Abstract
The current coronavirus disease 2019 (COVID-19) pandemic highlights the need for broad-spectrum antiviral therapeutics. Here we describe a new class of self-assembling immunostimulatory short duplex RNAs that potently induce production of type I and type III interferon (IFN-I and IFN-III). These RNAs require a minimum of 20 base pairs, lack any sequence or structural characteristics of known immunostimulatory RNAs, and instead require a unique sequence motif (sense strand, 5′-C; antisense strand, 3′-GGG) that mediates end-to-end dimer self-assembly. The presence of terminal hydroxyl or monophosphate groups, blunt or overhanging ends, or terminal RNA or DNA bases did not affect their ability to induce IFN. Unlike previously described immunostimulatory small interfering RNAs (siRNAs), their activity is independent of Toll-like receptor (TLR) 7/8, but requires the RIG-I/IRF3 pathway that induces a more restricted antiviral response with a lower proinflammatory signature compared with immunostimulant poly(I:C). Immune stimulation mediated by these duplex RNAs results in broad-spectrum inhibition of infections by many respiratory viruses with pandemic potential, including severe acute respiratory syndrome coronavirus (SARS-CoV)-2, SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), human coronavirus (HCoV)-NL63, and influenza A virus in cell lines, human lung chips that mimic organ-level lung pathophysiology, and a mouse SARS-CoV-2 infection model. These short double-stranded RNAs (dsRNAs) can be manufactured easily, and thus potentially could be harnessed to produce broad-spectrum antiviral therapeutics.
Keywords: MT: Oligonucleotides, Therapies and Applications, immunostimulatory RNA, antiviral therapeutic, interferon, SARS-CoV-2, influenza, organ on a chip, RIG-I, Hoogsteen G-G base pairing
Graphical abstract
Si et al. describe the discovery of a new family of short duplex RNAs that potently induce production of type I/III interferons. These RNAs, when administered to cells and animals, can protect against viral infections, including influenza and COVID-19.
Introduction
Recognition of duplex RNAs by cellular RNA sensors plays a central role in host response to infections by initiating signaling cascades that induce secretion of interferon (IFN) and subsequent upregulation of hundreds of IFN-stimulated genes (ISGs). This pathway therefore also serves as a potent point of therapeutic intervention in a broad range of viral diseases. Duplex RNAs with various structural features have been identified that are recognized by the three cellular RNA sensors that are responsible for this innate immune response.1 One of these, Toll-like receptor 3 (TLR3), is located on the cell membrane and the endosomal membrane, while the other two, retinoic acid inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), are located in the cytosol. Long forms of duplex RNA are recognized by these sensors based on their length (i.e., independently of the structure of their 5′ ends) with TLR3 recognizing duplex RNAs >35 bp and MDA5 sensing duplex RNAs >300 bp.2 Short stretches of duplex RNA (>19 bp) can be recognized by RIG-I, but strong activation is achieved only when a triphosphate or a diphosphate is present at its 5′ end and if the end is blunt with no overhangs.1, 2, 3, 4
Duplex RNA-mediated innate immune stimulation is a two-edged sword. For example, in the case of respiratory infections, such as those caused by pandemic viruses (e.g., severe acute respiratory syndrome coronavirus [SARS-CoV]-2, SARS-CoV, Middle East respiratory syndrome coronavirus [MERS-CoV], and influenza virus), RNA-mediated activation of this innate immune response provides the first line of host defense against the invading pathogen. However, on the other hand, the use of duplex RNAs for RNA interference (RNAi) approaches can result in undesired immunological off-target effects and misinterpretation of experimental results.5, 6, 7, 8, 9, 10, 11 Thus, gaining greater insight into the mechanism by which cells sense and respond to duplex RNAs could have a broad impact in biology and medicine.
In this study, we serendipitously discovered a class of new immunostimulatory RNAs while using >200 small interfering RNAs (siRNAs) to identify influenza infection-associated host genes in human lung epithelial cells. These short duplex RNAs potently induce type I and type III IFNs (IFN-I/III) in a wide type of cells but lack any sequence or structure characteristics of known immunostimulatory RNAs. Systematic mechanistic analysis revealed that these immunostimulatory RNAs specifically activate the RIG-I/IRF3 pathway by binding directly to RIG-I, and that this only occurs when these short RNAs have a common overhanging sequence motif (sense strand, 5′-C; antisense strand, 3′-GGG) and a minimum length of 20 bases. Interestingly, the terminal motif is responsible for the self-assembly of end-to-end RNA dimers through Hoogsteen G-G base pairing. In addition, these immunostimulatory RNAs appear to be novel in that they are capable of inducing IFN production regardless of whether they have blunt or overhanging ends, terminal hydroxyl or monophosphate groups, or RNA base or DNA base ends, in contrast to previously described immunostimulatory RNAs that require 5′-diphosphates or triphosphates to activate cellular RNA sensors.2,3 The RNA-mediated IFN-I/III production resulted in significant inhibition of infections by multiple human respiratory viruses, including influenza viruses and SARS-CoV-2 in established cell lines, human lung airway and alveolus chips that have been previously shown to recapitulate human lung pathophysiology, and mouse model.12, 13, 14 These findings also should facilitate the development of siRNAs that avoid undesired immune activation and may pave the way for the development of a new class of RNA therapeutics for the prevention and treatment of respiratory virus infections.
Results
Discovery of IFN-I pathway-activating immunostimulatory RNAs
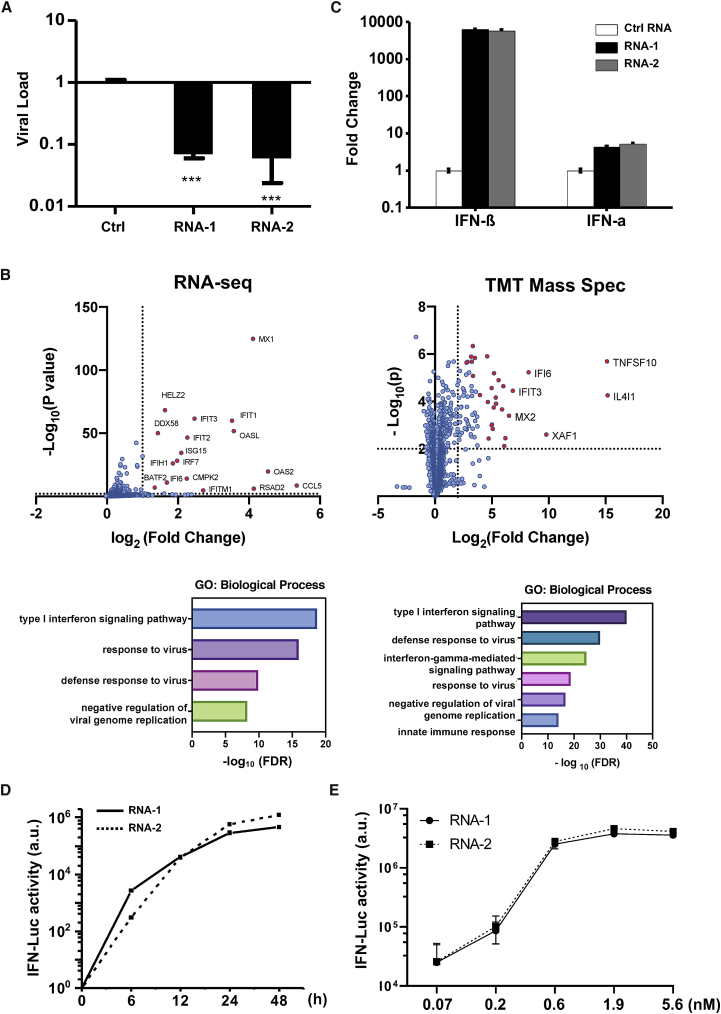
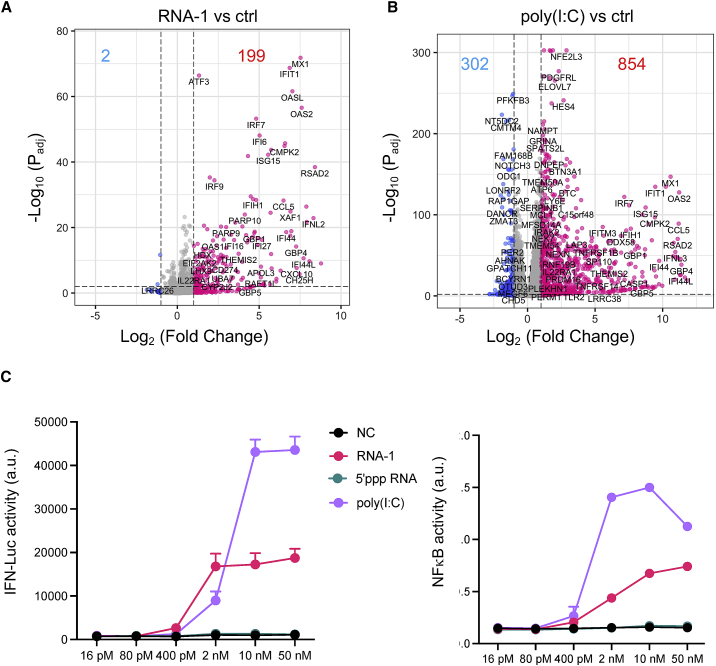
While using >200 siRNAs to identify host genes that mediate human A549 lung epithelial cell responses to influenza A/WSN/33 (H1N1) infection, we found that transfection of two siRNAs (RNA-1 and RNA-2) inhibited H1N1 replication by more than 90% (Figure 1A). To explore the mechanism of action of these siRNAs, we profiled the transcriptome and proteome of A549 cells transfected with RNA-1 (Figure 1B) and RNA-2 (Figure S1), which respectively target the long non-coding RNAs (lncRNAs) DGCR5 and LINC00261, and a scrambled siRNA was used as a control. RNA sequencing (RNA-seq) analysis showed that RNA-1 upregulates the expression of 21 genes by more than 2-fold (p value threshold of 0.01) (Figure 1B left and Figure S2A left). Gene Ontology (GO) enrichment analysis revealed that these genes are involved in IFN-I signaling pathway and host defense response to viral infections (Figure 1B, left), including MX1, OASL, IFIT1, and ISG15 (Figure S2A, left). In parallel, tandem mass tag (TMT) mass spectrometry quantification demonstrated upregulation of 73 proteins by more than 4-fold (p value threshold of 0.01), including IL4I1, TNFSF10, XAF1, IFI6, and IFIT3 (Figures 1B, right, and S2B). GO enrichment analysis of these upregulated proteins also confirmed an association between treatment of RNA-1 and induction of the IFN-I/III pathway (Figures 1B, right, and S3A). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay independently validated that RNA-1 preferentially activates the IFN-I/III pathway relative to the type II IFN pathways (Figure S3B), with IFN-β being induced to much higher levels (>1,000-fold) compared with IFN-α (Figure 1C). This potent induction of IFN-β by RNA-1 was verified at the protein level using enzyme-linked immunosorbent assay (ELISA) (Figure S4), and similar patterns of gene and protein expression were also observed for RNA-2 (Figures 1C, S1, and S2).
Figure 1.
Discovery of new immunostimulatory RNAs
(A) Viral load of A549 cells transfected with RNA-1, RNA-2, or a scrambled duplex RNA control, and 24 h later infected with influenza A/WSN/33 (H1N1) virus (MOI = 0.01). Titers of progeny viruses in medium supernatants collected at 48 h post infection were determined by quantifying plaque-forming units (PFU); data are shown as percentage viral infection measured in the cells treated with the control RNA (data shown are mean ± SD; N = 3; ∗∗∗p < 0.001). (B) Transcriptome by RNA-seq (left) and proteome by TMT mass spectrometry (Mass Spec) of A549 cells after RNA-1 transfection. Differentially expressed genes (DEGs) from RNA-seq or proteins from TMT Mass Spec are shown in volcano plots (top) and results of GO enrichment analysis performed for the DEGs are shown at the bottom (N = 3). (C) qPCR analysis of cellular IFN-β and IFN-α RNA levels at 48 h after A549 cells were transfected with RNA-1, RNA-2, or scrambled dsRNA control (N = 3). (D) RNA-mediated production kinetics of IFN production in wild-type A549-Dual cells that were transfected with RNA-1, RNA-2, or scramble RNA control measured using a Quanti-Luc assay. Optical density (OD) values from cells transfected with the scrambled RNA control were subtracted as background (N = 6). (E) Induction of IFN by different concentrations of RNA-1 and RNA-2 in A549-Dual cells compared with scrambled RNA control measured at 48 h post transfection (control OD values were subtracted as background; N = 6).
Interestingly, when we carried out studies with additional siRNAs to further validate the function of the lncRNAs they target, we found that knockdown of DGCR5 or LINC00261 by these other siRNAs did not induce IFN production (data not shown). This was surprising because, since the inception of RNAi technology, short duplex (double-stranded) siRNAs have been known to induce IFN-I,6,8 and thus subsequent design of these molecules, including the ones used in our study, were optimized to avoid this action and potential immunomodulatory side effects.15 siRNAs synthesized by phage polymerase that have a 5′-triphosphate end can trigger potent induction of IFN-α and -β,6 and siRNAs containing nine nucleotides (5′-GUCCUUCAA-3′) at the 3′ end can induce IFN-α through TLR7.7 Notably, RNAs with a 5′-diphosphate end can induce IFN-I as well,16 but our chemically synthesized duplex RNAs do not have any of these sequence or structural properties. Thus, our data suggested that the two specific RNAs we found to be potent IFN-I/III inducers (RNA-1 and RNA-2) may represent new immunostimulatory RNAs.
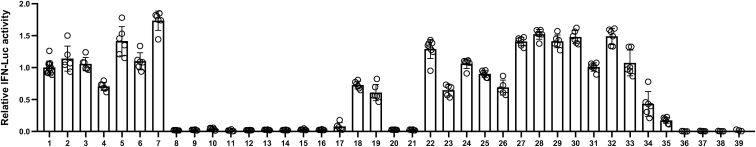
To explore this further, we assessed IFN production induced by the two putative immunostimulatory RNAs using an A549-Dual IFN reporter cell line, which stably expresses luciferase genes driven by promoters containing IFN-stimulated response elements.17 These studies revealed that both RNA-1 and -2 induce IFN production beginning as early as 6 h post transfection, consistent with IFN-I/III being an early-response gene in innate immunity, and high levels of IFN expression were sustained for at least 24 to 48 h (Figure 1D). We also observed induction of IFN production by these duplex RNAs over the nanomolar range (Figure 1E). In addition, we observed similar effects when we tested RNA-3, which was originally designed as a siRNA to knock down another lncRNA, LINC00885 (Figure 2; Table 1). Notably, all three immunostimulatory double-stranded RNAs (dsRNAs) that specifically upregulate strong IFN-I/III responses with high efficiency share a common motif (sense strand, 5′-C; antisense strand, 3′-GGG).
Figure 2.
Comparison of the immunostimulatory activities of different RNAs
IFN luciferase reporter activity of A549-Dual cells transfected with indicated duplex RNAs for 48 h. Data are shown as fold change relative to RNA-1, and the immunostimulatory activity of RNA-1 was set as 1 (N = 6). Data are shown as means ± SD.
Table 1.
Oligonucleotides of RNA monomers
| RNA ID | Sequence (5′–3′) | Note | Relative activity (%) | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNA-1 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | siRNA targeting DGCR5 | 100 ± 10 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-2 | C | U | G | A | G | G | U | U | A | C | U | G | A | A | U | C | U | A | A | C | A | A | U | G | A | siRNA targeting LINC00261 | 114 ± 20 | ||
| G | G | G | A | C | U | C | C | A | A | U | G | A | C | U | U | A | G | A | U | U | G | U | U | A | C | U | |||
| RNA-3 | C | C | A | G | U | G | G | A | A | U | C | A | U | G | G | G | G | A | U | U | U | C | U | T | A | siRNA targeting LINC00885 | 105 ± 10 | ||
| G | G | G | G | U | C | A | C | C | U | U | A | G | U | A | C | C | C | C | U | A | A | A | G | A | A | U | |||
| RNA-4 | C | U | G | A | C | A | U | C | G | U | C | U | C | G | C | A | U | U | U | A | U | G | A | G | C | keep 5′ motif and shuffle remaining | 70 ± 6 | ||
| G | G | G | A | C | U | G | U | A | G | C | A | G | A | G | C | G | U | A | A | A | U | A | C | U | C | G | |||
| RNA-5 | C | A | C | C | G | C | C | A | C | G | A | C | C | A | A | G | U | A | A | A | U | A | U | G | U | random sequence containing 5′ motif | 141 ± 23 | ||
| G | G | G | U | G | G | C | G | G | U | G | C | U | G | G | U | U | C | A | U | U | U | A | U | A | C | A | |||
| RNA-6 | C | U | A | G | U | C | A | C | C | A | C | U | U | C | U | U | A | U | G | G | U | C | U | C | U | random sequence containing 5′ motif | 110 ± 13 | ||
| G | G | G | A | U | C | A | G | U | G | G | U | G | A | A | G | A | A | U | A | C | C | A | G | A | G | A | |||
| RNA-7 | C | C | G | U | C | A | G | A | C | A | A | U | G | U | C | A | A | G | C | U | G | A | A | G | U | random sequence containing 5′ motif | 173 ± 15 | ||
| G | G | G | G | C | A | G | U | C | U | G | U | U | A | C | A | G | U | U | C | G | A | C | U | U | C | A | |||
| RNA-8 | A | C | A | C | U | G | G | C | C | C | C | U | G | A | U | A | G | U | U | C | A | C | C | T | T | move motif to center | 2 ± 0.3 | ||
| A | C | U | G | U | G | A | C | C | G | G | G | G | A | C | U | A | U | C | A | A | G | U | G | G | A | A | |||
| NA-9 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | Deleting G | 2 ± 0.6 | ||
| G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | ||||
| RNA-10 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | Deleting GG | 4 ± 2 | ||
| G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||||
| RNA-11 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | GG to CC | 1 ± 1 | ||
| C | C | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-12 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | GG to AA | 2 ± 0.1 | ||
| A | A | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-13 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | G to A | 2 ± 0.4 | ||
| G | A | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-14 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | G to A | 2 ± 0.3 | ||
| A | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-15 | U | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | G to A | 3 ± 0.9 | ||
| G | G | A | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-16 | G | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | G to C | 2 ± 0.5 | ||
| G | G | C | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-17 | A | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | G to U | 8 ± 6 | ||
| G | G | U | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-18 | Cm | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | 2′-O-methyl | 72 ± 6 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-19 | Cm | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | 2′-O-methyl | 61 ± 13 | ||
| Gm | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-20 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | Um | 2′-O-methyl | 3 ± 0.2 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | Am | |||
| RNA-21 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | 2′-O-methyl | 2 ± 0.6 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | Am | |||
| RNA-22 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | Um | 2′-O-methyl | 129 ± 15 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-23 | pC | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | Tp | monophosphate | 64 ± 9 | ||
| pG | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | Ap | |||
| RNA-24 | pC | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | monophosphate | 106 ± 7 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | Ap | |||
| RNA-25 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | Tp | monophosphate | 90 ± 5 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | Ap | |||
| RNA-26 | pC | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | monophosphate | 69 ± 12 | ||
| pG | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-27 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | change AA into DNA | 141 ± 7 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-28 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | U | U | change TT into RNA | 152 ± 8 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-29 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | U | T | change one T into U | 141 ± 10 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-30 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | U | change one T into U | 148 ± 10 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-31 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | overhang | 101 ± 7 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | |||||
| RNA-32 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | U | U | overhang | 149 ± 13 | ||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | |||||
| RNA-33 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | overhang | 107 ± 20 | ||||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | |||
| RNA-34 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | T | T | 6 nt deletion at 3′ | 43 ± 19 | ||||||||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | |||||||||
| RNA-35 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | T | 7 nt deletion at 3′ | 17 ± 5 | |||||||||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | ||||||||||
| RNA-36 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | 8 nt deletion at 3′ | 0 ± 0.4 | ||||||||||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | |||||||||||
| RNA-37 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | T | A | 9 nt deletion at 3′ | 0 ± 0.4 | |||||||||||
| G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | ||||||||||||
| RNA-38 | C | U | G | A | U | G | A | C | A | C | U | G | G | C | U | A | G | U | U | C | A | C | C | T | T | single sense strand | 0 ± 0.5 | ||
| RNA-39 | G | G | G | A | C | U | A | C | U | G | U | G | A | C | C | G | A | U | C | A | A | G | U | G | G | A | A | single antisense strand | 1 ± 1.5 |
The sense strand (left, 5′ end; right, 3′ end) is positioned on top, while the antisense strand (left, 3′ end; right, 5′ end) is below. If not indicated otherwise, both 5′ and 3′ ends of sense and antisense contain terminal hydroxyl groups. The underlined bases indicate DNA bases, and the bolded bases indicate the common IFN-inducing motif; p, monophosphate group; m, N1-2-O-methyl group. +++, high activity; ++, middle activity; +, low activity; -, no activity.
These short duplex RNAs bind directly to RIG-I
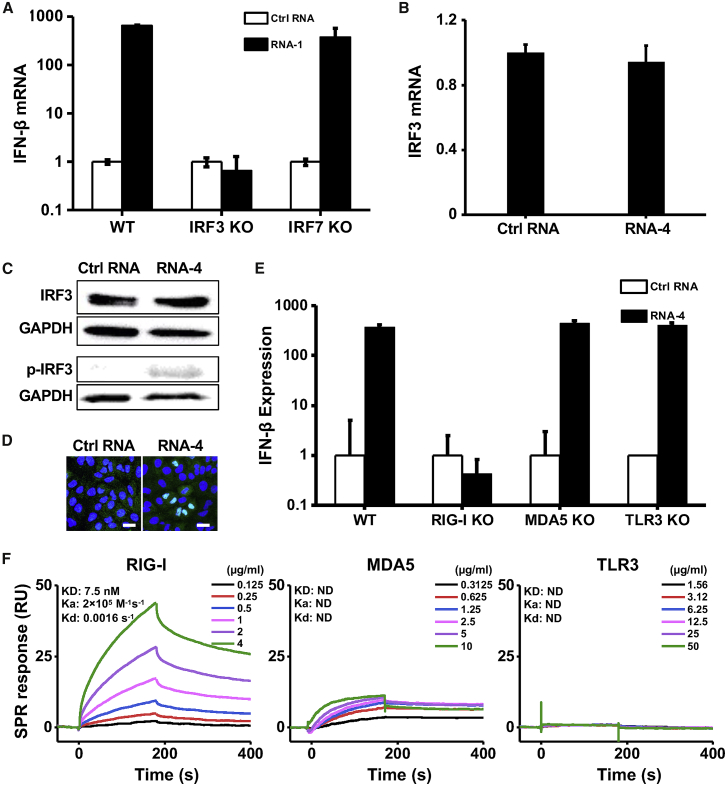
Transcription factor IFN regulatory factor 3 (IRF3) and 7 (IRF7) play vital roles in IFN-I production.18,19 Using IRF3 knockout (KO) and IRF7 KO cells, we found that loss of IRF3, but not IRF7, completely abolished the ability of RNA-1 to induce IFN-β (Figure 3A) and downstream ISGs, including STAT1, IL4L1, TRAIL, and IFI6 (Figure S5). IRF3 is the master and primary transcriptional activator of IFN-I and its induction of IFN-I involves a cascade of events, including IRF3 phosphorylation, dimerization, and nuclear translocation.20,21 To alleviate potential interference from host gene knockdown by RNA-1 that was developed as an siRNA, we performed further mechanistic studies using RNA-4, which contains the common motif of RNA-1, -2, and -3 that we hypothesized and proved to be involved in the immunostimulatory activity but does not target (silence) any host genes because its other nucleotides were randomized (Figure 2; Table 1). Although RNA-4 had no effect on IRF3 mRNA or total protein levels (Figures 3B and 3C), it increased IRF3 phosphorylation (Figure 3C), which is essential for its transcriptional activity18 and subsequent translocation to the nucleus (Figure 3D), where IRF3 acts as transcription factor that induces IFN-I expression.20,21
Figure 3.
Immunostimulatory RNAs induce IFN-I production through RIG-I-IRF3 pathway
(A) IFN-β mRNA levels in wild-type (WT) HAP1 cells, IRF3 knockout HAP1 cells, or IRF7 knockout HAP1 cells after transfection with RNA-1 or scrambled RNA control for 48 h. Data are shown as fold change relative to the scrambled RNA control (N = 3). Note that IRF3 knockdown completely abolished the IFN-β response. (B) IRF3 mRNA levels measured in A549 cells transfected with immunostimulatory RNA-4 or a scrambled RNA control (data are shown as fold change relative to the control RNA; N = 3). (C) Total and phosphorylated IRF3 protein levels in A549 cells at 48 h after transfection with RNA-4 or scrambled RNA control. GAPDH was used as a loading control. (D) Immunofluorescence micrographs showing the distribution of phosphorylated IRF3 in A549 cells transfected with RNA-4 or scrambled RNA control at 48 h post transfection (green, phosphorylated IRF3; blue, DAPI-stained nuclei; arrowheads, nuclei expressing phosphorylated IRF3). Scale bar, 20 μm. (E) IFN-β expression in WT A549-Dual cells, RIG-I knockout A549-Dual cells, MDA5 knockout A549-Dual cells, or TLR3 knockout A549 cells at 48 h after transfection with immunostimulatory RNA-4 or a scrambled RNA control. Data are shown as fold change relative to the scrambled RNA control; N = 6. Note that RIG-I knockout abolished the ability of the immunostimulatory RNAs to induce IFN-β. (F) SPR characterization of the binding affinity between cellular RNA sensors (RIG-I, MDA5, and TLR3) and RNA-1. Equilibrium dissociation constant (KD), association rate constant (Ka), and dissociation rate constant (Kd) are labeled on the graphs. (A, B, and E) Data are shown as means ± SD.
RIG-I, MDA5, and TLR3 are the main sensors upstream of IRF3 that recognize RNA.22 To investigate which of them detect the immunostimulatory short duplex RNAs, we quantified RNA-mediated production of IFN-I in RIG-I, MDA5, or TLR3 KO cells. Knockout of RIG-I completely suppressed the ability of RNA-4 (Figure 3E), as well as RNA-1 and -2 (Figure S6), to induce IFN-I, whereas loss of MDA5 or TLR3 had no effect on RNA-mediated IFN-I production (Figures 3E and S6). Importantly, surface plasmon resonance (SPR) analysis revealed that RNA-1 interacts directly with the RIG-I cellular RNA sensor, rather than MDA5 or TLR3 (Figure 3F). In addition, knockout or overexpression of other RNA sensors, such as TLR7 or TLR8, which sense RNA degradation products mediated by RNase 2 or RNase T2,23,24 did not affect the ability of these duplex RNAs to induce IFN production (Figure S7). Thus, these short duplex RNAs stimulate IFN-I production specifically via the RIG-I/IRF3 pathway.
Terminal GGG motif mediates IFN activation via duplex RNA dimerization
The active RNAs-1, -2, and -3 are chemically synthesized 27-mer RNA duplexes that include terminal hydroxyl groups, two DNA bases at the 3′ end of sense strands, and two-base overhangs at the 3′ end of antisense strands (Table 1). Importantly, their sequence and structure features do not conform to any characteristics of existing immunostimulatory RNA molecules (Table S1), suggesting that previously unknown elements must be responsible for this immunostimulatory activity. Remarkably, even though they were designed to target different host genes, sequence alignment revealed that RNA-1, -2, and -3 contained one identical motif at their 5′ ends (sense strand, C; antisense strand, 3′-GGG-5′) (Table 1). Because all the three RNAs were potent inducers of IFN, we hypothesized that this common motif may mediate their immunostimulatory activities.
To test this hypothesis, we systematically investigated IFN production induced by different sequence variants of RNA-1 (Table 1) using the IFN reporter-expressing A549 cell line. Maintaining the common motif while shuffling remaining nucleotides or replacing them with a random sequence (RNA-4 or RNA-5, -6, and -7, respectively, versus RNA-1, -2, and -3) did not affect the immunostimulatory activity of the duplex RNA (Figure 2; Table 1). However, moving the motif from 3′ GGG end to the middle region completely abolished the RNA’s immunostimulatory activity (RNA-8 versus RNA-1) (Figure 2; Table 1). Furthermore, the immunostimulatory activity was completely eliminated by any changes, including deletion or substitution, at the common motif (RNA-9 to -17 versus RNA-1) (Figure 2; Table 1). These data indicate that the common terminal 5′ GGG motif is necessary for IFN-I/III induction, and that this effect is sensitive to alterations in its position and sequence.
To determine whether this shared motif mediates binding to RIG-I, we evaluated the immunostimulatory activity of duplex RNAs bearing an N1-2′O-methyl group, which has been shown to block RIG-I activation by RNA when the modification occurs at the 5′-terminus.25 Surprisingly, the N1-2′O-methylation of the 5′ end of sense strand (RNA-18) or both simultaneously in the same duplex RNA (RNA-19) did not block RIG-I activation by RNA-1 (Figure 2; Table 1). In contrast, N1-2′O-methylation of the 5′ end of the antisense strand, but not the 3′ end of the sense strand, completely blocked RIG-I activation by RNA (RNA-20 to -22 versus RNA-1) (Figure 2; Table 1), indicating that RNA-1 binds to RIG-I via the 5′ end of its antisense strand.
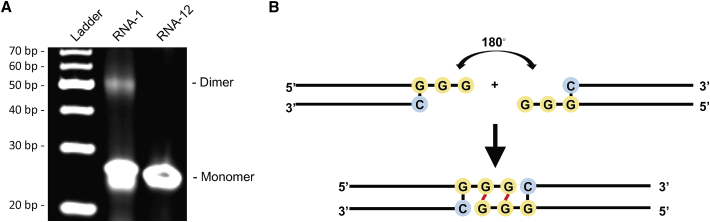
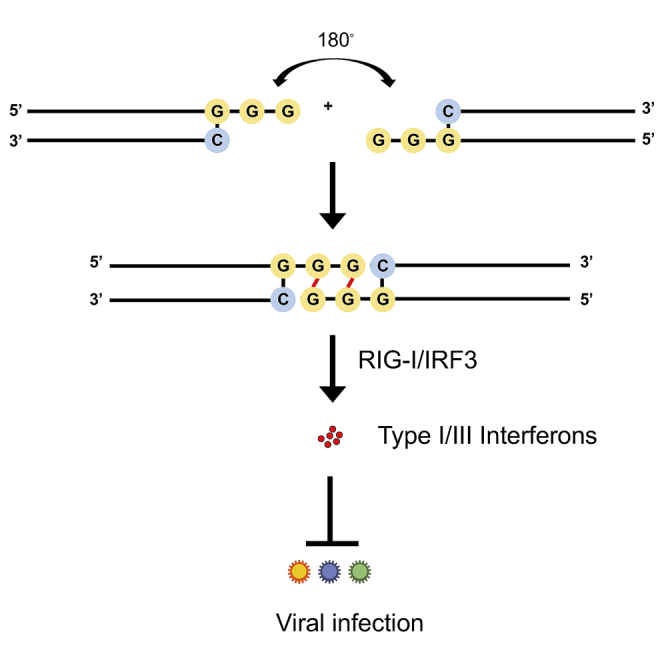
Given the critical role and high conservation of the common motif in this form of duplex RNA-mediated immunostimulation, we also explored whether this common motif could mediate the formation of higher-order structure of duplex RNA via an intramolecular G-quadruplex, a secondary structure that is held together by non-canonical G-G Hoogsteen base pairing.26 Interestingly, native gel electrophoresis revealed the formation of an RNA-1 dimer, while no dimer was detected when the GG overhang was replaced with AA bases (RNA-12 versus RNA-1) (Figure 4A). These data suggest that the common motif (sense strand, 5′-C; antisense strand, 3′-GGG-5′) may mediate formation of an end-to-end RNA-1 dimer via Hoogsteen G-G base pairing,26 which doubles the length of the dsRNA, thereby promoting efficient binding to RIG-I via the exposed 5′ antisense strand ends of each RNA and subsequently inducing IFN production (Figure 4B).
Figure 4.
The common motif mediates the formation of duplex RNA dimers via intramolecular G-quadruplex formed by GG overhang
(A) The image of native gel electrophoresis showing the formation of RNA-1 dimer; 1 μL of 10 μM RNA samples was loaded. RNA-12 was used as negative control. (B) The structure of end-to-end RNA-1 dimer due to terminal G-G Hoogsteen paring.
As chemically synthesized RNAs contain terminal hydroxyl groups, we tested whether adding a monophosphate at these sites affects the IFN-inducing activity. This is important to investigate because host RNAs contain a 5′-monophosphate, which has been reported to suppress RIG-I recognition.2 However, we found that RNA-1 containing terminal monophosphates exhibited immunostimulatory activity to a similar level as RNA-1 containing a hydroxyl groups (RNA-23 to -26 versus RNA-1) (Figure 2; Table 1), suggesting that a terminal monophosphate in these short duplex RNAs is neither required for, nor does it interfere with, their immunostimulatory activity.
As our dsRNAs contain two DNA bases at the 3′ end of their sense strand, we also tested whether the types of nucleosides affect the IFN-inducing activity. Interestingly, the duplex RNAs exhibited comparable immunostimulatory activity with RNA-1 regardless of whether DNA bases or RNA bases are inserted at the 3′ end of the sense strand and/or 5′ end of the antisense strand (RNA-27 to -30 versus RNA-1) (Figure 2; Table 1).
We then tested whether introduction of an overhang affects the IFN-inducing activity, because previous reports revealed that RIG-I can be activated by blunt duplex RNAs, and that almost any type of 5′ or 3′ overhang can prevent RIG-I binding and eliminate signaling.3 However, we found that the overhang did not affect the IFN-inducing activity of our duplex RNAs (RNA-31 to -33 versus RNA-1) (Figure 2; Table 1).
Finally, we analyzed the effects of RNA length on IFN production by gradually trimming bases from the 3′ end of RNA-1. Removal of increasing numbers of bases resulted in a gradual decrease in immunostimulatory activity (RNA-34 and -35 versus RNA-1) with complete loss of activity when eight bases or more were removed from the 3′ end of RNA-1 (RNA-36 and -37) (Figure 2; Table 1). Therefore, the minimal length of this new form of immunostimulatory RNA required for IFN induction is 20 bases on the antisense strand that can result in the formation of an RNA dimer containing ∼38 bases via Hoogsteen base pairing of their 5′ GG ends. Consistent with the proposed mechanism of action, RIG-I knockout also abolished the IFN-inducing ability of these RNA variants (RNA-1 to -7, -18, −19, −22 to -30, and -31 to -35) (Figure S8). Also, in a final control experiment, we found that neither the single sense strand nor the single antisense strand of RNA-1 alone is sufficient to induce IFN production (RNA-38 and -39) (Figure 2; Table 1), indicating that the dsRNA structure is absolutely required for its immunostimulatory activity.
Finally, given that the overhanging motif (sense strand. C; antisense strand. 3′-GGG-5′) is also found in the termini of many siRNAs that can be immunostimulatory, we evaluated its frequency in both human mRNAs and lncRNAs. Genome-wide sequence analysis revealed that the CCC motif is abundant in both mRNAs and lncRNAs sequences: 99.96% of human mRNAs contain CCC with an average distance of 75.45 bp between adjacent motifs and 98.08% of human lncRNAs contain CCC with an average distance of 75.93 bp between adjacent motifs (Figure S9). Thus, this indicates that the GGG motif that mediates short duplex RNA dimerization should be avoided when an siRNA’s immunostimulatory effect is undesired.
Self-assembling dsRNAs induce less proinflammatory genes than poly(I:C)
Polyinosinic:polycytidylic acid (poly(I:C)) is an immunostimulant used to simulate viral infections, which interacts with multiple pattern recognition receptors, including TLR3, RIG-I, and MDA5. To compare the immunostimulatory landscape induced by RNA-1 with poly(I:C), we performed bulk RNA-seq analysis of A549 cells transfected with the sample amounts of scrambled dsRNA as control, RNA-1, or poly(I:C) for 48 h. Principal-component analysis shows that RNA-1 and poly(I:C) induce distinct transcriptomic changes (Figure S10A). Similar to earlier results (Figure 1B), RNA-1 upregulated many genes that are involved in antiviral IFN response genes, such as MX1, OASL, IRF7, IFIT1 (Figure 5A), as well as both type I and type III IFN genes (Figure S10B). In contrast, poly(I:C) induces much broader changes in gene expression: 302 genes have decreased expression, while only two decrease when treated with RNA-1 (Figure 5B). A heatmap also shows that many proinflammatory cytokines and chemokines, such as CXCL11, TNF, CCL2, and IL1A, have much higher expression in cells transfected with poly(I:C) (Figure S10C). In addition, a number of genes involved in ion transport and cell adhesion are decreased by poly(I:C) but not by RNA-1. Notably, many of these genes (e.g., MYO1A, NEB, ADH6, H19, ELN) were also downregulated in SARS-CoV-2 infection.27 We further compared immune responses elicited by RNA-1 and poly(I:C). IFN reporter assay shows that poly(I:C) induce stronger IFN responses than RNA-1 (Figure 5C, left) despite comparable responses at the protein level (Figure S4). However, this is also accompanied by stronger induction of nuclear factor κB (NF-κB) activity (Figure 5C, right), which corroborates the RNA-seq results and the observation that poly(I:C) binds to many cellular RNA sensors in additional to RIG-I, such as TLR3 and MDA-5, which can also induce IFN. Interestingly, 5′-triphosphate RNA does not induce IFN under the tested concentrations (Figure 5C), which is consistent with a recent report that this type of RNA is unable to induce strong RIG-I signaling.28 These results indicate that, compared with poly(I:C), our dsRNAs induce a more targeted antiviral response and a lower level of tissue-damaging proinflammatory responses, while having no effect on critical biological processes, such as ion transport and cell adhesion, which should make them more suitable for antiviral therapeutic applications.
Figure 5.
Immunostimulatory RNAs elicit responses with a stronger antiviral component and a lower proinflammatory component
(A and B) Volcano plots showing significant upregulated genes (red) or downregulated genes (blue) in RNA-1 transfected (A) or poly(I:C) transfected (B) A549 cells. Threshold for fold change = 2, threshold for Padj = 0.01. (C) Comparison of the immunostimulatory activities of different RNAs. A549-Dual cells were transfected with indicated duplex RNAs at a 5-fold serial dilution from 50 nM to 16 pM for 24 h, and then activation of the IFN pathway and NF-kB pathway was measured by quantifying luciferase reporter activity or alkaline phosphatase activity, respectively. N = 4. Data are shown as means ± SD.
Broad-spectrum inhibition of multiple coronaviruses and influenza A viruses
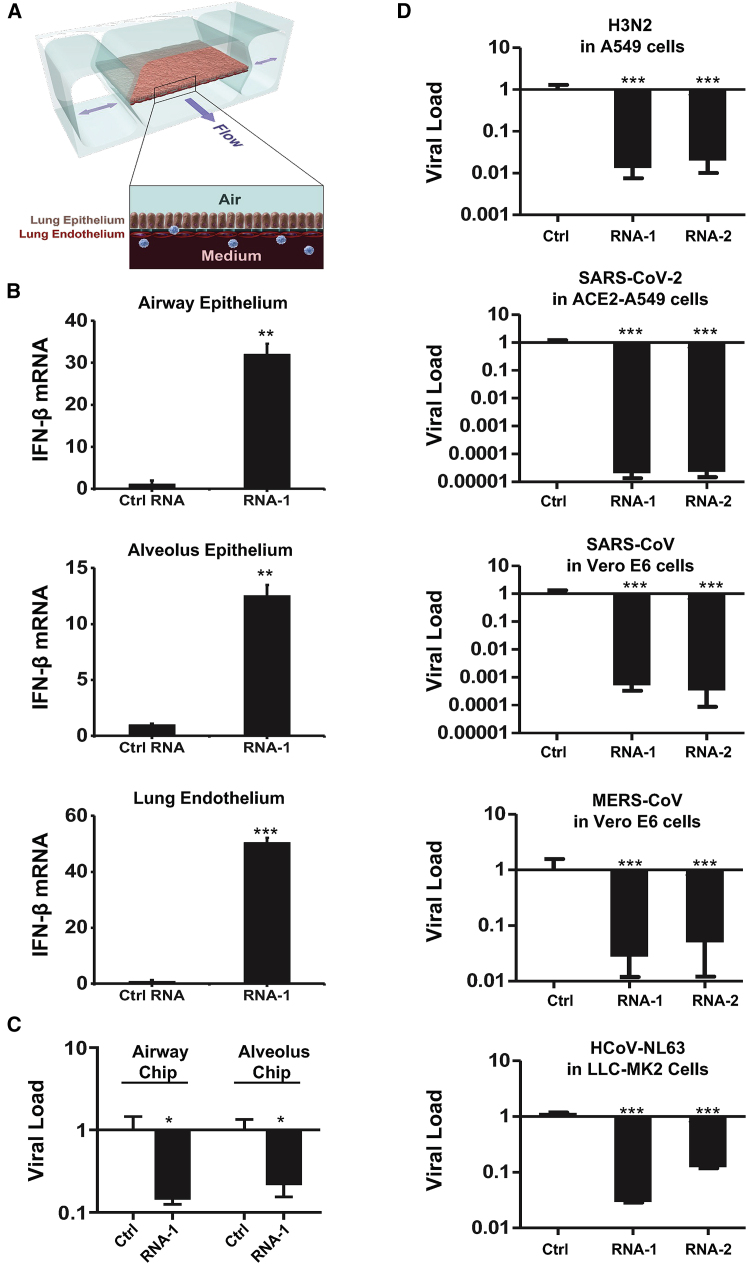
To explore the potential physiological and clinical relevance of these new RNAs that demonstrated immunostimulatory activities in established cell lines, we investigated whether they can trigger IFN-I responses in human lung airway and alveolus chip microfluidic culture devices lined by human primary lung bronchial or alveolar epithelium grown under an air-liquid interface in close apposition to a primary pulmonary microvascular endothelium cultured under dynamic fluid flow (Figure 6A), which have been demonstrated to faithfully recapitulate human organ-level lung physiology and pathophysiology.12,29,30 We observed 12- to 30-fold increases in IFN-β expression compared with a scrambled duplex RNA control when we transfected RNA-1 into human bronchial or alveolar epithelial cells through the air channels of the human lung chips (Figure 6B). In addition, treatment with RNA-1 induced robust (>40-fold) IFN-β expression in human primary lung endothelium on chip (Figure 6B) when it was introduced through the vascular channel. Given our initial finding that RNA-1 and -2 inhibit infection by H1N1 (Figure 1A) along with the known antiviral functions of IFN-I/III,31 we next explored the generalizability of these effects. First, we examined the potential of these IFN-inducing RNAs to block infection by influenza A/Hong Kong [HK]/8/68 (H3N2) virus in which cells were transfected with RNAs 1 day prior to infection, and then, with the advent of the coronavirus disease 2019 (COVID-19) pandemic, we extended this work by carrying out similar studies with SARS-CoV-2 and related coronaviruses, SARS-CoV, MERS-CoV, and HCoV-NL63. Analysis with qPCR for viral mRNA revealed that treatment with the immunostimulatory duplex RNAs significantly suppressed infections by H3N2 influenza virus in human lung airway and alveolus chips (80%–90% inhibition) and in A549 cells (>95% inhibition) (Figures 6C and 6D), as it did with H1N1 influenza virus in A549 cells (Figure 1A). Importantly, these same duplex RNAs inhibited MERS-CoV in Vero E6 cells and HCoV-NL63 in LLC-MK2 cells by >90% (Figure 6D), as well as SARS-CoV in Vero E6 cells by > 1,000-fold (>99.9%) (Figure 6D). Impressively, they were even more potent inhibitors of SARS-CoV-2 infection, reducing viral load in angiotensin-converting enzyme (ACE) 2 receptor-overexpressing A549 cells by over 10,000-fold (>99.99%) (Figures 6D and S11), which is consistent with the observation that SARS-CoV-2 regulates IFN-I/III signaling differently and fails to induce its expression relative to influenza virus and other coronaviruses.32,33 Finally, we assessed the in vivo antiviral efficacy of RNA-1 against SARS-CoV-2 in a K18-hACE2 mouse model.34 Intravenous delivery of 45 μg of RNA-1 using a commercial RNA delivery reagent resulted in >1,000-fold reduction of SARS-CoV-2 viral titers in the infected mouse lungs, while administration of the vehicle alone or with a scrambled control RNA has no effect (Figure 7).
Figure 6.
Immunostimulatory RNAs induce IFN-β production in differentiated human lung epithelial and endothelial cells in organ chips and exhibit broad-spectrum inhibition of infection by H3N2 influenza virus, SARS-CoV-2, SARS-CoV-1, MERS-CoV, and HCoV-NL63
(A) Schematic diagram of a cross section through the human lung on chip, which faithfully recapitulate human lung physiology and pathophysiology. (B) IFN-β mRNA in the epithelial or endothelial cells on human lung airway and alveolus chips at 48 h after transfection with RNA-1 or scrambled RNA control by perfusion through both channels of the chip. Data are presented as fold change relative to the RNA control; N = 3; ∗p < 0.05, ∗∗∗p < 0.001. (C) Effects of treatment with RNA-1 or a scrambled control on viral nucleoprotein (NP) mRNA levels in the human lung airway chips or human lung alveolus chips infected with influenza A/HK/8/68 (H3N2) (MOI = 0.1) at 24 h after RNA-1 treatment. Results are shown as fold change relative to RNA control; N = 3; ∗p < 0.05. (D) Viral load of indicated cells at 48 h after infection after transfection with RNA-1, RNA-2, or a scrambled control for 24 h. For infection, influenza A/HK/8/68 (H3N2) (MOI = 0.1), SARS-CoV-2 (MOI = 0.05), SARS-CoV-1 (MOI = 0.01), MERS-CoV (MOI = 0.01), and HCoV-NL63 (MOI = 0.002), respectively. qPCR in cell lysates was used to quantify viral NP gene for H3N2, and the N gene for SARS-CoV-2 and HCoV-NL63, and plaque-forming assay for SARS-CoV and MERS-CoV. All results are shown as fold change relative to RNA control; N = 3; ∗p < 0.05, ∗∗∗p < 0.001. Data are shown as means ± SD.
Figure 7.
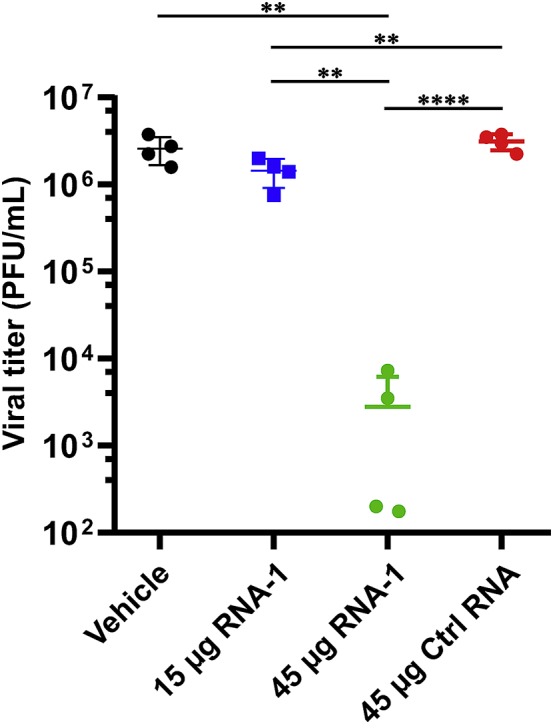
Viral titers at day 3 after challenge with SARS-CoV-2 virus in the lungs of K18-hACE2 mice treated with indicated RNAs or vehicle (n = 4)
Data are plotted for individual mice and overlaid with mean ± SD; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Discussion
In this study, we observed potent stimulation of IFN-I/III signaling by a new class of short duplex RNAs that contain an overhanging sequence motif and terminal monophosphate or hydroxyl groups in a broad spectrum of human cells. Mechanistic exploration revealed that these immunostimulatory RNAs specifically activate the RIG-I/IRF3 pathway by binding directly to RIG-I, even though duplex RNAs with monophosphate groups have previously been shown to antagonize IFN signaling by RNAs with 5′-diphosphates or -triphosphates.2,16 By systematically investigating the effects of various sequences and lengths of these RNAs on IFN-I induction, we identified that the immunostimulatory activity requires a minimal length of 17 bases, in addition to the overhanging sequence motif (sense strand, C; antisense strand, 3′-GGG-5′). This motif mediates the formation of end-to-end duplex RNA dimers via Hoogsteen base pairing, which enable its binding to RIG-I. In addition, the RNA-mediated IFN-I production that we observed resulted in significant inhibition of infections by multiple human respiratory viruses, including H1N1 and H3N2 influenza viruses, as well as coronaviruses SARS-CoV-2, SARS-CoV-1, MERS-CoV, and HCoV-NL63. Notably, these new immunostimulatory RNAs significantly reduced SARS-CoV-2 viral loads in cell lines and in human lung airway and alveolus chips containing primary lung epithelial and endothelial cells. These findings raise the possibility that these IFN-inducing immunostimulatory RNAs could offer alternative prophylactic and therapeutic strategies for the current COVID-19 pandemic, in addition to providing potential broad-spectrum protection against a wide range of respiratory viruses that might emerge in the future. In particular, this new duplex RNA approach provides a clear advantage over the commonly used pattern recognition receptor (PRR) agonist poly(I:C), as it is fully chemically defined, easier to synthesize, and exerts a more targeted antiviral effect with less proinflammatory activity.
While much has been learned about the molecular features of viral RNAs that drive RIG-I activation, considerably less is known about the conditions under which host-derived RNAs or other exogenous RNAs activate host innate immunity in the absence of infection, as well as the mechanistic basis for this activation. Indeed, the minimally required and exclusionary features of dsRNA for RIG-I activation have proved to be complex and sometimes contradictory. Some studies suggest that 5′ diphosphate group is the minimum feature required for RIG-I binding and activation, while 5′ monophosphate or 5′ hydroxyl groups can antagonize the process.2,3,35,36 However, other studies have shown that RIG-I can interact with dsRNA with 5′ monophosphate or hydroxyl groups and induce an innate immune response to a certain degree.37, 38, 39 A possible explanation for the discrepancy between these studies is that higher-order RNA structures may compensate for less-than-optimal ends.39
Our study clearly supports this hypothesis. The common overhanging motif we identified that contains 5′-C and 3′-GGG ends on the sense and antisense strands, respectively, appears to mediate end-to-end dimerization of the duplex RNAs via formation of an intramolecular G-quadruplex generated by the GG overhang, as any changes to this motif led to complete loss of immunostimulatory activity. The remaining exposed 5′ ends of the resultant longer dimers, in turn, appear to be responsible for binding directly to RIG-I, which thereby triggers IFN production. Consistent with this hypothesis, N1-2′O-methylation at the 5′ end of antisense strand, but not the other ends, of the original short dsRNA led to complete loss of the immunostimulatory activity. All these findings are consistent with previous research demonstrating that RIG-I recognizes the 5′ ends of longer duplex RNAs.2 Notably, similar Hoogsteen-like pairing has been identified between trans U-U base pairs in 5′-UU overhang dsRNA fragments40; however, our research establishes for the first time that Hoogsteen base pairing can lead to generation of duplex RNAs that are highly effective RIG-I agonists. To exploit this feature for designing siRNAs that have both gene silencing and IFN-inducing properties, it is necessary to first identify a GGG motif in the 3′ end of antisense strand. While chemical modification is not required, N1-2′O-methylation should be avoided. Importantly, while our research identified the minimum feature of immunostimulatory dsRNAs, new designs should be validated experimentally to ensure that both RNAi and immunostimulatory activities are preserved.
siRNA has been a common laboratory tool for gene silencing in biomedical research for almost two decades and a class of drugs that has recently been approved in clinics.10,11 However, the activation of innate immune responses by siRNAs is challenging their uses in both settings.10,11,41 A number of features that may elicit immune responses by siRNA have been identified (Table S1); for example, the presence of 5′ triphosphate in siRNA synthesized by phage polymerase6 or specific sequence motifs in the sense strand of siRNA.7 However, these features do not cover all possible scenarios, including the new immunostimulatory RNAs identified in our study. While optimally designed siRNAs may not have this motif in the overhang because of the potential for the siRNA to be cleaved by RNase at single-stranded G residues,42 our results further highlight the importance of excluding this feature in future siRNA design to alleviate unwanted activation of innate immune responses.
While immune stimulation by siRNAs is undesired in some gene silencing applications, it can be beneficial in others, such as treatment of viral infections or cancer. The IFN response constitutes the major first line of defense against viruses, and these infectious pathogens, including SARS-CoV-2, have evolved various strategies to suppress this response.32,43 In particular, transcriptomic analyses in both human cultured cells infected with SARS-CoV-2 and COVID-19 patients revealed that SARS-CoV-2 infection produces a unique inflammatory response with very low IFN-I, IFN-III, and associated ISG responses, while still stimulating chemokine and pro-inflammatory cytokine production,32,43 and this imbalance likely contributes to the increased morbidity and mortality seen in late-stage COVID-19 patients. Type I and type III IFN proteins are therefore being evaluated for their efficacy as therapeutics in preclinical models and clinical trials.44, 45, 46, 47 Pretreatment with IFN proteins has been shown to reduce viral titers, suggesting that induction of IFN-I responses may represent a potentially effective approach for prophylaxis or early treatment of SARS-CoV-2 infections.48,49 Triple combination of IFN-β1b, lopinavir-ritonavir, and ribavirin also has recently been reported to shorten the duration of viral shedding and hospital stay in patients with mild to moderate COVID-19.50
Consistent with these observations, our results showed that pretreatment with our immunostimulatory RNAs resulted in a dramatic decrease in infection by SARS-CoV-2, as well as SARS-CoV, MERS-CoV, HCoV-NL63 (common cold virus), and H1N1 and H3N2 influenza viruses. Importantly, our immunostimulatory RNAs specifically activate the RIG-I/IFN-I pathway but are not recognized by other cellular RNA sensors, such as TLR7, TLR8, MDA5, or TLR3. This is interesting because recent studies show that SARS-CoV-2 inhibits RIG-I signaling and clearance of infection via expression of nsp1.51 Importantly, the emerging SARS-CoV-2 Omicron subvariants BA.4 and BA.5 are notable for their ability to enhance innate immune suppression.52 Thus, in addition to demonstrating potent antiviral effects in the COVID-19 mouse model, our results demonstrate that these duplex RNAs can overcome viral antagonism of human innate immunity, at least in human lung epithelial and endothelial cells maintained in organ chips that have been previously shown to recapitulate human lung pathophysiology with high fidelity.53,54
Material and methods
Cell culture
A549 cells (ATCC CCL-185), A549-Dual cells (InvivoGen), RIG-I KO A549-Dual cells (InvivoGen), MDA5 KO A549-Dual cells (InvivoGen), TLR3 KO A549 cells (Abcam), HEK-Blue Null-k cells (InvivoGen, hkb-null1k), HEK-Blue hTLR7 cells (InvivoGen, htlr7), THP1-Dual cells (InvivoGen, thpd-nifs), THP1-Dual KO-TLR8 cells (InvivoGen, kotlr8), MDCK cells (ATCC CRL-2936), and LLC-MK2 cells (ATCC CCL-7.1) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Life Technologies) and penicillin-streptomycin (Life Technologies). HAP1 cells, IRF3 KO HAP1 cells, and IRF7 KO HAP1 cells were purchased from Horizon Discovery and cultured in Iscove’s modified Dulbecco’s medium (IMDM) (Gibco) supplemented with 10% FBS (Life Technologies) and penicillin-streptomycin (Life Technologies). All cells were maintained at 37°C and 5% CO2 in a humidified incubator. All cell lines used in this study were free of mycoplasma, as confirmed by the LookOut Mycoplasma PCR Detection Kit (Sigma). Cell lines were authenticated by the American Type Culture Collectionn (ATCC), InvivoGen, Abcam, or Horizon Discovery. Primary human lung airway epithelial basal stem cells (Lonza, USA) were expanded in 75-cm2 tissue culture flasks using airway epithelial cell growth medium (Promocell, Germany) until 60%–70% confluent. Primary human alveolar epithelial cells (Cell Biologics, H-6053) were cultured using alveolar epithelial growth medium (Cell Biologics, H6621). Primary human pulmonary microvascular endothelial cells (Lonza, CC-2527, P5) were expanded in 75-cm2 tissue culture flasks using human endothelial cell growth medium (Lonza, CC-3202) until 70%–80% confluent.
Viruses
Viruses used in this study include SARS-CoV-2, human coronavirus HCoV-NL63, influenza A/WSN/33 (H1N1), and influenza A/HK/8/68 (H3N2). SARS-CoV-2 isolate USA-WA1/2020 (NR-52281) was deposited by the Centers for Disease Control and Prevention, obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), and NIH, and propagated as described previously.32 HCoV-NL63 was obtained from the ATCC and expanded in LLC-MK2 cells. Influenza A/WSN/33 (H1N1) was generated using reverse genetics technique and influenza A/HK/8/68 (H3N2) was obtained from the ATCC. Both influenza virus strains were expanded in MDCK cells. HCoV-NL63 was titrated in LLC-MK2 cells by the Reed-Muench method. Influenza viruses were titrated by plaque formation assay.29 All experiments with native SARS-CoV-2, SARS-CoV, and MERS-CoV were performed in a BSL3 laboratory and approved by the respective institutional biosafety committees.
Stimulation of cell lines by transfection
All RNAs and negative control dsRNA were synthesized by Integrated DNA Technologies (IDT). The poly(I:C) was purchased from InvivoGen (catalog no. tlrl-picw), which specifically confirmed the absence of contamination by bacterial lipoproteins or endotoxins. The 5′ triphosphate dsRNA (catalog no. tlrl-3prna) was purchased from InvivoGen. Cells were seeded into a six-well plate at 3 × 105 cells/well or 96-well plate at 104 cells/well and cultured for 24 h before transfection. Transfection was performed using TransIT-X2 Dynamic Delivery System (Mirus) according to the manufacturer’s instructions, with some modifications. If not indicated otherwise, 6.8 μL of 10 μM RNA stock solution and 5 μL of transfection reagent were added in 200 μL of Opti-MEM (Invitrogen) to make the transfection mixture. For transfection in the six-well plate, 200 μL of the transfection mixture was added to each well; for transfection in the 96-well plate, 10 μL of the transfection mixture was added to each well. At indicated times after transfection, cell samples were collected and subjected to RNA-seq (Genewiz), TMT mass spectrometry, qRT-PCR, western blot, Quanti-Luc assay, and Quanti-Blue assay (InvivoGen).
RNA-seq and GO analysis
RNA-seq was processed by Genewiz using a standard RNA-seq package that includes poly(A) selection and sequencing on an Illumina HiSeq with 150-bp paired-end reads. Sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the Homo sapiens GRCh38 reference genome using the STAR aligner v.2.5.2b. Unique gene hit counts were calculated by using feature counts from the Subread package v.1.5.2 followed by differential expression analysis using DESeq2. GO analysis was performed using DAVID.55 Volcano plots and heat maps were generated using the EnhancedVolcano R package.56 Raw sequencing data files were deposited on NCBI GEO : GSE181827.
Proteomics analysis by TMT mass spectrometry
Cells were harvested on ice. Cells pellets were syringe lysed in 8 M urea and 200 mM EPPS pH 8.5 with protease inhibitor. BCA (bicinchoninic acid assay) was performed to determine protein concentration of each sample. Samples were reduced in 5 mM Tris(2-carboxyethyl)phosphine (TCEP), alkylated with 10 mM iodoacetamide, and quenched with 15 mM DTT, and 100 μg of protein was chloroform-methanol precipitated and re-suspended in 100 μL of 200 mM EPPS pH 8.5. Protein was digested by Lys-C at a 1:100 protease-to-peptide ratio overnight at room temperature with gentle shaking. Trypsin was used for further digestion for 6 h at 37°C at the same ratio with Lys-C. After digestion, 30 μL of acetonitrile (ACN) was added into each sample to 30% final volume, and 200 μg of TMT reagent (126, 127N, 127C, 128N, 128C, 129N, 129C, 130N, 130C) in 10 μL of ACN was added to each sample. After 1 h of labeling, 2 μL of each sample was combined, desalted, and analyzed using mass spectrometry. Total intensities were determined in each channel to calculate normalization factors. After quenching using 0.3% hydroxylamine, 11 samples were combined in 1:1 ratio of peptides based on normalization factors. The mixture was desalted by solid-phase extraction and fractionated with basic pH reversed-phase (BPRP) high-performance liquid chromatography (HPLC), collected onto a 96 six-well plates, and combined for 24 fractions in total. Twelve fractions were desalted and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).57
Mass spectrometric data were collected on an Orbitrap Fusion Lumos mass spectrometer coupled to a Proxeon NanoLC-1200 UHPLC. The 100-μm capillary column was packed with 35 cm of Accucore 50 resin (2.6 μm, 150Å; Thermo Fisher Scientific). The scan sequence began with an MS1 spectrum (Orbitrap analysis, resolution 120,000, 375–1,500 Th, automatic gain control [AGC] target 4E5, maximum injection time 50 ms). SPS-MS3 analysis was used to reduce ion interference.58,59 The top 10 precursors were then selected for MS2/MS3 analysis. MS2 analysis consisted of collision-induced dissociation (CID), quadrupole ion trap analysis, AGC 2E4, normalized collision energy (NCE) 35, q value 0.25, maximum injection time 35 ms), and isolation window at 0.7. Following acquisition of each MS2 spectrum, we collected an MS3 spectrum in which multiple MS2 fragment ions are captured in the MS3 precursor population using isolation waveforms with multiple frequency notches. MS3 precursors were fragmented by higher-energy collisional dissociation (HCD) and analyzed using the Orbitrap (NCE 65, AGC 1.5E5, maximum injection time 120 ms, resolution was 50,000 at 400 Th).
Mass spectra were processed using a Sequest-based pipeline.60 Spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included all entries from the Human UniProt database (downloaded 2014-02-04) This database was concatenated with one composed of all protein sequences in the reversed order. Searches were performed using a 50-ppm precursor ion tolerance for total protein-level analysis. The product ion tolerance was set to 0.9 Da. TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification.
Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR).61,62 PSM filtering was performed using linear discriminant analysis (LDA), as described previously,60 while considering the following parameters: XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy. For TMT-based reporter ion quantitation, we extracted the summed signal-to-noise (S:N) ratio for each TMT channel and found the closest matching centroid to the expected mass of the TMT reporter ion. For protein-level comparisons, PSMs were identified, quantified, and collapsed to a 1% peptide FDR and then collapsed further to a final protein-level FDR of 1%, which resulted in a final peptide level FDR of <0.1%. Moreover, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides. Proteins were quantified by summing reporter ion counts across all matching PSMs, as described previously.60 PSMs with poor quality, and MS3 spectra with TMT reporter summed signal-to-noise of less than 100, or having no MS3 spectra, were excluded from quantification.63 Each reporter ion channel was summed across all quantified proteins and normalized assuming equal protein loading of all tested samples. Raw data were submitted to ProteomeXchange : PXD027838.
qRT-PCR
Total RNA was extracted from cells using RNeasy Plus Mini Kit (QiaGen, catalog no. 74134) according to the manufacturer’s instructions. cDNA was then synthesized using AMV reverse transcriptase kit (Promega) according to the manufacturer’s instructions. To detect gene levels, real-time qPCR was carried out using the GoTaq qPCR Master Mix kit (Promega) with 20 μL of reaction mixture containing gene-specific primers or the PrimePCR assay kit (Bio-Rad) according the manufacturers’ instructions. The expression levels of target genes were normalized to GAPDH.
Antibodies and western blotting
The antibodies used in this study were anti-IRF3 (Abcam, ab68481), anti-IRF3 (Phospho S396) (Abcam, ab138449), anti-GAPDH (Abcam, ab9385), and goat anti-rabbit IgG H&L (HRP) (Abcam, ab205718). Cells were harvested and lysed in radio-immunoprecipitation assay (RIPA) buffer (Thermo Scientific, catalog no. 89900) supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Scientific, catalog no. 78440) on ice. The cell lysates were subject to western blotting. GAPDH was used as a loading control.
Confocal immunofluorescence microscopy
Cells were rinsed with PBS, fixed with 4% paraformaldehyde (Alfa Aesar) for 30 min, permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) in PBS (PBST) for 10 min, blocked with 10% goat serum (Life Technologies) in PBST for 1 h at room temperature, and incubated with anti-IRF3 (Phospho S396) (Abcam, ab138449) antibody diluted in blocking buffer (1% goat serum in PBST) overnight at 4°C, followed by incubation with Alexa Fluor 488 conjugated secondary antibody (Life Technologies) for 1 h at room temperature; nuclei were stained with DAPI (Invitrogen) after secondary antibody staining. Fluorescence imaging was carried out using a confocal laser-scanning microscope (SP5 X MP DMI-6000, Germany) and image processing was done using Imaris software (Bitplane, Switzerland).
Surface plasmon resonance
The interactions between duplex RNA-1 and cellular RNA sensor molecules (RIG-I (Abcam, catalog no. ab271486), MDA5 (Creative-Biomart, catalog no. IFIH1-1252H), and TLR3 (Abcam, catalog no. ab73825)) were analyzed by SPR with the Biacore T200 system (GE Healthcare) at 25°C (Creative-Biolabs). RNA-1 conjugated with biotin at 3′ end of sense strand (synthesized by IDT) was immobilized on an SPR Series S Sensor Chip SA (GE Healthcare, catalog no. BR100531) by flowing 2 nM RNA-1 conjugated with biotin diluted in running buffer (10 × HBS electrophoresis buffer, HBS-EP+); GE Healthcare, catalog no. BR100669) on the surface of SPR chip, with final levels of ∼50 response units (RU). Indicated concentrations of the RNA sensors (RIG-I, MDA5, or TLR3) diluted in running buffer (10 × HBS-EP+; GE Healthcare, catalog no. BR100669) were injected as analytes at a flow rate of 30 μL/min, a contact time of 180 s, and a dissociation time of 420 s. The surface was regenerated with 2 M NaCl for 30 s. Data analysis was performed on the Biacore T200 computer with the Biacore T200 evaluation software.
Organ chip culture
Microfluidic two-channel organ chip devices and automated ZOE instruments used to culture them were obtained from Emulate Inc (Boston, MA, USA). Our methods for culturing human lung airway chips29,30 and lung alveolus chips have been described previously.64 In this study, we slightly modified the alveolus chip method by coating the inner channels of the devices with 200 μg/mL of Collagen IV (5022-5MG, Advanced Biomatrix) and 15 μg/mL of laminin (L4544-100UL, Sigma) at 37°C overnight, and the next day (day 1) sequentially seeding primary human lung microvascular endothelial cells (Lonza, CC-2527, P5) and primary human lung alveolar epithelial cells (Cell Biologics, H-6053) in the bottom and top channels of the chip at a density of 8 × 106 and 1.6 × 106 cells/mL, respectively, under static conditions. On day 2, the chips were inserted into Pods (Emulate), placed within the ZOE instrument, and the apical and basal channels were respectively perfused (60 μL/h) with epithelial growth medium (Cell Biologics, H6621) and endothelial growth medium (Lonza, CC-3202). On day 5, 1 μM dexamethasone was added to the apical medium to enhance barrier function. On day 7, an air-liquid interface (ALI) was introduced into the epithelial channel by removing all medium from this channel while continuing to feed all cells through the medium perfused through the lower vascular channel, and this medium was changed to EGM-2MV with 0.5% FBS on day 9. Two days later, the ZOE instrument was used to apply cyclic (0.25 Hz) 5% mechanical strain to the engineered alveolar-capillary interface to mimic lung breathing on chip. RNAs were transfected on day 15.
RNA transfection in human lung airway and alveolus chips
Human airway or alveolus chips were transfected with duplex RNAs by adding the RNA and transfection reagent (Lipofectamine RNAiMAX) mixture into the apical and basal channels of the organ chips and incubating for 6 h at 37°C under static conditions before reestablishing an ALI. Tissues cultured on chip were collected by RNeasy Micro Kit (QiaGen) at 48 h post transfection by first introducing 100 μL of lysis buffer into the apical channel to lyse epithelial cells and then 100 μL into the basal channel to lyse endothelial cells. Lysates were subjected to qPCR analysis of IFN-β gene expression.
Native SARS-CoV-2 infection and inhibition by RNA treatment
ACE2-expressing A549 cells (a gift from Brad Rosenberg) were transfected with indicated RNAs. At 24 h post transfection, the transfected ACE2-A549 cells were infected with SARS-CoV-2 (MOI = 0.05) for 48 h. Cells were harvested in Trizol (Invitrogen) and total RNA was isolated and DNAse-I treated using Zymo RNA Miniprep Kit according to the manufacturer’s protocol. qRT-PCR for α-tubulin (forward, 5′-GCCTGGACCACAAGTTTGAC-3′; reverse, 3′-TGAAATTCTGGGAGCATGAC-5′) and SARS-CoV-2 N mRNA (forward, 5′-CTCTTGTAGATCTGTTCTCTAAACGAAC-3′; reverse, 3′-GGTCCACCAAACGTAATGCG-5′) were performed using KAPA SYBR FAST ONE-STEP qRT-PCR kits (Roche) according to manufacturer’s instructions on a Lightcycler 480 Instrument-II (Roche).
Native SARS-CoV-1 and MERS-CoV infection and inhibition by RNA treatment
Vero E6 cells (ATCC# CRL 1586) were cultured in DMEM (Quality Biological), supplemented with 10% (v/v) FBS (Sigma), 1% (v/v) penicillin/streptomycin (Gemini Bio-products), and 1% (v/v) L-glutamine (2 mM final concentration, Gibco). Cells were maintained at 37°C (5% CO2). Vero E6 cells were plated at 1.5 × 105 cells per well in a six-well plate 2 days prior to transfection. The RNA-1, RNA-2, and scrambled control RNA were transfected into each well using the Transit X2 delivery system (MIRUS; MIR6003) in OptiMEM (Gibco 31985-070). SARS-CoV (Urbani strain, BEI#NR-18925) and MERS-CoV (Jordan strain, provided by NIH) were added at MOI 0.01. At 72 h post infection, medium was collected and used for a plaque assay to quantify plaque-forming units (PFU) per milliliter of virus.
Efficacy study in K18-hACE2 mice
Sixteen 6-week-old female K18-hACE2 mice were purchased from Jackson Laboratory. Mice were randomized into four groups (n = 4) with vehicle, and vehicle plus treatment: 15 μg of RNA-1, 45 μg of RNA-1, or 45 μg of negative control RNA. Vehicle or treatment were delivered by intravenous administration through the tail in a total volume of 100 μL. Treatment was conducted at −24 h and 2 h prior to viral infection (two doses in total). Mice were weighed daily and physically assessed for signs of morbidity, anesthetized with isoflurane, and intranasally challenged with 2 × 104 PFU per mouse (25 μL/naris) using the SARS-CoV-2 WA1/2020 strain (20 × lethal dose 50 [LD50]). All animals were sacrificed in each group at 3 days post infection, and the lungs were collected for analysis. The left lobe of the lung tissue was placed into a bead mill tube (1.4-mm ceramic beads) containing a 1-mL solution of protease inhibitors (Halt Protease Inhibitor Cocktail) in PBS, homogenized using the Bead Mill 4 (Fisher) for one or two cycles of 10 s (5 m/s), centrifuged at 16,000 × g, and the supernatant was aliquoted and flash frozen in liquid nitrogen before being placed in a −80°C freezer. The viral titers were determined using plaque assay as described previously.65
Compliance
Experiments with SARS-CoV-2 were conducted at the Regional Biocontainment Laboratory at BSL-3 or ABSL-3. Studies were conducted with the approval of the Institutional Animal Care and Use Committee of University of Tennessee Health Science Center (protocol #20-0132).
Quantification and statistical analysis
All data are expressed as mean ± standard deviation (SD). N represents biological replicates. Statistical significance of differences in the in vitro experiments was determined by employing the paired two-tailed Student t test when comparing the difference between two groups and one-way ANOVA with multiple comparison when comparing the samples among groups with more than two samples. For all experiments, differences were considered statistically significant for p < 0.05 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; n.s., not significant).
Acknowledgments
We thank IDT Inc. for synthesizing the RNA oligonucleotides, X. Song and H. Queen at Creative-Biolabs Inc. for carrying SPR experiments, and Genewiz Inc. for carrying out the RNA-seq work. We thank the animal care staff at the University of Tennessee Health Science Center, Regional Biocontainment Laboratory for their support of these studies. This work was supported by the NIH (NCATS UH3-HL-141797 to D.E.I.) and the Defense Advanced Research Projects Agency under Cooperative Agreements (HR00111920008 and HR0011-20-2-0040 to D.E.I.).
Author contributions
L.S., H.B., and D.E.I. conceived this study. L.S. and H.B. conducted in vitro experiments and analyzed data with assistance from C.O., A.J., C.B., W.C., K.C., and R.P. T.Z. and S.P.G. performed TMT mass spectrometry and data analysis. F.H. performed the native gel electrophoresis experiments. Y.Y. performed the analysis of CCC sequence distribution in human mRNAs and lncRNAs. T.J., J.L., M.M., M.F., and B.R.t. performed the experiments on SARS-CoV-2, SARS-CoV, and MERS-CoV viruses. A.N. performed western blotting experiments. R.P.-B. assisted in the propagation and characterization of HCoV-NL63 virus. K.E.C. and C.B.J. designed and coordinated experiments to test RNA-1 activities in K18-ACE2 mouse models, which were carried out by D.Y. The manuscript was written by L.S., H.B., and D.E.I. with input from other authors.
Declaration of interest
D.E.I. is a founder, board member, SAB chair, and equity holder in Emulate Inc. D.E.I., L.S., H.B., C.O., and R.P. are inventors on relevant patent applications held by Harvard University.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.08.031.
Supplemental information
Data availability
Sharing of materials will be subject to standard material transfer agreements. The raw source data of RNA-seq have been deposited in the GEO: GSE181827 and TMT mass spectrometry in ProteomeXchange : PXD027838. Additional data are presented in the supplemental information.
References
- 1.Schlee M., Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren X., Linehan M.M., Iwasaki A., Pyle A.M. RIG-I Selectively Discriminates against 5'-monophosphate RNA. Cell Rep. 2019;26:2019–2027.e4. doi: 10.1016/j.celrep.2019.01.107. [DOI] [PubMed] [Google Scholar]
- 3.Ren X., Linehan M.M., Iwasaki A., Pyle A.M. RIG-I recognition of RNA targets: the influence of terminal base pair sequence and overhangs on affinity and signaling. Cell Rep. 2019;29:3807–3815.e3. doi: 10.1016/j.celrep.2019.11.052. [DOI] [PubMed] [Google Scholar]
- 4.Jiang F., Ramanathan A., Miller M.T., Tang G.Q., Gale M., Jr., Patel S.S., Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marques J.T., Williams B.R.G. Activation of the mammalian immune system by siRNAs. Nat. Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 6.Kim D.H., Longo M., Han Y., Lundberg P., Cantin E., Rossi J.J. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 7.Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 8.Sledz C.A., Holko M., de Veer M.J., Silverman R.H., Williams B.R.G. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 9.Robbins M., Judge A., Ambegia E., Choi C., Yaworski E., Palmer L., McClintock K., MacLachlan I. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum. Gene Ther. 2008;19:991–999. doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- 10.Setten R.L., Rossi J.J., Han S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 11.Meng Z., Lu M. RNA interference-induced innate immunity, off-target effect, or immune Adjuvant? Front. Immunol. 2017;8:331. doi: 10.3389/fimmu.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benam K.H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H.H., Alves S.E., Salmon M., Ferrante T.C., Weaver J.C., et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods. 2016;13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 13.Huh D., Matthews B.D., Mammoto A., Montoya-Zavala M., Hsin H.Y., Ingber D.E. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Si L., Bai H., Rodas M., Cao W., Oh C.Y., Jiang A., Moller R., Hoagland D., Oishi K., Horiuchi S., et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat. Biomed. Eng. 2021;5:815–829. doi: 10.1038/s41551-021-00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D.H., Behlke M.A., Rose S.D., Chang M.S., Choi S., Rossi J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 16.Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J., et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5'-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tissari J., Sirén J., Meri S., Julkunen I., Matikainen S. IFN-alpha enhances TLR3-mediated antiviral cytokine expression in human endothelial and epithelial cells by up-regulating TLR3 expression. J. Immunol. 2005;174:4289–4294. doi: 10.4049/jimmunol.174.7.4289. [DOI] [PubMed] [Google Scholar]
- 18.Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 19.Wang P., Xu J., Wang Y., Cao X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017;358:1051–1055. doi: 10.1126/science.aao0409. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y., Li M., Xue Y., Li Z., Wen W., Liu X., Ma Y., Zhang L., Shen Z., Cao X. Interferon-inducible cytoplasmic lncLrrc55-AS promotes antiviral innate responses by strengthening IRF3 phosphorylation. Cell Res. 2019;29:641–654. doi: 10.1038/s41422-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 22.Chow K.T., Gale M., Jr., Loo Y.M. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 2018;36:667–694. doi: 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- 23.Ostendorf T., Zillinger T., Andryka K., Schlee-Guimaraes T.M., Schmitz S., Marx S., Bayrak K., Linke R., Salgert S., Wegner J., et al. Immune sensing of Synthetic, bacterial, and Protozoan RNA by toll-like receptor 8 requires coordinated processing by RNase T2 and RNase 2. Immunity. 2020;52:591–605.e6. doi: 10.1016/j.immuni.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Greulich W., Wagner M., Gaidt M.M., Stafford C., Cheng Y., Linder A., Carell T., Hornung V. TLR8 is a sensor of RNase T2 degradation products. Cell. 2019;179:1264–1275.e13. doi: 10.1016/j.cell.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.M., Zillinger T., Goldeck M., Schmidt T., Schmid-Burgk J.L., Kerber R., Wolter S., et al. A conserved Histidine in the RNA sensor RIG-I controls immune tolerance to N1-2'O-Methylated self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyu K., Chow E.Y.C., Mou X., Chan T.F., Kwok C.K. RNA G-quadruplexes (rG4s): genomics and biological functions. Nucleic Acids Res. 2021;49:5426–5450. doi: 10.1093/nar/gkab187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;185:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linehan M.M., Dickey T.H., Molinari E.S., Fitzgerald M.E., Potapova O., Iwasaki A., Pyle A.M. A minimal RNA ligand for potent RIG-I activation in living mice. Sci. Adv. 2018;4:e1701854. doi: 10.1126/sciadv.1701854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Si L., Bai H., Rodas M., Cao W., Oh C.Y., Jiang A., Nurani A., Zhu D.Y., Goyal G., Gilpin S.E., et al. Human organs-on-chips as tools for repurposing approved drugs as potential influenza and COVID19 therapeutics in viral pandemics. bioRxiv. 2020 doi: 10.1101/2020.04.13.039917. Preprint at. [DOI] [Google Scholar]
- 30.Si L., Prantil-Baun R., Benam K.H., Bai H., Rodas M., Burt M., Ingber D.E. Discovery of influenza drug resistance mutations and host therapeutic targets using a human airway chip. bioRxiv. 2019 doi: 10.1101/685552. Preprint at. [DOI] [Google Scholar]
- 31.Mesev E.V., LeDesma R.A., Ploss A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019;4:914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galani I.E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E., Koukaki E., Fragkou P.C., Panou V., Rapti V., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- 34.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramanathan A., Devarkar S.C., Jiang F., Miller M.T., Khan A.G., Marcotrigiano J., Patel S.S. The autoinhibitory CARD2-Hel2i Interface of RIG-I governs RNA selection. Nucleic Acids Res. 2016;44:896–909. doi: 10.1093/nar/gkv1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hausmann S., Marq J.B., Tapparel C., Kolakofsky D., Garcin D. RIG-I and dsRNA-induced IFNbeta activation. PLoS One. 2008;3:e3965. doi: 10.1371/journal.pone.0003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brisse M., Ly H. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front. Immunol. 2019;10:1586. doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahl M.C., Rao S.T., Sundaralingam M. The structure of r(UUCGCG) has a 5'-UU-overhang exhibiting Hoogsteen-like trans U.U base pairs. Nat. Struct. Biol. 1996;3:24–31. doi: 10.1038/nsb0196-24. [DOI] [PubMed] [Google Scholar]
- 41.Bartoszewski R., Sikorski A.F. Editorial focus: understanding off-target effects as the key to successful RNAi therapy. Cell. Mol. Biol. Lett. 2019;24:69. doi: 10.1186/s11658-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., Clementi N., De Santis M., Mancini N., Granucci F., et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park A., Iwasaki A. Type I and type III interferons - induction, signaling, evasion, and application to Combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wadman M. Can interferons stop COVID-19 before it takes hold? Science. 2020;369:125–126. doi: 10.1126/science.2020.6500.369_125. [DOI] [PubMed] [Google Scholar]
- 47.Wadman M. Can boosting interferons, the body's frontline virus fighters, beat COVID-19? Science. 2020 doi: 10.1126/SCIENCE.ABD7137. [DOI] [Google Scholar]
- 48.Lokugamage K.G., Hage A., Schindewolf C., Rajsbaum R., Menachery V.D. SARS-CoV-2 is sensitive to type I interferon pretreatment. bioRxiv. 2020 doi: 10.1101/2020.03.07.982264. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hung I.F.N., Lung K.C., Tso E.Y.K., Liu R., Chung T.W.H., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reuschl A.-K., Thorne L.G., Whelan M.V.X., Mesner D., Ragazzini R., Dowgier G., Bogoda N., Turner J.L.E., Furnon W., Cowton V.M., et al. Enhanced innate immune suppression by SARS-CoV-2 Omicron subvariants BA.4 and BA.5. bioRxiv. 2022 doi: 10.1101/2022.07.12.499603. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain A., Barrile R., van der Meer A.D., Mammoto A., Mammoto T., De Ceunynck K., Aisiku O., Otieno M.A., Louden C.S., Hamilton G.A., et al. Primary human lung Alveolus-on-a-chip model of Intravascular Thrombosis for Assessment of therapeutics. Clin. Pharmacol. Ther. 2018;103:332–340. doi: 10.1002/cpt.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huh D., Leslie D.C., Matthews B.D., Fraser J.P., Jurek S., Hamilton G.A., Thorneloe K.S., McAlexander M.A., Ingber D.E. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 56.Blighe K., Rana S., Lewis M. 2020. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. [Google Scholar]
- 57.Navarrete-Perea J., Yu Q., Gygi S.P., Paulo J.A. Streamlined tandem mass tag (SL-TMT) protocol: an efficient strategy for quantitative (Phospho)proteome profiling using tandem mass Tag-Synchronous precursor selection-MS3. J. Proteome Res. 2018;17:2226–2236. doi: 10.1021/acs.jproteome.8b00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gygi J.P., Yu Q., Navarrete-Perea J., Rad R., Gygi S.P., Paulo J.A. Web-based search tool for Visualizing instrument performance using the Triple knockout (TKO) proteome standard. J. Proteome Res. 2019;18:687–693. doi: 10.1021/acs.jproteome.8b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paulo J.A., O'Connell J.D., Gygi S.P. A Triple knockout (TKO) proteomics standard for Diagnosing ion interference in isobaric labeling experiments. J. Am. Soc. Mass Spectrom. 2016;27:1620–1625. doi: 10.1007/s13361-016-1434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., Gygi S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elias J.E., Gygi S.P. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 63.McAlister G.C., Huttlin E.L., Haas W., Ting L., Jedrychowski M.P., Rogers J.C., Kuhn K., Pike I., Grothe R.A., Blethrow J.D., et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bai H., Si L., Jiang A., Belgur C., Plebani R., Oh C., Rodas M., Nurani A., Gilpin S., Powers R.K., et al. Mechanical control of innate immune responses against viral infection revealed in a human Lung Alveolus Chip. bioRxiv. 2021 doi: 10.1101/2021.04.26.441498. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue Y., Yang D., Vogel P., Stabenow J., Zalduondo L., Kong Y., Ravi Y., Sai-Sudhakar C.B., Parvathareddy J., Hayes E., et al. Cardiopulmonary Injury in the Syrian Hamster model of COVID-19. Viruses. 2022;14:1403. doi: 10.3390/v14071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sharing of materials will be subject to standard material transfer agreements. The raw source data of RNA-seq have been deposited in the GEO: GSE181827 and TMT mass spectrometry in ProteomeXchange : PXD027838. Additional data are presented in the supplemental information.