Summary
Seed size is one of the most important agronomic traits determining the yield of crops. Cloning the key genes controlling seed size and pyramiding their elite alleles will facilitate yield improvement. To date, few genes controlling seed size have been identified in soybean, a major crop that provides half of the plant oil and one quarter of the plant protein globally. Here, through a genome‐wide association study of over 1800 soybean accessions, we determined that natural allelic variation at GmST05 (Seed Thickness 05) predominantly controlled seed thickness and size in soybean germplasm. Further analyses suggested that the two major haplotypes of GmST05 differed significantly at the transcriptional level. Transgenic experiments demonstrated that GmST05 positively regulated seed size and influenced oil and protein contents, possibly by regulating the transcription of GmSWEET10a. Population genetic diversity analysis suggested that allelic variations of GmST05 were selected during geographical differentiation but have not been fixed. In summary, natural variation in GmST05 determines transcription levels and influences seed size and quality in soybean, making it an important gene resource for soybean molecular breeding.
Keywords: soybean, GmST05, natural allelic variation, GWAS, seed size, oil and protein contents
Introduction
Seeds serve as one of the most important organs for plant propagation, and they also provide the major source for crop food. In crop breeding, seed size is one of the most important agronomic traits that need to be considered. For instance, seeds of cultivated crops are usually larger than those of their corresponding wild ancestors, showing parallel selection (Doebley et al., 2006). How seed size is determined is not only an important fundamental question in developmental biology but also important for understanding the fitness, adaptation and yield determination of crops (Li et al., 2019). The global human population is continuously increasing and the demand for food will be higher in the future (Bodirsky et al., 2015). Therefore, the identification of key genes controlling important agronomic traits, such as seed size, and their utilization in molecular breeding are essential for crop yield improvement.
Seed size can be described by three main dimensions: length, width, and thickness (Xing and Zhang, 2010). In recent decades, a number of seed size regulatory genes have been cloned and characterized in cereal crops, particularly in rice (Chen et al., 2022; Duan and Li, 2021; Li and Li, 2016; Li et al., 2019; Xing and Zhang, 2010; Zuo and Li, 2014). Some of these characterized genes have been widely utilized in rice breeding, such as GS3, which functions as a negative regulator of grain and organ size (Fan et al., 2006; Mao et al., 2010); OsBG1, which functions as a positive regulator of grain length, grain width and 1000‐grain weight (Liu et al., 2015); and GW2, which can significantly enhance grain width, weight and yield via loss‐of‐function alleles (Song et al., 2007). Moreover, the orthologous genes of GW2 or OsBG1 showed conserved functions in controlling seed size in other cereals (Bednarek et al., 2012; Li et al., 2010; Sestili et al., 2019; Simmons et al., 2020; Su et al., 2011; Zhang et al., 2018). In addition, it was found that some grain size controlling genes could also affect grain quality. For instance, GW8/OsSPL16 and GW7/SLG7, two important regulators of grain size and shape in rice also strongly influence grain appearance quality (Wang et al., 2012; Wang et al., 2015a, 2015b, 2015c, 2015d; Zhou et al., 2015a, 2015b).
Soybean (Glycine max L. Merr.) is a major oilseed crop that supplies the plant protein and oil for humans and animals. Compared with that of other major crops, such as rice, wheat and maize, the yield of soybean is approximately two‐ to threefold lower. Increasing yield is an important and urgent task in soybean breeding (Liu et al., 2020a, 2020b). The yield of soybean is a complex trait that is determined by many components, among which seed size is one of the primary indexes. To date, although a number of quantitative trait loci (QTLs) controlling seed size and seed weight have been identified in soybean, few genes have been isolated and characterized (Zhang et al., 2022). Of the characterized genes, some were found to have conserved functions relative to those of their orthologs from other species, such as the GmCYP78A gene family (Du et al., 2017; Wang et al., 2015a, 2015b, 2015c, 2015d; Zhao et al., 2016a, 2016b), GmBS1 (Ge et al., 2016), GmCWI (Tang et al., 2016) and GmKIX8‐1 (Nguyen et al., 2021). To pyramid elite alleles in molecular breeding, the identification of functional genes with natural allelic variations is essential (Tian et al., 2021). Several seed size and quality regulatory genes with natural allelic variations in the soybean germplasm were cloned, such as GmPP2C‐1 (Lu et al., 2017), GmOLEO1 (Zhang et al., 2019), GmSWEET10a and GmSWEET10b (Miao et al., 2020; Wang et al., 2020; Zhang et al., 2020a, 2020b), GmST1 (Li et al., 2022) and cqSeed protein‐003 (Fliege et al., 2022; Marsh et al., 2022). Moreover, GmSWEET10a and GmSWEET10b were also found to have an effect on oil content and protein content simultaneously (Wang et al., 2020). The identification of more genes related to seed size will facilitate molecular breeding for yield improvement in soybean.
In this study, through a genome‐wide association study (GWAS) of over 1800 soybean accessions, we identified a significant association locus on chromosome 5 that predominantly controlled seed thickness. Further haplotype analyses revealed that allelic variation in a candidate gene, GmST05 (Seed Thickness on Chromosome 5), served as a major contributor to seed size in soybean germplasm. Molecular and transgenic analyses demonstrated that GmST05 not only positively regulated seed size but also influenced oil and protein content. Our study provides a useful genetic resource for soybean yield and quality improvement.
Results
GWAS revealed GmST05 as a regulator of seed thickness in soybean
The soybean seed morphology can be described from three dimensions: thickness (diameter vertical to hilum), length (diameter parallel to hilum) and width (diameter from hilum to the back of the seed; Figure S1a). To identify the genes controlling the seed thickness in soybean germplasm, we phenotyped 1853 accessions from our previous resequencing panel (Liu et al., 2020a, 2020b) in 2016 and 2017. We found that seed thickness of the phenotyped accessions was strongly correlated between the two independent years (Figure S1b), indicating that seed thickness is largely genetically controlled in soybean. The GWAS performing using a mixed linear model revealed a stable association signal across the 2 years in a 180‐kb interval block (from 43.58 to 43.76 Mb) on chromosome 5 (Figure 1a‐c; Figure S1c‐h). This region overlapped with several previously reported QTLs for seed weight, seed size, and oil and protein contents (Figure 1c) (Brummer et al., 1997; Jun et al., 2014; Kabelka et al., 2004; Pathan et al., 2013; Specht et al., 2001; Wang et al., 2014).
Figure 1.
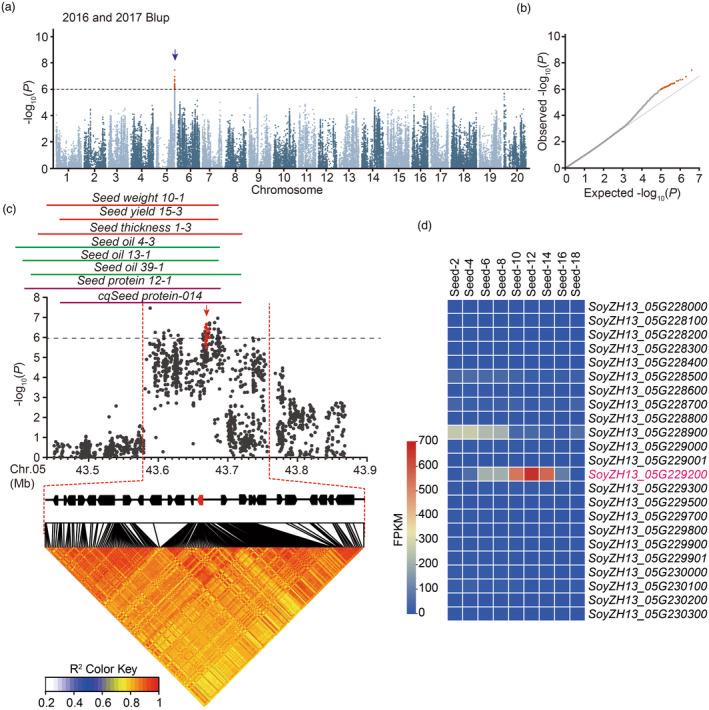
GWAS of seed thickness in the soybean germplasm. (a) Manhattan plot of GWAS results for seed thickness from 2016 and 2017 best linear unbiased prediction (BLUP) data. (b) Quantile–quantile plot of the GWAS results under a mixed linear model (MLM). (c) Local Manhattan plot (top) and linkage disequilibrium plot (bottom) for SNPs surrounding the peak on chromosome 5. The red dashed lines indicate the candidate region for the peak. The red plot indicates the nucleotide variation of GmST05. The solid lines above the plot represent the genomic locations of quantitative trait loci (QTLs) retrieved from SoyBase (https://soybase.org/). The red, green and orange lines are QTLs for seed size (yield), seed oil content and protein content, respectively. (d) A heat map for candidate genes located in the candidate region. The colour key (blue to red) represents gene expression (fragments per kilobase per million mapped reads, FPKM).
In this interval, 23 protein‐coding genes were annotated (Figure 1c) according to the reference genome of ZH13 (Shen et al., 2018; Shen et al., 2019). Transcriptome data demonstrated that among the 23 genes, SoyZH13_05G229200 exhibited significantly higher expression during seed development (Figure 1d), particularly in the suspensor, embryonal axis, embryo and endosperm regions (Figure S2a). SoyZH13_05G229200 encodes a protein homologue of Arabidopsis Mother of TFL1 and FT (MFT), which belongs to the phosphatidylethanolamine‐binding protein (PEBP) family (Figure S2b). Based on the association polymorphisms, SoyZH13_05G229200 could be classified into two major haplotypes in the studied accessions (Figure 2a; Figure S3). Haplotype I exhibited a significantly greater seed thickness than haplotype II (Figure 2b). The two haplotypes of the SoyZH13_05G229200 protein had the same localization in the nucleus and cytoplasm (Figure S2c). SoyZH13_05G229200 was considered a candidate gene for seed thickness in soybean and named GmST05.
Figure 2.
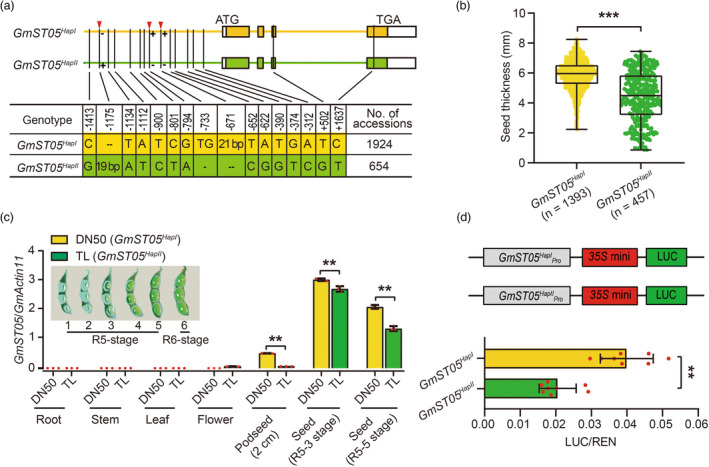
Two GmST05 haplotypes exhibiting differential expression levels. (a) Schematic representation of the two haplotypes of GmST05. (b) Comparison of seed thickness between two different haplotypes in 1853 soybean accessions. Each dot represents the seed thickness of an accession. All values represent the means ± SDs. Student's t test; *** P < 0.001. (c) qRT‐PCR results of GmST05 expression levels in various organs in DN50 and TL, which contained different haplotypes of GmST05. Values are means ± SDs (n = 3). Student's t test; ** P < 0.01. (d) Transient expression assays of the two GmST05 promoter types in Arabidopsis protoplasts. Values are means ± SDs (n = 8). Student's t test; ** P < 0.01.
Variations in the promoter resulted in differential expression of GmST05
Real‐time quantitative reverse transcription PCR (qRT‐PCR) investigation revealed that although GmST05 genes from Dongnong 50 (DN50, a cultivar harbouring haplotype I) and Tianlong 1 (TL, a cultivar harbouring haplotype II) exhibited similar transcriptional profile patterns, DN50 (GmST05 HapI ) showed significantly higher transcriptional levels than TL (GmST05 HapII ) at different seed developmental stages (Figure 2c), indicating that the allelic variations of the two haplotypes may affect the expression of GmST05.
To verify the transcriptional differences between the two haplotypes, we randomly selected 32 soybean accessions that harboured either GmST05 HapI or GmST05 HapII and then examined the expression levels of GmST05 in these accessions. The expression levels of GmST05 were obviously higher in GmST05 HapI accessions than in those carrying GmST05 HapII (Figure S4a). Together with the fact that the soybean accessions carrying GmST05 HapI had greater seed thickness values than the GmST05 HapII accessions (Figure 2b; Figure S4b‐d), the results suggested that GmST05 functioned as a positive regulator of seed thickness in soybean.
Moreover, we investigated the association between the expression level of GmST05 and seed thickness using 17 independent recombinant lines from a recombinant inbred line (RIL) population derived from Jindou 23 (harbouring GmST05 HapI ) and Huibuzhi (harbouring GmST05 HapII ). Consistent with the above results, the lines carrying GmST05 HapI exhibited significantly higher expression level than the lines carrying GmST05 HapII (Figure S5a, b), and the GmST05 expression levels were significantly positively correlated with seed thickness (Figure S5c, e). The expression levels of GmST05 were also significantly positively correlated with 100‐seed weight (Figure S5d, f).
We then compared the promoter activity of GmST05 from different haplotypes by transient expression assays, revealing that the promoter activity of GmST05 HapI was significantly higher than that of GmST05 HapII (Figure 2d) and confirming that natural genetic variation in GmST05 resulted in differential expression of GmST05 in different haplotypes.
Functional validation of GmST05 by transgenic experiments
To verify the function of GmST05, we introduced a 10.3‐kb genomic fragment of GmST05 HapI (including 5.6 kb upstream of the start codon, the sequences from the start codon to the stop codon and 3 kb downstream from the stop codon) into TL, a soybean cultivar with the GmST05 HapII haplotype. Compared with the parental line, each of the two independent homozygous overexpression (OE) transgenic lines exhibited higher expression and significantly greater seed thickness (Figure 3a‐c). Moreover, seed length and seed width were also higher in the OE lines (Figure 3d, e), which resulted in an approximately 5% greater 100‐seed weight in these OE lines compared with the parental line (Figure 3f). Interestingly, the plant height and pod number per plant of the OE lines were also higher (Figure 3g, h). As a result, compared with those of the parental line, the seed weight per plant and yield per plot increased approximately 12% and 10%, respectively (Figure 3i, j). The 10.3‐kb genomic fragment of GmST05 HapI was also introduced into DN50, a soybean cultivar with the GmST05 HapI haplotype, and results similar to those for TL were observed (Figure S6).
Figure 3.
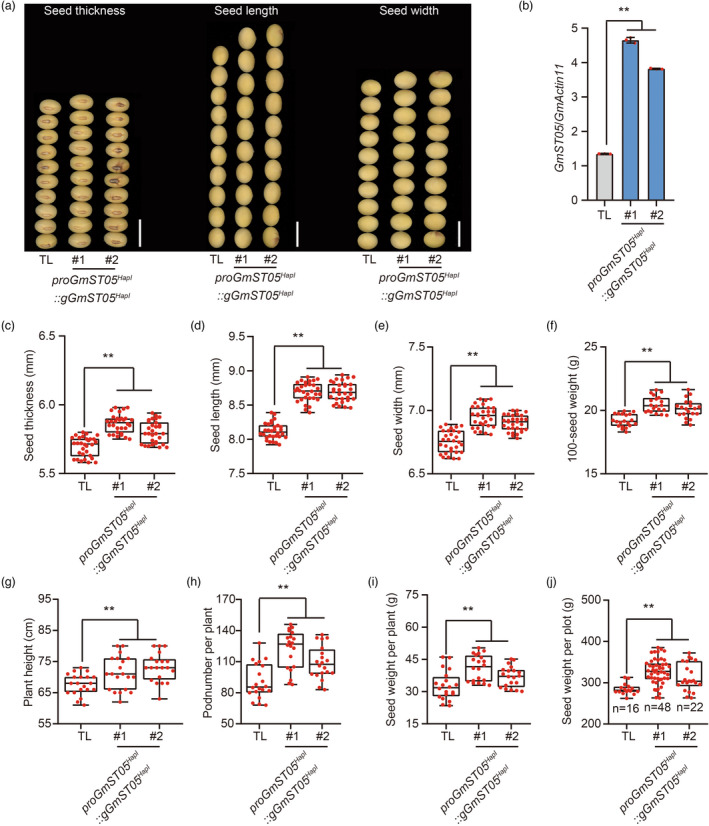
Functional validation of GmST05 in controlling seed size. (a) Comparison of the seed thickness, seed length and seed width of Tianlong (TL, background GmST05 HapII ) and overexpression transgenic plants. Scale bars, 1 cm. (b) Expression levels of GmST05 in TL and overexpression transgenic plants. Total RNA was isolated from soybean seeds in the R5‐4 stage. Values are means ± SDs (n = 3). (c‐j) Statistical analysis of seed thickness (c), seed length (d), seed width (e), 100‐seed weight (f), plant height (g), pod number per plant (h), seed weight per plant (i) and seed weight per plot (j) of TL and overexpression transgenic plants. Values are means ± SDs (for g, h, and i, n = 20; for others, n = 30; one plot indicates 1 m2; Student's t test; ** P < 0.01).
To further verify the function of GmST05, we disrupted GmST05 in DN50 using the CRISPR‐Cas9 genome editing system and obtained two independent homozygous knockout (CR) transgenic lines that led to premature terminations of GmST05 translation (Figure 4a). The CR lines showed significantly reduced seed thickness compared with that of the seeds from the wild type (Figure 4b, c). We found that seed length, seed width and 100‐seed weight were also lower in the CR lines (Figure 4d‐f). Compared with the wild type, the CR lines exhibited significantly decreased plant heights and pod numbers per plant (Figure 4g, h). As a result, the seed weight per plant decreased by approximately 19% compared with that of the wild type (Figure 4i).
Figure 4.
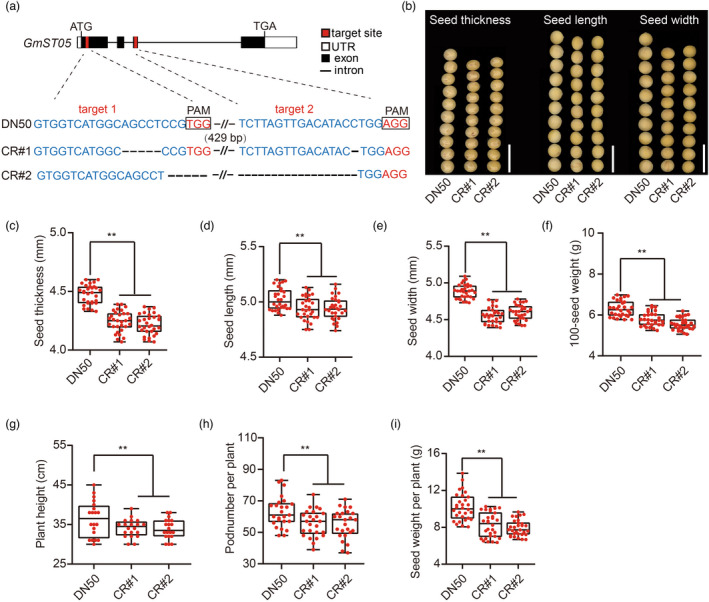
Knockout of GmST05 using CRISPR‐Cas9. (a) Schematics illustrating sgRNA (red lines) targets in the GmST05 coding region. sgRNA targets are highlighted in red. Sequences of the CRISPR‐Cas9‐induced mutant site are shown. (b) Comparison of seed thickness, seed length and seed width between the DN50 and GmST05 HapI knockout lines. Scale bars, 1 cm. (c‐i). Statistical analysis of seed thickness (c), seed length (d), seed width (e), 100‐seed weight (f), plant height (g), pod number per plant (h) and seed weight per plant (i) of DN50 and GmST05 HapI knockout transgenic plants. Values are means ± SDs (for g, n = 20; for others, n = 30; Student's t test; ** P < 0.01).
We also performed RNA interference (RNAi) for GmST05 in DN50 and found that the transgenic lines exhibited decreased GmST05 expression and lower seed thickness, seed length, seed width and 100‐seed weight values (Figure S7). Collectively, these results demonstrated that GmST05 could positively regulate seed thickness and seed size in soybean.
GmST05 affected oil content and protein content by influencing GmSWEET10a expression
It has been found that some genes can affect seed size and quality simultaneously (Wang et al., 2020). A study also suggested that TFL1, another PEBP family member, can modulate seed development except for regulating flowering (Zhang et al., 2020a, 2020b). We found that GmST05 was also located in several QTL regions related to seed oil and protein contents (Figure 1c), indicating that, in addition to regulating seed size, GmST05 may affect the oil and protein contents simultaneously. We then compared the oil and protein contents of GmST05 transgenic lines with those of their parental wild‐type lines and found that the seeds from GmST05 OE lines exhibited significantly higher oil contents and lower protein contents than those from the wild type. By contrast, GmST05 CR lines had significantly lower oil contents and higher protein contents than the wild type (Figure 5a‐d). We also investigated the oil content and protein content of different haplotypes in different soybean accessions and found that GmST05 HapI accessions had higher oil contents and lower protein contents than GmST05 HapII accessions (Figure 5e, f). These results demonstrated that GmST05 also modulates oil and protein contents in soybean.
Figure 5.
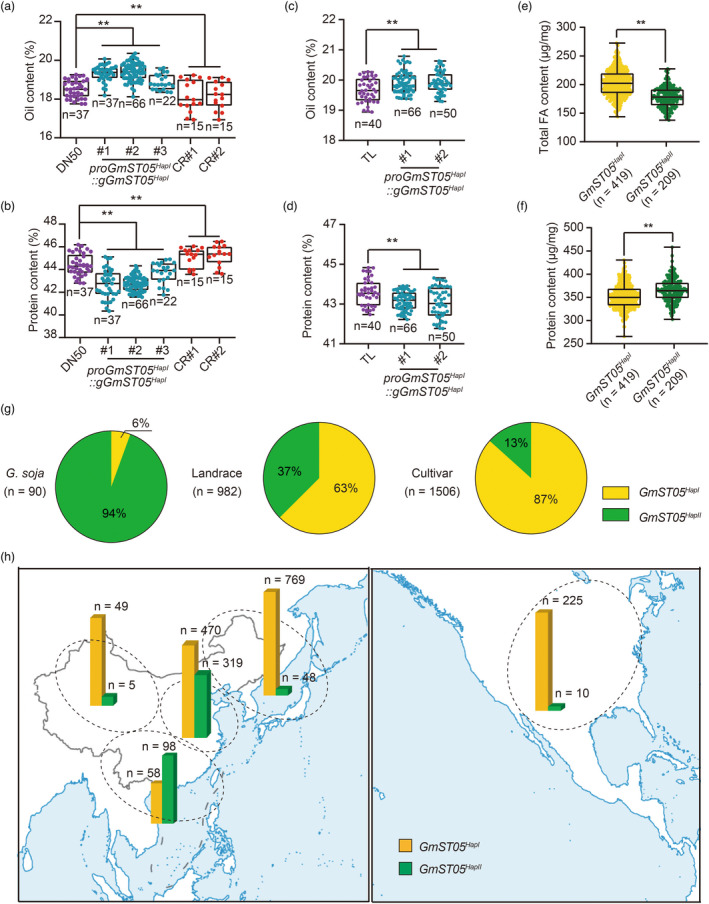
Geographic differentiation of two GmST05 haplotypes and their effects on soybean protein and oil contents. (a‐d) Comparison of the oil content and protein content of mature seeds from DN50 (or Tianlong, TL), GmST05 HapI overexpression transgenic plants and GmST05 HapI knockout transgenic plants. Student's t test; ** P < 0.01. (e, f) Total fatty acid (FA) content and protein content from the accessions of different GmST05 haplotypes. (g) Haplotype frequency distribution of GmST05 in soybean germplasm. (h) The geographical distribution of GmST05 alleles (landraces and cultivars) in Asia and North America.
To clarify the molecular basis of GmST05, we performed transcriptome sequencing analyses (RNA‐seq) and investigated the differentially expressed genes (DEGs) between the wild type and GmST05 OE and CR lines. We identified a total of 2539 DEGs between GmST05 OE lines and TL, 2301 DEGs between GmST05 OE lines and DN50, and 1533 DEGs between GmST05 CR lines and DN50 (Figure S8a). Gene Ontology (GO) analysis suggested that the DEGs were enriched in lipid transport and catabolic processes, amino acid biosynthetic and catabolic processes, sucrose transport and polysaccharide biosynthetic processes, cell cycle and cell proliferation, and phytohormone response pathways (Figure S8e). Of the DEGs, 57 genes were detected for all three pairs (Figure S8a). The SWEET gene family has been found to regulate seed size and seed quality in soybean (Miao et al., 2020; Wang et al., 2020; Zhang et al., 2020a, 2020b). We found that among the 57 overlapping DEGs, GmSWEET10a showed significantly increased expression in the OE lines and significantly decreased expression in the CR line (Figure S8b), which was further confirmed by qRT‐PCR (Figure S8c, d). The results indicated that GmST05 regulates seed size and oil and protein contents by influencing the transcription of GmSWEET10a.
Selection on GmST05 in geographical differentiation
Cultivated soybean was domesticated from wild soybean in China approximately 5000–8000 years ago (Nguyen and Bhattacharyya, 2017). Artificial selection and subsequent local breeding played a crucial role in soybean domestication and improvement (Zhou et al., 2015a, 2015b). We found that the ratio of GmST05 HapI to GmST05 HapII exhibited a continuously increasing pattern from wild soybean to landraces and then cultivars (Figure 5g). However, F ST , reduction of diversity (ROD) and cross‐population extended haplotype homozygosity (XP‐EHH) tests showed that GmST05 did not exceed the threshold for selective sweeps (Figure S9).
We then investigated the geographical distribution of homozygous GmST05 Hap1 and GmST05 HapII alleles in the cultivated soybean accessions. The analyses revealed that the majority of soybean accessions from high‐latitude regions (including north‐eastern Asia, north‐western China and North America) harboured the GmST05 HapI allele. As latitude decreased, the soybean accessions changed to predominantly carrying GmST05 HapII from GmST05 HapI (Figure 5h), which was consistent with the fact that the soybeans from high‐latitude regions usually had a higher oil content but lower protein content, whereas the soybeans from low‐latitude regions usually had a lower oil content but higher protein content. The results indicated that selection on GmST05 may have played important roles in local breeding and geographical differentiation.
Discussion
Improving crop yield has long been an important breeding task in human evolutionary history and will be a continuous never‐ending effort. Although the food supply has significantly increased, crop production still faces unprecedented challenges due to the booming global population, shortage of arable land and unpredicted changes in climate (Tian et al., 2021). Benefitting from biological development, knowledge‐based molecular design may accelerate the breeding process and help to overcome these challenges (Shen et al., 2022; Tian et al., 2021; Wallace et al., 2018). The production of soybean, predominant crop that provides plant oil and protein for the world, needs to be doubled by 2050 (Bodirsky et al., 2015). The identification of key genes regulating important agronomic traits, such as seed size, will facilitate soybean yield improvement. In this study, GmST05 was found to play an important role in controlling seed size and yield in soybean, making it a valuable gene for soybean molecular breeding.
GmST05 is a homologue of Arabidopsis AtMFT, which belongs to the PEBP family. The PEBP family is a very interesting gene family, with members exhibiting divergent and essential functions (Wang et al., 2015a, 2015b, 2015c, 2015d). The members of the PEBP family can be classified into three major subfamilies: TFL1, FT and MFT. Previous studies on Arabidopsis demonstrated that TFL1 members mainly function as flowering time repressors and FT members function as flowering time activators (Bradley et al., 1997; Corbesier et al., 2007; Hanano and Goto, 2011; Hengst et al., 2001; Kardailsky et al., 1999; Shannon and Meeks‐Wagner, 1991). Recent reports suggested that the different members of FT in soybean, which underwent two rounds of whole‐genome duplication and display multiple copies homologous to those in Arabidopsis, showed functional divergence in controlling flowering time (Cai et al., 2018; Fan et al., 2014; Guo et al., 2015; Kong et al., 2010; Lin et al., 2021; Samanfar et al., 2017; Wang et al., 2015a, 2015b, 2015c, 2015d; Zhai et al., 2014; Zhao et al., 2016a, 2016b). Moreover, a study demonstrated that TFL1 can also modulate endosperm cellularization and seed size in addition to regulating flowering (Zhang et al., 2020a, 2020b), suggesting that the genes involved in flowering time may also affect seed development. In Arabidopsis, MFT was first identified as a key gene in regulating seed germination and fertility. However, recently, it was found that it can also accelerate flowering (Xi et al., 2010; Xi and Yu, 2010; Yoo et al., 2004). Ectopic expression of GmMFT, the orthologous gene of MFT in soybean, in Arabidopsis moderately inhibited seed germination (Li et al., 2014), indicating a conserved function of MFT in soybean and Arabidopsis in regulating seed germination. In this study, we demonstrated that GmST05, a homologue of AtMFT, plays an important role in regulating both seed size and seed quality in soybean. Therefore, PEBP family members may have more pleiotropic roles in regulating plant development than previously thought.
The seed morphology of different soybean accessions may vary. In this study, we found that the host varieties used for transgenic experiments, TL and DN50, exhibited a significant difference in seed morphology (Figures 3 and 4). TL and DN50 had GmST05 HapII and GmST05 HapI haplotypes, respectively. The seed morphology differences between TL and DN50 further suggested that GmST05 plays an important role in controlling seed thickness in soybean. In addition to seed thickness, seed morphology is also determined by seed length and seed width (Figure S1a). A further dissection of the genes controlling seed length and seed width will give us more insights into how seed morphology is controlled in soybean.
Increasing evidence suggests that the natural variations present in promoter regions also play critical roles in altering agronomic traits by regulating gene expression levels (Springer et al., 2019; Swinnen et al., 2016). For instance, segmental deletion in the promoter region of qSW5/GW5 alters its expression level and results in slender grains (Duan et al., 2017; Liu et al., 2017), a 6‐bp tandem‐repeat sequence in the 5′ UTR of GLW7/OsSPL13 represses its expression level and enhances rice grain length and yield (Si et al., 2015), an 18‐bp fragment duplication in the 5.3 kb upstream of FZP represses its expression and prolongs the panicle branching period (Bai et al., 2017), and tandem duplication of a 17.1‐kb segment at the GL7 locus leads to upregulation of GL7 and results in increased grain length and improved grain quality (Wang et al., 2015a, 2015b, 2015c, 2015d). These findings provide important insights into the molecular mechanisms and functional significance of natural variation that regulates gene expression levels to alter cereal yield. In this study, we revealed that the functional differences of the two major GmST05 haplotypes resulted from natural variations in the promoter regions that affected their transcriptional level differences. Modulating the expression of important genes using genome editing will be a powerful approach for accelerating crop breeding (Hendelman et al., 2021; Wang et al., 2021; Song et al., 2022). Therefore, a modification of the transcription‐regulating regions of GmST05 may help us improve soybean yield. Taken together, our findings provide significant insight into the genetic basis for seed size in soybean and will be helpful for improving soybean yield through molecular breeding.
Methods
Plant material and phenotyping
The 1853 soybean accessions used for the GWAS were planted at the experimental station of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing (40°22′N and 116°23′E), during the summer season in 2016 and 2017. The 12 lines of the F15:16 RIL population derived from a cross between soybean accessions JinDou23 and HuiBuZhi were harvested in 2019. T3 transgenic plants of the GmST05 HapI genome fragment were planted at the experimental station during the summer season in 2021. The T3 transgenic seeds were planted in three‐row plots in a randomized complete block design with three replications for each environment. The length of each plot was 5 m, and the row spacing was 0.4 m. The space between two plots was 0.4 m. After 3 weeks, the seedlings were manually thinned to achieve an equal density of 120 000 individuals per hectare.
All above seeds were stored for 1 month after harvest and then used for seed trait determination. For seed thickness, seed length and seed width phenotyping, at least 20 representative dry seeds per accession were measured. For 100‐seed weight, at least 6 replicates were examined.
GWAS for the seed thickness trait
A total of 31 870 983 single‐nucleotide polymorphisms (SNPs) from our previously resequenced 1853 soybean accessions were used to perform GWAS for the seed thickness trait (Liu et al., 2020a, 2020b). The population structure was calculated using the Bayesian clustering program fastStructure (Raj et al., 2014). Only SNPs with a MAF ≥ 0.05 and missing rate <0.1 in the population were used in the GWAS. An association analysis was performed by a mixed linear model (MLM) implemented in the efficient mixed‐model association expedited (EMMAX) software package (Kang et al., 2010). The matrix of pairwise genetic distances, which were derived from the simple matching coefficients of the variance–covariance matrix of the random effects, was also calculated by EMMAX. The threshold for GWAS was determined based on a previous report (Fang et al., 2017); the brief procedure is as follows: First, we randomly shuffled the observed phenotypes to break the connections between these phenotypes and their corresponding genotypes. Then, we applied the GWAS to the permuted phenotypes by using the same model that was used for the observed phenotypes. The most significant P value across the whole genome was recorded. This random process was repeated 1000 times. The distribution of the most significant P values across the 1000 replicates was used to determine the threshold, which was the P value corresponding to a 5% chance of a type I error.
DNA and RNA extraction, PCR, and qRT‐PCR
Genomic DNA was isolated from fresh tender leaves by the CTAB method (Murray and Thompson, 1980). Total RNA was extracted from seeds at the R5 growth stage with the HUAYUEYANG Quick RNA Isolation Kit v1.0 (HUAYUEYANG, Beijing). The removal of genomic DNA and first‐strand synthesis were performed with TransScript® One‐Step gDNA Removal and cDNA Synthesis SuperMix (Transgene) according to the manufacturer's instructions. DNA fragment amplification was performed using KOD FX neo (Toyobo). qRT‐PCR was performed using LightCycler 480 SYBR Green I Master Mix (Roche) on a LightCycler 480 instrument (Roche). Gene expression was normalized to the expression of GmActin11. The primers used for PCR and RT‐PCR are listed in Table S1.
Vector construction and transformation
For functional validation in soybean, a 10.3‐kb genomic sequence of GmST05 (SoyZH13_05G229200), including 5.6 kb upstream of the start codon, the sequences from the start codon to the stop codon, and 3 kb downstream of the stop codon, was amplified from DN50 and ligated into the pTF101 vector. To generate Gmst05 mutants, sgRNAs targeting different positions of GmST05 were designed according to a previous study (Xie et al., 2014) and cloned into the CRISPR‐Cas9 vector pMDC123‐Cas9 (Addgene). To construct the GmST05 interference plasmid, an inverted repeat sequence harbouring the 213‐bp GmST05 cDNA fragment was ligated into the pFGC5941 vector. These constructs were introduced into Agrobacterium tumefaciens strain EHA101 (or EHA105) and then transformed into DN50 (or TL) as previously reported (Li et al., 2017). All primers used to construct vectors are listed in Table S1.
Subcellular localization and phylogenetic tree construction
To infer the subcellular localization of the GmST05 protein, the full‐length coding sequences (CDSs) of two GmST05 haplotypes were cloned into the pUC19‐35 s‐eGFP vector with green fluorescent protein (GFP) at the C‐terminus. We transformed the fusion protein into mesophyll protoplasts of four‐week‐old seedlings according to a previous study (Yoo et al., 2007). After 12–16 h of incubation at 22 °C under dark conditions, protoplasts were examined using confocal microscopy. GmST05 orthologous protein searching was performed with Phytozome 13 (https://phytozome‐next.jgi.doe.gov/). Duplicates and orthologs with high similarity to GmST05 from soybean and other representative species were downloaded and used to construct a maximum likelihood tree in MEGA6.0 software with 1000 bootstrap replicates.
Transient expression assays of promoter activity
The promoter activity analysis was performed using Arabidopsis protoplasts as previously described with some modifications (Li et al., 2016). The 3‐kb promoter fragment upstream of the GmST05 translation start site was amplified from DN50 and TL and used to construct the pGmST05 Hap1 ‐Luc and pGmST05 HapII ‐Luc plasmids. Then, both fragments were inserted into the pGreenII 0800‐LUC vector. Each of the GmST05 promoter‐Luc gene fusion constructs was used for transient transformation into Arabidopsis protoplasts. Luc and Ren activities were measured by the Dual‐Luciferase Reporter Assay System (Promega), and Luc activity was normalized to Ren activity.
Measurements of oil content and protein content
Measurements of oil content and protein content were performed at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. Uniform dry seeds were collected from DN50, TL and transgenic plants and measured in a multifunctional near‐infrared system (NIRS™ DS 2500F, FOSS, USA) to determine the oil content and protein content according to a previous study (Liu et al., 2018). Briefly, NIRS™ DS 2500F was preheated at least 30 min before use, and the instrument remained closed during the experiment to ensure the stability of the scan. During spectral acquisition, the uniform dry seeds were evenly distributed in the sample container to ensure that no spaces were generated, which can cause interference of objective factors such as spectral scattering. Each sample was scanned three times to obtain three sets of spectral data. These three sets of spectral data were averaged, and the average spectrum was used as the feature spectrum for this sample.
RNA‐seq and GO enrichment analysis
Total RNA was extracted from seeds of DN50, TL and transgenic plants at the R5 stage with three biological replicates. Paired‐end libraries were constructed and sequenced using an Illumina NovaSeq 6000 instrument at BerryGenomics Company (China). Cuffdiff was used to calculate FPKM (fragments per kilobase of exon per million mapped reads) values for each gene and identify differentially expressed genes (DEGs) with a fold change ≥2 and a false discovery rate <0.05. GO annotation was performed by PANNZER2 (Törönen et al., 2018). The enrichment test was performed by ClusterProfiler (Yu et al., 2012).
Genetic diversity analysis
SNP data from our previous study (Liu et al., 2020a, 2020b) were used to analyse the genetic diversity of GmST05 in soybean. SNPs with >10% missing data or a minor allele frequency (MAF) < 5% were excluded. The soybean accessions were divided into three populations: G. soja, landrace and cultivar.
F ST values were calculated with a 20‐kb to 10‐kb sliding window using VCFtools to calculate the pairwise genomic differentiation of wild and cultivated populations of soybean (Danecek et al., 2011). We used ROD values to define selective sweeps in the G. soja and cultivar populations. ROD is derived from π and estimates the reduction in genetic diversity in the derived group in comparison to the control group (Xu et al., 2011). XP‐EHH was used to detect signatures of recent selection by comparing EHH between G. soja and cultivars regardless of whether the favoured allele had reached fixation (Sabeti et al., 2007).
Conflict of interest
The authors declare no competing interests.
Author contributions
Z.T. designed and supervised this project; Z.D., M.Z., S.L., L.F., X.Y., Y.Y., Y.P. and G.Z. performed the experiments; Z.D., Z.Z., S.L. and Z.T. analysed the data; Z.D. and Z.T. wrote the manuscript.
Supporting information
Table S1 Primers used in this study.
Figure S1 GWAS of seed thickness using data from 1853 soybean accessions harvested in 2016 and 2017.
Figure S2 Characterization of GmST05.
Figure S3 Sequence of GmST05 from two different haplotypes.
Figure S4 The expression level of GmST05 was positively correlated with seed thickness in different soybean accessions.
Figure S5 Association analysis of GmST05 expression levels with seed thickness and seed weight in the recombinant inbred line (RIL) population.
Figure S6 Transgenic experiment test using genomic DNA of GmST05.
Figure S7 RNA interference test of GmST05.
Figure S8 GmST05 is involved in oil synthesis in soybean.
Figure S9 Selection of GmST05 during soybean domestication.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32090064, 32172053, 31788103), the National Key Research & Development Program of China (2021YFF1001201), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA24030501) and the Seed‐Industrialized Development Program in Shandong Province (2021LZGC003).
References
- Bai, X. , Huang, Y. , Hu, Y. , Liu, H. , Zhang, B. , Smaczniak, C. , Hu, G. et al. (2017) Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants, 3, 885–893. [DOI] [PubMed] [Google Scholar]
- Bednarek, J. , Boulaflous, A. , Girousse, C. , Ravel, C. , Tassy, C. , Barret, P. , Bouzidi, M.F. et al. (2012) Down‐regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. J. Exp. Bot. 63, 5945–5955. [DOI] [PubMed] [Google Scholar]
- Bodirsky, B.L. , Rolinski, S. , Biewald, A. , Weindl, I. , Popp, A. and Lotze‐Campen, H. (2015) Global food demand scenarios for the 21st century. PLoS One, 10, e0139201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, D. , Ratcliffe, O. , Vincent, C. , Carpenter, R. and Coen, E. (1997) Inflorescence commitment and architecture in Arabidopsis . Science, 275, 80–83. [DOI] [PubMed] [Google Scholar]
- Brummer, E. , Graef, G. , Orf, J. , Wilcox, J. and Shoemaker, R. (1997) Mapping QTL for seed protein and oil content in eight soybean populations. Crop. Sci. 37, 370–378. [Google Scholar]
- Cai, Y. , Chen, L. , Liu, X. , Guo, C. , Sun, S. , Wu, C. , Jiang, B. et al. (2018) CRISPR/Cas9‐mediated targeted mutagenesis of GmFT2a delays flowering time in soybean. Plant Biotechnol. J. 16, 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R. , Deng, Y. , Ding, Y. , Guo, J. , Qiu, J. , Wang, B. , Wang, C. et al. (2022) Rice functional genomics: decades' efforts and roads ahead. Sci. China Life Sci. 65, 33–92. [DOI] [PubMed] [Google Scholar]
- Corbesier, L. , Vincent, C. , Jang, S. , Fornara, F. , Fan, Q. , Searle, I. , Giakountis, A. et al. (2007) FT protein movement contributes to long‐distance signaling in floral induction of Arabidopsis . Science, 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Danecek, P. , Auton, A. , Abecasis, G. , Albers, C.A. , Banks, E. , MA, D.P. , Handsaker, R.E. et al. (2011) The variant call format and VCFtools. Bioinformatics (Oxford, England), 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J.F. , Gaut, B.S. and Smith, B.D. (2006) The molecular genetics of crop domestication. Cell, 127, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Du, J. , Wang, S. , He, C. , Zhou, B. , Ruan, Y.L. and Shou, H. (2017) Identification of regulatory networks and hub genes controlling soybean seed set and size using RNA sequencing analysis. J. Exp. Bot. 68, 1955–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, P. and Li, Y. (2021) Size matters: G protein signaling is crucial for grain size control in rice. Mol. Plant, 14, 1618–1620. [DOI] [PubMed] [Google Scholar]
- Duan, P. , Xu, J. , Zeng, D. , Zhang, B. , Geng, M. , Zhang, G. , Huang, K. et al. (2017) Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant, 10, 685–694. [DOI] [PubMed] [Google Scholar]
- Fan, C. , Hu, R. , Zhang, X. , Wang, X. , Zhang, W. , Zhang, Q. , Ma, J. et al. (2014) Conserved CO‐FT regulons contribute to the photoperiod flowering control in soybean. BMC Plant Biol. 14, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. , Xing, Y. , Mao, H. , Lu, T. , Han, B. , Xu, C. , Li, X. et al. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. [DOI] [PubMed] [Google Scholar]
- Fang, C. , Ma, Y. , Wu, S. , Liu, Z. , Wang, Z. , Yang, R. , Hu, G. et al. (2017) Genome‐wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 18, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliege, C.E. , Ward, R.A. , Vogel, P. , Nguyen, H. , Quach, T. , Guo, M. , Viana, J. et al. (2022) Fine mapping and cloning of the major seed protein quantitative trait loci on soybean chromosome 20. Plant J. 110, 114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, L. , Yu, J. , Wang, H. , Luth, D. , Bai, G. , Wang, K. and Chen, R. (2016) Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc. Natl. Acad. Sci. 113, 12414–12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, G. , Xu, K. , Zhang, X. , Zhu, J. , Lu, M. , Chen, F. , Liu, L. et al. (2015) Extensive analysis of GmFTL and GmCOL expression in northern soybean cultivars in field conditions. PLoS One, 10, e0136601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano, S. and Goto, K. (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell, 23, 3172–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendelman, A. , Zebell, S. , Rodriguez‐Leal, D. , Dukler, N. , Robitaille, G. , Wu, X. , Kostyun, J. et al. (2021) Conserved pleiotropy of an ancient plant homeobox gene uncovered by cis‐regulatory dissection. Cell, 184, 1724–1739. [DOI] [PubMed] [Google Scholar]
- Hengst, U. , Albrecht, H. , Hess, D. and Monard, D. (2001) The phosphatidylethanolamine‐binding protein is the prototype of a novel family of serine protease inhibitors. J. Biol. Chem. 276, 535–540. [DOI] [PubMed] [Google Scholar]
- Jun, T.H. , Freewalt, K. , Michel, A.P. and Mian, R. (2014) Identification of novel QTL for leaf traits in soybean. Plant Breed. 133, 61–66. [Google Scholar]
- Kabelka, E.A. , Diers, B.W. , Fehr, W.R. , LeRoy, A.R. , Baianu, I.C. , You, T. , Neece, D.J. et al. (2004) Putative alleles for increased yield from soybean plant introductions. Crop. Sci. 44, 784–791. [Google Scholar]
- Kang, H.M. , Sul, J.H. , Service SK , Zaitlen, N.A. , Kong, S.Y. , Freimer, N.B. , Sabatti, C. et al. (2010) Variance component model to account for sample structure in genome‐wide association studies. Nat. Genet. 42, 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I. , Shukla, V.K. , Ahn, J.H. , Dagenais, N. , Christensen, S.K. , Nguyen, J.T. , Chory, J. et al. (1999) Activation tagging of the floral inducer FT. Science, 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kong, F. , Liu, B. , Xia, Z. , Sato, S. , Kim, B.M. , Watanabe, S. , Yamada, T. et al. (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Cong, Y. , Liu, Y. , Wang, T. , Shuai, Q. , Chen, N. , Gai, J. et al. (2017) Optimization of agrobacterium‐mediated transformation in soybean. Front. Plant Sci. 8, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Fan, C. , Zhang, X. , Wang, X. , Wu, F. , Hu, R. and Fu, Y. (2014) Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PLoS One, 9, e99642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Fang, C. , Duan, Z. , Liu, Y. , Qin, H. , Zhang, J. , Sun, P. et al. (2016) Functional conservation and divergence of GmCHLI genes in polyploid soybean. Plant J. 88, 584–596. [DOI] [PubMed] [Google Scholar]
- Li, N. and Li, Y. (2016) Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Li, L. , Yang, X. , Warburton, M.L. , Bai, G. , Dai, J. , Li, J. et al. (2010) Relationship, evolutionary fate and function of two maize co‐orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Xu, R. and Li, Y. (2019) Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463. [DOI] [PubMed] [Google Scholar]
- Li, J. , Zhang, Y. , Ma, R. , Huang, W. , Hou, J. , Fang, C. , Wang, L. et al. (2022) Identification of ST1 reveals a selection involving hitchhiking of seed morphology and oil content during soybean domestication. Plant Biotechnol. J. 20, 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X. , Liu, B. , Weller, J.L. , Abe, J. and Kong, F. (2021) Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 63, 981–994. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Chen, J. , Zheng, X. , Wu, F. , Lin, Q. , Heng, Y. , Tian, P. et al. (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants, 3, 17043. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Du, H. , Li, P. , Shen, Y. , Peng, H. , Liu, S. , Zhou, G. et al. (2020b) Pan‐genome of wild and cultivated soybeans. Cell, 182, 162–176. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Sun, L. , Li, Y. , Zhang, D. , Song, Z. , Zhang, C. , Pan, X. et al. (2018) Quality evaluation of fried soybean oil base on near infrared spectroscopy. J. Food Process Eng. 41, e12887. [Google Scholar]
- Liu, L. , Tong, H. , Xiao, Y. , Che, R. , Xu, F. , Hu, B. , Liang, C. et al. (2015) Activation of big grain 1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. 112, 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Zhang, M. , Feng, F. and Tian, Z. (2020a) Toward a “green revolution” for soybean. Mol. Plant, 13, 688–697. [DOI] [PubMed] [Google Scholar]
- Lu, X. , Xiong, Q. , Cheng, T. , Li, Q. , Liu, X. , Bi, Y. , Li, W. et al. (2017) A PP2C‐1 allele underlying a quantitative trait locus enhances soybean 100‐seed weight. Mol. Plant, 10, 670–684. [DOI] [PubMed] [Google Scholar]
- Mao, H. , Sun, S. , Yao, J. , Wang, C. , Yu, S. , Xu, C. , Li, X. et al. (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. 107, 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, J.I. , Hu, H. , Petereit, J. , Bayer, P.E. , Valliyodan, B. , Batley, J. , Nguyen, H.T. et al. (2022) Haplotype mapping uncovers unexplored variation in wild and domesticated soybean at the major protein locus cqProt‐003. Theor. Appl. Genet. 135, 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, L. , Yang, S. , Zhang, K. , He, J. , Wu, C. , Ren, Y. , Gai, J. et al. (2020) Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol. 225, 1651–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H.T. and Bhattacharyya, M.K. (2017) The Soybean Genome. New York: Springer. 10.1007/978-3-319-64198-0 [DOI] [Google Scholar]
- Nguyen, C.X. , Paddock, K.J. , Zhang, Z. and Stacey, M.G. (2021) GmKIX8‐1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17‐1 . New Phytol. 229, 920–934. [DOI] [PubMed] [Google Scholar]
- Pathan, S.M. , Vuong, T. , Clark, K. , Lee, J.D. , Shannon, J.G. , Roberts, C.A. , Ellersieck, M.R. et al. (2013) Genetic mapping and confirmation of quantitative trait loci for seed protein and oil contents and seed weight in soybean. Crop. Sci. 53, 765–774. [Google Scholar]
- Raj, A. , Stephens, M. and Pritchard, J.K. (2014) fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics, 197, 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti, P.C. , Varilly, P. , Fry, B. , Lohmueller, J. , Hostetter, E. , Cotsapas, C. , Xie, X. et al. (2007) Genome‐wide detection and characterization of positive selection in human populations. Nature, 449, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanfar, B. , Molnar, S.J. , Charette, M. , Schoenrock, A. , Dehne, F. , Golshani, A. , Belzile, F. et al. (2017) Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 130, 377–390. [DOI] [PubMed] [Google Scholar]
- Sestili, F. , Pagliarello, R. , Zega, A. , Saletti, R. , Pucci, A. , Botticella, E. , Masci, S. et al. (2019) Enhancing grain size in durum wheat using RNAi to knockdown GW2 genes. Theor. Appl. Genet. 132, 419–429. [DOI] [PubMed] [Google Scholar]
- Shannon, S. and Meeks‐Wagner, D.R. (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell, 3, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. , Du, H. , Liu, Y. , Ni, L. , Wang, Z. , Liang, C. and Tian, Z. (2019) Update soybean Zhonghuang 13 genome to a golden reference. Sci. China Life Sci. 62, 1257–1260. [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Liu, J. , Geng, H. , Zhang, J. , Liu, Y. , Zhang, H. , Xing, S. et al. (2018) De novo assembly of a Chinese soybean genome. Sci. China Life Sci. 61, 871–884. [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Zhou, G. , Liang, C. and Tian, Z. (2022) Omics‐based interdisciplinarity is accelerating plant breeding. Curr. Opin. Plant Biol. 66, 102167. [DOI] [PubMed] [Google Scholar]
- Si, L. , Chen, J. , Huang, X. , Gong, H. , Luo, J. , Hou, Q. , Zhou, T. et al. (2015) OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. [DOI] [PubMed] [Google Scholar]
- Simmons, C.R. , Weers, B.P. , Reimann, K.S. , Abbitt, S.E. , Frank, M.J. , Wang, W. , Wu, J. et al. (2020) Maize BIG GRAIN 1 homolog overexpression increases maize grain yield. Plant Biotechnol. J. 18, 2304–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X. , Huang, W. , Shi, M. , Zhu, M. and Lin, H. (2007) A QTL for rice grain width and weight encodes a previously unknown RING‐type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Song, X. , Meng, X. , Guo, H. , Cheng, Q. , Jing, Y. , Chen, M. , Liu, G. et al. (2022) Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol. 10.1038/s41587-022-01281-7 [DOI] [PubMed] [Google Scholar]
- Specht, J. , Chase, K. , Macrander, M. , Graef, G. , Chung, J. , Markwell, J. , Germann, M. et al. (2001) Soybean response to water: a QTL analysis of drought tolerance. Crop. Sci. 41, 493–509. [Google Scholar]
- Springer, N. , de León, N. and Grotewold, E. (2019) Challenges of translating gene regulatory information into agronomic improvements. Trends Plant Sci. 24, 1075–1082. [DOI] [PubMed] [Google Scholar]
- Su, Z. , Hao, C. , Wang, L. , Dong, Y. and Zhang, X. (2011) Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 122, 211–223. [DOI] [PubMed] [Google Scholar]
- Swinnen, G. , Goossens, A. and Pauwels, L. (2016) Lessons from domestication: targeting cis‐regulatory elements for crop improvement. Trends Plant Sci. 21, 506–515. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Su, T. , Han, M. , Wei, L. , Wang, W. , Yu, Z. , Xue, Y. et al. (2016) Suppression of extracellular invertase inhibitor gene expression improves seed weight in soybean (Glycine max). J. Exp. Bot. 68, 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Z. , Wang, J. , Li, J. and Han, B. (2021) Designing future crops: challenges and strategies for sustainable agriculture. Plant J. 105, 1165–1178. [DOI] [PubMed] [Google Scholar]
- Törönen, P. , Medlar, A. and Holm, L. (2018) PANNZER2: a rapid functional annotation web server. Nucleic Acids Res. 46, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, J.G. , Rodgers‐Melnick, E. and Buckler, E.S. (2018) On the road to breeding 4.0: unraveling the good, the bad, and the boring of crop quantitative genomics. Annu. Rev. Genet. 52, 421–444. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Aguirre, L. , Rodríguez‐Leal, D. , Hendelman, A. , Benoit, M. and Lippman, Z.B. (2021) Dissecting cis‐regulatory control of quantitative trait variation in a plant stem cell circuit. Nat. Plants, 7, 419–427. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Jiang, G.L. , Green, M. , Scott, R.A. , Song, Q. , Hyten, D.L. and Cregan, P.B. (2014) Identification and validation of quantitative trait loci for seed yield, oil and protein contents in two recombinant inbred line populations of soybean. Mol. Genet. Genomics, 289, 935–949. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Li, S. , Liu, Q. , Wu, K. , Zhang, J. , Wang, S. , Wang, Y. et al. (2015d) The OsSPL16‐GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Li, Y. , Zhang, H. , Sun, G. , Zhang, W. and Qiu, L. (2015a) Evolution and association analysis of GmCYP78A10 gene with seed size/weight and pod number in soybean. Mol. Biol. Rep. 42, 489–496. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Liu, S. , Wang, J. , Yokosho, K. , Zhou, B. , Yu, Y.C. , Liu, Z. et al. (2020) Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 7, 1776–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Wu, K. , Yuan, Q. , Liu, X. , Liu, Z. , Lin, X. , Zeng, R. et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Xiong, G. , Hu, J. , Jiang, L. , Yu, H. , Xu, J. , Fang, Y. et al. (2015b) Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Zhou, Z. , Liu, Y. , Liu, T. , Li, Q. , Ji, Y. , Li, C. et al. (2015c) Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis . Plant Cell, 27, 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, W. , Liu, C. , Hou, X. and Yu, H. (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis . Plant Cell, 22, 1733–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, W. and Yu, H. (2010) MOTHER OF FT AND TFL1 regulates seed germination and fertility relevant to the brassinosteroid signaling pathway. Plant Signal. Behav. 5, 1315–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, S. , Shen, B. , Zhang, C. , Huang, X. and Zhang, Y. (2014) sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off‐target cleavage sites. PLoS One, 9, e100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y. and Zhang, Q. (2010) Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. [DOI] [PubMed] [Google Scholar]
- Xu, X. , Liu, X. , Ge, S. , Jensen, J.D. , Hu, F. , Li, X. , Dong, Y. et al. (2011) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30, 105–111. [DOI] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoo, S.Y. , Kardailsky, I. , Lee, J.S. , Weigel, D. and Ahn, J.H. (2004) Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Mol. Cells, 17, 95–101. [PubMed] [Google Scholar]
- Yu, G. , Wang, L. , Han, Y. and He, Q. (2012) ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, H. , Lu, S. , Liang, S. , Wu, H. , Zhang, X. , Liu, B. , Kong, F. et al. (2014) GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS One, 9, e89030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Goettel, W. , Song, Q. , Jiang, H. , Hu, Z. , Wang, M. and An, Y. (2020a) Selection of GmSWEET39 for oil and protein improvement in soybean. PLoS Genet. 16, e1009114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Li, C. , Li, Y. and Yu, H. (2020b) Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis . Nat. Plants 6, 1146–1157. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Li, D. , Zhang, D. , Zhao, X. , Cao, X. , Dong, L. , Liu, J. et al. (2018) Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 94, 857–866. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Liu, S. , Wang, Z. , Yuan, Y. , Zhang, Z. , Liang, Q. , Yang, X. et al. (2022) Progress in soybean functional genomics over the past decade. Plant Biotechnol. J. 20, 256–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Zhang, H. , Hu, Z. , Chu, S. , Yu, K. , Lv, L. , Yang, Y. et al. (2019) Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet. 15, e1008267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. , Dai, A. , Wei, H. , Yang, S. , Wang, B. , Jiang, N. and Feng, X. (2016b) Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 90, 33–47. [DOI] [PubMed] [Google Scholar]
- Zhao, C. , Takeshima, R. , Zhu, J. , Xu, M. , Sato, M. , Watanabe, S. , Kanazawa, A. et al. (2016a) A recessive allele for delayed FLOWERING at the soybean maturity LOCUS E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z. , Jiang, Y. , Wang, Z. , Gou, Z. , Lyu, J. , Li, W. , Yu, Y. et al. (2015a) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33, 408–414. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Miao, J. , Gu, H. , Peng, X. , Leburu, M. , Yuan, F. , Gu, H. et al. (2015b) Natural variations in SLG7 regulate grain shape in rice. Genetics, 201, 1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J. and Li, J. (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers used in this study.
Figure S1 GWAS of seed thickness using data from 1853 soybean accessions harvested in 2016 and 2017.
Figure S2 Characterization of GmST05.
Figure S3 Sequence of GmST05 from two different haplotypes.
Figure S4 The expression level of GmST05 was positively correlated with seed thickness in different soybean accessions.
Figure S5 Association analysis of GmST05 expression levels with seed thickness and seed weight in the recombinant inbred line (RIL) population.
Figure S6 Transgenic experiment test using genomic DNA of GmST05.
Figure S7 RNA interference test of GmST05.
Figure S8 GmST05 is involved in oil synthesis in soybean.
Figure S9 Selection of GmST05 during soybean domestication.


