Summary
Water is crucial for plant growth and survival. The transcellular water movement is facilitated by aquaporins (AQPs) that rapidly and reversibly modify water permeability. The abundance of AQPs is regulated by its synthesis, redistribution and degradation. However, the molecular mechanism of proteasomal degradation of AQPs remains unclear. Here, we demonstrate that a novel E3 ligase, OsRINGzf1, mediated the degradation of AQPs in rice. OsRINGzf1 is the candidate gene from a drought‐related quantitative trait locus (QTL) on the long arm of chromosome 4 in rice (Oryza sativa) and encodes a Really Interesting New Gene (RING) zinc finger protein 1. OsRINGzf1 possesses the E3 ligase activity, ubiquitinates and mediates OsPIP2;1 degradation, thus reducing its protein abundance. The content of OsPIP2;1 protein was decreased in OsRINGzf1 overexpression (OE) plants. The degradation of OsPIP2;1 was inhibited by MG132. The OsRINGzf1 OE plants, with higher leaf‐related water content (LRWC) and lower leaf water loss rate (LWLR), exhibited enhanced drought resistance, whereas the RNAi and knockout plants of OsRINGzf1 were more sensitive to drought. Together, our data demonstrate that OsRINGzf1 positively regulates drought resistance through promoting the degradation of OsPIP2;1 to enhance water retention capacity in rice.
Keywords: rice, ubiquitination, aquaporins, drought resistance
Introduction
Rice (Oryza sativa) is one of the most important cereals in the world. Compared with other cereal crops such as wheat and maize, rice is more sensitive to soil water deficiency because they are historically grown under flood irrigation conditions (Huang et al., 2014). Water resources are becoming one of the most limiting factors for rice production (Kumar et al., 2014). Therefore, there is an urgent demand for improved rice varieties with enhanced drought resistance.
Really Interesting New Gene (RING) finger proteins, defined by the presence of a RING finger motif with highly conserved Cys and His residues, are one of the most studied ubiquitin E3 ligases. The canonical RING domains can be subcategorized into RING‐H2 and RING‐HC subtypes based on the fifth coordination Cys or His residues, respectively. Studies over the past decade have revealed the crucial role of RING‐type E3 ligase in plant abiotic stress regulation. OsSDIR1 improves drought resistance by promoting the closing of stomata and reducing the loss of leaf water (Gao et al., 2011). OsDSG1, OsCTR1 and OsiSAP7 are involved in ABA signalling or biosynthesis, and play roles in drought stress response (Lim et al., 2014; Park et al., 2010; Sharma et al., 2015). OsPUB67 improved drought tolerance by enhancing reactive oxygen scavenging ability and stomatal closure (Qin et al., 2019). OsHCI1 and OsHTAS enhance heat tolerance of plant (Lim et al., 2013; Liu et al., 2016). OsSIRP2 and OsSIRH2‐14 act as a positive regulator of salt tolerance (Chapagain et al., 2018; Park et al., 2019), while OsMAR1, OsSIRP1, OsSIRP4 and OsSADR1 negatively regulate the response to salinity stress (Hwang et al., 2016; Kim and Jang, 2020; Park et al., 2018a,b). However, compared with the 425 predicted RING E3 ligase in rice (Lim et al., 2010), their regulatory role in abiotic stress, as well as their target proteins, remains largely elusive.
Aquaporins (AQPs) are intrinsic membrane protein channels and function in water movement across the cell membranes for water homeostasis. Increasing evidence in various crop plants indicates that AQPs are responsible for water deficit and play a crucial role in drought resistance (Ayadi et al., 2019; Grondin et al., 2015). The transcellular water movement is tightly controlled by the amount and activity of AQPs in the membrane, which is concerned with the trafficking, gating and degradation of AQPs. A profound understanding of trafficking and gating had been established (Maurel et al., 2015). Yet, the mechanism of the degradation is still poorly understood. MtCAS31 facilitated the autophagic degradation of MtPIP2;7 and reduced root hydraulic conductivity, thus reducing water loss and improving drought resistance (Li et al., 2019). AtPIP2;7 was shown to interact with a TSPO membrane protein to form a complex that was directed towards vacuolar degradation, using the autophagosome pathway (Hachez et al., 2014). Recently, researches on AQP degradation by the autophagic pathway have made big progress and proposed that PIP autophagy may be initiated at ER‐PM contact sites (EPCSs) through its interaction with TSPO and VAP27s (Fox et al., 2020). Few studies were concerned with the UPS‐dependent degradation of AQPs (Lee et al., 2009). In rice, there are total 33 AQPs (Sakurai et al., 2005). Some of them had been found to respond to drought (Bai et al., 2021; Lian et al., 2004), while the underlying regulation mechanism is unclear. Since AQPs are closely related to plant water status, regulation of rice AQPs might play a role in drought resistance.
Previously, we mapped a rice drought QTL interval on chromosome 4 (Liu et al., 2005; Zou et al., 2005). In this study, the gene OsRINGzf1 encoding a RING‐H2‐type E3 ligase was identified from this QTL interval. The expression of OsRINGzf1 was induced by drought stress. E3 ligase activity of OsRINGzf1 was confirmed in vitro and in vivo. OsRINGzf1 interacted with several AQPs and ubiquitinated OsPIP2;1, and OsSIP1;1. OsRINGzf1 and OsPIP2;1 were colocalized on endoplasmic reticulum (ER) and plasma membrane (PM). OsPIP2;1 protein abundance was significantly reduced both in vitro and in vivo with the existence of OsRINGzf1. Compared with the wild‐type (WT) plants, OsRINGzf1 overexpression (OE) plants exhibited enhanced drought resistance by reducing water loss, while the silencing of OsRINGzf1 leads to increased water loss, thereby reducing drought resistance. The OsRINGzf1 OE plants and the OsPIP2;1 mutant plants showed similar lower leaf water loss rate (LWLR) under water deficit conditions. Our study demonstrated that OsRINGzf1 positively regulated drought resistance through modulating the ubiquitin‐proteasome system (UPS)‐dependent degradation of OsPIP2;1 to reduce the water loss in rice.
Results
Identification of the potential Drought‐Regulating gene OsRINGzf1
A drought‐related QTL cluster on the long arm of chromosome 4 in rice was found to harbour QTL genes related to plant height (PH), spikelet fertility (SF), panicle number (PN), spikelet number per panicle (SNP), and biomass (BM) (Liu et al., 2005; Zou et al., 2005). The QTLs for PH, SF, and tiller number (TN) under drought conditions were fine‐mapped to a 237‐kb marker interval (M60‐RM17363) (Figure 1a, Table S1), which contains 29 predicted genes (Table S2). Considering that the QTLs were detected under drought conditions, we compared the drought resistance of recombinant plants of M60‐RM17363 interval. Line 326 (L326), possessing the QTL interval from IRAT109, the male parent, displayed significantly higher survival rates (SR) and showed better drought resistance (Figure 1b). Then, we analysed the expression of genes located in this interval under drought stress except for 5 retrotransposon proteins (Figure S1). Among them, six genes were not expressed. Interestingly, a novel gene, OsRINGzf1, annotated with RING zinc finger protein (LOC_Os04g48050), showed persistent up‐regulation. Furthermore, the expression of OsRINGzf1 was significantly up‐regulated in leaf, sheath and stem under drought conditions (Figure 1c). These results imply its potential role in the regulation of drought resistance.
Figure 1.
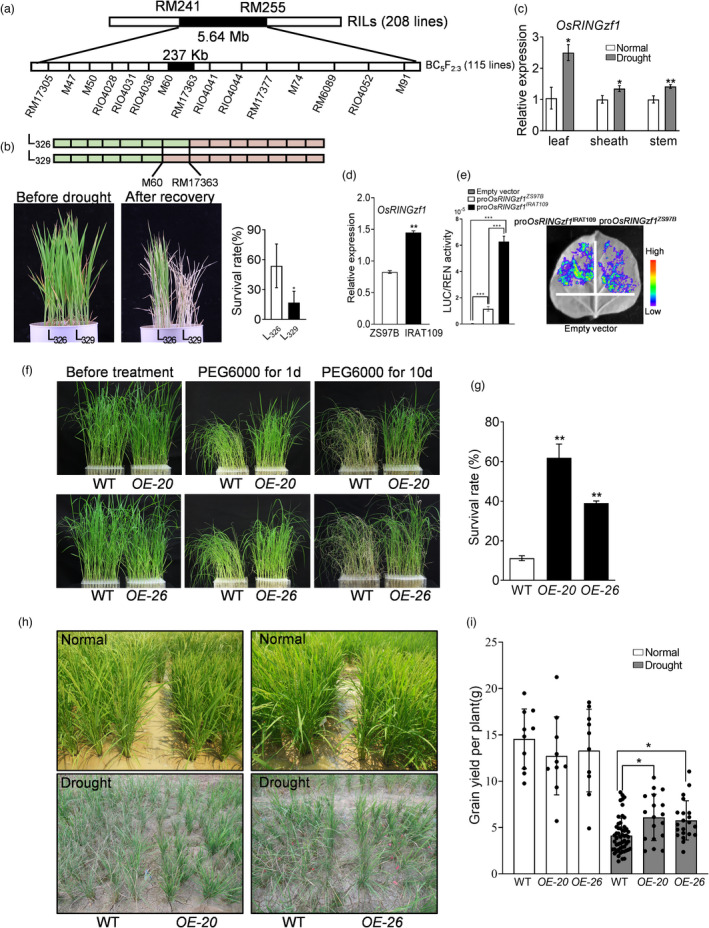
Functional identification of OsRINGzf1 gene on rice drought resistance. (a) The target QTL interval was narrowed down with a backcrossing population (BC5F2:3). (b) Drought stress response of the recombinant plants containing the target QTL interval. Genome schematic of two recombinant plants was indicated. Red represents the genome from receptor parent (Zhenshan97B), and green represents the genome from donor parent (IRAT109). Survival rate (SR) was calculated after 14 days of drought stress and 7 days of rehydration. (c) Expression profile of OsRINGzf1 under normal and drought conditions. (d) Differences in OsRINGzf1 expression between parents under normal conditions. (e) Relative luciferase activity of proOsRINGzf1‐LUC in N. benthamiana plants. The promoter fragment of Zhenshan97B was fused in proOsRINGzf1 ZS97B, and the promoter fragment of IRAT109 was fused in proOsRINGzf1 IRAT109. Relative reporter activity (LUC/REN) was calculated. (f) OsRINGzf1 improves the adaptation of rice to PEG‐simulated drought stress at the seedling stage. (g) The survival rate (SR) of WT, OE plants indicated in (f). (h) OsRINGzf1 improves the drought resistance of rice at the reproductive stage. (i) Grain yield of WT and OE plants under normal and drought stress conditions. Values in c, d, e and g are means ± SD based on three independent biological replicates. Asterisks represent the statistically significant differences according to a two‐tailed Student's t‐test. *P < 0.05, **P < 0.01 and ***P < 0.001.
Notably, there are 18 SNPs, including 16 substitutions and two indels in the promoter region, 1500 bp upstream of OsRINGzf1 between Zhenshan97B and IRAT109, two parents of the mapping population. In IRAT109, substitutions at −242 bp, −490 bp and −774 bp formed three TATA boxes, and substitution at −656 bp formed a CAAT box (Table S3). No differences were found in the coding sequence of OsRINGzf1 between the two parents. We proposed that these SNPs might affect the expression of OsRINGzf1 in the parents. Quantitative PCR showed that the expression level of OsRINGzf1 in IRAT109 was higher than that in Zhenshan97B (Figure 1d). The promoter of Zhenshan97B and IRAT109 was individually fused with luciferase (LUC) report gene and infiltrated into N. benthamiana leaves. Fluorescence signal with the promoter from IRAT109 was significantly higher than that of Zhenshan97B, implying that the promoter from IRAT109 possesses higher expression activity (Figure 1e).
We further sequenced the OsRINGzf1 promoter of 108 rice landraces, eight cultivars, and 17 wild species mainly from South China and South‐East Asia (Table S4). Among the 29 haplotypes, haplotype 3 group and 13 group contained 52 accessions including Zhenshan97B and 51 accessions including IRAT109, respectively (Table S4). There are 15 substitutions between haplotypes 3 and 13, which are the same as those of Zhenshan97B and IRAT109 (Table S3). The SNPs involved in TATA box and CAAT box related to expression regulation are included in these 15 SNPs. Phylogenetic analysis revealed that the promoter sequence of OsRINGzf1 has evolved from wild species into two main types, which is consistent with haplotype analysis (Figure S2). Together with Zhenshan 97B, IRAT109, and Nipponbare, the expression of OsRINGzf1 gene was examined in accessions randomly selected from haplotypes 3 and 13, respectively. The significant analysis between each accession and Nipponbare was analysed. The results showed that, in the haplotype 13 group, the expression levels of several accessions including IRAT109 were higher than those of Nipponbare. While for the haplotype 3 group, OsRINGzf1 expression in each accession was significantly lower than that of Nipponbare (Figure S3). These results collectively indicated that the sequence variation of OsRINGzf1 promoter could influence its expression.
To understand the function of OsRINGzf1 under drought stress, we investigated the response of OsRINGzf1 overexpression (OE), RNA interference (Ri) and knockout (KO) plants together with WT plants against drought stress. WT plants showed severe leaf rolling and wilting under 15% polyethylene glycol (PEG) 6000 treatment at the seedling stage (Figure 1f). The SR of WT plants is significantly lower than that of OsRINZzf1 OE plants (Figure 1g). With earlier and more severe leaf rolling, Ri plants of OsRINGzf1 are more sensitive to drought stress simulated by PEG (Figure S4a); the LRWC of OE plants was significantly higher than that of WT and Ri plants (Figure S4b, Table S5). In addition, KO plants were more sensitive to drought stress and have lower SR compared with WT plants (Figure S5). Similarly, OE plants with higher SR exhibited enhanced salt tolerance compared with WT plants, whereas KO plants exhibited the opposite (Figure S6). Furthermore, both the germination and the relative growth of KO plants were significantly inhibited under mannitol and NaCl stress during germination and budding stages. Under mannitol and NaCl stress, the relative growth of OE plants at the budding stage was significantly better than that of the WT plant (Figure S7). These results indicated that OsRINGzf1 plays a positive role in drought, salt and osmotic tolerance in rice.
We further examined the resistance of WT, OE and Ri plants to drought stress at the reproductive stage. The water supply was withdrawn at the early stage of panicle differentiation. The results showed that, compared with WT plants, Ri plants appeared earlier and more severe leaf rolling, while the OE plants exhibited delayed leaf rolling (Figure S8a). Similarly, the LRWC of OE plants was significantly higher than that of WT plants, whereas the LRWC of Ri plants performed the opposite (Figure S8b). The accumulation of proline, soluble protein, soluble sugar and the activities of catalase presented the same trend of change as LRWC (Figure S8c–f). In the field, OE plants exhibited better resistance than WT plants, with less dried‐up leaves and significantly less reduction in grain yield under drought conditions (Figure 1h–i). These results collectively demonstrated that OsRINGzf1 positively regulates rice drought resistance.
OsRINGzf1 has E3 ubiquitin ligase activity and is localized on ER and PM
Amino acid sequence analysis showed that OsRINGzf1 protein has 187 amino acids, with a RING domain in the C‐terminus and 3 transmembrane domains in the N‐terminus. RING domain alignment indicated that OsRINGzf1 is a RING‐H2‐type E3 ligase (Figure 2a). To investigate whether OsRINGzf1 is a functional E3 ligase enzyme, OsRINGzf1‐GST fusion protein was expressed in E. coli and the purified OsRINGzf1‐GST protein was used for in vitro self‐ubiquitination assay. The result showed that in the presence of E1, E2 and the ubiquitin protein, the self‐ubiquitinated form of OsRINGzf1‐GST was detected by both anti‐Ub and anti‐GST antibodies (Figure 2b –c). Meanwhile, when E1 or E2 was omitted, we could not detect obvious ubiquitination (Figure 2b–c). These results indicated that OsRINGzf1 possessed E3 ligase activity.
Figure 2.
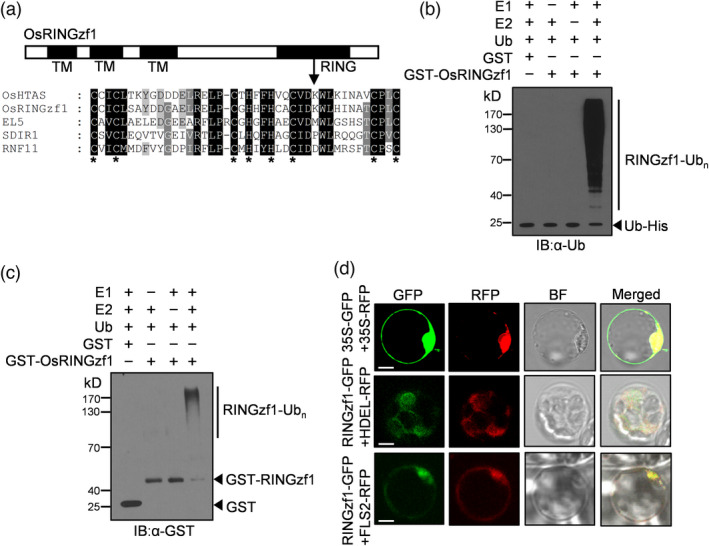
OsRINGzf1 possesses ubiquitin ligase activity. (a) Domain analysis of OsRINGzf1 and multiple alignment of RING domain. TM, transmembrane domain. RING, Really Interesting New Gene domain. The orthologs of RING‐H2‐type proteins including OsHTAS (ALN98169) and EL5 (BAA96874) from rice, SDIR1 (NP_191112) from Arabidopsis and RNF11 (NP_055187.1) from humans. Asterisk indicates conserved cysteine and histidine residues. (b) E3 ubiquitin ligase activity of OsRINGzf1. OsRINGzf1‐GST fusion protein was assayed for ubiquitin ligase activity in the presence of E1, E2 and ubiquitin (Ub). The numbers on the left indicate the molecular masses of marker proteins in kD. Ubiquitinated proteins were resolved by 10% (w/v) SDS‐PAGE, and then immunoblotted with anti‐ubiquitin antibodies. (c) Ubiquitinated proteins were immunoblotted with anti‐GST antibodies. (d) Subcellular localization of OsRINGzf1 in rice protoplasts. Green fluorescence is an indicative of OsRINGzf1‐GFP, whereas red fluorescence shows as indicator of HDEL‐RFP and FLS2‐RFP. BF, bright field. Scale bars, 10 μm.
OsRINGzf1 protein could be a membrane protein due to the three predicted transmembrane domains in its N‐terminus (Figure 2a). To identify the subcellular localization of OsRINGzf1, OsRINGzf1‐GFP fusion protein was introduced into rice protoplasts along with HDEL‐RFP (HDEL, as the endoplasmic reticulum marker) and FLS2‐RFP (FLS2, flagellin‐sensitive 2, as the plasma membrane marker), respectively. The green fluorescence signals of OsRINGzf1‐GFP overlapped with the red fluorescence signal of HDEL‐RFP and FLS2‐RFP marker (Figure 2d), suggesting that OsRINGzf1 is localized on the ER and PM of rice cells.
OsRINGzf1 interacts with OsPIP2;1
RING‐type E3 ligases were known to ubiquitinate specific target proteins and trigger their degradation via 26S proteasome. To decipher the molecular mechanism of OsRINGzf1 in response to drought stress, we attempted to identify the target proteins of OsRINGzf1. We firstly screened a rice cDNA library using OsRINGzf1 as the bait in the split‐ubiquitin membrane yeast two‐hybrid system. Ten positive colonies were identified, all of which were AQPs, including six plasma membrane intrinsic proteins (PIPs), two tonoplast intrinsic proteins (TIPs), one nodulin26‐like intrinsic protein (NIP) and one small basic intrinsic protein (SIP) (Table S6). We successfully cloned eight of the ten AQPs and performed the yeast two‐hybrid assay to test the interactions between OsRINGzf1 and these eight potential targets. Yeast strain containing OsPIP2;1 (LOC_Os07g26690.1), OsPIP2;2 (LOC_Os02g41860.1), OsPIP1;3 (LOC_Os02g57720.1), OsSIP1;1 (LOC_Os01g08660.1), OsPIP1;1 (LOC_Os02g44630.1) and a putative TIP protein (LOC_Os05g14240.1) could grow well on SD/−Leu/−Trp/‐His/−Ade selective medium (Figure S9a), implying their interactions with OsRINGzf1. We further examined the interaction between OsRINGzf1 and OsPIP2;1, which was known to have high water transport activities (Sakurai et al., 2008). Firstly, the yeast strain containing OsRINGzf1‐Cub and OsPIP2;1‐NubG turned blue on X‐gal indicator plate (Figure S9b). Secondly, a semi‐in vitro pull‐down experiment was conducted. The mixture of the protein extraction including the OsPIP2;1‐GFP fusion protein expressed in N. benthamiana leaves and the GST‐OsRINGzf1 fusion protein expressed in E. coli was separated by GFP attached beads, and then detected by anti‐GST and anti‐GFP antibodies. Western blot results showed that GST‐OsRINGzf1 could be pulled down in the presence of OsPIP2;1‐GFP (Figure 3a). Thirdly, the LUC complementation imaging assay showed that the fluorescence signal of LUC could be detected when OsRINGzf1‐Nluc and Cluc‐OsPIP2;1 were coinfiltrated into N. benthamiana leaves (Figure 3b). Furthermore, we asked whether OsRINGzf1 had similar subcellular localization with OsPIP2;1. To answer this question, we constructed 35S:OsPIP2;1‐RFP and 35S:OsRINGzf1‐GFP fusion vectors, and cointroduced the two constructs into rice protoplasts. Both red and green fluorescence signals displayed a similar pattern and overlapped with each other (Figure 3c). All these data demonstrated that OsRINGzf1 and OsPIP2;1 colocalized on ER and PM of rice cells and OsRINGzf1 could interact with OsPIP2;1.
Figure 3.
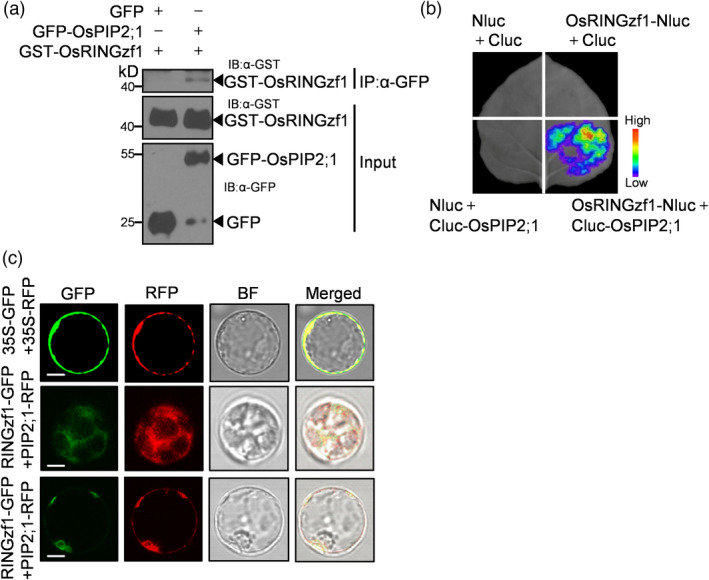
OsRINGzf1 interacts with OsPIP2;1. (a) OsRINGzf1 interacts with OsPIP2;1 in a semi‐in vitro pull‐down assay. Proteins were pulled down by GFP‐Trap agarose beads and detected by Western blots with antibody against GFP or GST. GFP was used as negative control. The numbers on the left indicate the molecular masses of marker proteins in kD. (b) OsRINGzf1 interacts with OsPIP2;1 in a luciferase (LUC) complementation imaging assay. The indicated constructs were transiently expressed in N. benthamiana for 48 h. LUC activity is depicted with false colour from low (blue) to high (red). Nluc, N‐terminal half of firefly LUC; Cluc, C‐terminal half of firefly LUC. (c) Subcellular colocalization of OsRINGzf1 and OsPIP2;1. 35S:OsRINGzf1‐GFP was transformed into rice protoplasts along with 35S: RFP‐OsPIP2;1. Scale bars, 10 μm.
OsRINGzf1 mediates the ubiquitination and UPS‐Dependent degradation of OsPIP2;1
The above findings that OsRINGzf1 exhibited E3 ligase activity and interacted with OsPIP2;1 prompted us to test whether OsPIP2;1 might be the ubiquitination target of OsRINGzf1. To validate this hypothesis, we first examined whether OsPIP2;1 is ubiquitinated in plants. The protein extraction of N. benthamiana leaves infiltrated with Agrobacterium containing OsPIP2;1‐GFP fusion protein was separated by GFP attached beads, and OsPIP2;1‐GFP was detected by immunoblot with anti‐GFP and anti‐Ub antibodies. The smear bands with higher molecular weight indicate that OsPIP2;1 was polyubiquitinated in vivo (Figure 4a). Furthermore, in vitro ubiquitination assay was conducted to examine whether OsPIP2;1 is the ubiquitination substrate of OsRINGzf1. As shown in Figure 4b, in the presence of E1, E2, ubiquitin and OsRINGzf1, OsPIP2;1 was ubiquitinated. These results collectively demonstrated that OsPIP2;1 is ubiquitinated by OsRINGzf1. Besides, our in vitro ubiquitination assay verified that OsSIP1;1 could also be ubiquitinated by OsRINGzf1 (Figure S10).
Figure 4.
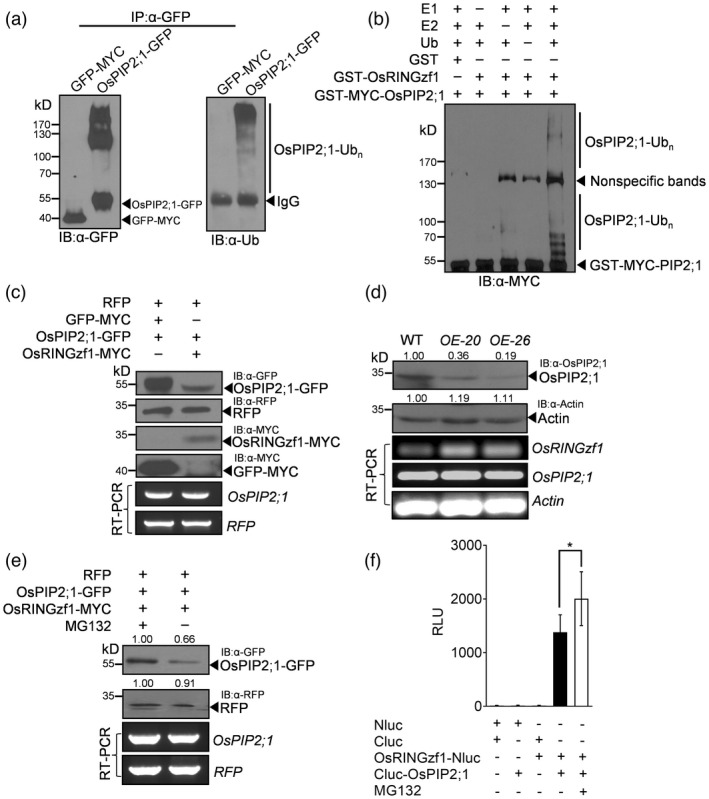
OsPIP2;1 is the ubiquitination substrate of OsRINGzf1, which mediates its UPS‐dependent degradation. (a) In vivo ubiquitination of OsPIP2;1 protein. GFP‐MYC was used as negative control. The numbers on the left indicate the molecular masses of marker proteins in kD. (b) In vitro ubiquitination of OsPIP2;1 protein. GST was used as a negative control. The numbers on the left indicate the molecular masses of marker proteins in kD. (c) OsRINGzf1‐MYC decreased OsPIP2;1 protein level. GFP‐MYC and RFP proteins were used as negative control. (d) OsPIP2;1 proteins were reduced in OsRINGzf1 OE plants. Actin was used as a loading control. (e) MG132 inhibited the reduction in OsPIP2;1‐GFP protein. (f) Fluorescent signal of luciferase of OsRINGzf1‐OsPIP2;1 interaction increased in the presence of MG132. RLU, relative luminescence unit. Value are means ± SD based on five independent biological replicates. Asterisks represent the statistically significant differences according to a two‐tailed Student's t‐test, *P < 0.05.
To investigate whether OsRINGzf1 mediates the degradation of OsPIP2;1, the Agrobacterium containing OsRINGzf1‐MYC fusion protein and the OsPIP2;1‐GFP fusion protein were coinfiltrated into N. benthamiana leaves. Western blot analysis revealed that the abundance of OsPIP2;1‐GFP was much lower in the coinfiltration of OsRINGzf1‐MYC and OsPIP2;1‐GFP combination (Figure 4c). Meanwhile, we detected the transcription of OsRINGzf1 and OsPIP2;1, and the protein abundance of OsPIP2;1 in WT and OsRINGzf1 OE plants (Figure 4d). The expression of OsRINGzf1 in OE plants was remarkably higher than that in WT plants, while for the transcription level of OsPIP2;1, there was no obvious difference between WT and OE plants (Figure 4d). However, the abundance of OsPIP2;1 protein in OE plants was significantly lower than that in WT plants (Figure 4d). According to these results, we concluded that OsRINGzf1 promotes OsPIP2;1 degradation in vivo. To examine whether 26S proteasome was involved in the degradation of OsPIP2;1 or not, we compared the change of OsPIP2;1 protein content treated with or without MG132, a proteasome inhibitor. The results displayed that in the protein extraction of N. benthamiana leaves coinfiltrated with OsPIP2;1‐GFP and OsRINGzf1‐MYC, the abundance of OsPIP2;1 protein was obviously higher in the presence of MG132 but lower without MG132 (Figure 4e). Moreover, the LUC fluorescence signal of OsRINGzf1‐OsPIP2;1 interaction in MG132‐treated N. benthamiana leaves was stronger than that without MG132 (Figure 4f). As the ubiquitin–proteasome pathway is an ATP‐dependent process, we further investigated whether the in vivo degradation of OsPIP2;1 is ATP‐dependent. We extracted protein from N. benthamiana leaves infiltrated with OsPIP2;1‐GFP and treated the protein extraction with or without ATP. Then, the abundance of OsPIP2;1‐GFP fusion protein was detected by immunoblot with anti‐GFP antibody. We found that the degradation of OsPIP2;1 is ATP‐dependent (Figure S11). Together, we concluded that OsRINGzf1 mediates the UPS‐dependent degradation of OsPIP2;1.
The interaction between OsRINGzf1 and OsPIP2;1 reduces the water loss under drought stress
In plants, AQPs are membrane channels that facilitate water movement across cell membranes. Among different AQPs families, PIPs and TIPs are found to be involved in water transport (Hove and Bhave, 2011). Downregulation of some AQPs decreased the membrane water permeability to avoid excessive water loss (Zargar et al., 2017). In this study, we found that OsPIP2;1 protein was remarkably reduced in OsRINGzf1 OE plant (Figure 4d), and the leaf rolling of OsRINGzf1 OE plants was delayed under drought stress compared with the WT and RNAi plants (Figure 1f, Figures S4 and S8). Furthermore, LRWC of OsRINGzf1 OE plant was significantly higher than that of WT plants under drought stress (Figures S4b and S8b). These results suggested that the reduction in OsPIP2;1 mediated by OsRINGzf1 leads to a better leaf water status. The better leaf water status is mainly attributed to a better root system or reduced leaf water loss. To investigate the underlying mechanism for the better water status of OE plants, we first observed the roots of WT, OE and Ri plants and found that under normal and drought conditions, the root length, volume, and dry weight of OE plants were smaller than those of WT plants, especially under drought conditions, the root weight and volume were significantly reduced. The root length, volume, and dry weight of Ri plants did not differ from WT under normal conditions but decreased under drought conditions (Figure S12). These results indicated that the better water status of OsRINGzf1 OE plant was not due to water absorption by the roots.
To further determine the role of OsRINGzf1 and OsPIP2;1 in the leaf water retention, we generated OsPIP2;1 OE plants and OsPIP2;1 mutants (pip2;1) (Figure 5d and Figure S13a). Three independent experiments were conducted to detect the LWLR of the detached leaves of OsPIP2;1 OE, pip2;1, OsRINGzf1 OE, and WT plants, respectively. Compared with WT plants, LWLR of OsRINGzf1 OE and pip2;1 plants was significantly lower, whereas the LWLR of OsPIP2;1 OE plants was remarkably higher (Figure 5a–c). OsPIP2;1 OE plants with significantly reduced SR were more sensitive to drought stress, while pip2;1 plants exhibited higher drought resistance than WT (Figure 5e–f and Figure S13b–c). These data indicated that excessive OsPIP2;1 might lead to loss of more leaf water, and the degradation of OsPIP2;1 mediated by OsRINGzf1 enables plants to enhance drought resistance through reducing leaf water loss.
Figure 5.
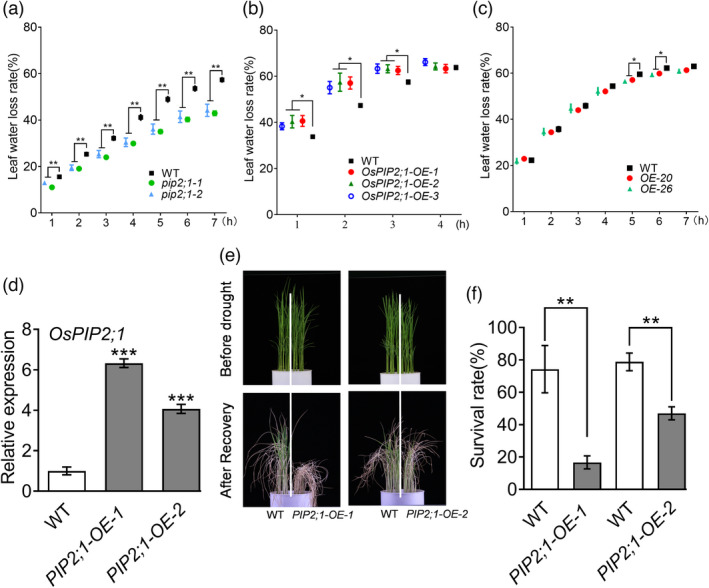
Leaf water loss rate (LWLR) of OsRINGzf1 and OsPIP2;1 transgenic plants and drought resistance of OsPIP2;1‐OE plants. (a) The LWLR of pip2;1 and WT plants. Values are means ± SE (n = 17). (b) The LWLR of OsPIP2;1‐OE and WT plants. Values are means ± SE (n = 5). (c) The LWLR of OsRINGzf1‐OE and WT plants. Values are means ± SE (n = 7). The WT and transgenic plants were grown in the field for 2 months. Fresh weights of detached leaves of wild‐type and the transgenic plants at the indicated time were determined. (d) Expression level of OsPIP2;1 gene in OsPIP2;1 OE plants. Values are means ± SD (n ≥ 10). (e) Phenotype of OsPIP2;1‐OE and WT plants under soil drought stress. (f) Survival rate of OsPIP2;1‐OE and WT plants under soil drought stress. Values are means ± SD based on three independent biological replicates. Asterisks represent the statistically significant differences according to a two‐tailed Student's t‐test. *P < 0.05, **P < 0.01 and ***P < 0.001.
Discussion
OsRINGzf1 is a novel gene involving the aquaporin ubiquitination
OsPIP2;1 is a homologue protein of AtPIP2;1 with 83% identity (Figure S14; Maurel et al., 2015). CaRma1H1, a RING‐type E3 ubiquitin ligase from hot pepper, and its homologues AtRma1, AtRma2 and AtRma3 in Arabidopsis were reported to ubiquitinate and degrade the AtPIP2;1, and significantly increase the drought tolerance of plants (Lee et al., 2009). Homologous alignment revealed the homologous protein OsRDCP1 of CaRma1H1 in rice, which can also improve the tolerance of rice to drought stress (Bae et al., 2011). However, the substrate of OsRDCP1 is still unclear. CaRma1H1 and OsRINGzf1 belong to two different E3 ligase subfamilies. OsRINGzf1 is RING‐H2 type E3 ligase, while CaRma1H1, AtRma1 and OsRDCP1 are RING‐HC‐type E3 ligases and were clustered into the same group (Figure S15). CaRma1H1 localizes on ER and prevents the trafficking of PIP2;1 from ER to PM (Lee et al., 2009), while OsRINGzf1 localizes at ER and PM, which is consistent with the localization of OsPIP2;1(Figures 2d and 3c). What's more, CaRma1H1 cannot enhance the tolerance of plant to high salinity (100 mm NaCl) (Lee et al., 2009), while OsRINGzf1 can improve the resistance to high salinity (150 mm NaCl) and osmotic stress (150 mm mannitol) (Figure S6 and S7). All these results imply that OsRINGzf1 is a novel E3 ligase that participates in the ubiquitination of AQPs and responds to various abiotic stresses.
OsRINGzf1 positively regulates drought resistance through modulating the Water Channel to enhance leaf water retention capacity
Drought resistance refers to the ability of plants to maintain high water status by absorbing water or reducing water loss in dry conditions (Luo, 2010). Aquaporins, especially for PIPs with high water transport activity, play an important role in water transcellular flow by regulating membrane water permeability (Chaumont and Tyerman, 2014; Heinen et al., 2009; Laloux et al., 2018). Overexpression of OsPIP2s, especially OsPIP2;1, increased yeast membrane water permeability, while the root hydraulic conductivity was decreased by approximately fourfold in OsPIP2;1 RNAi knock‐down plants (Ding et al., 2019). Transcriptional regulation and post‐translational modifications influence the abundance and activity of AQPs, which determines the flux of water within and across the cells under drought (Fox et al., 2017; Martre et al., 2002). However, the molecular regulatory mechanism of OsPIP2;1 under drought is still unclear. In Arabidopsis, a hot pepper E3 ligase CaRma1H1 and its homologues Rma1, Rma2 and Rma3 modulate the UPS‐dependent degradation of AtPIP2;1 (Lee et al., 2009). Recently, the endoplasmic reticulum‐associated degradation (ERAD) component ubiquitin‐conjugating enzyme UBC32 was found to act together with Rma1 to ubiquitinate a phosphorylated form of PIP2;1 and to promote its degradation (Chen et al., 2021). In this study, we identified a RING‐H2 E3 ligase, OsRINGzf1, and elucidated the positive drought resistance regulation mechanism of OsRINGzf1 (Figure 6). The up‐regulated expression of OsRINGzf1 under drought conditions promoted the ubiquitination and degradation by 26S proteasomes of OsPIP2;1, causing decreased water diffusion. As a result, the plant water loss decreased under drought.
Figure 6.
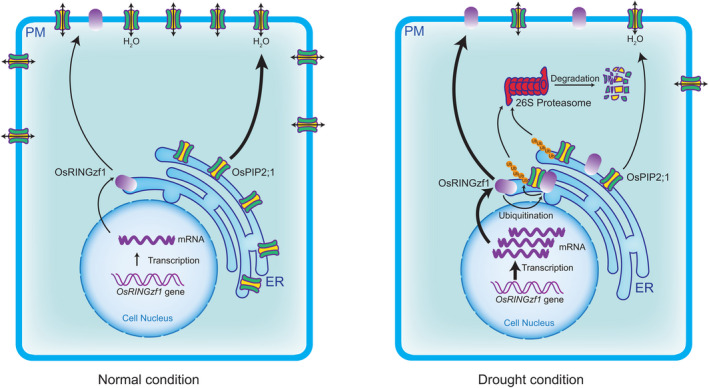
Proposed working model for OsRINGzf1‐mediated mechanism of drought resistance. The expression of OsRINGzf1 is increased under drought stress. OsRINGzf1 interacts with, ubiquitinates and targets OsPIP2;1 for 26S proteasome‐mediated degradation under drought in rice. ER, endoplasmic reticulum; PM, plasma membrane.
In addition, we found that the other three AQPs, OsPIP1;3, OsPIP2;2 and OsSIP1;1, interact with OsRINGzf1 (Figure S9). The in vitro ubiquitination assay directly confirmed that OsSIP1;1 can be ubiquitinated by OsRINGzf1 (Figure S10). OsPIP1;3, responds strongly to drought stress and can be recruited to the plasma membrane by OsPIP2;2, to be a water‐transporting facilitator (Lian et al., 2004; Liu et al., 2020). Recent research has revealed the highest water transporting activity of OsPIP2;2 among the 11 PIPs in rice, which is beneficial to enhance the drought tolerance in rice protoplasts and seedlings (Bai et al., 2021). SIPs had a moderate water transport activity (Maurel et al., 2015). VvSIP1, AtSIP1;1 and AtSIP1;2 are located at ER and possessed water transport activity (Ishikawa et al., 2005; Noronha et al., 2014). These results suggest that OsRINGzf1 enhances the water retention capacity under drought stress by interacting with multiple AQPs. Further research should be carried out around these findings.
The multiple roles of OsRINGzf1 in plants
In a network analysis of rice gene, OsRINGzf1 was proposed to be related to seed development and abiotic stress (Cooper et al., 2003). Our results indicated that OsRINGzf1 functions on plant agronomy, yield and resistance to abiotic stresses. Thus, we propose that OsRINGzf1 may play multiple roles through interacting with multiple substrates, including AQPs confirmed in this study. Plant AQPs are a large family and have been found to be not only water channel but also channels for H2O2 (Dynowski et al., 2008; Rodrigues et al., 2017), CO2 (Uehlein et al., 2003), NH3 (Loqué et al., 2005), glycerol (Biela et al., 1999) and micronutrients (boron, silicon, arsenic) (Ma et al., 2006, 2008; Takano et al., 2006). Therefore, the physiological role of AQPs might be involved in a diversity of functions such as the transport of micronutrients, signalling molecules and photosynthetic substrates, as well as water channels. In this study, we elucidate the effect of interaction between OsRINGzf1 and OsPIP2;1 on plant water status. Actually, PIP2;1 was found to be a water channel, as well as a channel for H2O2 and CO2 in Arabidopsis. The phosphorylation of PIP2;1 by OST1 not only contributed to ABA‐triggered stomatal closure (Grondin et al., 2015) but also involved CO2 permeability (Wang et al., 2016). Four PIP2 AQPs of barley, namely HvPIP2;1, HvPIP2;2, HvPIP2;3 and HvPIP2;5, were reported to facilitate CO2 transport across the oocyte cell membrane (Mori et al., 2014). The transportation of CO2 may be directly related to photosynthesis, which is a basic bioprocess and has a widespread influence on plants. In rice, OsPIP2;1 can be phosphorylated by OsCPK17 in a calcium‐dependent manner in response to cold stress (Almadanim et al., 2017). OsPIP2;1 was also found to have a distinct role in developing grains (Hayashi et al., 2015). OsPIP2;1 and OsPIP2;2 are involved in the rapid internode elongation of deep‐water rice (Muto et al., 2011). Furthermore, OsPIP1;3, another interacting protein of OsRINGzf1, could enhance tolerance to boron toxicity (Mosa et al., 2016) and chill (Matsumoto et al., 2009). OsPIP1;3 responds to exogenous nitric oxide (NO) stimulation and plays a role in NO signalling pathway in seed germination (Liu et al., 2007). All these findings support the hypothesis that OsRINGzf1 plays multiple regulatory roles in plant growth and stress responses through recruiting multiple AQPs (Figure S16).
In conclusion, OsRINGzf1 is a novel E3 ligase and plays a crucial role in the regulation of AQPs. OsRINGzf1 can ubiquitinate OsPIP2;1 and lead it to be degraded by 26S proteasome. This process reduces the abundance of water channel in the membrane system, intensifies the cell capacity of water retention and ultimately improves the drought resistance of rice. These results enrich the understanding of the regulation mechanism of AQPs and plant water status.
Materials and methods
Fine mapping
A recombinant line containing the target QTL interval from IRAT109 was selected for backcross with Zhenshan97B. Zhenshan97B and IRAT109 are the maternal and paternal plant of the mapping population used previously (Liu et al., 2005; Zou et al., 2005). Zhenshan97B is an indica variety, and IRAT109 is a japonica variety. A BC5F2:3 population containing 115 lines was developed for fine mapping. The population was planted in a drought resistance facility, and the drought stress was supplied as described previously (Zou et al., 2005). PH, TN, and the yield‐related traits under both normal water and drought stress were investigated. The QTLs were detected by mixed linear‐model composite interval mapping method at LOD >2.5 with ICIMAPPING. The primers used for fine mapping are shown in Table S7.
qPCR assay
For the expression‐level analysis of OsRINGzf1 and OsPIP2;1 in rice and in the infiltrated N. benthamiana leaves, total RNA was extracted from rice leaves of varieties indicated and N. benthamiana leaves using TRIzol (TaKaRa) according to the manufacturer's protocol. The full‐length cDNAs were reverse‐transcribed using a cDNA synthesis kit (TRANSGEN, AE311‐03). The reaction products were used as the template for qPCR according to the manufacturer's instructions (TRANSGEN, Beijing, China, AQ601‐04). Reaction mixtures include 50–500 ng template, 0.2 μm forward primer, 0.2 μm reverse primer, 10 μL of 2X PerfectStart Green qPCR SuperMix, and nuclease‐free water to a final volume of 20 μL. The qPCR was performed with the Bio‐Rad CFX96 system with the following reaction conditions: 95 °C for 30s, then 40 cycles of 95 °C for 5 s and 60 °C for 31 s. The primer sequences used for qPCR are shown in Table S7.
Plant growth conditions, stress treatment and trait measurement
To test the resistance of OsRINGzf1 transgenic plants to osmotic and salt stress at the germination and budding stage, seeds (for germination test) and germinated seeds (for budding test) were sown on 1/2MS medium containing 150 mm NaCl or 150 mm mannitol, respectively. The germination rates were calculated 5 days after sowing. Ten days after sowing, the relative growth, that is the ratio of the fresh weight of plants under stress conditions and normal conditions, was calculated. To test the resistance of OsRINGzf1 transgenic plants to drought and salt stress at the seedling stage, seeds were sowed on 96‐well plates in Hoagland hydroponic culture medium. The 4‐leaf seedlings were transferred into the nutrient solution containing 15% PEG6000 or 150 mm NaCl, respectively. The leaf relative water content (LRWC) was measured at 0, 12, 24 and 48 h after stress. The survival rate (SR) was calculated 10 days (for PEG) and 3 days (for NaCl) after stress, respectively. Dehydration stress was induced by transferring 4‐leaf seedlings to filter papers, and leaves were sampled at 0, 0.5, 2, 4, 6, and 8 h for expression analysis of genes located in the fine mapped QTL region. To test the resistance of OsRINGzf1 and OsPIP2;1 transgenic plants to soil drought stress at the seedling stages, seeds were sowed in the bucket, and drought stress was applied to seedlings at four‐leaf age. SR was calculated after 2 weeks of drought stress and 7 days of rehydration. For the experiment at the germination, budding and seedling stage, plants were grown in a climate chamber under a 14 h‐light/10‐h dark photoperiod at 30 °C in the day and 25 °C at night.
Resistance test experiment of OsRINGzf1 transgenic plants to soil drought stress at the reproductive stage was conducted in a rainproof shelter. Thirty‐day seedlings were transplanted in the buckets and field. The water supply was withdrawn at the early stage of panicle differentiation and lasted for 2 weeks for the plants in buckets and 3 weeks for the plants in the field. For the plants in the bucket, the first fully expanded leaves at the top were sampled for measurement of LRWC, catalase activity, and the content of proline, soluble sugar and soluble protein 10 days after stress. For the plants in the field, the yield‐related traits were measured after harvest.
The N. benthamiana, used as host plants for Agrobacterium‐mediated transient expression analysis, were grown in a growth chamber at 25 °C under a 16‐h light/8‐h dark cycle for 6 weeks before infiltration.
Haplotype analysis and phylogenetic analysis
Alignment of OsRINGzf1 promoter sequence of the materials indicated in Table S4 was performed by ClustalW (Thompson et al., 1994) using the MEGA5.1 software (Tamura et al., 2011). Then, the phylogenetic tree was constructed from the aligned sequences by neighbour‐joining tree method with 1000 bootstrap. The phylogenetic tree was visualized and annotated by the EvolView (www.evolgenius.info). The alignment result was used for haplotype analysis using DnaSP v5.
Plasmid construction and rice transformation
To generate constructs for recombinant proteins, coding sequence of OsRINGzf1 was inserted into pGEX‐6P‐1 with BamHI and EcoRI sites, and OsPIP2;1 was inserted into pGEX‐4 T‐2‐Myc with KpnI and XhoI sites. To generate constructs for transient expression, coding sequence of OsRINGzf1 was inserted into pCambia1300‐221‐Myc with XbaI and AscI sites, OsPIP2;1 was inserted into pCambia1300‐221‐GFP.1 with KpnI and BamHI sites. To generate constructs for OsRINGzf1 OE, coding sequence of OsRINGzf1 was inserted into pCAMBIA 1321 with BamHI and SacI sites. To generate constructs for OsRINGzf1 RNAi, the 382‐bp cDNA fragment of OsRINGzf1 was amplified by PCR and inserted as inverted repeats into the RNAi vector PTCK303 with KpnI /SpeI and BamHI/SacI sites, to generate hairpin RNAi constructs. The CRISPR/Cas9 vector system was used to generate constructs for OsPIP2;1 KO. The CRISPR/Cas9 vectors were constructed according to the protocol described previously (Ma et al., 2015). All these vectors were introduced into Nipponbare via Agrobacterium‐mediated transformation.
Subcellular localization
Coding sequence of OsRINGzf1 was cloned into the transient expression vector pGFP‐2, and then cotransformed with the ER marker HDEL‐RFP (De et al., 2011) or the PM marker FLS2‐RFP (Lal et al., 2018) into rice protoplasts via PEG‐mediated transformation. The fluorescence signal was observed with a confocal microscopy (Leica) about 16 h after transformation.
Yeast strains and growth conditions
Saccharomyces cerevisiae strain NMY51 was grown in yeast extract–peptone–dextrose media at the indicated temperatures. The presence of transgene was confirmed by growth on SD/−Leu/−Trp/‐His/−Ade plates (Clontech, Moutain view, CA, USA). The transformed yeasts were suspended in 0.9% NaCl to OD600 = 1.0. Then, 2 μL of suspension yeasts was spotted onto the indicated selection medium.
Bacterial strains and growth conditions
The Agrobacterium strain GV3101 was incubated overnight and resuspended in the activation buffer (10 mm MES, pH 5.6, 10 mm MgCl2 and 150 μm acetosyringone). The desired Agrobacterium combinations were mixed and incubated at room temperature for at least 2 h by gentle shaking at a final concentration of OD600 = 1 for each construct. The activated Agrobacterium strains were then infiltrated into fully expanded N. benthamiana leaves through a 1‐mL needleless syringe. E. coli BL21 (DE3) transformed with the indicated constructs were grown to a density OD600 of 0.4–0.5 on LB supplemented with 100 μg/mL ampicillin or 50 μg/mL kanamycin. Protein expression was then induced by adding 0.1 mm IPTG and incubated 3 h at 37 °C with agitation.
Yeast Two‐Hybrid screening
The dual‐membrane system (Dualsystems Biotech, Zurich, Switzerland) was used to screen interacting proteins according to the manufacturer's instructions. The coding region of OsRINGzf1 was cloned into the bait vector pBT3‐SUC and then transformed into yeast strain NMY51. The cDNA library from the callus of IRAT109 roots, stems, leaves, sheath, and young panicles, which was constructed by fusing cDNAs in the pPR3‐N prey vector, was cotransformed into the strain NMY51 with pBT3‐SUC‐OsRINGzf1. After two rounds of cDNA library screening, positive transformants were identified. To retest the interaction, OsPIP1;3, OsPIP2;1, OsPIP2;2, OsSIP1;1, and LOC_Os05g14240.1 were constructed in pPR3‐N prey vector, respectively, and cotransformed with pBT3‐SUC‐OsRINGzf1 into NMY51. The cotransformed mixtures were plated on synthetic dropout medium SD/−Leu/−Trp/–His/−Ade to detect the interactions.
E3 ubiquitin ligase activity assay
The full‐length cDNA of OsRINGzf1 was cloned into the pGEX‐6P‐1 vector, fused to the C‐terminal GST, and expressed in Escherichia coli strain BL21 (DE3). The OsRINGzf1‐GST fusion protein was prepared according to the instruction manual (Amersham Biosciences, Piscataway, NJ, USA). For E3 ubiquitin ligase activity assay of the fusion proteins, the reaction containing recombinant wheat (Triticum aestivum) E1 (GI:136632), human E2 (UBCH5b; approximately 40 ng), purified E3 OsRINGzf1‐GST (approximately 1 μg) and purified Arabidopsis (Arabidopsis thaliana) ubiquitin (a ubiquitin monomer of UBQ14; At4g02890; approximately 2 mg) fused to a His‐tag. The reaction was incubated at 30 °C for 1.5 h. In vitro E3 ligase assays were performed as previously described (Xie et al., 2002; Zhang et al., 2007). For protein expression, E. coli BL21 (DE3) transformed with the indicated constructs were grown to a density OD600 of 0.4–0.5 on LB supplemented with 100 μg/mL of ampicillin or 50 μg/mL kanamycin. Protein expression was then induced by adding 0.1 mm IPTG and incubated at 37 °C for 3 h with agitation.
In vitro ubiquitination assay
For in vitro ubiquitination assay, crude extracts containing recombinant wheat (Triticum aestivum) E1 (GI: 136632), human E2 (UBCH5b; approximately 40 ng), purified OsRINGzf1(1 mg) fused to GST tag, purified Arabidopsis ubiquitin (UBQ14; At4g02890; approximately 2 μg) fused to His‐tag and purified OsPIP2;1‐GST‐MYC (2 mg) fused to both GST and MYC tags were used for the assay. GST was used as the negative control. Reactions were performed following the protocol described previously (Xie et al., 2002). The reaction was stopped by the addition of 4× protein loading sample buffer (258 mm Tris–HCl, pH 6.8, 8% SDS, 40% glycerol, 0.4% Coomassie Brilliant Blue and 0.4 m β‐mercaptoethanol) and boiling for 10 min. Samples were detected with anti‐MYC antibody.
Semi‐in vitro pull‐down assay
Agrobacterium strains GV3101 containing OsPIP2;1‐GFP or GFP were infiltrated into N. benthamiana leaves, respectively. The infiltrated leaves were ground into powder in liquid nitrogen and dissolved in protein extraction buffer (50 mm Tris‐MES, pH 8.0, 0.5 m sucrose, 1 mm MgCl2, 10 mm EDTA, 5 mm DTT, protease inhibitor cocktail complete Mini Tablets); then, the extracts were centrifuged thrice at 13200 g at 4 °C for 10 min. The supernatants were incubated with GFP beads at 4 °C for 1 h. The beads were washed 5 times with PBS buffer and incubated with OsRINGzf1‐GST at 4 °C for 1 h. The beads were washed 5 times and boiled in 4 × protein loading sample buffer for 10 min, separated in an SDS‐PAGE gel and detected with anti‐GST antibodies.
In vivo degradation assay and the MG132 treatment
For in vivo degradation assay, we followed the protocol by Liu et al. (2010). We coinfiltrated the Agrobacterium strains carrying the OsPIP2;1‐GFP and OsRINGzf1‐MYC plasmids into N. benthamiana leaves. Meanwhile, GFP‐MYC and OsPIP2;1‐GFP were also coinfiltrated into N. benthamiana and act as a negative control, and the RFP was infiltrated and acts as an internal control. Three days after infiltration, samples were collected for protein and RNA extraction. For protein‐level analysis, the extracts were analysed using anti‐GFP antibody, anti‐MYC antibody and anti‐RFP antibody. For RNA‐level expression analysis, RT‐PCR was performed. For MG132 treatment, MG132 was infiltrated into N. benthamiana leaves at a final concentration of 50 mm 24 h before sampling, and DMSO was used as a control. The harvested tissues were used for Western blot and RT‐PCR analyses.
Split‐luciferase complementation imaging assay
Split‐LUC complementation assay was performed as previously described (Chen et al. (2008). The coding regions of OsRINGzf1 and OsPIP2;1 were cloned into pCAMBIA1300‐35S‐Nluc and pCAMBIA1300‐35S‐Cluc, respectively. Agrobacterium strains GV3101 containing the indicated constructs were infiltrated into Nicotiana benthamiana leaves. After infiltration, plants were incubated at 25 °C for 48 h, and the LUC fluorescence signal was detected by a NightSHADE LB 985 in vivo Plant Imaging System (Berthold Technology, Bad Wildbad, Germany). To detect LUC activity, 1 mm D‐luciferin solution (Promega, Madison, WI, USA, Cat. No: E1602) was sprayed onto the leaves of N. benthamiana, followed by incubation in darkness for 5–10 min. Image was captured using a low‐light cooled CCD imaging apparatus (NightOWL II LB983 with Indigo software). Images was taken with an exposure time of 3 min.
To quantify protein–protein interaction intensity, leaf discs infiltrated with the desired Agrobacterium combinations were taken and incubated with 50 μL water containing 1 mm D‐luciferin in a Costar® 96‐Well White Flat Bottom Polystyrene High Bind Microplate (Corning, Corning, NY, USA, Cat. No: 7400) for 5 min. Luminescence activity was measured using the GLOMAX 96 microplate luminometer (Promega). Protein–protein interaction intensity was shown as relative luminescence unit (RLU). For MG132 treatment, MG132 was infiltrated into N. benthamiana leaves at a final concentration of 50 mm 24 h before sampling, and DMSO was used as a control. To avoid the bias of transfection efficiency, we used at least 12 individual leaves of six tobacco plants for each combination.
Antibodies
The antibodies used in this assay were as follows: anti‐Ub monoclonal antibody produced by Xie's laboratory, anti‐GST (1 : 2000 diluted, EASY‐BIO, Seoul, Korea, Cat# BE2013), anti‐GFP (1 : 2000 diluted, Roche, Basel, Switzerland, Cat#11814460001), anti‐Myc (1 : 3000 diluted, EASY‐BIO, Seoul, Korea, Cat#BE2073), anti‐RFP (1 : 2000 diluted, EASY‐BIO, Seoul, Korea, Cat# BE2023), anti‐actin (1 : 5000 diluted, EASY‐BIO, Seoul, Korea, Cat#BE0027), anti‐PIP2;1 (1 : 2000 diluted, COSMO‐BIO, Tokyo, Japan, Cat#COP‐080024), goat anti‐rabbit (1 : 3000 diluted, Proteintech, Chicago, IL, USA, Cat# SA00001‐2) and goat anti‐mouse (1 : 3000 diluted, Proteintech, Chicago, IL, USA, Cat# SA00001‐1).
Accession number
The cDNA and protein sequences of OsRINGzf1 were deposited in NCBI GenBank under the accession No. KC004026. Other sequence data from this article can be found in NCBI database under following accession number: CaRma1H1 (AAR99376), AtRma1 (NP_974506), AtRma2 (AEE85462.1), AtRma3 (NP_194477.2), OsRDCP1 (BAS90208), HsRma1 (BAB39359), OsHTAS (ALN98169), EL5 (BAA96874), SDIR1 (NP_191112), RNF11 (NP_055187), OsPIP2;1(XP_015646828) and AtPIP2;1 (NP_001030851).
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Chen SJ designed and performed the experiments, conducted gene cloning, expression profile analysis, pull down assay, ubiquitination assay and LUC complementation imaging assays, and wrote the manuscript; Xu K constructed the vectors and wrote the manuscript; Kong DY conducted the yeast two‐hybrid; Wu LY performed LUC complementation imaging assays; Chen Q performed the E3 ligase assay in vitro; Ma XS fine‐mapped the gene and investigated the phenotype of materials; Ma SQ identified the Subcellular localization of proteins; Li TF generated transgenic rice plants; Xie Q designed the study; Liu HY designed and supervised the study, and wrote the manuscript; Luo LJ conceived the project, supervised the study and wrote the manuscript. All the authors carefully read and discussed the manuscript.
Supporting information
Figure S1 Expression profiles of candidate genes in the fine mapped QTL interval under dehydration stress.
Figure S2 Phylogenetic analysis of OsRINGzf1 promoter sequence.
Figure S3 Expression of OsRINGzf1 in two main haplotypes.
Figure S4 OsRINGzf1 positively regulates rice drought resistance under simulated drought stress at the seedling stage.
Figure S5 Knockout of OsRINGzf1 reduces the resistance of rice to drought stress.
Figure S6 OsRINGzf1 improves the salt tolerance of rice.
Figure S7 OsRINGzf1 enhances the tolerance to osmotic and salt stress at the germination and budding stage.
Figure S8 OsRINGzf1 enhances the drought resistance in rice at the reproductive stage.
Figure S9 Yeast two‐hybrid assays to test the interaction between OsRINGzf1 and AQPs.
Figure S10 In vitro ubiquitination of OsSIP1; 1 by OsRINGzf1.
Figure S11 In vivo degradation of OsPIP2;1 is an ATP dependent process.
Figure S12 Roots phenotype of OsRINGzf1 OE, Ri and WT plants under normal and drought conditions.
Figure S13 Knockout of OsPIP2;1 increases the resistance of rice to drought stress.
Figure S14 Amino acid sequence alignment of PIP2;1 in rice (Oryza sativa japonica cv Nipponbare) and Arabidopsis thaliana.
Figure S15 Clustering analysis and homology alignment of OsRINGzf1, CaRma1H1 and its homologues.
Figure S16 A proposed model for multiple roles of OsRINGzf1 in rice abiotic stresses resistance and development through interacting with rice AQPs.
Table S1 QTLs detected by mixed linear‐model composite interval mapping across two soil water level.
Table S2 Putative genes located in the fine mapped QTL interval.
Table S3 SNPs Between Zhenshan97B and IRAT109, and Haplotype 3 and 13.
Table S4 Germplasm for phylogenetic analysis.
Table S5 Significance analysis of LRWC in WT, OsRINGzf1 OE and Ri seedlings under drought stress.
Table S6 Genes encoding for proteins that interact with OsRINGzf1 screened by split‐ubiquitin membrane yeast two hybrid system (DUAL membrane system).
Table S7 Primers used for fine mapping, gene expression analysis, cloning and vector construction.
Acknowledgements
We thank Prof. Yaoguang Liu for providing the CRISPR/Cas9 vector system. This study was supported by the National Key Research and Development Program of China (No. 2018YFE0106200), National Natural Science Foundation of China (Grant No. 31930080, 31100237, 31100862), Natural Science Foundation of Shanghai (19ZR1446600), International S&T Cooperation Program of Shanghai (19310711700), SAAS Program for Runup Talent Project (ZP21231) and National Program of Key Basic Research Project (973 Program) (2012CB114305).
Contributor Information
Qi Xie, Email: qxie@genetics.ac.cn.
Hongyan Liu, Email: lhy@sagc.org.cn.
Lijun Luo, Email: lijun@sagc.org.cn.
References
- Almadanim, M.C. , Alexandre, B.M. , Rosa, M.T.G. , Sapeta, H. , Leitão, A.E. , Ramalho, J.C. , Lam, T.T. et al. (2017) Rice calcium‐dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose‐phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 40, 1197–1213. [DOI] [PubMed] [Google Scholar]
- Ayadi, M. , Brini, F. and Masmoudi, K. (2019) Overexpression of a wheat aquaporin gene, TdPIP2;1, enhances salt and drought tolerance in transgenic durum wheat cv. Maali. Int. J. Mol. Sci. 20, 2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, H. , Kim, S.K. , Cho, S.K. , Kang, B.G. and Kim, W.T. (2011) Overexpression of OsRDCP1, a rice RING domain‐containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.). Plant Sci. 180, 775–782. [DOI] [PubMed] [Google Scholar]
- Bai, J.Q. , Wang, X. , Yao, X.H. , Chen, X.C. , Lu, K. , Hu, Y.Q. , Wang, Z.D. et al. (2021) Rice aquaporin OsPIP2;2 is a water‐transporting facilitator in relevance to drought‐tolerant response. Plant Direct, 5, e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biela, A. , Grote, K. , Otto, B. , Hoth, S. , Hedrich, R. and Kaldenhoff, R. (1999) The Nicotiana tabacum plasma membrane aquaporin NtAQP1 is mercury‐insensitive and permeable for glycerol. Plant J. 18, 565–570. [DOI] [PubMed] [Google Scholar]
- Chapagain, S. , Park, Y.C. , Kim, J.H. and Jang, C.S. (2018) Oryza sativa salt‐induced RING E3 ligase 2 (OsSIRP2) acts as a positive regulator of transketolase in plant response to salinity and osmotic stress. Planta, 247, 925–939. [DOI] [PubMed] [Google Scholar]
- Chaumont, F. and Tyerman, S.D. (2014) Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol. 164, 1600–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Zou, Y. , Shang, Y. , Lin, H. , Wang, Y. , Cai, R. , Tang, X. , et al. (2008) Firefly luciferase complementation imaging assay for protein‐protein interactions in plants. Plant Physiol., 146(2), 368–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. , Liu, R.J. , Wu, Y.R. , Wei, S.W. , Wang, Q. , Zheng, Y.N. , Xia, R. et al. (2021) ERAD‐related E2 and E3 enzymes modulate the drought response by regulating the stability of PIP2 aquaporins. Plant Cell, 33, 2883–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, B. , Clarke, J.D. , Budworth, P. , Kreps, J. , Hutchison, D. , Park, S. , Guimil, S. et al. (2003) A network of rice genes associated with stress response and seed development. Proc. Natl Acad. Sci. USA, 100, 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De, C.M. , Lenucci, M.S. , Di Sansebastiano, G.P. , Dalessandro, G. , De Lorenzo, G. and Piro, G. (2011) Protein trafficking to the cell wall occurs through mechanisms distinguishable from default sorting in tobacco. Plant J. 65, 295–308. [DOI] [PubMed] [Google Scholar]
- Ding, L. , Uehlein, N. , Kaldenhoff, R. , Guo, S. , Zhu, Y. and Kai, L. (2019) Aquaporin PIP2;1 affects water transport and root growth in rice (Oryza sativa L.). Plant Physiol. Biochem. 139, 152–160. [DOI] [PubMed] [Google Scholar]
- Dynowski, M. , Schaaf, G. , Loque, D. , Moran, O. and Ludewig, U. (2008) Plant plasma membrane water channels conduct the signalling molecule H2O2 . Biochem. J. 414, 53–61. [DOI] [PubMed] [Google Scholar]
- Fox, A.R. , Maistriaux, L.C. and Chaumont, F. (2017) Toward understanding of the high number of plant aquaporin isoforms and multiple regulation mechanisms. Plant Sci. 264, 179–187. [DOI] [PubMed] [Google Scholar]
- Fox, A.R. , Scochera, F. , Laloux, T. , Filik, K. , Degand, H. , Morsomme, P. , Alleva, K. et al. (2020) Plasma membrane aquaporins interact with the endoplasmic reticulum resident VAP27 proteins at ER–PM contact sites and endocytic structures. New Phytol. 228, 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, T. , Wu, Y. , Zhang, Y. , Liu, L. , Ning, Y. , Wang, D. , Tong, H. et al. (2011) OsSDIR1 overexpression greatly improves drought tolerance in transgenic rice. Plant Mol. Biol. 76, 145–156. [DOI] [PubMed] [Google Scholar]
- Grondin, A. , Rodrigues, O. , Verdoucq, L. , Merlot, S. , Leonhardt, N. and Maurel, C. (2015) Aquaporins contribute to ABA‐triggered stomatal closure through OST1‐mediated phosphorylation. Plant Cell, 27, 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez, C. , Veljanovski, V. , Reinhardt, H. , Guillaumot, D. , Vanhee, C. , Chaumont, F. and Batoko, H. (2014) The Arabidopsis abiotic stress‐induced TSPO‐related protein reduces cell‐surface expression of the aquaporin PIP2;7 through protein‐protein interactions and autophagic degradation. Plant Cell, 26, 4974–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, H. , Ishikawa‐Sakurai, J. , Murai‐Hatano, M. , Ahamed, A. and Uemura, M. (2015) Aquaporins in developing rice grains. Biosci. Biotechnol. Biochem. 79, 1422–1429. [DOI] [PubMed] [Google Scholar]
- Heinen, R.B. , Ye, Q. and Chaumont, F. (2009) Role of aquaporins in leaf physiology. J. Exp. Bot. 60, 2971–2985. [DOI] [PubMed] [Google Scholar]
- Hove, R.M. and Bhave, M. (2011) Plant aquaporins with non‐aqua functions: deciphering the signature sequences. Plant Mol. Biol. 75, 413–430. [DOI] [PubMed] [Google Scholar]
- Huang, L.Y. , Zhang, F. , Wang, W.S. , Zhou, Y.L. , Fu, B.Y. and Li, Z.K. (2014) Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genomics, 15, 1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, S.G. , Kim, J.J. , Lim, S.D. , Park, Y.C. , Moon, J.C. and Jang, C.S. (2016) Molecular dissection of Oryza sativa salt‐induced RING finger protein 1 (OsSIRP1): possible involvement in the sensitivity response to salinity stress. Physiol. Plant. 158, 168–179. [DOI] [PubMed] [Google Scholar]
- Ishikawa, F. , Suga, S. , Uemura, T. , Sato, M.H. and Maeshima, M. (2005) Novel type aquaporin SIPs are mainly localized to the ER membrane and how cell‐specific expression in Arabidopsis thaliana . FEBS Lett. 579, 5814–5820. [DOI] [PubMed] [Google Scholar]
- Kim, J.H. and Jang, C.S. (2020) E3 ligase, the Oryza sativa salt‐induced RING finger protein 4(OsSIRP4), negatively regulates salt stress responses via degradation of the OsPEX11‐1 protein. Plant Mol. Biol. 105, 231–245. [DOI] [PubMed] [Google Scholar]
- Kumar, A. , Dixit, S. , Ram, T. , Yadaw, R.B. , Mishra, K.K. and Mandal, N.P. (2014) Breeding high‐yielding drought‐tolerant rice: genetic variations and conventional and molecular approaches. J. Exp. Bot. 65, 6265–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal, N.K. , Nagalakshmi, U. , Hurlburt, N.K. , Flores, R. , Bak, A. , Sone, P. , Ma, X. et al. (2018) The receptor‐like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe, 23, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloux, T. , Junqueira, B. , Maistriaux, L.C. , Ahmed, J. , Jurkiewicz, A. and Chaumont, F. (2018) Plant and mammal aquaporins: same but different. Int. J. Mol. Sci. 19, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.K. , Cho, S.K. , Son, O. , Xu, Z. , Hwang, I. and Kim, W.T. (2009) Drought stress‐induced Rma1H1, a RING membrane‐anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell, 21, 622–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Liu, Q.W. , Feng, H. , Deng, J. , Zhang, R.X. , Wen, J.Q. , Dong, J.L. et al. (2019) Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy, 16, 862–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, H.L. , Yu, X. , Ye, Q. , Ding, X. , Kitagawa, Y. , Kwak, S.S. , Su, W.A. et al. (2004) The role of aquaporin RWC3 in drought avoidance in rice. Plant Cell Physiol. 45, 481–489. [DOI] [PubMed] [Google Scholar]
- Lim, S.D. , Cho, H.Y. , Park, Y.C. , Ham, D.J. , Lee, J.K. and Jang, C.S. (2013) The rice RING finger E3 ligase, OsHCI1, drives nuclear export of multiple substrate proteins and its heterogeneous overexpression enhances acquired thermotolerance. J. Exp. Bot. 64, 2899–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S.D. , Lee, C. and Jang, C.S. (2014) The rice RING E3 ligase, OsCTR1, inhibits trafficking to the chloroplasts of OsCP12 and OsRP1, and its overexpression confers drought tolerance in Arabidopsis. Plant Cell Environ. 37, 1097–1113. [DOI] [PubMed] [Google Scholar]
- Lim, S.D. , Yim, W.C. , Moon, J.C. , Kim, D.S. , Lee, B.M. and Jang, C.S. (2010) A gene family encoding RING finger proteins in rice: their expansion, expression diversity, and co‐expressed genes. Plant Mol. Biol. 72, 369–380. [DOI] [PubMed] [Google Scholar]
- Liu, H.Y. , Yu, X. , Cui, D.Y. , Sun, M.H. , Sun, W.N. , Tang, Z.C. , Kwak, S.S. et al. (2007) The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 17, 638–649. [DOI] [PubMed] [Google Scholar]
- Liu, H.Y. , Zou, G.H. , Liu, G.L. , Hu, S.P. , Li, M.S. , Yu, X.Q. , Mei, H.W. et al. (2005) Correlation analysis and QTL identification for canopy temperature, leaf water potential and spikelet fertility in rice under contrasting moisture regimes. Chin. Sci. Bull. 50, 317–326. [Google Scholar]
- Liu, J. , Zhang, C. , Wei, C. , Liu, X. , Wang, M. , Yu, F. , Xie, Q. et al. (2016) The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2‐induced stomatal closure in Rice. Plant Physiol. 170, 429–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.Y. , Fukumoto, T. , Gena, P. , Feng, P. , Sun, Q. , Li, Q. , Matsumoto, T. et al. (2020) Ectopic expression of a rice plasma membrane intrinsic protein (OsPIP1;3) promotes plant growth and water uptake. Plant J. 102, 779–796. [DOI] [PubMed] [Google Scholar]
- Luo, L.J. (2010) Breeding for water‐saving and drought‐resistance rice (WDR) in China. J. Exp. Bot. 61, 3509–3517. [DOI] [PubMed] [Google Scholar]
- Liu, L.J. , Zhang, Y.Y. , Tang, S.Y. , Zhao, Q.Z. , Zhang, Z.H. , Zhang, H.W. , Dong, L. et al. (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana . Plant J. 61, 893–903. [DOI] [PubMed] [Google Scholar]
- Loqué, D. , Ludewig, U. , Yuan, L. and von Wirén, N. (2005) Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol. 137, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.F. , Tamai, K. , Yamaji, N. , Mitani, N. , Konishi, S. , Katsuhara, M. , Ishiguro, M. et al. (2006) A silicon transporter in rice. Nature, 440, 688–691. [DOI] [PubMed] [Google Scholar]
- Ma, J.F. , Yamaji, N. , Mitani, N. , Xu, X.Y. , Su, Y.H. , McGrath, S.P. and Zhao, F.J. (2008) Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl Acad. Sci. USA, 105, 9931–9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martre, P. , Morillon, R. , Barrieu, F. , North, G.B. , Nobel, P.S. and Chrispeels, M.J. (2002) Plasma membrane aquaporins play a significant role during recovery from water deficit. Plant Physiol. 130, 2101–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al. (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Matsumoto, T. , Lian, H.L. , Su, W.A. , Tanaka, D. , CW, L. , Iwasaki, I. and Kitagawa, Y. (2009) Role of the aquaporin PIP1 subfamily in the chilling tolerance of rice. Plant Cell Physiol. 50, 216–229. [DOI] [PubMed] [Google Scholar]
- Maurel, C. , Boursiac, Y. , Luu, D.T. , Santoni, V. , Shahzad, Z. and Verdoucq, L. (2015) Aquaporins in plants. Physiol. Rev. 95, 1321–1358. [DOI] [PubMed] [Google Scholar]
- Mori, I.C. , Rhee, J. , Shibasaka, M. , Sasano, S. , Kaneko, T. , Horie, T. and Katsuhara, M. (2014) CO2 transport by PIP2 aquaporins of barley. Plant Cell Physiol. 55, 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosa, K.A. , Kumar, K. , Chhikara, S. , Musante, C. , White, J.C. and Dhankher, O.P. (2016) Enhanced Boron tolerance in plants mediated by bidirectional transport through plasma membrane intrinsic proteins. Sci. Rep. 6, 21640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto, Y. , Segami, S. , Hayashi, H. , Sakurai, J. , Murai‐Hatano, M. , Hattori, Y. , Ashikari, M. et al. (2011) Vacuolar proton pumps and aquaporins involved in rapid internode elongation of Deepwater rice. Biosci. Biotechnol. Biochem. 75, 114–122. [DOI] [PubMed] [Google Scholar]
- Noronha, H. , Agasse, A. , Martins, A.P. , Berny, M.C. , Gomes, D. , Zarrouk, O. , Thiebaud, P. et al. (2014) The grape aquaporins VvSIP1 transports water across the ER membrane. J. Exp. Bot. 65, 981–993. [DOI] [PubMed] [Google Scholar]
- Park, G.G. , Park, J.J. , Yoon, J. , Yu, S.N. and An, G. (2010) A RING finger E3 ligase gene, Oryza sativa delayed seed germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol. Biol. 74, 467–478. [DOI] [PubMed] [Google Scholar]
- Park, Y.C. , Chapagain, S. and Jang, C.S. (2018a) A negative regulator in response to salinity in Rice: Oryza sativa salt‐, ABA‐ and Drought‐induced RING finger protein 1 (OsSADR1). Plant Cell Physiol. 59, 575–589. [DOI] [PubMed] [Google Scholar]
- Park, Y.C. , Chapagain, S. and Jang, C.S. (2018b) The microtubule‐associated RING finger protein 1 (OsMAR1) acts as a negative regulator for salt‐stress response through the regulation of OCPI2 (O. sativa chymotrypsin protease inhibitor 2). Planta, 247, 875–886. [DOI] [PubMed] [Google Scholar]
- Park, Y.C. , Lim, S.D. , Moon, J.C. and Jang, C.S. (2019) A rice really interesting new gene H2‐type E3 ligase, OsSIRH2‐14, enhances salinity tolerance via ubiquitin/26S proteasome‐mediated degradation of salt‐related proteins. Plant Cell Environ. 42, 3061–3076. [DOI] [PubMed] [Google Scholar]
- Qin, Q. , Wang, Y.X. , Huang, L.Y. , Du, F.P. , Zhao, X.Q. , Li, Z.K. , Wang, W.S. et al. (2019) A U‐box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 102, 89–107. [DOI] [PubMed] [Google Scholar]
- Rodrigues, O. , Reshetnyak, G. , Grondin, A. , Saijo, Y. , Leonhardt, N. , Maurel, C. and Verdoucq, L. (2017) Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA‐ and pathogen‐triggered stomatal closure. Proc. Natl Acad. Sci. USA, 114, 9200–9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai, J. , Ahamed, A. , Murai, M. , Maeshima, M. and Uemura, M. (2008) Tissue and cell‐specific localization of rice aquaporins and their water transport activities. Plant Cell Physiol. 49, 30–39. [DOI] [PubMed] [Google Scholar]
- Sakurai, J. , Ishikawa, F. , Yamaguchi, T. , Uemura, M. and Maeshima, M. (2005) Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 46, 1568–1577. [DOI] [PubMed] [Google Scholar]
- Sharma, G. , Giri, J. and Tyagi, A.K. (2015) Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water‐deficit stress in Arabidopsis. Plant Sci. 237, 80–92. [DOI] [PubMed] [Google Scholar]
- Takano, J. , Wada, M. , Ludewig, U. , Schaaf, G. , von Wirén, N. and Fujiwara, T. (2006) The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell, 18, 1498–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol., 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehlein, N. , Lovisolo, C. , Siefritz, F. and Kaldenhoff, R. (2003) The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature, 425, 734–737. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Hu, H. , Qin, X. , Zeise, B. , Xu, D. , Rappel, W.J. , Boron, W.F. et al. (2016) Reconstitution of CO2 regulation of SLAC1 Anion Channel and function of CO2‐permeable PIP2;1 aquaporin as CARBONIC ANHYDRASE4 interactor. Plant Cell, 28, 568–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q. , Guo, H.S. , Dallman, G. , Fang, S.Y. , Weissman, A.M. and Chua, N.H. (2002) SINAT5 promotes ubiquitin‐related degradation of NAC1 to attenuate auxin signals. Nature, 419, 167–170. [DOI] [PubMed] [Google Scholar]
- Zargar, S.M. , Nagar, P. , Deshmukh, R. , Nazir, M. , Wani, A.A. , Masoodi, K.Z. , Agrawal, G.K. et al. (2017) Aquaporins as potential drought tolerance inducing proteins: towards instigating stress tolerance. J. Proteomics, 169, 233–238. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.Y. , Yang, C.W. , Li, Y. , Zheng, N.Y. , Chen, H. , Zhao, Q.Z. , Gao, T. et al. (2007) SDIR1 Is a RING Finger E3 Ligase That Positively Regulates Stress‐Responsive Abscisic Acid Signaling in Arabidopsis. Plant Cell, 19, 1912–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, G.H. , Mei, H.W. , Liu, H.Y. , Liu, G.L. , Hu, S.P. , Yu, X. , Li, M.S. et al. (2005) Grain yield responses to moisture regimes in a rice population: association among traits and genetic markers. Theor. Appl. Genet. 112, 106–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Expression profiles of candidate genes in the fine mapped QTL interval under dehydration stress.
Figure S2 Phylogenetic analysis of OsRINGzf1 promoter sequence.
Figure S3 Expression of OsRINGzf1 in two main haplotypes.
Figure S4 OsRINGzf1 positively regulates rice drought resistance under simulated drought stress at the seedling stage.
Figure S5 Knockout of OsRINGzf1 reduces the resistance of rice to drought stress.
Figure S6 OsRINGzf1 improves the salt tolerance of rice.
Figure S7 OsRINGzf1 enhances the tolerance to osmotic and salt stress at the germination and budding stage.
Figure S8 OsRINGzf1 enhances the drought resistance in rice at the reproductive stage.
Figure S9 Yeast two‐hybrid assays to test the interaction between OsRINGzf1 and AQPs.
Figure S10 In vitro ubiquitination of OsSIP1; 1 by OsRINGzf1.
Figure S11 In vivo degradation of OsPIP2;1 is an ATP dependent process.
Figure S12 Roots phenotype of OsRINGzf1 OE, Ri and WT plants under normal and drought conditions.
Figure S13 Knockout of OsPIP2;1 increases the resistance of rice to drought stress.
Figure S14 Amino acid sequence alignment of PIP2;1 in rice (Oryza sativa japonica cv Nipponbare) and Arabidopsis thaliana.
Figure S15 Clustering analysis and homology alignment of OsRINGzf1, CaRma1H1 and its homologues.
Figure S16 A proposed model for multiple roles of OsRINGzf1 in rice abiotic stresses resistance and development through interacting with rice AQPs.
Table S1 QTLs detected by mixed linear‐model composite interval mapping across two soil water level.
Table S2 Putative genes located in the fine mapped QTL interval.
Table S3 SNPs Between Zhenshan97B and IRAT109, and Haplotype 3 and 13.
Table S4 Germplasm for phylogenetic analysis.
Table S5 Significance analysis of LRWC in WT, OsRINGzf1 OE and Ri seedlings under drought stress.
Table S6 Genes encoding for proteins that interact with OsRINGzf1 screened by split‐ubiquitin membrane yeast two hybrid system (DUAL membrane system).
Table S7 Primers used for fine mapping, gene expression analysis, cloning and vector construction.


