Abstract
The gram-negative marine bacterium Pseudoalteromonas atlantica produces extracellular polysaccharide (EPS) that is important in biofilm formation by this bacterium. Insertion and precise excision of IS492 at a locus essential for extracellular polysaccharide production (eps) controls phase variation of EPS production in P. atlantica. Examination of IS492 transposition in P. atlantica by using a PCR-based assay revealed a circular form of IS492 that may be an intermediate in transposition or a terminal product of excision. The DNA sequence of the IS492 circle junction indicates that the ends of the element are juxtaposed with a 5-bp spacer sequence. This spacer sequence corresponds to the 5-bp duplication of the chromosomal target sequence found at all IS492 insertion sites on the P. atlantica chromosome that we identified by using inverse PCR. IS492 circle formation correlated with precise excision of IS492 from the P. atlantica eps target sequence when introduced into Escherichia coli on a plasmid. Deletion analyses of the flanking host sequences at the eps insertion site for IS492 demonstrated that the 5-bp duplicated target sequence is essential for precise excision of IS492 and circle formation in E. coli. Excision of IS492 in E. coli also depends on the level of expression of the putative transposase, MooV. A regulatory role for the circular form of IS492 is suggested by the creation of a new strong promoter for expression of mooV by the joining of the ends of the insertion sequence element at the circle junction.
IS492 is a 1.2-kb multicopy insertion sequence (IS) found in the gram-negative marine bacterium Pseudoalteromonas atlantica (3). Insertion and precise excision of IS492 at an essential locus for extracellular polysaccharide (EPS) synthesis (eps) is associated with phase variation of EPS production in P. atlantica (2). The ability of P. atlantica to switch the adhesin EPS on and off is important in the life cycle of this biofilm-forming bacterium, which moves from solid surfaces (such as seaweed or sand) to the open ocean and then back to a solid substrate to initiate a new biofilm (6, 7). Southern blot analysis performed by Bartlett et al. (2) on chromosomal DNA from EPS phase variants of P. atlantica indicated that excision of IS492 from the eps locus does not result in insertion of IS492 at a new site in the chromosome. Therefore, precise excision of IS492 from eps is not associated with replicative transposition of IS492 or with a cut-and-paste mechanism for transposition in which excision is directly linked to insertion at a new site. Instead, the result of the Southern analysis suggests that precise excision results in loss of the element or that the element moves into an extrachromosomal form.
IS492 is a member of the IS110-IS492 family of insertion elements, a group recognized as atypical in several aspects (for a review, see reference 19). Its members have either extremely small or no terminal inverted repeats, and many do not create target site duplications upon insertion. The transposases for this family, including MooV (for mover of IS492 in oceanic variants) from IS492, are also unusual. These transposases have homology with each other and with the novel site-specific DNA invertase Piv from Moraxella lacunata and Moraxella bovis (16). Despite the homology with Piv, there is no shared catalytic amino acid motif with the site-specific DNA recombinases of the λ Int and Hin-resolvase families (for a review, see reference 23). In addition, it is not apparent that the DNA recombinases of the IS110-IS492 family contain the DDE motif, which is the most common catalytic motif of the characterized transposases-phosphoryltransferases from eukaryotes and prokaryotes (for reviews, see references 8 and 19). These combined unique aspects suggest that the transposases from the IS110-IS492 family of insertion elements use a different catalytic mechanism than previously characterized for transposases and warrant further investigation.
In the work presented here, we have focused on the excision reaction of IS492 transposition. A circular product of IS492 excision can be detected by PCR analysis at low levels in P. atlantica and in an Escherichia coli model system. Both excision and formation of the circular product are dependent on the transposase MooV. Determination of the requirements for and consequences of IS492 precise excision and circle formation suggests that circularization of the element plays a regulatory role in IS492 transposition. Surprisingly, these analyses also revealed that the target sequence that is duplicated upon insertion of IS492 is required for excision of IS492 in E. coli.
MATERIALS AND METHODS
Bacterial strains.
P. atlantica DB27 was a gift from D. Bartlett (2, 3). P. atlantica 19262, 43666, and 43667 (7, 36); Pseudoalteromonas espejiana 29659 (5); and Pseudoalteromonas haloplanktis 14393 (26) were obtained from the American Type Culture Collection (ATCC). E. coli InvαF′ [F′ (φ80lacZΔM15) endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF)U169] (Invitrogen), TOP10 [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG] (Invitrogen), DH5α [(φ80lacZΔM15) supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (obtained from C. Moran), and HMS174(DE3) [(λDE3) recA1 hsdR rifR] (Novagen) were used as hosts for IS492-containing plasmids and MooV expression vectors. E. coli MC1061(recA) [hsdR araD139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi recA] (ATCC) and DH5α were used for circle promoter studies.
Media, reagents, and enzymes.
All E. coli strains were grown on Luria-Bertani (LB) agar (Difco). Pseudoalteromonas strains were cultured on Difco 2216 marine agar at 75% of the recommended concentration (4). The following drug concentrations were used for plasmid selection and maintenance: ampicillin at 80 or 100 μg/ml, chloramphenicol at 50 or 80 μg/ml, tetracycline at 12.5 μg/ml, and streptomycin at 25 μg/ml. When indicated, 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) (40 μl of a 40-mg/ml solution) was spread onto plates. All chemicals were purchased from Sigma Chemical Co. Restriction enzymes, T4 DNA ligase, and Klenow DNA polymerase were purchased from New England Biolabs. Sequenase, dideoxynucleotides, and deoxynucleotides were purchased from Amersham, and [α-32P]dATP was purchased from Dupont NEN. Pfu DNA polymerase used in PCR was obtained from Stratagene. Oligonucleotides were purchased from Gibco-Bethesda Research Laboratories.
Plasmids and plasmid constructions.
The sequences of all oligonucleotides utilized in this work are listed in Table 1. All constructs created with PCR-amplified DNA fragments were sequenced to determine that there were no PCR-generated mutations. Cosmids pDB200 (eps+) and pDB440 (eps::IS492) were gifts from D. Bartlett and were previously described (2). IS492 was amplified by PCR from pDB440 by using oligonucleotides 1 and 2 and cloned into the TA cloning vector pCR2.1 (Invitrogen), resulting in pAG949. A mooV mutant derivative of pAG949, pAG957, was constructed by removing the 357-bp MfeI fragment of IS492 and replacing it with an 884-bp DNA fragment containing the chloramphenicol acetyltransferase gene (cam); the cam fragment was PCR amplified from pACYC184 with oligonucleotides 3 and 4, encoding the MfeI sites to facilitate cloning. Both IS492 and IS492ΔmooV::cam were subcloned from pAG949 and pAG957, respectively, as BamHI-to-XhoI fragments into the BamHI-to-XhoI region of low-copy-number plasmid pACYC177, to generate pAG956 and pAG959.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| 1 | 5′ GGCTCTCGAGCGGTACTGTCTTATCATCCTAATCG 3′ |
| 2 | 5′ CTCGCTCGAGCAGGAGGCTCTCTATTGTACAGC 3′ |
| 3 | 5′ GCGCAATTGAGGGCACCAATAACTGCCTT 3′ |
| 4 | 5′ CCTACAATTGACGGAAGATCACTTCGCAGA 3′ |
| 5 | 5′ GGCCCATATGAATCCAAAAACAAATCAAAGCGTTAAC 3′ |
| 6 | 5′ GCGCCTCGAGATTTGTAGTCTTCGGCGCTTC 3′ |
| 7 | 5′ ATGAATCCAAAAACAAATCAAAGCG 3′ |
| 8 | 5′ CTCGAAGCTTAGACTATGGCGTCAATAGCTAA 3′ |
| 9 | 5′ GGCCGGATCCGCATTATGCAAGCAAGAGTT 3′ |
| 10 | 5′ GATCCAAATTAGCTATTGACGCCATAGTCTCTTGTATGGATAATAACTCAGGT 3′ |
| 11 | 5′ CTAGACCTGAGTTATTATCCATACAAGAGACTATGGCGTCAATAGCTAATTTG 3′ |
| 12 | 5′ AGACCCAGTAGACTATTCGAGTC 3′ |
| 13 | 5′ TGT ACA TGG CGA TGA TGT CAG CG 3′ |
| 14 | 5′ GGCCCTCGAGACCTAACAAGATGGATAATAACTCAG 3′ |
| 15 | 5′ TTTGGTGGCATTGAAAACCTTGT 3′ |
| 16 | 5′ GAGAGCTTGATAGACCTAACAAG 3′ |
| 17 | 5′ AAAACCTTGTATGGATAATAACTCAG 3′ |
| 18 | 5′ ACCTAACAAGAGACTATGGC 3′ |
| 19 | 5′ CTTGTATGGATAATAACTCAG 3′ |
| 20 | 5′ ACAAGAGACTATGGCGTCAA 3′ |
| 21 | 5′ ATGGATAATAACTCAGGTAAATCAT 3′ |
| 22 | 5′ TTAGCTATTGACGCCATAGTC 3′ |
| 23 | 5′ AATCACTCATAGCCTGCATAG 3′ |
| 24 | 5′ GCGCTGCAGCAGCTATTTTGTAATCACGTACTCTC 3′ |
| 25 | 5′ GGCACGTAGTGAGAATCCATACATAACGGAAAGAGCC 3′ |
| 26 | 5′ GGCATTGAAAACCTTGTATGG 3′ |
| 27 | 5′ TTGATAGACCTAACAAAGAGACT 3′ |
| 28 | 5′ GGCTCTCGAGCGGTACTGTCTTATCATCCTAATCG 3′ |
| 29 | 5′ CTC GCTCGAGCAGGAGGCTCTCTATTGTACAGC 3′ |
| 30 | 5′ ACCCCAGGCTTTACACTTTATGCTTCCGGCTCGTATAATGTGTGGA 3′ |
| 31 | 5′ TCCACACATTATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGT 3′ |
| 32 | 5′ GAGAGACCGATAACAAGAAAGAAC 3′ |
| 33 | 5′ GCTGTTGGATTTTTTTAATGGCTTC 3′ |
| 34 | 5′ CATTTTAGCTTCCTTAAGCTCCTGAAAATC 3′ |
| 35 | 5′ GATTTTCAGAGCTTAAGGAAGCTAAAATG 3′ |
| 36 | 5′ GATCTTAAGTTTGGTGGCATTGAAAACCTTGT 3′ |
| 37 | 5′ CTGCTTAAGAGAGCTTGATAGACCTAACAAG 3′ |
| 38 | 5′ CCGGCAATTGTTAATGAAACGCCGAGTTAACGCCATC 3′ |
| 39 | 5′ GCTACAATTGAGGAAACAGCTATGACCATGATTAC 3′ |
Multiple plasmids were constructed for MooV overexpression. The MooV protein with a carboxyl-terminal His6 tag was created by PCR amplifying mooV from pDB440 with oligonucleotides 5 and 6, containing NdeI and XhoI restriction sites, respectively, for cloning into the polylinker of pET21-a (Novagen), forming pAG921. Derivatives of pAG921 were made in order to change antibiotic resistance or to obtain compatibility with other plasmids used in this study: (i) pAG922 was constructed by deleting the PstI-to-DraIII region of pAG921, containing the β-lactamase gene (bla), followed by replacement with the tetM gene from Tn916, which was PCR amplified from pAM120 (12), with oligonucleotides 24 and 25, and (ii) pAG954 was constructed by removing the PstI-to-BspLUII1 region of pAG922, containing the ColE1 ori, and replacing it with the PstI-to-BspHI region of pACYC177, containing the p15a ori. A pET21-a derivative, pAG1300 (32), containing the site-specific recombinase gene, piv, from M. lacunata was modified in the same fashion as pAG921 to create pAG955. pMalE-MooV, which expresses a translational fusion of malE (maltose binding protein [MBP] gene) to the 5′ end of mooV, was created by cloning a PCR-generated fragment from pDB440 (by using oligonucleotides 7 and 8), containing mooV and the correct cloning sites, into the XmnI-to-HindIII region of pMal-C2 (New England Biolabs). The wild-type MooV (nonfusion) protein was expressed from pAG900, which has mooV under control of the tac promoter and with its own Shine-Dalgarno sequence in plasmid pKH197 (generously provided by K. Hughes; pKH197 is a derivative of pKH66 [14], having one BamHI site upstream from the tac promoter filled in and religated); the BamHI-to-HindIII region of pKH197 containing the hin recombinase gene was replaced by a fragment containing mooV, generated by PCR with oligonucleotides 8 and 9 and pDB440.
Two circle junction lacZ fusions were constructed: (i) the multicopy plasmid pDV6 is a derivative of pCB267 (28) with the BamHI-to-XbaI region replaced by annealed oligonucleotides 10 and 11, which span the IS492 junction, and (ii) the single-copy plasmid pAG967, derived from pNN387 (9), contains the IS492 circle junction flanked by 203 bp of the right-end sequence and 66 bp of the left-end sequence of IS492; this 274-bp sequence was obtained as a HindIII-to-NotI restriction fragment from pAG952.1 (described below) and cloned into the HindIII-to-NotI region of pNN387. Plasmid pAG952.1 is a pCR2.1 (Invitrogen) derivative containing a 400-bp PCR fragment insert which was amplified from P. atlantica lysates with IS492-complementary oligonucleotides (oligonucleotides 12 and 13) with their 3′ extendable ends in opposite directions (see Results). Constitutive lacZ control plasmids were also constructed. The constitutive lacUV5 promoter was cloned into the SmaI site of pCB267 by using annealed oligonucleotides 30 and 31, generating pDV5. A constitutive tac promoter, with all regulatory elements deleted, was subcloned from pNN396 (9) as a NotI-to-HindIII fragment into the NotI-to-HindIII region of pNN387 (9), to generate pAG620.
An IS492 target site mutant was created by first generating a DNA fragment containing the full IS492 sequence with the left end flanked by 12 bp of target eps sequence identical to the right-end target sequence and the right end flanked by 76 bp of eps right-end target sequence. This fragment was PCR amplified from pDB440 by using oligonucleotides 2 and 14 and then inserted into the unique XhoI site of pACYC177, generating pAG903. A series of target site deletion plasmids were constructed by cloning into the TA cloning site of pCR2.1 the PCR products generated from pAG949, using the following primer pairs (the number of base pairs of original target sequence remaining on each side of IS492 is indicated): oligonucleotides 15 and 16, pAG970 (24 bp on left and 23 bp on right); oligonucleotides 26 and 27, pAG976 (18 bp on left and 17 bp on right); oligonucleotides 17 and 18, pAG971 (10 bp on left and 10 bp on right); oligonucleotides 19 and 20, pAG972 (5 bp on left and 5 bp on right); and oligonucleotides 21 and 8, pAG973 (1 bp on left and 0 bp on right).
The reporter plasmids pAG993 and pAG994 were constructed to examine IS492 excision in vivo. Wild-type IS492 and an excision-deficient IS492, IS492ΔmooV::lacZα, were subcloned from pAG949 and pAG989, respectively, as AflII fragments (approximately 1.2 kb each) and cloned into the unique AflII site between the chloramphenicol acetyltransferase gene and its promoter on plasmid pAG987. The QuikChange site-directed mutagenesis kit (Stratagene) and oligonucleotides 34 and 35 were used to generate pAG987, which is a pACYC184 derivative containing a unique AflII site, created by a single-base insertion (A/T) at position 232. pAG989 was constructed by inserting the 1,248-bp PCR product resulting from amplification of pAG963 with oligonucleotides 36 and 37 into pCR2.1. pAG963 is a derivative of pAG956 (described above) containing a 357-bp deletion between distal MfeI sites within mooV, which was replaced by a 453-bp promoterless lacZα fragment. This lacZα fragment was generated as a PCR product from the amplification of pEU730 (10) by using oligonucleotides 38 and 39.
All PCR products created for cloning purposes were amplified by using Pfu DNA polymerase (Stratagene). The thermal cycling conditions were as follows: 95°C for 3 min followed by 25 cycles of 95°C for 1 min (denaturing), the Tm of the oligonucleotide with the lesser Tm value minus 5°C for 1 min (annealing), and 72°C for 2 min (extension). The products were digested with the indicated restriction enzymes, or, if ligated into pCR2.1, they were incubated for an additional 10 min at 72°C with Taq DNA polymerase (Sigma). PCR products were purified away from reaction components with the QIAquick kit (Quiagen) or by phenol extraction followed by ethanol precipitation. All ligations were done at 16°C overnight with 400 U of T4 DNA ligase or, for pCR2.1 cloning, with topoisomerase from the Invitrogen kit. Transformation of ligation mixtures into host strains was generally performed with competent cells prepared by CaCl2 treatment (27) or by using the One Shot (InvαF′ or TOP10) competent cells from Invitrogen and then plating on LB agar containing the appropriate antibiotics.
Southern blotting.
Standard techniques for transfer of DNA onto a nylon support with alkaline buffer were performed as described by Ausubel et al. (1). Ethidium bromide staining of the agarose gel before and after transfer of the DNA for the Southern blot showed that approximately equal amounts of chromosomal DNA from each strain had transferred to the blot filter. Hybridizations were performed with Rapid-hyb buffer (Amersham) according to the manufacturer’s recommendations: the hybridization temperature was 65°C, and hybridization was followed by one low-stringency wash in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) sodium dodecyl sulfate (SDS) for 20 min at 65°C and two high-stringency washes in 0.5× SSC–0.1% (wt/vol) SDS for 15 min at 50°C. Following the final washes after hybridization, the blots were visualized by using a Molecular Dynamics 445SI PhosphorImager.
The DNA probe for detection of IS492 on blots containing chromosomal DNAs from various Pseudoalteromonas species (see Fig. 1) was prepared by end labeling the EcoRI fragment of pAG949 containing IS492 with [α-32P]dATP and Klenow fragment. The DNA probe for detection of circle junctions was prepared by PCR, incorporating [α-32P]dATP into newly synthesized 405-bp circle junction fragments. The PCR mixture (50 μl) contained 0.1 μg of oligonucleotide 12, 0.1 μg of oligonucleotide 13, 0.1 μg of circle junction fragment (see “PCR assay to detect IS492 circles” below), 50 μM each deoxynucleoside triphosphate (dNTP) (dATP, dCTP, dTTP, and dGTP), 1× Pfu reaction buffer, 0.33 μM [α-32P]dATP, and 1 U of Pfu polymerase (Stratagene). The thermal cycling conditions were as follows: 95°C for 2 min, followed by 25 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min. The probe was boiled for 10 min, and one-fifth of the reaction mixture was added to 20 ml of hybridization buffer.
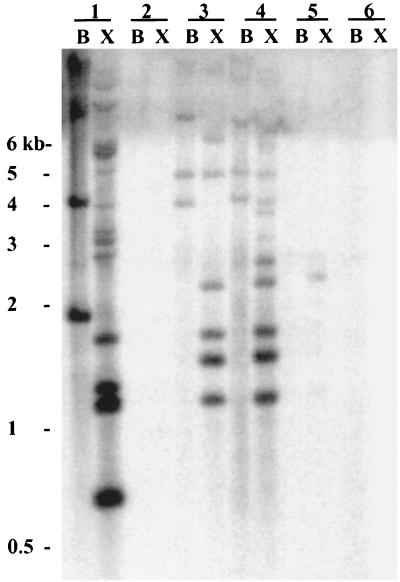
FIG. 1.
Southern analysis of P. atlantica and closely related marine bacteria with 32P-labeled IS492 DNA as a probe. Chromosomal DNAs from the following strains were digested with either BamHI alone (lanes B) or BamHI and XmnI (lanes X) and subjected to agarose gel electrophoresis: P. atlantica DB27 (lanes 1), P. atlantica ATCC 19262 (lanes 2), P. atlantica ATCC 43666 (lanes 3), P. atlantica ATCC 43667 (lanes 4), P. espejiana ATCC 29659 (lanes 5), and P. haloplanktis ATCC 14393 (lanes 6). After transfer to a nylon membrane, the DNA was probed with a 1-kb [32P]dATP-labeled IS492 DNA fragment. The positions and sizes of the molecular size markers on the agarose gel are indicated to the left.
PCR assay to detect IS492 circles.
IS492 circle junctions were detected by using either of two sets of PCR primers directed in opposing orientations (with 3′ extendable ends directed outwardly): oligonucleotides 12 and 13 or oligonucleotides 22 and 23, with 405- and 648-bp products, respectively. For initial circle detection, colony PCR was used; template DNA was prepared from single colonies by resuspending the colony in 30 μl of Tris-EDTA (pH 8.0), boiling to lyse the cells, and centrifuging to remove insoluble debris. The colonies were grown overnight on solid medium, with the appropriate antibiotics and various concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG) as indicated, at 30°C (for P. atlantica) or 37°C (for E. coli), followed by incubation at 4°C for 2 to 24 h. The incubation at 4°C was essential for circle detection. Comparative PCR with wild-type IS492 and mutant strains was done with template DNA prepared by plasmid minipreps (Qiagen) on cells grown as described above; the DNA concentration was normalized for all samples, first by UV spectroscopy at 260 nm and then by finding the concentration of template which gave equal-intensity control PCR products as determined by electrophoresis of dilutions of the products on agarose gels, staining with ethidium bromide, image acquisition on a fluorescence transilluminator with a digital camera, and quantitative analysis using the image TIFF file and ImageQuant Software Package from Molecular Dynamics. Control PCR products were generated in triplicate by using twofold template dilutions with primers which amplify a 603-bp region of the β-lactamase resistance marker found on the IS492-containing plasmids; 30 cycles of PCR amplification were used, whereby the linear range of the standard curve extended from 9.3 × 104 to 9.3 × 108 pAG949 (IS492) molecules, and differences between product intensities could be reproducibly determined. Reaction conditions for the experimental and control products were identical: 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min.
For assays in which complementation of MooV activity for circle formation was determined by using increasing concentrations of IPTG to induce expression of mooV from pAG954, it was noted that the colony appearance and size changed with increasing [IPTG]. Growth curves were generated for HMS174(DE3) containing pAG954 and pAG957 in LB broth containing 0, 0.01, 0.05, or 0.1 mM IPTG (higher concentrations of IPTG resulted in nonviable cells) and the appropriate antibiotics. Overnight cultures were used to inoculate LB broth containing the different levels of IPTG to a final optical density at 600 nm (OD600) of 0.1. As the cultures were aerated at 37°C, the OD600 was determined every 30 min, and dilutions were plated to determine viable cell counts.
PCR assay to detect repaired donor plasmid.
The repaired donor plasmid was detected by PCR with primers complementary to known eps sequence, oligonucleotides 28 and 29. Template DNA was prepared and normalized as described above for the detection of IS492 circles, using HMS174(DE3) carrying either pCR2.1, pAG949, pAG957, or pAG957 and pAG954. Reactions were done in 1× Pfu buffer with 50 μM dNTPs and 2.5 U of Pfu polymerase. Cycling conditions were 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, 57°C for 1 min, and 72°C for 1.5 min. Products were detected on a 2% agarose gel (1% NuSieve, 1% SeaKem [FMC BioProducts]).
Genetic assay for excision of IS492.
Competent TOP10 cells were transformed with the excision reporter plasmid pAG993 (wild-type IS492) or pAG994 (IS492ΔmooV::cam), selecting for tetracycline resistance. These transformants were then made competent by the CaCl2 method (27), transformed with pMal-C2 or pMalE-MooV, and then plated onto LB agar plates containing 12.5 μg of tetracycline per ml and 100 μg of ampicillin per ml. Two primary transformant colonies were each grown to an OD600 of 0.2 to 0.4, and 10-fold serial dilutions were plated in duplicate. Acquisition of chloramphenicol resistance was scored as colonies which were visible after overnight growth at 37°C on LB agar containing 80 μg of chloramphenicol per ml. The number of viable cells containing the excision reporter plasmid was determined by plating dilutions of the same cultures on LB agar containing 12.5 μg of tetracycline per ml. To physically determine if excision of the IS492 element had occurred, the plasmid DNA was prepared from chloramphenicol-resistant cells, cut with NcoI, and examined by agarose gel electrophoresis in comparison with the original plasmids. The Cmr Tetr and Cms Tetr colonies from these platings were also checked by the PCR assay for circle junction formation.
Electrophoretic gel fractionation for detection of IS492 circular form.
HMS174(DE3) containing pAG949 was streaked for confluent growth on three standard (90-mm-diameter) LB agar plates containing ampicillin at 100 μg/ml. The cells were grown at 37°C overnight, followed by a 2-h incubation at 4°C. Plasmid DNA was prepared from total scraped cells by using a Quiagen Plasmid Maxi kit, and 0.25 μg of this DNA was loaded onto a 1% agarose gel. The gel was lightly stained with ethidium bromide. DNA was size fractionated by dividing the gel into segments relative to a linear DNA marker ladder. DNA was eluted from the agarose, phenol-chloroform extracted, and ethanol precipitated by standard protocols described by Sambrook, et al. (27). Two nanograms of a control artificial IS492 circle was fractionated similarly from the same agarose gel. The control circle was constructed with IS492 plus 150 bp of surrounding sequence which was removed from pAG949 by EcoRI restriction digestion. The 1,352-bp EcoRI fragment was gel purified from 1% agarose, followed by phenol-chloroform extraction and ethanol precipitation. T4 DNA ligase was added to DNA diluted to 0.3 μg/ml, a concentration at which intramolecular ligation predominates. The covalently closed circular form was then supercoiled by using DNA gyrase according to the specifications of the manufacturer (Gibco-BRL).
DNA isolated in different fractions was used as the template in PCRs with mixtures containing the circle junction primers, oligonucleotides 12 and 13, 50 μM dNTPs, 1× Pfu buffer, and 2.5 U of Pfu polymerase. Cycling conditions were as follows: 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min. Products were detected on a 2% agarose gel (1% NuSieve and 1% SeaKem).
Identification and characterization of IS492 target sites in P. atlantica.
Inverse PCR was performed with chromosomal DNA isolated from EPS+ P. atlantica DB27 by the protocol of Ochman et al. (24). P. atlantica chromosomal DNA (0.5 μg) was digested with either RsaI or HaeIII (which do not have recognition sites within the IS492 sequence). Restriction enzymes were removed by phenol extraction followed by ethanol precipitation. The digested DNA was diluted to 5 μg/ml, and T4 DNA ligase was added; these conditions promote intramolecular ligation. After overnight incubation at 16°C, the ligase was inactivated by heat. Various dilutions of the ligated products were used as templates in PCRs with mixtures containing primers complementary to IS492 with their 3′ ends directed outwardly (oligonucleotides 12 and 13); therefore, these primers could be used to amplify DNA from either side of an IS492 insertion. PCR products were generated by Pfu (turbo) polymerase (Stratagene) under the following thermal cycling conditions and cloned into the plasmid vector pCR2.1 (Invitrogen) for subsequent sequencing: 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, 57°C for 1 min, and 72°C for 6 min.
Circle junction promoter activity assay.
For measurement of β-galactosidase activity, cultures of MC1061(recA) carrying either pDV6, pCB267, or pDV5 and cultures of DH5α carrying either pAG967, pNN387, or pAG620 were grown overnight at 37°C in LB medium with the appropriate antibiotics and diluted 1:50 into fresh medium. β-Galactosidase activity was measured when cells reached mid-exponential phase (OD600 = 0.4 to 0.6), according to the method of Miller (21) with an SDS-chloroform lysis modification. Lysates were centrifuged prior to measurements of absorbance at 420 nm.
RESULTS
Prevalence of IS492 in P. atlantica and related marine bacteria.
Bartlett et al. (2) previously reported that P. atlantica DB27 contains several copies of IS492. It has not been reported that IS492 appears in other isolates of P. atlantica or in other gram-negative bacteria. To determine if other P. atlantica strains or related marine bacteria (11) also contain IS492, Southern blot analysis (Fig. 1) was performed with chromosomal DNAs from P. atlantica DB27 (3, 7), P. atlantica ATCC 19262 (36), P. atlantica ATCC 43666 (7) and ATCC 43667 (7), Pseudoalteromonas (Alteromonas) espejiana ATCC 29659 (5), and Pseudoalteromonas (Alteromonas) haloplanktis ATCC 14393 (26). The chromosomal DNAs were digested with BamHI alone (Fig. 1, lanes B) or with BamHI and XmnI (Fig. 1, lanes X). BamHI has no recognition site within the known IS492 DNA sequence, and XmnI has a single recognition site within IS492 at 565 bp from the left end of the 1,202-bp element. Figure 1 shows that in addition to P. atlantica DB27, two of the ATCC P. atlantica strains (ATCC 43666 and ATCC 43667) have IS492 insertions in the chromosome. As expected, the BamHI fragments that hybridized with IS492 (Fig. 1, lanes B) were cleaved by XmnI, thus generating twice as many hybridizing bands (Fig. 1, lanes X). The differences in the hybridization profiles for the multicopy IS492 element may reflect movement of the IS element or restriction fragment polymorphisms. There is at least one weakly hybridizing band in the P. espejiana DNA (Fig. 1, lane 5X); this may indicate that a cryptic or diverged IS element, related to IS492, is present on the chromosome of P. espejiana. No hybridization of the IS492 probe with the DNA from P. atlantica 19262 or P. haloplanktis was seen (Fig. 1, lanes 2 and 6). Although strain 19262 is considered the type strain for P. atlantica, it differs from the other P. atlantica isolates in growth rate (higher), colony appearance (more mucoid and no phase variation), and odor, as well as in its lack of IS492 on the chromosome.
IS492 forms circles in P. atlantica and E. coli.
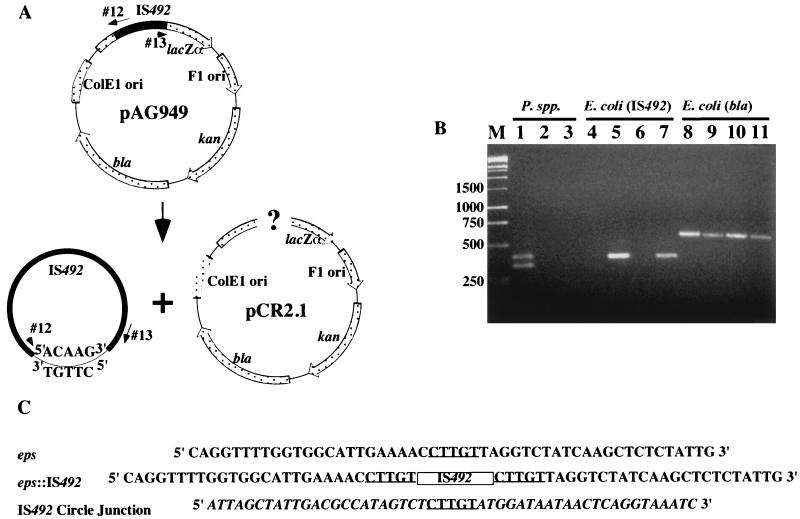
Bartlett et al. (2) performed Southern blot analysis of the chromosomes from P. atlantica DB27 that had switched from EPS− to EPS+; the results indicated that precise excision of IS492 from the eps locus is not linked to insertion of the element at a new site on the chromosome. A possible explanation for the unlinking of IS492 excision and insertion is that IS492 may form a circular, extrachromosomal element as an intermediate in transposition or as an end product of precise excision. Indeed, the related IS element IS117, from the IS110 family, has a circular, extrachromosomal form that is proposed to be an intermediate in transposition (13). To assay for a circular form of IS492 in P. atlantica DB27, PCR was used to detect the junction of the left and right ends of IS492. Primers that are complementary to the DNA sequences proximal to the ends of IS492 were designed to have their 3′ ends directed away from each other, such that the predicted PCR product generated with this primer set would represent amplification from a circular form of the IS (Fig. 2A). Two independent primer sets were used to generate PCR products. PCR amplification of crude DNA isolated from EPS+ colonies yielded products of the appropriate size for both primer sets, i.e., a 405-bp product for primers 12 and 13 (Fig. 2B, lane 1) and a 648-bp product for primers 22 and 23 (data not shown), suggesting that a circular IS492 element was the template in the reaction. An additional, smaller PCR product was present only for primers 12 and 13 (Fig. 2B, lane 1). The related Pseudoalteromonas species P. espejiana and P. haloplanktis, which showed no bands hybridizing to IS492 in the Southern assay, did not yield a PCR product in this assay (Fig. 2B, lanes 2 and 3).
FIG. 2.
Detection of IS492 circles in vivo by PCR analysis. (A) Diagram of IS492-containing plasmid pAG949 and locations of primers (oligonucleotides 12 and 13) used in this PCR analysis. Potential products of excision are drawn below the diagram. (B) Agarose gel electrophoresis of products generated from colony PCR with the circle junction primers (oligonucleotides 12 and 13) and total DNA from P. atlantica DB27 (lane 1), P. espejiana (lane 2), P. haloplanktis (lane 3), E. coli InvαF′ containing pCR2.1 (lane 4), InvαF′ containing pAG949 (lane 5), E. coli HMS174(DE3) containing pCR2.1 (lane 6), or HMS174(DE3) containing pAG949 (lane 7). Lanes 8 to 11 have the same template DNAs as lanes 4 to 7, respectively, but PCR was performed with the bla primers (see Materials and Methods), which amplify the β-lactamase gene of pAG949, as a control for template addition. Lane M, molecular size markers in base pairs. (C) Sequences of the eps locus prior to and after IS492 insertion and the circle junction sequence with flanking IS492 termini. The 5-bp target duplication-circle junction sequence is underlined, the IS492 insertion in eps is represented by a box, and the IS492 sequence is italicized in the circle junction sequence.
The PCR products obtained for P. atlantica with each primer set were ligated into the plasmid pCR2.1 and sequenced (see Methods and Materials). The primary products for both primer sets had the IS492 right- and left-end sequences separated by a 5-bp junction sequence (Fig. 2C). The 5-bp junction sequence, 5′-CTTGT-3′, matches the 5-bp sequence of eps that is duplicated upon IS492 insertion into the eps target site (Fig. 2C). The sequence for the secondary product from primers 12 and 13 revealed that this product resulted from annealing of primer 12 to a partially homologous sequence within the eps locus.
To determine if formation of this IS492 excision product requires any host factors or conditions specifically provided by P. atlantica, we used the same PCR assay as used for P. atlantica to detect IS492 circle junction formation in E. coli. Plasmids pAG949 and pAG956, containing the 1.2-kbp IS492 element and 134 bp of surrounding eps target sequence (59- and 76-bp flanking sequences from the left and right ends, respectively) within a ColE1 ori plasmid and a p15a ori plasmid, respectively, were introduced into E. coli InvαF′ and HMS174(DE3). The circle junction PCR product was detected in the transformant colonies (Fig. 2B, lanes 5 and 7). The DNA sequences of the PCR products derived from the E. coli strains containing pAG949 and pAG956 contained the same circle junction sequence as had been identified for IS492 in P. atlantica (Fig. 2C).
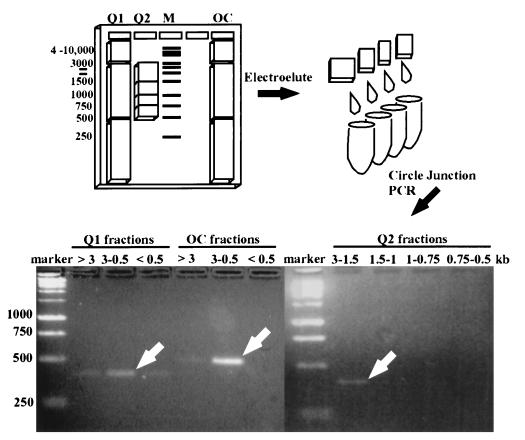
Although the circle junction PCR products support the hypothesis that IS492 forms a circle upon excision in P. atlantica and E. coli, confirmation of an extrachromosomal, circular form of IS492 was needed because the same PCR products could be generated from tandem copies of IS492 in the chromosome (or on a plasmid) and not from a circular form. To address this possibility, we assayed for the presence of a circular form of IS492 in purified extrachromosomal DNA isolated from an E. coli strain containing the 5.2-kb IS492-carrying plasmid pAG949. Extrachromosomal circular DNA, purified from total cellular DNA by using the anion-exchange resin, was fractionated by molecular size by agarose gel electrophoresis (Fig. 3). PCR analysis of the fractionated DNA indicated that fractions containing DNA migrating between 1,500 and 3,000 bp (linear DNA standards) yielded the circle junction PCR product (Fig. 3). As a control, a 1.35-kb DNA fragment from pAG949, containing the 1.2-kb IS element and flanking sequence from the plasmid, was ligated to give an open circular form. This control circle migrated a similar distance relative to the DNA markers as the IS492 circle. The PCR product for the control circle is slightly larger than the circle junction for IS492 circular form, since it has an additional 150 bp of adjacent plasmid DNA (Fig. 3). The apparent molecular size for the circular form of IS492 in this fractionation was slightly higher than the molecular size of the linear form of IS492 (1,200 bp), suggesting that it is an open circle. These results confirm that the circle junction PCR product does indeed correlate with a circular form of IS492 and not tandem copies of the element.
FIG. 3.
Demonstration of circular product from IS492 excision by electrophoretic fractionation of circular DNA from E. coli containing IS492. The illustration shows the 1% agarose gel used to fractionate the open circular IS492 control DNA (OC) and the Qiagen plasmid (circular) DNA preparations (Q1 and Q2) from E. coli containing pAG949, which carries wild-type IS492. Slices were cut from the gel based on comigration with the DNA molecular size standards (1-kb ladder; Promega). DNAs electroeluted from these slices were used as template DNAs for PCR analysis to detect the IS492 circle junction. The PCR products from the wide-range molecular size gel slices (Q1 and OC, >3, 3 to 0.5, and <0.5 kb) and from the narrow-range molecular size slices (Q2, 3 to 1.5, 1.5 to 1, 1 to 0.75, and 0.75 to 0.5 kb) were separated on a 2% agarose gel. The white arrows indicate the primary circle junction PCR product for each group of fractions.
It is important to note that detection of IS492 circles in P. atlantica and in E. coli required incubation of the cells at 4°C for at least 2 h before isolation of the DNA (data not shown). These results suggest that, unlike the circular form of IS117 (13), the IS492 circle is not persistent, and perhaps the lower temperature permits isolation of a transient intermediate in transposition.
Circle formation is transposase dependent.
The predicted transposase for IS492 transposition is encoded by the only long open reading frame of the element (957 bp), designated mooV. To determine if IS492 circle formation is dependent on expression of mooV, we replaced 357 bp of the mooV sequence with an 884-bp DNA fragment containing the chloramphenicol acetyltransferase (cam) gene. This ΔmooV::cam derivative of IS492 was introduced into E. coli on a ColE1-ori plasmid, and transformants were assayed for circle formation by using the colony PCR assay for the circle junction (see Materials and Methods). The IS492 ΔmooV::cam construct gave no circle junction product (Fig. 4, lane 3), indicating no circle formation, whereas the wild-type IS492 construct generated circles that served as a template for producing the circle junction PCR fragment (Fig. 4, lane 2). Several control measures were performed to confirm the negative result for the IS492ΔmooV::cam construct, including determining the template concentration in the reaction mixture and Southern blotting to increase the sensitivity for detection of the PCR product (see Materials and Methods) (data not shown).
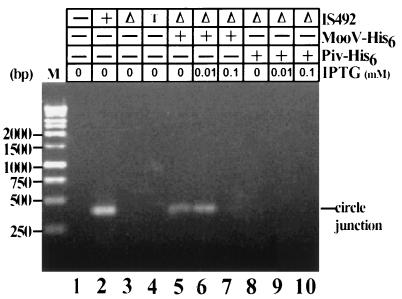
FIG. 4.
Effect of MooV-His6 and Piv-His6 on IS492 circle formation. IS492 circle formation was examined in E. coli HMS174(DE3) by the PCR assay diagrammed in Fig. 1. The circle junction PCR products after agarose gel electrophoresis are shown. MooV-His6 and Piv-His6 (presence or absence is indicated by + or −, respectively) were provided in trans from pAG954 and pAG955, respectively, to the IS492ΔmooV::cam-carrying plasmid, pAG957 (Δ). The levels of MooV-His6 and Piv-His6 were varied by using different concentrations of IPTG for induction. The following plasmids in HMS174(DE3) were included as controls: pCR2.1 (− for IS492), pAG949 (+ for IS492), and pAG903 (T, IS492 target site mutant [see Materials and Methods]). Lane M, Promega 1-kb markers.
Complementation analysis was performed to examine whether the mooV deletion had affected expression of mooV only or the function of a cis-acting site as well. In strain HMS174(DE3), MooV-His6 was supplied in trans to IS492ΔmooV::cam from a compatible plasmid, pAG954, which has mooV translationally fused to six histidine codons at its 3′ end and expressed from the T7 promoter. In this strain the T7 polymerase gene is expressed from the lacUV5 promoter on the λDE3 prophage. Western blot analysis showed that IPTG-induced expression of mooV-his6 produced a 35.7-kDa protein, consistent with the predicted amino acid sequence of the MooV-His6 fusion protein (data not shown). Supplying MooV-His6 in trans resulted in a detectable IS492ΔmooV::cam circle junction PCR product (Fig. 4, lane 6); therefore, the deletion affected only expression of MooV and not the function of a cis-acting site.
To determine how the formation of circle product corresponds to the level of MooV, we quantitated the response of IS492ΔmooV::cam to various concentrations of MooV-His6 by using increasing levels of IPTG to induce expression of mooV-his6. After induction, the DNA was isolated and its concentration was normalized, as described in Materials and Methods; this was followed by PCR analysis for the circle junction. Circle formation did vary with different levels of MooV (Fig. 4, lanes 5 to 7). A low level of circle formation is detected when no IPTG is added; this is likely due to leaky expression of T7 polymerase from the lacUV5 promoter in the absence of IPTG. Optimal circle formation was obtained at approximately 0.01 mM IPTG, and no circle formation could be detected at or above 0.1 mM IPTG. The lack of circle formation when mooV-his6 expression was induced at 0.1 mM IPTG (or higher) could be explained either by repression of IS492 excision at high concentrations of MooV or by a decrease in cell viability with higher levels of MooV-His6. The latter explanation is supported by growth rate and cell viability assays, which showed that induction of mooV-his6 expression at 0.1 or 0.5 mM IPTG results in decreased growth rate or cell inviability, respectively (data not shown).
We also performed the complementation assay for circle formation with an MBP-MooV fusion protein and the wild-type MooV protein supplied in trans from compatible plasmids (see Materials and Methods). Both MBP-MooV and wild-type MooV supported circle formation by IS492ΔmooV::cam (data not shown). Since the site-specific recombinase Piv from M. lacunata has significant homology with the transposases of the IS110-IS492 family elements, we determined the ability of Piv-His6, MBP-Piv, and wild-type Piv (32) to complement for IS492 excision. The complementation assays were performed as with MooV; however, Piv did not mediate detectable circle formation at any level of expression tested (Fig. 4, lanes 8 to 10). Western blot analysis confirmed that Piv-His6, MBP-Piv, and wild-type Piv were expressed in these assays (data not shown).
Precise excision of IS492 and repair of the donor molecule.
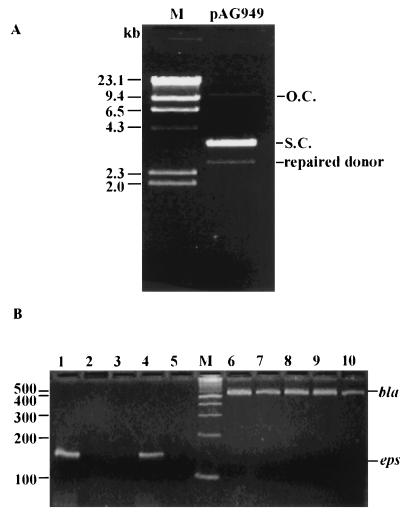
In P. atlantica, excision of IS492 from the eps locus restores expression of eps, indicating that either there is precise exchange of DNA strands in the excision process or the eps target sequence is frequently repaired upon excision of IS492. If circle formation by IS492 corresponds to precise excision from eps, then the repaired donor DNA molecule should be detectable. In the assays for circle formation by IS492 in E. coli, the donor molecules are multicopy plasmids carrying the IS492 element within the eps target sequence. Examination of plasmid DNA carrying wild-type IS492 (pAG949) by electrophoresis on an agarose gel revealed an additional, faster-migrating DNA species. The slower-migrating plasmid DNA was electroeluted from the agarose gel and transformed into naive E. coli cells at a DNA concentration at which the likelihood of cotransformation was negligible. Plasmid preparations from these transformants again yielded the additional faster-migrating DNA on agarose gels (Fig. 5A). PCR amplification was performed on this plasmid preparation by using primers to the eps target sequence; DNA sequencing of this PCR product (Fig. 5B, lane 4) revealed that the sequence matches the eps locus with no IS492 insertion. This result indicates that the faster-migrating species was the donor molecule after precise excision of IS492. The eps target sequence PCR product was not amplified from the same E. coli strain containing the plasmid with IS492ΔmooV::cam within the eps sequence or from the E. coli strain containing the parent plasmid pCR2.1 (Fig. 5B, lanes 3 and 5, respectively). Repair of the donor molecule can be detected for IS492ΔmooV::cam when MooV-His6 is supplied in trans and its expression is induced by IPTG (Fig. 4B, lanes 1 and 2). Thus, precise excision was detected only under conditions that also gave IS492 circles. These results indicate that excision of IS492 and repair of the donor correlate with circle formation. However, every excision event that results in circle formation may not also give a repaired donor molecule; the results given in Fig. 4, lane 5, show that circle formation can be detected for IS492::cam at very low levels of mooV-his6 expression (in the absence of IPTG), but the repaired eps target sequence is not detected by PCR (Fig. 5B, lane 2) until induction of mooV-his6 expression at 0.01 mM IPTG.
FIG. 5.
Detection of the repaired donor DNA after IS492 excision. (A) Plasmid preparation of pAG949 after agarose gel electrophoresis. The following forms of the plasmid are indicated to the right of the gel: open circular (O.C.), supercoiled (S.C.), and the repaired pAG949 after excision of IS492 (repaired donor). The DNA molecular size marker (lane M) is the Promega λ/H3 markers, with sizes indicated to the left. (B) PCR assay for the repaired donor DNA, using primers complementary to eps (lanes 1 to 5), and controls for addition of template to the reaction mixtures, using primers complementary to bla (lanes 6 to 10). Strains tested for the repaired donor DNA were HMS174(DE3) containing pCR2.1 (no IS492) (lanes 5 and 6), pAG949 (wild-type IS492) (lanes 4 and 7), pAG957 (IS492ΔmooV::cam) (lanes 3 and 8), pAG957 and pAG954 (IS492ΔmooV::cam plus mooV-his6) (lanes 2 and 9), and pAG957 and pAG954 (inducing conditions [0.01 mM IPTG]) (lanes 1 and 10). The molecular size marker (lane M) is the New England Biolabs 100-bp marker with sizes (in base pairs) indicated to the left.
To further examine excision of IS492 from the eps sequence, we used reporter plasmids in which IS492 or IS492ΔmooV::lacZ (with 23-bp flanking eps sequences on the left and right ends of the element) is inserted between the chloramphenicol acetyltransferase gene (cam) and its promoter (pcam) on the plasmid pACYC184 (pAG993 or pAG994, respectively). Transformants with pAG993 gave a mixed population of plasmids containing the IS492 insertion or the target eps sequence from which IS492 had been precisely excised (data not shown); therefore, all transformant colonies exhibited chloramphenicol resistance (Cmr) (Table 2). Insertion of IS492ΔmooV::lacZ (pAG994) downstream of pcam resulted in termination of transcription before the cam gene and chloramphenicol sensitivity (Cms) in the absence of MooV (Table 2). When MooV was provided in trans as an MBP-MooV fusion protein from pMalE-MooV, the frequency of excision, as determined by the number of Cmr colonies versus the total number of Tetr colonies that contain the excision reporter plasmid, was approximately 10−5 for IS492ΔmooV::lacZ (Table 2). Excisant plasmids were sequenced for both the wild-type excision substrate and the IS492ΔmooV::lacZ substrate and were found to have undergone precise excision of the IS element, thus restoring the eps sequence. The 23-bp eps sequences flanking the left and right ends of the IS element contain the 5-bp duplication; therefore, after excision, the eps sequence is the total 46-bp flanking sequences minus one copy of the 5-bp duplication. This 41-bp eps sequence did not affect cam expression, as evidenced by the value of 1 for the ratio of Cmr to Tetr colonies for one of the excisants (pAG992.2) that was derived from pAG993 (Table 2). Colony PCR assays for circle junctions were performed on colonies for all strains used in the excision assays. Again, circle formation correlated with excision of IS492; however, unlike the results of the PCR assays for excision compared to circle formation under noninducing conditions (Fig. 5B, lane 2, and Fig. 4, lane 5), the genetic assay indicated that both excision and circle formation occurred at the low level of MooV expression in the absence of IPTG (Table 2). This result probably reflects the fact that different levels of MooV-His6 or MBP-MooV are expressed in the absence of IPTG due to the different expression systems (see Materials and Methods).
TABLE 2.
IS492 excision from the eps target sequence in E. coli
| Plasmid(s) | IS492 genotype (source of MooVa) | Cells/ml
|
Frequency of excision (Cmr/Tetr) | Circle formation (PCR assay) | |
|---|---|---|---|---|---|
| Tetrb | Cmrc | ||||
| pAG993 | Wild type (IS492+) | 6.0 × 107 | 5.7 × 107 | 0.95 | + |
| pAG994 | ΔmooV (IS492ΔmooV::lacZ) | 1.4 × 108 | 0 | 0 | − |
| pAG994, pMalC-2 | ΔmooV | 2.6 × 108 | 0 | 0 | − |
| pAG994, pMal-MooV | ΔmooV (malE-mooV) | 2.3 × 108 | 2.0 × 103 | 0.87 × 10−5 | + |
| pAG992.2 | Excisant (IS492−)d | 1.3 × 108 | 1.3 × 108 | 1.00 | − |
Plasmids were maintained in E. coli TOP10 cells (see Materials and Methods).
Cultures were grown in LB broth without IPTG, and dilutions were plated on LB agar containing 12.5 μg of tetracycline per ml.
The same culture dilutions were plated on LB agar containing 80 μg of chloramphenicol per ml.
pAG992.2 is the product of precise excision of IS492 from pAG993.
IS492 excision requires flanking eps sequences.
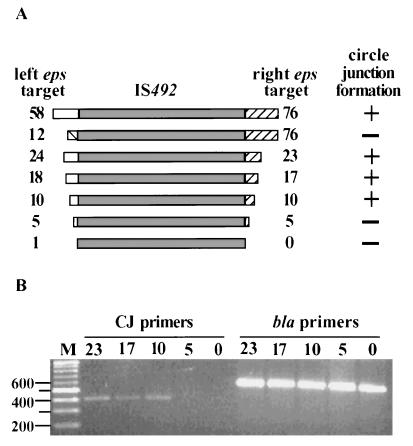
The ability of IS492 to be excised precisely and restore the donor sequence is a very unusual feature for a transposable element (8, 19). Another unusual aspect of IS492 is that it does not have inverted-repeat DNA sequences at the ends of the element; for most IS elements, the short terminal inverted-repeat DNA sequences are required for transposase recognition and cleavage of the DNA at the ends of the element (for a review, see reference 19). These features of IS492 suggest that the sequence flanking the IS element at its insertion sites may play an essential role in transposase binding and in the excision reaction. For example, the 5-bp sequence that is duplicated upon insertion of IS492 may function very much like the attL and attR sequences in excisive site-specific recombination for bacteriophage λ (for a review, see reference 15). To determine if flanking DNA sequences affected circle formation of IS492, a deletion analysis of the eps target sequence was performed. Our original plasmid constructs containing the wild-type IS492 (pAG949 and pAG956) carried 58 and 76 bp of eps target sequence flanking the left and right ends of the inserted element, respectively. This eps target sequence was reduced systematically on either side of IS492 in a series of plasmids (see Materials and Methods), resulting in IS492 flanked by 24 and 23 bp, 18 and 17 bp, 10 and 10 bp, 5 and 5 bp, or 1 and 0 bp of eps target sequence (Fig. 6A). PCR amplification to detect the circle junction, using DNA from E. coli carrying each of these derivative plasmids as a template, indicated that having between 5 and 10 bp of the flanking eps target sequence is essential for detection of the circle junction fragment and, therefore, IS492 excision (Fig. 6B).
FIG. 6.
Target site requirements for IS492 excision. (A) Schematic diagram showing the extent of eps target sequence on either side of IS492 (left flanking sequence, open box; right flanking sequence, hatched box). The number at either end of IS492 indicates the number of nucleotides of eps sequence. Whether a circle junction PCR product could be detected is indicated as + or −. (B) Agarose gel electrophoresis of circle junction (CJ primers) PCR products and bla (bla primers) control products generated from plasmids carrying IS492 sequence with the flanking sequences shown in panel A. Lane M, Promega 1-kb marker, with the sizes of some of the markers (in base pairs) shown to the left.
The 5-bp eps target sequence (5′-CTTGT-3′) is normally in direct repeats at the ends of the element. To determine whether excision of IS492 required that this sequence be in direct repeats at the ends of the element, we replaced the eps sequence at the left end of the element with 12 bp of flanking sequence from the right end, but in an inverted orientation. The resulting plasmid, pAG903, carried the IS492 element with the flanking 5 bp in an inverted repeat, i.e., 5′-ACAAG/IS492/CTTGT-3′ (Fig. 6A). This IS492 mutant is not excised to form circles, as demonstrated by PCR analysis (Fig. 4, lane 4). A minor, approximately 1,600-bp PCR product is detected instead when primers which normally generate the 405-bp product in our standard assay and used. Upon sequencing, it appears that this product might be the result of an aberrant one-ended excision event within pAG903 (data not shown).
Other insertion sites for IS492 in P. atlantica DB27.
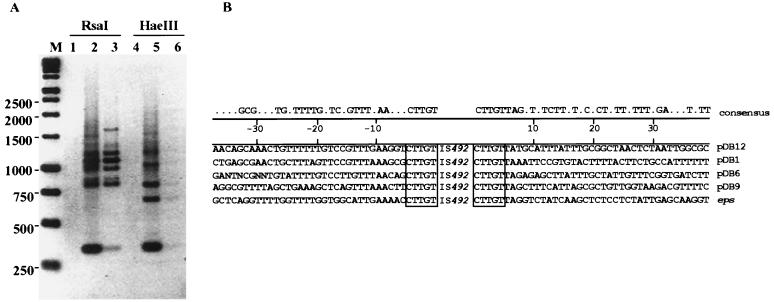
When multiple circle junction PCR products were sequenced for the circular form of IS492 in P. atlantica, all of the junctions had the identical 5-bp sequence that is duplicated at the eps locus upon insertion of IS492. That same 5-bp sequence is critical in excision of the IS element in E. coli, based on our deletion analysis described above. Therefore, it was of interest to determine whether this 5-bp sequence is found at the other insertion sites for IS492 on the chromosome of P. atlantica. We used the inverse PCR method described by Ochman et al. (24) for amplifying flanking DNA to determine the sequences of other IS492 target sites in P. atlantica DB27. Digestion of total cellular DNA with RsaI or HaeIII, which have no recognition sequences within the IS492 element, results in linear restriction fragments from the chromosomal DNA containing the full IS element with some of its flanking sequence from both the left and right ends at each of the IS492 insertion sites. Dilution and ligation of the linear fragments result in open circles that are then used as templates in PCRs with the primer set (primers 12 and 13) used in the circle junction assays (Fig. 2A). Thus, the PCR products contained the ends of IS492 separated by the flanking DNA sequence from either end of the element up to the nearest RsaI or HaeIII site, where ligation occurred. The different-sized PCR products generated from the ligated dilutions of the RsaI- and HaeIII-restricted chromosomal DNA (Fig. 7A) should each represent a different insertion site for IS492 on the chromosome of P. atlantica; as expected, the 405-bp circle junction fragment is also seen from each amplification with the inverse PCR primers.
FIG. 7.
Identification of other target sites in P. atlantica DB27. (A) Agarose gel electrophoresis of PCR products generated from inverse PCR (described in Materials and Methods) of diluted and ligated P. atlantica DB27 chromosomal DNA following digestion with either RsaI (lanes 1 to 3) or HaeIII (lanes 4 to 6). Either 0.25 μg (lanes 1 and 4), 0.025 μg (lanes 2 and 5), or 0.0005 μg (lanes 3 and 6) of P. atlantica chromosomal DNA was used as a template in inverse PCRs. Lane M, Promega 1-kb marker. The inverse image of the UV-illuminated ethidium bromide-stained gel is shown. (B) Alignment of target site sequences. The duplicated target sequence is indicated with a box.
The PCR products shown in lanes 3 and 5 of Fig. 7A were cloned in pCR2.1 and sequenced, as described in Materials and Methods. The DNA sequences for different IS492 target sites are shown in Fig. 7B. All of these target sites have the same 5-bp duplication as seen for the eps target site (5′-CTTGT-3′); therefore, any of these copies of IS492 could be the source of the circular form of IS492 detected in P. atlantica. Consistent with this result, we found that the IS492 circle junction could be detected in EPS+ cells which lack the eps::IS492 insertion (data not shown). Comparison of the flanking sequence beyond the 5-bp duplication for the eps and four other insertion sites indicates that there is limited sequence conservation at the left and right ends of the inserted elements (Fig. 7B). Comparison searches of the databases with the insertion site sequences (by using the Wisconsin Package, version 9.1 [Genetics Computer Group]) indicate that one of the other insertion sites (pDB12) appears to be within an IS10-like transposase gene. No significant homology or similarity to other genes in prokaryotes or eukaryotes could be found for the other insertion sites.
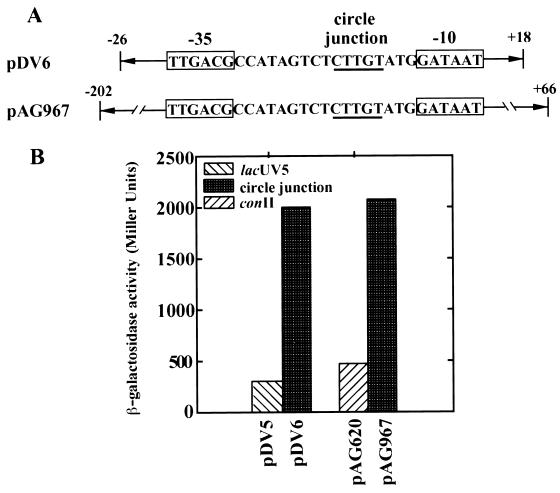
Assembly of a strong promoter by circle junction formation.
A possible advantage for an insertion element to have a circular intermediate has been proposed based on studies of IS911 transposition (33, 34). For IS911, it has been shown that inverse repeat left- and right-end joining results in the formation of a strong promoter which can drive transposase expression during a brief time when the presence of the transposase is essential for continued survival. We examined whether IS492, too, might possess such a regulatory mechanism for transposase expression. The 5-bp circle junction flanked by 26 bp of the left end and 18 bp of the right end of IS492 was inserted into a multicopy vector used for promoter identification in front of a promoterless lacZ (pDV6) (Fig. 8A). Positive transformants were dark blue on LB agar plates containing X-Gal, thus indicating that the circle junction of IS492 does form a promoter driving β-galactosidase expression. β-Galactosidase assays were performed with these cells as well as a reference strain containing the same plasmid but with a lacUV5 constitutive promoter driving β-galactosidase expression (pDV5). β-Galactosidase levels were 6.7-fold higher for the IS492 circle junction promoter, with 2,000 Miller units of activity versus 300 Miller units for the lacUV5 control promoter (pDV6 versus pDV5 in Fig. 8B). The circle junction promoter was also cloned in single copy on pNN387 (9) to estimate its strength. A larger fragment containing the circle junction was cloned, with 66 bp of the left end and 202 bp of the right end of IS492, in front of a promoterless lacZY; this plasmid construction was designated pAG967 (Fig. 8A). Positive transformants were dark blue on LB agar containing X-Gal, suggesting the circle junction represents a strong promoter. β-Galactosidase assays were performed with these cells, and the activity levels were compared to those in control cells containing the same plasmid with a constitutive tac promoter driving lacZα expression; this control tac promoter, conII, has all regulatory elements deleted (9). The IS492 circle junction promoter demonstrated 4.3-fold higher β-galactosidase activity than the constitutive tac promoter, with 1,700 versus 400 Miller units (Fig. 8B).
FIG. 8.
Promoter activity associated with the IS492 circle junction sequence. (A) The potential promoter sequence created when IS492 ends are joined at the circle junction is indicated by boxes. Relevant regions of plasmids pDV6 and pAG967, used to evaluate circle junction promoter activity, are schematically shown. Arrows and numbers (base pairs) are used to indicate the region of IS492 included on either side of the circle junction on these plasmids. The circle junction sequence is underlined. (B) Promoter activity measured as β-galactosidase activity (Miller units) in plasmids having constitutive control promoters (lacUV5 [pDV5] or conII [pAG620]) or the circle junction and surrounding IS492 sequence (pDV6 or pAG967). Besides having different amounts of surrounding IS492 sequence, pDV6 and pAG967 have different copy numbers (multiple and single copy, respectively). The relative β-galactosidase activities for these promoter constructions remained constant in multiple experiments.
DISCUSSION
IS492 is a member of an atypical family of insertion elements that do not have inverted repeats at their ends and whose transposition is mediated by an apparently novel group of DNA recombinases (16). Transposition of IS492 controls phase variation of EPS production in the marine bacterium P. atlantica. The frequency of switching from EPS− to EPS+ and, apparently, the frequency of IS492 precise excision from a locus that is essential for EPS production (eps) varies more than 104-fold depending on the growth conditions (2). These unique features of the IS element and its transposase, along with the control of IS492 excision frequency by environmental conditions, suggest that there are likely to be many interesting features to the regulation of IS492 transposition. In this study we have begun to address the mechanism for IS492 transposition.
In PCR-based assays we detected a circular form of IS492 in P. atlantica DB27. The same circular form of IS492 was also detected in E. coli when we introduced wild-type IS492 on multi- or single-copy plasmids by transformation. DNA sequencing of the junction of the ends of the element in the circular form showed that a 5-bp sequence linked the ends of IS492. This 5-bp sequence corresponds to the eps sequence that is duplicated at the insertion site for IS492. In previous Southern analyses of IS492 transposition (2), only the eps-associated copy of the element appeared to have been excised. This suggested to us that perhaps the 5-bp eps sequence defined the circular form as being generated only from the copy of IS492 in the eps site. We identified the other insertion sites of IS492 on the P. atlantica chromosome by inverse PCR (24) and found that all of the target sites had this same 5-bp sequence; therefore, any of these copies of the element could be the source for the IS492 circle.
The conservation of this 5-bp sequence at all of the target sites for IS492 suggests an interesting possibility for the mechanism of transposition of IS492. Perhaps the transposases for IS492 and the related IS110 family of insertion elements utilize a recombination reaction more like the site-specific recombinases such as the integrases of the λ-Int family. In this scenario, the 5-bp target sequence would function similarly to the core sequences of the attB/attP and attR/attL sites, where DNA cleavage and precise strand exchange occur in bacteriophage λ integration and excision, respectively (for a review, see reference 15). The homology of these transposases with the site-specific invertase of M. lacunata lends some support to this hypothesis. If the 5-bp sequence does function in this manner, then it would be essential for the excision process. A stepwise deletion analysis of the flanking DNA sequence showed that, indeed, between 5 and 10 bp of the flanking sequence is required for excision and circle formation. Inverting the orientation of 12 bp of the flanking sequence at one end of the element also affected circle formation; no circles could be detected, but instead of a site-specific inversion of the element, an apparent one-ended transposition event was detected by PCR. We are currently undertaking a more detailed analysis of the essential sequences at the ends of IS492 and in the flanking DNA to address their role in the excisive recombination reaction.
While some transposable elements, such Tn7, Tn10, and bacteriophage Mu, show various extents of target site selectivity for insertion, a critical role for flanking host DNA sequences in transposition is exceptional. Mu transposase shows a conditional effect of flanking host sequences on cleavage at the donor site in vitro (35), and the transposases of conjugative transposons, which share the catalytic amino acid motif of the λ-Int site-specific recombinases, exhibit dependence on flanking host sequences for frequency of transposition (14a). The requirement for the 5-bp duplicated target sequence for excision of IS492 is truly an unusual feature for an IS element. Another unusual aspect of IS492 excision is that it appears to frequently result in restoration of the target DNA sequence. In both P. atlantica (2, 3) and E. coli (Fig. 5), the donor DNA is repaired after excision of IS492. This ability to excise precisely is not necessarily linked to circle formation. The related IS element IS117 from Streptomyces coelicolor has been shown to form a minicircle which may be an intermediate in transposition; however, precise excision of IS117 is never seen (18). In fact, it has been suggested that all IS117 transposition events are replicative in S. coelicolor and Streptomyces lividans (31). Thus, although our results show that circle formation and precise excision require the same trans-acting factor, MooV, and at least some of the same cis-acting DNA sequences in the flanking host DNA, the questions of whether precise excision and circle formation by IS492 are directly linked and of what role circle formation plays in the transposition reaction still remain.
Formation of circular products in transposition is not unique to the IS110-IS117 family of insertion elements; circles have been detected in a number of other systems, including IS3 (29), IS2 (17), IS150 (19), IS10 (22), IS1 (19, 30), IS91 (20), and IS911 (19, 25). IS911 circles have been shown to actively integrate in vivo and in vitro, supporting their possible role as transposition intermediates (33, 34). Strong evidence supporting a functional role of the IS911 circle in transposition comes from regulation studies of the IS911 transposase. The assembly of IS911 inverse repeat right and left ends as a circle junction creates a strong promoter which is stronger than the indigenous promoter driving transposase transcription; thus, it has been suggested that in a postexcision situation where a transposon circle has been formed, the upregulation of transposase transcription may increase the likelihood of integration occurring (33).
IS492, like IS911, forms a strong promoter from the left and right ends when joined by the 5-bp host target sequence at the circle junction (Fig. 8). The high level of transcription from this newly assembled promoter upon excision of IS492 could either positively or negatively regulate reinsertion of the element. The new promoter is appropriately positioned to initiate increased expression of mooV. In E. coli, the amount of circle formation by a ΔmooV derivative of IS492 was dependent on the level of mooV expression from a plasmid in trans (Fig. 4). In general, for most transposable elements, there are multiple levels of regulation to keep transposition frequencies low (for a review, see reference 8). The decrease in circle formation at high levels of MooV (Fig. 4) may reflect autoregulation of transposase expression, as is seen for IS1 and Tn7 (8, 30), although the decrease in cell viability upon induction of MooV complicates this analysis. Alternatively, a negative effect of the high level of transcription from the promoter overlapping the ends of the element may be that RNA polymerase then prevents binding of MooV to the ends. An assay for insertion of IS492 is needed to address the role of the IS492 circle and the circle junction promoter. We have not observed insertion of IS492 into the eps sequence or a new locus in E. coli (data not shown), which suggests that regulatory or accessory recombination factors that are involved in transposition of IS492 in P. atlantica are absent in E. coli. We are pursuing the characterization of the mechanism for IS492 transposition and its regulation genetically in P. atlantica and E. coli and biochemically in an in vitro recombination system.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM 49794-05 from the National Institutes of Health and Presidential Young Investigator award MCB-9396003 from the National Science Foundation to A.C.G.
We thank Russell Karls, Mick Chandler, Gordon Churchward, and June Scott for helpful discussions and critical review of the manuscript. We appreciate the contribution of Wendy Mahler to the construction of pAG956.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1993. [Google Scholar]
- 2.Bartlett D H, Wright M E, Silverman M. Variable expression of extracellular polysaccharide in the marine bacterium Pseudomonas atlantica is controlled by genome rearrangement. Proc Natl Acad Sci USA. 1988;85:3923–3927. doi: 10.1073/pnas.85.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett D H, Silverman M. Nucleotide sequence of IS492, a novel insertion sequence causing variation in extracellular polysaccharide production in the marine bacterium Pseudomonas atlantica. J Bacteriol. 1989;171:1763–1766. doi: 10.1128/jb.171.3.1763-1766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belas R, Bartlett D, Silverman M. Cloning and gene replacement mutagenesis of a Pseudomonas atlantica agarase gene. Appl Environ Microbiol. 1988;54:30–37. doi: 10.1128/aem.54.1.30-37.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan K Y, Baumann L, Garza M M, Baumann P. Two new species of Alteromonas: Alteromonas espejiana and Alteromonas undina. Int J Syst Bacteriol. 1978;28:217–222. [Google Scholar]
- 6.Corpe W A. Attachment of marine bacteria to solid surfaces. In: Manley R S, editor. Adhesion in biological systems. New York, N.Y: Academic Press; 1970. pp. 73–87. [Google Scholar]
- 7.Corpe W A. Proceedings of the 3rd International Congress on Marine Corrosion and Fouling. Evanston, Ill: Northwestern University Press; 1973. Microfouling: the role of primary film forming marine bacteria; pp. 598–609. [Google Scholar]
- 8.Craig N. Transposition. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2339–2362. [Google Scholar]
- 9.Elledge S J, Davis R W. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 1989;3:185–197. doi: 10.1101/gad.3.2.185. [DOI] [PubMed] [Google Scholar]
- 10.Froehlich B J, Scott J R. A single promoter-cloning vector for use in E. coli. Gene. 1991;108:99–101. doi: 10.1016/0378-1119(91)90492-t. [DOI] [PubMed] [Google Scholar]
- 11.Gauthier G, Gauthier M, Christen R. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int J Sys Bacteriol. 1995;45:755–761. doi: 10.1099/00207713-45-4-755. [DOI] [PubMed] [Google Scholar]
- 12.Gawron-Burke C, Clewell D B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984;159:214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson D J, Lydiate D J, Hopwood D A. Structural and functional analysis of the mini-circle, a transposable element of Streptomyces coelicolor A3(2) mini-circle to and from a cloned target site and into secondary chromosomal sites. Mol Gen Genet. 1989;224:65–71. [Google Scholar]
- 14.Hughes K T, Youderian P, Simon M I. Phase variation in Salmonella, analysis of Hin recombinase and hix recombination site interaction in vivo. Genes Dev. 1988;2:937–948. doi: 10.1101/gad.2.8.937. [DOI] [PubMed] [Google Scholar]
- 14a.Jaworski D D, Clewell D B. Evidence that coupling sequences play a frequency-determining role in conjugative transposition of Tn916 in Enterococcus faecalis. J Bacteriol. 1994;176:3328–3335. doi: 10.1128/jb.176.11.3328-3335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landy A. Mechanistic and structural complexity in the site-specific recombination pathway of Int and Flp. Curr Opin Genet Dev. 1993;3:699–707. doi: 10.1016/s0959-437x(05)80086-3. [DOI] [PubMed] [Google Scholar]
- 16.Lenich A G, Glasgow A C. Amino acid sequence homology between Piv, an essential protein in site-specific DNA inversion in Moraxella lacunata, and transposases of an unusual family of insertion elements. J Bacteriol. 1994;176:4160–4164. doi: 10.1128/jb.176.13.4160-4164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis L A, Grindley N D F. Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol Microbiol. 1997;25:517–529. doi: 10.1046/j.1365-2958.1997.4871848.x. [DOI] [PubMed] [Google Scholar]
- 18.Lydiate D J, Ikeda H, Hopwood D A. A 2.6 kb DNA sequence of Streptomyces coelicolor A3(2) which functions as a transposable element. Mol Gen Genet. 1986;203:79–88. doi: 10.1007/BF00330387. [DOI] [PubMed] [Google Scholar]
- 19.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendiola M V, Bernales I, de la Cruz F. Differential roles of the transposon termini in IS91 transposition. Proc Natl Acad Sci USA. 1994;91:1922–1926. doi: 10.1073/pnas.91.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Morisato D, Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987;51:101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- 23.Nash H. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2363–2376. [Google Scholar]
- 24.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking DNA by inverse PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press, Inc.; 1990. pp. 219–227. [Google Scholar]
- 25.Polard P, Prere M F, Fayet O, Chandler M. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 1992;11:5079–5090. doi: 10.1002/j.1460-2075.1992.tb05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reichelt J L, Baumann P. Change of the name Alteromonas marinopraesens (ZoBell and Upham) Baumann et al. to Alteromonas haloplanktis (ZoBell and Upham) comb. nov. and assignment of strain ATCC 23821 (Pseudomonas enali) and strain c-A1 of DeVoe and Oginsky to this species. Int J Syst Bacteriol. 1973;23:438–441. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Schneider K, Beck C F. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene. 1986;42:37–48. doi: 10.1016/0378-1119(86)90148-4. [DOI] [PubMed] [Google Scholar]
- 29.Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- 30.Sekine Y, Eisaki N, Kobayashi K, Ohtsubo E. Isolation and characterization of IS1 circles. Gene. 1997;191:183–190. doi: 10.1016/s0378-1119(97)00057-7. [DOI] [PubMed] [Google Scholar]
- 31.Smokvina T, Henderson D J, Melton R E, Brolle D, Kieser T, Hopwood D. Transposition of IS117, the 2.5 kb Streptomyces coelicolor A3(2) ‘minicircle’: roles of open reading frames and origin of tandem insertions. Mol Microbiol. 1994;12:459–468. doi: 10.1111/j.1365-2958.1994.tb01034.x. [DOI] [PubMed] [Google Scholar]
- 32.Tobiason D M, Lenich A G, Glasgow A C. Multiple DNA binding activities of the novel site-specific recombinase, Piv, from Moraxella lacunata. J Biol Chem. 1999;274:9698–9706. doi: 10.1074/jbc.274.14.9698. [DOI] [PubMed] [Google Scholar]
- 33.Ton-Hoang B, Betermier M, Polard P, Chandler M. Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. EMBO J. 1997;16:3357–3371. doi: 10.1093/emboj/16.11.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ton-Hoang B, Polard P, Chandler M. Efficient transposition of IS911 circles in vitro. EMBO J. 1998;17:1169–1181. doi: 10.1093/emboj/17.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Z, Chaconas G. Flanking host sequences can exert an inhibitory effect on the cleavage step of the in vitro Mu DNA strand transfer reaction. J Biol Chem. 1992;267:9552–9558. [PubMed] [Google Scholar]
- 36.Yaphe W. The use of agarase from Pseudomonas atlantica in the identification of agar in marine algae (Rhodophycae) Can J Microbiol. 1957;3:987–993. doi: 10.1139/m57-109. [DOI] [PubMed] [Google Scholar]