Abstract
Insertional inactivation of the yrbA gene of Bacillus subtilis reduced the resistance of the mutant spores to lysozyme. The yrbA mutant spores lost their optical density at the same rate as the wild-type spores upon incubation with l-alanine but became only phase gray and did not swell. The response of the mutant spores to a combination of asparagine, glucose, fructose, and KCl was also extremely poor; in this medium yrbA spores exhibited only a small loss in optical density and gave a mixture of phase-bright, -gray, and -dark spores. Northern blot analysis of yrbA transcripts in various sig mutants indicated that yrbA was transcribed by RNA polymerase with ςE beginning at 2 h after the start of sporulation. The yrbA promoter was localized by primer extension analysis, and the sequences of the −35 (TCATAAC) and −10 (CATATGT) regions were similar to the consensus sequences of genes recognized by ςE. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of proteins solubilized from intact yrbA mutant spores showed an alteration in the protein profile, as 31- and 36-kDa proteins, identified as YrbA and CotG, respectively, were absent, along with some other minor changes. Electron microscopic examination of yrbA spores revealed changes in the spore coat, including a reduction in the density and thickness of the outer layer and the appearance of an inner coat layer-like structure around the outside of the coat. This abnormal coat structure was also observed on the outside of the developing forespores of the yrbA mutant. These results suggest that YrbA is involved in assembly of some coat proteins which have roles in both spore lysozyme resistance and germination.
Many gram-positive soil microorganisms, such as Bacillus subtilis, develop dormant spores when nutrients are exhausted. Spore formation is the result of a complex process of macromolecular assembly that is controlled at different stages of sporulation. For example, RNA polymerase sigma factors are activated sequentially in the mother cell or forespore compartment and regulate the expression of sporulation-related genes (10, 38). The bacterial spores are metabolically dormant and have a unique thick protein shell known as the spore coat (3). The coat is composed of dozens of proteins (38) arranged in an electron-dense thick outer layer and a thinner, lamella inner layer (6). These layers provide a protective barrier against bactericidal enzymes and chemicals, such as lysozyme and organic solvents (9). SpoIVA is synthesized from 2 h after the start of sporulation (T2) in the mother cell compartment and assembles around the outer membrane of the forespore in B. subtilis (39). This protein is thought to be required for the formation of a basement layer on which spore coat proteins assemble (8, 29, 39). One of the coat protein components, CotE, is also a morphogenic protein required for the assembly of the outer coat (47). cotE mutant spores are refractile and resistant to heat and chemicals but are lysozyme sensitive and germinate slower and less efficiently than wild-type spores (47). The CotT protein of B. subtilis is synthesized as a 10.1-kDa precursor, which is processed to a coat polypeptide of 7.8 kDa, and insertional inactivation of the cotT gene resulted in spores with an altered appearance of the inner coat layers and slow germination in response to a solution containing fructose, glucose, and asparagine (4). Thus, the coat components may play an important role in responding to germinants and also in preventing access of lysozyme to the peptidoglycan of the spore cortex.
We identified a DNA fragment containing three deduced open reading frames, orf1, orf2, and orf3 (42) (DDBJ accession no. D50551) near the nadA gene in the 243° region (nadC, nadA, yrbA, yrbB, and yrbC, in that order); orf1, orf2, and orf3 in our work correspond to yrbA, yrbB, and yrbC, respectively, as determined by the B. subtilis genome sequencing project (19). In a previous paper (42), we demonstrated by immunoelectron microscopy that the YrbB protein was located in spores, primarily in the cortex layer. In this work, we have analyzed the function and expression of yrbA and found that yrbA expression was dependent on ςE-containing RNA polymerase. The yrbA mutant spores had abnormal coat layers, had lost their response to a germination solution containing asparagine, glucose, fructose, and KCl (AGFK) and resistance to lysozyme, and were deficient in some coat proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and general techniques.
The B. subtilis strains used in this study are listed in Table 1 and were all grown in DS medium at 37°C (34). Escherichia coli was grown in Luria-Bertani medium. The conditions for sporulation of B. subtilis and the method for purification of mature spores have been described previously (2, 41). Recombinant DNA methods were as described by Sambrook et al. (33). Methods for preparing competent cells for transformation and for preparing chromosomal DNA from B. subtilis were as described by Cutting and Vander Horn (5).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Source (reference) |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| SC1159 | spoIIAC1 (SigF mutant) | S. Cutting (7) |
| 1S60 | leuB8 tal-1 spoIIG41 (SigE mutant) | BGSCa |
| spoIIIGΔ1 | trpC2 spoIIIGΔ1 (SigG mutant) | J. Sekiguchi (36) |
| 1S38 | trpC2 spoIIIC94 (SigK mutant) | BGSC |
| TB711 | trpC2 yrbB::cat | This work |
| TB713 | trpC2 yrbA::cat | This work |
| E. coli JM109 | relA supE44 endA1 hsdR17 gyrA96 mcrA mcrB+ thiΔ(lac-proAB)/F′ (traD36 proAB+ lacIq lacZΔM15) | Sambrook et al. (33) |
| Plasmids | ||
| pDH88 | Cmr AmprPspac-1 | D. Henner (12) |
| pCOXA5 | Cmr AmprPspac-1 yrbB′ | This work |
| pYRBA5 | Cmr AmprPspac-1 yrbA′ | This work |
BGSC, Bacillus Genetic Stock Center.
Preparation and purification of spores.
B. subtilis spores prepared in DS medium were harvested after incubation for 48 h and washed several times with deionized water. The spores obtained were then purified by a urografin gradient procedure as described by Nicholson and Setlow (27).
Construction of yrbA and yrbB mutants.
Oligonucleotide primers YRBA158 (5′-CGTCTAGAAAAGAGCCAAAAGCGG-3′) and YRBA562R (5′-TTAGATCTTCTACACCGCCTACCT-3′) were used to amplify a DNA fragment from nucleotide (nt) +158 to +562 of yrbA, and primers CRB1 (5′-AATCTAGAGAACTGACACGCTTAA-3′) and CRB2 (5′-TAGATCTGTGGTGTTTCGGTTACC-3′) were used to amplify a DNA fragment from nt −93 to +234 of yrbB. The PCR products were cleaved at the XbaI and BglII sites and inserted between the XbaI and BglII sites of pDH88 (12) to construct plasmids pYRBA5 and pCOXA5, respectively. The resulting plasmids were introduced into strain 168 by a single crossover with selection for resistance to chloramphenicol (5 μg/ml) to give strains TB713 (yrbA mutant) and TB711 (yrbB mutant).
RNA preparation and Northern analysis.
B. subtilis cells were grown in DS medium, and 20-ml samples were harvested every hour throughout sporulation. RNA for Northern blots was then prepared by a modification of the procedure described by Igo and Losick (15). Aliquots (10 μg) of the RNA preparation were analyzed by size fractionation through a 1% (wt/vol) agarose gel containing 2.2 M formaldehyde and were transferred to a positively charged Hybond-N+ membrane (Amersham). The membrane was stained with 0.04% methylene blue in 0.5 M sodium acetate (pH 5.2) to measure the concentrations of 16S and 23S RNAs in the preparations as described previously (13). The RNA on the membrane was hybridized to probe 1 and probe 2 DNAs, which are specific for yrbA and yrbB, respectively. The 0.5-kb probe 1, corresponding to nt 22 to 526 downstream of the putative translation initiation codon of yrbA, was prepared by PCR with primers COXRA10 (5′-AAAGGCGATTCGCTCTGG-3′) and YRBA500T (5′ - TAATACGACTCAC TATAGGGCGAGGGCATAT TAGGCATAT TCGG -3′). The 0.4-kb probe 2, corresponding to nt −170 to +216 relative to the putative translation initiation codon of yrbB, was prepared by PCR with primers CRB1 (5′-AATCTAGAGAACTGACACGCTTAA-3′) and COX200T (5′-TAATACGACTCACTATAGGGCGAGTTCCGTCAGTTGCCAAAGG-3′). The underlined regions in the primers represent the T7 promoter sequence. The RNA probes were prepared by using the Boehringer Mannheim digoxigenin labeling system, and hybridization was performed by the procedure recommended by Boehringer Mannheim.
Mapping of the 5′ terminus of yrbA mRNA.
Cells were grown in DS medium, and 20-ml samples were harvested at T5 of sporulation. RNA for primer extension analyses were prepared by a modification of the procedure described by Igo and Losick (15) and Cutting et al. (7). Primer extension was performed with a cDNA synthesis kit (Pharmacia Biotech) and a 5′-end digoxigenin-labeled primer, COXPM1R (5′-TTCCCCTCCTATGCAAAACG-3′), which was complementary to nt+5 to nt+24 downstream of the translational start point of yrbA. The reaction was carried out as recommended by Boehringer Mannheim, except that the reaction mixture was incubated at 42°C. Oligonucleotide primers COXRAM400 (5′-TCGACACAATCAACCAGGCT-3′) and COXRA320R (5′-ACATCAGCTTCAGGGTACAC-3′) were used to amplify a DNA fragment from nt −394 to +374 of yrbA. The 5′-end-labeled primer was also used to generate a sequence ladder by the dideoxy chain termination method with the DNA fragment as a template. The products of primer extension were then subjected to electrophoresis in a 5% (wt/vol) polyacrylamide slab gel containing 8 M urea, and products were detected as recommended by Boehringer Mannheim.
Spore germination.
Purified spores were heat activated at 65°C for 15 min and suspended in 50 mM Tris-HCl (pH 7.5) buffer at an optical density at 660 nm of 0.5. Either 10 mM l-alanine, AGFK (3.3 mM l-asparagine, 5.6 mM d-glucose, 5.6 mM d-fructose, and 10 mM KCl), or 10 mM Tris-HCl (pH 7.5) was then added. Germination was monitored by measurement of the decrease in absorbance (660 nm) of the spore suspension at 37°C for up to 90 min as described previously (2).
Spore resistance.
Cells were grown in DS medium at 37°C for 18 h after the end of exponential growth (to T18), and spore resistance was assayed as follows. The cultures were either heated at 80°C for 30 min, treated with lysozyme (250 μg/ml [final concentration]) at 37°C for 10 min, or treated with 10% (vol/vol) chloroform at room temperature for 10 min as described previously (27). After the cultures were serially diluted 100-fold in distilled water, appropriate volumes of the dilutions were spread on Luria-Bertani agar plates, which were incubated overnight at 37°C. The proportion of survivors was determined by counting the colonies.
Solubilization of proteins from spores.
Cultures (5 ml) were harvested at T18 of sporulation and washed with 10 mM sodium phosphate buffer (pH 7.2) containing 0.5 M sodium chloride. The pellets were suspended in 0.1 ml of lysozyme solution (10 mM sodium acetate [pH 7.2], 1% lysozyme) and incubated for 15 min at 37°C. After addition of 1.0 ml of 10 mM sodium phosphate buffer (pH 7.2) containing 0.5 M sodium chloride, the suspensions were centrifuged to remove soluble proteins from mother cells and spores. The spores in the pellet fraction were suspended in 100 μl of buffer containing 2% (wt/vol) sodium dodecyl sulfate (SDS), 5% (vol/vol) 2-mercaptoethanol, 10% (vol/vol) glycerol, 62.5 mM Tris-HCl (pH 6.8), and 0.05% (wt/vol) bromophenol blue and boiled for 5 min.
SDS-PAGE and immunoblotting.
Protein samples were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (12% acrylamide) as described previously (1). For immunoblotting, proteins were transferred onto a polyvinylidene difluoride membrane (0.45-μm pore size) (Immobilon; Millipore) and detected by using rabbit immunoglobulin G against YrbB (42) as the first antibody and donkey anti-rabbit immunoglobulin G–horseradish peroxidase conjugate as the second antibody (Amersham).
NH2-terminal sequence analysis.
Samples were subjected to SDS-PAGE, electroblotted onto a polyvinylidene difluoride membrane as described above, and briefly stained with Coomassie brilliant blue. After extensive washing, the protein bands of interest were excised and applied to a Procise 492 gas-phase sequencer (Applied Biosystems Division, Perkin-Elmer), and sequences of NH2-terminal amino acids were determined as described previously (22).
Electron microscopy.
Purified spores and sporulating cells were fixed with 2.5% glutaraldehyde and then 2% OsO4 as described by Ryter et al. (31) and embedded in Quetol 653 by the method of Kushida (20). Thin sections of spores and sporulating cells were observed with a JEM-1200EX electron microscope operating at 80 kV.
RESULTS
Expression of yrbA during growth and sporulation.
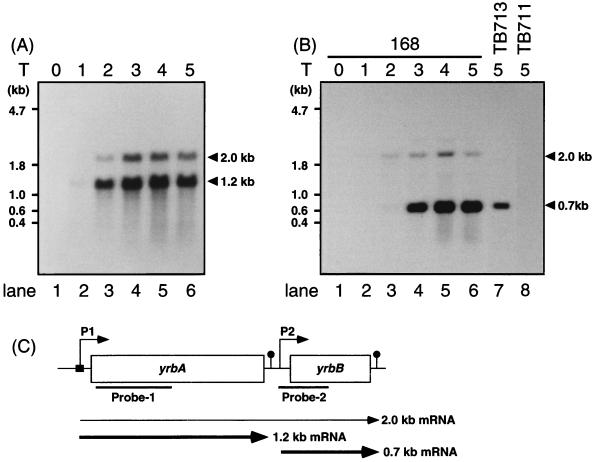
The yrbA-yrbC region contains at least three complete open reading frames, designated yrbA, yrbB, and yrbC. We first determined the sizes and times of appearance of yrbA and yrbB mRNAs during growth and sporulation by Northern blot analysis of samples containing essentially the same amounts of 16S and 23S RNAs (Fig. 1). Both 1.2- and 2.0-kb transcripts containing yrbA mRNA were detected beginning at T2 of sporulation by using probe 1 (Fig. 1A). By using probe 2, which is specific for yrbB mRNA, a 0.7-kb mRNA was found to be transcribed from yrbB beginning at T3 of sporulation (Fig. 1B, lane 4), while the 2.0-kb mRNA found with the yrbA probe also hybridized to the yrbB probe. The origins of these mRNAs were confirmed by additional Northern blot analysis of RNAs from yrbA or yrbB mutants. As shown in Fig. 1B, only the 0.7-kb yrbB mRNA was present in the yrbA mutant (TB713 cells), while this mRNA was not found in the yrbB mutant (TB711 cells); the 2.0-kb mRNA was not detected in either mutant. These results suggest that at least two putative promoters are used for expression of these genes and that the 2.0-kb mRNA is most probably a read-through product from yrbA to yrbB.
FIG. 1.
Northern blot analysis of yrbA mRNA. The gene products of B. subtilis 168 trpC2 (wild-type) were analyzed by using probes specific for yrbA (probe 1) (A) and yrbB (probe 2) (B). The loci, probes used in these experiments, and prospective regions of promoters for yrbA and yrbB are shown in panel C. Total RNA (10 μg) was prepared from cells of strains TB713 (yrbA) and TB711 (yrbB) at T5 of sporulation and analyzed by hybridization with probe 2 (lanes 7 and 8 in panel B). Arrowheads indicate the positions of mRNAs hybridized with the probes. The number of hours after the end of the exponential phase of growth is shown at the top.
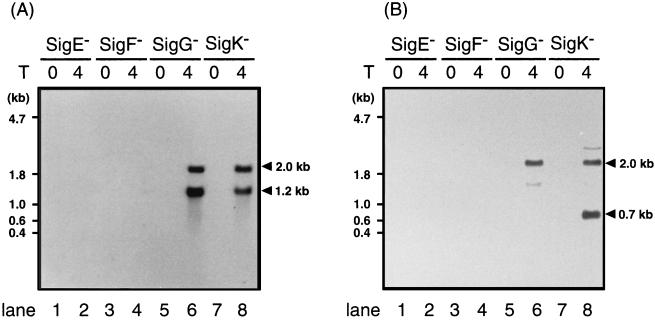
We then examined dependency of yrbA expression on various sigma factors. After the onset of sporulation of B. subtilis, ςE is the first of the sigma factors to appear in the mother cell, with ςF as its counterpart in the forespore; ςF is essential for the activation of pro-ςE (17, 21, 40). Northern blot analysis showed that RNA from either sigE or sigF mutant cells failed to hybridize with the yrbA-specific probe, whereas RNA from sigG or sigK mutant cells gave the same two hybridizing bands as did wild-type cells when analyzed at T4 of sporulation (Fig. 2A). The results in Fig. 2B allow a similar conclusion; probe 2 hybridized to a 2.0-kb mRNA species in the RNAs from sigG and sigK mutants, while the 0.7-kb mRNA was detectable only in the sample prepared from sigK cells (Fig. 2B). SpoIIID was also not essential for the transcription of yrbA (data not shown). These results strongly suggest that expression of yrbA starts at T2 and is dependent on E-ςE RNA polymerase in the mother cell compartment and that the 0.7-kb yrbB mRNA was transcribed by RNA polymerase containing the forespore-specific ςG.
FIG. 2.
Northern blot analysis of yrbA and yrbB transcripts in various sigma factor-deficient cells. RNAs (10 μg) of cells of spoIIG41 (sigE), spoIIAC1 (sigF), spoIIIGΔ1 (sigG), and spoIIIC94 (sigK) mutant strains were subjected to Northern blot analysis with probes specific for yrbA (A) and yrbB (B). The times of harvest of the cells are shown at the top. Arrowheads indicate the positions of RNAs hybridized with the probes.
Location of the yrbA promoter.
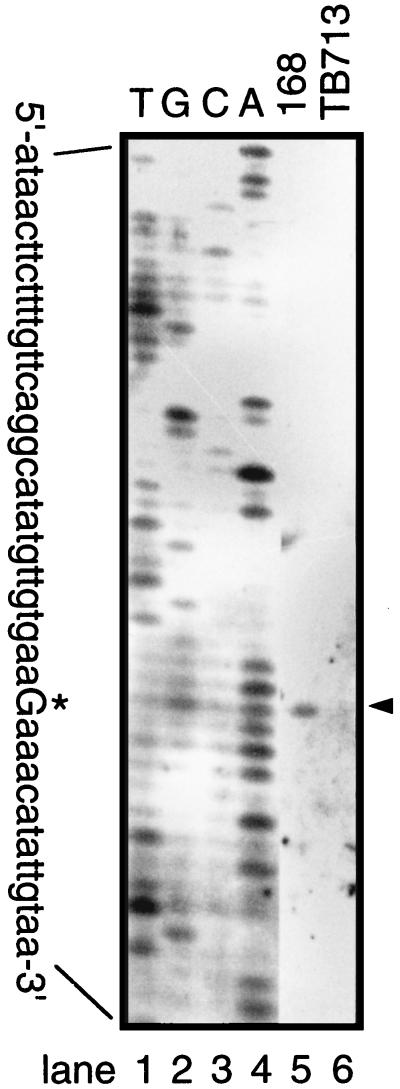
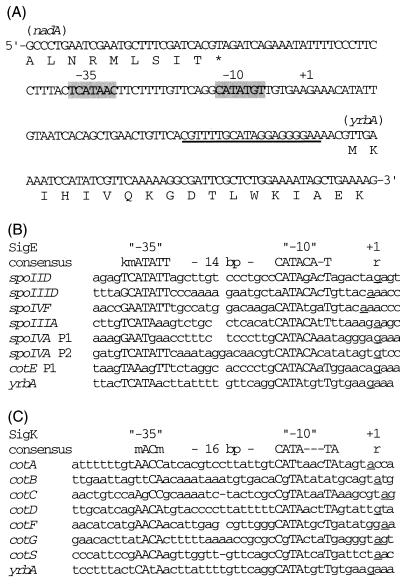
To further analyze the dependence of yrbA expression on sigma factors, the start point of yrbA transcription was mapped by primer extension (Fig. 3). No extension product was detected when RNA of yrbA mutant cells at T5 of sporulation was analyzed; in contrast, a single extension product was seen with RNA isolated from the wild-type cells at T5 of sporulation. The size of this transcript indicated that transcription of yrbA starts at a G residue 56 nt upstream from the TTG translation initiation codon (Fig. 4A), and regions centered 10 and 35 bp upstream of the apparent yrbA transcription start site are very similar to −10 and −35 regions of promoters utilized by ςE-containing RNA polymerase (Fig. 4B).
FIG. 3.
Identification of the yrbA promoter. RNAs (20 μg) extracted from wild-type (168) (lane 5) or yrbA (lane 6) cells at T5 of sporulation were used for primer extension. The sizes of the extended products were compared with a DNA sequencing ladder of the adjacent sequence of the 5′ region of yrbA. The yrbA transcription start site is shown by an arrowhead and an asterisk on the sequence.
FIG. 4.
Genomic structure of the region upstream of yrbA. (A) Nucleotide sequence of the yrbA promoter, showing putative −35 and −10 regions and the transcription start site (+1). The nucleotide sequence complementary to the synthetic oligonucleotide used in primer extension is underlined. The asterisk indicates a translation stop codon. (B) Sequences near the transcription start sites of genes transcribed by RNA polymerase containing ςE. The underlined nucleotides denote the transcription start site. The consensus sequence proposed by Roels et al. (29) is shown at the top (K = G or T; m = C or A; r = G or A). References for the sequences of these promoters are as follows: spoIID, 30; spoIIID, 18 and 43; spoIVF 7; spoIIIA, 8; spoIVA P1 and spoIVA P2, 29; and cotE P1, 48.
Properties of mutant spores.
The resistance of yrbA spores was also examined to learn whether YrbA played any role in spore properties. The yrbA mutation had no effect on vegetative growth (data not shown) or on spore resistance to heat and chloroform (Table 2). However, the yrbA mutation reduced spore resistance to lysozyme. The sensitivity of yrbA spores to lysozyme was confirmed by observation by phase-contrast microscopy, as lysozyme treatment generated lysed yrbA spores (data not shown).
TABLE 2.
Resistance of mutant spores
| Genotype | Viability (CFU/ml) after the following treatmenta:
|
|||
|---|---|---|---|---|
| None | Heat | Lysozyme | Chloroform | |
| Wild type | 4.4 × 108 | 3.4 × 108 | 3.3 × 108 | 2.5 × 108 |
| yrbA | 2.3 × 108 | 1.9 × 108 | 2.7 × 107 | 1.4 × 108 |
| yrbB | 3.9 × 108 | 3.3 × 108 | 2.9 × 108 | 2.4 × 108 |
Spores were spread onto L-agar medium after the following treatment: heating at 80°C for 30 min, incubation with lysozyme (250 μg/ml [final concentration] at 37°C for 10 min, or incubation with 10% (vol/vol) chloroform at room temperature for 10 min. The proportion of survivors was determined by counting colonies after 12 h of incubation at 37°C.
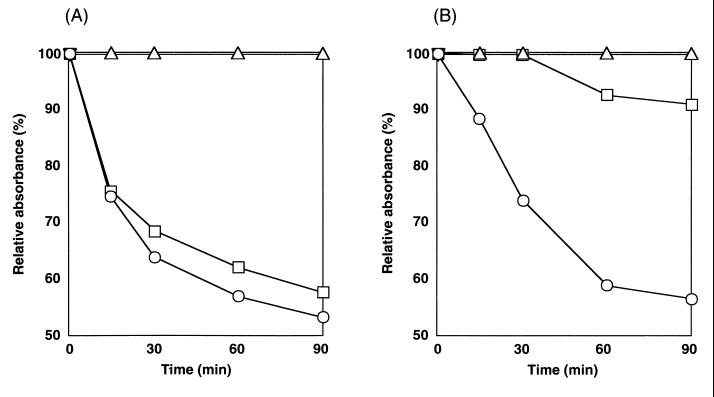
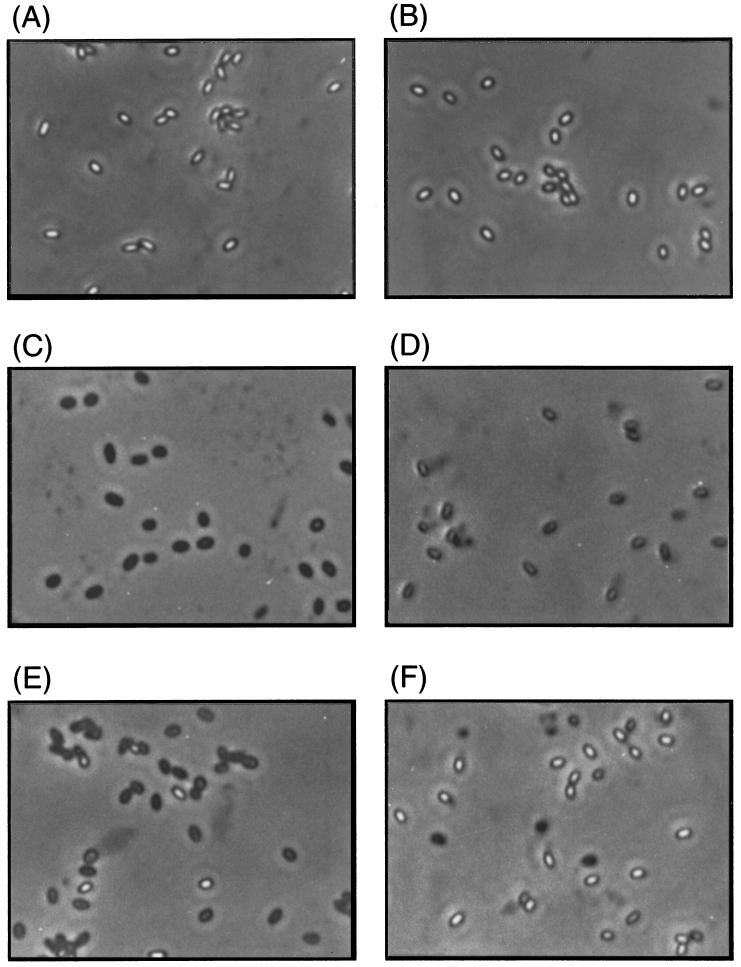
The yrbA mutant spores lost most of their optical density in germination with l-alanine (Fig. 5), and dipicolinic acid was released from the yrbA spores to almost the same extent as from wild-type spores after incubation with l-alanine for 90 min (data not shown). However, microscopic observation revealed that the germinated wild-type spores were phase dark and swollen, while the mutant spores were only phase gray and not swollen (Fig. 6C and D). Dormant-spore properties disappear sequentially during germination, with dipicolinic acid released in the first minutes of germination, followed by loss of spore refractility (35, 37, 44). Consequently, our results suggest that in l-alanine, yrbA mutant spores have a defect at a late stage of spore germination. The most notable property of the yrbA spores was their germination response to AGFK. With AGFK the yrbA spores showed only an extremely small change in optical density (Fig. 5), and after 90 min the population had some phase-bright spores, some phase-gray spores, and some phase-dark spores (Fig. 6F). Since yrbA spores lost no significant heat resistance or dipicolinic acid content after incubation with AGFK for 90 min (data not shown), this suggests that spores with the yrbA defect have an additional early defect in germination with AGFK.
FIG. 5.
Germination of wild-type and yrbA mutant spores. Wild-type (A) and TB713 (yrbA) (B) spores were heat activated at 65°C for 15 min and either germinated in 10 mM l-alanine (○) or AGFK (□) or incubated with 10 mM Tris-HCl (▵) at 37°C.
FIG. 6.
Phase-contrast microscopy of wild-type and yrbA spores. Wild-type spores (A, C, and E) and TB713 (yrbA) spores (B, D, and F) were incubated with 10 mM Tris-HCl (pH 7.5) (A and B), 10 mM l-alanine (C and D), or AGFK (E and F) at 37°C for 90 min.
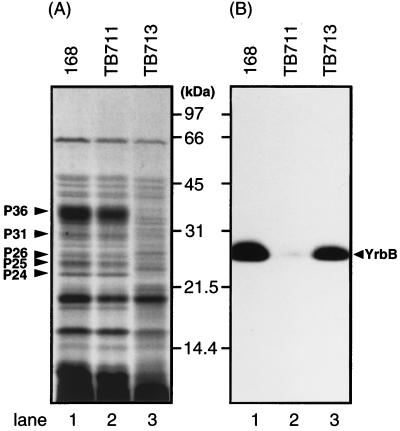
Since the reduction in lysozyme resistance of the yrbA spores implied that yrbA is involved in spore coat morphogenesis, we analyzed coat proteins by SDS-PAGE. The coat protein profile of the yrbA (TB713) mutant spores on SDS-PAGE was significantly different from that of the wild-type spores (Fig. 7A), as proteins of 24 (P24), 25 (P25), 26 (P26), 31 (P31), and 36 (P36) kDa were absent from yrbA spores. Analysis of the NH2-terminal sequence of protein P36 gave HYSHYDIEEAV, corresponding to the sequence from His-3 to Val-13 of CotG (32), while the NH2-terminal sequence of P31 was MENANYPNM, corresponding to the sequence from Met-164 to Met-172 of YrbA. YrbA is deduced to be a 43-kDa protein from its nucleotide sequence; therefore, we assume that P31 was generated by proteolysis of YrbA. We could not determine a unique N-terminal sequence for P24, P25, or P26 because these bands contained several different polypeptides.
FIG. 7.
SDS-PAGE analysis of proteins solubilized from spores. Spores were prepared from T18 sporulating cells. The protein samples were solubilized from the spores by boiling with SDS and 2-mercaptoethanol and analyzed by SDS-PAGE (12% gel). (A) Coomassie brilliant blue stain; (B) immunoblotting with anti-YrbB antibody. The protein samples were from wild-type spores (lanes 1), TB711 (yrbB) spores (lanes 2), and TB713 (yrbA) spores (lanes 3). The arrowhead in panel B indicates the migration position of YrbB.
Since yrbA is upstream of yrbB, it was possible that the yrbA mutation might affect YrbB synthesis. However, YrbB is present in yrbA mutant spores (Fig. 7B), and no visible difference between the coat protein samples prepared from wild-type and yrbB spores was seen upon SDS-PAGE (Fig. 7A, lanes 1 and 2).
Morphology of yrbA mutant spores.
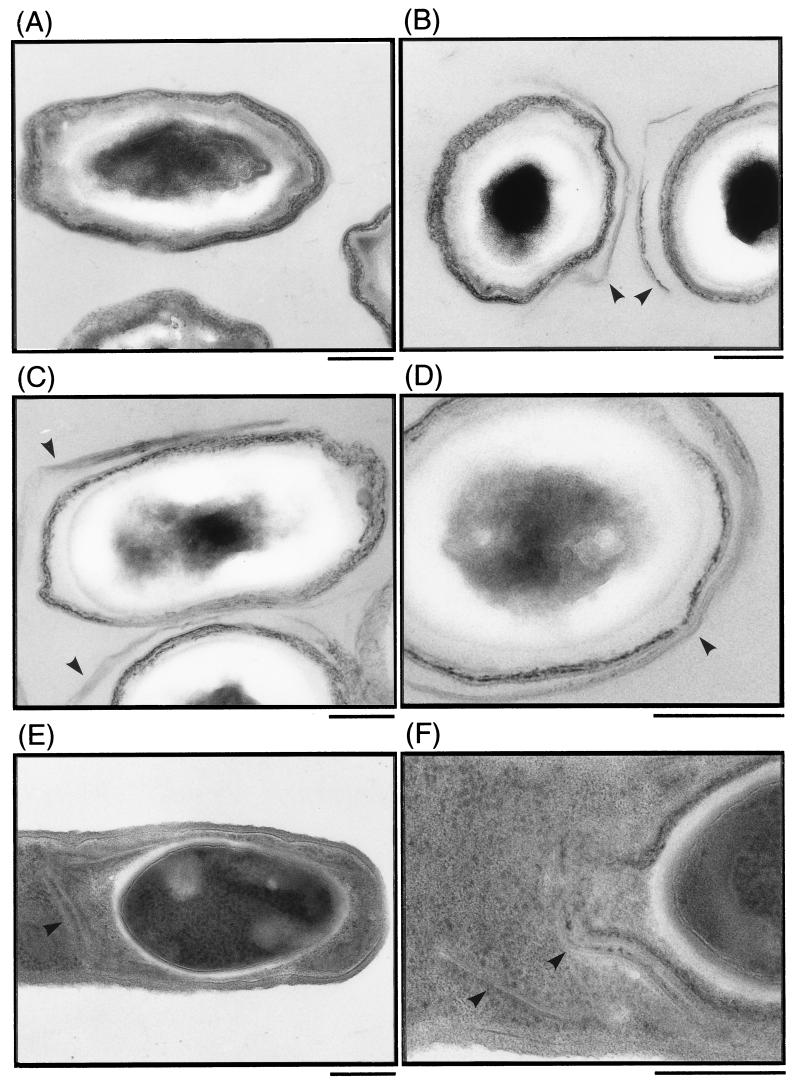
Given the spore coat defects in yrbA spores, we analyzed the ultrastructure of these spores by electron microscopy (Fig. 8). The coat of the wild-type spores has two major layers, a highly electron-dense and thicker outer coat layer and a fine lamellar inner coat layer (Fig. 8A). Some changes in coat morphology were observed in the yrbA mutant spores (Fig. 8B, C, and D); the outer layer was less electron dense and less thick (Fig. 8B and C), and a separate layer-like structure loosely surrounded the outside of the coat (Fig. 8D). The abnormal coat structure was also observed outside the developing forespores of the mutant at T8 of sporulation, and the spore coat layer(s) was partially detached from the surface of the developing forespores (Fig. 8E and F). An inner coat layer-like structure was found around the outside of an electron-dense layer of the developing forespores (Fig. 8E and F).
FIG. 8.
Ultrastructure of yrbA spores. Wild-type (A) and yrbA (B, C, and D) spores were collected at T18 of sporulation and analyzed by electron microscopy as described in Materials and Methods. Sporulating cells of the yrbA mutant were also collected at T8 of sporulation and analyzed similarly (E and F). Arrowheads indicate abnormal spore coat. Bars, 0.2 μm.
DISCUSSION
The control of yrbA by ςE is supported by two types of experiments. First, Northern blot analysis showed that transcription of yrbA mRNA was dependent on ςE and ςF but not on ςG or ςK (Fig. 2). Since activation of ςE is regulated by ςF during sporulation, expression of yrbA is under ςE or ςF control. Second, examination of the yrbA promoter revealed −35 (TCATAAC) and −10 (CATATGT) sequences separated by 14 bp; these sequences conform well to the −10 and −35 consensus sequences of many ςE-dependent promoters (7, 11, 18, 26, 28–30, 43, 48) (Fig. 3 and 4). These results indicated that transcription of yrbA is most likely regulated by RNA polymerase with ςE in the mother cell compartment and also suggest that YrbA may be a coat protein made in the mother cell. However, it is possible that RNA polymerase with ςK also recognizes the yrbA promoter to a small degree, because the yrbA promoter is similar to those of the ςK-dependent genes (Fig. 4C).
In order to assess YrbA function, we prepared an insertional yrbA mutant and purified the mutant spores by our standard method with lysozyme digestion to remove vegetative cell debris. However, during the lysozyme treatment, the yrbA spores lost their refractility, becoming phase gray after the treatment (data not shown). Lysozyme’s target site in spores is the cortex, and digestion of this structure results in loss of spore refractility. However, dormant spores generally resist lysozyme digestion because of impermeability of their complex coat structure, which is exterior to the cortex. Consequently, the lysozyme sensitivity of the yrbA spores suggested that YrbA has some effect on expression or assembly of some coat components. This was confirmed by SDS-PAGE analysis of yrbA spores (Fig. 7), as yrbA spores lacked not only YrbA but also CotG, even though Northern blot analysis showed that cotG was transcribed normally from T4 of sporulation in yrbA cells (data not shown). These data suggest that YrbA is involved in spore coat assembly but not in regulation of cot gene transcription. CotE is another protein required for morphogenesis of the coat layer of B. subtilis, and a mutation in the cotE gene results in the loss of some proteins from spore coats, with a resultant decrease of spore resistance to lysozyme (47). Electron microscopic observation also revealed that the coat layers of yrbA spores were less electron dense and less thick than coat layers in wild-type spores, and a coat layer(s) was partially detached from the surface of yrbA mutant forespores and extended into the mother cell compartment (Fig. 8). This morphological change is somewhat similar to that of a spoIVA mutant, in which abnormally assembled coat layers develop in the mother cell compartment (8, 29, 39). These results further imply that YrbA is a morphogenetic protein that is required for the assembly of protein components into the spore coats and the development of lysozyme resistance.
The yrbA mutant spores were also defective in some late step of l-alanine-induced germination and had a defect early in germination with AGFK (Fig. 5). The process of spore germination in B. subtilis requires the action of a germinant on a trigger site within the spores. B. subtilis spores germinate with l-alanine alone or with AGFK, none of whose components is germinative on its own (45, 46). Spores of mutants with mutations in the gerA operon are defective specifically in the response to l-alanine but germinate normally in AGFK (24). In contrast gerB, gerK, and fruB mutant spores germinate normally in l-alanine but are defective in germination in AGFK (16, 24). This difference suggests that the spore has two different systems for detecting these two germination signals (25). Both cotE and gerE spores also lack some spore coat proteins and have defects in spore germination, as do yrbA mutant spores (23, 47). CotE is required for the assembly of outer coat proteins, and GerE is a DNA binding protein essential for expression of some cot genes (14, 47, 49), but these proteins are thought to be neither germinant receptors nor factors which directly control the process of spore germination. We assume that YrbA is also involved in assembly of some spore coat components required for l-alanine- or AGFK-stimulated germination.
ACKNOWLEDGMENTS
We are grateful to Anne Moir for critical review and discussions, and we thank Michael G. Bramucci for critical reading of the manuscript. We thank JEOL Datum Co. (Tokyo, Japan) for technical support for electron microscopy.
This work was supported by grant JPSP-RFTF96L00105 from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Abe A, Ogawa S, Kohno T, Watabe K. Purification of Bacillus subtilis spore coat protein by electrophoretic elution procedure and determination of NH2-terminal amino acid sequences. Microbiol Immunol. 1993;37:809–812. doi: 10.1111/j.1348-0421.1993.tb01709.x. [DOI] [PubMed] [Google Scholar]
- 2.Abe A, Koide H, Kohno T, Watabe K. A Bacillus subtilis spore coat polypeptide gene, cotS. Microbiology. 1995;141:1433–1442. doi: 10.1099/13500872-141-6-1433. [DOI] [PubMed] [Google Scholar]
- 3.Aronson A I, Fitz-James P C. Structure and morphogenesis of the bacterial spore coat. Bacteriol Rev. 1976;40:360–402. doi: 10.1128/br.40.2.360-402.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne N, Fitz-James P C, Aronson A I. Structural and germination defects of Bacillus subtilis spores with altered contents of a spore coat protein. J Bacteriol. 1991;173:6618–6625. doi: 10.1128/jb.173.20.6618-6625.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutting S M, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. pp. 27–74. [Google Scholar]
- 6.Cutting S, Zheng L B, Losick R. Gene encoding two alkali-soluble components of the spore coat from Bacillus subtilis. J Baceteriol. 1991;173:2915–2919. doi: 10.1128/jb.173.9.2915-2919.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 8.Driks A, Roels S, Beall B, Moran C P, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 9.Gould G W. Mechanisms of resistance and dormancy. In: Hurst A, Gould G W, editors. The bacterial spore. Vol. 2. London, United Kingdom: Academic Press; 1983. pp. 173–210. [Google Scholar]
- 10.Haldenwang W. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay R E, Tatti K M, Vold B S, Green C J, Moran C P., Jr Promoter used by sigma-29 RNA polymerase from Bacillus subtilis. Gene. 1986;48:301–306. doi: 10.1016/0378-1119(86)90090-9. [DOI] [PubMed] [Google Scholar]
- 12.Henner D J. Inducible expression of regulatory gene in Bacillus subtilis. Methods Enzymol. 1990;185:223–228. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- 13.Herrin D L, Schmidt G W. Rapid, reversible staining of Northern blots prior to hybridization. Bio/Technology. 1988;6:196–200. [PubMed] [Google Scholar]
- 14.Holland S K, Cutting S, Mandelstam J. The possible DNA-binding nature of the regulatory proteins, encoded by spoIID and gerE, involved in the sporulation of Bacillus subtilis. J Gen Microbiol. 1987;133:2381–2391. doi: 10.1099/00221287-133-9-2381. [DOI] [PubMed] [Google Scholar]
- 15.Igo M M, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- 16.Irie R, Okamoto T, Fujita Y. A germination mutant of Bacillus subtilis deficient in response to glucose. J Gen Appl Microbiol. 1982;28:345–354. [Google Scholar]
- 17.Karow M L, Glaser P, Piggot P J. Identification of gene, spoIIR, that links the activation of sigma E to the transcriptional activity of sigma F during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel B, Kroos L, Poth H, Youngman P, Losick R. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev. 1989;3:1735–1744. doi: 10.1101/gad.3.11.1735. [DOI] [PubMed] [Google Scholar]
- 19.Kunst F, Ogasawara N, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 20.Kushida H. An improved embedding method using ERL 4206 and Quetol 653. J Electron Microsc. 1980;29:193–194. [Google Scholar]
- 21.London-Vallejo J A, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;15:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- 22.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 23.Moir A. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J Bacteriol. 1981;146:1106–1116. doi: 10.1128/jb.146.3.1106-1116.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moir A, Lafferty E, Smith D A. Genetic analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype with map location. J Gen Microbiol. 1979;111:165–180. doi: 10.1099/00221287-111-1-165. [DOI] [PubMed] [Google Scholar]
- 25.Moir A, Smith D A. The genetics of bacterial spore germination. Annu Rev Microbiol. 1990;44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- 26.Moran C P., Jr . RNA polymerase and transcription factors. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 653–667. [Google Scholar]
- 27.Nicholson W L, Setlow P. Sporulation, germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular microbiological method for Bacillus. Chichester, United Kingdom: John Wiley & Sons, Ltd.; 1990. pp. 391–450. [Google Scholar]
- 28.Rather P N, Hay R E, Ray G L, Haldenwang W G, Moran C P., Jr Nucleotide sequences that define promoters that are used by Bacillus subtilis sigma-29 RNA polymerase. J Mol Biol. 1986;192:557–565. doi: 10.1016/0022-2836(86)90276-7. [DOI] [PubMed] [Google Scholar]
- 29.Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rong S, Rosenkrantz M S, Sonenshein A L. Transcriptional control of the Bacillus subtilis spoIID gene. J Bacteriol. 1986;165:771–779. doi: 10.1128/jb.165.3.771-779.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryter A, Kellenberger E, Birch-Andersen A, Maaloe O. Etude au microscope electronique de plasmas contenant de l’acide desoxyribonucleique. I. Les nucleoides des bacteries en croissance active. Z Naturforsch. 1958;13B:597–605. [PubMed] [Google Scholar]
- 32.Sacco M, Ricca E, Losick R, Cutting S. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J Bacteriol. 1995;177:372–377. doi: 10.1128/jb.177.2.372-377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Schaefer P, Millet J, Aubert J P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott I R, Stewart G S A G, Koncewicz M J, Ellar D J, Crafts-Lighty A. Sequence of biochemical events during germination of Bacillus megaterium spores. In: Chambliss G, Vary J C, editors. Spores VII. Washington, D.C: American Society of Microbiology; 1978. pp. 95–103. [Google Scholar]
- 36.Sekiguchi J, Akeo K, Yamamoto H, Khasanov F K, Alonso J C, Kuroda A. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J Bacteriol. 1995;177:5582–5589. doi: 10.1128/jb.177.19.5582-5589.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Setlow P. Biochemistry of bacterial forespore development and spore germination. In: Levinson H S, Sonenshein A L, Tipper D J, editors. Sporulation and germination. Washington, DC: American Society for Microbiology; 1981. pp. 13–28. [Google Scholar]
- 38.Setlow P. Spore structure proteins. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 801–809. [Google Scholar]
- 39.Stevens C M, Daniel R, Illing N, Errington J. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J Bacteriol. 1994;174:586–594. doi: 10.1128/jb.174.2.586-594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stragier P, Bonamy C, Karamazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 41.Takamatsu H, Chikahiro Y, Kodama T, Koide H, Kozuka S, Tochikubo K, Watabe K. A spore coat protein, CotS, of Bacillus subtilis is synthesized under the regulation of sigmaK and GerE during development and is located in the inner coat layer of spores. J Bacteriol. 1998;180:2968–2974. doi: 10.1128/jb.180.11.2968-2974.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takamatsu H, Hiraoka T, Kodama T, Koide H, Kozuka S, Tochikubo K, Watabe K. Cloning of a novel gene yrbB, encoding a protein located in the spore integument of Bacillus subtilis. FEMS Microbiol Lett. 1998;166:361–367. doi: 10.1111/j.1574-6968.1998.tb13913.x. [DOI] [PubMed] [Google Scholar]
- 43.Taylor M. Undergraduate honors thesis. Cambridge, Mass: Harvard University; 1991. [Google Scholar]
- 44.Venkatasubramanian P, Johnstone E. Biochemical analysis of the Bacillus subtilis 1604 spore germination response. J Gen Microbiol. 1989;135:2723–2733. doi: 10.1099/00221287-135-10-2723. [DOI] [PubMed] [Google Scholar]
- 45.Wax R, Freese E, Cashel M. Separation of two functional roles of L-alanine in the initiation of Bacillus subtilis spore germination. J Bacteriol. 1967;94:522–529. doi: 10.1128/jb.94.3.522-529.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wax R, Freese E. Initiation of germination of Bacillus subtilis spores by a combination of compounds in place of L-alanine. J Bacteriol. 1968;95:433–438. doi: 10.1128/jb.95.2.433-438.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng L B, Donovan W P, Fitz-James P C, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988;2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]
- 48.Zheng L B, Losick R. Cascade regulation of spore coat gene expression in Bacillus subtilis. J Mol Biol. 1990;212:645–660. doi: 10.1016/0022-2836(90)90227-d. [DOI] [PubMed] [Google Scholar]
- 49.Zheng L, Halberg R, Roels S, Ichikawa H, Kroos L, Losick R. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J Mol Biol. 1992;226:1037–1050. doi: 10.1016/0022-2836(92)91051-p. [DOI] [PubMed] [Google Scholar]