual a score based on their tertile of exposure (e.g. a patient who did not receive external beam radiation received a score of zero for that particular agent. If the patient’s cumulative radiation dose, as expressed in EQD2, was within the first tertile, a score of one was assigned, and so forth). The twins were selected by NASA, since they were the only twin astronauts on Earth.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis was conducted in R environment (https://www.R-project.org; R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria). Figures were produced using the package ggplot2 also in R (Wickham, 2016). Statistical significance was defined as p < 0.05. Statistical tests were performed using R software.
As per the procedure in Bolton et al., we used a linear model for VAF changes. For each mutation in each individual with sequential sequencing data available, we modeled the growth rate of the mutation between the two time points according to the following formula: α = log (V / V0) / (T − T0) Where T and T0 indicates the age of the individual (in days) at the two measurement time points and V and V0 correspond to the VAF at T and T0 respectively. We also classified mutations as 600 having increased, decreased or remained constant during the follow-up period based on a binomial test comparing the two VAFs. Generalized estimating equations were used to test for an association between exposure to cytotoxic therapy and external beam radiation therapy and CH growth rate adjusting for age, gender and smoking status accounting for correlation between the growth rate of mutations in the same person. Among patients with at least one mutation in a 605 DDR CH gene and another non-DDR CH gene (N = 34), we calculated the difference in the growth rate between mutations. When patients had more than two mutations in the same gene category, we used the highest growth rate for that category. A paired t test was used to test for significance in the difference between growth rates of DDR mutations compared to non-DDR mutations within individuals who received cytotoxic therapy and/or external beam radiation 610 therapy and within those who were untreated during the follow-up period.
ADDITIONAL RESOURCES
Additional details are on NASA’s sites including http://www.nasa.gov/twins-study/.
List of Genes in CH Panel (n = 93)
ACTA2, ACTC1, ANGPTL4, ANKRD26, APC, APOA5, APOB, APOC3, ASXL1, ATM, BAP1, BARD1, BMPR1A, BRAF, BRCA1, BRCA2, BRIP1, CBL, CDH1, CDK4, CDKN2A, CEBPA, CHEK2, COL3A1, DDX41, DNMT3A, DSC2, DSG2, DSP, EPCAM, ETV6, FBN1, FLT3, GATA1, GATA2, GLA, GREM1, IDH1, IDH2, JAK2, KCNH2, KCNQ1, KDM1A, KRAS, LDLR, LMNA, LPA, LPL, MITF, MLH1, MPL, MSH2, MSH6, MUTYH, MYBPC3, MYH11, MYH7, MYL2, MYL3, NBN, NPC1L1, NPM1, NRAS, PALB2, PCSK9, PKP2, PMS2, POLD1, POLE, PPM1D, PRKAG2, PTEN, RAD51C, RAD51D, RUNX1, RYR2, SCN5A, SF3B1, SMAD3, SMAD4, SRP72, SRSF2, STK11, TET2, TGFBR1, TGFBR2, TMEM43, TNNI3, TNNT2, TP53, TPM1, U2AF1, ZRSR2
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| CD8 | Miltenyi Biotec | Cat# 130-097-057 |
| CD4 | Miltenyi Biotec | Cat# 130-097-048 |
| CD19 | Miltenyi Biotec | Cat# 130-097-055 |
| Biological Samples | ||
| NA12878 | Coriell | Cat # NA12878 |
| Critical Commercial Assays | ||
| 4 mL CPT vacutainers | BD Biosciences | Cat # 362760 |
| KAPA Hyper Prep Library Preparation | Roche | Cat # 07962347001 |
| Twist Hybridization and Wash Kit | Twist Biosciences | Cat# 101025 |
| Twist Binding and Purification Beads | Twist Biosciences | Cat# 100983 |
| 10x genomics Chromium Genome kit (2016) | 10X Genomics | N/A |
| NEBNext UltraTM II Directional RNA Library Prep Kit | New England Biolabs | Cat # E7760/E7765 |
| NEBNext rRNA Depletion Kit | New England Biolabs | Cat # E6310 |
| Deposited Data | ||
| Raw and analyzed data | This paper | NASA LSDA |
| RNA sequencing | Garrett-Bakelman et al., 2019 | NASA LSDA |
| Whole Genome sequencing | Garrett-Bakelman et al., 2019 | NASA LSDA |
| Analyzed mutation data on cancer patients treated with External Beam radiation or Untreated | Bolton et al., 2019b | https://github.com/papaemmelab/bolton_NG_CH |
| Human reference genome UCSC build hg38 (GRCh38) | UCSC | https://genome.ucsc.edu/cgi-bin/hgGateway?db=hg38 |
| Software and Algorithms | ||
| Gencore v0.13.0 | Chen et al., 2019 | https://github.com/OpenGene/gencore |
| fastp v0.20.1 | Chen et al., 2018 | https://github.com/lh3/bwa |
| BWA MEM v0.7.17 | Li, 2013 | https://github.com/lh3/bwa |
| VarDict-Java v1.7.0 | Lai et al., 2016 | https://github.com/AstraZeneca-NGS/VarDictJava |
| Bcftools v1.10.2 | Li, 2011 | http://samtools.sourceforge.net |
| Vcflib v1.0.1 | Garrison, 2016 | https://github.com/vcflib/vcflib |
| Variant-Effect-Predictor (VEP) v100.3 | McLaren et al., 2016 | https://github.com/Ensembl/ensembl-vep |
| LongRanger (1.0.0) | N/A | https://support.10xgenomics.com/genome-exome/software/pipelines/latest/installation |
| Genome Analysis ToolKit (GATK) (4.1.9.0) | N/A | https://github.com/broadinstitute/gatk/releases |
| Rsoftware | N/A | https://www.R-project.org/ |
Highlights.
Blood samples from twin astronauts were studied for clonal hematopoiesis (CH)
Distinct CH mutations and RNA variant trajectories were found across 4 years
CH was found almost two decades prior to the mean age at which it is typically detected
Longitudinal monitoring of CH is an important disease risk metric for astronauts.
ACKNOWLEDGMENTS
The study was supported by NASA/TRISH grants (NNX14AH51G, NNX17AB26G, NNX16AO69A:0107, NNX16AO69A:0061), as well as the Bert L and N Kuggie Vallee Foundation, Igor Tulchinsky and the WorldQuant Foundation, and The Pershing Square Sohn Cancer Research Alliance (Mason). We also want to thank Bill Ackman and Olivia Flatto for their support. We would also like to thank the Genomics, Epigenomics, and Applied Bioinformatics Core Facilities at Weill Cornell Medicine for sequencing and data services. We would like to thank Chaithanya Ponnaluri, Eileen Dimalanta, Brittany Sexton, and Bradley W. Langhorst from New England Biolabs for their awesome collaboration in conducting these experiments, sequencing, and analysis.
Footnotes
DECLARATION OF INTERESTS
No relevant conflicts apply to this work, but C.E.M. is a cofounder of Onegevity and Biotia. D.C.H. and C.E.M. also have positions at Tempus Labs.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108458.
REFERENCES
- Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, Barda N, Zuzarte PC, Heisler L, Sundaravadanam Y, et al. (2018). Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaev VR, Fazio S, Gleaves LA, Carter KJ, Semenkovich CF, and Linton MF (1999). Macrophage lipoprotein lipase promotes foam cell formation and atherosclerosis in vivo. J. Clin. Invest. 103, 1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, Huch M, Boymans S, Kuijk E, Prins P, et al. (2016). Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton KL, Gillis NK, Coombs CC, Takahashi K, Zehir A, Bejar R, Garcia-Manero G, Futreal A, Jensen BC, Diaz LA Jr., et al. (2019a). Managing clonal hematopoiesis in patients with solid tumors. J. Clin. Oncol. 37, 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton KL, Ptashkin RN, Gao T, Braunstein L, Devlin SM, Kelly D, Patel M, Berthon A, Syed A, Yabe M, et al. (2019b). Oncologic therapy shapes the fitness landscape of clonal hematopoeisis. bioRxiv. 10.1101/848739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y, Maragh M, and Gilliland DG (1996). Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 88, 59–65. [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. (2012). Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 44, 1179–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvillo-Argüelles O, Jaiswal S, Shlush LI, Moslehi JJ, Schimmer A, Barac A, and Thavendiranathan P (2019). Connections between clonal hematopoiesis, cardiovascular disease, and cancer: A review. JAMA Cardiol. 4, 380–387. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, and Gu J (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Huang T, Liao W, Xu Y, Li Z, and Gu J (2019). Gencore: an efficient tool to generate consensus reads for error suppressing and duplicate removing of NGS data. BMC Bioinformatics 20 (Suppl 23), 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, Ng V, Xia B, Witkowski MT, Mitchell-Flack M, et al. (2017). Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 170, 1079–1095.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, Hyman DM, Solit DB, Robson ME, Baselga J, et al. (2017). Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 21, 374–382.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S (2011). Somatic mosaicism in healthy human tissues. Trends Genet. 27, 217–223. [DOI] [PubMed] [Google Scholar]
- Desai P, Mencia-Trinchant N, Savenkov O, Simon MS, Cheang G, Lee S, Samuel M, Ritchie EK, Guzman ML, Ballman KV, et al. (2018). Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24, 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu CL, Sano S, Muralidharan S, Rius C, et al. (2017). Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Bakelman FE, Darshi M, Green SJ, Gur RC, Lin L, Macias BR, McKenna MJ, Meydan C, Mishra T, Nasrini J, et al. (2019). The NASA Twins Study: A multidimensional analysis of a year-long human spaceflight. Science 364, eaau8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E (2016). Vcflib, a simple C++ library for parsing and manipulating VCF files (MIT). https://github.com/vcflib/vcflib.
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. (2014). Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz ML, Chin CR, Tomoiaga D, Mackay M, Butler D, Afshinnekoo E, Bezdan D, Schmidt MA, Mozsary C, Melnick A, et al. (2020). Multi-omic, Single-Cell, and Biochemical Profiles of Astronauts Guide Pharmacological Strategies for Returning to Gravity. Cell Rep Published online November 25, 2020. 10.1016/j.celrep.2020.108429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JW, Pedersen DA, Larsen LA, Husby S, Clemmensen SB, Hjelmborg J, Favero F, Weischenfeldt J, Christensen K, and Grønbæk K (2020). Clonal hematopoiesis in elderly twins: concordance, discordance, and mortality. Blood 135, 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iosim S, MacKay M, Westover C, and Mason CE (2019). Translating current biomedical therapies for long duration, deep space missions. Precis Clin Med 2, 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, and Ebert BL (2019). Clonal hematopoiesis in human aging and disease. Science 366, eaan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. (2014). Agerelated clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, et al. (2017). Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, and Majeti R (2012). Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 4, 149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, Johnson J, Dougherty B, Barrett JC, and Dry JR (2016). VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 44, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, 1303.3997v2. https://arxiv.org/abs/1303.3997. [Google Scholar]
- Luxton JJ, McKenna MJ, Lewis A, Taylor LE, George KA, Dixit SM, Moniz M, Benegas W, Mackay MJ, Mozsary C, et al. (2020a). Telomere Length Dynamics and DNA Damage Responses Associated with Long-Duration Spaceflight. Cell Rep Published online November 25, 2020. 10.1016/j.celrep.2020.108457. [DOI] [PubMed] [Google Scholar]
- Luxton JJ, McKenna MJ, Taylor LE, George KA, Zwart SR, Crucian B, Drel VR, Garrett-Bakelman FE, Mackay MJ, Butler D, et al. (2020b). Temporal Telomere and DNA Damage Responses in the Space Radiation Environment. Cell Rep Published online November 25, 2020. 10.1016/j.celrep.2020.108435. [DOI] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, et al. (2015). Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, and Cunningham F (2016). The Ensembl Variant Effect Predictor. Genome Biol. 17, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle SN, Wolfson MY, Hartsough L, Ma NJ, Mason CE, Merighi M, Nathan V, Silver PA, Simon M, Swett J, et al. (2020). The case for biotech on Mars. Nat. Biotechnol. 38, 401–407. [DOI] [PubMed] [Google Scholar]
- Sano S, Oshima K, Wang Y, MacLauchlan S, Katanasaka Y, Sano M, Zuriaga MA, Yoshiyama M, Goukassian D, Cooper MA, et al. (2018). Tet2-Mediated Clonal Hematopoiesis Accelerates Heart Failure Through a Mechanism Involving the IL-1β/NLRP3 Inflammasome. J. Am. Coll. Cardiol. 71, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma DP (2018). Clinical implications of clonal hematopoiesis. Mayo Clin. Proc. 93, 1122–1130. [DOI] [PubMed] [Google Scholar]
- Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, and Ebert BL (2015). Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattathil S, and Scheet P (2016). Extensive hidden genomic mosaicism revealed in normal tissue. Am. J. Hum. Genet. 98, 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilkki S, Tsao JL, Loukola A, Pöyhönen M, Vierimaa O, Herva R, Aaltonen LA, and Shibata D (2001). Extensive somatic microsatellite mutations in normal human tissue. Cancer Res. 61, 4541–4544. [PubMed] [Google Scholar]
- Wang Y, Sano S, Yura Y, Ke Z, Sano M, Oshima K, Ogawa H, Horitani K, Min KD, Miura-Yura E, et al. (2020). Tet2-mediated clonal hematopoiesis in nonconditioned mice accelerates age-associated cardiac dysfunction. JCI Insight 5, e135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; ). [Google Scholar]
- Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, Thorgeirsson TE, Sigurdsson A, Gudjonsson SA, Gudmundsson J, et al. (2017). Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130, 742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NASA Life Sciences Data Archive (LSDA) is the repository for all human and animal research data, including that associated with this study. LSDA has a public facing portal where data requests can be initiated (https://lsda.jsc.nasa.gov/Request/dataRequestFAQ). The LSDA team provides the appropriate processes, tools, and secure infrastructure for archival of experimental data and dissemination while complying with applicable rules, regulations, policies, and procedures governing the management and archival of sensitive data and information. The LSDA team enables data and information dissemination to the public or to authorized personnel either by providing public access to information or via an approved request process for information and data from the LSDA in accordance with NASA Human Research Program and Johnson Space Center (JSC) Institutional Review Board direction.


