Abstract
The SARS-CoV-2 virus has been identified as a causative agent for COVID-19 pandemic. About more than 6.3 million fatalities have been attributed to COVID-19 worldwide to date. Finding a viable cure for the illness is urgently needed in light of the present pandemic. The prominence of main protease in the life cycle of virus shapes the main protease as a viable target for design and development of antiviral agents to combat COVID-19. The current study presents the fragment linking strategy to design the novel Mpro inhibitors for COVID-19. A total of 293,451 fragments from diversified libraries have been screened for their binding affinity towards Mpro enzyme. The best 1600 fragment hits were subjected to fragment joining to achieve 100 new molecules using Schrödinger software. The resulting molecules were further screened for their Mpro binding affinity, ADMET, and drug-likeness features. The best 13 molecules were selected, and the first 6 compounds were investigated for their ligand-receptor complex stability through a molecular dynamics study using GROMACS software. The resulting molecules have the potential to be further evaluated for COVID-19 drug discovery.
Keywords: COVID-19, SARS-CoV-2, Mpro, Fragment-based drug design, Molecular docking, Molecular dynamics
Introduction
The ongoing global pandemic of coronavirus 2019 (COVID-19) is caused by SARS-CoV-2. According to the world health organization (WHO), as of 27 July 2022, there have been 570,005,017 confirmed cases of COVID-19, including 6,384,128 deaths globally [1]. During a public health emergency, like COVID-19, the Food and Drug Administration (FDA) can issue emergency use authorizations (EUAs) to make new medications and medical products available without full FDA approval [2]. Many medications have been authorized for emergency use to treat COVID-19, including oral treatments like Paxlovid (nirmatrelvir and ritonavir) and molnupiravir [3]. In the past time, doctors used to prescribe existing drugs notably, antimalarials (chloroquine, hydroxychloroquine) [4], antibacterials (azithromycin, doxycycline) [5], antivirals (remdesivir, ribavirin, favipiravir, lopinavir, oseltamivir) [6], antiparasitic (ivermectin) [7], monoclonal antibody (bebtelovimab, bamlanivimab, etesavimab, tosulizumab) [8], corticosteroids (dexamethasone, prednisolone, methyl prednisolone) [9], 2-deoxy-d-glucose [10], and even convalescent plasma [11] as a treatment option for COVID-19. Many phytochemicals (polyphenols—rutin, acetoside, procyanidins, solanine, hypericin) have also been recorded to be useful to combat COVID-19 [12]. As there is no other treatment option at an early stage of infection, the WHO emergency use authorization (EUA) has qualified the use of COVID-19 vaccines notably, Covaxin (whole inactivated coronavirus), Covishield (adenovirus vector), BNT162b2/COMIRNATY (mRNA vaccine), mRNA-1273 (mRNA vaccine), AZD1222 Vaxzevria (adenovirus vector), Ad26.COV2.5 (adenovirus type 26 vector), Covilo/BBIBP-CorV (whole inactivated coronavirus), CoronaVac (whole inactivated coronavirus), Nuvaxovid, and Covovax (protein subunit) [13]. Despite the fact that vaccination campaigns have continued and have provided protection against the virus, it is still necessary to identify novel treatments that could treat the infection while also acting to prevent potential mutations. The SARS-CoV-2 viral life cycle could be crucial to study the various targets for COVID-19 drug discovery. The main protease (Mpro) is mainly involved in virus replication and thus is the target of choice for COVID-19 drug discovery.
SARS-CoV-2 main protease (Mpro)
SARS-CoV-2 Mpro, a cysteine protease is an attractive target for the development of therapeutics because of its critical role in the viral life cycle and its high conservation amongst the family of coronaviruses [14]. Mpro consists of three main domains with a 306 amino acids chain. Domain 1 consists of 8–101 residues, domain 2 of 102–184 residues, and the domain 3 of 201–203 amino acid residues, linked to domain 2 by residues 185–200 loop. The substrate-binding site is present in a cleft between domains 1 and 2 with a His41-Cys145 catalytic dyad [15]. The S1 subsite is formed by Phe140, Leu141, Asn142, His163, Glu166, and His172 amino acids. Hydrophobic S2 subsite involves His41, Met49, Tyr54, Met165, and Asp187. The S4 binding subsite is made up of Met165, Leu167, Phe185, Gln189, and Gln192 amino acids [16].
Fragment-based drug discovery (FBDD) principles
FBDD is an imperative strategy for drug design for industry and academia. In FBDD method, a group of low molecular weight polar fragments/compounds is screened against a specific target [17]. Usually, the screening methods are biophysical methods including X-ray crystallography, nuclear magnetic resonance, differential scanning fluorimetry, isothermal titration calorimetry, and surface plasmon resonance [18]. One of the critical factors favoring the FBDD’s success is the smaller size of the fragment-like compound compared to the size of the drug-like compounds.
The recent advancement of computational tools and methods for fragment-based approaches improves the identification of promising fragment hits. This approach generally begins with target protein structure determination followed by virtual fragment library preparation, docking, and hit confirmation through docking and molecular dynamics simulation [19]. With current technology advancements, a library with a huge number of fragments can be easily screened, which offers a high hit rate. The fragments can be further grown, merged or scaffold hopped, or linked to develop a new molecule. Software like LUDI, HOOK, CAVEAT, RECORE, Schrödinger, CCG, AGFIS server, and many others have been developed for this purpose [20].
The traditional de novo drug discovery requires an ample amount of time and money to provide a single clinical candidate. Faster approaches involve the usage of computer-aided drug design (CADD) tools like structure-based drug design (SBDD), ligand-based drug design (LBDD), and fragment-based drug design (FBDD) [21]. FBDD has been proven a flourishing design strategy for many compounds. The other computational methods may experience a low hit rate and narrow coverage of drug-like chemical space to some extent. It is widely considered that a chemical space of 109 diversified molecules can be sampled with 103 fragments [22]. Thus, the coverage of chemical space can be improved with FBDD approach. It is also easier and more convenient to modify the chemical structure of fragment hits. Therefore, the FBDD strategy becomes the complimentary choice for drug discovery and development.
In continuation of our research for unraveling potential novel therapeutics to combat COVID-19, herein we report the screening of diversified fragments from natural and synthetic categories against Mpro of SARS-CoV-2 through molecular docking. The top-ranked fragments were linked to get the novel compounds as Mpro inhibitors.
Materials and methods
Data collection and preparation
The in silico fragment-based drug design study was carried out using Schrödinger maestro v 12.2 (Schrödinger, LLC, NY, 2020) [23]. The crystal structure of SARS-CoV-2 Mpro was retrieved from the RCSB PDB database. The fragment structures were retrieved from different commercial fragment screening libraries.
Preparation of protein structure
The 3D X-ray crystal structure of SARS-CoV-2 main protease (PDB ID: 6LU7) was obtained from RCSB protein databank [24]. Then, it was processed in protein preparation wizard of Schrödinger maestro using OPLS3e force field [25]. The structural correctness was ensured for bond orders, hydrogen consistency, steric clashes, and charges during protein processing. The prepared structure was then utilized for receptor grid generation.
Fragment libraries
A total of 293,451 fragment structures were downloaded from the diversified fragment databases available publicly. These are the fragment libraries from Life Chemicals General and Natural Product-Like (https://lifechemicals.com/screening-libraries/fragment-libraries), Aurora fine chemicals (https://aurorafinechemicals.com/targeted-library.html), Otava general and natural product-like (https://otavachemicals.com/products/fragment-libraries), Enamine natural product-like (https://enamine.net/compound-libraries/fragment-libraries), Schrodinger Glide (https://www.schrodinger.com/Glide), ZINC (https://zinc15.docking.org/), and ChemBridge (https://www.chembridge.com/screening_libraries/fragment_library/). Some of the library’s fragments were not following “Rule of 3.” So, all the fragments were first screened for Rule of 3 using ChemAxon tools. After applying this filter, the resulted number of fragments was 210,123. They were prepared at physiological pH conditions using the LigPrep module of Schrödinger suite v 12.2 (Schrödinger, LLC, NY, 2020) where 5 conformers of each fragment were generated [26]. OPLS3e force field algorithm was applied for the minimization of ligand geometry. The minimum energy conformer from each fragment was then selected for molecular docking.
Preparation of main protease crystal structure for molecular docking
PDB ID 6LU7 is an X-ray crystal structure of Mpro in a complex with an N3 inhibitor having a 2.16 Å resolution. Firstly, the covalent bond between co-crystallized ligand N3 and amino acid residue Cys145 was cleaved. The Cys145 and ligand molecule were rebuilt by making necessary changes, and the ligand-receptor complex was generated and refined by Protein Preparation Wizard in Schrödinger. This prepared complex was employed for receptor grid generation. The receptor grid was generated on the active site of Mpro by taking the centroid of ligand molecule N3 as a centre of the grid. The X, Y, and Z grid coordinates were −10.47, 12.23, and 68.7, respectively [27].
Fragment screening
All the prepared fragments after applying the rule of three were used for docking against active site of Mpro. Glide module of Schrödinger v 12.2 (Schrödinger, LLC, NY, 2020) [28] was used for molecular docking using standard precision (SP) mode. The validation of docking protocol was done by calculating the all-atom RMSD value of the re-docked N3 ligand with the co-crystallized ligand.
Fragment linking
The top 1600 fragments with SP docking score ≤ −7.00 were selected for the fragment linking. The direct joining of the fragments prepositioned at various regions of the Mpro binding site was carried out by the “combine fragments” tool from the library design module of Schrodinger to design novel compounds. The combine fragment tool usually joins the fragments by picking out the feasible bonds that can be formed between the fragments. The default parameters of the tool were selected where the maximum output of the newly formed structures was 100. The fragments were also checked for Van der Waals clashes before joining. For direct joining, maximum atom–atom distance from different fragments was kept 1 Å, minimum fragment centroid distance was kept 2 Å, minimum bond angle deviation was set to 15°, and maximum number of fragment atoms was set to 200. All bonds attached to halogens or hydrogen in a fragment were selected for breaking and re-joining with another fragment. All atoms present in the newly built molecule were subsequently minimized. Total three rounds of fragment joining were carried out [29]. In the first round, pairs of fragments were joined, and in the subsequent round, the outputs of the previous round were used as inputs to join up to 4 fragments and so on.
Molecular docking and visualization of receptor-ligand interaction
The 100 new molecules were generated after fragment linking. They were first prepared using LigPrep, similarly as done for the fragments, and then docked within the active site of Mpro (6LU7) using extra precision (XP) mode of Glide docking module of Maestro. The same grid coordinates were used here for docking of new molecules as used for the fragments screening. The obtained docking scores of the compounds were used to rank them. The ligand-receptor interactions within the active sites of the protein were visualized using PyMOL. The docked ligand-receptor complexes were further analyzed by Molecular Mechanics-Generalized Born Surface Area (MM-GBSA)-based-free binding energy (ΔGbind) calculation with the help of the prime tool. The same docking protocol and MM-GBSA-free binding energy calculation were also been carried out for the approved SARS-CoV-2 Mpro inhibitor, Nirmaltrevir as a standard compound.
In silico physicochemical and pharmacokinetic parameter prediction
The new molecules with predicted good binding affinities towards Mpro were explored for their virtual physicochemical and pharmacokinetic parameters. The prediction was performed by the QikProp module of Maestro [30].
Molecular dynamic simulations
The molecular dynamic (MD) simulation study of the compounds (1-6) and Nirmatrelvir with Mpro was carried out for a period of 100 ns using GROMACS 2020.1 software [31]. The ligand-receptor complex generated in the molecular docking study was used as the initial point for simulation. CHARMM36, an all-atom force field was employed to find the ligand-receptor complex stability [32]. Ligand topology was built by CGenFF server [33, 34]. The ligand-receptor complex was built and solvated by the tip3p/SPC216 water model [35]. Neutralization of the solvate system having charged protein was performed by using Na + or Cl − ions. The energy minimization of the complex was carried out by the steepest descent method. It was followed by two sequential equilibration simulation phases using canonical (NVT) and isobaric-isothermic (NPT) ensemble for 100 ns each. The production MD simulation was carried out using NPT group, and long-range electrostatic interactions were identified using the particle mesh Ewald (PME) method [36]. The MD calculations were performed using the GROMACS 2020.1 simulation package at 300 K temperature and 1 bar pressure, and the resulting data were analyzed by plotting RMSD, RMSF, radius of gyration, and solvent accessible surface area (SASA).
Results and discussion
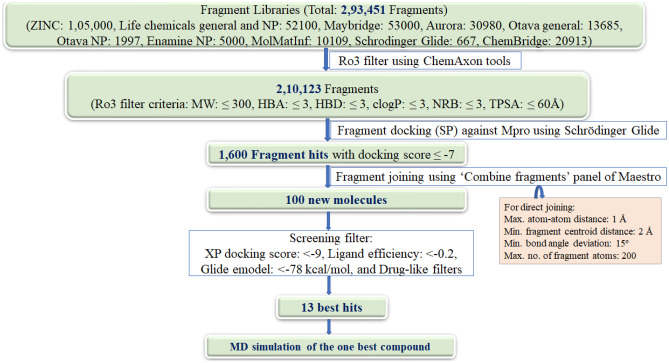
The stated fragment-based drug discovery investigation has been carried out in an effort to create a molecule that combines the properties of synthetic and natural compounds. The publicly available fragment libraries from a set of diversified synthetic chemicals and natural product like compounds were chosen for the present study. Figure 1 represented the schematic diagram for the workflow of the current work.
Fig. 1.
Schematic diagram of workflow
Fragment database screening and linking
A total of 293,451 diversified fragments have been retrieved from the aforementioned libraries. These fragments have been screened initially for the rule of three (Ro3) [37]. The Ro3 filter criteria were molecular weight: ≤ 300, number of H-bond acceptors (HBA): ≤ 3, number of H-bond donors (HBD): ≤ 3, clogP: ≤ 3, number of rotatable bonds (NRB): and ≤ 3, and total polarizable surface area (TPSA): ≤ 60Å. After applying this filter, 210,123 fragments have been obtained. All the filtered fragments were then prepared for energy minimization using LigPrep module. Four low-energy conformers have been generated for each ligand at physiological pH using OPLS3e force field.
These fragments were screened using the receptor grid generated by retaining the co-crystallized ligand N3 as a centre of grid. For the validation of generated grid, the co-crystallized ligand (N3) of PDB 6LU7 was first drained out, re-built, and re-docked into the active site of the protein against the prepared grid. Here, the N3 molecule exhibited an analogous pattern of orientation and interactions, maintaining the hydrogen bonding with Glu166, Gln189, and Thr190 residues of the active site of the protein [27]. The XP docking score of N3 for Mpro was −7.93 kcal/mol. This docking was further analysed by considering all-atom RMSD value of the re-docked N3 ligand with the co-crystallized ligand, and it was found to be 0.095 Å, which validates the docking protocol.
The standard precision (SP) algorithm of Glide was applied to screen the fragments through molecular docking. The SP docking scores of all the fragments were in the range of −9.0 to −5.6 kcal/mol. The top 1600 fragments have been found to have high docking score ≤ −7 kcal/mol. These fragment hits are linked through the “combine fragments panel” of Schrödinger to afford 100 new compounds. These new compounds are then prepared by LigPrep tool, keeping all the parameters same as done for the preparation of fragments.
Molecular docking of newly designed compounds
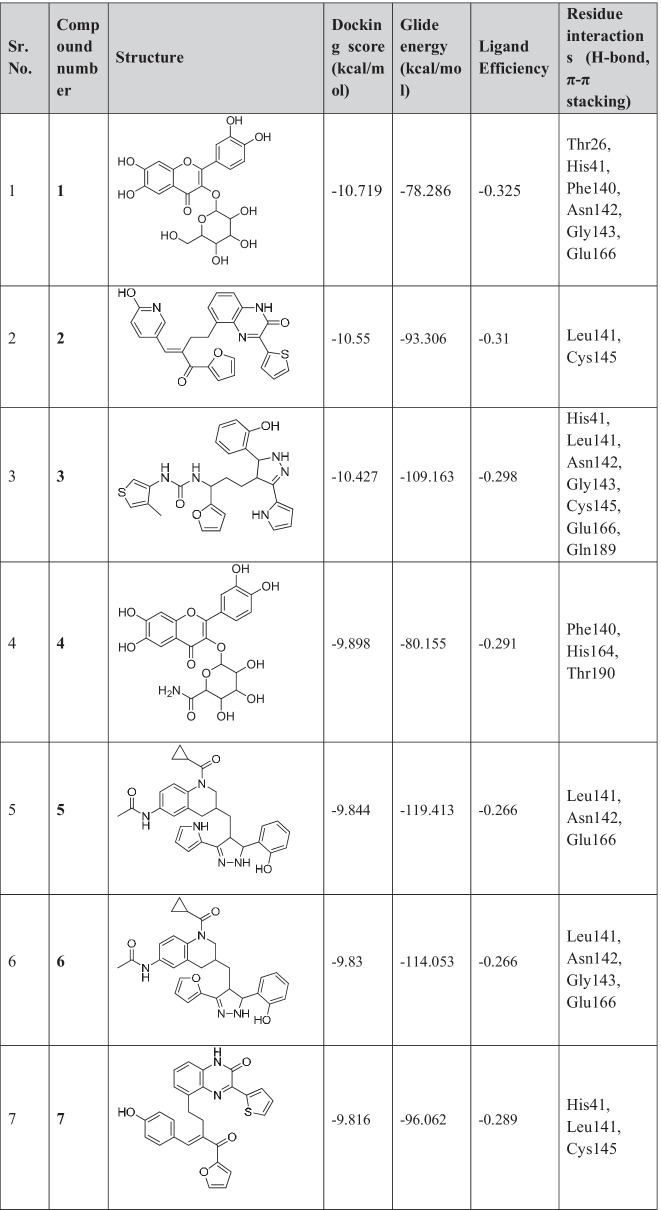
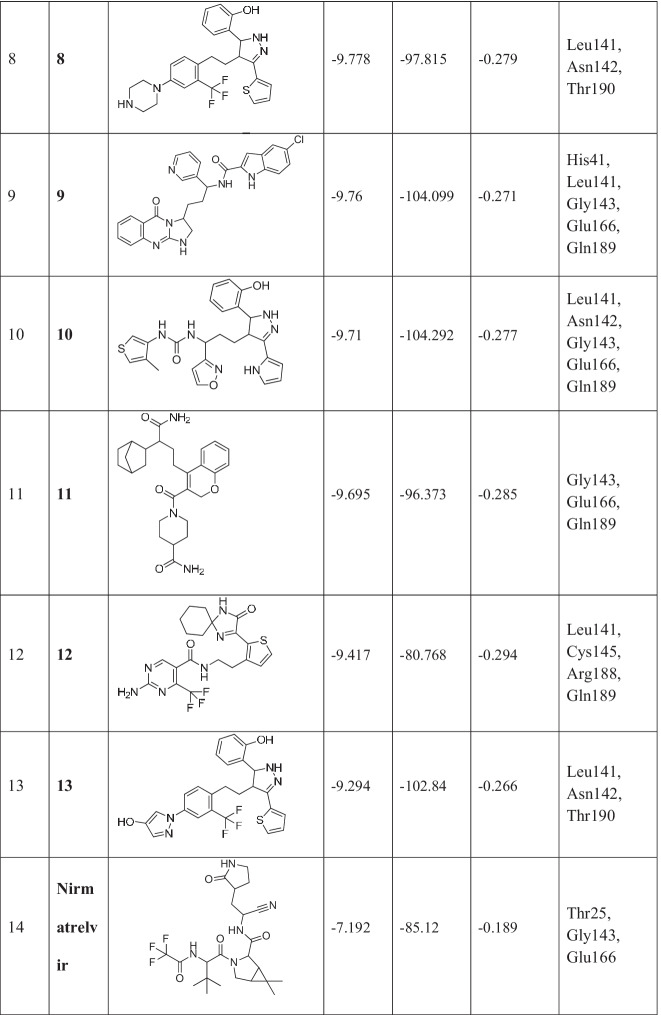
Newly designed compounds were docked using extra precision (XP) algorithm of Glide. The same grid was used for the docking of newly designed compounds as was prepared for fragments docking. Top-scoring 13 compounds with docking scores ranging from −10.71 to −9.29 kcal/mol were obtained (Table 1).
Table 1.
Chemical structures and docking results of newly formed compounds
These compounds were also screened based on their ligand efficiency and Glide emodel. Ligand efficiency is an estimate of the binding energy per atom of a ligand to its receptor. It is defined as the ratio of Gibb’s free energy to the number of hydrogen atoms present in the compound. It is directly proportional to the docking score. The ligand efficiencies of top-scored compounds were in the range of −0.36 to −0.26, which suggests that these compounds have high binding affinity with receptor. Glide utilizes emodel to identify the best pose of ligand and then ranks these best poses against one another with GlideScore. The shortlisted 13 compounds have Glide emodel values ranging from −119.14 to −80.15 kcal/mol.
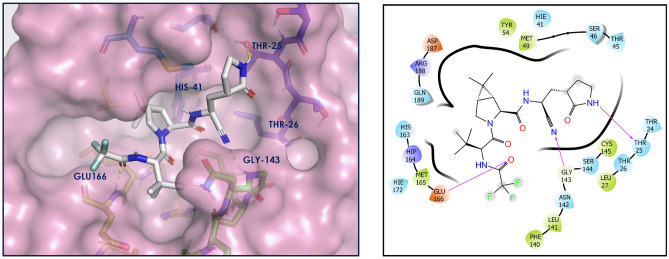
The top docked 13 compounds have better docking scores compared to the co-crystallized ligand N3 of PDB ID 6LU7 (−7.93 kcal/mol). The detailed molecular interaction study between Mpro and N3 has been described in our earlier articles [12, 27]. As a control/reference, nirmatrelvir, an approved Mpro inhibitor, has also been taken in the present study and the docking of it has been carried out using the same docking protocol before the docking of designed compounds. The interactions with Mpro have been shown in Fig. 2. Nirmatrelvir showed key interactions (H-bonding) with Thr25, Gly143, and Glu166, having a docking score of −7.19 kcal/mol.
Fig. 2.
Nirmatrelvir-Mpro interaction diagram. Nirmatrelvir is depicted as white sticks, and Mpro residues are presented as atom type color sticks
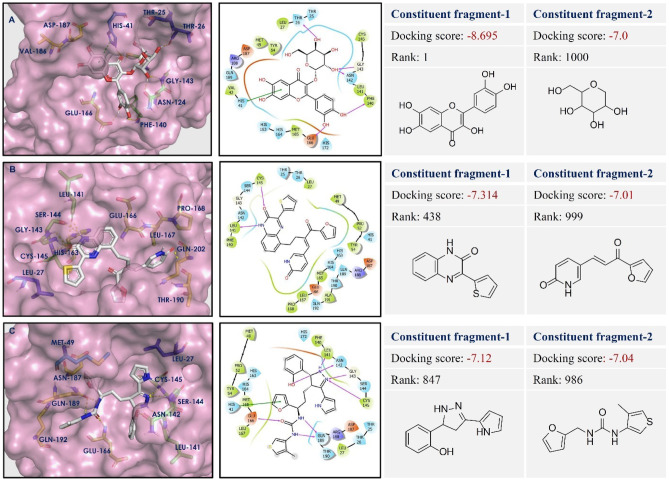
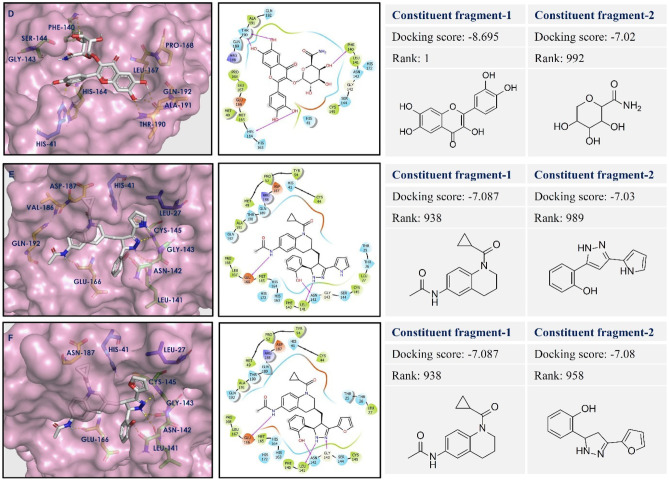
The designed 13 compounds showed the key interactions with active site of main protease. The 2D and 3D ligand-receptor interaction diagrams of the 6 top-scored compounds (1-6) have been shown in Figs. 3 and 4. All these compounds were prepared by linking of fragments using Schrödinger. The top 1600 best docked fragments have been selected for the same. These fragments were ranked from 1 to 1600 according to their docking scores. Compound (1) was constituted by the linking of rank 1 (docking score − 8.69 kcal/mol) and rank 1000 (docking score − 7.0 kcal/mol) fragments by ether linkage. Compound (1) has a structural similarity with natural glycosides. The aromatic ring of chromene of compound (1) formed π-π stacking with His41, the key amino acid residue of the active site of Mpro. The hydroxyl groups present in compound (1) exhibited H-bonding with Thr26, Phe140, Asn142, Gly143, and Glu166 amino acid residues. These amino acids are the active site residues of Mpro. Compound (2) was constituted by the linking of rank 438 (docking score − 7.31 kcal/mol) and rank 999 (docking score − 7.01 kcal/mol) fragments by ethyl chain linkage. The oxygen of amide group in quinoxalin-2(1H)-one ring of compound (2) showed hydrogen bonding with Cys145 residue of Mpro. This interaction is particularly important as the Cys145 amino acid residue is present in the catalytic dyad and acts as a nucleophile in the proteolytic cleavage of the natural substrate of Mpro [38]. The nitrogen of amide group in quinoxalin-2(1H)-one ring of compound (2) formed hydrogen bond with Leu141 of Mpro. Compound (3) was generated by the ethyl chain linking of rank 847 (docking score − 7.12 kcal/mol) and rank 986 (docking score − 7.04 kcal/mol) fragments. In compound (3), the oxygen of hydroxyl group, present in fragment 1 portion showed hydrogen bonding with Asn142 residue of Mpro. The NH- group and nitrogen of the pyrazole ring showed hydrogen bonding with Leu141 and Gly143, Cys145, respectively. The nitrogens of urea group present in fragement-2 portion of compound (3) showed hydrogen bonding with Gln189 of Mpro. H-bonding between the oxygen of urea and Glu166 residue provided stability to the ligand-receptor complex. The other compounds (4-13) also showed similar types of interactions with active site residues of the protein. The constituent fragments for compounds (4-6) have been shown in Fig. 4.
Fig. 3.
Ligand-receptor interaction diagram of compounds (1-3: A–C) at the active sites of Mpro protein (PDB ID: 6LU7). The two constituent fragments are shown on right side, ligands are depicted as white sticks, and Mpro residues are presented as atom type color sticks
Fig. 4.
Ligand-receptor interaction diagram of compounds (4-6: D–F) at the active sites of Mpro protein (PDB ID: 6LU7). The two constituent fragments are shown on right side, ligands are depicted as white sticks, and Mpro residues are presented as atom type color sticks
MM-GBSA-free binding energy calculations
The binding energy has been considered as a more accurate screening parameter than any other molecular docking scoring parameter. Thus, the docked ligand-receptor complexes were further analyzed for Molecular Mechanism-Generalized Born Surface Area (MM-GBSA) analysis to predict the free binding energy of ligand-receptor complexes [39]. The total free binding energy (MM-GBSA ΔGBind) was estimated as follows using the software:
where GComplex represents the energies of optimized ligand-receptor complex and GReceptor and GLigand represent the energies of optimized receptor and optimized ligand, respectively.
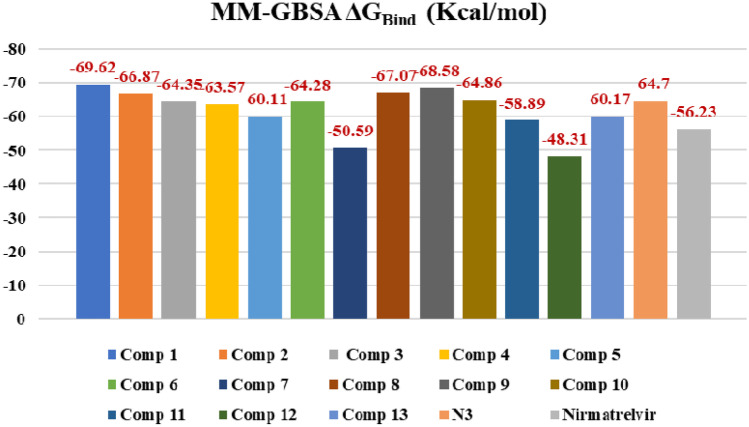
Prime MM-GBSA uses the VSGB 2.0 solvation model. MM-GBSA ΔGBind values of top-scored 13 compounds, co-crystal, N3 and standard Mpro inhibitor, and nirmatrelvir have been demonstrated in Fig. 5. As predicted by the MM-GBSA protocol, compound (1) has the most negative binding free energy (−69.62 kcal/mol), which is in agreement with the molecular docking results and showed the best binding affinity with Mpro. Compounds (9), (8), and (2) showed −68.58, −67.07, and −66.87 kcal/mol binding free energy, respectively, which also suggest better binding with the receptor. N3 showed −64.7 kcal/mol binding free energy which is similar to the experimental compounds but nirmatrelvir showed less binding affinity (−56.23 kcal/mol) compared to the experimental compounds.
Fig. 5.
MM-GBSA free binding energy values of compounds 1–13, N3, and Nirmatrelvir
In silico prediction of physicochemical and pharmacokinetics parameters
The 13 novel compounds screened based on docking score and ligand efficiency were then further screened for their physicochemical and pharmacokinetic parameters using QikProp tool (Tables 2 and 3). These parameters have been predicted to check the drug likeliness of the obtained novel compounds.
Table 2.
Predicted physicochemical parameters of compounds (1-13)
| Sr. No. | Compound number | MW (g/mol) | HBD | HBA | QPlogPo/w | PSA | Rotor |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 464.38 | 8 | 12 | −0.79 | 206.6 | 4 |
| 2 | 2 | 471.53 | 2 | 4 | 3.89 | 109.93 | 7 |
| 3 | 3 | 489.59 | 5 | 4 | 4.64 | 114.68 | 8 |
| 4 | 4 | 477.38 | 8 | 12 | −1.28 | 229.46 | 4 |
| 5 | 5 | 497.60 | 4 | 5 | 3.47 | 109.82 | 6 |
| 6 | 6 | 498.58 | 3 | 5 | 3.53 | 107.17 | 6 |
| 7 | 7 | 469.53 | 2 | 4 | 5.89 | 96.48 | 7 |
| 8 | 8 | 501.59 | 3 | 4 | 5.13 | 64.47 | 7 |
| 9 | 9 | 499.98 | 4 | 5 | 3.53 | 103.68 | 6 |
| 10 | 10 | 490.58 | 5 | 5 | 4.18 | 127.57 | 8 |
| 11 | 11 | 465.59 | 2 | 4 | 1.83 | 115.72 | 7 |
| 12 | 12 | 466.48 | 3 | 6 | 3 | 122.36 | 6 |
| 13 | 13 | 498.52 | 3 | 5 | 6.03 | 82.67 | 7 |
| Range as per QikProp module of Schrödinger | 130–725 | 0–6 | 2–20 | −2 to 6.5 | 7–200 | 0–15 | |
MW molecular weight, HBD hydrogen-bond donor atoms, HBA hydrogen-bond acceptor atoms, QPlogPo/w predicted octanol/water partition coefficient, PSA polar surface area, Rotor number of rotatable bonds
Table 3.
Predicted pharmacokinetic parameters of compounds (1–13)
| Sr. No. | Compound number | Volume | QPlogS | QPlogHERG | QPPCaco | QPlogBB |
|---|---|---|---|---|---|---|
| 1 | 1 | 1295.116 | −5.629 | −5.123 | 132.622 | −1.36 |
| 2 | 2 | 1212.159 | −2.265 | −5.052 | 3.027 | −3.602 |
| 3 | 3 | 1369.935 | −5.737 | −7.32 | 187.076 | −1.747 |
| 4 | 4 | 1490.377 | −6.773 | −6.153 | 363.682 | −1.213 |
| 5 | 5 | 1224.429 | −1.469 | −3.398 | 0.844 | −3.738 |
| 6 | 6 | 1466.536 | −5.961 | −5.694 | 286.681 | −1.318 |
| 7 | 7 | 1475.018 | −6.032 | −5.873 | 460.519 | −1.12 |
| 8 | 8 | 1376.969 | −6.218 | −7.439 | 425.858 | −1.396 |
| 9 | 9 | 1485.017 | −7.665 | −7.856 | 234.735 | −0.241 |
| 10 | 10 | 1478.488 | −7.049 | −7.472 | 298.099 | −1.401 |
| 11 | 11 | 1481.394 | −6.474 | −5.727 | 70.037 | −2.052 |
| 12 | 12 | 1443.777 | −3.591 | −2.7 | 31.12 | −2.024 |
| 13 | 13 | 1295.116 | −5.629 | −5.123 | 132.622 | −1.36 |
| Range as per QikProp module of Schrödinger | 500–2000 | −6.5 to 0.5 | Above −5 | < 25 poor, > 500 great | −3 to 1.2 | |
QPlogS predicted aqueous solubility, QPlogHERG, predicted IC50 value for blockage of HERG K + channels, QPPCaco caco-2 cell permeability, QPlogBB brain/blood partition coefficient
As per Lipinski’s rule of five [40], the most drug-like molecules should possess molecular weight ≤ 500, LogP ≤ 5, number of hydrogen bond donors ≤ 5, and number of hydrogen bond acceptors ≤ 10. Topological polar surface area (TPSA) and number of rotatable bonds are the other two parameters introduced by Veber and co-workers [41]. Except for a very small number of exceptions, all the compounds, as indicated in Table 2, adhere to the aforementioned two principles.
As shown in Table 3, pharmacokinetic indicators of the compounds, such as volume, QPlogS, QPlogHERG, QPPCaco, and QPlogBB, were also predicted. QPlogS is an indicator of aqueous solubility. QPlogHERG gives the predicted value for blockage of HERG K+ channels, an indicator for cardiotoxicity. It should be above −5. All the compounds complied with this parameter except compounds (5) and (12). QPlogBB is the predicted brain/blood barrier coefficient. QPPCaco is the indicator for oral absorption of a compound. It estimates the alleged gut-blood barrier permeability. A value of less than 25 predicts poor oral absorption. All the compounds exhibited better oral absorption, except compounds (2) and (5).
Molecular dynamics (MD)
The MD study was performed to comprehend the time-dependent stability of the complex between the most favourable compounds (1–6) and Mpro. The same MD protocol was also run for nirmatrelvir and Mpro complex as a reference. The study was carried out over a period of 100 ns using GROMACS2020.1 package. The docked pose of the ligand in complex was contemplated as the initial frame for MD study, and some statistical parameters such as root mean square deviation of protein (RMSD-P), root mean square deviation of ligand (RMSD-L), root mean square fluctuation of protein (RMSF-P), radius of gyration, and solvent accessible surface area (SASA) were determined.
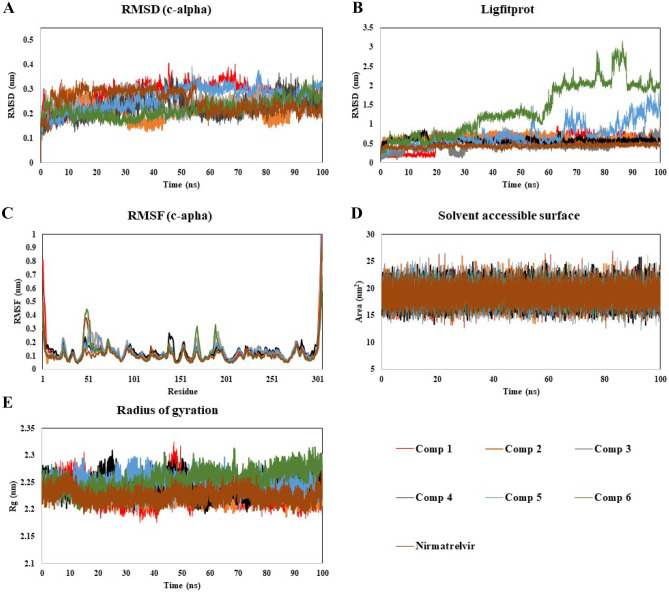
The protein RMSD is measured to determine the extent of movement of protein or atoms while ligand is present in the active site and indicating the extent of stability, deviation, and protein conformations during the simulation time. RMSD of protein Mpro in all the systems indicated that the simulations have equilibrated (Fig. 6A). The RMSD-P value for Mpro in complexation with compound (1) was in the range of 0.05–0.4 nm with an average of 0.28 nm for 100 ns. The range of the RMSD values of protein in all the ligand–protein complexes, including reference nirmatrelvir complex was found to be about 0.04–0.4 nm. This signifies the stability of the protein having designed compounds present in the active site of protein throughout the analysis time. To characterize the local changes of protein chain further, the RMSF of each residue in each ligand–protein complex was also analyzed throughout the simulation time (Fig. 6C). The RMSF-P value describes the residual mobility and integrity of the protein structure. For most of the systems, the obtained RMSF-P value for the residues up to 300 was below 0.38 nm, whereas the protein tail with residue number from 300 onwards displayed a fluctuation, but it was not participating in binding with the ligands. Thus, RMSD-P and RMSF-P results indicate the stability of protein during simulation, which proves the basis for using the protein structure further for ligand–protein interaction studies.
Fig. 6.
Stability analysis of compounds (1-6) and Nirmatrelvir with Mpro throughout 100 ns MD simulation A RMSD-P, B RMSD-L, C RMSF-P, and D SASA and E radius of gyration plots
The RMSD of ligands (compounds (1–6) including nirmatrelvir) with respect to protein has been investigated (Fig. 6B). The RMSD-L values for all the systems were found in the overall range of 0.04–1.8 nm (except some fluctuation found in the case of compound (6)), which implies the ligand-receptor complex stability without a significant change in orientation of the ligand in the active site during the analysis time.
To analyze the stability of ligand further in the binding pocket of protein, the surface area of the ligands accessible by the water molecules was calculated and analysed (Fig. 6D). The solvent accessible surface area by ligands for all the systems was found in the range of 13–26 nm2 which indicates that the ligands remained embedded in the binding pocket throughout the simulation time. The radius of gyration measures the compactness of ligand. The radius of gyration values for all the systems were observed in the range of 2.17–2.32 nm (Fig. 6E), suggesting the stable ligand profile and thus advocating the stable binding of designed compounds and nirmatrelvir as well.
To further validate the ligand-receptor interactions, two important energy terms have been extracted from 100 ns MD simulation trajectories: (A) short-range electrostatic/coulomb energy (Coul-SR) and (B) van der Waals/hydrophobic Lennard–Jones energy. The average values of observed Coul-SR were −139.25 ± 3.4, −42.17 ± 7.9, −126.26 ± 1.2, −148.59 ± 1.7, −44.21 ± 3.5, −40.16 ± 1.2, and −95.43 ± 2.5 kJ/mol for compounds (1), (2), (3), (4), (5), (6), and nirmatrelvir complexes, respectively; and the average values of observed LJ-SR were −116.13 ± 7.8, 114.09 ± 6.6, −139.39 ± 3.3, −223.88 ± 2.6, −118.48 ± 7.5, −116.29 ± 1.6, and 110.84 ± 2.6 kJ/mol for compounds (1), (2), (3), (4), (5), (6), and nirmatrelvir complexes, respectively. These numbers suggested that hydrophobic contacts contribute more to the complex’s stabilization than electrostatic interactions.
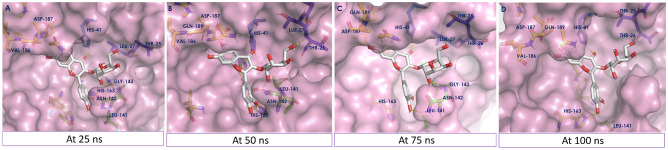
To acquire more information regarding the newly adopted ligand–protein conformations by compound (1) throughout the 100 ns MD simulation run time, the selected frames of the system were extracted and the conformational ligand–protein interactions have been visualized using PyMOL (Fig. 7). The interactions at 0 ns have been shown in Fig. 2A. During the analysis of interactions at 25, 50, 75, and 100 ns, no drastic fluctuation has been observed. These results were in accordance with RMSD-P and RMSD-L.
Fig. 7.
Ligand–protein complex conformations by compound (1) through selected trajectories A at 25 ns, B at 50 ns, C at 75 ns, and D at 100 ns
Conclusion
Fragment-based drug design has proven to be an efficacious tool for drug development in recent times. The present study attempts to design SARS-CoV-2 main protease inhibitors through an in silico fragment-based drug design approach. A huge library of diversified fragments of natural as well as chemical origin was used for the study. The rule of three was applied to the fragments for initial screening. The resulting fragments were screened against SARS-CoV-2 Mpro through molecular docking. The top-scored fragments were used for fragment linking to design new compounds. The 13 such molecules were secured by molecular docking against the Mpro, applying Lipinski’s rule of five and prediction of pharmacokinetic parameters. The 6 top-scored compounds, along with nirmatrelvir as a reference compound, were also checked for their ligand–protein complex stability using molecular dynamics, which gave promising results. As per the present study, the designed compounds were found to be more effective Mpro inhibitors compared to the reference compound. Thus, we obtained potential SARS-CoV-2 main protease inhibitors using in silico fragment-based drug design which can be further explored for COVID-19 drug discovery process.
Acknowledgements
The authors acknowledge the Drug Discovery Hackathon 2020 (DDH2020), MHRD, Government of India, and the proprietary software partners Schrödinger and ChemAxon for providing the training and software support to complete the present work.
Author contributions
The manuscript was written through the contributions of all authors. Ms. Divya M. Teli and Ms. Bansari Patel collected the data, performed the computational study, and analyzed the data. Ms. Divya M. Teli wrote the manuscript. Dr. Mahesh T. Chhabria conceptualized and reviewed the whole study and edited the manuscript. The authors have given approval to the final version of the manuscript.
Funding
The authors declare that there were no funds or grants received for the present work.
Availability of data and materials
Not applicable.
Declarations
Code availability
Not applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World health organization (2022) https://covid19.who.int/ Accessed on 28 Jul 2022
- 2.Moshkovits I, Shepshelovich D. Emergency use authorizations of COVID-19-related medical products. JAMA Intern Med. 2022;182:228–229. doi: 10.1001/jamainternmed.2021.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen W, Chen C, Tang J, Wang C, Zhou M, Cheng Y, Zhou X, Wu Q, Zhang X, Feng Z, Wang M, Mao Q. Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID-19: a meta-analysis. Ann Med. 2022;54:516–523. doi: 10.1080/07853890.2022.2034936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh R, Vijayan V. Chloroquine: a potential drug in the COVID-19 scenario. Trans Indian Natl Acad Eng. 2020;5:399–410. doi: 10.1007/s41403-020-00114-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel J, Berezowski I, Abdelmonem A, Taylor D, Pourmand A. Azithromycin for mild-to-moderate COVID-19. Lancet Respir Med. 2021;9:e99. doi: 10.1016/S2213-2600(21)00379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costanzo M, De Giglio M, Roviello G. SARS-CoV-2: Recent reports on antiviral therapies based on lopinavir/ritonavir, darunavir/umifenovir, hydroxychloroquine, remdesivir, favipiravir and other drugs for the treatment of the new coronavirus. Curr Med Chem. 2020;27:4536–4541. doi: 10.2174/0929867327666200416131117. [DOI] [PubMed] [Google Scholar]
- 7.Bryant A, Lawrie T, Dowswell T, Fordham E, Mitchell S, Hill S, Tham T. Ivermectin for prevention and treatment of COVID-19 infection: a systematic review, meta-analysis, and trial sequential analysis to inform clinical guidelines. Am J Ther. 2021;28:e434–e460. doi: 10.1097/MJT.0000000000001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougan M, Nirula A, Azizad M, Mocherla B, Gottlieb RL, Chen P, Hebert C, Perry R, Boscia J, Heller B, Morris J, Crystal C, Igbinadolor A, Huhn G, Cardona J, Shawa I, Kumar P, Adams AC, Van Naarden J, Custer KL, Durante M, Oakley G, Schade AE, Holzer TR, Ebert PJ, Higgs RE, Kallewaard NL, Sabo J, Patel DR, Dabora MC, Klekotka P, Shen L, Skovronsky DM. Bamlanivimab plus etesevimab in mild or moderate Covid-19. N Engl J Med. 2021;385:1382–1392. doi: 10.1056/nejmoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Annane D. Corticosteroids for COVID-19. J Intensive Med. 2021;1:14–25. doi: 10.1016/j.jointm.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahu K, Kumar R. Role of 2-deoxy-d-glucose (2-DG) in COVID-19 disease: a potential game-changer. J Fam Med Prim Care. 2021;10:3548. doi: 10.4103/jfmpc.jfmpc_1338_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agarwal A, Mukherjee A, Kumar G, Chatterjee P, Bhatnagar T, Malhotra P (2020) Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial). BMJ 371. 10.1136/bmj.m3939 [DOI] [PMC free article] [PubMed]
- 12.Teli DM, Shah MB, Chhabria MT. In silico screening of natural compounds as potential inhibitors of SARS-CoV-2 main protease and spike RBD: targets for COVID-19. Front Mol Biosci. 2021;7:429. doi: 10.3389/fmolb.2020.599079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forni G, Mantovani A, Forni G, Mantovani A, Moretta L, Rappuoli R, Rezza G, Bagnasco A, Barsacchi G, Bussolati G, Cacciari M, Cappuccinelli P, Cheli E, Guarini R, Bacci ML, Mancini M, Marcuzzo C, Morrone MC, Parisi G, Pasquino G, Patrono C, Curzio AQ, Remuzzi G, Roncaglia A, Schiaffino S, Vineis P. COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ. 2021;28:626–639. doi: 10.1038/s41418-020-00720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ullrich S, Nitsche C (2020) The SARS-CoV-2 main protease as drug target. Bioorg Med. Chem Lett 30. 10.1016/j.bmcl.2020.127377 [DOI] [PMC free article] [PubMed]
- 15.Huff S, Kummetha IR, Tiwari SK, Huante MB, Clark AE, Wang S, Bray W, Smith D, Carlin AF, Endsley M, Rana TM. Discovery and mechanism of SARS-CoV-2 main protease inhibitors. J Med Chem. 2022;65:2866–2879. doi: 10.1021/acs.jmedchem.1c00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui W, Yang K, Yang H. Recent progress in the drug development targeting SARS-CoV-2 main protease as treatment for COVID-19. Front Mol Biosci. 2020;7:398. doi: 10.3389/fmolb.2020.616341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall RJ, Mortenson PN, Murray CW. Efficient exploration of chemical space by fragment-based screening. Prog Biophys Mol Biol. 2014;116:82–91. doi: 10.1016/j.pbiomolbio.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad S, Mirza MU, Yean Kee L, Nazir M, Abdul Rahman N, Trant JF, Abdullah I. Fragment-based in silico design of SARS-CoV-2 main protease inhibitors. Chem Biol Drug Des. 2021;98:604–619. doi: 10.1111/cbdd.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortier J, Rakers C, Frederick R, Wolber G. Computational tools for in silico fragment-based drug design. Curr Top Med Chem. 2012;12:1935–1943. doi: 10.2174/156802612804547371. [DOI] [PubMed] [Google Scholar]
- 20.Bian Y, Xie XQ (2018) Computational fragment-based drug design: current trends, strategies, and applications. AAPS J 20. 10.1208/s12248-018-0216-7 [DOI] [PMC free article] [PubMed]
- 21.Gurung AB, Ali MA, Lee J, Farah MA, Al-Anazi KM (2021) An updated review of computer-aided drug design and its application to COVID-19. Biomed Res Int 2021. 10.1155/2021/8853056 [DOI] [PMC free article] [PubMed]
- 22.Sheng C, Zhang W. Fragment informatics and computational fragment-based drug design: an overview and update. Med Res Rev. 2013;33(3):554–598. doi: 10.1002/med.21255. [DOI] [PubMed] [Google Scholar]
- 23.Schrödinger (2020) Release 2020–1, LLC, New York
- 24.Protein data bank https://www.rcsb.org/ Accessed on 22 Oct 2020
- 25.Protein Preparation Wizard (2020) Epik, Schrödinger, LLC, New York, NY, 2020; Impact, Schrödinger, LLC, New York, NY, 2020; Prime, Schrödinger, LLC, New York, NY, 2020.
- 26.LigPrep, Schrödinger, LLC, NY, (2020)
- 27.Kanhed AM, Patel DV, Teli DM, Patel NR, Chhabria MT, Yadav MR. Identification of potential Mpro inhibitors for the treatment of COVID-19 by using systematic virtual screening approach. Mol Divers. 2021;25:383–401. doi: 10.1007/s11030-020-10130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glide, Schrödinger, LLC, NY (2020)
- 29.Choudhury C. Fragment tailoring strategy to design novel chemical entities as potential binders of novel corona virus main protease. J Biomol Struct Dyn. 2021;39:3733–3746. doi: 10.1080/07391102.2020.1771424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.QikProp, Schrödinger, LLC, NY (2020)
- 31.Lindahl, Abraham, Hess, van der Spoel, GROMACS (2020.1) Source code (Version 2020.1). Zenodo. 10.5281/zenodo.3685919. Accessed on 15 Nov 2020
- 32.Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, De Groot BL, Grubmüller H, MacKerell AD. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2016;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD (2009) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 9999. 10.1002/jcc.21367 [DOI] [PMC free article] [PubMed]
- 34.Yu W, He X, Vanommeslaeghe K, MacKerell AD. Extension of the CHARMM general force field to sulfonyl-containing compounds and its utility in biomolecular simulations. J Comput Chem. 2012;33:2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mark P, Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem A. 2001;105:9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- 36.Darden T, York D, Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 37.Jhoti H, Williams G, Rees DC, Murray CW. The “rule of three” for fragment-based drug discovery: where are we now? Nat Rev Drug Discov. 2013;12:644. doi: 10.1038/nrd3926-c1. [DOI] [PubMed] [Google Scholar]
- 38.Patel DV, Teli DM, Kanhed AM, Patel NR, Shah BS, Vora AK, Chhabria MT, Yadav MR. Identification of potential Mpro inhibitors for the treatment of COVID-19 by targeted covalent inhibition. Int J Quant Struct Relationships. 2021;6:58–77. doi: 10.4018/ijqspr.20210401.oa1. [DOI] [Google Scholar]
- 39.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2012;64:4–17. doi: 10.1016/j.addr.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 41.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.