Abstract
STUDY QUESTION
How does the human embryo breach the endometrial epithelium at implantation?
SUMMARY ANSWER
Embryo attachment to the endometrial epithelium promotes the formation of multinuclear syncytiotrophoblast from trophectoderm, which goes on to breach the epithelial layer.
WHAT IS KNOWN ALREADY
A significant proportion of natural conceptions and assisted reproduction treatments fail due to unsuccessful implantation. The trophectoderm lineage of the embryo attaches to the endometrial epithelium before breaching this barrier to implant into the endometrium. Trophectoderm-derived syncytiotrophoblast has been observed in recent in vitro cultures of peri-implantation embryos, and historical histology has shown invasive syncytiotrophoblast in embryos that have invaded beyond the epithelium, but the cell type mediating invasion of the epithelial layer at implantation is unknown.
STUDY DESIGN, SIZE, DURATION
Fresh and frozen human blastocyst-stage embryos (n = 46) or human trophoblast stem cell (TSC) spheroids were co-cultured with confluent monolayers of the Ishikawa endometrial epithelial cell line to model the epithelial phase of implantation in vitro. Systems biology approaches with published transcriptomic datasets were used to model the epithelial phase of implantation in silico.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Human embryos surplus to treatment requirements were consented for research. Day 6 blastocysts were co-cultured with Ishikawa cell layers until Day 8, and human TSC spheroids modelling blastocyst trophectoderm were co-cultured with Ishikawa cell layers for 48 h. Embryo and TSC morphology was assessed by immunofluorescence microscopy, and TSC differentiation by real-time quantitative PCR (RT-qPCR) and ELISA. Single-cell human blastocyst transcriptomes, and bulk transcriptomes of TSC and primary human endometrial epithelium were used to model the trophectoderm–epithelium interaction in silico. Hypernetworks, pathway analysis, random forest machine learning and RNA velocity were employed to identify gene networks associated with implantation.
MAIN RESULTS AND THE ROLE OF CHANCE
The majority of embryos co-cultured with Ishikawa cell layers from Day 6 to 8 breached the epithelial layer (37/46), and syncytiotrophoblast was seen in all of these. Syncytiotrophoblast was observed at the embryo-epithelium interface before breaching, and syncytiotrophoblast mediated all pioneering breaching events observed (7/7 events). Multiple independent syncytiotrophoblast regions were seen in 26/46 embryos, suggesting derivation from different regions of trophectoderm. Human TSC spheroids co-cultured with Ishikawa layers also exhibited syncytiotrophoblast formation upon invasion into the epithelium. RT-qPCR comparison of TSC spheroids in isolated culture and co-culture demonstrated epithelium-induced upregulation of syncytiotrophoblast genes CGB (P = 0.03) and SDC1 (P = 0.008), and ELISA revealed the induction of hCGβ secretion (P = 0.03). Secretory-phase primary endometrial epithelium surface transcriptomes were used to identify trophectoderm surface binding partners to model the embryo-epithelium interface. Hypernetwork analysis established a group of 25 epithelium-interacting trophectoderm genes that were highly connected to the rest of the trophectoderm transcriptome, and epithelium-coupled gene networks in cells of the polar region of the trophectoderm exhibited greater connectivity (P < 0.001) and more organized connections (P < 0.0001) than those in the mural region. Pathway analysis revealed a striking similarity with syncytiotrophoblast differentiation, as 4/6 most highly activated pathways upon TSC-syncytiotrophoblast differentiation (false discovery rate (FDR < 0.026)) were represented in the most enriched pathways of epithelium-coupled gene networks in both polar and mural trophectoderm (FDR < 0.001). Random forest machine learning also showed that 80% of the endometrial epithelium-interacting trophectoderm genes identified in the hypernetwork could be quantified as classifiers of TSC-syncytiotrophoblast differentiation. This multi-model approach suggests that invasive syncytiotrophoblast formation from both polar and mural trophectoderm is promoted by attachment to the endometrial epithelium to enable embryonic invasion.
LARGE SCALE DATA
No omics datasets were generated in this study, and those used from previously published studies are cited.
LIMITATIONS, REASONS FOR CAUTION
In vitro and in silico models may not recapitulate the dynamic embryo-endometrial interactions that occur in vivo. The influence of other cellular compartments in the endometrium, including decidual stromal cells and leukocytes, was not represented in these models.
WIDER IMPLICATIONS OF THE FINDINGS
Understanding the mechanism of human embryo breaching of the epithelium and the gene networks involved is crucial to improve implantation success rates after assisted reproduction. Moreover, early trophoblast lineages arising at the epithelial phase of implantation form the blueprint for the placenta and thus underpin foetal growth trajectories, pregnancy health and offspring health.
STUDY FUNDING/COMPETING INTEREST(S)
This work was funded by grants from Wellbeing of Women, Diabetes UK, the NIHR Local Comprehensive Research Network and Manchester Clinical Research Facility, and the Department of Health Scientist Practitioner Training Scheme. None of the authors has any conflict of interest to declare.
Keywords: implantation, embryo development, trophoblasts, gene expression, cell culture
Introduction
Pregnancy is established as the embryo implants into the endometrium, whereupon placentation enables foetal development. Trophectoderm (TE) forms the outer cell layer of the blastocyst-stage embryo, attaching to the receptive luminal epithelium of the endometrium to initiate implantation before TE-derived trophoblast penetrates the epithelium and invades the endometrial stroma (Aplin and Ruane, 2017). Recent progress with in vitro culture has revealed details of the cellular choreography of peri-implantation human embryos, up to Day 14, in isolation (Deglincerti et al., 2016; Shahbazi et al., 2016, 2017; Popovic et al., 2019; West et al., 2019; Zhou et al., 2019; Xiang et al., 2020), and the effects of maternal cell interactions on peri-implantation development up to Day 10 have been explored using endometrial stromal cells (Lv et al., 2019). However, breaching of the endometrial epithelium in the first phase of human embryo implantation remains largely uncharacterized.
Human embryo implantation in situ has been observed only from the early stromal invasion phase in historic samples (Hertig et al., 1956). Many studies have reported interaction of the human blastocyst with endometrial epithelial cells (EEC) in vitro (Lindenberg et al., 1985; Bentin-Ley et al., 2000; Galan et al., 2000; Meseguer et al., 2001; Petersen et al., 2005; Lalitkumar et al., 2007, 2013; Kang et al., 2014; Berger et al., 2015; Boggavarapu et al., 2016; Aberkane et al., 2018; Ruane et al., 2020), however, these have predominantly focussed on blastocyst attachment with limited analysis of cellular morphology. Ultrastructural analysis of six blastocysts attached to EEC in vitro demonstrated shared desmosomal junctions mediating TE-EEC attachment, TE invasion between EEC and bi-nucleated TE cells potentially representing syncytiotrophoblast (STB) (Bentin-Ley et al., 2000). Trophoblast develops from TE during implantation, and STB forms by cell fusion to produce multinucleated cells. STB was seen to mediate early stromal invasion at human implantation in vivo (Hertig et al., 1956), and in earlier implantation samples in non-human primates it was observed to penetrate the endometrial epithelium (Enders et al., 1983; Smith et al., 1987). STB has been seen in human embryos developing beyond the blastocyst stage in isolation (Deglincerti et al., 2016; Shahbazi et al., 2016; West et al., 2019) and in culture with endometrial cells (Aberkane et al., 2018; Lv et al., 2019), but whether it mediates embryo breaching of the endometrial epithelium in human remains unknown. Our previous study in mouse embryos suggested that interaction with EEC stimulates trophoblast differentiation (Ruane et al., 2017), implicating the epithelial phase of implantation as important for trophoblast development.
The maternal-facing cells of the placenta derive from the TE, and as such a subpopulation of trophoblast must retain multipotency during implantation (Knofler et al., 2019), while post-mitotic trophoblast invades the endometrium (Chuprin et al., 2013; Lu et al., 2017; Velicky et al., 2018). Multipotent human trophoblast stem cells (TSCs) have recently been derived from blastocysts and first trimester placenta (Okae et al., 2018), and from naïve embryonic stem cells (Dong et al., 2020), providing promising systems to study implantation. In mice, the pioneering invasive trophoblast derives from the subset of the TE not in contact with the inner cell mass (ICM) of the blastocyst, termed mural TE, while the ICM-adjacent TE, termed polar TE, gives rise to multipotent trophoblast of the ectoplacental cone (Sutherland, 2003). In vitro studies have demonstrated that human blastocysts orient such that polar TE may mediate attachment to EEC or adhesive surfaces (Bentin-Ley et al., 2000; Deglincerti et al., 2016; Shahbazi et al., 2016; Aberkane et al., 2018), while it is not clear from specimens of later stages of human implantation in vivo whether polar TE initiates invasion into the endometrium (Hertig et al., 1956). Differences between human polar and mural TE have been described at the transcriptomic level (Petropoulos et al., 2016; Meistermann et al., 2021), but little is understood about differential function of these TE subpopulations.
We and others have recently characterized human blastocyst attachment to the endometrial epithelial Ishikawa cell line. This revealed that both clinical embryology grading of TE morphology and hatching from the zona pellucida affect blastocyst attachment (Ruane et al., 2020), while adhesion-related genes are upregulated in attached embryos (Aberkane et al., 2018). Here, we further interrogate embryo-EEC interactions in vitro and show that STB mediates embryonic breaching of the epithelial layer. Moreover, our TSC spheroid and in silico models indicate that attachment to EEC promotes STB differentiation.
Materials and methods
Human embryos
Embryos generated for IVF treatment but not used by patients were obtained with informed written consent at Old Saint Marys Hospital, Manchester, or other IVF units in England. Embryos from both current and previous (cryopreserved) treatment cycles were used in accordance with ethics approval from the NRES committee south central (Berkshire; REC reference: 12/SC/0649), and a research licence from the Human Fertilisation and Embryology Authority (R0026; Person Responsible: Daniel Brison), centre 0067 (Old Saint Mary’s Hospital; fresh embryo research) and University of Manchester (0175; frozen-thawed embryo research). Day 6 blastocysts were artificially hatched using acid Tyrode (pH 2.5), as described (Ruane et al., 2020), before use. Forty-six blastocysts (expanded morphology at Day 6 and of unknown clinical grade) were used in this study; four of the embryos were thawed from cryopreservation at pro-nuclear stage or on Day 2 after fertilization (clinical morphology grades were not assessed). The remaining 42 embryos were donated as blastocysts fresh from IVF cycles and were unsuitable for freezing, with grades below 2/5 for expansion, 4/5 for ICM and 2/3 for TE (Cutting et al., 2008; ASiRMaESIGo, 2011).
Ishikawa cell culture
Ishikawa EEC (ECACC 99040201) were cultured at 37°C, 95% air and 5% CO2 in growth medium (1:1 Dulbecco’s modified Eagle’s medium: Ham’s-F12 (Sigma) containing 10% v/v foetal bovine serum (Gibco) and supplemented with 2 mM L-glutamine, 100 µg/ml streptomycin and 100 IU/ml penicillin (Sigma)). Ishikawa cells were grown to confluency to form layers in 24-well plates (Greiner) on 13 mm glass coverslips for co-culture with blastocysts and TSC spheroids and subsequent microscopy, or in 48-well plates without coverslips for co-culture with TSC spheroids for subsequent gene expression and ELISA analysis.
Human blastocyst implantation assay
Confluent Ishikawa EEC layers were washed and replenished with serum-free growth medium immediately prior to the transfer of single hatched Day 6 blastocysts to individual wells. After 48 h, all co-cultures were washed in PBS and fixed with 4% w/v paraformaldehyde (PFA) in PBS for 20 min.
TSC culture and spheroid formation
TSC (line CT27, derived from first trimester placenta) were acquired as a gift from Dr Hiroake Okae, and cultured and differentiated to adherent STB as described (Okae et al., 2018). TSC were seeded into Aggrewell™ plates (Stem Cell Technologies) at ∼200 cells/microwell and cultured in serum- and BSA-free TSC medium for 48 h to produce spheroids.
TSC spheroid implantation assay
TSC spheroids were collected from Aggrewell™ plates and resuspended in co-culture medium (1:1 DMEM: Ham’s-F12, 1% ITS-X (Gibco), 5 µM Y27632 (Adooq Bioscience), 2 mM L-glutamine and 100 µg/ml streptomycin and 100 IU/ml penicillin). Approximately 20 spheroids, pre-treated with CellTracker™ (Life Technologies), were co-cultured per well of Ishikawa cells in 24-well plates for 48 h before fixation with 4% PFA. Attachment was monitored over the first 6 h of co-culture by agitating the culture plate to determine which spheroids were attached to the Ishikawa EEC layer. Alternatively, ∼400 spheroids per well of Ishikawa cells in 48-well plates were co-cultured for 48 h, before lysis and RNA extraction.
Staining and fluorescence imaging
Fixed samples were quenched with 50 mM ammonium chloride solution before permeabilization with 0.5% Triton-X 100 PBS. Samples were incubated with primary antibody in PBS for 2 h or overnight at 4°C, followed by alexa568-phalloidin (Life Technologies), 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) and alexa488/649 secondary antibodies (Life Technologies) for 1 h. Human embryo samples were mounted in a chamber of 3% 1,4-diazabicyclo[2.2.2]octane (DABCO) (Sigma) in PBS. TSC samples were mounted on a glass slide with 3% DABCO Mowiol (Sigma). Fluorescence microscopy was carried out using a Zeiss Axiophot microscope equipped with an Apotome module for optical sectioning. Images were processed using Zeiss Zen software and ImageJ. Antibodies: rabbit anti-E-cadherin (EP700Y, Abcam), rabbit anti-CDX2 (D11D10, Cell Signalling Technologies), mouse anti-GATA3 (MAB6330, R&D Systems), mouse anti-hCGβ (5H4-E2, Abcam) and mouse anti-HLA-G (4H84, Abcam).
RNA extraction and PCR
RNA was extracted using the RNeasy Mini Kit (Qiagen), according to the manufacturer’s instructions. Reverse transcription using random 9mer primers (Agilent) followed protocols for Sensiscript Reverse Transcription kit (Qiagen). Real-time quantitative PCR (RT-qPCR) were performed using reverse transcription reactions together with 0.25 µM gene-specific primers (SDC1 CTGCCGCAAATTGTGGCTAC, TGAGCCGGAGAAGTTGTCAGA; CGB CAGCATCCTATCACCTCCTGGT, CTGGAACATCTCCATCCTTGGT; OVOL1 TGAACCGCCACATGAAGTGTC, GACGTGTCTCTTGAGGTCGAA; ACTB AAGCCACCCCACTTCTCTCT, CTATCACCTCCCCTGTGTGG) and QuantiTect SYBR green PCR kit (Qiagen) on StepOne Plus machines; 40 cycles, 60°C annealing temperature. Raw data were analysed with StepOne Plus software to yield cycle threshold (Ct) values, which were expressed as 2−ct relative to housekeeping gene, ACTB. In order to normalize total transcript levels of TSC spheroids cultured in isolation to that of transcripts from TSC spheroids cultured with Ishikawa cells, reverse transcribed RNA from samples of isolated culture TSC spheroids were pooled with reverse transcribed RNA extracts from samples of isolated Ishikawa cultures before analysis by RT-qPCR.
ELISA
Media from TSC spheroid cultures and co-cultures were collected and subject to hCGβ ELISA using kit EIA-1496 (DRG Instruments GmbH), according to the manufacturer’s instructions.
Modelling the TE-EEC interface in silico
All in silico modelling and analysis was performed in R version 3.4.2 (R Foundation for Statistical Computing). Differentially expressed genes (DEGs) were defined by ANOVA (cut-off P < 0.01) between proliferative phase (n = 4) and mid-secretory-phase (n = 4) endometrial epithelial RNA sequencing (RNAseq) transcriptomes (GEO accession number GSE132711) (Chi et al., 2020). Localization of the protein products of these genes was investigated using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang da et al., 2009a,b). Proteins defined as localized to any of ‘extracellular space’, ‘extracellular matrix’, ‘proteinaceous extracellular matrix’ or ‘cell surface’ were refined from the list of DEGs. Putative binding partners were identified using the Biological General Repository for Interaction Datasets (BioGRID) (Stark et al., 2006) and refined to those which were localized as above in Day 6 TE single cell-RNAseq transcriptomes (n = 331) (Array Express Accession E-MTAB-3929) (Petropoulos et al., 2016). Transcriptomic differences between samples were visualized by principal component analysis (PCA) and partial least-squares-discriminant analysis (PLS-DA). All pathway analyses were performed using Webgestalt (Zhang et al., 2005), and word clouds were generated using WocEA (Ning et al., 2018).
Hypernetworks
Hypernetworks were used to quantify shared correlations between EEC-interacting TE surface genes across the rest of the transcriptome. Correlation matrices were generated using the Day 6 TE single cell-RNAseq transcriptomes. All analyses were repeated for polar and mural TE separately, modifying the samples included in each experiment, using previously inferred TE subpopulations (Petropoulos et al., 2016). Correlation matrices were binarized using the distribution of r-values in order to include only the strongest present correlations. Correlations further from the mean r-value than ±1 SD in each set were assigned ‘1’, while those between the SD boundaries was assigned ‘0’. Hypernetworks were generated by multiplying the binary matrix () by the transpose of that matrix () to describe the number of shared correlations between a pair of TE genes. The central cluster of the hypernetworks, arranged using hierarchical clustering, represent a subset of EEC-interacting TE genes whose expression patterns are correlated with a large number of genes across the transcriptome. The connectivity (mean number of shared connections) and organization (Shannon entropy) of the central clusters were quantified. These metrics were also calculated by permuting 1000 hypernetworks of random genes within the transcriptome.
Classification of genes using random forest
EEC-interacting TE genes present in TSC and TSC-derived STB (Okae et al., 2018) were used for random forest (RF) machine learning (Breiman, 2001), which was applied over 1000 iterations to test whether the genes could accurately classify STB differentiation. Noise within the RF was modelled using Boruta (Kursa and Rudnicki, 2010).
RNA velocity
RNA velocity analysis was performed on TE cells of Day 5 embryo cells, and mural TE and polar TE cells of Day 6 and 7 embryos (Petropoulos et al., 2016), using the scVelo package (La Manno et al., 2018; Bergen et al., 2020). The inferred velocity of genes identified in the hypernetwork analysis were then compared between these TE cells, with velocity (v) plotted as .
Statistical comparisons
Statistical significance was measured by Chi-squared and Mann–Whitney analyses using Prism (GraphPad). ANOVA and Wilcoxon rank sum analyses were performed using R.
Results
Gross and cellular morphology of embryos attached to Ishikawa EEC layers
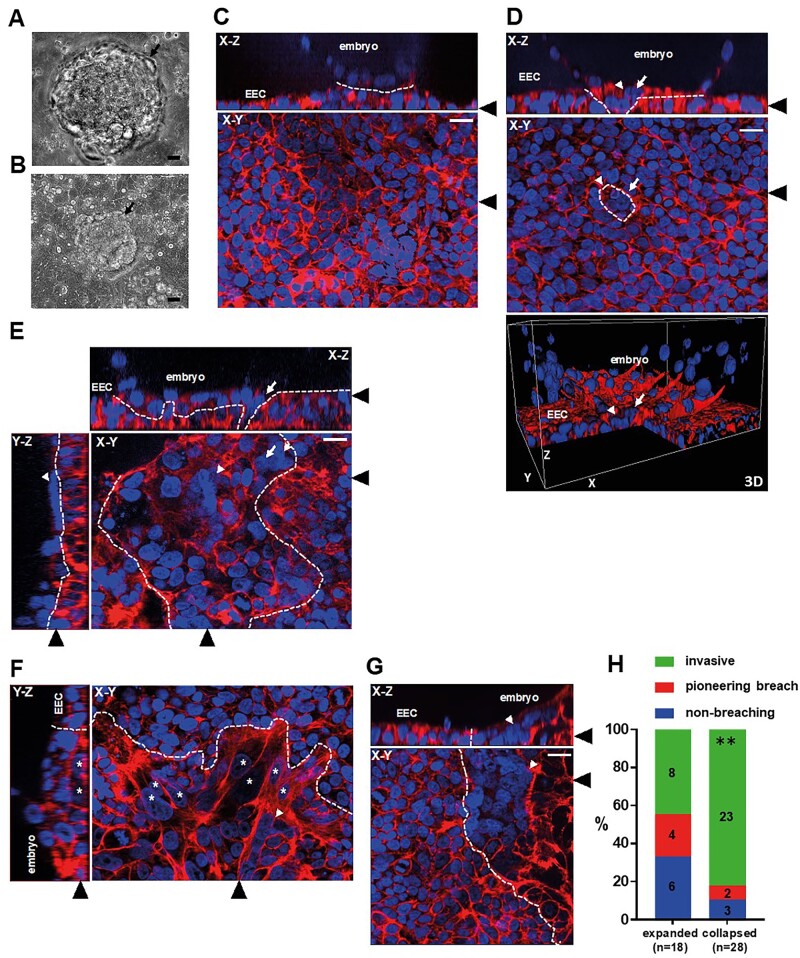
Forty-six blastocysts attached to Ishikawa EEC layers were analysed in this study, all after co-culture for 48 h, from Day 6 post-fertilization to Day 8. Attached embryos exhibited either expanded (18/46, 39%) (Fig. 1A) or collapsed (28/46, 61%) blastocyst morphology (Fig. 1B). We were able to ascertain whether embryos attached in a polar TE- or mural TE-oriented fashion in seven instances, with 5/7 exhibiting polar TE-oriented attachment. Phalloidin labelling of cortical actin filaments combined with DAPI labelling of nuclei enabled single cell resolution of embryos attached to EEC layers. Attached embryos with no clear breaching of the epithelium were observed (Fig. 1C). Embryos in which a defined region of multinuclear STB was mediating initial, pioneering breaching of the epithelium and contacting the underlying substrate were also seen (Fig. 1D and E; Supplementary Movies S1 and S2). In addition, STB was observed in contact with the apical EEC surface (Fig. 1E; Supplementary Movie S2). The majority of embryos had progressed beyond these initial stages to invade laterally into the EEC layer, with morphologically distinct mononuclear and multinuclear outgrowths present (Fig. 1F and G, respectively; Supplementary Movies S3 and S4).
Figure 1.
Syncytiotrophoblast (STB) pioneers embryo invasion of the Ishikawa endometrial epithelial cell (EEC) layer. (A, B) Phase contrast images of embryos attached to Ishikawa EEC layers, showing expanded blastocyst and collapsed blastocyst morphology. Black arrows indicate embryos. Scale bars 20 µm. (C–G) Attached embryos were stained with phalloidin (red) and DAPI (blue) to label actin and nuclei, respectively, and imaged by optical sectioning fluorescence microscopy. Panels show X–Y, X–Z and Y–Z planes and a 3D image, as labelled. Ishikawa EEC and embryos are indicated in X–Z and Y–Z planes and the 3D image. Black arrowheads indicate the region of the section for the adjoining panels. White arrows indicate trophoblast breaching of the Ishikawa EEC layer, white arrowheads indicate STB, and white asterisks indicate invasive mononuclear trophoblast. Dotted lines indicate embryo-EEC interface. Scale bars 20 µm. (H) Bar graph showing proportions of embryo morphologies (n = 46). **P < 0.01 Chi-squared. DAPI, 4′,6-diamidino-2-phenylindole.
It was observed that 10/18 embryos with expanded blastocyst morphology (56%) were superficially attached to the EEC layer or displayed only pioneering breaching of the epithelium, while 8/18 (44%) had invaded laterally and often extensively into the epithelium. In comparison, 5/28 collapsed embryos (18%) had not breached or exhibited only pioneering breaching, with 23/28 (82%) found to be invasive (Fig. 1H). Collapsed embryos were therefore more likely to have extensively invaded the EEC layer (P < 0.01). Importantly, each of the seven pioneering breaching events, observed in six embryos as cells that were in the EEC plane but continuous with an embryonic cell above, consisted of STB protruding through the EEC layer (Figs 1D and E and 2B; Supplementary Movies S1 and S2).
Figure 2.
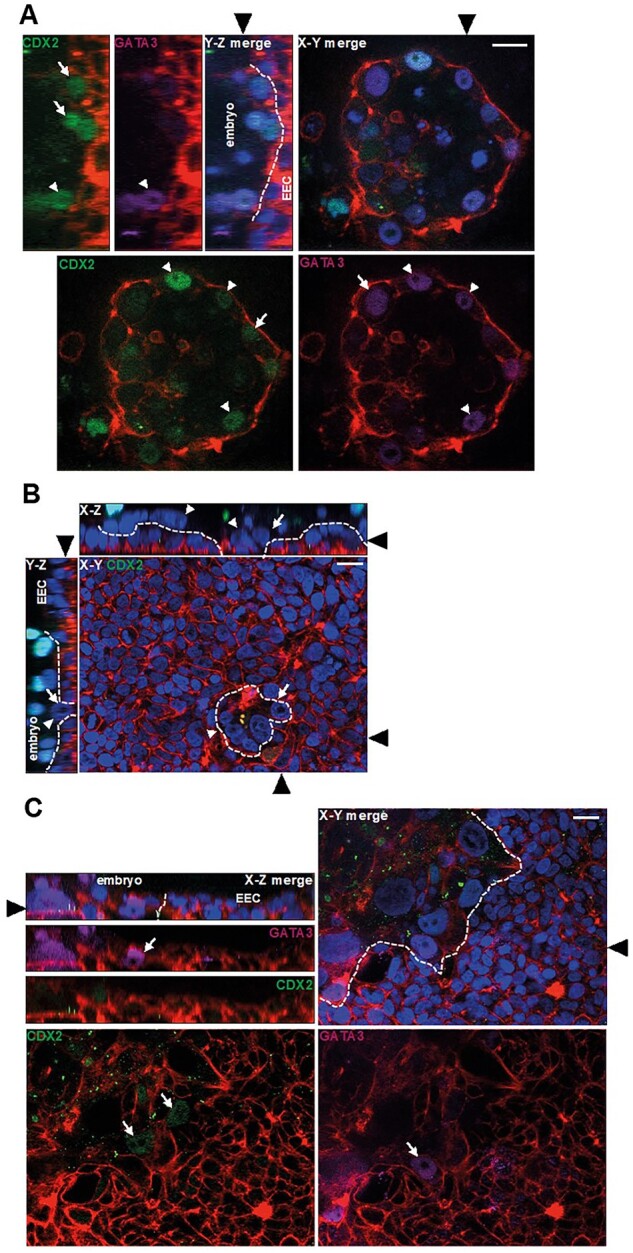
Trophectoderm (TE) transcription factors at the embryo- endometrial epithelial cell (EEC) interface. (A) Fluorescence optical sections of an attached embryo labelled with phalloidin (red), DAPI (blue), anti-CDX2 (green) and anti-GATA3 antibody (magenta). Y–Z and X–Y planes are shown, and Ishikawa EEC and embryos are indicated Y–Z panel. Black arrowheads indicate the location of the section for the adjoining panels. White arrows show CDX2 or GATA3-positive TE and white arrowheads indicate double-positive CDX2–GATA3-positive TE. Dotted lines indicate embryo-EEC interface. Scale bar 20 µm. (B) An attached embryo labelled with phalloidin (red), DAPI (blue) and anti-CDX2 antibody (green) and imaged by optical sectioning fluorescence microscopy. The planes shown in each panel are indicated (X–Z, X–Y) and black arrowheads indicate the location of the section for the adjoining panels. White arrows point to trophoblast breaching of the Ishikawa EEC layer and white arrowheads indicate STB. Dotted lines indicate embryo-EEC interface. Scale bar 20 µm. (C) An invasive embryo labelled with phalloidin (red), DAPI (blue), anti-CDX2 (green) and anti-GATA3 antibody (magenta), optical planes indicated on the panels. White arrows indicate CDX2- or GATA3-positive mononuclear trophoblast. Black arrowheads indicate the location of the section for the adjoining panels. Dotted lines indicate embryo-EEC interface. Scale bar 20 µm. DAPI, 4′,6-diamidino-2-phenylindole.
TE markers in EEC-attached embryos
To characterize the TE/trophoblast cell types in attached embryos, we labelled samples with antibodies to the transcription factors CDX2 and GATA3, which are expressed in this lineage (Hemberger et al., 2020). Co-staining for CDX2 and GATA3 in expanded blastocysts that had not breached the epithelium revealed heterogeneity amongst TE both in contact with, and distal to, EEC; some nuclei were positive only for CDX2, some only for GATA3 and yet others were co-labelled (Fig. 2A). In embryos which had just breached the epithelium, CDX2 was present only in embryonic nuclei above the plane of the EEC layer (Fig. 2B). In collapsed embryos, CDX2 and GATA3 labelling was faintly present in mononuclear trophoblast that had invading extensively into the epithelium (Fig. 2C). Neither transcription factor was detected in STB.
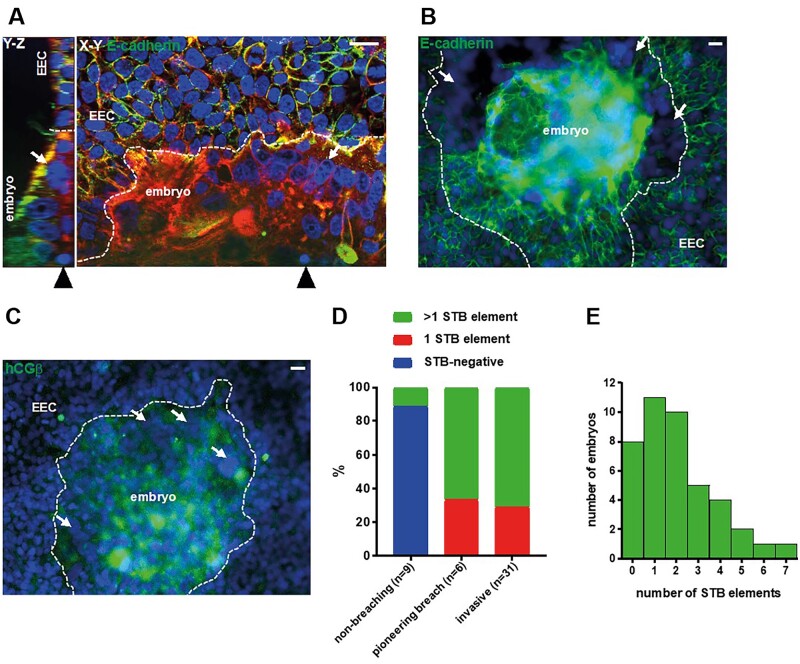
Multiple STB regions invade into the EEC layer
E-cadherin is present at intercellular junctions in human TE and mononuclear trophoblasts (Alikani, 2005; Aplin et al., 2009), and was used to demarcate mononuclear cells. Multinuclear elements lacking inter-nuclear E-cadherin labelling were observed in contact with EEC at the invading front of embryonic outgrowths (Fig. 3A). This STB contained densely packed nuclei and often occurred at multiple, distinct sites (Fig. 3B). STB and the central area of the embryo were also positive for hCG subunit β (Fig. 3C), the maternal recognition of pregnancy hormone that is secreted by STB from the onset of pregnancy. HLA-G, the canonical marker for invasive mononuclear extravillous trophoblast, was not detected in embryos implanting in this model (Supplementary Fig. S1).
Figure 3.
Multiple regions of syncytiotrophoblast (STB) form at the embryo- endometrial epithelial cell (EEC) interface. (A) Fluorescence optical sections of an invasive embryo labelled with phalloidin (red), DAPI (blue) and anti-E-cadherin (green), panels show Y–Z and X–Y planes. Black arrowheads point to the location of the section for the adjoining panel. White arrows indicate STB. Scale bars 20 µm. (B) Fluorescence micrograph of an invasive embryo labelled with DAPI (blue) and E-cadherin (green). White arrows point to distinct regions of STB. Dotted lines indicate embryo-EEC interface. Scale bar 20 µm. (C) Fluorescence microscopy image of an invasive embryo labelled with DAPI (blue) and anti-hCGβ (green). White arrows point to distinct regions of STB. Dotted lines indicate embryo-EEC interface. Scale bar 20 µm. (D) Bar graph relating STB quantities to embryo invasiveness (n = 46). (E) Bar graph of STB quantities in embryos (n = 42; the number of STB elements could not be discerned in four samples). DAPI, 4′,6-diamidino-2-phenylindole.
STB, defined as multiple DAPI-stained nuclei within E-cadherin- or phalloidin-stained cell borders, was seen in only 1/9 non-invasive embryos (11.1%). All breaching and invasive embryos contained STB, with approximately two-thirds containing more than one STB element (Fig. 3D). Notably, STB was never observed in TE not in contact with EEC. Analysis of the number of STB elements (multinuclear structures bound by E-cadherin and/or phalloidin staining) present in each embryo demonstrated that up to seven could be found and >70% embryos had one to four distinct STB elements (Fig. 3E).
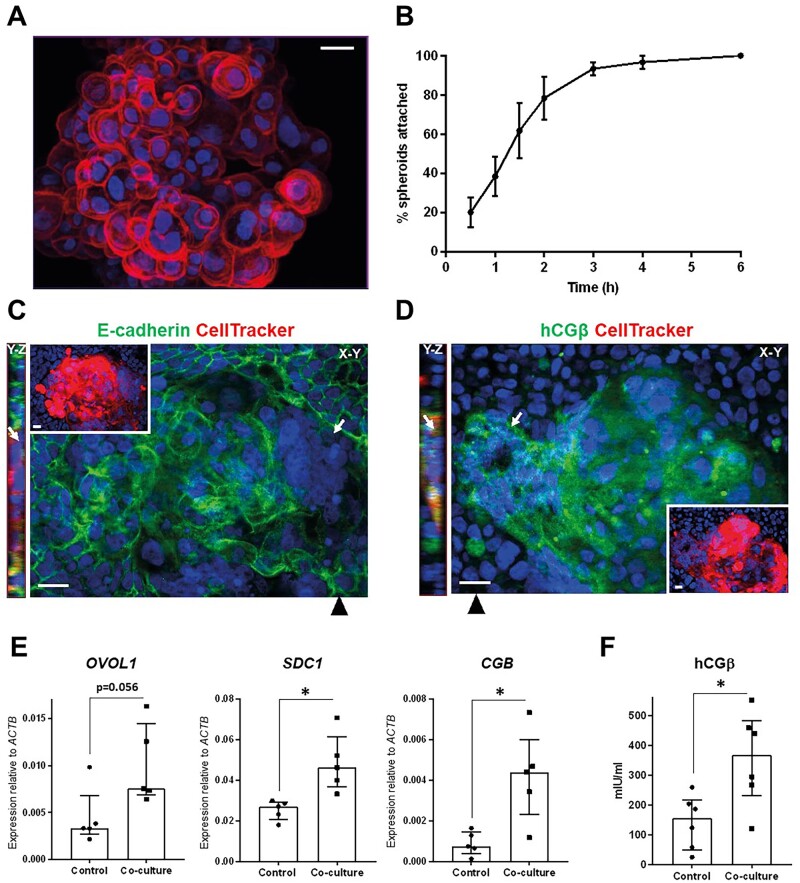
Ishikawa EEC layer stimulates STB differentiation in TSC spheroids
Human TSC can differentiate to form the major trophoblast subtypes, including STB (Okae et al., 2018), and thus offer a model with greater experimental scope than single embryos for interrogating trophoblast differentiation at implantation. Treatment of TSCs with the previously reported STB differentiation medium induced syncytialization and upregulation of hCGβ at the mRNA and protein level (Supplementary Fig. S2A and B). TSC culture in non-adherent conditions produced blastocyst-sized spheroids of mostly mononuclear cells (Fig. 4A). These were co-cultured in basal medium with Ishikawa EEC layers to model human embryo implantation in vitro. TSC spheroids attached to EEC layers within 6 h (Fig. 4B), reflecting the kinetics of blastocyst attachment (Ruane et al., 2020). After 48 h, spheroids had invaded the epithelium with STB at the invasion front (Fig. 4C). Invasive trophoblast was hCGβ-positive, although this was not localized only to invasive multinuclear cells but was seen also in mononuclear trophoblast (Fig. 4D). The expression of STB markers in spheroids attached to EEC layers (co-culture) and those cultured in isolation and attached to tissue culture plastic (control) was compared with determine whether interaction with EEC stimulated STB differentiation. STB genes CGB and SDC1 were significantly upregulated by co-culture with EEC layers (Fig. 4E), while secreted hCGβ levels were also higher in co-cultured TSC spheroids (Fig. 4F). These data suggest that EEC interactions stimulate STB differentiation from multipotent trophoblast.
Figure 4.
Syncytiotrophoblast (STB) formation from trophoblast stem cell (TSC) spheroids is promoted by Ishikawa endometrial epithelial cell (EEC) layers. (A) TSC spheroid labelled with phalloidin (red) and DAPI (blue), imaged by optical sectioning fluorescence microscopy and displayed as maximum intensity projection. Scale bar 20 µm. (B) TSC spheroid attachment to Ishikawa EEC layers was monitored over 6 h. Three independent repeats were performed (∼20 spheroids per repeat), mean ± SEM plotted on line graph. CellTracker-loaded TSC spheroids attached to Ishikawa EEC layers after 48 h co-culture were labelled with DAPI (blue) and (C) anti-E-cadherin (green) or (D) anti-hCGβ (green), and optically sectioned by fluorescence microscopy. X–Y and Y–Z planes are shown as indicated, with black arrowheads indicating the location of the Y–Z plane. CellTracker (red) and DAPI labelling is shown in insets. STB is indicated by white arrows. Scale bars 20 µm. (E) TSC spheroid-Ishikawa EEC co-cultures and isolated TSC and Ishikawa EEC cultures were lysed after 48 h for real-time quantitative PCR (RT-qPCR) analysis. Expression of STB markers OVOL1, SDC1 and CGB was assessed relative to ACTB expression. Five experimental repeats, median ± IQR plotted, *P < 0.05 Mann–Whitney. (F) Medium was collected from TSC spheroid-Ishikawa EEC co-cultures and isolated TSC and Ishikawa EEC cultures after 48 h for hCGβ ELISA. Six experimental repeats, median ± IQR plotted, * P < 0.05 Mann–Whitney. DAPI, 4′,6-diamidino-2-phenylindole.
Gene networks at the TE-EEC interface
To identify candidate gene networks functioning at the TE-EEC interface, we used single-cell transcriptomic datasets from blastocyst TE (Petropoulos et al., 2016), and bulk transcriptomic data from EEC directly isolated from endometrium (Chi et al., 2020). Cell surface or secreted genes, differentially expressed in receptive (mid-secretory phase) versus non-receptive (proliferative phase) endometrial epithelium (Supplementary Fig. S3A), were used to identify 39 cognate genes in Day 6 TE. The resulting in silico TE-EEC interface (Table I), was enriched for extracellular matrix components, peptidases, cell adhesion molecule-binding proteins, signalling receptor-binding proteins and glycosaminoglycan-binding proteins (Supplementary Fig. S3B).
Table I.
Summary of in silico trophectoderm (TE)-endometrial epithelial cell (EEC) interface, hypernetwork analysis and random forest-defined syncytiotrophoblast (STB) classifier status.
| EEC gene | Cognate TE gene | TE subset hypernetwork cluster | Classifier of STB differentiation |
|---|---|---|---|
| LOXL1 | FBLN5* | Polar and mural | Yes |
| IGF1 | IGFBP7* | Polar and mural | Yes |
| CRISPLD2 | HSP90AB1* | Polar | Yes |
| NOG | NOG* | Polar | Yes |
| CP | LTF | Polar | Yes |
| TNC | NCAN | Polar | Yes |
| NRXN1 | NLGN1* | Mural | Yes |
| ALPP | COL2A1* | Mural | Yes |
| SERPINB8 | FURIN* | Mural | Yes |
| CST7 | POTEE* | Mural | Yes |
| FLNB | PSEN1* | Mural | Yes |
| TGFBR3 | TGFB1* | Mural | Yes |
| VCAN | HAPLN1* | Mural | Yes |
| HEXB | HEXB | Mural | Yes |
| CSTB | CTSH | Mural | Yes |
| SLPI | PLSCR1 | Mural | Yes |
| AMBP | AMBP | Polar and mural | No |
| KLKB1 | KNG1 | Mural | No |
| GUSB | CES1 | Mural | No |
| THBD | F2 | Mural | No |
| C4BPA | APCS | Polar and mural | ND† |
| IFNG | IFNG | Polar and mural | ND† |
| IFNAR2 | IFNA2 | Polar and mural | ND† |
| SERPING1 | SELE | Polar and mural | ND† |
| FGA | FGB | Mural | ND† |
| SFRP1 | FZD6* | Neither | ND |
| DKK3 | UBA52* | Neither | ND |
| LAMA1 | FBLN2* | Neither | ND |
| DCN | EGFR* | Neither | ND |
| F2 | SERPINE1* | Neither | ND |
| CD55 | CD55* | Neither | ND |
| PPFIBP2 | APP | Neither | ND |
| ANXA2 | CTSB | Neither | ND |
| PROS1 | F5 | Neither | ND |
| ANK3 | KCNC1 | Neither | ND |
| CPE | ROBO2 | Neither | ND |
| GPLD1 | APOA4 | Neither | ND |
| FGG | FGA | Neither | ND |
| APOD | APOD | Neither | ND |
TE gene differentially expressed in TSC-STB differentiation; ND, not determined; ND†, not determined as not expressed in TSC/STB.
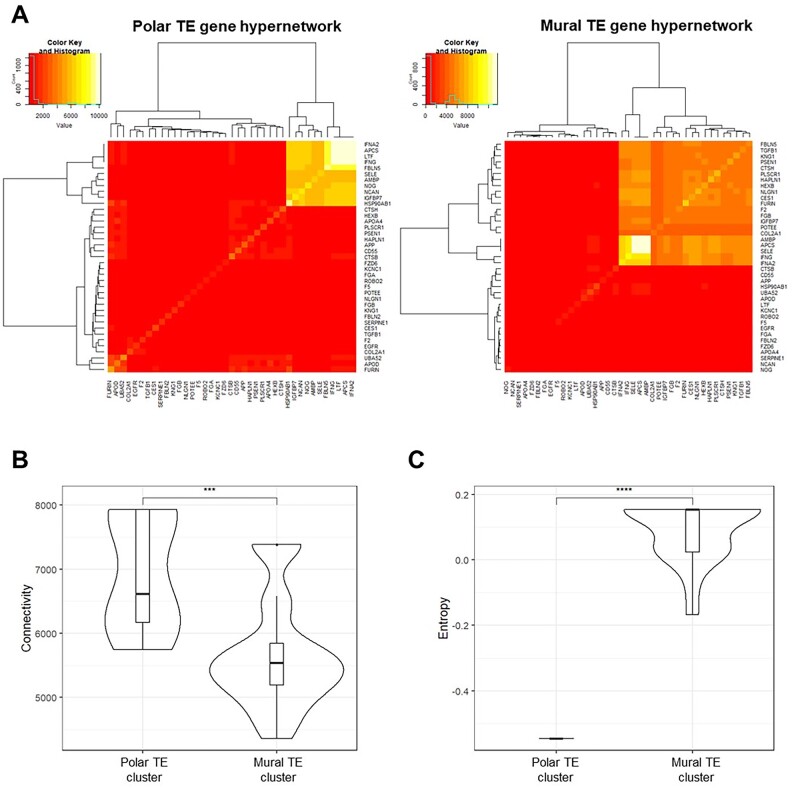
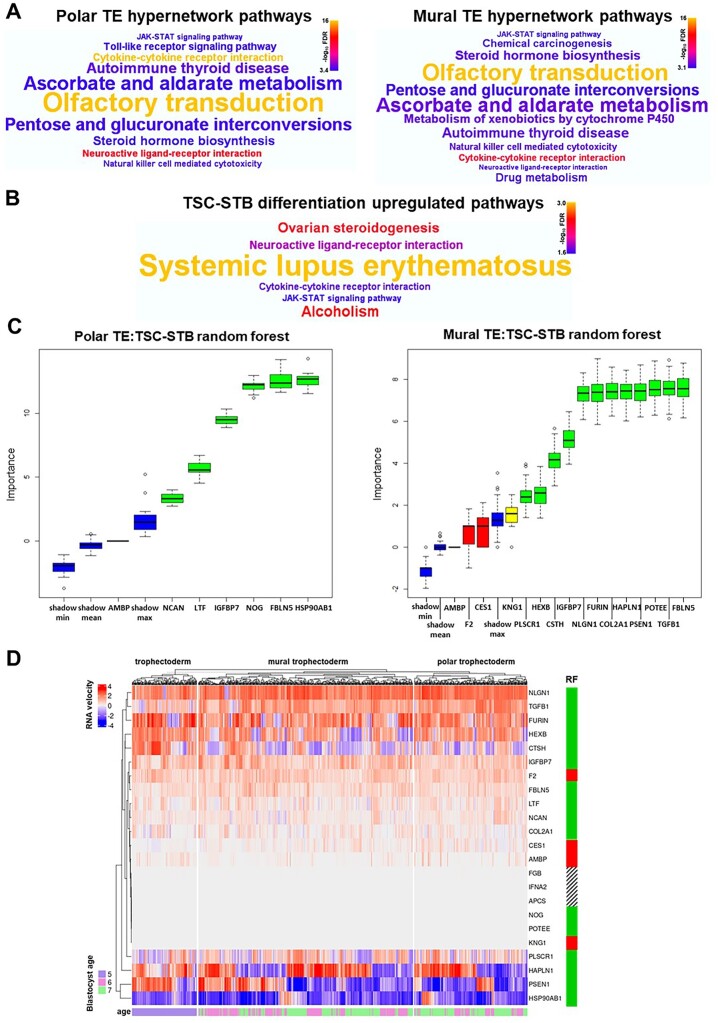
Hypernetwork analysis enables clustering of genes based on co-expression within the transcriptome, providing an indication of connectivity in functional gene networks (Pearcy et al., 2016; Gaudelet et al., 2018). Day 6 TE subsets previously defined as polar and mural (Supplementary Fig. S3C) (Petropoulos et al., 2016), enabled us to use hypernetwork analysis to detect differences between polar and mural TE interactions with EEC. A total of 25 distinct genes were identified in the hypernetworks; a cluster of 11 genes from analysis of EEC-interacting polar TE genes, while a 21-gene cluster was found from EEC-interacting mural TE genes (Fig. 5A), with seven genes common between the two clusters (Table I).
Figure 5.
Polar and mural trophectoderm (TE) gene networks are differentially connected to endometrial epithelial cell (EEC) surface genes in an in silico model of the TE-EEC interface. (A) Hypernetwork analysis was performed on EEC-interacting TE genes (n = 39) in polar TE cells (n = 86) and mural TE cells (n = 245), and presented as a heatmap. Eleven genes form the highly connected central cluster in the polar TE hypernetwork and 21 genes form that in the mural TE hypernetwork. (B) Connectivity of hypernetwork clusters (the number of pairwise shared correlations) in polar and mural TE is presented as a violin plot. Median line and interquartile range are illustrated in each box, and whiskers illustrate the range with outliers as points. ***P < 0.001 Wilcoxon rank sum test. (C) Entropy from the hypernetwork clusters of polar and mural TE (a measure of the organization of gene connections), relative to entropy generated by permuting 1000 hypernetworks of random genes within the respective transcriptomes. Comparison of absolute entropy values between gene networks and subsets therein is not informative connectivity organization. Data are presented in violin plot form as above. ****P < 0.0001 Wilcoxon rank sum test.
The hypernetwork heatmaps suggest enhanced transcriptome connectivity in polar TE compared with mural TE. Quantification of the hypernetwork properties revealed a higher connectivity (number of shared transcriptome correlations) between EEC-interacting polar TE genes compared with those in mural TE (Fig. 5B), and this reflected differences between background transcriptome connectivity in polar and mural TE (Supplementary Fig. S3D). Entropy is a measure of the organization of gene connections within the hypernetwork and is presented relative to background levels. A markedly lower entropy, indicative of a more co-ordinated organization of gene connectivity, was found for EEC-interacting polar TE genes than for those in mural TE (Fig. 5C).
EEC-coupled TE gene networks are linked to STB differentiation
Pathway analysis of the EEC-coupled polar and mural TE gene hypernetworks (3541 and 2447 genes, respectively) uncovered similar pathways (Fig. 6A; Supplementary Tables SI and SII, respectively). The most enriched pathways consisted of olfactory receptors (olfactory transduction) and cytokines (autoimmune thyroid disease, cytokine–cytokine receptor interaction and natural killer cell-mediated cytotoxicity). Glucuronosyltransferases were also highly prevalent in enriched pathways including steroid hormone biosynthesis, within which central progesterone metabolism genes HSD3B2, CYP17A1, CYP11B2 and AKR1C1 were present in the hypernetworks. Steroid hormone biosynthesis, especially progesterone synthesis, is a key pathway for endocrine activity in STB (Okae et al., 2018). Remarkably, comparing EEC-coupled TE pathways to a gene set enrichment analysis of TSC differentiation to STB (9728 genes; Okae et al., 2018) demonstrated that 4/6 most highly upregulated pathways in STB differentiation (false discovery rate (FDR < 0.026)) were highly significantly enriched in both polar and mural hypernetworks (FDR < 0.001): steroid hormone biosynthesis/ovarian steroidogenesis (overlapping pathways), cytokine–cytokine receptor interaction, neuroactive ligand-receptor interaction and the JAK-STAT signalling pathway (Fig. 6B; Supplementary Table SIII).
Figure 6.
Endometrial epithelial cell (EEC)-interacting trophectoderm (TE) gene networks are coupled to syncytiotrophoblast (STB) differentiation pathways. (A) Word clouds representing highly enriched (FDR < 0.001) KEGG pathways from over-representation analysis of genes connected within the polar and mural TE hypernetwork clusters, respectively. Word size reflects enrichment ratio, colour relates to significance level. Full list of significantly enriched pathways (FDR < 0.05) in Supplementary Tables SI and SII, respectively. (B) Word cloud representing the six most upregulated (FDR < 0.026) KEGG pathways from gene set enrichment analysis of genes differentially expressed upon trophoblast stem cell (TSC) differentiation to STB (P < 0.01, TSC n = 4, STB n = 4; Okae et al., 2018). Word size relates to enrichment ratio, colour to significance level. Full list of significantly enriched pathways (FDR < 0.05) in Supplementary Table SIII. (C) Boruta plots of random forest machine learning (1000 iterations) showing STB classification importance (TSC n = 4, STB n = 4; Okae et al., 2018) of hypernetwork-clustered EEC-interacting polar and mural TE genes expressed in TSC/STB (n = 7 and n = 16, respectively). Box-whisker plots represent importance Z-scores, a metric of informativity of a variable for classification. Green confirms statistical significance in the classification of STB, red indicates a non-informative gene in the classification of STB, yellow indicates a failure to either confirm or reject a gene as informative within the allotted number of random forest runs, and blue demonstrates the background variation of predictive value within the data. (D) RNA velocity was measured for each distinct hypernetwork-clustered EEC-interacting TE gene (n = 23, IFNG and SELE velocity data not available) in each cell of Day 5 TE (polar and mural distinction not available) and Day 6 and 7 polar and mural TE (Petropoulos et al., 2016). These velocity values were clustered within TE cell types in a heatmap, with blastocyst age in days indicated for each TE cell and random forest (RF) classification also indicated for each gene (green, variable of importance in the classification of STB; red, non-informative variable in the classification of STB; barred, importance not determined due to lack of expression in TSC/STB). FDR, false discovery rate.
To test whether the TE genes in our in silico interface were linked to STB differentiation, we applied RF machine learning. This quantitative approach revealed that 80% (16/20) of the distinct EEC-interacting genes in polar and mural TE could be considered variables of importance in classifying STB (Fig. 6C and Table I), with FBLN5 and IGFBP7 appearing as significant in both polar and mural TE. RNA velocity was assessed for TE genes in the in silico model as a direct measure of the rate of change of expression (La Manno et al., 2018). The genes classified as important variables by RF exhibited dynamic gene expression in TE cells from Day 5 to 7, clustering at the upper and lower regions of the heatmap compared with genes that were not classified as variables of importance that generally clustered in the middle with little dynamic expression (Fig. 6D), therefore, further implicating functionality for these genes in TE development. Hypernetworks, pathway analysis, RF machine learning and RNA velocity analysis in our in silico model together suggest that EEC-interacting genes in both polar and mural TE are linked to functional gene networks involved in STB differentiation.
Discussion
Embryo implantation into the uterine endometrium is essential for human development, and this process begins with blastocyst attachment to the endometrial epithelium and breaching of this barrier (Aplin and Ruane, 2017). Here, in vitro and in silico models of the epithelial phase of human embryo implantation reveal for the first time that interactions with EEC promote TE differentiation to STB, which then goes on to breach the endometrial epithelium. These findings establish the importance of EEC interactions to the development of trophoblast lineages at implantation. To further investigate the pattern of trophoblast development at implantation, sequential interactions with endometrial epithelium, subjacent stromal matrix and decidual cell types must be considered. Understanding these processes will illuminate infertility and obstetric disease mechanisms thought to be underpinned by defective trophoblast development at implantation (Aplin et al., 2020).
Blastocysts that had attached to Ishikawa EEC layers after 48 h co-culture, from Day 6 to 8, revealed a range of implantation stages, from those attached to the EEC apical surface to those that had extensively invaded into the epithelium. Blastocyst cavity collapse is consistently observed by Day 8 in embryos cultured in isolation (Deglincerti et al., 2016; Shahbazi et al., 2016), and consistent with this, most embryos here displayed collapsed cavities and invasive morphology. Those embryos that remained expanded and did not breach the epithelium may reflect slower implantation kinetics or implantation failure. STB was observed in all invasive embryos and consistently pioneered breaching of the epithelium, with these breaching events appearing to form between cells of the EEC layer. STB may negotiate invasion between EEC via the formation of junctional complexes between trophoblast and lateral EEC membranes, which have been seen in ultrastructural studies in both human and monkey (Enders et al., 1983; Smith et al., 1987; Bentin-Ley et al., 2000). Both mononuclear trophoblast and STB were present at the leading edge of embryos invading laterally into the EEC layer, suggesting that the initial stromal phase of invasion may not be driven solely by STB (James et al., 2012; Knofler et al., 2019; Turco and Moffett, 2019). However, the 2D nature of the in vitro EEC model and the absence of underlying matrix and stromal cell types may have influenced invasive cell behaviour. The well-differentiated endometrial adenocarcinoma-derived Ishikawa cell line provides a consistent, polarized, receptive 2D epithelial model that is difficult to generate using primary EEC (Campbell et al., 2000; Singh and Aplin, 2015; Ruane et al., 2017), nevertheless 3D cultures of primary endometrial cells are required to better recapitulate implantation. Moreover, using high grade blastocysts may also improve in vitro models of implantation.
Our observational evidence from embryo-EEC cultures and experimental evidence from TSC spheroid-EEC cultures suggest that interactions with the apical surface of EEC promotes extensive STB formation before this cell type invades between cells of the EEC layer. Formation of STB has been seen in Day 8 embryos cultured in the absence of endometrial cells (Deglincerti et al., 2016; Shahbazi et al., 2016), while syncytialization is well established to occur spontaneously in primary placental trophoblast (Kliman et al., 1986). Our in silico models provide independent evidence that EEC interactions stimulate differentiation pathways in TE, and this may act to promote STB formation and ensure timely breaching of the epithelium. The STB we observed in both implanting embryos and the TSC spheroid implantation model was invasive, and this primary STB present at implantation contrasts with the canonical STB that forms from villous cytotrophoblasts and bounds the villi at later stages of placentation (Aplin, 2010; James et al., 2012; Yabe et al., 2016). TSC spheroid interactions with Ishikawa cell layers produced invasive STB that was morphologically very similar to that formed from embryos, thus leading us to favour a model in which the primary STB phenotype is induced by interactions with EEC. Conversely, formation directly from TE as opposed to from cytotrophoblast could also contribute to the unique phenotype of primary STB. EEC-induced invasive trophoblast differentiation from TE was previously shown in mouse blastocysts (Ruane et al., 2017), implicating evolutionary conservation of this mechanism at the onset of interstitial implantation.
We modelled the TE-EEC interface using network biology techniques as a discovery-phase approach to identify candidate functional gene networks that could be assessed in future in vitro studies. A prior network biology approach used whole embryo and whole endometrium microarray transcriptomes to identify key gene networks, including adhesion, extracellular matrix and cytokine genes (Altmae et al., 2012). While we found some of the same genes (APOD, VCAN, FBLN2, LAMA1, SERPINE1, TGFB1), our focus on EEC interactions with polar and mural TE subpopulations using RNAseq data provided a model with enhanced resolution. Furthermore, hypernetwork analysis enabled enrichment of functional gene networks, which we have previously used to investigate ICM and TE lineage allocation from the eight-cell blastomere stage in human embryos (Smith et al., 2019). The expression correlation matrices underlying hypernetworks capture gene networks that relate to functional units within the transcriptome, with the sensitivity of this correlation approach lending itself to highly dynamic biological systems (Pearcy et al., 2016; Gaudelet et al., 2018; Chantzichristos et al., 2021). Increased resolution of in silico implantation models could be obtained by stratifying EEC populations, as have been recently characterized using spatial transcriptomics (Garcia-Alonso et al., 2021). Indeed, comparison with that dataset revealed that all but one (NRXN1) of the EEC genes identified as candidates to promote TE differentiation to STB were expressed in the luminal EEC populations to which blastocysts attach in vivo.
Accordingly, pathway analysis of the hypernetworks linked to EEC-interacting TE genes uncovered a striking resemblance to the pathways activated upon STB differentiation from TSC (Okae et al., 2018), a process that we directly observed when TSC spheroids implanted into Ishikawa EEC layers. Hormone production, including progesterone, is a classical function of STB (Costa, 2016), so the coupling of EEC-interacting TE genes to steroid hormone biosynthetic pathways is strongly suggestive of STB differentiation. The presence of cytokines, predominantly chemokines and interleukins, in the EEC-coupled hypernetworks may also be a marker of STB, since immunomodulation is another key STB attribute (Du et al., 2014). The common signalling pathways seen in TSC-STB differentiation and the TE hypernetworks consisted of JAK-STAT and G-protein-coupled receptors of the neuroactive ligand-receptor interaction pathway, and both of these have been implicated in transducing STB differentiation signals (Knerr et al., 2005; Leduc et al., 2012). RF machine learning was used as a quantitative measure to test the hypothesis that the functionally enriched TE genes at the interface with EEC were linked to STB differentiation, and this analysis yielded a strong signal favouring the hypothesis. Furthermore, dynamic expression indicative of function was demonstrated for many of these genes by RNA velocity analysis (La Manno et al., 2018). Delineating the exact pathways signalling from the interface to differentiation processes was not possible with these approaches, however, the regulation of integrin and insulin-like growth factor (IGF) signalling is hinted at through the robust identification of integrin ligand FBLN5 and IGF-binding protein IGFBP7 as common functional factors in both polar and mural TE. Prior in vitro studies have shown these pathways to function during blastocyst-EEC attachment at implantation (Kang et al., 2014; Green et al., 2015), indeed IGF can signal through JAK-STAT (Zhou et al., 2021) and there is evidence of co-regulation between IGF and integrin signalling (Takada et al., 2017).
Single cell transcriptomic discrimination of TE subpopulations have linked polar TE with upregulation of STB differentiation genes (Petropoulos et al., 2016; Lv et al., 2019). Here, we used these TE subpopulations and found increased levels and organization of connectivity in EEC-interacting polar TE gene networks. Gene network properties of connectivity and organization are indicative of cell differentiation state; high network organization (measured as low entropy) characterizes reduced regulatory cross-talk and pathway redundancy in more differentiated cells (Banerji et al., 2013; Teschendorff and Enver, 2017). Our analysis, therefore, corroborates polar TE as more differentiated than mural TE and suggests that interactions with EEC may be more strongly linked to cell differentiation networks in polar TE than in mural TE. Polar TE-oriented attachment to endometrial epithelium was observed here and in previous studies (Bentin-Ley et al., 2000; Aberkane et al., 2018), and embryonic pole-oriented stromal invasion was observed in vivo (Hertig et al., 1956). The more differentiated state of EEC-attached polar TE could therefore lead to polar-oriented STB invasion. In contrast, our pathway, RF and RNA velocity analyses found overall similarities between polar and mural TE. Moreover, in embryos we observed multiple independent STB regions in contact with Ishikawa cells. Together, our findings lead to a model whereby STB forms from EEC-attached polar and polar-proximal mural TE before initiating invasion into the endometrium. The corollary of this model could be that distal mural TE not engaged with EEC gives rise to TSCs. This scenario is opposite to that in mice, where mural TE initiates invasion and polar TE gives rise to multipotent trophoblast that go on to form the placenta (Sutherland, 2003). Studies of differential phagocytosis activity between polar and mural TE support this inversion of embryonic pole functionality between mouse and human blastocysts (Rassoulzadegan et al., 2000; Li et al., 2016).
Further work with sophisticated in vitro models consisting of multiple maternal cell types are required to pursue some of the outcomes of this study and to fully characterize the establishment of trophoblast populations at implantation. Failure of this process is likely behind many of the recurrent implantation failures seen in assisted reproduction patients and may also contribute to cases of recurrent miscarriage (Koot et al., 2011). Furthermore, there is increasing awareness that later stage obstetric pathologies have their origin in implantation and early pregnancy (Rabaglino et al., 2015; Garrido-Gomez et al., 2017; Than et al., 2018; Aplin et al., 2020), and understanding endometrial shaping of trophoblast development is central to developing new treatments.
Data availability
Much of the data underlying this article are available within the article and in its online supplementary material. The totality of the data underlying this article will be shared on reasonable request to the corresponding author.
Supplementary Material
Acknowledgements
We would like to thank the assisted reproduction patients who kindly donated their embryos to this research programme, and the clinic staff at the Department of Reproductive Medicine, St Mary’s Hospital, Manchester, UK; Manchester Fertility, Manchester, UK and the Hewitt Centre, Liverpool Women’s Hospital, Liverpool, UK, who made this possible. We also thank Dr Hiroake Okae for the kind gift of the TSC line used in this study.
Authors’ roles
P.T.R., S.J.K., A.S., M.W., D.R.B. and J.D.A. designed the study, and S.J.K., D.R.B., M.W. and J.D.A. obtained funding. P.T.R., T.G., L.P., P.A.B. and I.W. carried out the experimental and analytical work. P.T.R. wrote the manuscript, which was edited by T.G., S.J.K., A.S., M.W., D.R.B. and J.D.A.
Funding
This work was supported by funds from the charities Wellbeing of Women (RG1442) and Diabetes UK (15/0005207), and by the NIHR Local Comprehensive Research Network and Manchester Clinical Research Facility, and the Department of Health Scientist Practitioner Training Scheme.
Conflict of interest
None declared.
References
- Aberkane A, Essahib W, Spits C, De Paepe C, Sermon K, Adriaenssens T, Mackens S, Tournaye H, Brosens JJ, Van de Velde H.. Expression of adhesion and extracellular matrix genes in human blastocysts upon attachment in a 2D co-culture system. Mol Hum Reprod 2018;24:375–387. [DOI] [PubMed] [Google Scholar]
- Alikani M. Epithelial cadherin distribution in abnormal human pre-implantation embryos. Hum Reprod 2005;20:3369–3375. [DOI] [PubMed] [Google Scholar]
- Altmae S, Reimand J, Hovatta O, Zhang P, Kere J, Laisk T, Saare M, Peters M, Vilo J, Stavreus-Evers A. et al. Research resource: interactome of human embryo implantation: identification of gene expression pathways, regulation, and integrated regulatory networks. Mol Endocrinol 2012;26:203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin JD. Developmental cell biology of human villous trophoblast: current research problems. Int J Dev Biol 2010;54:323–329. [DOI] [PubMed] [Google Scholar]
- Aplin JD, Jones CJ, Harris LK.. Adhesion molecules in human trophoblast—a review. I. Villous trophoblast. Placenta 2009;30:293–298. [DOI] [PubMed] [Google Scholar]
- Aplin JD, Myers JE, Timms K, Westwood M.. Tracking placental development in health and disease. Nat Rev Endocrinol 2020;16:479–494. [DOI] [PubMed] [Google Scholar]
- Aplin JD, Ruane PT.. Embryo-epithelium interactions during implantation at a glance. J Cell Sci 2017;130:15–22. [DOI] [PubMed] [Google Scholar]
- ASiRMaESIGo. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod 2011;26:1270–1283. [DOI] [PubMed] [Google Scholar]
- Banerji CR, Miranda-Saavedra D, Severini S, Widschwendter M, Enver T, Zhou JX, Teschendorff AE.. Cellular network entropy as the energy potential in Waddington's differentiation landscape. Sci Rep 2013;3:3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentin-Ley U, Horn T, Sjogren A, Sorensen S, Falck Larsen J, Hamberger L.. Ultrastructure of human blastocyst-endometrial interactions in vitro. J Reprod Fertil 2000;120:337–350. [DOI] [PubMed] [Google Scholar]
- Berger C, Boggavarapu NR, Menezes J, Lalitkumar PG, Gemzell-Danielsson K.. Effects of ulipristal acetate on human embryo attachment and endometrial cell gene expression in an in vitro co-culture system. Hum Reprod 2015;30:800–811. [DOI] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ.. Generalizing RNA velocity to transient cell states through dynamical modelling. Nat Biotechnol 2020;38:1408–1414. [DOI] [PubMed] [Google Scholar]
- Boggavarapu NR, Berger C, von Grothusen C, Menezes J, Gemzell-Danielsson K, Lalitkumar PG.. Effects of low doses of mifepristone on human embryo implantation process in a three-dimensional human endometrial in vitro co-culture system. Contraception 2016;94:143–151. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- Campbell S, Larsen J, Seif MW, Allen TD, Knox F, Jones CJ, Aplin JD.. Mosaic characteristics of human endometrial epithelium in vitro: analysis of secretory markers and cell surface ultrastructure. Mol Hum Reprod 2000;6:41–49. [DOI] [PubMed] [Google Scholar]
- Chantzichristos D, Svensson PA, Garner T, Glad CA, Walker BR, Bergthorsdottir R, Ragnarsson O, Trimpou P, Stimson RH, Borresen SW. et al. Identification of human glucocorticoid response markers using integrated multi-omic analysis from a randomized crossover trial. Elife 2021;10:e62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi RA, Wang T, Adams N, Wu SP, Young SL, Spencer TE, DeMayo F.. Human endometrial transcriptome and progesterone receptor cistrome reveal important pathways and epithelial regulators. J Clin Endocrinol Metab 2020;105:1419–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuprin A, Gal H, Biron-Shental T, Biran A, Amiel A, Rozenblatt S, Krizhanovsky V.. Cell fusion induced by ERVWE1 or measles virus causes cellular senescence. Genes Dev 2013;27:2356–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MA. The endocrine function of human placenta: an overview. Reprod Biomed Online 2016;32:14–43. [DOI] [PubMed] [Google Scholar]
- Cutting R, Morroll D, Roberts SA, Pickering S, Rutherford A; BFS and ACE. Elective single embryo transfer: guidelines for practice British fertility society and association of clinical embryologists. Hum Fertil (Camb) 2008;11:131–146. [DOI] [PubMed] [Google Scholar]
- Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED, Brivanlou AH.. Self-organization of the in vitro attached human embryo. Nature 2016;533:251–254. [DOI] [PubMed] [Google Scholar]
- Dong C, Beltcheva M, Gontarz P, Zhang B, Popli P, Fischer LA, Khan SA, Park KM, Yoon EJ, Xing X. et al. Derivation of trophoblast stem cells from naive human pluripotent stem cells. Elife 2020;9:52504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du MR, Wang SC, Li DJ.. The integrative roles of chemokines at the maternal–fetal interface in early pregnancy. Cell Mol Immunol 2014;11:438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders AC, Hendrickx AG, Schlafke S.. Implantation in the rhesus monkey: initial penetration of endometrium. Am J Anat 1983;167:275–298. [DOI] [PubMed] [Google Scholar]
- Galan A, Herrer R, Remohi J, Pellicer A, Simon C.. Embryonic regulation of endometrial epithelial apoptosis during human implantation. Hum Reprod 2000;15:74–80. [PubMed] [Google Scholar]
- Garcia-Alonso L, Handfield LF, Roberts K, Nikolakopoulou K, Fernando RC, Gardner L, Woodhams B, Arutyunyan A, Polanski K, Hoo R. et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. bioRxiv 2021. doi: 10.1101/2021.01.02.425073. [DOI] [PMC free article] [PubMed]
- Garrido-Gomez T, Dominguez F, Quinonero A, Diaz-Gimeno P, Kapidzic M, Gormley M, Ona K, Padilla-Iserte P, McMaster M, Genbacev O. et al. Defective decidualization during and after severe preeclampsia reveals a possible maternal contribution to the etiology. Proc Natl Acad Sci U S A 2017;114:E8468–E8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelet T, Malod-Dognin N, Przulj N.. Higher-order molecular organization as a source of biological function. Bioinformatics 2018;34:i944–i953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CJ, Fraser ST, Day ML.. Insulin-like growth factor 1 increases apical fibronectin in blastocysts to increase blastocyst attachment to endometrial epithelial cells in vitro. Hum Reprod 2015;30:284–298. [DOI] [PubMed] [Google Scholar]
- Hemberger M, Hanna CW, Dean W.. Mechanisms of early placental development in mouse and humans. Nat Rev Genet 2020;21:27–43. [DOI] [PubMed] [Google Scholar]
- Hertig AT, Rock J, Adams EC.. A description of 34 human ova within the first 17 days of development. Am J Anat 1956;98:435–493. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009a;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009b;4:44–57. [DOI] [PubMed] [Google Scholar]
- James JL, Carter AM, Chamley LW.. Human placentation from nidation to 5 weeks of gestation. Part I: What do we know about formative placental development following implantation? Placenta 2012;33:327–334. [DOI] [PubMed] [Google Scholar]
- Kang YJ, Forbes K, Carver J, Aplin JD.. The role of the osteopontin-integrin alphavbeta3 interaction at implantation: functional analysis using three different in vitro models. Hum Reprod 2014;29:739–749. [DOI] [PubMed] [Google Scholar]
- Kliman HJ, Nestler JE, Sermasi E, Sanger JM, Strauss JF III. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology 1986;118:1567–1582. [DOI] [PubMed] [Google Scholar]
- Knerr I, Schubert SW, Wich C, Amann K, Aigner T, Vogler T, Jung R, Dotsch J, Rascher W, Hashemolhosseini S.. Stimulation of GCMa and syncytin via cAMP mediated PKA signaling in human trophoblastic cells under normoxic and hypoxic conditions. FEBS Lett 2005;579:3991–3998. [DOI] [PubMed] [Google Scholar]
- Knofler M, Haider S, Saleh L, Pollheimer J, Gamage T, James J.. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell Mol Life Sci 2019;76:3479–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koot YE, Boomsma CM, Eijkemans MJ, Lentjes EG, Macklon NS.. Recurrent pre-clinical pregnancy loss is unlikely to be a ‘cause’ of unexplained infertility. Hum Reprod 2011;26:2636–2641. [DOI] [PubMed] [Google Scholar]
- Kursa MB, Rudnicki WR.. Feature selection with the Boruta package. J Stat Softw 2010;36:1–13. [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A. et al. RNA velocity of single cells. Nature 2018;560:494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalitkumar PG, Lalitkumar S, Meng CX, Stavreus-Evers A, Hambiliki F, Bentin-Ley U, Gemzell-Danielsson K.. Mifepristone, but not levonorgestrel, inhibits human blastocyst attachment to an in vitro endometrial three-dimensional cell culture model. Hum Reprod 2007;22:3031–3037. [DOI] [PubMed] [Google Scholar]
- Lalitkumar S, Boggavarapu NR, Menezes J, Dimitriadis E, Zhang JG, Nicola NA, Gemzell-Danielsson K, Lalitkumar LP.. Polyethylene glycated leukemia inhibitory factor antagonist inhibits human blastocyst implantation and triggers apoptosis by down-regulating embryonic AKT. Fertil Steril 2013;100:1160–1169. [DOI] [PubMed] [Google Scholar]
- Leduc K, Bourassa V, Asselin E, Leclerc P, Lafond J, Reyes-Moreno C.. Leukemia inhibitory factor regulates differentiation of trophoblastlike BeWo cells through the activation of JAK/STAT and MAPK3/1 MAP kinase-signaling pathways. Biol Reprod 2012;86:54. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu J, Zhou CQ, Zhang CL, Zhuang GL.. Nonprofessional phagocytosis in trophectoderm cells of human preimplantation blastocysts. Syst Biol Reprod Med 2016;62:243–248. [DOI] [PubMed] [Google Scholar]
- Lindenberg S, Nielsen MH, Lenz S.. In vitro studies of human blastocyst implantation. Ann N Y Acad Sci 1985;442:368–374. [DOI] [PubMed] [Google Scholar]
- Lu X, Wang R, Zhu C, Wang H, Lin H-Y, Gu Y, Cross JC, Wang H.. Fine-tuned and cell-cycle-restricted expression of fusogenic protein syncytin-2 maintains functional placental syncytia. Cell Rep 2017;21:1150–1159. [DOI] [PubMed] [Google Scholar]
- Lv B, An Q, Zeng Q, Zhang X, Lu P, Wang Y, Zhu X, Ji Y, Fan G, Xue Z.. Single-cell RNA sequencing reveals regulatory mechanism for trophoblast cell-fate divergence in human peri-implantation conceptuses. PLoS Biol 2019;17:e3000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistermann D, Bruneau A, Loubersac S, Reignier A, Firmin J, François-Campion V, Kilens S, Lelièvre Y, Lammers J, Feyeux M. et al. Integrated pseudotime analysis of human pre-implantation embryo single-cell transcriptomes reveals the dynamics of lineage specification. Cell Stem Cell 2021;28:1625–5909. [DOI] [PubMed] [Google Scholar]
- Meseguer M, Aplin JD, Caballero-Campo P, O'Connor JE, Martin JC, Remohi J, Pellicer A, Simon C.. Human endometrial mucin MUC1 is up-regulated by progesterone and down-regulated in vitro by the human blastocyst. Biol Reprod 2001;64:590–601. [DOI] [PubMed] [Google Scholar]
- Ning W, Lin S, Zhou J, Guo Y, Zhang Y, Peng D, Deng W, Xue Y.. WocEA: the visualization of functional enrichment results in word clouds. J Genet Genomics 2018;45:415–417. [DOI] [PubMed] [Google Scholar]
- Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T.. Derivation of human trophoblast stem cells. Cell Stem Cell 2018;22:50–63. [DOI] [PubMed] [Google Scholar]
- Pearcy N, Chuzhanova N, Crofts JJ.. Complexity and robustness in hypernetwork models of metabolism. J Theor Biol 2016;406:99–104. [DOI] [PubMed] [Google Scholar]
- Petersen A, Bentin-Ley U, Ravn V, Qvortrup K, Sorensen S, Islin H, Sjogren A, Mosselmann S, Hamberger L.. The antiprogesterone Org 31710 inhibits human blastocyst-endometrial interactions in vitro. Fertil Steril 2005;83:1255–1263. [DOI] [PubMed] [Google Scholar]
- Petropoulos S, Edsgard D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F.. Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 2016;165:1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M, Bialecka M, Gomes Fernandes M, Taelman J, Van Der Jeught M, De Sutter P, Heindryckx B, Chuva De Sousa Lopes SM.. Human blastocyst outgrowths recapitulate primordial germ cell specification events. Mol Hum Reprod 2019;25:519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaglino MB, Post Uiterweer ED, Jeyabalan A, Hogge WA, Conrad KP.. Bioinformatics approach reveals evidence for impaired endometrial maturation before and during early pregnancy in women who developed preeclampsia. Hypertension 2015;65:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Rosen BS, Gillot I, Cuzin F.. Phagocytosis reveals a reversible differentiated state early in the development of the mouse embryo. EMBO J 2000;19:3295–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruane PT, Berneau SC, Koeck R, Watts J, Kimber SJ, Brison DR, Westwood M, Aplin JD.. Apposition to endometrial epithelial cells activates mouse blastocysts for implantation. Mol Hum Reprod 2017;23:617–627. [DOI] [PubMed] [Google Scholar]
- Ruane PT, Buck CJ, Babbington PA, Aboussahoud W, Berneau SC, Westwood M, Kimber SJ, Aplin JD, Brison DR.. The effects of hyaluronate-containing medium on human embryo attachment to endometrial epithelial cells in vitro. Hum Reprod Open 2020;2020:hoz033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty NM, Campbell A, Devito LG, Ilic D. et al. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol 2016;18:700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi MN, Scialdone A, Skorupska N, Weberling A, Recher G, Zhu M, Jedrusik A, Devito LG, Noli L, Macaulay IC. et al. Pluripotent state transitions coordinate morphogenesis in mouse and human embryos. Nature 2017;552:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Aplin JD.. Endometrial apical glycoproteomic analysis reveals roles for cadherin 6, desmoglein-2 and plexin b2 in epithelial integrity. Mol Hum Reprod 2015;21:81–94. [DOI] [PubMed] [Google Scholar]
- Smith CA, Moore HD, Hearn JP.. The ultrastructure of early implantation in the marmoset monkey (Callithrix jacchus). Anat Embryol (Berl) 1987;175:399–410. [DOI] [PubMed] [Google Scholar]
- Smith HL, Stevens A, Minogue B, Sneddon S, Shaw L, Wood L, Adeniyi T, Xiao H, Lio P, Kimber SJ. et al. Systems based analysis of human embryos and gene networks involved in cell lineage allocation. BMC Genomics 2019;20:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M.. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006;34:D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland A. Mechanisms of implantation in the mouse: differentiation and functional importance of trophoblast giant cell behavior. Dev Biol 2003;258:241–251. [DOI] [PubMed] [Google Scholar]
- Takada Y, Takada YK, Fujita M.. Crosstalk between insulin-like growth factor (IGF) receptor and integrins through direct integrin binding to IGF1. Cytokine Growth Factor Rev 2017;34:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Enver T.. Single-cell entropy for accurate estimation of differentiation potency from a cell's transcriptome. Nat Commun 2017;8:15599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than NG, Romero R, Tarca AL, Kekesi KA, Xu Y, Xu Z, Juhasz K, Bhatti G, Leavitt RJ, Gelencser Z. et al. Integrated systems biology approach identifies novel maternal and placental pathways of preeclampsia. Front Immunol 2018;9:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco MY, Moffett A.. Development of the human placenta. Development 2019;146:163428. [DOI] [PubMed] [Google Scholar]
- Velicky P, Meinhardt G, Plessl K, Vondra S, Weiss T, Haslinger P, Lendl T, Aumayr K, Mairhofer M, Zhu X. et al. Genome amplification and cellular senescence are hallmarks of human placenta development. PLoS Genet 2018;14:e1007698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RC, Ming H, Logsdon DM, Sun J, Rajput SK, Kile RA, Schoolcraft WB, Roberts RM, Krisher RL, Jiang Z. et al. Dynamics of trophoblast differentiation in peri-implantation-stage human embryos. Proc Natl Acad Sci U S A 2019;116:22635–22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Yin Y, Zheng Y, Ma Y, Li Y, Zhao Z, Guo J, Ai Z, Niu Y, Duan K. et al. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 2020;577:537–542. [DOI] [PubMed] [Google Scholar]
- Yabe S, Alexenko AP, Amita M, Yang Y, Schust DJ, Sadovsky Y, Ezashi T, Roberts RM.. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci U S A 2016;113:E2598–E2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J.. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005;33:W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Lv M, Wang P, Guo C, Ni Z, Bao H, Tang Y, Cai H, Lu J, Deng W. et al. Sequential activation of uterine epithelial IGF1R by stromal IGF1 and embryonic IGF2 directs normal uterine preparation for embryo implantation. J Mol Cell Biol 2021;13:646–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Wang R, Yuan P, Ren Y, Mao Y, Li R, Lian Y, Li J, Wen L, Yan L. et al. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature 2019;572:660–664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Much of the data underlying this article are available within the article and in its online supplementary material. The totality of the data underlying this article will be shared on reasonable request to the corresponding author.