Abstract
Objectives
Antigen rapid diagnostic tests (RDTs) for SARS coronavirus 2 (SARS-CoV-2) are quick, widely available, and inexpensive. Consequently, RDTs have been established as an alternative and additional diagnostic strategy to quantitative reverse transcription polymerase chain reaction (RT-qPCR). However, reliable clinical and large-scale performance data specific to a SARS-CoV-2 virus variant of concern (VOC) are limited, especially for the Omicron VOC. The aim of this study was to compare RDT performance among different VOCs.
Methods
This single-centre prospective performance assessment compared RDTs from three manufacturers (NADAL, Panbio, MEDsan) with RT-qPCR including deduced standardized viral load from oropharyngeal swabs for detection of SARS-CoV-2 in a clinical point-of-care setting from November 2020 to January 2022.
Results
Among 35 479 RDT/RT-qPCR tandems taken from 26 940 individuals, 164 of the 426 SARS-CoV-2 positive samples tested true positive with an RDT corresponding to an RDT sensitivity of 38.50% (95% CI, 34.00–43.20%), with an overall specificity of 99.67% (95% CI, 99.60–99.72%). RDT sensitivity depended on viral load, with decreasing sensitivity accompanied by descending viral load. VOC-dependent sensitivity assessment showed a sensitivity of 42.86% (95% CI, 32.82–53.52%) for the wild-type SARS-CoV-2, 43.42% (95% CI, 32.86–54.61%) for the Alpha VOC, 37.67% (95% CI, 30.22–45.75%) for the Delta VOC, and 33.67% (95% CI, 25.09–43.49%) for the Omicron VOC. Sensitivity in samples with high viral loads of ≥106 SARS-CoV-2 RNA copies per mL was significantly lower in the Omicron VOC (50.00%; 95% CI, 36.12–63.88%) than in the wild-type SARS-CoV-2 (79.31%; 95% CI, 61.61–90.15%; p 0.015).
Discussion
RDT sensitivity for detection of the Omicron VOC is reduced in individuals infected with a high viral load, which curtails the effectiveness of RDTs. This aspect furthert: limits the use of RDTs, although RDTs are still an irreplaceable diagnostic tool for rapid, economic point-of-care and extensive SARS-CoV-2 screening.
Keywords: Antigen rapid diagnostic test, Clinical performance evaluation, Omicron, PCR, SARS-CoV-2, Virus variants of concern
Graphical abstract
Introduction
Owing to their independence of diagnostic infrastructure, short analysis time, self-testing option, and affordability, SARS coronavirus 2 (SARS-CoV-2) rapid diagnostic tests (RDTs), technically based on lateral flow enzyme-linked immunosorbent assays, have been established as an important diagnostic alternative to quantitative reverse transcription polymerase chain reaction (RT-qPCR) as a reference standard [1,2]. With respect to different SARS-CoV-2 variants of concern (VOCs) and the variable VOC dominance during the course of the coronavirus disease 2019 (COVID-19) pandemic, the available RDT performance assessments present heterogeneous results regarding a potential limitation of RDT sensitivity because of a particular VOC. Comparable data from real-life, extensive point-of-care usage are still lacking [[3], [4], [5], [6]].
In this prospective performance evaluation study, the accuracy of RDTs with that of normalized RT-qPCR in the daily clinical routine was compared, with the main focus being on SARS-CoV-2 VOC–dependent test performance and sensitivity in highly infectious individuals and specificity in broad screening use.
Methods
Study setting
Clinical performance assessment was performed at a tertiary care hospital in Bavaria, Germany, as a single-centre study. Presented data were collected from 12 November 2020 to 31 January 2022, which included the second to the fifth wave of the COVID-19 pandemic in Germany caused by the wild-type SARS-CoV-2, Alpha VOC, Delta VOC, and Omicron VOC [7]. The study continued a previous RDT evaluation up to February 2021 including 5068 samples [8].
During the data collection period, the spread of SARS-CoV-2 infection in the Federal State of Bavaria was quantified, with an average weekly incidence of 191.61 per 100 000 inhabitants, which reached unprecedented maximum incidence values in January 2022 owing to the increasing number of Omicron VOC infections (Fig. S1) [7,9,10].
Test enrolment
As part of the measures in place to prevent and reduce the intrahospital spread of SARS-CoV-2, RDT testing in tandem with RT-qPCR was implemented in key situations for prevention of nosocomial SARS-CoV-2 transmission chains. As part of the local SARS-CoV-2 screening, conception RDT diagnostics were established for patients, accompanying individuals staying overnight with underage and otherwise dependent patients on admission, and employees in case of the onset of symptoms potentially resulting from COVID-19.
Although tandem testing was performed on all patients on admission during high SARS-CoV-2 incidence periods (1 February 2021 to 30 June 2021 and 4 November 2021 to 31 January 2022), during low incidence periods (12 November 2020 to 31 January 2021 and 1 July 2021 to 3 November 2021), RDTs were only performed in critical areas, such as emergency departments and the delivery room. This reflects in the weekly number of RDTs included in the study, as shown in Fig. S1. In case of more than one documented RDT per day per person, only the first RDT was considered for data analysis. Patients who fulfilled the inclusion criteria on multiple days of the study period were tested and included once per visit. Individuals with a recent SARS-CoV-2 infection and subsequent deisolation were also excluded owing to the potential persistent RT-qPCR positivity that is not related to a risk of viral spread [8].
Antigen RDTs
To ensure continuous supply, the following RDTs from three different manufacturers were selected out of 23 products listed by the German Federal Institute for Drugs and Medical Devices in October 2020 for implementation to clinical point-of-care diagnostics: (a) NADAL COVID-19 Ag Test (nal von minden GmbH, Regensburg, Germany), (b) PANBIO COVID-19 Ag Rapid Test (Abbott Laboratories, Abbott Park IL, USA), and (c) MEDsan SARS-Cov-2 Antigen Rapid Test (MEDsan GmbH, Hamburg, Germany) [8,11].
The selected three RDTs were maintained for the complete study period, with randomly varying distribution across the hospital departments over the study period for infrastructural and availability reasons.
In case of an invalid RDT result, the RDT was repeated. Because only the first RDT per day per person was included, these repeated RDTs were excluded from the analysis.
RDT and RT-qPCR specimens were gathered in successive, paired oropharyngeal swabs by trained medical staff in accordance with manufacturers' instructions; all the used RDTs target the SARS-CoV-2 nucleoprotein antigen. Because RDTs were performed at point-of-care, RT-qPCR results were not available to the performers.
RT-qPCR tests and analytical devices
The RT-qPCR tests, analytical devices, and the viral load determination are described in detail in the Supplementary Methods. To prioritize RDT positive samples, RDT results were made available to the RT-qPCR–conducting staff.
VOC-specific polymerase chain reaction and determination of SARS-CoV-2 VOC prevalence levels
Between 3 February 2021 and 19 January 2022, all new RT-qPCR positive samples underwent spike protein variant–specific polymerase chain reaction (PCR) to differentiate between present spike protein VOC using the VirSNiP SARS-CoV-2 Spike N501Y, del 69/70, E484K, N501Y, L452R, T478K, and 371L 373P 452R kits (TIB molbiol, Berlin, Germany) on a cobas z 480 analyser (Roche Diagnostics, Rotkreuz, Switzerland). The determination of the VOC by multiple PCR tests has shown a high agreement with VOC determination by whole genome sequencing [12]. In case of combinations not corresponding to a frequent VOC, the described analytical procedure was followed using spike protein sequencing. Owing to the high COVID-19 prevalence and Omicron VOC in 95% of all COVID-19 cases in Germany, VOC determination was interrupted as of 19 January 2022. All positive samples before 3 February 2021 were assumed to be of wild-type SARS-CoV-2, and all samples after the termination of virus variant determination were assumed to be Omicron VOC. In case VOC determination was not possible owing to low viral loads, the VOC was, if possible, derived from the known SARS-CoV-2 infection source. Otherwise, samples were assumed to belong to the VOC responsible for more than 90 % of prevalent SARS-CoV-2 cases in that calendar week in Germany: wild-type SARS-CoV-2 (12 November 2020 to 24 January 2021), Alpha VOC (19 April 2021 to 2 May 2021 and 17 May 2021 to 30 May 2021), Delta VOC (12 July 2021 to 19 December 2021), and Omicron VOC (17 January 2022 to 31 January 2022) [10].
Data collection
Documented RDT and RT-qPCR results and demographic data were collected from the local hospital information system SAP ERP 6.0 (SAP, Walldorf, Germany).
Ethical approval
The Ethics committee of the University of Wuerzburg considered the study protocol and waived the need to formally apply for ethical clearance because of the study design (file number 20210813 01).
Statistics
Data analysis was performed with Excel 2019 (Microsoft, Redmond, WA, USA) and GraphPad Prism 9.0.2 (GraphPad Software, San Diego, CA, USA). CIs were calculated using the Wilson/Brown method [13], statistical significance levels were calculated using Fisher's exact test, chi-squared test, Mann-Whitney U test, and Kruskal-Wallis test. The two-tailed significance level α was set to 0.05. The limit of detection (LOD) analysis was performed in R, version 4.2.1. A logistic regression model was fitted to test results in dependence of log10 viral load; 50% and 95% LOD values with 95% CIs were calculated from the regression curve.
Results
Test enrolment
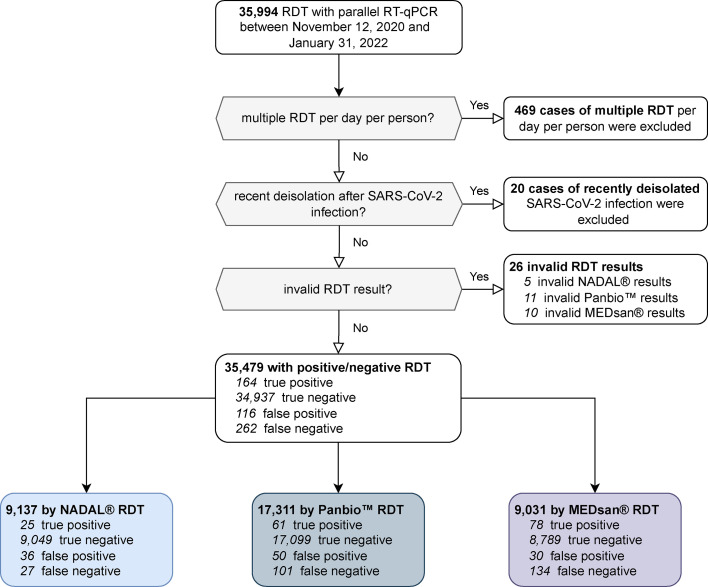
Between 12 November 2020 and 31 January 2022, a total of 35 994 RDTs with parallel RT-qPCR were conducted, out of which 35 479 RDTs with parallel RT-qPCR taken from 26 940 individuals were taken into consideration for performance assessment. A total of 9137 (25.75%) RDTs were carried out with NADAL, 17 311 (48.79%) RDTs were carried out with Panbio, and 9031 (25.45%) RDTs were carried out with MEDsan (Fig. 1 ).
Fig. 1.
Distribution of enrolled rapid diagnostic test (RDT) results. Four hundred sixty-nine cases of multiple RDT performances per day per person and 20 cases of recent deisolation after SARS coronavirus 2 (SAR-CoV-2) infection were excluded from data analysis. Twenty-six RDTs with invalid test results (missing positive control or interfering lines, 5 NADAL, 11 Panbio, and 10 MEDsan) were also not taken into account. RT-qPCR, quantitative reverse transcription polymerase chain reaction.
Study population
Enrolled study participants were aged from 0 to 101 years (median age, 50.5 years; interquartile range, 30–69 years). A total of 17 906 (50.43%) RDTs were performed on female individuals, 17 597 (49.56%) RDTs were performed on male individuals, and one RDT was performed on an individual with allocation to a diverse gender. A total of 30 588 (86.15%) RDTs were performed on patients, 598 (1.68%) RDTs were performed on employees, and 4319 (12.16%) RDTs were performed on accompanying individuals staying overnight with underage and otherwise dependent patients.
Performance of RDT compared with that of RT-qPCR
In 426 specimens among 35 479 enrolled RDT/RT-qPCR tandems, SARS-CoV-2 positivity could be confirmed, corresponding to a SARS-CoV-2 prevalence of 1.20%. Of all tandem samples, 164 (0.46%) were tested positive by an RDT (true positive), and 262 (0.74%) were tested negative (false negative). Of the 35 053 RT-qPCR-negative samples, 34 937 (98.47%) tested negative by an RDT (true negative), and 116 (0.33%) tested positive (false positive). Three of the 26 invalid assessed RDTs tested RT-qPCR positive.
The overall RDT sensitivity was calculated as 38.5% (95% CI, 34.0–43.2%), and the overall specificity was calculated as 99.67% (95% CI, 99.60–99.72%). The positive predictive value was 58.57% (95% CI, 52.57–64.19%), and the negative predictive value was 99.26% (95% CI, 99.16–99.34%).
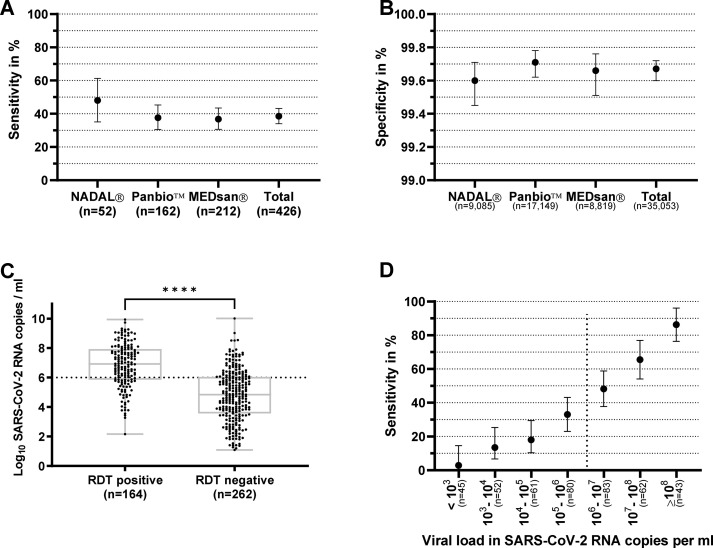
The differences among manufacturers in sensitivity (p 0.31) (Fig. 2 [a] and Supplementary Results) and specificity (p 0.37; chi-squared test) (Fig. 2[b]) were not significant.
Fig. 2.
Sensitivity of antigen rapid diagnostic testing (RDT) compared with that of quantitative reverse transcription polymerase chain reaction by manufacturer and viral load (a) Sensitivity and (b) specificity of antigen rapid diagnostic tests from three manufacturers (nal von minden NADAL, Abbott Panbio, and MEDsan) compared with those of quantitative reverse transcription polymerase chain reaction (RT-qPCR) as reference standard (n = 35 479). (c) Viral load of an RT-qPCR–positive specimen that tested positive and negative by RDTs. (d) Sensitivity of RDT compared with that of RT-qPCR in relation to viral load determined from Ct values. Sensitivity is sharply increasing with higher viral loads (n = 426). The dotted lines in (c) and (d) represent the viral load of 106 SARS coronavirus 2 (SARS-CoV-2) RNA copies per mL, assumed as infectivity threshold [14]. The asterisks represent a p value of <0.0001.
Relation of RDT sensitivity to viral load
Median viral loads were significantly higher in RDT positive (median, 8.22 × 106 SARS-CoV-2 RNA copies per mL) than in RDT negative samples (median, 6.84 × 104 copies per mL; p < 0.0001; Mann-Whitney U test) (Fig. 2[c]). Sensitivity increased with rising viral loads (Fig. 2[d]) [14].
Relation of RDT performance to molecularly determined SARS-CoV-2 VOC
In 257 (60.32%) of all RT-qPCR–positive samples, VOCs were determined by PCR tests. A total of 16 (6.23%) specimens were assigned as containing wild-type SARS-CoV-2, 70 (27.24%) specimens were assigned as containing Alpha VOC, 132 (51.36%) specimens were assigned as containing Delta VOC, and 36 (14.01%) specimens were assigned as containing Omicron VOC. In two samples (0.78%) Iota variant of interest (VOI) was detected; in one case results of variant-specific PCR tests and spike protein sequencing did not conform to any commonly described VOC.
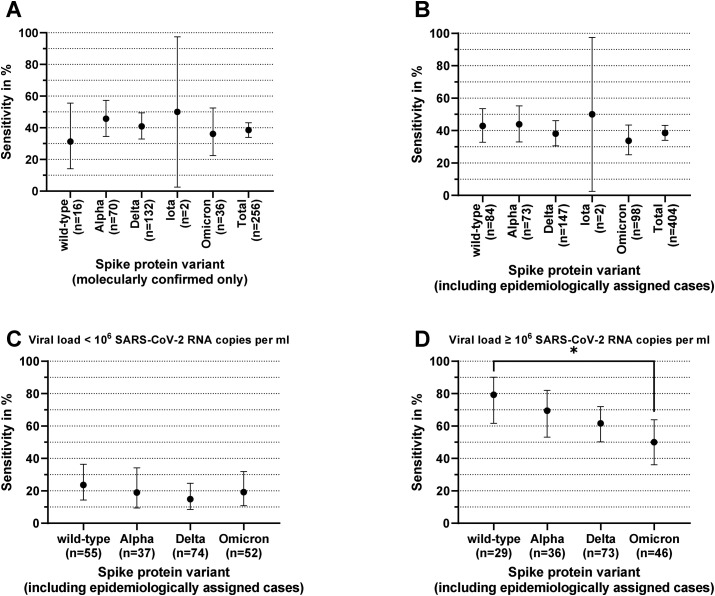
Including all RDT/RT-qPCR tandems with molecularly determined VOC, RDT sensitivity was 31.25% (5/16; 95% CI, 14.16–55.60%) for wild-type SARS-CoV-2, 45.71% (32/70; 95% CI, 34.57–57.30%) for Alpha VOC, 40.91% (54/132; 95% CI, 32.89–49.44%) for Delta VOC, 36.11% (13/36; 95% CI, 22.48–52.42%) for Omicron VOC, and 50.00% (1/2; 95% CI, 2.56–97.44%) for Iota VOI (Fig. 3 [a]).
Fig. 3.
Variant of concern (VOC) depending on antigen rapid diagnostic test (RDT) sensitivity. Antigen rapid diagnostic test sensitivity compared with that of quantitative reverse transcription polymerase chain reaction as reference standard by VOC. (a) Included 257 samples with molecularly confirmed VOC. (b) Included 407 samples with either molecularly confirmed VOC or epidemiologically assigned VOC (in case no VOC was determined molecularly and the VOC of the infection source was known or a VOC was responsible for more than 90% of all coronavirus disease 2019 cases in Germany at the time of sampling). (c) Included 218 samples with a viral load of <106 SARS coronavirus 2 (SARS-CoV-2) RNA copies per mL and an either molecularly or epidemiologically assigned wild-type, Alpha VOC, Delta VOC, or Omicron VOC. (d) Included 184 samples with a viral load of ≥106 SARS-CoV-2 RNA copies per mL and an either molecularly or epidemiologically assigned wild-type SARS-CoV-2, Alpha VOC, Delta VOC, or Omicron VOC. The asterisk represents a p value of <0.05.
Relation of RDT performance to SARS-CoV-2 VOC, including epidemiological assignment
Using epidemiological data, 148 further samples could be assigned to a presumed SARS-CoV-2 VOC, resulting in 405 RT-qPCR–positive samples with confirmed or presumed VOC: 84 (20.74%) specimens to wild-type SARS-CoV-2, 73 (18.02%) specimens to Alpha VOC, 147 (36.30%) specimens to Delta VOC, 98 (24.20%) specimens to Omicron VOC, and 2 (0.49%) specimens to Iota VOI. One sample showed an uncommon VOC, and 21 remaining specimens without VOC allocation could not be epidemiologically identified because of VOC prevalence levels below 90% and were consequently not considered for VOC-dependent performance evaluation [10].
On the basis of this assignment [10], the following VOC-dependent sensitivities were observed: 42.86% (36/84; 95% CI, 32.82–53.52%) for wild-type virus, 43.84% (32/73; 95% CI, 33.05–55.24%) for Alpha VOC, 38.10% (56/147; 95% CI, 30.64–46.15%) for Delta VOC, 33.67% (33/98; 95% CI, 25.09–43.49%) for Omicron VOC, and 50.00% (1/2; 95% CI, 2.56–97.44%) for Iota VOI (Fig. 3[b]).
The median viral load was significantly lower in wild-type samples (7.71 × 104 SARS-CoV-2 RNA copies per mL) than in VOI and VOC samples (p < 0.001, Mann-Whitney U test). No significant difference was observed in comparison of Alpha VOC (median, 8.28 × 105 copies per mL), Delta VOC (median: 7.43 × 105 copies per mL), Iota VOI (6.84 × 105 copies per mL), and Omicron VOC (5.30 × 105 copies per mL; p 0.71, Kruskal-Wallis test).
Although no significant differences in RDT sensitivity could be detected in samples with a viral load of <106 SARS-CoV-2 RNA copies per mL (Fig. 3[c]), differences in sensitivity were observed in samples with a viral load greater than this threshold: sensitivity was 79.31% (23/29; 95% CI, 61.61–90.15%) in wild-type SARS-CoV-2–containing samples, whereas it decreased in other VOCs to 69.44% (25/36; 95% CI, 53.14–82.00%; p 0.41) in Alpha VOC, 61.64% (45/73; 95% CI, 50.17–71.95%; p 0.11) in Delta VOC, and 50.00% (23/46; 95% CI, 36.12–63.88%; p 0.015, Fisher's exact test) in Omicron VOC (Fig. 3[d]).
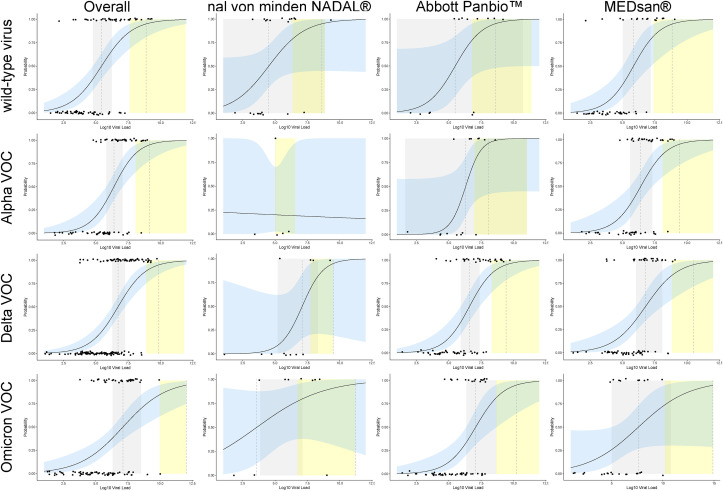
Further, 50% LOD increased from 2.75 × 105 (95% CI, 6.03 × 104–1.74 × 106) SARS-CoV-2 RNA copies per mL for wild-type virus over 2.51 × 106 (95% CI, 6.46 × 105–1.17 × 107) copies per mL for Alpha VOC and 5.13 × 106 (95% CI, 1.86 × 106–1.82 × 107) for Delta VOC to 1.32 × 107 (95% CI, 2.09 × 106–3.39 × 108) for Omicron VOC. LOD (95%) increased from 7.59 × 108 (95% CI, 4.17 × 107–8.71 × 1011) SARS-CoV-2 RNA copies per mL for wild-type virus over 1.38 × 109 (95% CI, 1.20 × 108–1.74 × 1012) copies per mL for Alpha VOC and 7.41 × 109 (95% CI, 7.76 × 108–6.76 × 101) for Delta VOC to 2.34 × 1012 (95% CI, 9.33 × 109–2.00 × 1018) for Omicron VOC (Fig. 4 ).
Fig. 4.
Variant of concern (VOC) depending level of detection (LOD), overall and by manufacturer. Included 402 samples either molecularly confirmed or epidemiologically assigned to wild-type virus, Alpha VOC, Delta VOC, or Omicron VOC (in case no VOC was determined molecularly and the VOC of the infection source was known or a VOC was responsible for more than 90% of all coronavirus disease 2019 cases in Germany at the time of sampling). Further, 50% and 95% LODs were marked by dashed vertical lines with 95% CIs visualized in grey (50% LOD) and yellow (95%), respectively. The regression curve is shown as a straight line with 95% CI visualized in blue. The difference in overall 50% LOD was significantly higher for Omicron VOC than for wild-type virus, whereas a nonsignificant increase in 50% and 95% LOD was observed from wild-type virus over Alpha VOC, Delta VOC in the overall data, and for Abbott Panbio (n = 156) and MEDsan (n = 197). Owing to the limited case numbers, no reliable VOC-specific LOD calculation could be obtained for nal von minden NADAL (n = 49).
Discussion
With an overall sensitivity of 38.5%, the clinical sensitivity matches the range of a laboratory analysis of the used products (36–64%) [15]. The overall sensitivity is on the lower end of the range reported by other studies, which can be explained by the inclusion of asymptomatic individuals, whereas most of the previously published clinical evaluation studies are limited to symptomatic individuals [[16], [17], [18], [19]]. Among the subgroup of specimens with a viral load of ≥106 SARS-CoV-2 RNA copies per mL, suggested as the viral load threshold for infectivity [14], RDT sensitivity was statistically significantly impaired in Omicron VOC samples compared with wild-type SARS-CoV-2. The 50% LOD for Omicron VOC increased by the factor 48 compared with that for wild-type SARS-CoV-2, and the 95% LOD increased by the factor 1425. This is in line with the previously published small-cohort or laboratory studies that claim Omicron VOC as curtailing RDT sensitivity when compared with wild-type SARS-CoV-2 and Delta VOC. The present data support the previous studies and provide a translation of data to real-life and large-scale clinical conditions [[3], [4], [5], [6]].
Data from a laboratory evaluation suggest that the decrease in sensitivity may be caused by a lower nucleoprotein-to-RNA ratio in Omicron VOC–infected individuals [5]. A possible explanation for the fact that this difference was not significant among individuals with a viral load of ≤106 RNA copies per mL may be the higher medium viral load in Omicron VOC than in wild-type SARS-CoV-2 samples, which may compensate for the restriction in sensitivity. Another possible bias might be the differences in immunization status of general public: while nearly no wild-type SARS-CoV-2 and only a few Alpha VOC infections occurred in vaccinated or convalescent individuals enrolled to this study, most adults were vaccinated twice or thrice during the Delta and Omicron VOC–dominated interval [7,20]. Recently, this potential influence of immunization status, including vaccination status, on RDTy among the selected RDT manufacturers. In a large-scale laboratory performance evaluation, the three RDTs used belonged to a moderate sensitivity group. RDTs with sensitivity was discussed in a preprint analysis [21].
The presented results are limited in several aspects. Because RDT point-of-care usage was implemented under real-life clinical conditions and differing product availabilities, absolute numbers and proportions of used RDT products varied among several clinical departments and in course of the study period. The participating clinical departments differ regarding patient structure and morbidity. Enrolled study participants were only tested using one of the three chosen RDTs, which restricts direct comparability among the selected RDT manufacturers. In a large-scale laboratory performance evaluation, the three RDTs used belonged to a moderate sensitivity group. RDTs with higher sensitivity have since become available, although their performance regarding Omicron VOC is still unclear [15]. In acceptance of these limitations, the study allowed an analysis of real-life data in a large cohort with a broad demographic structure and consequently high reliable transferability in in vivo conditions, including VOC dependence. Because sample collection for RDT and RT-qPCR was performed by a multitude of skilled, professional operators, the influence of potential inhomogeneity in sampling, test execution, and interpretation could not be avoided. Further, the role of preanalytical quality and accurate specimen collection must be highlighted. Compared with RT-qPCR, RDTs are more prone to incorrect swabbing. Consequent lower viral loads of the gained samples may in part be responsible for increasing numbers of false negative results. Although NADAL and MEDsan RDTs were recommended for nasopharyngeal and oropharyngeal sampling, the Panbio RDT was used with oropharyngeal sampling, in contrast to the manufacturer's instructions recommending nasopharyngeal specimen collection, which might limit the comparability to studies based on nasopharyngeal sampling.
VOC determination was only performed between January 2021 and January 2022. Therefore, a relevant proportion of wild-type SARS-CoV-2 and Omicron VOC samples could only be epidemiologically assigned. Omicron VOC sublineages could not be differentiated. Owing to the inability of VOC determination in several low viral load samples, sensitivity in molecularly determined VOCs is biased towards higher values. With only two specimens of Iota VOI, no precise sensitivity data could be determined for this VOI. Compared with previously published studies, this study represents a low SARS-CoV-2 prevalence setting of 1.20% but corresponds to a real-life scenario including RDT testing on asymptomatic individuals [[16], [17], [18], [19],22].
Omicron VOC reduces RDT sensitivity even among individuals with high viral loads, which limits the use of RDTs in the group most relevant for SARS-CoV-2 transmission. However, an RDT is still an important strategic tool for rapid identification of highly infectious individuals before the availability of RT-qPCR results and for regular screening of large cohorts. The self-testing option and straightforward feasibility emphasize these aspects. This points out the importance of further large clinical studies on RDTs in large populations not only in times of Omicron VOC prevalence but also for future possible VOCs and VOC sublineages.
Author contributions
All authors had unlimited access to all data. IW and MK take responsibility for the integrity of the data and the accuracy of the data analysis. OA, TW, RIE, JF, DW, BW, LD, JL, OK, UV, and MK conceived and designed the study. Quantitative reverse transcription polymerase chain reaction testing and a standardized Ct quantification were performed by KK, DH, and BW. Antigen rapid diagnostic test use and documentation instruction in different departments were performed by VR, ME, SF, OA, MST, MP, RVF, AS, DZ, HB, AG, AKS, MK, MG, RT, TW, DW, and MK. User support was provided by VR, ME, UV, and MK. IW, VR, ME, NP, MM, and MK collected clinical data from patients' files. Limit of detection calculation was performed by LK. Statistical analysis was performed by IW, JR, AG, NP, and MK. LD, OK, and UV obtained funding. The first draft of the manuscript was prepared by IW and MK. KK, DH, VR, ME, JR, AG, SF, OA, NP, MST, MP, MM, RVF, AS, DZ, HB, AG, AKS, MK, MG, RT, TW, RIE, JF, DW, BW, LD, JL, LK, OK, and UV reviewed and modified the manuscript and approved its final version.
Transparency declaration
The authors declare that they have no conflicts of interest. This study was funded by the Federal Ministry for Education and Science via a grant provided to the University Hospital of Wuerzburg by the Network University Medicine on COVID-19 (B-FAST, grant-No 01KX2021) as well as by the Free State of Bavaria with COVID-research funds provided to the University of Wuerzburg, Germany. This work was supported by the Bavarian Staten Ministry of Health and Care via Bay-VOC. Nils Petri is supported by the German Research Foundation (DFG) funded scholarship UNION CVD. This study was initiated by the investigators. The sponsoring institutions had no role in study design, data collection, analysis, and interpretation of data as well as in writing of the manuscript. All authors had unlimited access to all data. The first and the corresponding author had final responsibility for the decision to submit for publication.
Acknowledgements
We thank all the hospital staff for conducting antigen rapid diagnostic test testing and documenting test results and the entire laboratory staff in the virological diagnostic laboratories for performing quantitative reverse transcription polymerase chain reaction analyses. We thank the accounting department from medical controlling for SAP support. The study has been published as a preprint (https://doi.org/10.2139/ssrn.4075840) at Social Science Research Network. Results of the study have been presented at the 31st Annual Meeting of the German Society for Virology and the 32nd European congress of clinical microbiology and infectious diseases. The graphical abstract was created using biorender.com.
Editor: J. M. Hübschen
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2022.08.006.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Diel R., Nienhaus A. Point-of-care COVID-19 antigen testing in German emergency rooms–a cost-benefit analysis. Pulmonology. 2022;28:164–172. doi: 10.1016/j.pulmoe.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization Use of SARS-CoV-2 antigen-detection rapid diagnostic tests for COVID-19 self-testing. https://www.who.int/publications/i/item/WHO-2019-nCoV-Ag-RDTs-Self_testing-2022.1 Available at:
- 3.Barrera-Avalos C., Luraschi R., Vallejos-Vidal E., Mella-Torres A., Hernández F., Figueroa M., et al. The rapid antigen detection test for SARS-CoV-2 underestimates the identification of covid-19 positive cases and compromises the diagnosis of the SARS-CoV-2 (K417N/T, E484K, and N501Y) variants. Front Public Health. 2021;9 doi: 10.3389/fpubh.2021.780801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bekliz M., Perez-Rodriguez F., Puhach O., Adea K., Melancia S.M., Baggio S., et al. Sensitivity of SARS-CoV-2 antigen-detecting rapid tests for Omicron variant. medRxiv. https://www.medrxiv.org/content/10.1101/2021.12.18.21268018v2 [DOI] [PMC free article] [PubMed]
- 5.Osterman A., Badell I., Basara E., Stern M., Kriesel F., Eletreby M., et al. Impaired detection of omicron by SARS-CoV-2 rapid antigen tests. Med Microbiol Immunol. 2022;211:105–117. doi: 10.1007/s00430-022-00730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao A., Bassit L., Lin J., Verma K., Bowers H.B., Pachura K., et al. Assessment of the Abbott BinaxNOW SARS-CoV-2 rapid antigen test against viral variants of concern. iScience. 2022;25 doi: 10.1016/j.isci.2022.103968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayerisches Landesamt für Gesundheit und Lebensmittelsicherheit: Übersicht der Fallzahlen von Coronavirusinfektionen in Bayern. https://www.lgl.bayern.de/gesundheit/infektionsschutz/infektionskrankheiten_a_z/coronavirus/karte_coronavirus/ Available at:
- 8.Wagenhäuser I., Knies K., Rauschenberger V., Eisenmann M., McDonogh M., Petri N., et al. Clinical performance evaluation of SARS-CoV-2 rapid antigen testing in point of care usage in comparison to RT-qPCR. EBioMedicine. 2021;69 doi: 10.1016/j.ebiom.2021.103455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayerisches Landesamt für Statistik Bevölkerung: Gemeinden, amtliche Einwohnerzahl aktuell (jährlich, vierteljährlich) https://www.statistikdaten.bayern.de/genesis/online?operation=abruftabelleBearbeiten&levelindex=1&levelid=1647519965087&auswahloperation=abruftabelleAuspraegungAuswaehlen&auswahlverzeichnis=ordnungsstruktur&auswahlziel=werteabruf&code=12411-000&auswahltext=&nummer=7&variable=7&name=GEMEIN&werteabruf=Werteabruf#abreadcrumb Available at:
- 10.Koch-Institut Robert. Anzahl und anteile von VOC und VOI in Deutschland. https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Daten/VOC_VOI_Tabelle.html Available at:
- 11.Bundesinstitut für Arzneimittel und Medizinprodukte Antigen-tests zum direkten erregernachweis des coronavirus SARS-CoV-2. 2020. https://antigentest.bfarm.de/ords/f?p=101:100:13556100388097 Previously available at:
- 12.Lind A., Barlinn R., Landaas E.T., Andresen L.L., Jakobsen K., Fladeby C., et al. Rapid SARS-CoV-2 variant monitoring using PCR confirmed by whole genome sequencing in a high-volume diagnostic laboratory. J Clin Virol. 2021;141 doi: 10.1016/j.jcv.2021.104906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown L., Cai T., DasGupta A. Interval estimation for a binomial proportion. Stat Sci. 2001;16:101–133. doi: 10.1214/ss/1009213286. [DOI] [Google Scholar]
- 14.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 15.Scheiblauer H., Filomena A., Nitsche A., Puyskens A., Corman V.M., Drosten C., et al. Comparative sensitivity evaluation for 122 CE-marked rapid diagnostic tests for SARS-CoV-2 antigen, Germany, September 2020 to April 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.44.2100441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gremmels H., Winkel B.M.F., Schuurman R., Rosingh A., Rigter N.A.M., Rodriguez O., et al. Real-life validation of the Panbio™ COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalmedicine. 2021;31 doi: 10.1016/j.eclinm.2020.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhi S., Tayler N., Hoang T., Ballard S.A., Graham M., Rojek A., et al. Multi-site assessment of rapid, point-of-care antigen testing for the diagnosis of SARS-CoV-2 infection in a low-prevalence setting: a validation and implementation study. Lancet Reg Health West Pac. 2021;9 doi: 10.1016/j.lanwpc.2021.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olearo F., Nörz D., Heinrich F., Sutter J.P., Roedl K., Schultze A., et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. J Clin Virol. 2021;137 doi: 10.1016/j.jcv.2021.104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres I., Poujois S., Albert E., Colomina J., Navarro D. Evaluation of a rapid antigen test (Panbio™ COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clin Microbiol Infect. 2021;27 doi: 10.1016/j.cmi.2020.12.022. 636e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bundesministerium für Gesundheit Impfdashboard. https://impfdashboard.de Available at:
- 21.Meiners L., Horn J., Mühlemann B., Schmidt M.L., Walper F., Menzel P., et al. Social Science Research Network (SSRN); 2022. SARS-CoV-2 rapid antigen test sensitivity and viral load in freshly symptomatic hospital employees, December 2020 to February 2022.https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4099425 Available at: [DOI] [PubMed] [Google Scholar]
- 22.Schildgen V., Demuth S., Lüsebrink J., Schildgen O. Limits and opportunities of SARS-CoV-2 antigen rapid tests: an experienced-based perspective. Pathogens. 2021;10:38. doi: 10.3390/pathogens10010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.