Introduction
What do drug transporters and drug metabolizing enzymes (DMEs)--as well as the various regulatory proteins involved in the absorption, distribution, metabolism and excretion (ADME) of drugs--really do (1)? To those coming from a non-pharmaceutical background, this seems a very reasonable, if not urgent, question. What do they really do? What is their true biological function?
To understand this, we must go beyond the classic multi-specific “drug” transporters and “drug” metabolizing enzymes (2, 3). We must consider the oligo-specific and mono-specific transporters, enzymes and regulatory proteins (e.g., nuclear receptors, kinases) they often work with--in the contexts of both pharmacokinetics and endogenous metabolism. The list of proteins is quite large; a rough estimate is 500–1000 (4). In other words, perhaps as much as 5% of the human genome is dedicated to handling endogenous small molecules that bear a molecular resemblance to numerous pharmaceuticals.
Many of these proteins appear to be part of a large endogenous network, where multi-specific transporters, enzymes and nuclear receptors are hubs (4). Many gene families well-known for their roles in drug ADME (e.g., ABCC, SLC22) are evolutionarily conserved in organisms as diverse as mouse, flies and fish (5). Nevertheless, these “model” organisms, along with pre-industrial humans, are not known to wait in line at the pharmacy for their prescriptions of small molecule drugs to be filled. Moreover, it is now clear--based on human genome wide association studies (GWAS), knockout mouse metabolomics, in vitro assays, and systems biology studies--that these proteins work in concert to regulate metabolism and signaling pathways involving endogenous small molecules. Some well-established examples are the physiology of bile acids, sex steroids, urate, and uremic toxins derived from gut microbes (6). Urate, for example, is handled by a number of multi-, oligo-, and monospecific transporters in different organs. While most have very high expression in the kidney (e.g., OAT1, OAT3, URAT1, SLC2A9), the liver and intestine also express transporters that handle urate (e.g., ABCG2). This broad expression, when coupled with shared substrate specificity--and the activity of gut microbial and liver enzymes involved in purine metabolism--creates a cross-tissue network of proteins that work together to tightly regulate urate levels.
What to make of all this? Over 15 years ago--based on data from the SLC22 transporter family which contains some of the best-known “drug transporters” (e.g., OAT1, OAT3, OCT1, OCT2)--what is now known as the Remote Sensing and Signaling Theory began to be formulated (1, 6). Among the initial prompts were the discovery of alterations in endogenous metabolites in the serum of the Oat1 knockout mouse (7). These mice also had defects in the renal elimination of diuretics and other drugs. While much of the field was characterizing drugs handled by multi-specific transporters of the family, other oligo-specific and (relatively) mono-specific SLC22 family members were discovered; many of these have now been shown to be central to metabolism and signaling (8). For example, a close relative of OAT1 in mice that is mainly expressed in the olfactory system binds odorants with much higher affinity than OAT1 (9). These and other data led us to consider the endogenous functions not only of SLC22 transporters, but of all other transporters, enzymes and regulatory protein families implicated in ADME of small molecule drugs--particularly in the context of remote organ communication (1, 4).
Essential aspects of the Remote Sensing and Signaling Theory (RSST) are summarized in Table 1. The RSST is a multi-faceted general biological theory that is concerned with the roles of transporters, enzymes, nuclear receptors and other regulatory proteins in optimizing inter-organ crosstalk (e.g., gut-liver-kidney-brain) and inter-organismal communication (e.g., gut microbes-host, mother-fetus) in the service of homeostasis (Figure 1A). In this context, we consider homeostasis from the viewpoint of the organism’s ability to maintain a relatively stable environment by ensuring that small molecules remain within a certain range of concentrations in any body fluids, tissues, and cells. The Remote Sensing and Signaling System achieves this by regulating the functional expression and/or activity of key proteins that mediate net flux in and out of tissues of numerous small, endogenous, molecules with “high informational content.” These high informational content molecules include rate-limiting metabolites (e.g., Krebs cycle intermediates), signaling molecules (e.g., bile acids), redox molecules (e.g., urate), gut microbe-derived products (e.g., indole derivatives), essential nutrients and natural products (e.g., flavonoids). Remote communication refers to the movement of these small molecules with high informational content between organisms, organs, cells, and organelles, where they can modulate distinct metabolic and signaling pathways.
Table 1:
Key tenets of the Remote Sensing and Signaling Theory.
| Remote Sensing and Signaling Theory Concepts |
|---|
| Proteins: Drug and other transporters, drug metabolizing enzymes (DMEs), nuclear receptors, kinases, and other regulatory proteins |
| Endogenous small molecules with “high informational content”: rate-limiting metabolites (e.g., dicarboxylates), signaling molecules (e.g., sex steroids, prostaglandins), nutrients (e.g., pantothenic acid), antioxidants (e.g., urate), gut microbiome products (e.g., indoxyl sulfate, secondary bile acids) |
| Regulated expression of transporters on apical and basolateral surfaces of polarized epithelial tissues, such as kidney, liver, intestine, choroid plexus, retina, and placenta (as well as endothelial cells) |
| Combinatorial possibilities of multi-, oligo-, and mono-specificity to (re)optimize metabolite levels at multiple scales (organelle, cell, tissue, organ, organ system, organism, multiple organisms) |
| Inter-organ small molecule remote communication (e.g., gut-liver-kidney) |
| Inter-organismal small molecule remote communication (e.g., gut microbiome-host, mother-fetus) |
| Representation as Remote Sensing and Signaling Network consisting of ~500–1000 proteins with hubs including multi-specific transporters, DMEs, and nuclear receptors |
| DMEs play a critical role by tagging small molecules with sulfate, glucuronide, and other groups for destinations in other cells/organs with specific drug transporter expression profiles |
| Evolutionary conservation of key gene families as well as network topology |
| The Remote Sensing and Signaling System functions in parallel and in tandem with other homeostatic systems (e.g., neuroendocrine, growth factor-cytokines, autonomic nervous system) |
| The Remote Sensing and Signaling System resets after perturbation (e.g., acute and chronic organ injury, metabolic disorders) to help restore homeostasis via transcriptional, post-translational and epigenetic mechanisms |
| Endogenous small molecule homeostasis by the Remote Sensing and Signaling System (Network) is viewed in terms of activity necessary to meet tens to hundreds of small, if sometimes conflicting, biological objectives. This in turn leads to optimization of levels of hundreds to thousands of metabolites and signaling molecules in cells, tissues, and body fluids |
| Major implications: biological basis for pharmacokinetics, drug-metabolite interactions, drug-induced metabolic diseases, treatment of complex metabolic disease, pharmacokinetic and other modeling |
Figure 1:
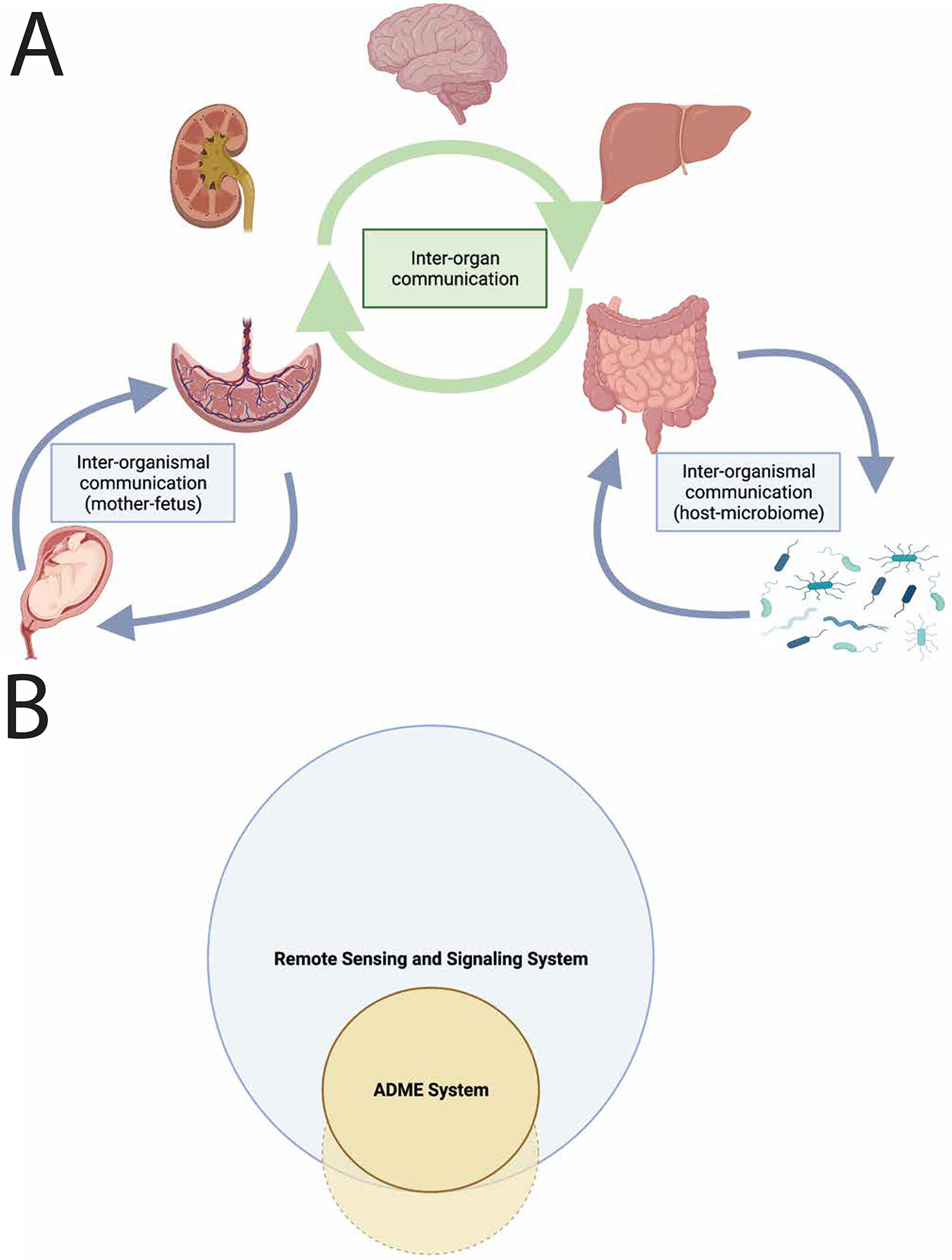
A) The Remote Sensing and Signaling System (Network) mediates inter-organ and inter-organismal communication through small molecules with “high informational content.” In healthy and diseased states, endogenous and other small molecules with key roles in metabolism, signaling and maintaining redox state are mediators of remote communication between organs. Important axes--like the brain-gut and gut-liver-kidney axes--are characterized by sets of transporters, enzymes and regulatory proteins (e.g., nuclear receptors) in each organ. Often, these multi-specific, oligo-specific and mono-specific proteins are dynamically regulated at the apical and basolateral surfaces of polarized epithelial tissues--thereby providing many combinatorial possibilities for regulating net flux between tissues, body fluids, and other interfaces. Small molecule communication mediated by transporters, enzymes and regulatory proteins of the Remote Sensing and Signaling System (Network) also occurs at the inter-organismal level, as in the case of mother-fetus and host-gut microbiome. For example, the gut microbiome-derived small molecules play important metabolic and signaling roles in the host through the activity of transporters, enzymes and other proteins of the Remote Sensing and Signaling System in the intestine, liver, kidney, brain and other remote tissues. B) The ADME system overlaps largely or completely with the greater Remote Sensing and Signaling System. The Remote Sensing and Signaling System consists of hundreds of proteins, including multi-specific, oligo-specific, and mono-specific transporter and enzymes. Within a subset of these proteins exists the ADME system, which includes what are often termed “drug” transporters and “drug” metabolizing enzymes. These systems exist simultaneously and have overlapping functionality, with drugs potentially perturbing the Remote Sensing and Signaling System. The ADME system likely also contains some proteins that are independent of the Remote Sensing and Signaling System.
The RSST emphasizes the roles of transporters, enzymes and regulatory proteins--in collaboration with other homeostatic systems like the neuroendocrine and autonomic nervous system--in the maintenance of small molecule homeostasis at multiple scales: organelle, cell, organ, multi-organ system, multiple interacting organisms. Small molecule homeostasis in this context means the regulation within a certain range of concentrations of hundreds to thousands of endogenous organic molecules (~100–1500 Da). While many of these molecules are essential for signaling, cellular metabolism, and maintenance of redox state--and have a range in which they are beneficial at one or more scales (e.g. organism, organ, organelle) --beyond this range they can cause disease. A high urate level is strongly associated with gout; the signaling molecule indoxyl sulfate is toxic in the setting of poor kidney function; carnitine deficiency can impact muscle and heart function. The RSST also emphasizes the role of the Remote Sensing and Signaling System (Network) in correcting disrupted small molecule homeostasis in pathophysiological states (e.g., acute or chronic kidney injury). This can occur, for instance, by taking advantage of the many possibilities for resetting the system through the combinatorial action of multi-, oligo-, and mono-specificity across different scales (e.g., organism, organ, organelle) in an effort to restore endogenous small molecule homeostasis (Table 1). The theory was not developed with man-made small molecule pharmaceuticals in mind, but that said, many of the proteins considered central to ADME are part of the Remote Sensing and Signaling System (Network) (4).
That begets the following questions. What is the biological basis of pharmacokinetics? Can the Remote Sensing and Signaling Theory provide such a biological basis?
One system or two?
At this point, we hope the reader sees the value in asking these questions. We hasten to emphasize that a major “focus” of the Remote Sensing and Signaling System is optimizing the levels of hundreds to thousands of endogenous small molecules necessary for proper biological functioning at one or more scales--from organism to organ to organelle. In contrast, the applied science of pharmacokinetics generally focuses on the ADME of small molecule drugs. These drugs are generally meant to be completely eliminated from the body.
Having made those distinctions, the RSST takes the view that most of those small molecule drugs are, in actuality, “probes” of the endogenous Remote Sensing and Signaling System (Network). Indeed, were it not for this relentless probing of the (endogenous) system with drugs--the careful work of numerous scientists--we might not have such a large “parts list” of the Remote Sensing and Signaling System. Or, for that matter, so much converging data on the role of “drug” transporters and DMEs in endogenous metabolism. For example, the application of multi-omics profiling to OAT1 and other “drug” transporter knockout animals probably would not have occurred so early were not these transporters of such high interest to the fields of pharmacy and pharmacology, as well as funding/regulatory agencies. It should, however, be noted that these pharmaceutical “probes” are not harmless. Drugs make use of many proteins in the Remote Sensing and Signaling System and, by doing so, can disrupt endogenous function.
If one views small molecule drugs as probes of a largely endogenous Remote Sensing and Signaling System (Network), how do we reconcile the operational pharmacokinetic data on drug ADME--which heavily emphasizes pharmaceutical metabolism/detoxification and elimination--with the essential role of the Remote Sensing and Signaling System in organ cross-talk, inter-organismal communication, and endogenous small molecule homeostasis at multiple scales?
One potential answer is that ADME of small molecules is an essential part of a larger homeostatic system concerned with the influx, transformation and efflux of endogenous small molecules. For example, indole originating from tryptophan-consuming bacteria of the gut microbiome is absorbed across the intestine and into the circulation, whereupon it is converted into indoxyl sulfate by Phase 2 DMEs in the liver. Indoxyl sulfate has the interesting property of being a deleterious uremic toxin in the setting of chronic kidney disease and a signaling molecule that, once taken up by OAT1, activates the aryl hydrocarbon receptor (AHR)--and thereby regulates metabolism (10). Assuming this type of scenario is quite common, it is conceivable that the protein network involved in small molecule drug ADME is a subset of the larger Remote Sensing and Signaling Network concerned with homeostasis of endogenous small molecules at levels ranging from organelles to multiple interacting organisms.
Another possibility is that the ADME network and Remote Sensing and Signaling Network only partly overlap (Figure 1B). That raises the question of the endogenous function of the non-overlapping part of the ADME network. If we consider pre-industrial humans, mice, flies, and fish, we will probably agree that the function is not to anticipate the development of the biopharmaceutical industry. One obvious possibility is the handling of toxic natural products. Indeed, the Remote Sensing and Signaling System handles many such products, like aristolochic acid, a kidney toxin found in certain food grains that is transported by OAT1 and OAT3 (11).
Leaving aside the question of one network or two overlapping networks, it seems likely that the need to eliminate toxic natural products, cohabit with various microbiomes--as well as the requirement of certain biochemical pathways for exogenous essential nutrients--heavily shaped a Remote Sensing and Signaling System focused on organ crosstalk and small molecule homeostasis at many scales. The RSS System is also shaped by its multiple interactions with other homeostatic systems like the neuroendocrine system (e.g., transport of thyroxine and sex steroids) and the autonomic nervous system (e.g., neurotransmitter transport).
So then, if we consider the Remote Sensing and Signaling Theory as the biological basis of small molecule drug ADME, what does that do for us? What is--if only in the sense that William James might have used--its “cash value”?
Drug-metabolite interactions (DMI) and drug-induced metabolic disease resulting from perturbation of the RSS system
The most immediate consequence of two overlapping networks may well be the competition that occurs between them. The RSST, being based in metabolism, may be particularly useful for understanding drug-metabolite interactions (DMI) not only at the transporter level but also downstream effects (1, 6). Understanding the basis of these downstream effects--with an aim toward minimizing potentially deleterious ones and maximizing beneficial ones--can influence drug development and drug repurposing.
Certain conditions require long-term drug treatment. For example, HIV antivirals must be taken for the rest of the patient’s life following diagnosis, and many of these drugs are handled by different kinds of organic anion transporters which also regulate metabolism (e.g., OATs, MRPs). HIV antiviral treatments have been associated with metabolic disorders, such as dyslipidemia, fatty liver, and others (12), suggesting that chronic disruption of key hubs in the Remote Sensing and Signaling System may be a root cause of the dysregulation. Long-term drug-metabolite interactions occurring at the site of a transporter like OAT1 may lead to a permanent shift in metabolism.
For instance, probenecid, a well-established inhibitor of three SLC22 transporters--OAT1, OAT3 and URAT1--has long been used to treat hyperuricemia as well as to therapeutically increase serum levels of certain antibiotics and antivirals. Even short-term treatment with probenecid leads to a vast number of metabolic alterations, including lipids and metabolites regulating redox state as well as energy metabolism (13). Since many other drugs, including antivirals, are known to inhibit OAT1 and OAT3, it is conceivable that, over the long term, they could play a role in drug-induced metabolic syndrome. With the current focus on the development of antiviral drugs for COVID, improving our understanding of the overlap between the Remote Sensing and Signaling System and the ADME system may help predict metabolic issues likely to arise from new antivirals drugs.
Disruptions to the Remote Sensing and Signaling System can exacerbate diseases characterized by altered endogenous metabolite handling (e.g., chronic kidney disease)
Many diseases are characterized by imbalances in metabolite handling. Take the example of chronic kidney disease (CKD), which leads to elevations in small molecule uremic toxins and urate--the latter being associated with gout, metabolic syndrome, kidney stones, and hypertension. Urate handling involves multi-, oligo- and mono-specific transporters, mainly in the kidney. When urate levels rise in CKD, intestinal ABCG2 (BCRP), a multi-specific “drug” transporter that facilitates urate movement into the gut lumen, becomes much more important (14). This example of inter-organ crosstalk--with a multi-specific intestinal transporter seeming to “rescue” the loss of multi-, oligo- and mono-specific transporters in the diseased kidney--is consistent with key concepts of the RSST.
The Remote Sensing and Signaling Theory can lead to more consistent modeling with underlying biology
Pharmacokinetic models should benefit from the inclusion of endogenous molecules and RSST concepts--if for no other reason than the constant competition (DMI) for access to transporters and enzymes in the body. However, the point of this article is not to add epicycles. If one seriously takes the RSST as a biological basis of small molecule drug ADME, a very substantial revision of pharmacokinetic models and, even more, a revision of pharmacokinetics theory, is in order.
FUNDING
This work was supported by a grant from the National Institutes of Health (NIH) to S. K. N. from the National Institute of General Medical Sciences (NIGMS) (R01GM132938). Support for J. C. G. comes from a supplement to R01GM132938. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
CONFLICT OF INTEREST
The authors report no conflicts of interest.
References
- (1).Nigam SK What do drug transporters really do? Nature Reviews Drug Discovery 14, 29–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Giacomini KM et al. Membrane transporters in drug development. Nature Reviews Drug Discovery 9, 215–36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).You G & Morris ME Drug transporters : molecular characterization and role in drug disposition. Third edition. edn. (Wiley: Hoboken, NJ, 2022). [Google Scholar]
- (4).Rosenthal SB, Bush KT & Nigam SK A Network of SLC and ABC Transporter and DME Genes Involved in Remote Sensing and Signaling in the Gut-Liver-Kidney Axis. Scientific Reports 9, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Engelhart DC et al. Systems Biology Analysis Reveals Eight SLC22 Transporter Subgroups, Including OATs, OCTs, and OCTNs. Int J Mol Sci 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Nigam SK, Bush KT, Bhatnagar V, Poloyac SM & Momper JD The Systems Biology of Drug Metabolizing Enzymes and Transporters: Relevance to Quantitative Systems Pharmacology. Clin Pharmacol Ther 108, 40–53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Eraly SA et al. Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. Journal of Biological Chemistry 281, 5072–83 (2006). [DOI] [PubMed] [Google Scholar]
- (8).Yee SW & Giacomini KM Emerging Roles of the Human Solute Carrier 22 Family. Drug Metab Dispos, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kaler G et al. Olfactory mucosa-expressed organic anion transporter, Oat6, manifests high affinity interactions with odorant organic anions. Biochem Biophys Res Commun 351, 872–6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jansen J et al. Remote sensing and signaling in kidney proximal tubules stimulates gut microbiome-derived organic anion secretion. Proc Natl Acad Sci U S A 116, 16105–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Xue X et al. Critical role of organic anion transporters 1 and 3 in kidney accumulation and toxicity of aristolochic acid I. Mol Pharm 8, 2183–92 (2011). [DOI] [PubMed] [Google Scholar]
- (12).Jain RG, Furfine ES, Pedneault L, White AJ & Lenhard JM Metabolic complications associated with antiretroviral therapy. Antiviral Res 51, 151–77 (2001). [DOI] [PubMed] [Google Scholar]
- (13).Granados JC, Bhatnagar V & Nigam SK Blockade of organic anion transport in humans after treatment with the drug probenecid leads to major metabolic alterations in plasma and urine. Clin Pharmacol Ther, In press (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bhatnagar V et al. Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: potential role of remote sensing and signaling. Clinical Kidney Journal 9, 444–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]


