Abstract
The widespread transmission and evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) call for rapid nucleic acid diagnostics that are easy to use outside of centralized clinical laboratories. Here we report the development and performance benchmarking of Cas13-based nucleic acid assays leveraging lyophilised reagents and fast sample inactivation at ambient temperature. The assays, which we named SHINEv.2 (for ‘streamlined highlighting of infections to navigate epidemics, version 2’), simplify the previously reported RNA-extraction-free SHINEv.1 technology by eliminating heating steps and the need for cold storage of the reagents. SHINEv.2 detected SARS-CoV-2 in nasopharyngeal samples with 90.5% sensitivity and 100% specificity (benchmarked against the reverse transcription quantitative polymerase chain reaction) in less than 90 min, using lateral-flow technology and incubation in a heat block at 37 °C. SHINEv.2 also allows for the visual discrimination of the Alpha, Beta, Gamma, Delta and Omicron SARS-CoV-2 variants, and can be run without performance losses by using body heat. Accurate, easy-to-use and equipment-free nucleic acid assays could facilitate wider testing for SARS-CoV-2 and other pathogens in point-of-care and at-home settings.
Frequent and widespread testing is critical to prevent and respond to infectious disease outbreaks. For example, large-scale testing to track the prevalence and transmission of SARS-CoV-2 has been essential in managing the ongoing coronavirus disease 2019 (COVID-19) pandemic1,2. Ubiquitous and frequent diagnostic testing leads to the rapid identification of new cases and permits the swift treatment or isolation of infected individuals, thereby preventing further viral spread3. However, the gold standard for COVID-19 diagnosis—reverse transcription quantitative polymerase chain reaction (RT-qPCR)—remains suboptimal for orchestrating this response. RT-qPCR requires specialized equipment and expertise rarely found outside of centralized laboratories. In addition, an insufficient testing infrastructure coupled with reagent shortages and high testing demand have led to slow sample-to-answer times (often of several days) for RT-qPCR4,5. Alternative diagnostic technologies that enable rapid and decentralized testing are vital to respond to the current and future pandemics.
Lateral-flow antigen-capture tests and isothermal nucleic acid diagnostics with visual readouts represent promising alternatives for SARS-CoV-2 testing outside of centralized laboratories. The effectiveness of such tests in curbing the spread of SARS-CoV-2 has been demonstrated in multiple settings, ranging from nursing homes to whole countries2,6. Antigen-capture tests are quick and user-friendly, but their moderate sensitivity means they can miss potentially infectious individuals7-9. Isothermal nucleic acid amplification methods, such as loop-mediated isothermal amplification (LAMP), are more sensitive than antigen-capture tests and operate at a single temperature10,11. The deployment of LAMP-based tests has been facilitated by the use of auxiliary devices to eliminate the need for intensive nucleic acid purification and maintain a stable temperature during amplification12. However, these devices are often too costly for single use and are difficult to manufacture at the scale required for population-wide distribution, which limits their utility for frequent and widespread testing13. Hence, the continued development and deployment of user-friendly, sensitive and equipment-free diagnostics are key to enhancing the public health response to COVID-19.
CRISPR-based diagnostics (CRISPR-Dx) are promising technologies for SARS-CoV-2 testing with minimal equipment requirements. CRISPR-Dx usually combine isothermal nucleic acid amplification methods (often LAMP or recombinase polymerase amplification (RPA)) with an RNA-guided CRISPR-Cas nuclease (usually Cas12 or Cas13) for pathogen detection14-17. Detection of an amplified target nucleic acid triggers cleavage of a reporter molecule by the CRISPR-Cas enzyme. This reporter cleavage can be detected by the appearance of visual bands on a lateral-flow strip, among other methods17,18. Pairing isothermal nucleic acid amplification methods with Cas nuclease detection considerably enhances specificity and sensitivity, but at the expense of increasing assay complexity. Some groups have simplified the workflow by eliminating the need for amplification using more active Cas enzymes, although they rely on custom-built equipment and nucleic acid extraction19,20. Several groups have combined multiple enzymatic steps into individual reactions to streamline CRISPR-Dx21,22. We previously developed SHINE, henceforth SHINEv.1, a diagnostic assay that does not require nucleic acid extractions or custom equipment22. However, SHINEv.1 involves multiple heating steps, and uses reagent mixes that require cold temperature storage and manual preparation by trained personnel. Further simplifications and technical improvements to CRISPR-Dx would greatly facilitate test distribution and expand their use cases.
Frequent and widespread testing is even more important with the rise of highly transmissible SARS-CoV-2 variants. In recent months, the evolution of SARS-CoV-2 has led to the emergence and sustained transmission of viral variants with sets of mutations that can increase infectivity or reduce neutralization by antibodies, making the virus harder to contain23. The World Health Organization (WHO) has designated some of the SARS-CoV-2 variants with increased transmissibility, virulence and/or antigenicity as Variants of Concern (VOCs). Notably, the Alpha, Beta, Gamma, Delta and Omicron VOCs (designated B.1.1.7, B.1.351, P.1, B.1.617.2 and BA.1 using the PANGO nomenclature system, respectively) are widely circulating in many countries around the world and have placed unprecedented strain on several healthcare systems24,25. Surveillance of SARS-CoV-2 VOCs is vital to inform public health and clinical decisions, such as for prioritizing testing or vaccination in the populations most heavily impacted by the pandemic. Currently, the identification of viral mutations and the tracking of circulating SARS-CoV-2 variants are performed by viral genomic sequencing and a small number of mutation-specific RT-qPCR tests26-28. However, the lack of infrastructure and expertise have made routine genomic surveillance difficult to perform outside of specialized genomic centres29.
CRISPR-Dx are uniquely suited to detect VOCs for routine surveillance applications. In contrast to available point-of-need tests, CRISPR-Dx can easily detect individual SARS-CoV-2 mutations. They do so by measuring relative signal intensities using a set of two CRISPR RNAs (crRNAs) designed against the variant region14,18,30. So far, only a few CRISPR-Dx have been developed for the identification of single or multiple nucleotide substitutions present in various SARS-CoV-2 VOCs, but none of them are suitable for point-of-care (POC) deployment31-33. These tests consist of multiple liquid-handling steps or rely on custom-built equipment32,33. The development of equipment-free and user-friendly CRISPR-Dx assays to distinguish SARS-CoV-2 VOCs would fill a major unmet need in COVID-19 diagnostics.
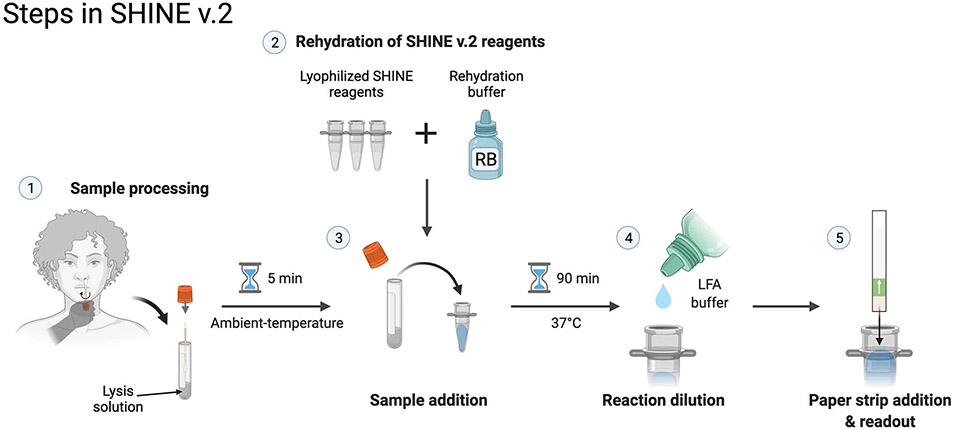
Here we develop SHINEv.2, a fast, user-friendly and widely deployable technology for the detection of SARS-CoV-2 and its variants in clinical samples. We improve upon SHINEv.1 by incorporating a fast and ambient-temperature sample lysis procedure and enabling the lyophilisation of the assay reagents. These improvements considerably reduce assay complexity and facilitate its distribution. We demonstrate that SHInEv.2 can detect SARS-CoV-2 RNA in unextracted patient samples with 90.5% sensitivity and 100% specificity compared with the gold standard RT-qPCR. Moreover, we design and validate SHINEv.2 assays to discriminate the Alpha, Beta, Gamma, Delta and Omicron VOCs in clinical samples. Finally, we introduce a Cas12-based human RNase P control, further simplify the lateral-flow assay and examine the performance of SHINEv.2 at ambient temperature. These additional advances could expand the use cases of SHINEv.2 to virtually any location, including at-home settings.
Results
Design of SHINE assay for SARS-CoV-2 detection – S gene assay.
We previously developed a SHINEv.1 assay to detect SARS-CoV-2 open reading frame 1a (ORF1a) directly from clinical samples22. We wondered whether the performance of SHINE could be further enhanced through the selection of target sites with more active RPA primers and crRNAs. Using ADAPT34—a genomics-informed computational design tool for nucleic-acid-based diagnostics—on 308,315 SARS-CoV-2 genomes collected before February 2021, we identified RPA primers and 3 crRNAs with high predicted activity in detecting the S gene of SARS-CoV-2. We selected the most active crRNA by measuring Cas13 cleavage activity on synthetic RNA targets without previous amplification (Supplementary Fig. 1) and optimized the RPA primer concentration to create the S gene assay. We performed bioinformatic analysis to ensure that the crRNA and RPA primer sequences were highly conserved across SARS-CoV-2 genomes. The crRNA sequence was fully conserved in 99.79% of the 308,315 SARS-CoV-2 genomes analysed and the RPA primers were predicted to amplify 99.77% of the genomes. Compared with the ORF1a assay, the S gene assay was found to be 1–2× faster and 10× more sensitive (Supplementary Fig. 2). Therefore, given its increased sensitivity and favourable kinetics, we selected the S gene assay for further development.
Improving the accessibility of SHINEv.1.
For SARS-CoV-2 diagnostic testing to occur in virtually any location, improvements were needed to increase user-friendliness and facilitate test distribution. Towards this goal, we developed SHINEv.2, which incorporates ambient-temperature sample processing and lyophilised testing reagents, thereby greatly simplifying reagent transport and storage and decreasing the overall complexity of the assay.
To be an effective approach, extraction-free sample processing must inactivate nucleases and infectious viral particles, and must be compatible with the assay, allowing for good diagnostic performance. Previously, we circumvented the need for clinical sample extraction using HUDSON, a nuclease inactivation and viral lysis method that relies on heat and chemical reduction18,22. To eliminate HUDSON’s need for heating elements and incubation at two temperatures, we sought to find an ambient temperature alternative for sample processing. FastAmp Viral and Cell Solution for COVID-19 testing (Intact Genomics) functions at ambient temperatures and has been reported to be compatible with isothermal nucleic acid amplification methods35. Therefore, we investigated whether this lysis reagent was also compatible with Cas13-based detection methods.
We assessed the ability of the FastAmp lysis reagent to inactivate RNases in contrived nasal samples. Our contrived samples consisted of 10% (v/v) healthy nasal fluid in universal transport medium (UTM). After treatment, we added RNaseAlert, a reporter that fluoresces in the presence of RNase activity (Fig. 1a). Without treatment, contrived nasal samples contained high levels of RNases as indicated by the high fluorescence. Treatment with FastAmp lysis reagent supplemented with 5% RNase inhibitors decreased the RNase activity by over 85% in contrived nasal samples (Fig. 1a). In addition, the ratio of nasal fluid to UTM in clinical samples derived from nasal swabs is probably considerably lower than 10% and therefore, the residual RNase activity in lysed patient samples is expected to be even lower. Moreover, we assessed the ability of the FastAmp lysis reagent to inactivate RNases in saliva. Similar to the results in contrived nasal samples, treatment of pooled healthy saliva samples with FastAmp lysis reagent supplemented with 5% RNase inhibitors decreased the RNase activity by over 85% (Supplementary Fig. 3).
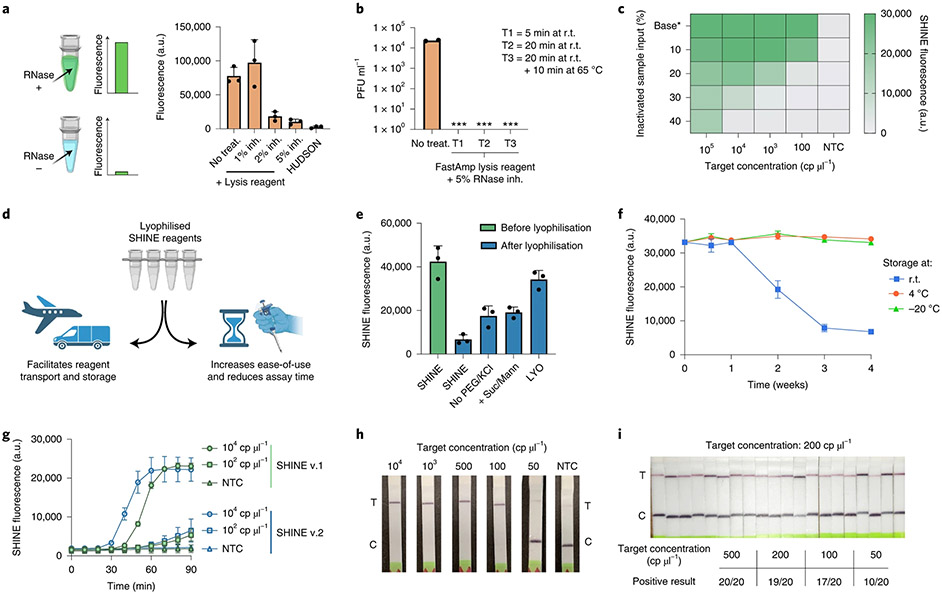
Fig. 1 ∣. Increasing the ease-of-use and deployability of SHINE.
a, RNase activity in nasal fluid mixed with UTM untreated or treated with FastAmp lysis reagent supplemented with RNase inhibitor (inh.) or treated with HUDSON (a heat- and chemical-treatment). Activity measured using RNaseAlert at r.t. for 30 min. b, SARS-CoV-2 seedstock titre without treatment or after being incubated with lysis reagent (+5% RNase inh.) at r.t. for 5 min, 20 min or 20 min+10 min at 65 °C. ***, infection not detected; PFU, plaque forming units. c, SHINE fluorescence with different proportions of blank inactivated sample input (that is, FastAmp lysis reagent, RNase inh. and UTM) after a 90 min incubation. Base*, baseline (that is, no inactivated sample added). d, Schematic of the advantages of lyophilising SHINE. e, SHINE fluorescence on synthetic RNA target (104 cp μl−1) before and after lyophilisation using different buffers. Fluorescence measured after 90 min. For buffer composition, see Methods. f, SHINE fluorescence after a 90 min incubation using lyophilised (LYO) reagents stored at r.t., 4 °C or −20 °C over time. Target concentration: 104 cp μl−1. g, Fluorescence kinetics for SHINEv.1 and SHINEv.2 using synthetic RNA targets; NTC, no target control. h, Lateral-flow detection of SARS-CoV-2 RNA in lysis buffer-treated viral seedstocks using SHINEv.2 after a 90 min incubation. C, control band; T, test band. i, Determination of analytical limit of detection with 20 replicates of SHINEv.2 at different concentrations of SARS-CoV-2 RNA from lysis reagent-treated contrived samples incubated for 90 min. Mean ± s.d. for 3 technical replicates for a, e and g; 2 technical replicates for b; 3 biological replicates with 3 technical replicates each for f. In c, heatmap values are the means of 3 technical replicates.
Having determined the nuclease inactivation capacity of this lysis solution, we tested its ability to inactivate SARS-CoV-2 viral seedstocks. For each treatment, we quantified the amount of viable virus after two passages using a plaque-based assay (Fig. 1b). Incubation of SARS-CoV-2 with the lysis solution for as little as 5 min at room temperature (r.t., 25 °C) yielded no discernible plaques, thereby confirming the ability of our lysis solution to swiftly inactivate the virus.
We then examined the compatibility of this lysis solution with good SHINE assay performance. Solution-based sample processing methods often introduce an upper bound of inactivated sample input that can be added, which is dictated by the balance between increased sample volume and decreased assay performance caused by the lysis reagents. Therefore, we evaluated the inhibitory effect of the lysis reagents by supplementing these SHINE reactions with different proportions of blank inactivated sample inputs (that is, contrived nasal samples treated with lysis solution) while controlling for target RNA concentration (Fig. 1c). An input of 10% (v/v) inactivated sample into SHINE was able to retain full activity (despite a 3–12% decrease in assay kinetics) while still doubling the amount of sample added with respect to the previous instantiation of SHINE, which used 5% (v/v) inactivated sample22 (Supplementary Fig. 4). Larger input volumes, which would increase the volume of clinical sample added, led to a marked decrease in the sensitivity of SHINE. (Fig. 1c and Supplementary Fig. 4). Thus, our lysis procedure—10% (v/v) input of sample treated by FastAmp lysis reagent with 5% RNase inhibitor for 5 min—is compatible with SHINE and can effectively inactivate nucleases and SARS-CoV-2 virions, making it ideal for rapid and equipment-free sample processing at ambient temperature.
Lyophilisation can facilitate the transport and storage of point-of-care CRISPR-Dx by eliminating the need for a cold chain and increasing shelf life (Fig. 1d). Lyophilisation also simplifies reaction setup by allowing the full reaction to be easily reconstituted by the end user. Initially, lyophilisation with the original SHINE buffer led to a profound drop in activity post-lyophilisation (Fig. 1e). We sought to improve the lyophilisation process through the addition of non-reducing disaccharides (such as sucrose), which can act as stabilizers, the addition of bulking agents (such as mannitol) and the removal of potentially destabilizing components, such as polyethylene glycol (PEG) and potassium chloride (KCl). Specifically, the removal of 3.5% PEG-8000 and 60 mM KCl from the original SHINE buffer or the addition of 5% (w/v) sucrose and 150 mM mannitol each moderately increased SHINE’s activity post-lyophilisation. With these modifications combined (that is, LYO buffer), SHINE retained its limit of detection (LoD) after lyophilisation despite a reduction in maximal fluorescence at lower target RNA concentrations (Fig. 1e and Supplementary Fig. 5).
Having optimized the lyophilisation excipients and procedure, we conducted a month-long experiment to determine the stability of the lyophilised SHINE reagents at different temperatures over time. After being stored at r.t., 4 °C or −20 °C for 4, 7, 14, 21 or 28 d, SHINE pellets were rehydrated and tested on full-genome synthetic RNA standards (Twist Bioscience). SHINE pellets retained full activity for >1 week at r.t. and at least a month at 4 °C/−20 °C (Fig. 1f). To evaluate the performance of SHINEv.2 after long-term storage, we performed SHINEv.2 tests 5 months after lyophilisation. When tested over a range of SARS-CoV-2 RNA concentrations in triplicate, we found the sensitivity of lyophilised SHINEv.2 pellets to be unaffected after 5 months in storage (Supplementary Fig. 6). By enabling transport and storage at above-freezing temperatures (4 °C), lyophilised SHINE greatly simplifies distribution logistics.
Our SHINEv.2 technology, which incorporates a new sample lysis procedure and lyophilisation into SHINEv.1, can sensitively detect nucleic acids with a simple workflow. SHINEv.2 substantially reduced the hands-on time and liquid-handling steps needed from the user, from over 45 min and 20 pipetting steps in SHINEv.1 to less than 10 min and 5 user operations in SHINEv.2 (Extended Data Fig. 1). When tested side-by-side with SHINEv.1, we found the sensitivity and reaction kinetics of the two methods to be equivalent at multiple target concentrations (Fig. 1g). SHINEv.2 detected down to 100 copies per microliter (cp μl−1) within 90 min, using either a plate-based fluorescence readout or a colorimetric lateral-flow-based readout (Fig. 1g,h). Moreover, we evaluated the impact of transportation and storage on SHINEv.2 by comparing its performance over time in a second laboratory in Nigeria. Six weeks after production and transportation to a different continent, SHINEv.2 was shown to detect down to 100 cp μl−1 of full-genome synthetic RNA standards using a paper-based readout (Supplementary Fig. 6).
We sought to determine the analytical LoD of SHINEv.2 following the guidance released by the US Food and Drug Administration (FDA)36. Under this guidance, the LoD is defined as the lowest concentration at which at least 19/20 technical replicates are detected. Using SHINEv.2 in lateral-flow format, we tested 20 replicates of a dilution series of heat-inactivated SARS-CoV-2 viral seedstock in UTM. We determined the analytical LoD of SHINEv.2 to be 200 cp μl−1 (Fig. 1i and Supplementary Fig. 7), which is over an order of magnitude lower than that of state-of-the-art antigen-capture tests and comparable to that of some equipment-based isothermal nucleic acid tests37,38. Given the optimal performance of SHINEv.2 on contrived SARS-CoV-2 samples, we sought to determine its performance on clinical samples.
Assessing the clinical performance of SHINEv.2 on nasopharyngeal swab samples.
We assessed the clinical performance of SHINEv.2 on unextracted nasopharyngeal swab samples, using the US Center for Disease Control and Prevention (CDC)-recommended RT-qPCR protocol for SARS-CoV-2 as the gold-standard reference test39. We conducted side-by-side testing on a set of 72 nasopharyngeal swab samples from SARS-CoV-2-positive and negative patients, with a viral load distribution (median viral load of ~107 cp ml−1) representative of the general population40,41 (Supplementary Table 1). SHINEv.2 detected SARS-CoV-2 RNA in 38 of the 42 RT-qPCR-positive samples (90.5% sensitivity) and in none of the RT-qPCR-negative samples (100% specificity) (Fig. 2a,b and Supplementary Fig. 8).
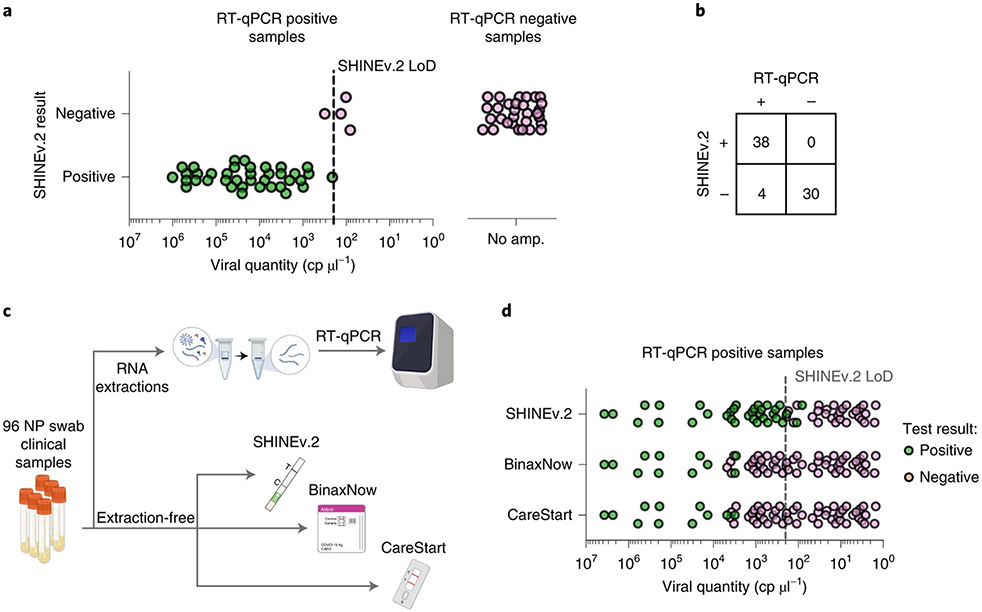
Fig. 2 ∣. Performance of SHINEv.2 on clinical samples.
a, Positive and negative SHINEv.2 test results for RT-qPCR-positive and negative clinical samples relative to viral RNA concentration. b, Concordance between SHINEv.2 and RT-qPCR for 72 nasopharyngeal swab samples. c, Schematic of side-by-side clinical sample testing using SHINEv.2, BinaxNow, CareStart and RT-qPCR. NP, nasopharyngeal swab. d, Positive and negative test results for SHINEv.2, BinaxNow and CareStart tests for RT-qPCR-positive clinical samples relative to viral RNA concentration.
While equipment-reliant RT-qPCR is the gold standard for sensitivity, we also wanted to compare the clinical performance of SHINEv.2 with two widely used FDA emergency use authorized (EUA) antigen-capture tests, as the gold standards in terms of user-friendliness and deployability. First, Abbott’s BinaxNow COVID-19 antigen self-test (BinaxNow), which is the most widely used rapid test in the United States and represents the most user-friendly of antigen-capture tests42. Second, Access Bio’s CareStart COVID-19 antigen test (CareStart), which has the most similar workflow to SHINEv.2 and comparable performance to BinaxNow43.
We performed SHINEv.2 testing side-by-side with the RT-qPCR, BinaxNow and CareStart tests in an additional set of 96 unextracted nasopharyngeal swab samples (Fig. 2c). RT-qPCR detected SARS-CoV-2 RNA in 63 samples, with cycle threshold (Ct) values ranging from 14.7 to 37.2 (Supplementary Table 1). SHINEv.2 was 50 times more sensitive than either antigen-capture test, albeit with a slower sample-to-answer time42-45 (Fig. 2d and Supplementary Figs. 9-11). SHINEv.2 excelled at detecting samples with moderate viral loads (25–30 Ct), thereby allowing the identification of potentially infectious individuals that antigen-capture tests would miss7-9. Notably, the viral load distribution of this sample set was 50–100 times lower than expected in the general population (median viral load of ~1.9 × 105 cp ml−1), which was useful to ascertain the clinical LoD40,41 of SHINEv.2. Our assay correctly identified all positive samples with titres above our analytical LoD of 200 cp μl−1 and several samples below this LoD, which confirmed that the clinical performance of SHINEv.2 is equivalent to its analytical performance.
SHINEv.2, BinaxNow and CareStart were all highly specific, each having no false-positive results in any of the 33 RT-qPCR-negative samples (100% specificity, Supplementary Figs. 10-13). This is consistent with previously reported results for these antigen-capture tests42,43 and is also expected for CRISPR-Dx, given that the RPA primers during amplification and the crRNA during Cas13-based detection provide two layers of specificity14. Consequently, the sensitivity and specificity of SHINEv.2 on patient samples, together with its user-friendliness, make this test especially valuable for community surveillance testing.
Design and testing of SHINEv.2 assays for SARS-CoV-2 VOCs.
The emergence of SARS-CoV-2 variants with increased transmissibility and virulence, and the specificity and flexibility of SHINEv.2 prompted us to develop assays to detect and distinguish five widely circulating VOCs. Initially, we focused on detecting the 69/70 deletion in the S gene, a hallmark of the Alpha VOC23,46. Then, we expanded to the K417N and K417T mutations found in the Beta and Gamma VOCs, respectively, as well as the L452R mutation and 156/157 deletion + R158G mutation present in the Delta VOC23,46,47. Finally, in light of the rampant spread of the Omicron VOC in many countries around the world, we sought to rapidly develop new SHiNev.2 assays to detect the 142–144 deletion + Y145D mutation present in this variant48. We designed sets of crRNAs with differential activities on the reference RNA genome (ancestral target) with respect to the RNA genome containing a given mutation (derived target), or vice versa (Fig. 3a).
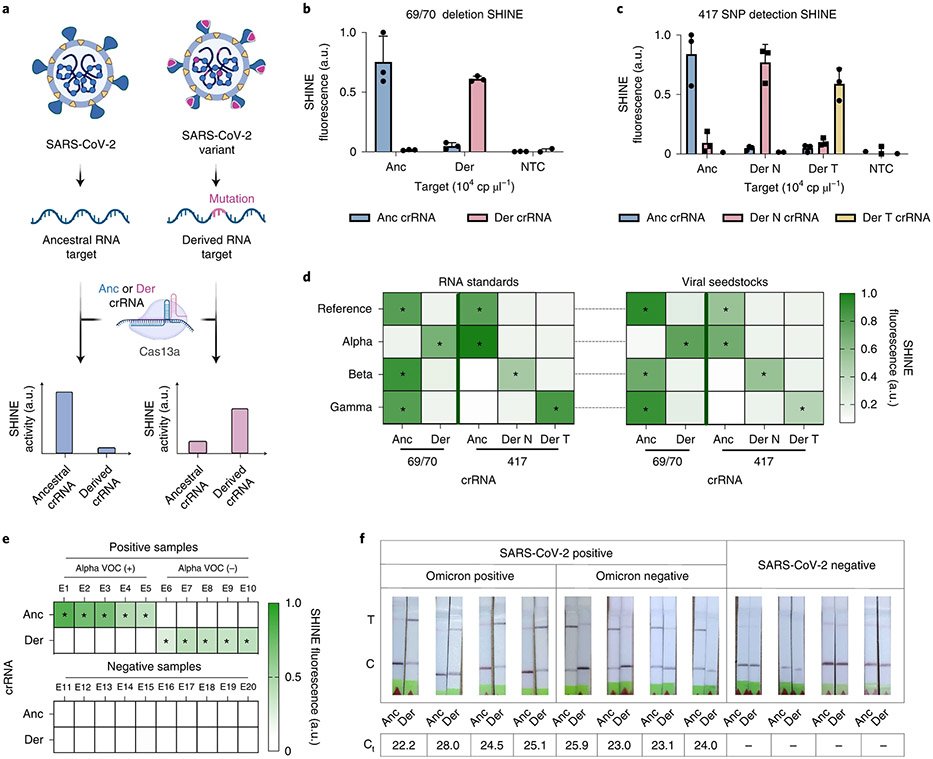
Fig. 3 ∣. Development of SHINEv.2 assays for the detection of SARS-CoV-2 VOCs.
a, Schematic of Cas13a-based detection of mutations in SARS-CoV-2 using a fluorescent readout; anc, ancestral; der, derived. b,c, SHINE fluorescence of the ancestral and derived crRNAs for the 69/70 deletion assay (b) and the 417 mutation assay (c) on synthetic RNA targets after 90 min; der N, derived N (K417N); der T, derived T (K417T). d, Identification of SARS-CoV-2 VOCs using SHINE fluorescence on full-genome synthetic RNA standards and RNA extracted from viral seedstocks; target RNA concentration: 104 cp μl−1. e, Mean fluorescence of 69/70 SHINEv.2 assay on SARS-CoV-2 RNA extracted from clinical samples after 90 min. f, Lateral-flow detection of SARS-CoV-2 RNA and VOC discrimination with SHINEv.2 Omicron assay on unextracted clinical samples after a 90 min incubation. RT-qPCR cycle threshold (Ct) specified for each sample; –, no amplification detected (SARS-CoV-2-negative sample). For b and c, mean ± s.d. for 3 technical replicates. In d, e and f, the heatmap values represent the mean for 3 technical replicates; *, positive signal expected.
For the Alpha VOC, we manually designed a set of 10 ancestral and derived crRNAs to detect the 69/70 deletion. On the basis of Cas13 cleavage activity and the ability to discriminate between ancestral and derived RNA targets, we selected Anc 1 and Der 2 as the ancestral and derived crRNAs for identifying the 69/70 deletion (Supplementary Fig. 12). The 69/70 deletion falls within the region amplified by the RPA primers from the S gene assay we previously developed (Supplementary Fig. 13). Using these primers, these assays successfully detected the 69/70 deletion in synthetic RNA targets (Fig. 3b and Supplementary Fig. 14). For example, on the ancestral RNA target, using the ancestral crRNA led to 7× greater fluorescence than using the derived crRNA (Fig. 3b). Conversely, on the derived RNA target, we observed 5× higher fluorescence with the derived crRNA than with the ancestral crRNA.
Next, we sought to design additional assays for the identification of the Beta and Gamma VOCs. These variants contain distinct single nucleotide polymorphisms (SNPs) at amino acid position 417 in the S gene (that is, K417N for Beta and K417T for Gamma), none of which are present on the reference genome or the Alpha VOC23,46. We used a computational design technique (see Methods; Mantena S. et al., manuscript in preparation) to generate crRNAs that maximize predicted SNP discrimination at position 417, and used ADAPT34 to help in designing RPA primers for these crRNAs. After assay optimization, we demonstrated that our 417 assays can differentiate between the three genotypes with high accuracy (Fig. 3c and Supplementary Fig. 15). When tested on synthetic RNA targets, each 417 assay was shown to be at least 3× more active (that is, higher fluorescence) on its preferred target than on the non-preferred targets (Fig. 3c).
Having demonstrated the sensitivity and specificity of our 69/70 and 417 assays on short synthetic RNA targets, we next tested their performance on full-genome synthetic RNA standards and on RNA extracted from SARS-CoV-2 viral seedstocks. In each case, these assays correctly identified the presence or absence of their respective mutations (Fig. 3d). For the Alpha VOC, the fluorescence signal of the derived crRNA was over 3× higher than that of the ancestral crRNA, indicating the presence of the 69/70 deletion. Conversely, the fluorescence signal for the ancestral crRNA was 1.25–3.5× higher for the reference strain as well as the Beta and Gamma VOCs, which confirmed the absence of the S gene deletion in these cases. Similarly, the genotype of each variant was correctly identified with the 417 assays, based on which crRNA showed the highest activity (Fig. 3d). For example, for the Gamma VOC, the derived T crRNA led to the highest fluorescence level, thereby correctly identifying the presence of the K417T SNP in this variant. With a fluorescence-based readout in a plate reader, these VOC assays could bring rapid VOC identification capacity to primary healthcare facilities.
Given the promising performance of these assays, we sought to assess their capacity to detect SARS-CoV-2 VOCs in clinical samples. We assessed the clinical performance of the 69/70 assay on RNA extracted from 20 nasopharyngeal swab samples. SHINEv.2 correctly identified the presence of the 69/70 deletion in the Alpha VOC-positive samples (Ct values, 23–29), as indicated by the higher fluorescence with the derived crRNA in samples E6 to E10 (Fig. 3e). In addition, the 69/70 deletion assay showed excellent specificity (100%), detecting all RT-qPCR-positive samples and none of the RT-qPCR-negative samples (Fig. 3e).
We further assessed the performance of the 69/70 assay on contrived samples. To emulate unextracted nasopharyngeal swab samples, we spiked in heat-inactivated viral seedstocks (reference strain or Alpha VOC) into SARS-CoV-2-negative clinical samples. Using a colorimetric lateral-flow-based readout, SHINEv.2 correctly identified the presence or absence of the alpha VOC in the range of viral RNA concentrations (103–105 cp μl−1) tested (Supplementary Fig. 16). The presence of a test band for the ancestral crRNA and the lack of it for the derived crRNA confirmed the presence of the 69/70 deletion in Alpha VOC-positive contrived samples with lower viral RNA concentrations (103 and 104 cp μl−1). At higher viral RNA concentrations (105 cp μl−1), the presence of the deletion was determined by the difference in test band intensity between the ancestral and derived crRNAs. Altogether, these data indicate that the 69/70 SHINEv.2 assay is well poised to identify SARS-CoV-2 VOCs in clinical samples at the POC.
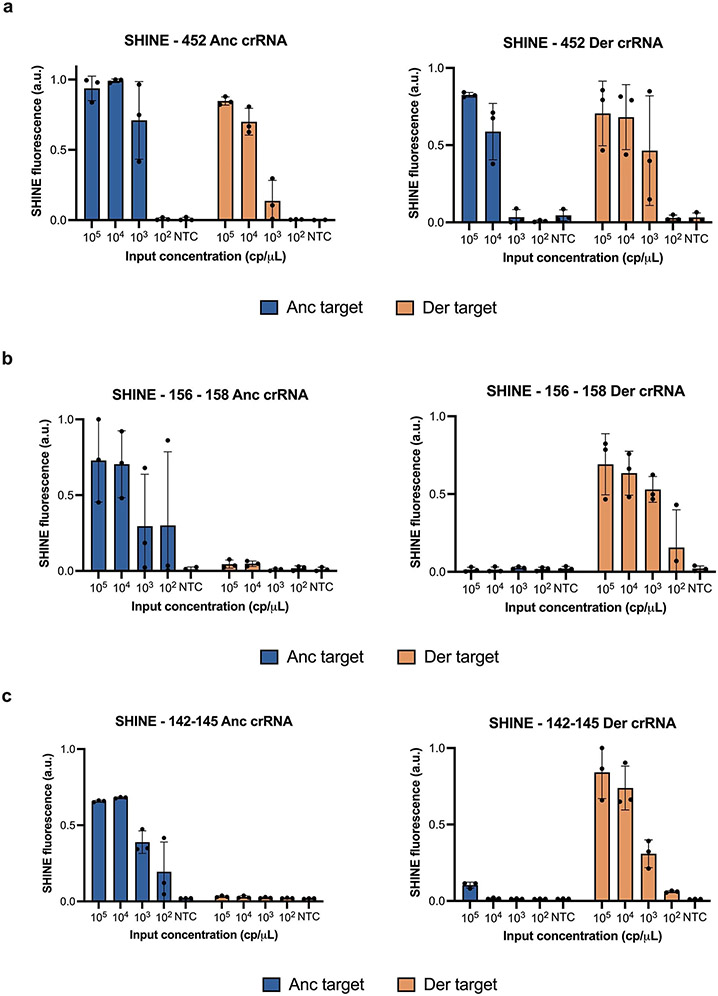
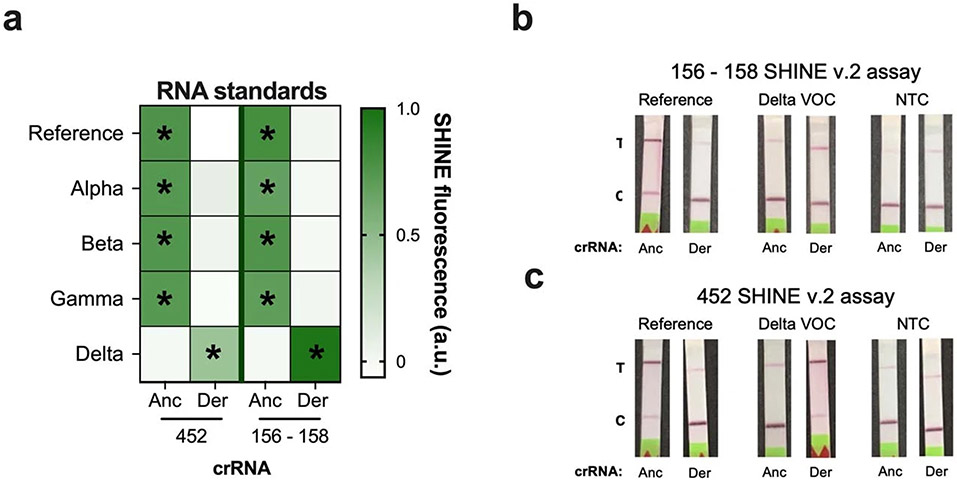
SHINEv.2 is a flexible technology that can be rapidly adapted to detect new viral variants. This allowed us to develop new SHINEv.2 assays for the 452 and 156–158 mutations present in the Delta VOC, a VOC that arose as we were preparing this manuscript for the initial submission. We designed a set of crRNAs and RPA primers for each assay as described above, and evaluated their performance on full-genome synthetic RNA standards. Both assays successfully detected the mutations in synthetic RNA targets with high sensitivity and specificity (Extended Data Fig. 2a,b). The 156–158 assay, in particular, displayed excellent specificity. Both the ancestral and derived crRNAs detected their respective targets with high accuracy while showing no detection for the opposite target (Extended Data Fig. 2b). Next, we examined the compatibility of these assays with lyophilisation and ambient-temperature sample processing. These SHINEv.2 assays showed excellent specificity on full-genome synthetic RNA standards, clearly distinguishing the Delta VOC from the reference strain as well as from the other VOCs (Extended Data Fig. 3a). The 452 and 156–158 assays correctly identified the genotype of each variant tested, on the basis of the differential fluorescence activity of each crRNA pair. Additionally, both SHINEv.2 assays correctly identified the presence or absence of each mutation using a colorimetric lateral-flow-based readout (Extended Data Fig. 3b,c).
As we were revising the manuscript for publication, a new SARS-CoV-2 VOC arose (Omicron) and quickly spread throughout the world, even among highly vaccinated populations. Given the fast transmission of Omicron, we sought to rapidly develop an Omicron-specific SHINEv.2 assay. Within 12 d, we designed a set of crRNAs and RPA primers for the 142–145 mutation present in the Omicron VOC, optimized the Omicron-specific assay and evaluated its performance on unextracted clinical samples (Fig. 3f and Extended Data Fig. 2c). When tested on 12 unextracted nasopharyngeal swab samples, the Omicron-specific SHINEv.2 assay correctly identified the presence of the 142–145 mutation in all the Omicron VOC-positive samples, as indicated by the darker band in the paper strip containing the derived crRNA (Fig. 3f). In addition, the Omicron-specific SHINEv.2 assay detected the absence of the 142–145 mutation in all Omicron-negative samples and showed excellent specificity (100%), detecting all RT-qPCR-positive samples and none of the RT-qPCR-negative samples (Fig. 3f). All in all, we have developed mutation-specific SHINEv.2 assays that are widely deployable and, in combination, can effectively discriminate between five widely circulating SARS-CoV-2 VOCs.
Towards at-home SHINEv.2 testing.
SHINEv.2 increases the accessibility of CRISPR-based SARS-CoV-2 diagnostics and enables VOC identification outside of laboratory settings. However, further improvements are necessary for SHINEv.2 to be used in virtually any environment. For example, the incorporation of internal controls, or further simplifying the assay readout, could greatly expand the use cases of SHINEv.2 and enable its use in at-home settings.
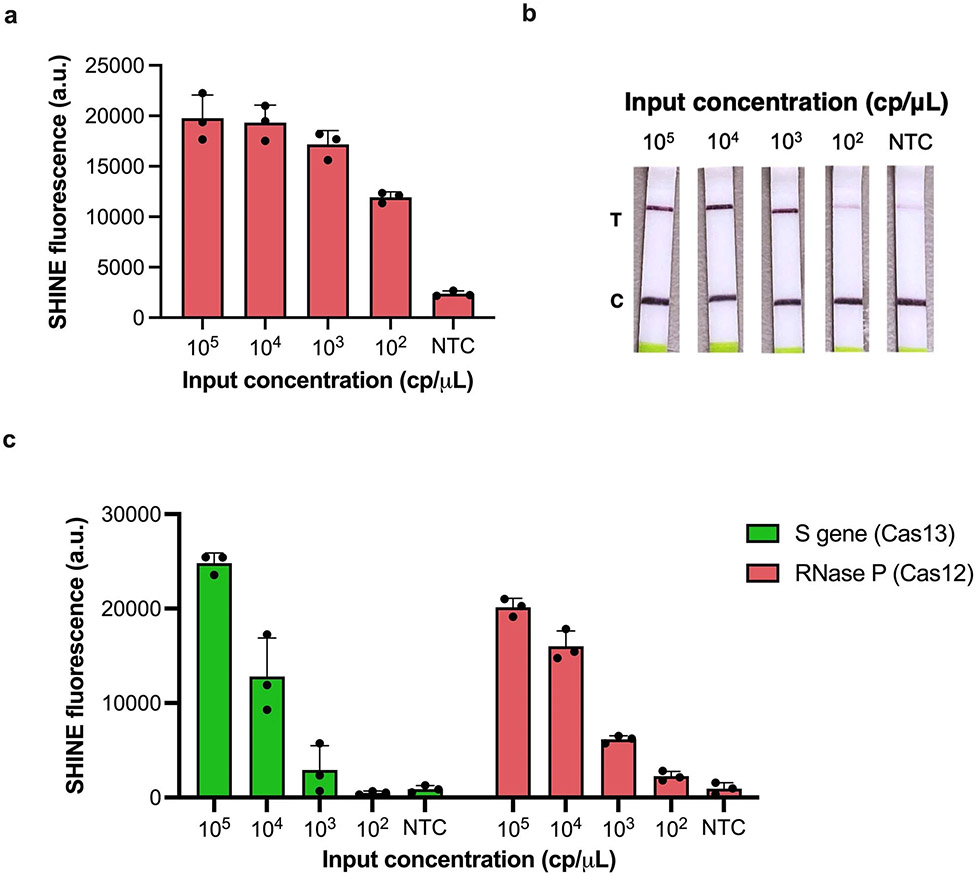
Nucleic acid diagnostic tests often require an internal control as part of the regulatory approval process49. The use of Cas enzymes with orthogonal recognition and cleavage properties can enable the detection of multiple targets from a single reaction17. Cas12, a Cas enzyme that detects and cleaves DNA, can be used as an orthogonal enzyme to Cas1315,16. Therefore, we designed and developed a Cas12-based assay for human RNase P to serve as an internal control for SHINEv.2. After optimizing the concentration of RPA primers, our RNase P assay was able to detect synthetic DNA down to 100 cp μl−1 with the fluorescence-based readout and 1,000 cp μl−1 with the paper-based format (Extended Data Fig. 4a,b). Next, we tested whether this Cas12-based assay was compatible with the FastAmp lysis solution and with lyophilisation. In this format, the Cas12-based RNase P assay detected synthetic DNA target down to 100 cp μl−1 using either fluorescence-based or lateral-flow-based readouts, which supports its potential use as an internal control for SHINEv.2 (Fig. 4a,b). Our results also demonstrate the use of an ambient-temperature lysis solution for Cas13- and Cas12-based diagnostics. Therefore, we sought to determine the compatibility of the Cas12-based RNase P assay with the Cas13-based S gene assay and evaluate the performance of this duplex assay on synthetic nucleic acid targets. The duplex SHINEv.2 assay successfully detected both targets, with an LoD of 103 cp μl−1 for each target (Extended Data Fig. 4c). Although additional improvements will be required to achieve clinically relevant sensitivity with the duplex SHINEv.2 assay, we have taken steps towards incorporating an internal control for SHINEv.2. Moreover, these results confirmed that the improvements made to SHINEv.2 are applicable to other systems, including in Cas12-based assays and potentially other CRISPR-Dx.
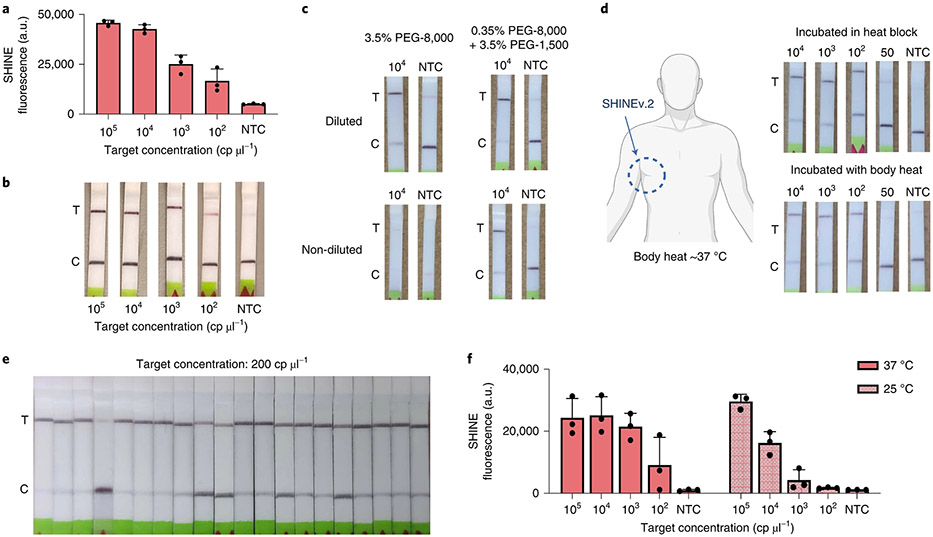
Fig. 4 ∣. Enhancing the accessibility of SHINEv.2.
a,b, RNase P SHINEv.2 assay results on synthetic DNA target after 90 min using a plate-based readout (a) and a lateral-flow-based readout (b). c, Lateral-flow-based detection of SARS-CoV-2 RNA using SHINEv.2 with different PEG compositions with or without dilution after a 90 min incubation. d, Lateral-flow-based SHINEv.2 detection of SARS-CoV-2 RNA after a 90 min incubation in a heat block or using body heat (underarm incubation). e, Determination of analytical LoD with 20 replicates of SHINEv.2 at 200 cp μl−1 of SARS-CoV-2 RNA from lysis solution-treated contrived samples incubated for 90 min. f, SHINE fluorescence on SARS-CoV-2 RNA after 90 min at 37°C or 25°C. For a and f, mean ± s.d. for 3 technical replicates.
Moreover, the user-friendliness of SHINEv.2 could be enhanced by further simplifying the assay. The lateral-flow-based readout for CRISPR-Dx, the preferred option for point-of-need implementation, requires a buffer to be added after the detection reaction is performed, adding a liquid-handling step for the user and increasing the risk of sample contamination. Directly inserting the paper strip into the SHINEv.2 reaction would substantially reduce this risk. However, undiluted SHINEv.2 reactions block the flow of nanoparticles through the paper strip, resulting in a failed test (Fig. 4c). We reasoned that high molecular weight PEG—an indispensable crowding agent for RPA—could be responsible for the lack of flow. We sought to identify a combination of PEGs with different molecular weights that enables flow while retaining the activity of SHINEv.2. In the absence of RNA targets, lowering the concentration and molecular weight of PEG in SHINEv.2 reactions facilitated the flow of nanoparticles through the paper strip (Supplementary Fig. 17a). However, the sensitivity of SHINEv.2 was markedly reduced when lowering the concentration of PEG-8000 in the reaction (Supplementary Fig. 18). Using a combination of 0.35% PEG- 8000 and 3.5% PEG-1500, undiluted SHINEv.2 was able to detect SARS-CoV-2 RNA without a drop in performance while retaining flow (Fig. 4c and Supplementary Fig. 17b). Thus, we managed to eliminate a liquid-handling step in SHINEv.2 and simplify the test readout.
In addition, SHINEv.2 can be run without equipment due to its minimal heating requirements. SHINE’s single 90 min incubation step at 37 °C can be easily maintained with low-cost and energy-efficient heat blocks. However, the removal of all heating elements would enable the unrestricted deployment of SHINEv.2. Using body temperature, which is approximately 37 °C, could provide an alternative for fully equipment-free SHINEv.2 testing50. Indeed, the underarm incubation of SHINEv.2 was successful at detecting down to 100 cp μl−1 of SARS-CoV-2 RNA, and with a similar efficiency to SHINEv.2 incubated in a heat block (Fig. 4d). We then sought to combine body-heat incubation with the elimination of the liquid-handling step to develop a further simplified SHINEv.2 technology, achieving a fully equipment-free CRISPR-based POC diagnostic. Following FDA guidance, we determined the analytical LoD of the simplified and equipment-free SHINEv.2 to be 200 cp μl−1 (19/20 replicates were positive, Fig. 4e). This is the same LoD as we achieved for instrument-based SHINEv.2, which remains lower than other equipment-free methods for detecting SARS-CoV-2.
An ideal test would perform with similar efficiency at ambient temperature as well. We examined the performance of SHINEv.2 at 25 °C using both a plate-based fluorescence readout and a colorimetric paper-based readout. SHINEv.2 was able to detect SARS-CoV-2 RNA at 25 °C with the plate reader, albeit with lower speed and sensitivity than at 37 °C (Fig. 4f). We observed a larger drop in sensitivity with the lateral-flow-based readout (Supplementary Fig. 19). We believe the drop in sensitivity was caused by a decline in the performance of RPA at lower temperatures51, since Cas13 detection alone performed well at 25 °C and 37 °C (Supplementary Fig. 20). Collectively, these improvements increase the accessibility of SHINEv.2 and enable its use in almost any situation.
Discussion
SHINEv.2 is a widely deployable CRISPR-Dx for SARS-CoV-2 RNA detection and VOC identification from unextracted samples with a straightforward workflow. Previously developed CRISPR-Dx are not well poised for widespread deployment, as they are often too complex or require a cold chain and auxiliary equipment14,19,33. SHINEv.2 addresses these limitations. Lyophilisation considerably simplifies the assay and facilitates its transportation and storage. SHINEv.2 can be distributed overseas without a loss in performance. Moreover, the use of an equipment-free and ambient-temperature sample lysis method further increases the user-friendliness of the assay. SHINEv.2 involves as few steps from the user as antigen-capture tests while providing a 50-fold boost in sensitivity6,42,43. Importantly, SHINEv.2 demonstrates perfect (100%) concordance with RT-qPCR, the gold standard for SARS-CoV-2 diagnosis, in samples with RNA levels above our analytical LoD of 200 cp μl−1.
SHINEv.2 can accurately identify several clade-specific mutations in the Alpha, Beta, Gamma, Delta and Omicron SARS-CoV-2 VOCs, and it can be rapidly adapted to respond to emerging viral variants as well as other viruses in current and future outbreaks. Thus, SHINEv.2 can provide critical information to inform public health responses, and it fills a major gap in point-of-need diagnostics. At the population level, SHINEv.2 could be used to prioritize testing and vaccine rollout in highly affected communities or to select subsets of samples for further viral sequencing. SHINEv.2 could also assist clinicians in selecting the right treatment (such as monoclonal-antibody cocktails) for patients with severe COVID-19. Altogether, we believe that SHINEv.2 will be especially valuable for community surveillance testing, as it combines user-friendly and equipment-free preparation methods with sufficient sensitivity and the capacity to discriminate VOCs.
Further advances will be required for CRISPR-based diagnostic testing to take place in any location, including in at-home settings. Ideally, such a test would involve a few simple, ambient-temperature steps and provide a fast and accurate visual readout without the need for specialized equipment. To our knowledge, current nucleic acid diagnostics are not capable of meeting all these requirements simultaneously. The combination of sample processing, nucleic acid amplification and CRISPR-based detection into a single ambient-temperature reaction could reduce liquid-handling steps. The incorporation of solution-based colorimetric readouts could further simplify the assay and reduce the risk of contamination52. Moreover, with SHINEv.2, we have taken steps to eliminate all equipment needs, although additional advances will be required to boost the performance of SHINEv.2 at ambient temperature. The speed and sensitivity of ambient-temperature CRISPR-Dx could be enhanced through the addition of auxiliary proteins or using alternative isothermal amplification methods17,20,53. Collectively, these improvements could greatly enhance the accessibility of SHINEv.2 and provide a critical tool in the fight against current and future outbreaks. By reducing assay complexity and simplifying test distribution without sacrificing sensitivity or specificity, we have taken steps towards the development of such a diagnostic tool.
Methods
Clinical samples and ethics statement.
To conduct this research, de-identified nasopharyngeal swab patient samples were purchased from Boca Biolistics (USA) and processed under a non-human subjects research determination from the Broad Institute Office of Research Subject Protections (NHSR-4318). Additional de-identified patient samples were obtained from the CDC. This was reviewed by the CDC and the study was conducted consistent with applicable federal law and CDC policy. Following the regulations for the protection of human subjects in research (Code of Federal Regulations 45 CFR 46.102(1)), the CDC determined that the collection of these samples was not considered to be human subject research. CDC samples were processed at the Broad Institute under an exempt determination issued by the Broad Institute Office of Research Subject Protections (EX-7209). SARS-CoV-2 clinical excess samples were obtained from the Rhode Island Department of Health under a waiver of consent and processed at the Broad Institute with approval from the MIT Institutional Review Board (Protocol No.1612793224).
SARS-CoV-2 assay design and synthetic template information.
RPA primers were designed using ADAPT34. The S gene, which contains SNPs of interest in several SARS-CoV-2 VOCs, was extracted from an alignment of 308,315 SARS-CoV-2 genomes downloaded from GISAID in January 202154,55. ADAPT was then run on the S gene alignment with the following parameters: 35–65% guanine-cytosine (GC) content, max. 1 mismatch between the primer and target, full coverage in >98% of the genomes and overlap allowed between amplicons but not primers. Forward RPA primers were ordered with a T7 promoter sequence (5’-GAAATTAATACGACTCACTATAGGG-3’) appended upstream. Both forward and reverse RPA primers were purchased from Integrated DNA Technologies (IDT) as single-stranded DNA oligos.
The crRNAs for SNP discrimination were designed using a generative sequence design algorithm (S.M. et al., manuscript in preparation). This approach uses ADAPT’s predictive model to predict the activity of candidate crRNA sequences against on-target and off-target sequences34. These predictions of candidate crRNA activity guide the generative algorithm’s optimization process, in which it seeks to design crRNA probes that have maximal predicted on-target activity and minimal predicted off-target activity. These crRNAs were purchased from IDT as Alt-R CRISPR guide RNAs.
Synthetic DNA targets with an upstream T7 promoter sequence were ordered as double-stranded DNA (dsDNA) gene fragments from IDT, and were in vitro transcribed to generate synthetic RNA targets. In vitro transcription was conducted as previously described22. In brief, the dsDNA template was transcribed using the HiScribe T7 high-yield RNA synthesis kit (New England Biolabs (NEB)) for 2 h at 37 °C. Transcribed RNA was then treated with RNase-free DNase I (Qiagen) according to the manufacturer’s instructions. Finally, DNase-treated RNA was purified using RNAClean SPRI XP beads using a 10:3:5 mixture of RNA solution, isopropanol and ethanol.
Sequence information for the synthetic targets, RPA primers, Cas13-crRNA and RNA reporters is listed in Supplementary Table 2.
Full-genome synthetic RNA controls and viral seedstocks.
Synthetic SARS-CoV-2 RNA controls (controls 2, 14, 16, 17 and 23) were purchased from Twist Biosciences and are referred to as full-genome synthetic RNA standards. Heat-inactivated viral stocks (2019-nCoV/USA-WA1/2020 as reference strain and USA/CA_CDC_5574/2020 as Alpha VOC) were purchased from ATCC.
Analysis of crRNA activity with Cas13a.
Initial measurements of crRNA activity were obtained under simplified SHINE conditions. That is, crRNA activity was measured using 1× optimized reaction buffer (20 mM HEPES pH 8.0 with 60 mM KCl and 3.5% PEG-8000), 45 nM LwaCas13a protein, 125 nM polyU (that is, 6 uracils (6U) in length) quenched FAM reporter, 1 U μl−1 murine RNase inhibitor, 22.5 nM crRNA and 14 mM magnesium acetate. Target RNA was added to each reaction at a ratio of 1:20 and fluorescence was measured on a Biotek Cytation 5 plate reader (excitation, 485 nm; emission, 520 nm) at 37 °C every 5 min for up to 3 h.
Nuclease inactivation with FastAmp lysis reagent.
A concentrated (5×) version of FastAmp viral and cell solution B for Covid-19 testing was purchased directly from Intact Genomics. This solution, referred to as FastAmp lysis reagent, was used at a final concentration of 1×. For nuclease inactivation experiments, 10% healthy nasal fluid (Lee BioSolutions) in UTM or pooled saliva samples (Lee BioSolutions) were mixed with 1× lysis reagent and either 1%, 2% or 5% (v/v) murine RNase inhibitor (NEB). HUDSON treatment was used as a control for successful RNase inactivation and performed as previously described22. In short, 100 mM tris(2-carboxyethyl)phosphine (TCEP) (Thermo Fisher), 1 mM ethylenediaminetetraacetic acid (EDTA) (Thermo Fisher) and 0.8 U μl−1 murine RNase inhibitor were added to the nasal fluid/UTM mix or pooled saliva sample and incubated at 40 °C for 5 min. PBS was used instead of HUDSON reagents as a no-treatment control. All treated products were then mixed 1:1 with 400 mM RNaseAlertv2 substrate (Thermo Fisher) in nuclease-free water and incubated at 25 °C while measuring the fluorescence (excitation, 485 nm; emission, 520 nm) every 5 min for up to 30 min using a Biotek Cytation 5 plate reader.
Viral inactivation with Intact Genomics lysis buffer.
An isolate of SARS-CoV-2 (2019-nCoV/USA-WA1-A12/2020) was obtained from the CDC. At the Integrated Research Facility (IRF) – Frederick, the virus was passaged by inoculating grivet kidney epithelial Vero cells (ATCC CCL-81) at a multiplicity of infection of 0.01 under high containment (BSL-3) conditions. Infected cells were incubated for 48 or 72 h in Dulbecco’s modified Eagle medium with 4.5 g l−1 d-glucose, l-glutamine and 110 mg l−1 sodium pyruvate (DMEM, Gibco) containing 2% heat-inactivated foetal bovine serum (SAFC Biosciences) in a humidified atmosphere at 37 °C with 5% CO2. The resulting viral seedstock was collected and quantified by plaque assay using Vero E6 cells (ATCC CRL-1586) with a 2.5% Avicel overlay, and stained after 48 h with a 0.2% crystal violet stain.
For testing the viral inactivation capacity of the FastAmp lysis reagent, the viral stock (107 PFU ml−1) was mixed with 1× lysis buffer supplemented with 5% (v/v) murine RNAse inhibitor and incubated at ambient temperature for either 5 min, 20 min or 20 min followed by 10 min at 65 °C. A no-treatment (PBS) control was also included. Treated viral stocks were then cleaned and quantified by plaque assay using Vero E6 cells (ATCC CRL-1586) with a 2.5% Avicel overlay, and stained after 48 h with a 0.2% crystal violet stain.
Single-step SARS-CoV-2 SHINE reactions.
RPA primer and crRNA optimizations were performed using the following SHINE conditions: 1× original SHINE buffer (20 mM HEPES pH 8.0 with 60 mM KCl and 3.5% PEG-8000), 45 nM LwaCas13a protein resuspended in 1× storage buffer (SB) (such that the resuspended protein is at 2.26 μM), 125 nM polyU quenched FAM reporter, 2 mM of each ribonucleoside triphosphate (rNTP), 1 U μl−1 murine RNase inhibitor, 1 U μl−1 NextGen T7 RNA polymerase, 0.1 U μl−1 RNase H (NEB), 2 U μl−1 Invitrogen SuperScript IV (SSIV) reverse transcriptase (Thermo Fisher), an assay-specific concentration of forward and reverse RPA primers (detailed below), and 22.5 nM crRNA. Once the master mix was created, it was used to resuspend the TwistAmp basic kit RPA pellets (1 lyophilised pellet per 102 μl final master mix volume). After that, magnesium acetate (as the only magnesium cofactor) was added at a final concentration of 14 mM to generate the final master mix. Finally, a synthetic RNA target or a sample was added to the complete master mix at a ratio of 1:19. Fluorescence kinetics were measured on a Biotek Cytation 5 plate reader (excitation, 485 nm; emission, 520 nm) at 37 °C every 5 min for up to 3 h.
For lateral-flow readout, the quenched FAM reporter was exchanged for a biotinylated FAM reporter at a final concentration of 1 μM. After incubation at 37 °C for 90 min, the sHiNE reaction was diluted 1:4 in 80 μl HybriDetect assay buffer (Milenia Biotec), and a HybriDetect 1 lateral-flow strip was added. Test images were collected with a smartphone camera 5 min after the addition of the strip.
RPA primer concentrations varied between assays, but the ratio of forward to reverse primers was always kept at 1:1. For each assay, the concentrations of forward and reverse RPA primers were as follows: 120 nM for ORF1a and 417 assays; 160 nM for 452 assay and 180 nM for S gene, 69/70 deletion and 156–158 assays.
SHINE tolerance to samples inactivated with lysis solution.
Blank contrived nasal samples were prepared by mixing 10% healthy nasal fluid (v/v) in UTM. These contrived samples were inactivated by incubating with the 5× FastAmp lysis reagent and RNase inhibitor (15:4:1 ratio) for 5 min at ambient temperature. SHINE reactions were generated as described above, except that a variable amount of water was replaced with inactivated contrived nasal samples. Synthetic RNA target (with the concentration specified in Fig. 1c) was added to the modified SHINE reactions at a ratio of 1:19. Plate-based fluorescence detection was performed as described above. In these reactions, the inactivated sample volume comprised 0%–40% of the total volume.
Lyophilisation optimization for SHINE.
The original SHINE buffer was optimized to retain SHINE’s activity after lyophilisation. For lyophilisation, we generated the master mix as described above, except that the 1× SHINE buffer was substituted for 1× lyophilisation buffer (composition described below) and magnesium acetate was omitted. The master mix was aliquoted, flash frozen in liquid nitrogen and lyophilised overnight. The lyophilisation buffers tested had the following composition: (1) SHINE (20 mM HEPES pH 8.0 with 60 mM KCl and 3.5% PEG-8000), (2) SHINE without PEG and KCl (20 mM HEPES pH 8.0), (3) SHINE with sucrose and mannitol (20 mM HEPES pH 8.0 with 60 mM KCl, 3.5% PEG-8000, 5% (w/v) sucrose and 150 mM mannitol), (4) LYO (20 mM HEPES pH 8.0 with 5% w/v sucrose and 150 mM mannitol). Lyophilised pellets were resuspended with a buffer composed of magnesium acetate (14 mM final concentration) and whichever reagent was left out of the original SHINE buffer (for example, 60 mM KCl, 3.5% PEG-8000 and 14 mM magnesium acetate for the LYO resuspension buffer). After resuspension, synthetic RNA target was added to the resuspended SHINE reactions at a ratio of 1:19. Plate-based fluorescence detection or lateral-flow-based detection was performed as described above.
Lyophilised SHINE pellets for stability testing were prepared using the LYO buffer as described above. SHINE pellets were stored at room temperature, 4 °C or −20 °C for 4, 7, 14, 21, 28 d or 5 months before resuspension and testing. For SHINEv.2 testing in Nigeria, SHINE pellets were lyophilised in the United States, shipped in dry ice to Nigeria and then resuspended and tested after a total of 6 weeks in storage.
Side-by-side clinical sample testing.
Clinical samples were thawed on ice. SHINEv.2, BinaxNow and CareStart testing, as well as RNA extractions and RT-qPCR, were performed side-by-side according to the manufacturer’s instructions (see details below). Nasopharyngeal swab samples (168) were tested in batches of 24 samples. In addition, unextracted VOC samples were also tested side-by-side with SHINEv.2 and RT-qPCR.
Information on clinical samples, including Ct values, can be found in Supplementary Table 1. Clinical sample test results are in Supplementary Figs. 8-11.
Clinical sample extractions and RT-qPCR.
Clinical sample extractions and RT-qPCR were performed according to the CDC’s recommended protocol39. Nucleic acid extractions for samples S1–S168 were performed using the QIAamp viral RNA mini kit (Qiagen). The starting volume for extraction was 100 μl and extracted nucleic acid was eluted into 100 μl of nuclease-free water. Extracted RNA was used immediately or stored at −80 °C. Extracted RNA was tested for the presence of SARS-CoV-2 RNA using the CDC’s SARS-CoV-2 RT-qPCR assay (2019-nCoV CDC EUA kit, IDT). RT-qPCR cycling conditions were as follows: hold at 25 °C for 2 min, reverse transcription at 50 °C for 15 min, polymerase activation at 95 °C for 2 min and 45 cycles with a denaturing step at 95 °C for 3 s, followed by annealing and elongation steps at 55 °C for 30 s. RT-qPCR was run on a QuantStudio 6 (Applied Biosystems) and data were analysed using the Standard Curve module of the Applied Biosystems analysis software.
SHINEv.2 testing for clinical samples.
SHINEv.2 reactions were prepared the day before testing and consist of 1× optimized lyophilisation buffer (20 mM HEPES pH 8.0 with 5% w/v sucrose and 150 mM mannitol), 45 nM LwaCas13a, 1 μM lateral-flow biotinylated FAM reporter, 2 mM of each rNTP, 1 U μl−1 murine RNase inhibitor, 1 U μl−1 T7 RNA polymerase, 0.1 U μl−1 RNase H, 2 U μl−1 SSIV, 180 nM forward and reverse RPA primers and 22.5 nM crRNA. Once the master mix was created, it was used to resuspend the TwistAmp basic kit lyophilised reaction components (1 lyophilised pellet per 102 μl final master mix volume). The master mix was aliquoted into individual reactions (20 μl final volume), flash frozen in liquid nitrogen and lyophilised overnight.
The following day, clinical samples were mixed with lysis reagent with 5% RNase inhibitor and incubated at ambient temperature for 5 min. In the meantime, the lyophilised SHINEv.2 pellets were resuspended with resuspension buffer to 1× (60 mM KCl, 3.5% PeG-8000 and 14 mM magnesium acetate). Inactivated clinical samples were added to the resuspended SHINEv.2 reactions at a ratio of 1:9. After incubation at 37 °C for 90 min, the SHINEv.2 reaction was diluted in 80 μl HybriDetect assay buffer, and a HybriDetect 1 lateral-flow strip was added. Test images were collected 5 min after the addition of the strip. Tests were considered positive when the intensity of the test band was higher than that of the negative control.
BinaxNow COVID-19 antigen self-test for clinical samples.
BinaxNow COVID-19 antigen self-tests (Abbott) were purchased from Walmart. Since we were not able to procure dry swabs, the test was performed according to the manufacturers’ protocol for determining the analytical sensitivity with a liquid input (Instructions For Use submitted for FDA EUA)56. Briefly, 6 drops of BinaxNow liquid were added to the top hole of each test card. Clinical samples (20 μl) were absorbed onto a clinical swab. The swab was then inserted into the bottom hole of the test card and turned 3 times. Without removing the swab, the test card was closed and sealed. The test was incubated at room temperature for at least 15 min before imaging with a smartphone camera. Any faint coloured lines in the test region were considered as positive.
CareStart COVID-19 antigen testing for clinical samples.
CareStart COVID-19 antigen tests (Access Bio) were purchased from Medek Health. The test was performed according to the manufacturer’s instructions for use57. Briefly, clinical samples were mixed 1:1 with the extraction buffer, then 3 drops of the mix were added onto the sample well in a test cartridge and incubated at ambient temperature for at least 10 min before imaging with a smartphone camera. As suggested by the manufacturer, any faint coloured line in the test region was considered as positive.
Contrived samples testing (VOCs).
Contrived samples were created by diluting a known concentration of heat-inactivated viral seedstocks (reference strain or Alpha VOC) 1:9 in UTM. Contrived samples were then inactivated with lysis solution for 5 min at ambient temperature. Lyophilised SHINEv.2 pellets were prepared and resuspended as described above. Inactivated samples were added to the resuspended SHINEv.2 reactions at a ratio of 1:9. After incubation at 37 °C for 90 min, the SHINEv.2 reaction was diluted in 80 μl HybriDetect assay buffer, and a Milenia HybriDetect 1 lateral-flow strip was added. Test images were collected 5 min after the addition of the strip.
Extracted clinical samples testing (VOCs).
Clinical sample extractions and RT-qPCR were performed according to the CDC’s recommended protocol39. Nucleic acid extraction for samples E6–E10 was performed with the MagnaPure 96 instrument using the DNA and viral SV kit (Roche). The starting volume for extraction was 100 μl and extracted nucleic acid was eluted into 100 μl of nuclease-free water. Extracted RNA was used immediately or stored at −80 °C. Extracted RNA was tested for the presence of SARS-CoV-2 RNA using the CDC’s SARS-CoV-2 RT-qPCR assay (2019-nCoV CDC EUA kit, IDT). RT-qPCR cycling conditions were as follows: hold at 25 °C for 2 min, reverse transcription at 50 °C for 15 min, polymerase activation at 95 °C for 2 min and 45 cycles with a denaturing step at 95 °C for 3 s, followed by annealing and elongation steps at 55 °C for 30 s. RT-qPCR was run on a QuantStudio 6 (Applied Biosystems) and data were analysed using the Standard Curve module of the Applied Biosystems analysis software. RNA extracted from clinical samples was added to resuspended SHINEv.2 reactions at a ratio of 1:9 as described above. Plate-based fluorescence detection was performed as described above.
Cas12-based RNase P assay.
Cas12-based SHINEv.2 reactions were prepared as follows: 1× original SHINE buffer or LYO buffer, 20 nM LbaCas12a, 250 nM polyC (that is, 5 cytosines (5C) in length) quenched HEX reporter, 1 U μl−1 murine RNase inhibitor, 90 nM of forward and reverse RPA primers, and 22.5 nM Cas12a crRNA were combined to create a master mix. The master mix was used to resuspend the TwistAmp basic kit RPA pellets (1 lyophilised pellet per 102 μl final master mix volume). For lyophilisation, the magnesium acetate was omitted, and individual reactions were aliquoted, flash frozen and lyophilised overnight. Otherwise, magnesium acetate (as the only magnesium cofactor) was added at a final concentration of 14 mM. Lyophilised Cas12 reactions were reactivated with resuspension buffer as described above. Finally, synthetic DNA target was added to the complete master mix at a ratio of 1:19. Fluorescence kinetics were measured on a Biotek Cytation 5 plate reader (excitation, 530 nm; emission, 580 nm) at 37 °C every 5 min for up to 3 h. For lateral-flow readout, the quenched HEX reporter was exchanged for a 5C biotinylated FAM reporter at a final concentration of 1 μM. After incubation at 37 °C for 90 min, the Cas12-based reaction was diluted in 80 μl HybriDetect assay buffer, and a HybriDetect 1 lateral-flow strip was added. Test images were collected 5 min after the addition of the strip.
Duplex SHINEv.2.
Duplex SARS-CoV-2 and RNase P SHINEv.2 reactions were prepared as follows: 1× original SHINE buffer or 1× LYO buffer, 45 nM LwaCas13a, 20 nM LbaCas12a, 62.5 nM polyU quenched FAM reporter, 250 nM polyC quenched HEX reporter, 2 mM of each rNTP, 1 U μl−1 murine RNase inhibitor, 1 U μl−1 T7 RNA polymerase, 0.1 U μl−1 RNase H, 2 U μl−1 SSIV, 90 nM S gene RPA primers, 90 nM RNase P RPA primers, 22.5 nM Cas12a crRNA and 22.5 nM Cas13a crRNA were combined to create a master mix. The master mix was then used to resuspend the TwistAmp basic kit RPA pellets (1 lyophilised pellet per 102 μl final master mix volume). After that, pre- and post-lyophilisation reactions were performed as previously described. Fluorescent readout was measured on a Biotek Cytation 5 plate reader in two separate fluorescent channels (excitation, 485 nm; emission, 520 nm; excitation, 530 nm; emission, 580 nm).
PEG experiments.
To evaluate the effect of PEG composition and concentration on flow through the lateral-flow strip, SHINEv.2 reactions were prepared as described above, except that the 3.5% PEG-8000 present in the original SHINE buffer was exchanged for 0%–7% PEG-8000, PEG-1500 or a combination of the two. Without adding synthetic targets and with or without diluting in HybriDetect assay buffer, a Milenia HybriDetect 1 lateral-flow strip was added to these modified reactions. Test images were collected 5 min after the addition of the strip.
To evaluate the performance of SHINEv.2 with the modified PEG composition, SHINEv.2 reactions were performed as described above, except that the composition of the original SHINE and resuspension buffers was modified to be: SHINE (20 mM HEPES pH 8.0 with 60 mM KCl, 0.35% PEG-8000 and 3.5% PEG-1500) and resuspension buffer (60 mM KCl, 0.35% PEG-8000, 3.5% PEG-1500 and 14 mM magnesium acetate).
Body-heat experiments.
Lyophilised SHINEv.2 pellets were prepared and resuspended as described above. Full-genome synthetic RNA standards were added to individual SHINEv.2 reactions and incubated using body heat or a heat block for 90 min. For body-heat experiments, individual SHINEv.2 reactions were closed, inserted in a ziploc bag, taped to the underarm of one of the researchers and incubated for 90 min. The reactions were then diluted in 80 μl HybriDetect assay buffer, and a HybriDetect 1 lateral-flow strip was added to each reaction. Test images were collected 5 min after the addition of the strip.
Data analysis.
SHINE fluorescence values are reported as background-subtracted fluorescence, with the fluorescence value collected before reaction progression (usually t = 10 min) subtracted from the final fluorescence value (90 min, unless otherwise indicated). For VOC data analysis, we introduced an assay-specific correction factor before data normalization to account for differences in background signal across VOC assays and plate readers used. For each condition, the crRNA-specific mean fluorescent signal across negative sample replicates was subtracted from the fluorescence measured for that condition. For normalization, the maximal corrected fluorescence value in an experiment was set to 1 and the fluorescence values from that same experiment were set as ratios of the maximum fluorescence value.
Drawing of schematics.
The schematics shown in Figs. 1a, 1d, 2c, 3a, 4d and Extended Data Fig. 1 were created using www.biorender.com. The data panels were primarily generated using Prism 8 (GraphPad).
Extended Data
Extended Data Fig. 1 ∣. SHINEv.2 requires 5 user manipulations.
Schematic of the SHINEv.2 workflow, including sample inactivation, rehydration of pellets, sample addition, reaction dilution after incubation and result readout.
Extended Data Fig. 2 ∣. Performance of SARS-CoV-2 Delta and Omicron assays on synthetic RNA targets.
SHINE fluorescence of the (a) 452, (b) 156 - 158 and (c) 142 - 145 ancestral (anc) and derived (der) assays on (a,b) full genome synthetic RNA standards or (c) synthetic RNA targets after 90 minutes. NTC, no target control. Center = mean and error bars = s.d. for 3 technical replicates.
Extended Data Fig. 3 ∣. Validation of SHINEv.2 Delta assays.
a, Discrimination of SARS-CoV-2 VOCs using SHINE fluorescence of Delta assays on full-genome synthetic RNA standards, after 90 minutes. Target RNA concentration: 104 genomes/μL. (b,c), Colorimetric lateral flow based detection of full-genome synthetic RNA standards using the (b) 156 - 158 and (c) 452 SHINEv.2 assays. SHINEv.2 incubation time: 90 minutes. NTC, no target control. T, test line; C, control line.
Extended Data Fig. 4 ∣. Development of Cas12-based SHINE assay for RNase P detection.
a. SHINE fluorescence of the RNase P assay on synthetic DNA target after 90 minutes. b. Lateral flow detection of synthetic DNA target using Cas12-based SHINE assay, after 90 minute incubation; c. SHINE fluorescence of the duplex SARS-CoV-2 S gene and human RNase P assays on synthetic nucleic acid targets after 90 minutes. NTC = no target control; C = control band; T = test band. For (a,c), center = mean and error bars = s.d. for 3 technical replicates.
Supplementary Material
Acknowledgements
We thank the TIDE group at the Broad Institute for providing additional laboratory space to perform the work; all the researchers and laboratories who generously made SARS-CoV-2 sequencing data publicly available, to inform the design of our assay; H. Metsky for his contributions to the development of ADAPT, which guided the assay design; the Sabeti laboratory, notably S. Siddiqui, H. Metsky and E. Normandin for thoughtful discussions and reading of the manuscript; the personnel at the Rhode Island Department of Health for the samples they provided, in particular, E. King, Associate Director of Health, and R. C. Huard, Chief Clinical Laboratory Scientist, both at the Division of State Laboratories and Medical Examiner at Rhode Island Department of Health. Funding was provided by DARPA D18AC00006. This work was made possible by support from the Flu Lab and a cohort of generous donors through TED’s Audacious Project, including the ELMA Foundation, MacKenzie Scott, the Skoll Foundation and Open Philanthropy. J.A.-S. was supported by a fellowship from ‘la Caixa’ Foundation (ID 100010434, code LCF/BQ/AA18/11680098). C.M. was supported by start-up funds from Princeton University. For L.E.H., this work was funded under Agreement No. HSHQDC-15-C-00064 awarded to Battelle National Biodefense Institute (BNBI) by the Department of Homeland Security (DHS) Science and Technology (S&T) Directorate for the management and operation of the National Biodefense Analysis and Countermeasures Center (NBACC), a Federally Funded Research and Development Center. M.S. was also supported by the National Institutes of Health (RO1 GM120122-01). P.C.S. was supported by the Howard Hughes Medical Institute, Merck KGaA Future Insight Prize and NIH (U01AI151812 and U54HG007480). The views, opinions, conclusions and/or findings expressed should not be interpreted as representing the official views or policies, either expressed or implied, of the Department of Defense, US Government, National Institute of General Medical Sciences, DHS or the National Institutes of Health. The DHS does not endorse any products or commercial services mentioned in this presentation. In no event shall the DHS, BNBI or NBACC have any responsibility or liability for any use, misuse, inability to use or reliance upon the information contained herein. In addition, no warranty of fitness for a particular purpose, merchantability, accuracy or adequacy is provided regarding the contents of this document. Notice: this manuscript has been authored by Battelle National Biodefense Institute, LLC under Contract No. HSHQDC-15-C-00064 with the U.S. Department of Homeland Security. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
Footnotes
Competing interests
C.A.F., P.C.S. and C.M. are co-inventors on patent PCT/US2018/022764, which covers the SHERLOCK and HUDSON technology for viral RNA detection held by the Broad Institute. J.A.-S., C.A.F., P.C.S. and C.M. are co-inventors on patent PCT/US2021/049145, which covers the SHINE technology for viral RNA detection held by the Broad Institute. M.S. is a co-founder of Rhinostics, Inc., and a shareholder and advisor to the company. P.C.S. is a co-founder of Sherlock Biosciences, Inc., and a shareholder and advisor to the company, as well as a Board member and shareholder of Danaher Corporation. All other authors declare no competing interests.
Reporting summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended data is available for this paper at https://doi.org/10.1038/s41551-022-00889-z.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41551-022-00889-z.
Code availability
The code used in the design of primers and crRNAs are available at adapt.sabetilab.org.
Data availability
The data and detailed methods used in the design of primers and crRNAs are available at adapt.sabetilab.org. Raw data for the clinical samples are available in the Supplementary Information. All raw and analysed datasets generated during the study are available for research purposes from the corresponding author on reasonable request.
References
- 1.Summers J et al. Potential lessons from the Taiwan and New Zealand health responses to the COVID-19 pandemic. Lancet Reg. Health West Pac 4, 100044 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavelka M et al. The impact of population-wide rapid antigen testing on SARS-CoV-2 prevalence in Slovakia. Science 372, 635–641 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walensky RP & del Rio C From mitigation to containment of the COVID-19 pandemic. JAMA 323, 1889–1890 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Mina MJ & Andersen KG COVID-19 testing: one size does not fit all. Science 371, 126–127 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Mögling R et al. Delayed laboratory response to COVID-19 caused by molecular diagnostic contamination. Emerg. Infect. Dis 26, 1944–1946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKay SL et al. Performance evaluation of serial SARS-CoV-2 rapid antigen testing during a nursing home outbreak. Ann. Intern. Med 174, 945–951 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferguson J et al. Validation testing to determine the sensitivity of lateral flow testing for asymptomatic SARS-CoV-2 detection in low prevalence settings: testing frequency and public health messaging is key. PLoS Biol. 19, e3001216 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Kampen JJA et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat. Commun 12, 267 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pray IW et al. Performance of an antigen-based test for asymptomatic and symptomatic SARS-CoV-2 testing at two university campuses - Wisconsin, September–October 2020. MMWR Morb. Mortal. Wkly Rep 69, 1642–1647 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rabe BA & Cepko C SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl Acad. Sci. USA 117, 24450–24458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thi VLD et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med 12, eabc7075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LUCIRA HEALTH. Lucira™ COVID-19 Test Kit Instructions for Use (2021) https://www.fda.gov/media/147494/download
- 13.Land KJ, Boeras DI, Chen X-S, Ramsay AR & Peeling RW REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol 4, 46–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gootenberg JS et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JS et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S-Y et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 4, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gootenberg JS et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myhrvold C et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fozouni P et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184, 323–333.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu TY et al. Accelerated RNA detection using tandem CRISPR nucleases. Nat. Chem. Biol 17, 982–988 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joung J et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med 383, 1492–1494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arizti-Sanz J et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun 11, 5921 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harvey WT et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol 19, 409–424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konings F et al. SARS-CoV-2 Variants of Interest and Concern naming scheme conducive for global discourse. Nat. Microbiol 6, 821–823 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Volz E et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593, 266–269 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Lemieux JE et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science 371, eabe3261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borges V et al. Tracking SARS-CoV-2 lineage B.1.1.7 dissemination: insights from nationwide spike gene target failure (SGTF) and spike gene late detection (SGTL) data, Portugal, week 49 2020 to week 3 2021. Euro Surveill. 26, 1–6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogels CBF et al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 19, e3001236 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brito AF et al. Global disparities in SARS-CoV-2 genomic surveillance. Preprint at medRxiv 10.1101/2021.08.21.21262393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ackerman CM et al. Massively multiplexed nucleic acid detection with Cas13. Nature 582, 277–282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakraborty D, Agrawal A & Maiti S Rapid identification and tracking of SARS-CoV-2 variants of concern. Lancet 397, 1346–1347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Puig H et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv 7, eabh2944 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casati B et al. ADESSO: a rapid, adaptable and sensitive Cas13-based COVID-19 diagnostic platform. Preprint at medRxiv 10.1101/2021.06.17.21258371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metsky HC et al. Designing sensitive viral diagnostics with machine learning. Nat. Biotechnol 10.1038/s41587-022-01213-5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian J et al. An enhanced isothermal amplification assay for viral detection. Nat. Commun 11, 5920 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (U.S. Food and Drug Administration, 2021). https://www.fda.gov/media/135659/download [Google Scholar]
- 37.Perchetti GA, Huang M-L, Mills MG, Jerome KR & Greninger AL Analytical sensitivity of the Abbott BinaxNOW COVID-19 Ag card. J. Clin. Microbiol 59, e02880–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xun G, Lane ST, Petrov VA, Pepa BE & Zhao H A rapid, accurate, scalable, and portable testing system for COVID-19 diagnosis. Nat. Commun 12, 2905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel (Centers for Disease Control and Prevention, 2021). https://www.fda.gov/media/134922/download [Google Scholar]
- 40.Yang Q et al. Just 2% of SARS-CoV-2-positive individuals carry 90% of the virus circulating in communities. Proc. Natl Acad. Sci. USA 118, 1–6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teyssou E et al. The Delta SARS-CoV-2 variant has a higher viral load than the Beta and the historical variants in nasopharyngeal samples from newly diagnosed COVID-19 patients. J. Infect 83, e1–e3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollock NR et al. Performance and implementation evaluation of the Abbott BinaxNOW rapid antigen test in a high-throughput drive-through community testing site in Massachusetts. J. Clin. Microbiol 10.1128/JCM.00083-21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollock NR et al. Performance and operational evaluation of the Access Bio Carestart rapid antigen test in a high-throughput drive-through community testing site in Massachusetts. Open Forum Infect. Dis 10.1093/ofid/ofab243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allan-Blitz L-T & Klausner JD A real-world comparison of SARS-CoV-2 rapid antigen vs. polymerase chain reaction testing in Florida. J. Clin. Microbiol 10.1128/JCM.01107-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okoye NC et al. Performance characteristics of BinaxNOW COVID-19 antigen card for screening asymptomatic individuals in a university setting. J. Clin. Microbiol 59, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodcroft EB CoVariants: SARS-CoV-2 Mutations and Variants of Interest https://covariants.org/ (2021). [Google Scholar]
- 47.Planas D et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Viana R et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 603, 679–686 (2022). 10.1038/s41586-022-04411-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emmadi R et al. Molecular methods and platforms for infectious diseases testing. A review of FDA-approved and cleared assays. J. Mol. Diagn 13, 583–604 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crannell ZA, Rohrman B & Richards-Kortum R Equipment-free incubation of recombinase polymerase amplification reactions using body heat. PLoS ONE 9, e112146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh P et al. Evaluation of recombinase-based isothermal amplification assays for point-of-need detection of SARS-CoV-2 in resource-limited settings. Int. J. Infect. Dis 114, 105–111 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan C-Q et al. Universal and naked-eye gene detection platform based on CRISPR/Cas12a/13a system. Anal. Chem 10.1021/acs.analchem.9b05597 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Carter JG et al. Ultrarapid detection of SARS-CoV-2 RNA using a reverse transcription-free exponential amplification reaction, RTF-EXPAR. Proc. Natl Acad. Sci. USA 118, 1–6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elbe S & Buckland-Merrett G Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall 1, 33–46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shu Y & McCauley J GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 22, 30494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.BinaxNOW COVID-19 Antigen Self TEST (U.S. Food and Drug Administration, 2021). https://www.fda.gov/media/147254/download [Google Scholar]
- 57.CareStart COVID-19 Antigen Rapid Diagnostic Test for the Detection of SARS-CoV-2 Antigen (U.S. Food and Drug Administration, 2021). https://www.fda.gov/media/142919/download [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and detailed methods used in the design of primers and crRNAs are available at adapt.sabetilab.org. Raw data for the clinical samples are available in the Supplementary Information. All raw and analysed datasets generated during the study are available for research purposes from the corresponding author on reasonable request.