Abstract
We have developed a method for identifying essential genes by using an in vitro transposition system, with a small (975 bp) insertional element containing an antibiotic resistance cassette, and mapping these inserts relative to the deduced open reading frames of Haemophilus influenzae by PCR and Southern analysis. Putative essential genes are identified by two methods: mutation exclusion or zero time analysis. Mutation exclusion consists of growing an insertional library and identifying open reading frames that do not contain insertional elements: in a growing population of bacteria, insertions in essential genes are excluded. Zero time analysis consists of monitoring the fate of individual insertions after transformation in a growing culture: the loss of inserts in essential genes is observed over time. Both methods of analysis permit the identification of genes required for bacterial survival. Details of the mutant library construction and the mapping strategy, examples of mutant exclusion, and zero time analysis are presented.
The increasing incidence of antibiotic-resistant bacteria in clinical practice has stimulated renewed interest within the pharmaceutical industry in searching for and developing new classes of antibiotics. One approach used in this work is molecular screening against defined targets. Until recently, the identification of appropriate antibacterial targets has been a slow, laborious process and has been limited to a few well-defined bacterial functions. The availability of the complete nucleotide sequences of a number of bacterial species has stimulated global approaches (12, 23) to understanding and identifying previously undiscovered functions.
Even a simple analysis of genomic sequence from bacterial pathogens of commercial interest reveals a large fraction (∼40%) of open reading frames (ORFs) of unknown or hypothetical function. Among this collection are ORFs required for bacterial growth and survival—potential antibacterial targets. Accordingly, we have developed an experimental method to annotate a bacterial genome at a simple level: is the deduced ORF required for growth under the chosen conditions? The answer to this question would be one criterion for choosing an antibacterial target for development.
The minimum number of genes or functions required for autonomous bacterial growth has been variously estimated (17, 18). While it is clear that bacteria possess redundant, or backup, functions, there are individual genes that are absolutely required for growth or viability. We define essential genes as those for which an insertional mutation cannot be obtained in a growing bacterium. This definition provides the theoretical foundation for the experiments in this paper.
We describe an experimental, as opposed to computational (2), method for identifying essential genes in Haemophilus influenzae. The technique makes use of in vitro transposition to generate a large, random, insertional mutant library and a combination of PCR and Southern analysis to map the chromosomal location of the inserts. Our choice of H. influenzae was influenced by the quality of its genomic sequence (10), the ease and efficiency of DNA transformation in this organism, and its continued importance as a human pathogen. The details of the library construction, the insert mapping strategy, and the analysis used for identification of previously unknown essential genes are described.
MATERIALS AND METHODS
Strain construction.
H. influenzae BC200 (the kind gift of Jane Setlow) was cured of plasmid pDM2 by growth in brain heart infusion supplemented with NAD (10 μg/ml) and hemin (12 μg/ml) (sBHI) at 37°C without antibiotics. After serial passage, individual isolates were tested for sensitivity to ampicillin and chloramphenicol. A sensitive isolate was examined for plasmid content and transformation efficiency and designated NP200 (for no plasmid).
Competent cell preparation.
NP200 competent cells were prepared by using competence-inducing MIV medium (4). Cells were stored at −80°C in 1.0-ml aliquots.
Transformation of NP200 competent cells.
Frozen competent cells were thawed on wet ice, spun briefly, and resuspended in 1.0 ml of freshly prepared MIV medium (4). One microgram of DNA was added, and the cells were incubated at 37°C for 30 min. Fresh sBHI was then added (5 ml), and the cells were incubated for an additional 90 min (with shaking). Chloramphenicol was added to a final concentration of 1.5 μg/ml, and the cells were grown for an additional 90 min. The culture was then plated on sBHI agar containing 1.5 μg of chloramphenicol per ml.
Genomic DNA preparation.
The CTAB method (3) was used for the isolation of genomic DNA from H. influenzae with the addition of 10 μl of RNase A (50 μg/ml) and incubation at 37°C for 15 min, prior to the second phenol extraction.
DNA quantification.
DNA was quantified fluorometrically (Turner Designs) relative to lambda standards by using Pico green (Molecular Probes).
Generation of AT-Cm.
The region from bp 19 to bp 3757 from pACYC184 (New England Biolabs) was PCR amplified with primers containing XmnI restriction sites [AT-Cm (+), ATTAATGAACATGTTCTACCTGTGACGGAAGATCAC; AT-Cm (−), ATTAATGAACATGTTCACCGGGTCGAATTTGCTTTC]. The PCR product was purified by phenol-chloroform extraction, precipitation with NaOAc, and repeated ultrafiltration (Ultrafree CL; Millipore). The recognition sites for Ty-1 transposase (sequence in boldface type) were generated by XmnI digestion of the purified DNA (XmnI sites underlined).
In vitro transposition.
Primer Island transposition kits (Perkin-Elmer) were used essentially as outlined by the manufacturer. Briefly, 1 μg of H. influenzae genomic DNA was mixed with transposase buffer, 0.2 μg of AT-Cm, and 3 μl of transposase, in a final volume of 30 μl, for 3 h at 30°C. The reaction was terminated by the addition of proteinase K and EDTA. The DNA was precipitated with ammonium acetate, and single-stranded gaps, introduced by the in vitro insertion reaction, were subsequently repaired.
DNA repair reaction.
In vitro-mutagenized genomic DNA was repaired with 2.5 μl of Escherichia coli PolI (NEB), 1 μl of T4 DNA ligase (NEB), and 20 mM deoxynucleoside triphosphates (dNTPs) in 1× ligase buffer for 30 min at 37°C. The DNA was precipitated with sodium acetate, washed carefully in 70% ethanol, and stored at −20°C.
Mutant library construction.
In vitro-mutagenized genomic DNA was transformed into H. influenzae NP200, and the transformation mix was plated on sBHI agar containing 1.5 μg of chloramphenicol per ml. After 24 h, chloramphenicol-resistant colonies were pooled by the addition of sBHI (5 ml) to the plates and gently scraping the colonies together. The number of plates that were pooled determined the size of the mutant library. We routinely obtained 1,000 to 3,000 mutants from a single Ty-1 reaction.
PCRs.
TaKaRa Taq polymerase was used according to the manufacturer in 50-μl reaction mixtures with 50 ng of genomic DNA as template. A three-step PCR was used: 94°C for 5 min (1 cycle); 94°C for 1 min, 60°C for 0.5 min, and 68°C for 2.5 min (35 cycles); and 68°C for 10 min (1 cycle).
Long PCRs (LPCRs).
TaKaRa LA Taq was used according to the manufacturer in 100-μl reaction mixtures with 20 ng of DNA as template. A three-step PCR was used: 95°C for 4 min (1 cycle) and 98°C for 20 s, 50°C for 2 min, and 68°C for 18 min (35 cycles), with the oligonucleotides (5′ 0991)-CCATTATGAACAGAAAACATTTTTTTATTTTC and (3′ 0997)-CCAATTTCGAGATAAATTCTATTTTTATCATAAC.
Southern analysis.
Large-format (25 by 20 cm) agarose gels were soaked sequentially with 0.1 N HCl and 0.4 M NaOH and transferred to Hybond N+ membrane (Amersham) by vacuum blotting (Bio-Rad). Membranes were prehybridized for 1 h and hybridized overnight in 20 ml of hybridization solution (GIBCO) with [33P]dCTP random-labeled probes (20). Membranes were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (42°C), followed by two washes in 0.1× SSC (63°C), exposed overnight to a phosphor screen, and visualized by phosphoimaging (Molecular Dynamics).
Molecular weight markers.
A 1-kb ladder (Gibco) was used for both ethidium staining and, after random-primed [33P]dCTP labelling, as a probe for Southern analysis.
Oligonucleotides.
PCR primers specific for At-Cm and metE (AT-Cm 542, AAAGAAAAATAAGCACAAGTTTTATCCG) were designed by using OLIGO (MBInsights) with a calculated melting temperature of 70°C (metE, 5′-ATGACAACATCACATATTTTAGGCTTTC and 3′-CGCTAATTCCGCACGTAATTTT).
Genomic sequencing.
H. influenzae genomic DNA (3 to 5 μg) was used as a template for PCR cycle sequencing (Perkin-Elmer) with the oligonucleotide primers AT-Cm Seq (+) (ATTGGTGCCCTTAAACGCCTG) and AT-Cm Seq (−) (TTACGTGCCGATCAACGTCTC).
RESULTS
Characterization of in vitro transposon-mutagenized H. influenzae.
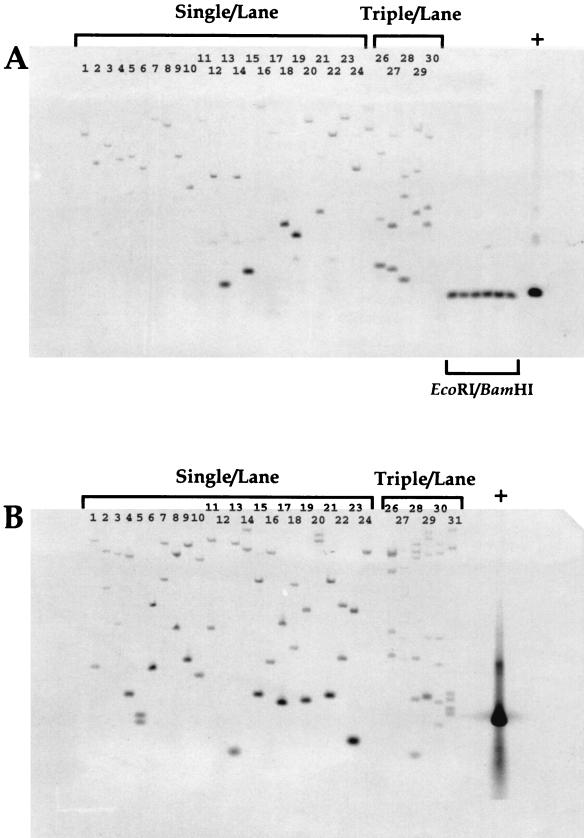
The in vitro transposition reaction catalyzed by Ty-1 randomly inserts a DNA fragment with defined ends into a DNA target (5, 7, 8). We tested this system with two antibiotic resistance cassettes (Fig. 1) and high-molecular-weight H. influenzae genomic DNA as a target. After in vitro reaction and repair (see Materials and Methods), the DNA was transformed into competent H. influenzae and the transformation mixture was plated on selective media (trimethoprim for AT-2 and chloramphenicol for AT-Cm). We examined the resultant antibiotic-resistant colonies for the number and randomness of insertions into the H. influenzae chromosome by Southern analysis (Fig. 2). Genomic DNA from overnight cultures inoculated from single colonies or three independently picked colonies was isolated, digested with EcoRI (Fig. 2A and B, lanes 1 to 23) or with EcoRI-BamHI (Fig. 2A, lanes 31 to 36), separated by agarose gel electrophoresis, and transferred to nylon membranes. These filters were probed with a random-primed 33P-labeled AT-2 (Fig. 2A) or AT-Cm (Fig. 2B) probe. The single Southern hybridizing band seen in each lane with the AT-2 probe is evidence that resistant clones contain a single AT-2 insertion (Fig. 2A, lanes 1 to 23). We interpret the size distribution of Southern hybridizing genomic EcoRI fragments as evidence for the randomness of insertion sites in the H. influenzae chromosome. The fidelity and integrity of the in vitro reactions were examined by digesting genomic DNA samples with restriction sites that are at each end of the AT-2 cassette (EcoRI-BamHI): the entire AT-2 insert should be released from high-molecular-weight DNA. A Southern hybridizing band can clearly be seen that migrates with the same apparent molecular weight as authentic AT-2 (Fig. 2A, lanes 30 to 35), confirming that the in vitro reaction, transformation, and selection proceed such that an entire antibiotic cassette is randomly inserted into high-molecular-weight DNA.
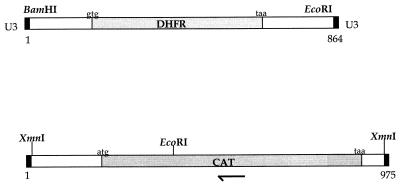
FIG. 1.
Features and partial restriction maps of in vitro transposition cassettes. Relevant restriction sites, positions of start and stop codons, and positions of ORFs coding for antibiotic resistance determinants are indicated. Solid bars indicate the positions of U3 termini recognized by Ty-1 transposase. (Top diagram) AT-2. (Bottom diagram) AT-Cm. The position of the AT-Cm-specific insert-anchored primer is indicated by the half arrow. DHFR, dihydrofolate reductase.
FIG. 2.
Southern analysis of antibiotic-resistant H. influenzae isolates. (A) Genomic Southern blot of trimethoprim-resistant colonies. (B) Genomic Southern blot of chloramphenicol-resistant colonies. Lanes: 1 to 24, 1 colony/lane; 25 to 30, three colonies/lane. (A) Lanes 1 to 31, EcoRI digest; 31 to 36, EcoRI-BamHI double digest. (B) Lanes 1 to 31, EcoRI digest; +, positive controls for Southern hybridization with AT-2 and AT-Cm, respectively.
A similar analysis was performed with chloramphenicol-resistant clones (Fig. 2B). The AT-Cm cassette contains a unique internal EcoRI site (Fig. 1); therefore, a single insertion will yield two Southern hybridizing bands when an EcoRI-digested genomic Southern blot is probed with a randomly primed 33P-labelled AT-Cm. We interpret the observed pattern to indicate that for the AT-Cm cassette, insertions are also randomly distributed in the H. influenzae chromosome. The results from the multiple isolate cultures (Fig. 2A and B, lanes 25 to 30) provide further evidence for the random nature of the insertion reaction and for the conclusion that each isolate contains a single insert: the number of observed bands can be accounted for by the number of colonies picked to grow the culture (1 band/colony for AT-2 and 2 bands/colony for AT-Cm).
Identification of insertion sites.
More precise localization of inserts in the H. influenzae chromosome was determined by direct sequencing. Oligonucleotide primers specific for either AT-2 or AT-Cm were designed (∼150 bp from the ends of the inserts [see Materials and Methods]) that permitted the junctions between the cassettes and the H. influenzae genome to be identified by comparing our sequencing results to the H. influenzae genomic sequence (10). The DNA template for the sequencing reactions was the genomic DNA used for Southern analysis (see above). The results (Table 1) show that the in vitro reaction can insert AT-2 and AT-Cm into a variety of DNA elements: ORFs, intergenic regions, and ribosomal operons. No sequence preferences for insertion sites were observed. Comparison of the sequence data derived from the outward-reading primers (appropriate to each cassette) with the published H. influenzae genome revealed no deletions or insertions near the transposon insertion sites. We interpret these results as further evidence that the in vitro reaction, repair, and subsequent transformation introduce no local DNA rearrangements or deletions near the insertion site. One isolate, AT-Cm10, contained an AT-Cm insert in metE (codon 603), and a strain bearing this mutation was reconstructed from isolated genomic DNA by standard techniques (see Materials and Methods).
TABLE 1.
AT insertion sites
| Isolate no. | ORF name | HI no. | Insertion site | Orientation | Location |
|---|---|---|---|---|---|
| At-2 insertions | |||||
| 1 | basS | 1707 | 1781335 | → | In coding region |
| 2 | ftsH | 1335 | 1410642 | ← | In coding region |
| 3 | pnp | 229 | 258896 | → | In coding region |
| 4 | prlC | 214 | 231467 | ← | In coding region |
| 5 | queA | 245 | 273497 | → | In coding region |
| 6 | metE | 1702 | 1773705 | → | In coding region |
| 7 | crp | 957 | 1014169 | → | Upstream of ORF |
| 8 | arcB | 220 | 241588 | → | Upstream of ORF |
| At-Cm insertions | |||||
| 9 | nifR3 | 979 | 1037244 | → | In coding region |
| 10 | metE | 1702 | 1772287 | ← | In coding region |
| 11 | ilvC | 682 | 724475 | → | In coding region |
| 12 | Hypothetical | 159 | 175595 | → | In coding region |
| 13 | ompA | 1164 | 1236029 | ← | In coding region |
| 14 | slyD | 699 | 745450 | → | Upstream of ORF |
| 15 | Hypothetical | 736 | 789499 | ← | Upstream of ORF |
| 16 | HIrrna | In rRNA operon |
Due to the redundancy of HIrrn operons in H. influenzae, the exact position of AT-Cm16 could not be determined.
PCR and Southern detection of chromosomal insertions.
Our strategy for identifying putative essential genes uses a technique for mapping the location of inserts, relative to deduced ORFs, in a population of growing bacteria. A pilot experiment using genomic DNA from a small AT-Cm insertional mutant library (∼5,000 inserts) was spiked with known quantities of metE mutant DNA and used as a template for PCR and Southern analysis. metE mutant DNA was serially diluted into genomic DNA prepared from the insertional library, and these dilutions were used in PCRs with a primer pair consisting of one primer specific for AT-Cm (see Materials and Methods) and another primer specific for the 5′ coding sequence of metE (Fig. 3). This primer combination (insert-anchored primers) was ∼104-fold more sensitive for detecting the metE insertions from the mixed template than ORF-specific primers (PCR primer pairs that spanned the coding region of metE [data not shown]). PCRs using the serially diluted templates were separated by agarose gel electrophoresis, transferred to a nylon membrane, and probed with a 33P-random-labeled AT-Cm probe. The results show a significant signal from as few as ∼10 copies of metE insert DNA in a background of ∼107 wild-type metE genes (Fig. 3, lane 7).
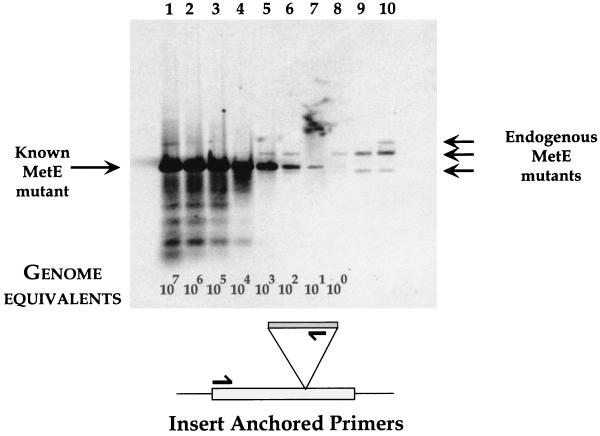
FIG. 3.
Detection of metE insert mutant by PCR and Southern analysis. A Southern blot of dilutions of metE mutant DNA with genomic DNA from a small insert library is shown. The positions of known metE insert and library mutants are shown. Genome equivalents indicate the calculated copies of PCR template in the reactions. Lanes: 1, 50 ng of metE mutant DNA and 50 ng of insert library DNA; 2, 5 ng of metE DNA and 50 ng of library DNA; 3, 0.5 ng of metE DNA and 50 ng of library DNA; 4, 50 pg of metE DNA and 50 ng of library DNA; 5, 5 pg of metE DNA and 50 ng of library DNA; 6, 0.5 pg of metE DNA and 50 ng of library DNA; 7, 50 fg of metE DNA and 50 ng of library DNA; 8, 5 fg of metE DNA and 50 ng of library DNA; 9, 0.5 fg of metE DNA and 50 ng of library DNA; 10, 50 ng of insert library DNA. The schematic shows the positions of the PCR primers relative to the metE coding region and AT-Cm insert.
When only genomic DNA from the insertional library was used as a PCR template, we observed several Southern hybridizing bands (Fig. 3, lane 10) with metE-specific, insert-anchored primers. We interpret this result as evidence for AT-Cm insertions in metE that are present in the mutant library. These endogenous inserts can be detected by PCR and Southern analysis in the presence of small numbers of competing metE mutant DNA templates (Fig. 3, lanes 5, 6, and 8). As the ratio of endogenous mutants to metE mutant DNA decreases, the signal from the library diminishes (Fig. 3, lanes 9 to 4). In order to identify chromosomal insertions, a combination of PCR and Southern analysis gave the required sensitivity and specificity: PCR and agarose gel-ethidium staining alone did not give reliable or reproducible results (data not shown). Because the positions of the PCR primers are precisely known (for both the AT-Cm cassette and the ORF of interest), the size of the Southern hybridizing fragments relates to the position of the insert relative to the ORF-specific primer, thereby identifying the chromosomal location of every insert. By varying the ORF-specific primer, a map of the locations of AT-Cm inserts relative to every ORF in H. influenzae can be derived. This mapping approach can be used to identify essential (by our definition of insertional inactivation) genes.
Zero time analysis.
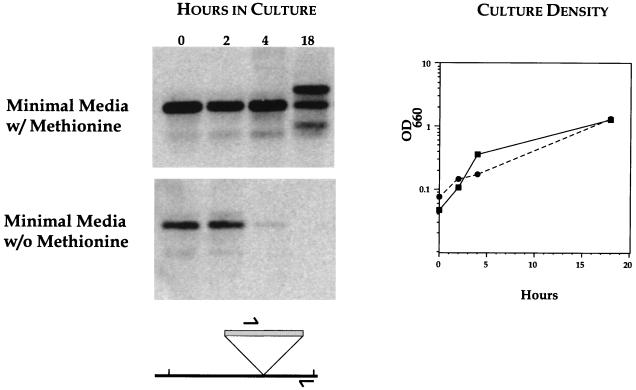
The in vitro transposition reaction can create insertional mutations in both essential and nonessential genes: potentially lethal events will only be manifest after transformation and subsequent expression. Inserts in essential genes will therefore be present in vitro (zero time) and should be lost from the population as the transformation culture grows. This hypothesis was first tested by using the defined metE mutant and a small AT-Cm insertional library. A culture in complete medium (sBHI) was seeded with the metE insert strain and with the small insertional library. This mixed culture was grown for 2 h, and the bacteria were then diluted into minimal medium containing all required amino acids or a defined medium lacking methionine (11, 21). Aliquots at the time of dilution (zero time) and 2, 4, and 18 h postdilution were removed and processed for PCR and Southern analysis (Fig. 4). The presence of the metE mutant strain in the culture can be deduced from the insert-anchor-derived Southern hybridizing band that is clearly visible at the beginning of the experiment (Fig. 4, both panels, lane t = 0). The metE insert strain persists throughout the growth of the culture in the samples derived from the minimal medium containing methionine (Fig. 4, upper panel). The samples from minimal medium lacking methionine clearly show the disappearance of the metE mutant strain over time (Fig. 4, lower panel). Under the conditions of the experiment, metE is an essential function, and cells bearing inserts in this gene are lost from the population. This loss is specific to a subset of mutants, because the growth rate and final cell density of the cultures in both media (with and without methionine) are essentially identical (Fig. 4, graph). We interpret the presence of the additional Southern hybridizing bands seen in the minimal medium with methionine at the 18-h time point as evidence for the outgrowth of endogenous metE mutants present in the insertional library. These mutants were identified previously (Fig. 3, lane 10). As expected, these Southern hybridizing bands derived from the insertional library mutants are not seen in the experimental samples derived from minimal medium lacking methionine. These data illustrate our ability to monitor the loss of specific insertional mutants in a growing population of cells, thus providing experimental justification for our definition of essential genes.
FIG. 4.
Zero time analysis of metE insertion loss. Aliquots from growing cultures were removed at the indicated times and processed for PCR and Southern analysis (see text). Results are from minimal media with (upper panel) and without (lower panel) methionine. The optical densities at 660 nm (OD660) of bacterial cultures (right panel) for minimal media with (solid line) and without (dashed line) methionine are shown. The schematic illustrates the positions of PCR primers used in the analysis.
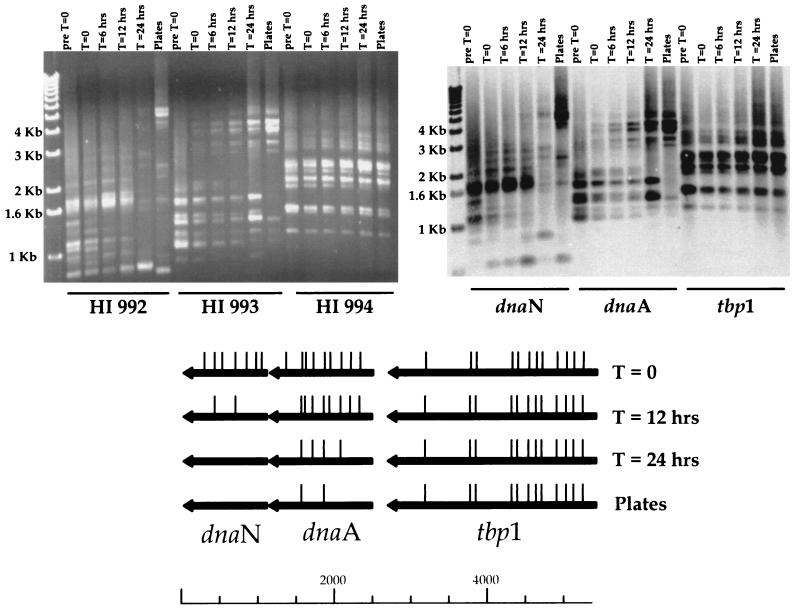
An additional example of zero time analysis illustrates differences in the rate of mutation loss in a dividing population of bacteria (Fig. 5). A defined region of H. influenzae (bp 1053737 to 1063605, covering the ORFs HI 991 to 997) was cloned in a multicopy vector and used as a template for in vitro transposition. The mutagenized plasmids were then transformed into E. coli, and a library of inserts was accumulated in this neutral host. This focused insertional library was used as a template for an LPCR that spanned the cloned insert, HI 991 to HI 997, and this LPCR DNA was then used to transform H. influenzae. Aliquots of the chloramphenicol-selected transformation mix were processed for insert-anchored PCR (Fig. 5) immediately after transformation (t = 0), at defined times in broth culture (t = 6, 12, and 24 h), and after selection on solid medium (plates). The presence of inserts in the transforming DNA could be verified by analyzing the LPCR DNA itself (Fig. 5, pre t = 0). The locations of inserts, mapped by insert-anchored PCR, in HI 992, HI 993, and HI 994 are shown (Fig. 5, bottom). The results illustrate that inserts obtained before selection (in E. coli) can be transferred to H. influenzae (compare pre t = 0 with t = 6 h for all three genes), and essential genes (by our definition) can be identified by the loss of inserts within a gene over time. This is shown by the pattern seen for HI 992 (Fig. 5), where inserts are lost after 12 h in culture. The rate of loss of inserts can clearly vary, because inserts in HI 993 are still seen after 24 h. There are genes that appear dispensable, because the pattern of inserts remains stable throughout the experiment (Fig. 5, HI 994, t = 0 to 24 h and plates). We interpret the insert pattern differences seen for HI 993 at t = 24 h and for the plates, as evidence for the different selection pressures exerted by liquid culture and solid media.
FIG. 5.
Zero time analysis of focused mutant library. (Top left panel) Ethidium-stained agarose gel of insert-anchored PCRs with primers specific for HI 992 to 994. (Top right panel) Southern analysis of gel probed with [33P]dCTP random-primed region probe. (Bottom) ORF map of chromosomal region. Arrows indicate the direction of transcription and relative sizes of ORFs. The deduced locations of inserts are indicated by the vertical bars above the ORF map for the individual time points. Size standards (1-kb ladder) are indicated.
Mutation exclusion.
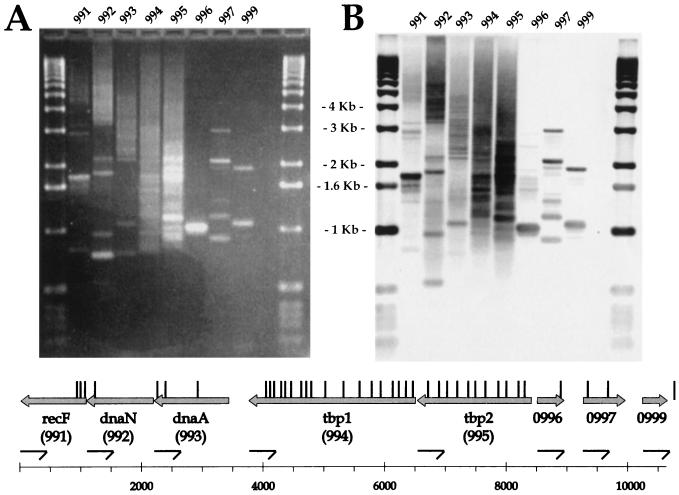
Our definition of gene essentiality states that inserts in essential functions will be lost from a growing population of bacteria. Mapping the positions of AT-Cm inserts in a large mutant library should identify regions of the chromosomes that do not contain inserts: AT-Cm cassettes will be excluded from regions of the chromosome required for bacterial survival. By using PCR and Southern analysis to map inserts in a large mutant library (∼40,000 inserts, or ∼20 inserts/gene), we examined a contiguous region of the H. influenzae genome, ORF by ORF, for genes that do not contain AT-Cm inserts. Genomic DNA isolated from the insertional library was used as a template for insert-anchored PCR. Each reaction mixture contained a primer pair consisting of a primer specific for AT-Cm and a primer specific for an ORF. For ease of analysis, the ORF-specific primers were chosen from a single strand of the chromosome. The ethidium-stained agarose gel (Fig. 6A) and resulting Southern analysis (Fig. 6B) were generated from these reactions. The positions of the AT-Cm inserts relative to the deduced ORFs in this region of the H. influenzae chromosome were mapped by calculating the size of the Southern hybridizing bands in each lane and are shown above the ORF map (Fig. 6, vertical bars). There are clearly regions that do not contain AT-Cm inserts: these areas map to both annotated and hypothetical ORFs. When the insert library was examined with PCR primers designed to map AT-Cm inserts present in the opposite orientation, the pattern of AT-Cm insertions in this region of the chromosome was preserved (data not shown). We interpret gaps in the AT-Cm insertion mapping data, which correspond to deduced ORFs, as defining putative essential genes. Under these experimental conditions, ORFs 991, 992, 996, and 999 have no At-Cm insertions and are therefore potentially essential for growth, while ORFs 993, 994, 995, and 997 clearly have insertions distributed throughout their length, and bacteria harboring inserts in these genes are well represented in the culture.
FIG. 6.
Mutation exclusion analysis of HI 991 to 999. Ethidium-stained agarose gel and Southern analysis of insert-anchored PCRs with primers specific for HI 991 to 999 are shown (see text for details), as is an ORF map of the chromosomal region. Arrows indicate the direction of transcription and relative sizes of the ORFs. (A) Ethidium-stained agarose gel. (B) Southern analysis of gel probed with [33P]dCTP random-primed region probe. The positions and orientations of ORF-specific primers are shown by half arrows. The deduced locations of inserts are indicated by the vertical bars above the ORF map.
Mutation exclusion analysis of HI 991 to 999 identifies a known essential gene, dnaN (HI 992) (9, 15), and several new essential gene candidates. We had anticipated that dnaA (HI 993) would also be devoid of AT-Cm inserts, but we consistently were able to find insertions in this gene (were insertions also seen by zero time analysis [Fig. 5]). The central region of dnaN is devoid of insertions, while the carboxy-terminal region of dnaN and the amino-terminal region of recF appear to tolerate insertions. The ability of putative essential genes to tolerate insertions at the extremities of the reading frame has previously been noted (1). The unannoted genes HI 996 and 999 are also essential by our analysis: they do not contain At-Cm insertions. HI 998 (ribosomal protein L34) was not directly tested, but inserts in this gene would have been revealed by the overlapping PCRs specific for HI 997 (and by exclusion analysis using ORF-specific primers derived from the opposite chromosomal strand for HI 999). No inserts are detected in this region, and HI 998 would therefore be annoted as putative essential. The genes coding for transferrin binding proteins (HI 994 and HI 995) clearly contain multiple insertions and would therefore be considered dispensable in rich media, although in an iron-limiting environment or in an animal host, these mutants might be nonviable, and H. influenzae strains bearing At-Cm inserts in these genes might disappear from the population (6).
By placing the insert-anchored PCRs in sequential order on the gel and manipulating the PCR conditions for longer extensions, overlapping insert mapping data can be generated. Thus, Southern hybridizing bands near the top of the gel in each lane represent AT-Cm inserts in the following ORF. This is mostly clearly seen in the repeated pattern of bands in lanes 991, 992, and 993. Southern hybridizing bands that are not observed in multiple lanes are presumed to be artifactual and are not included in the analysis. This mapping procedure can be continued for every deduced ORF in the H. influenzae genome for which a PCR primer can be synthesized.
DISCUSSION
We have developed a method of identifying regions of the H. influenzae chromosome that are required for viability, making use of an in vitro transposition reaction, complete and accurate genomic sequence data, and the sensitivity of PCR and Southern analysis to map the chromosomal locations of a selectable marker. This approach is generally applicable, although the efficiency of transformation, the accuracy of the genomic sequence, and the number of generated insertions will modulate the confidence in the results. Organisms that are naturally competent and whose genome sequences are available are clear candidates for extending this technique (e.g., Streptococcus pneumoniae, Helicobacter pylori, and Neisseria sp.).
Our analysis has concentrated on ORFs in H. influenzae. We have made no attempt to identify essential structural RNA genes or DNA structural elements or to analyze ORFs smaller than 300 bp (100 amino acids) in length. These elements could be discovered with a library of sufficient coverage and the appropriate genomic PCR primers. They are clearly biologically important, but they are not generally regarded as primary antibacterial drug targets, the initial impetus for our work.
The number of inserts we observed in individual ORFs by PCR and Southern analysis corresponds well with our estimate of the number of mutants obtained from colony counting (assuming ∼1,000 bp/ORF, random insertions, and 1.8 × 106 bp/genome). In analyzing several regions of the H. influenzae chromosome for essential genes, we have noted that the distribution of insert orientation is not random and could be influenced by the local DNA transcriptional environment. We interpret the observation that the number of antibiotic-resistant colonies recovered after in vitro transposition is strongly dependent on the chloramphenicol concentration (higher chloramphenicol concentration = fewer mutants) as evidence that the chloramphenicol acetyltransferase (CAT) promoter in AT-Cm is only weakly transcribed in H. influenzae. We believe that a weak CAT promoter will reduce the polar effects of transposase-generated insertions on surrounding chromosomal genes, simplifying our analysis.
We anticipate that by using our mutant library and searching for genes required for survival in animal models of infection, virulence determinants could be identified as well. This approach could be refined further, to identify genes required for survival in specific niches or organs (e.g., lung versus liver versus spleen) or in different animal models of infection (e.g., murine versus rat). Given the size of the mutant libraries that can be generated, we believe that genome scanning could give a more complete picture of the functions required for pathogenesis than other in vivo mutagenesis methods (13, 16).
Our initial goal was to develop a method that would identify genes required for bacterial viability; we have settled for a technique that can generate a list of genes that cannot be mutagenized by our in vitro insertional technique. As a matter of convenience, we chose rich medium (sBHI) as a growth condition for selection. The selective properties of solid medium versus broth culture were noted in initial experiments and shown by the zero time example, and we chose to use sBHI agar for generating our mutant libraries. Other culture conditions could be tested, including various minimal media, partial oxygen pressure, heat shock, cold shock, growth in serum, limiting iron, etc. Identification of functions required for survival in stationary phase could also be considered.
Several different approaches to identifying essential genes in microorganisms have been proposed, both before and after the availability of genomic sequences (18, 19). Postgenomic approaches include a systematic knockout strategy being undertaken by the yeast community, in silico analysis to determine common, shared and unique ORFs (2), systematic complementation of temperature-sensitive alleles, and a similar in vitro transposition mutagenesis strategy that has recently been described (1). We have used a well-characterized (5, 7, 8, 22) in vitro transposition system to generate a large mutant insert library and analyzed the library by mapping the location of inserts relative to ORFs and by monitoring the rate of loss of particular mutants. The ability to monitor the disappearance of a particular mutant over time provides both a positive control for the ORF of interest (that the in vitro transposition reaction targeted the ORF) and biological information concerning the ORF itself. The rate of gene loss will be modulated by a number of factors, including the steady-state level of expression of the protein, its half-life, the cell doubling time, and the cellular function that is abrogated. These additional data will be relevant to choosing targets for antibacterial drug discovery.
The recognition sequence for Ty-1 transposase is 4 bp, allowing for simple and efficient construction of translational fusions for structure-function studies. This, coupled with the focused mutant library approach, would allow for detailed topological analysis (using alkaline phosphatase fusions [14]) and protein functional domain identification (because loss of an enzymatic function could be correlated to the position of inserts). The 4-bp recognition sequence is also amenable to transcriptional fusions with reporter genes (e.g., green fluorescent protein or β-galactosidase) for cell sorting and identification.
Genome scanning provides an experimental technique for assigning a rudimentary annotation to the large fraction of bacterial genomes that have no known function. We hope this method, and its variations, will begin to provide solutions to understanding and predicting the minimal gene complement required for autonomous bacterial survival.
ACKNOWLEDGMENTS
We thank Ken Idler for sequencing, Paul Jung for oligonucleotide synthesis, Don Halbert for support, and Jane Setlow for the kind gift of strains and protocols.
REFERENCES
- 1.Akerley B J, Rubin E J, Camilli A, Lampe D J, Roberston H M, Mekalanos J J. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arigoni F, Talabot F, Peitsch M, Edgerton M D, Meldrum E, Allet E, Fish R, Jamotte T, Curchod M L, Loferer H. A genome based approach for the identification of essential bacterial genes. Nat Biotechnol. 1998;16:851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1991. pp. 2.4.1–2.4.2. [Google Scholar]
- 4.Barcak G J, Chandler M C, Redfield R J, Tomb J F. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 5.Braiterman L T, Monokian G M, Eichinger D J, Merbs S L, Gabriel A, Boeke J D. In frame linker insertion mutagenesis of yeast transposon Ty1: phenotypic analysis. Gene. 1994;139:19–26. doi: 10.1016/0378-1119(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 6.Cornelissen C N, Sparling P F. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol Microbiol. 1994;14:843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 7.Devine S E, Boeke J D. Efficient integration of artificial transposons into plasmid targets in vitro: a useful tool for DNA mapping, sequencing and genetic analysis. Nucleic Acids Res. 1994;22:3765–3772. doi: 10.1093/nar/22.18.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devine S E, Chissoe S L, Eby Y, Wilson R K, Boeke J D. A transposon-based strategy for sequencing repetitive DNA in eukaryotic genomes. Genome Res. 1997;7:551–563. doi: 10.1101/gr.7.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donachie W D. The cell cycle of Escherichia coli. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 10.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G G, FitzHugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.Herriott R M, Meyer E Y, Vogt M, Modan M. Defined medium for growth of Haemophilus influenzae. J Bacteriol. 1970;101:513–516. doi: 10.1128/jb.101.2.513-516.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hood D W, Deadman M E, Jennings M P, Bisercic M, Fleischmann R D, Venter J C, Moxon E R. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 15.Marians K J. Replication fork propagation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 749–763. [Google Scholar]
- 16.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 17.Mushegian A R, Koonin E V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid M B, Kapur N, Isaacson D R, Lindroos P, Sharpe C. Genetic analysis of temperature-sensitive lethal mutants of Salmonella typhimurium. Genetics. 1989;123:625–633. doi: 10.1093/genetics/123.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharetzsky S, Edlind T D, LiPuma J J, Stull T L. A novel approach to insertional mutagenesis of Haemophilus influenzae. J Bacteriol. 1991;173:1561–1564. doi: 10.1128/jb.173.4.1561-1564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 21.Talmadge M B, Herriott R M. A chemically defined medium for growth, transformation, and isolation of nutritional mutants of Haemophilus influenzae. Biochem Biophys Res Commun. 1960;2:203–206. [Google Scholar]
- 22.Westphal C H, Leder P. Transposon-generated ‘knock-out’ and ‘knock-in’ gene targeting constructs for use in mice. Curr Biol. 1997;7:R530–R533. doi: 10.1016/s0960-9822(06)00224-7. [DOI] [PubMed] [Google Scholar]
- 23.Wood D W, Deadman M E, Allen T, Masoud H, Martin A, Brisson J R, Fleischman R, Venter J C, Richards J C, Moxon E R. Use of the complete genome sequence information of Haemophilus influenzae Rd to investigate lipopolysaccharide biosynthesis. Mol Microbiol. 1996;22:951–965. doi: 10.1046/j.1365-2958.1996.01545.x. [DOI] [PubMed] [Google Scholar]