Abstract
Single cell RNA sequencing (scRNA-seq) is a novel high-throughput technique that enables the investigation of a single cell’s entire transcriptome. It elucidates intricate cellular networks and generates indices that will eventually enable the development of more targeted and personalized medications. The importance of scRNA-seq has been highlighted in complex biological systems such as cancer and the immune system, which exhibit significant cellular heterogeneity. Colorectal cancer (CRC) is the third most common type of cancer and the second leading cause of cancer-related death globally. Chemotherapy continues to be used to treat these patients. However, 5-FU has been utilized in chemotherapy regimens with oxaliplatin and irinotecan since the 1960s and is still used today. Additionally, chemotherapy-resistant metastatic CRCs with poor prognoses have been treated with immunotherapy employing monoclonal antibodies, immune checkpoint inhibitors, adoptive cell therapy and cancer vaccines. Personalized immunotherapy employing tumor-specific neoantigens allows for treating each patient as a distinct group. Sequencing and multi-omics approaches have helped us identify patients more precisely in the last decade. The introduction of modern methods and neoantigen-based immunotherapy may usher in a new era in treating CRC. The unmet goal is to better understand the cellular and molecular mechanisms that contribute to CRC pathogenesis and resistance to treatment, identify novel therapeutic targets, and make more stratified and informed treatment decisions using single cell approaches. This review summarizes current scRNA-seq utilization in CRC research, examining its potential utility in the development of precision immunotherapy for CRC.
Keywords: immune landscape, single cell, colorectal cancer, metastasis, tumor immune microenvironment, immunotherapy, precision medicine
Introduction
According to the International Agency for Research on Cancer, colorectal cancer (CRC) was the third most commonly diagnosed cancer and ranked second in global cancer-related mortality in 2020 (1). Approximately 20% of the newly diagnosed CRC patients developed distant metastases, while another 25% with localized tumors will eventually acquire metastases (2, 3). The expected 5-year survival decreases from 82%-90% in stage I or II CRC patients to a dismal 12% in those of stage IV (4). Currently, surgery alone is still the gold standard for stage I and II cancer, while for some stage II, III and early-stage IV CRC patients with minimal metastases, neoadjuvant, or adjuvant chemotherapy coupled with surgery is still an option (4). Although these conventional cancer therapies target the tumor cells and yield positive outcomes in the majority of CRC patients, the benefits are often offset by tumor reoccurrence and chemoresistance after prolonged treatment (5). In short, metastasis is still a major challenge in CRC’s effective treatment (6).
Conversely, immunotherapy exploits the immune system to induce a systemic response targeting malignant tumor cells (7–9). The discovery of first-generation antibody-based immunotherapy, also known as immune checkpoint inhibitor (ICI), altered the CRC therapeutic landscape. It functions via preventing anti-programmed cell death ligand 1 (PD-L1) and anti-cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) monoclonal antibodies (mAbs) from binding/interacting with the receptors during T cell activation, allowing the latter to recognize and destroy CRC cells (10, 11). Nivolumab (Opdivo®; PD-1 blocker) is the most extensively studied ICI in metastatic CRC (mCRC) (12–14). Additionally, combinational administration with Ipilimumab (Yervoy®; CTLA-4 blocker) has also shown improvement in overall survival and long-term treatment responses particularly in mCRC patients with microsatellite instability-high (MSI-H) and mismatch repaired (MMR) aberrations (15–17). Nevertheless, the success of CRC immunotherapy remains unsatisfactory since positive responses are confined to a minority of patients (18, 19).
Successful tumor control by immunotherapy is observed only in metastatic CRC (mCRC) of MMR and MSI-H with immune infiltration, indicating that CRC and its treatment response could be influenced by the tumor immune microenvironment (TIME) (20, 21). Interplay underlying immunotherapy and the TIME is therefore vital not only for deciphering the mechanisms of action but also for identifying advanced biomarkers and revising current immunotherapy strategies for better efficiency (22, 23). In this regard, single cell technology has quickly emerged as a potent technique to investigate the TIME in mCRC. Single cell RNA-sequencing (scRNA-seq) not only provides an unprecedented, detailed characterization of transcriptomes of cell diversity and heterogeneity in immune cell populations but also potentially discovers novel cellular or molecular factors involved in ICI, thereby allowing comprehensive assessment of the complexity of TIME (24, 25). Henceforth, scRNA-seq will be firmly embedded as a tool in oncology, with increasing incorporation of genes/neoantigens into the cancer panels, and the fusion of immunotherapy with scRNA-seq is expected to deliver truly precision treatment to an expanding number of mCRC patients.
Tumour immune microenvironment reshapes the current transcriptional landscape
Recent advances in cancer immunotherapy encounter a bottleneck due to the complex tumor microenvironment (TME) which provides a formidable barrier to immune infiltration and function (26–28). The TME consists of various cell types including transformed cancer cells from the epithelium of the tissue of origin; stromal cells/non-cancer cells (fibroblasts, adipocytes, endothelial, immune cells) and the extracellular matrix. As the heart of TME, cancer cells exploited non-malignant cells to prevent recognition and elimination by T cells, followed by establishing dormant tumor immune tolerance via ‘immunoediting’ their immunogenicity or antigen presentation as well as secreting favorable cytokines to exhaust T cells (29–31). Other consequences of such crosstalk are reflected in tumor growth maintenance, metastasis formation, deficient immunotherapeutic response and multi-drug resistance (32–35). Although single cell transcriptome analyses have been extensively used to study the relationship between TME and immunotherapy response, it remains unclear how cancer cells and host tissues differentially influence the immune composition within TME (36–38). Moreover, there is a lack of effective predictive biomarkers, making it difficult to accurately grasp the effect of immunotherapy (39, 40). As such, the transition towards tumor immune microenvironmental (TIME) transcriptional landscaping is rational given that a successful tumor control induced by immunotherapy requires the activation of the immune system, expansion of the effector cells, infiltration of activated effector cells to the tumor tissue, and elimination of the tumor cells (41). A deeper understanding of the crosstalk between cancer and immune cells not only permits dissection of the immune complexity of TME in CRC but also provides a comprehensive characterization of all immune cells that participate in modulation of TIME. Additionally, scRNA-seq studies on the immune landscape of CRC might unveil the underlying mechanisms modulating immune cells exhaustion, and the identification of advanced biomarkers, enabling the devising of novel personalized immunotherapy strategies. In the future, it is possible that single cell studies on the TIME could provide a snapshot of CRC tumor evolution since tumor cells interact with immune cells most frequently, tumor immunology and evolution are interwoven, and co-evolution has been proven to exist between them (42, 43). Thus, in this review, we aimed to summarize the current scRNA-seq studies on TIME of CRC and assess their potential utilities as immune-based therapeutic biomarkers in personalized immunotherapy illustrated in Figure 1 .
Figure 1.
Potential immune-based precision medicine targets governed by scRNA-seq.
Fundamental importance of T lymphocytes in TIME and immunotherapy responses
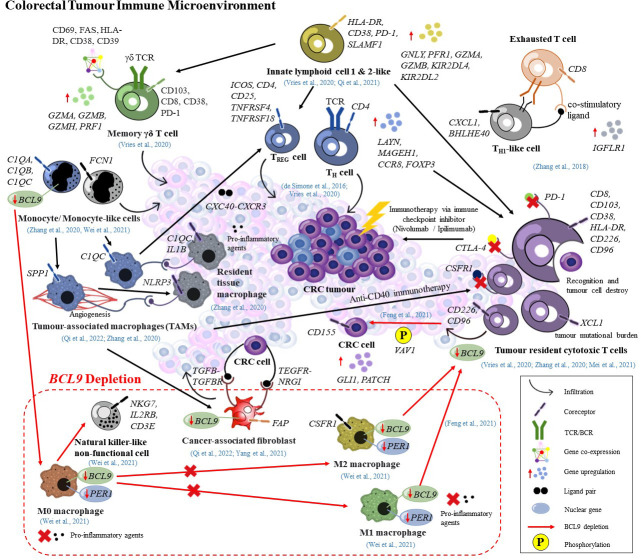
The colorectal tumor immune microenvironment (TIME) is a heterogeneous microenvironment containing a variety of immune cells and their products in tumor tissues (44). Many scRNA-seq studies have found a link between significant infiltration of CD8 + T cells (45), CD4 + T helper 1 (Th1) (46), regulatory T (Treg) cells (47), tumor-infiltrating exhausted T cells (48), CD45 + macrophages, dendritic cells, and myeloid cells (49), and a favourable outcome in mCRC. However, these immune cells, particularly T cells are always in a hyporesponsive state, a phenomenon often referred to as exhaustion (30). Since immunotherapy primarily relies on T cells to attack and kill tumor cells, the response of the former could be impaired if the latter’s capabilities fade, resulting in tumor immune escape, whereby CRC cells evade recognition and are not attacked by the human immune system (31, 50). Furthermore, a recent study by Sorrentino et al. has reported that exhaustion is not exclusive to T cells, but could also affect other immune cells including B lymphocytes and conventional natural killer (NK) cells from CRC patients (51). Restoring exhausted immune cells is currently an inspiring CRC therapeutic technique that is anticipated to yield promising results and mark a significant breakthrough in CRC immunotherapy. Hence, the rationale behind the shift of traditional scRNA-seq from targeting only tumor cells to neighboring infiltrated immune cells is understandable and is believed to reshape the current transcriptional landscape, redefine CRC classification, and shed insights on the tumor progression, restoration of immune cells exhaustion and immunotherapy response in CRC patients (31, 52).
The first scRNA-seq based immune transcriptional investigation was performed on CRC-infiltrating CD4 + T cells, where the authors confirmed the influence of TIME on specific gene expressions (LAYN, MAGEH1, and CCR8) in tumor-infiltrating Treg cells and their correlations with immunotherapy response, tumor suppressive activity and prognosis (53). Another attempt was also performed on T cells from primary human CRC, where cell heterogeneity was discovered, and the dynamic relationships between CD4 + and CD8 + T cell subpopulations were explained via integrated transcriptomic analysis and T cell receptor (TCR) (54). Concisely, TIME plays a vital role in reshaping CRC therapeutic landscape, and high throughput technologies permit the dissection of heterogeneity of immune cells at single cell resolution as well as cell-cell interaction. Nevertheless, a comprehensive understanding of the microenvironmental interactions between tumor cells and their surrounding immune cells is still lacking since both studies focus on T cell subpopulations only.
Following this, unbiased characterization of the immune contexture of CRC was performed via the inclusion of all CD45 + cells from both MMR-deficient and MMR-proficient tumors. This work explained the intricate immune landscape of primary CRC and healthy mucosa, including the discovery of a previously unappreciated innate lymphoid cells (ILCs) subpopulation and reveal a potential multitargeted immunotherapeutic response via modulating adaptive (cytotoxic and helper) and innate (γδ) T cells (55). In the same year, Lee and their co-workers further explored the TIME, including six immune cells (epithelial, stromal, myeloid, T, B, and mast cells) and their matched normal mucosa, followed by an illustration of an immune transcriptional landscape and reconstruction of putative interaction network between tumor cells and their surrounding microenvironment via the dominance of IgA-type humoral immunity in normal mucosa and γδ T-cell-driven innate immunity in CRC (56). In 2021, a significant link between B-cell and myeloid-cell signalling was revealed using two scRNA-seq approaches, while CCL8 + cycling B cells/CCR5 + T-cell interactions were identified as a potential antitumoral mechanism in advanced CRC tumors (57). In essence, these studies proved that T, B, and myeloid cells play a dominant role in cancer-associated immune surveillance. Profiling TIME in both healthy and neoplastic states allows uncovering of the underlying mechanisms as well as the identification of new therapeutic targets in CRC (58).
Tumour-associated macrophage is the second significant key component after T cells
Tumour-associated macrophages (TAMs) are critical in the establishment of the TIME through their production of cytokines and chemokines, representing the most abundant immune cell population infiltrating colorectal tumors (59, 60). A detrimental effect on cancer treatment is more likely with TAMs than a beneficial effect since they are known to promote tumor angiogenesis, growth, metastasis (via miRNA-containing exosome secretion, matrix metalloproteinase 9 expression, epithelial-to-mesenchymal transition) and immunosuppression (61, 62). In conjunction with prior research findings, Chinese researchers verified in 2020 that depletion of specific infiltrating-TAMs impacted CRC immunotherapy outcomes. Based on combined scRNA-seq methods, they tracked two distinct human C1QC + and SPP1 + TAM subpopulations, the latter of which exhibited inflammatory and angiogenic characteristics in human CRC and distant liver metastatic site. Moreover, they highlighted two murine TAM subsets that resembled human SPP1 + TAM, showed resistance to anti-CSF1R depletion, and described a previously unrecognized immunological mechanism upon anti-CD40 treatment. Interestingly, they were unable to explain the dichotomy of C1QC + and SPP1 + TAMs in the CRC TIME using genes related to M1- and M2-TAMs (49). In line with this, Qi et al. reported a dramatic increase in infiltrating SPP1+ macrophages in CRC tissue and a positive correlation with FAP+ fibroblasts, which impaired immunotherapeutic effect (63). Similarly, an expanded SPP1 + TAM subpopulation was recovered and proposed to have both pro- and anti-inflammatory signatures via scRNA-seq investigation (56). Mei and her colleagues also discovered polyfunctional SPP1 + TAM subsets in CRC that do not fit the M1 and M2 polarization paradigm (37).
Conversely, another study conducted by Hicks and her co-authors revealed that a combination of tumor-targeted interleukin-12 (IL12) and Entinostat therapy was capable of TAM reprogramming, resulting in a significant shift in M1/M2 TAMs balance favoring tumor resolution. Furthermore, polarization to M1-like TAMs was shown to be substantially linked with complete tumor eradication (41.7%-100%), triggered by combination therapy, correlating to antitumor efficacy (64). More recently, China experts conducted a single-cell and spatial transcriptomics analysis and identified highly metabolically active MRC1 + CCL18 + M2-like TAMs in colorectal liver metastasis site. They discovered that M2-like TAMs had increased metabolic activity, which might be reduced by effective neoadjuvant chemotherapy, implying the possibility of targeting metastatic metabolism pathways via TAM reprogramming (65). Intriguingly, Wei et al. believed that re-educating TAMs as M1 phenotype might be an efficient anticancer strategy since pro-tumor M0- and M1-TAMs were actively involved in CRC inflammation (60). Collectively, these findings indicated that targeting certain TAM subsets in conjunction with reprogramming could be beneficial in CRC treatment and immunotherapy response prediction. Unfortunately, knowledge of TAM cellular architecture and transcriptional profiles in the CRC TIME landscape remains limited.
Remodelling of TIME via nuclear B-cell lymphoma 9 expression in Wnt pathway
Apart from incorporation of a wider subsets of tumor-neighboring immune cell populations to provide a more comprehensive exploration of the underlying cellular interactions during tumor progression and immunotherapy treatment, the remodelled immune translational landscape could elucidate the role of diverse signalling pathways involved in the modulation of CRC TIME. Currently, wingless-related integration site-beta catenin (Wnt/β-catenin) signalling is the best-studied pathway, whereby B-cell lymphoma 9 (BCL9) is the critical transcription co-factor (66). For instance, Yang et al. detected that knock down of nuclear BCL9 promoted tumorigenesis in murine cancer-associated fibroblasts (CAFs), whereas aberrant inactivation of Wnt/β-catenin due to BCL9 suppression aided T-cell–mediated antitumor immune responses. Briefly, the authors illustrated cellular landscape and transcription differences in CAFs upon BCL9 depletion, as well as the reconfiguration of CRC immune surveillance in TIME via Wnt signalling blockage (67).
In agreement with the previous research finding, BCL9 depletion was reported to benefit CD8 + T cells infiltration into CRC tumor and improve anti-PD-1 immunotherapy response in murine models via increased VAV1 phosphorylation in CD8 + T cells and enhanced GLI1 and PATCH expression, promoting CD155 production in CRC cells. Moreover, BCL9 was linked to adenomatous polyposis coli (APC) mutation involved in patient survival following anti-PD-L1 treatment. Ultimately, this study proved that BCL9 inhibition altered cellular diversity within the tumor immune milieu and shed light on the role of BCL9 in regulating CD226 and CD96 checkpoints. Using identical mice xenograft models, a group of scientists from China described that BCL9 inhibition attenuated CRC growth via inhibiting TAM polarization from M0 to M2 phenotype that interfered with inflammatory actions of M0 and M1 TAMs. Based on the cellular landscape and transcription differences of TAMs after BCL9 suppression, they demonstrated that regulation of Wnt signalling via BCL9 suppression was expected to impair TAM-induced inflammation, CRC progression and immune surveillance (60). In a nutshell, unlike traditional immunotherapy, which targets solely CRC patients in late stages with MSI-H, scRNA-seq immune transcriptional studies offer an alternative option for precision medicine by targeting the Wnt signalling pathway via BCL9 depletion.
Immune transcriptomics identifies novel biomarker and revises immunotherapy strategies
The expanding pool of knowledge regarding the immunological complexity of the tumor microenvironment (TME) resulted in the discovery of a vast majority of previously unappreciated biomarkers, which played crucial roles in the modulation of CRC TIME. These biomarkers provide candidates for immunotherapy prediction, resulting in a paradigm shift in personalized immuno-oncology. The identification of new regulatory roles in neighboring immune cells via cancer biomarkers/gene expressions are not limited to T cells or TAMs as previously described. One such example is tumor specific innate lymphoid cells (ILCs). Single cell characterization of ILCs in healthy and CRC conditions was performed, with ILC1s, ILC3s, and ILC3/NKs subsets identified in the healthy gut; and ILC1-like and ILC2s subsets found to be tumor specific. Moreover, SLAMF1 expression in ILCs had been discovered as an anti-tumor biomarker in CRC (68).
Other biomarkers such as tumor mutational burden (TMB), inflammation, alteration in a specific gene or signalling pathway (e.g. BCL9 in Wnt pathway), APC mutation and MSI, allow stratification of patients for targeted immunotherapy and are consistent with several studies whereby TMB contributed to CRC immune landscape modelling (37); and BCL9 depletion in Wnt signalling reprogrammed TIME, promoting anti-tumoral immune response (60, 67, 69). On the other hand, the conversion of TIME into a functionally inflamed immune hub via IL12 and Entinostat combinational therapy promoted and sustained the clinical benefits of immune therapy to a wider proportion of CRC patients. This study provides a rationale for combination therapy in the clinical setting for tailored immunotherapy (64). Furthermore, Wu et al. pinpointed the presence of energetic MRC1 + CCL18 + M2-like macrophages under an immunosuppressive state and susceptible to neoadjuvant chemotherapy (NAC) that restored anti-tumoral immune balance in TIME, suggesting the possibility of personalized immunotherapy in combination with NAC (65). In summary, the remodelling of an immune translational landscape in CRC appears promising in terms of the development of personalized cancer treatment and improvement in the selection of patients who may benefit from immunotherapy. Despite ground-breaking discoveries in single cell immune transcriptional studies, a comprehensive exploration of infiltrating immune cells in CRC TIME is still inadequate for clinical applications. Table 1 .
Table 1.
The summary of key discoveries on the single-cell transcriptional CRC immune landscape.
| Type of Sample | Cell Infiltration | Key Finding | Number and Type of Cell | Sequencing Technology | Cells Identification/Screening | Reference |
|---|---|---|---|---|---|---|
|
|
|
|
|
Nine main cells:
|
(63) |
|
|
|
|
|
Seven main immune cells:
|
(65) |
|
|
b) CCL8 + cycling B-cell/CCR5 + T-cell and IgA + IGLC2 + plasma-cell/CCR5 + T-cell interactions
|
|
Two scRNA-seq methods:
|
Four immune cell types:
Three nonimmune cell types:
|
(57) |
|
|
|
|
|
Three ILC subsets in the healthy gut:
Two ILC subsets in CRC patients:
|
(68) |
|
|
|
|
|
Seven clusters from total CRC cells:
|
(37) |
|
|
|
|
|
14 clusters from EMT6 tumor infiltrating CD45
+ cells:
|
(64) |
|
• TAMs |
|
|
|
Six main cell clusters:
|
(60) |
|
Cells interacting with infiltrated leucocytes:
|
|
|
|
|
(67) |
|
|
|
|
|
Six main cell clusters:
|
(69) |
|
Three cells related to CRC tumor infiltration:
|
|
|
|
Six main cell clusters:
|
(56) |
|
|
|
|
|
Seven main cell clusters:
|
(55) |
|
|
b) Two distinct TAM subsets with differential sensitivity to CSF1R blockade
|
|
|
13 myeloid subsets:
|
(49) |
|
|
|
|
|
Three main subsets from primary human lung or CRC tumors and non-neoplastic counterparts:
|
(53) |
|
|
|
|
|
|
(54) |
Conclusion and future direction
Although conventional cancer transcriptomics focused on TME holds promise in cancer treatment, the treatment of advanced metastatic CRCs remains challenging. Hence, it is of paramount importance to explore innovative therapeutic targets/strategies. The transition towards single cell immune transcriptomics unveils the precise landscape of both immune and non-immune cells throughout CRC TIME. This immune-based approach not only translates gene signatures into a collective landscape, but it also investigates cellular interactions between immune cells and highlights their potential values as novel CRC classification systems as well as immunotherapy targets for personalized cancer treatment. With the growing number of studies on single-cell transcriptional profiling of cancer-associated immune cells, we believe that integrating and comprehensively characterizing these data would deepen our understanding of the immunobiology of CRC TIME. The new insights provided into cancer biology and metastasis may allow new applications in precision medicine.
Author contributions
N-SAM contributed the idea. N-SAM and FYFT prepared the manuscript. FYFT prepared the figure. L-HL provided critical insights and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The related research is funded by the Ministry of Higher Education Higher Institution Centre of Excellence Grant JJ-2021-002. The APC was funded by Monash University Malaysia.
Acknowledgments
The authors acknowledge the supports provided by the universities and Ministry of Higher Education Malaysia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. van der Pool AEM, Damhuis RA, Ijzermans JNM, de Wilt JHW, Eggermont AMM, Kranse R, et al. Trends in incidence, treatment and survival of patients with stage IV colorectal cancer: a population-based series. Colorectal Disease: Off J Assoc Coloproctol Great Britain Ireland (2012) 14(1):56–61. doi: 10.1111/j.1463-1318.2010.02539.x [DOI] [PubMed] [Google Scholar]
- 3. Väyrynen V, Wirta E-V, Seppälä T, Sihvo E, Mecklin JP, Vasala K, et al. Incidence and management of patients with colorectal cancer and synchronous and metachronous colorectal metastases: A population-based study. BJS Open (2020) 4(4):685–92. doi: 10.1002/bjs5.50299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistic. CA: Cancer J Clin (2019) 69(5):363–85. doi: 10.3322/caac.21565 [DOI] [PubMed] [Google Scholar]
- 5. Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer (2013) 13(10):714–26. doi: 10.1038/nrc3599 [DOI] [PubMed] [Google Scholar]
- 6. Chakraborty S, Rahman T. The difficulties in cancer treatment. ecancer (2012) 6(16):1–5. doi: 10.3332/ecancer.2012.ed16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schuster M, Nechansky A, Kircheis R. Cancer immunotherapy. Biotechnol J (2006) 1(2):138–47. doi: 10.1002/biot.200500044 [DOI] [PubMed] [Google Scholar]
- 8. Dillman RO. Cancer immunotherapy. Cancer Biotherapy Radiopharmaceuticals (2011) 26(1):1–64. doi: 10.1089/cbr.2010.0902 [DOI] [PubMed] [Google Scholar]
- 9. Sharma G, Harmanci AO. ‘Iterative estimation of structures of multiple rna homologs: Turbofold’. IEEE [Preprint] (2011) 2011:529–32. doi: 10.1109/ICASSP.2011.5946457 [DOI] [Google Scholar]
- 10. Ballas ZK. The 2018 Nobel prize in physiology or medicine: An exemplar of bench to bedside in immunology. J Allergy Clin Immunol (2018) 142(6):1752–3. doi: 10.1016/j.jaci.2018.10.021 [DOI] [PubMed] [Google Scholar]
- 11. Yi M, Jiao D, Xu H, Liu Q, Zhao W, Han X, et al. ‘Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors’. Mol Cancer (2018) 17(1):129. doi: 10.1186/s12943-018-0864-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz H-J, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol (2017) 18(9):1182–91. doi: 10.1016/S1470-2045(17)30422-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarshekeh AM, Overman MJ, Kopetz S. Nivolumab in the treatment of microsatellite instability high metastatic colorectal cancer. Future Oncol (2018) 14(18):1869–74. doi: 10.2217/fon-2017-0696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jácome AA, Eng C. Role of immune checkpoint inhibitors in the treatment of colorectal cancer: Focus on nivolumab. Expert Opin Biol Ther (2019) 19(12):1247–63. doi: 10.1080/14712598.2019.1680636 [DOI] [PubMed] [Google Scholar]
- 15. Overman MJ, Kopetz S, McDermott RS, Leach J, Lonardi S, Lenz H-J, et al. ‘Nivolumab ± ipilimumab in treatment (tx) of patients (pts) with metastatic colorectal cancer (mCRC) with and without high microsatellite instability (MSI-h): CheckMate-142 interim results.’. J Clin Oncol (2016) 34(15_suppl):3501–1. doi: 10.1200/JCO.2016.34.15_suppl.3501 [DOI] [Google Scholar]
- 16. Andre T, Lonardi S, Wong M, Lenz H-J, Gelsomino F, Aglietta M, et al. ‘Nivolumab + ipilimumab combination in patients with DNA mismatch repair-deficient/microsatellite instability-high (dMMR/MSI-h) metastatic colorectal cancer (mCRC): First report of the full cohort from CheckMate-142’. J Clin Oncol (2018) 36(4_suppl):553–3. doi: 10.1200/JCO.2018.36.4_suppl.553 [DOI] [Google Scholar]
- 17. Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA (2021) 325(7):669–85. doi: 10.1001/jama.2021.0106 [DOI] [PubMed] [Google Scholar]
- 18. Smith KM, Desai J. Nivolumab for the treatment of colorectal cancer. Expert Rev Anticancer Ther (2018) 18(7):611–8. doi: 10.1080/14737140.2018.1480942 [DOI] [PubMed] [Google Scholar]
- 19. Rotte A. ‘Combination of CTLA-4 and PD-1 blockers for treatment of cancer’. J Exp Clin Cancer Res (2019) 38(1):255. doi: 10.1186/s13046-019-1259-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, et al. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat Rev Gastroenterol Hepatol (2019) 16(6):361–75. doi: 10.1038/s41575-019-0126-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arrichiello G, Poliero L, Borrelli C, Paragliola F, Nacca V, Napolitano S, et al. ‘Immunotherapy in colorectal cancer: Is the long-awaited revolution finally happening?’. Cancer Treat Res Commun (2021) 28:100442. doi: 10.1016/j.ctarc.2021.100442 [DOI] [PubMed] [Google Scholar]
- 22. Tang H, Qiao J, Fu Y-X. Immunotherapy and tumor microenvironment. Cancer Lett (2016) 370(1):85–90. doi: 10.1016/j.canlet.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med (2018) 24(5):541–50. doi: 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods (2014) 11(1):41–6. doi: 10.1038/nmeth.2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol (2018) 18(1):35–45. doi: 10.1038/nri.2017.76 [DOI] [PubMed] [Google Scholar]
- 26. Ventola CL. ‘Cancer immunotherapy, part 3: Challenges and future trends’. Pharm Ther (2017) 42(8):514–21. [PMC free article] [PubMed] [Google Scholar]
- 27. Gaissmaier L, Elshiaty M, Christopoulos P. ‘Breaking bottlenecks for the TCR therapy of cancer’. Cells (2020) 9(9):2095. doi: 10.3390/cells9092095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bai R-L, Chen N-F, Li L-Y, Cui J-W. A brand new era of cancer immunotherapy: Breakthroughs and challenges. Chin Med J (2021) 134(11):1267–75. doi: 10.1097/CM9.0000000000001490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nurieva R, Wang J, Sahoo A. T-Cell tolerance in cancer. Immunotherapy (2013) 5(5):513–31. doi: 10.2217/imt.13.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang Y, Li Y, Zhu B. ‘T-cell exhaustion in the tumor microenvironment’. Cell Death Dis (2015) 6(6):e1792–2. doi: 10.1038/cddis.2015.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Z, Guo Y, Yang X, Chen C, Fan D, Wu X, et al. Immune landscape refines the classification of colorectal cancer with heterogeneous prognosis, tumor microenvironment and distinct sensitivity to frontline therapies. Front Cell Dev Biol (2022) 9:784199. doi: 10.3389/fcell.2021.784199Accessed:6 April 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsai M-J, Chang W-A, Huang M-S, Kuo P-L. ‘Tumor microenvironment: A new treatment target for cancer’. ISRN Biochem (2014) p:e351959. doi: 10.1155/2014/351959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. ‘Targeting tumor microenvironment for cancer therapy’. Int J Mol Sci (2019) 20(4):840. doi: 10.3390/ijms20040840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Communication Signaling (2020) 18(1):59. doi: 10.1186/s12964-020-0530-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The tumor microenvironment: A milieu hindering and obstructing antitumor immune responses. Front Immunol (2020) 11:940. doi: 10.3389/fimmu.2020.00940(Accessed: 18 July 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Merlano MC, Abbono A, Denaro N, Garrone O. Knowing the tumour microenvironment to optimise immunotherapy. Acta Otorhinolaryngologica Italica (2019) 39(1):2–8. doi: 10.14639/0392-100X-2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mei Y, Xiao W, Hu H, Lu G, Chen L, Sun Z, et al. ‘Single-cell analyses reveal suppressive tumor microenvironment of human colorectal cancer’. Clin Trans Med (2021) 11(6):e422. doi: 10.1002/ctm2.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caligola S, De Sanctis F, Canè S, Ugel S. Breaking the immune complexity of the tumor microenvironment using single-cell technologies. Front Genet (2022) 13:867880. doi: 10.3389/fgene.2022.867880(Accessed: 18 July 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spencer KR, Wang J, Silk AW, Ganesan S, Kaufman HL, Mehnert JM. ‘Biomarkers for immunotherapy: Current developments and challenges’. Am Soc Clin Oncol Educ Book (2016) 36):e493–503. doi: 10.1200/EDBK_160766 [DOI] [PubMed] [Google Scholar]
- 40. Sankar K, Ye JC, Li Z, Zheng L, Song W, Hu-Lieskovan S. ‘The role of biomarkers in personalized immunotherapy’. Biomark Res (2022) 10(1):32. doi: 10.1186/s40364-022-00378-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Valdes-Mora F, Handler K, Law AMK, Salomon R, Oakes SR, Ormandy CJ, et al. Single-cell transcriptomics in cancer immunobiology: The future of precision oncology. Front Immunol (2018) 9:2582. doi: 10.3389/fimmu.2018.02582(Accessed: 18 July 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galon J, Bruni D. Tumor immunology and tumor evolution: Intertwined histories. Immunity (2020) 52(1):55–81. doi: 10.1016/j.immuni.2019.12.018 [DOI] [PubMed] [Google Scholar]
- 43. Wu Y, Biswas D, Swanton C. Impact of cancer evolution on immune surveillance and checkpoint inhibitor response. Semin Cancer Biol (2022) 84:89–102. doi: 10.1016/j.semcancer.2021.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Sci (New York N.Y.) (2006) 313(5795):1960–4. doi: 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 45. Yang X, Qi Q, Pan Y, Zhou Q, Wu Y, Zhuang J, et al. Single-cell analysis reveals characterization of infiltrating T cells in moderately differentiated colorectal cancer. Front Immunol (2021) 11:620196. doi: 10.3389/fimmu.2020.620196(Accessed: 30 April 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Zheng L, Zhang L, Hu X, Ren X, Zhang Z. ‘Deep single-cell RNA sequencing data of individual T cells from treatment-naïve colorectal cancer patients’. Sci Data (2019) 6(1):1–15. doi: 10.1038/s41597-019-0131-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bhatt D, Kang B, Sawant D, Zheng L, Perez K, Huang Z, et al. STARTRAC analyses of scRNAseq data from tumor models reveal T cell dynamics and therapeutic targets. J Exp Med (2021) 218(6):e20201329. doi: 10.1084/jem.20201329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim M, Min YK, Jang J, Park H, Lee S, Lee CH. ‘Single-cell RNA sequencing reveals distinct cellular factors for response to immunotherapy targeting CD73 and PD-1 in colorectal cancer’. J Immunother Cancer (2021) 9(7):e002503. doi: 10.1136/jitc-2021-002503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O’Brien SA, et al. ‘Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer’. Cell (2020) , 181(2):442–459.e29. doi: 10.1016/j.cell.2020.03.048 [DOI] [PubMed] [Google Scholar]
- 50. Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol (2020) 20(11):651–68. doi: 10.1038/s41577-020-0306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sorrentino C, D’Antonio L, Fieni C, Ciummo SL, Di Carlo E. ‘Colorectal cancer-associated immune exhaustion involves T and b lymphocytes and conventional NK cells and correlates with a shorter overall survival’. Front Immunol (2021) 12:778329. doi: 10.3389/fimmu.2021.778329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet (2017) 49(5):708–18. doi: 10.1038/ng.3818 [DOI] [PubMed] [Google Scholar]
- 53. De Simone M, Arrigoni A, Rossetti G, Gruarin P, Ranzani V, Politano C, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity (2016) 45(5):1135–47. doi: 10.1016/j.immuni.2016.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature (2018) 564(7735):268–72. doi: 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- 55. Vries NL, de Unen V, van Ijsselsteijn ME, Abdelaal T, Breggen R, van der Sarasqueta AF, et al. High-dimensional cytometric analysis of colorectal cancer reveals novel mediators of antitumour immunity. Gut (2020) 69(4):691–703. doi: 10.1136/gutjnl-2019-318672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee H-O, Hong Y, Etlioglu HE, Cho YB, Pomella V, Van den Bosch B, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet (2020) 52(6):594–603. doi: 10.1038/s41588-020-0636-z [DOI] [PubMed] [Google Scholar]
- 57. Wang W, Zhong Y, Zhuang Z, Xie J, Lu Y, Huang C, et al. ‘Multiregion single-cell sequencing reveals the transcriptional landscape of the immune microenvironment of colorectal cancer’. Clin Trans Med (2021) 11(1):e253. doi: 10.1002/ctm2.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pérez-Romero K, Rodríguez RM, Amedei A, Barceló-Coblijn G, Lopez DH. ‘Immune landscape in tumor microenvironment: Implications for biomarker development and immunotherapy’. Int J Mol Sci (2020) 21(15):5521. doi: 10.3390/ijms21155521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li J, Li L, Li Y, Long Y, Zhao Q, Ouyang Y, et al. Tumor-associated macrophage infiltration and prognosis in colorectal cancer: Systematic review and meta-analysis. Int J Colorectal Dis (2020) 35(7):1203–10. doi: 10.1007/s00384-020-03593-z [DOI] [PubMed] [Google Scholar]
- 60. Wei Z, Yang M, Feng M, Wu Z, Rosin-Arbesfeld R, Dong J, et al. ‘Inhibition of BCL9 modulates the cellular landscape of tumor-associated macrophages in the tumor immune microenvironment of colorectal cancer’. Front Pharmacol (2021) 12:713331. doi: 10.3389/fphar.2021.713331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hao N-B, Lü M-H, Fan Y-H, Cao Y-L, Zhang Z-R, Yang S-M, et al. ‘Macrophages in tumor microenvironments and the progression of tumors’. Clin Dev Immunol (2012) 2012:948098. doi: 10.1155/2012/948098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu Y, Zhang Q, Xing B, Luo N, Gao R, Yu K, et al. ‘Immune phenotypic linkage between colorectal cancer and liver metastasis’. Cancer Cell (2022) 40(4):424–437.e5. doi: 10.1016/j.ccell.2022.02.013 [DOI] [PubMed] [Google Scholar]
- 63. Qi J, Sun H, Zhang Y, Wang Z, Xun Z, Li Z, et al. ‘Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer’. Nat Commun (2022) 13(1):1742. doi: 10.1038/s41467-022-29366-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hicks KC, Chariou PL, Ozawa Y, Minnar CM, Knudson KM, Meyer TJ, et al. ‘Tumour-targeted interleukin-12 and entinostat combination therapy improves cancer survival by reprogramming the tumour immune cell landscape’. Nat Commun (2021) 12(1):5151. doi: 10.1038/s41467-021-25393-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov (2022) 12(1):134–53. doi: 10.1158/2159-8290.CD-21-0316 [DOI] [PubMed] [Google Scholar]
- 66. Li X, Xiang Y, Li F, Yin C, Li B, Ke X. ‘WNT/β-catenin signaling pathway regulating T cell-inflammation in the tumor microenvironment’. Front Immunol (2019) 10:2293. doi: 10.3389/fimmu.2019.02293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yang M, Wei Z, Feng M, Zhu Y, Chen Y, Zhu D. Pharmacological inhibition and genetic knockdown of BCL9 modulate the cellular landscape of cancer-associated fibroblasts in the tumor-immune microenvironment of colorectal cancer. Front Oncol (2021) 11:603556. doi: 10.3389/fonc.2021.603556(Accessed: 6 April 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Qi J, Crinier A, Escalière B, Ye Y, Wang Z, Zhang T, et al. ‘Single-cell transcriptomic landscape reveals tumor specific innate lymphoid cells associated with colorectal cancer progression’, cell reports. Medicine (2021) 2(8):100353. doi: 10.1016/j.xcrm.2021.100353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Feng M, Wu Z, Zhou Y, Wei Z, Tian E, Mei S, et al. BCL9 regulates CD226 and CD96 checkpoints in CD8+ T cells to improve PD-1 response in cancer. Signal Transduction Targeted Ther (2021) 6(1):1–14. doi: 10.1038/s41392-021-00730-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Guo J-N, Chen D, Deng S-H, Huang J-R, Song J-X, Li X-Y, et al. ‘Identification and quantification of immune infiltration landscape on therapy and prognosis in left- and right-sided colon cancer’. Cancer Immunol Immunother: CII [Preprint] (2021) 71(6):1313–30. doi: 10.1007/s00262-021-03076-2 [DOI] [PMC free article] [PubMed] [Google Scholar]