Abstract
A 37-kb DNA fragment containing five fengycin synthetase genes, including fenC, fenD, fenE, fenA, and fenB, was cloned and sequenced. Among these genes, fenC encodes a fengycin synthetase 2,560 amino acids long with an estimated molecular mass of 287 kDa. This protein contains two amino acid activation modules, FenC1 and FenC2, which activate l-glutamic acid and l-ornithine, respectively. Primer extension, using mRNA isolated from the log-phase cells, identified a transcription start site located 86 nucleotides upstream from the initiation codon of fenC, implying that a promoter is located upstream from the start site. Primer extension using total RNA isolated from stationary-phase cells also identified a transcription start site located 61 nucleotides upstream from the initiation codon of fenC. Gene fusion studies demonstrated that in nHA medium, the cells transcribe the fengycin synthetase genes at two different stages of cell growth. The promoter is active during the log phase, and the activity reaches the highest level during the late log phase. The activity decreases sharply but is maintained at a low level for approximately 24 h after cells enter the early stationary phase. The results of this investigation also suggest that the transcription of fenC is positively regulated during the late log phase. Results presented herein provide further insight into fengycin synthesis by B. subtilis F29-3.
Fengycin, an antifungal antibiotic produced by Bacillus subtilis F29-3 (39), is a cyclic lipopeptide with the sequence fatty acid · l-Glu · d-Orn · l-Tyr · d-allo-Thr · l-Glu · d-Ala (d-Val) · l-Pro · l-Glu · d-Tyr · l-Ile, with a lactone bond connecting L-Tyr and L-Ile (19). Many peptide antibiotics produced by Bacillus spp., including fengycin, gramicidin S, surfactin, and tyrocidine, are synthesized nonribosomally by peptide synthetases (9, 14, 17, 18, 20, 27). These enzymes typically contain one or several amino acid activation modules approximately 1,000 amino acids long. In each peptide synthetase module, an adenylation domain recognizes and adenylates a specific amino acid (17, 31–33). The adenylated amino acid then forms a thioester bond with the cofactor 4′-phosphopantetheine at the thiolation domain (18, 32, 35, 40). A transpeptidation process then transfers the activated amino acid in the initiating module to the activated amino acid in the thiolation domain in the next module, ultimately forming a peptide. This process continues from one module to another until an antibiotic is completely synthesized (18). Kleinkauf and von Döhren postulated that peptide synthetases are connected to each other in a specific order in cells, thereby allowing the amino acids to link sequentially to form a peptide (18). In addition to the adenylation domain and the thiolation domain, each module, except for the module involved in initiation of peptide synthesis, also contains a region called the condensation domain (12, 18, 32–34), which is present in the N-terminal region upstream from the adenylation domain (20, 25, 41). The final module of a peptide synthetase, except for the one involved in the termination process, typically contains an epimerization domain (12) which converts an l-amino acid to a d-amino acid. The module involved in the termination of peptide synthesis contains a thioesterase domain in the C-terminal region (11, 24, 30, 32, 33).
In a previous study, we cloned a fengycin synthetase gene, fenB. This gene encodes a fengycin synthetase which activates l-isoleucine (24). This enzyme also contains a thioesterase domain, indicating that FenB participates in terminating fengycin synthesis (24). The entire chromosome of B. subtilis has recently been sequenced (21). According to this sequence, B. subtilis 168 contains a pps cluster. Among the five pps genes in this cluster, ppsE encodes a protein with 74% sequence identity to FenB (37, 38). Based on this sequence identity, Tosato et al. postulated that the proteins encoded by the pps genes are fengycin synthetases (38). Because the pps genes are not transcribed in B. subtilis 168 (38), the functions of these pps genes have not been demonstrated experimentally. Recently, Steller et al. (36) characterized the enzymes involved in the synthesis of plipastatin, an antibiotic similar to fengycin. They also showed that the genes encoding fengycin, plipastatin, and Pps synthetases are highly homologous (36). In light of above developments, we analyze the function of the first gene (fenC) of the fengycin synthetase operon and examine how this gene is transcribed in B. subtilis F29-3.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
Escherichia coli HB101 (5) was used for gene cloning. E. coli M15(pREP4) and pQE60 (Qiagen, Hilden, Germany) (6, 43) were used for overexpressing the adenylation domains of FenC. Plasmids pGEM-3Zf(+), pGEM-5Zf(+), and pGEM-7Zf(+) were purchased from Promega Corp. (Madison, Wis.). Plasmid pGHL6 (7.9 kb) is a B. subtilis-E. coli shuttle vector carrying a pC194 ori, a ColE1 ori, an ampicillin resistance gene, a chloramphenicol resistance gene, and luxAB genes of Vibrio harveyi. Plasmid pD917lux, a derivative of pD917, was constructed by inserting the luxAB genes into the NotI site in pD917 (8). This insertion created a transposon, Tn917lux, capable of generating transcriptional fusion on the chromosome of B. subtilis. LB medium (26) was used as a general-purpose medium; nHA medium (39) was used for analyzing the activity of the fenC promoter. An nHA-spore plate was prepared as previously described (9). Culture media were supplemented with ampicillin (100 μg/ml), chloramphenicol (5 μg/ml), and erythromycin (1 μg/ml) to select antibiotic-resistant colonies.
DNA techniques.
Plasmids in E. coli were screened as described by Kado and Liu (16) and purified by the alkaline lysis method described by Sambrook et al. (29). Plasmids in B. subtilis were screened by using an alkaline lysis method described elsewhere (9). A cosmid library of B. subtilis F29-3 was constructed with pHC79 in E. coli HB101 as previously described (9). Plasmids were isolated from each cosmid clone and blotted onto Zeta-Probe membrane (Bio-Rad, Richmond, Calif.) by using a dot blot apparatus (Bio-Rad) as recommended by the manufacturer. The DNA probe was prepared with [α-32P]dCTP (3,000 Ci/mmol; Amersham, Little Chalfont, Buckinghamshire, England), using a Rediprime labeling kit (Amersham). Finally, hybridization was performed as described elsewhere (29).
Transformation.
E. coli was transformed by the CaCl2 transformation method of Cohen and Chang (10). B. subtilis F29-3 was transformed by the protoplast transformation method of Imanaka et al. (15).
DNA sequencing.
DNA fragments to be sequenced were subcloned into pGEM-5Zf(+) or pGEM-7Zf(+). The sequencing reaction was performed with dye-labeled T7 and SP6 primers (Li-Cor, Lincoln, Neb.) and a SequiTherm Excel Long Read DNA sequencing kit (Epicentre Technologies, Madison, Wis.). The reaction mixtures were initially incubated at 95°C for 5 min; PCR was performed at 95°C for 30 s, 60°C for 30 s, and 70°C for 1 min, for 30 cycles. Finally, the DNA sequence was analyzed with an automated DNA sequencer (model 4000; Li-Cor).
Cloning of the adenylation domains of FenC1 and FenC2.
A DNA fragment encoding the adenylation domain (AD-FenC1) of FenC1 was amplified with primers fenCAN (5′-GCGCCATGGATATGGCAGAAAAACGTGAG) and fenCAC (5′-ACAGGATCCCTGGGAAAGCTTCATCTCTGTC). A DNA fragment encoding the adenylation domain (AD-FenC2) of FenC2 was amplified with primers fenCB5 (5′-TATCCATGGTTTCACAAGTGGATATACTC) and fenCBC (5′-ATAGGATCCCGCTGTCATTCCCTGCAA). A DNA fragment encoding ADS-FenC2 was amplified with fenCBN2 (5′-CCGGATCCATGGATATACTCAGCGAAAAAGAA) and fenCBC. PCR was performed at 94°C for 1 min, 50°C for 2 min, and 72°C for 3 min, for a total of 30 cycles. PCR fragments were digested with NcoI and BamHI and inserted into the NcoI-BamHI sites of pQE60. Finally, the plasmids were transformed into E. coli M15(pREP4).
Purification of AD-FenC1 and ADS-FenC2.
Cells were cultured at 37°C in LB-ampicillin broth to mid-log phase. Isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM was added to the culture medium to induce gene expression. Cells were cultured for 3 h, harvested by centrifugation, and lysed by a method previously described (24). Cell lysate was centrifuged at 15,000 rpm for 60 min with a Sorvall SS-34 rotor. Recombinant protein in the supernatant was purified by affinity chromatography with a His-Bind column (Novagen, Madison, Wis.) as previously described (24). Finally, proteins were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (22) and staining with Coomassie blue (Merck, Darmstadt, Germany).
ATP-PPi exchange assay.
Activities of AD-FenC1 and ADS-FenC2 were determined by an ATP-PPi exchange assay (23). Each reaction mixture contained 1 μCi of [32P]tetrasodium pyrophosphate (14.5 Ci/mmol) (NEN, Boston, Mass.), 2 mM amino acid, and 20 mM HEPES-morpholine ethanesulfonic acid buffer (pH 4.5). Reaction mixtures were incubated at 25°C for 10 min.
Primer extension.
B. subtilis F29-3 was inoculated in nHA broth at a density of 5 × 107 CFU/ml and cultured at 37°C for 6 h and for 20 h. Total RNA was prepared from the cells according to an acid-phenol extraction method described by Aiba et al. (1) except that the cells were homogenized with acid-treated glass beads (425 to 600-μm diameter; Sigma) in hot phenol. Primer fenp-4 (5′-CCCTCCAATTCTAATTTATAAGAGG) was end labeled with 0.3 μCi of [γ-32P]ATP (6,000 Ci/mmol; Amersham) and 8 U of T4 polynucleotide kinase (Promega) (29). Primer extension was performed with a kit purchased from Promega. Finally, labeled cDNA was analyzed on an 8% urea-polyacrylamide gel (29).
Generation of transcriptional fusion in pGHL6.
A 2,076-bp EcoRI fragment containing the sequence immediately upstream from the initiation codon of fenC was cloned into the EcoRI site of pGEM-3Zf(+). A fragment (2,083 bp) containing this region was then isolated from the plasmid by SacI digestion and cloned into the SacI site of pGHL6 to generate a transcriptional fusion between the promoter of fenC and luxAB.
Isolation of mutants containing Tn917lux insertion in fenC.
Plasmid pD917lux was transformed into B. subtilis F29-3. Transformants were selected on LB agar containing chloramphenicol. Cells were then cultured at 37°C for 3 h in LB broth containing 0.1 μg of erythromycin per ml to induce transposition (42). Cells were plated on LB-chloramphenicol agar and were incubated at 45°C overnight to eliminate pD917lux from the cells. Colonies resistant to chloramphenicol but sensitive to erythromycin were selected. The colonies were then spotted on an nHA-spore plate (9) to screen mutants incapable of synthesizing fengycin. Chromosomal DNA fragments adjacent to the transposons were cloned as previously described (8). Finally, the fragments adjacent to the transposon were sequenced to determine the locations of Tn917lux insertions.
Luciferase assay.
Cells expressing luxAB genes were cultured overnight in LB-ampicillin broth. Cells were inoculated into nHA-ampicillin broth at a density of 5 × 107 CFU/ml and cultured under constant shaking at 37°C. Luciferase activity exhibited by B. subtilis (measured in relative light units [RLU]) was monitored at different stages of cell growth with a luminometer (model LB953; Berthold, Bad Wildbad, Germany) as described elsewhere (7).
Nucleotide sequence accession numbers.
The nucleotide sequences of fenC, fenE, fenA, and fenB have been deposited in GenBank under accession no. AF087452, AF023465, AF023464, and L42523, respectively. The sequence of fenD was deposited in the EMBL database under accession no. AJ011849.
RESULTS
Isolation of cosmid clones containing fengycin synthetase genes.
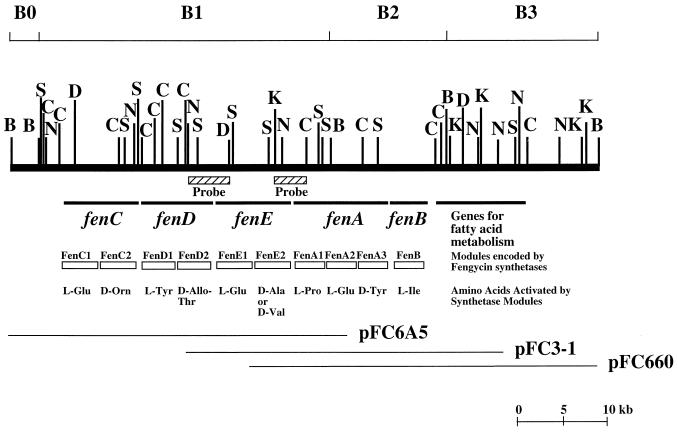
Genes for nonribosomal peptide synthesis are often clustered. If this is also the case for fengycin synthetase genes, it was estimated that a DNA fragment 37 kb long would be necessary to accommodate all of the fengycin synthetase genes. We have previously isolated a cosmid clone, pFC660, from a genomic library of B. subtilis F29-3 (9). Sequencing and Southern hybridization studies revealed that this cosmid contains only the 3′ region, approximately 17 kb in length, of the gene cluster (9). To isolate the cosmid clones containing the 5′ region of the gene cluster, we screened a genomic library of B. subtilis F29-3, using a 2.4-kb KpnI-ClaI fragment in pFC660 as a probe (Fig. 1). This screening identified an overlapping cosmid, pFC3-1 (Fig. 1). However, this clone did not extend far enough into the 5′ region to cover the entire gene cluster (Fig. 1). Therefore, the library was screened again, using a 4.2-kb NcoI-DraIII fragment in pFC3-1 as a probe (Fig. 1). This screening identified the cosmid clone pFC6A5. Restriction mapping depicted that the distance between the 5′ end of pFC6A5 and the end of the gene cluster (fenB) in pFC660 was approximately 45 kb (Fig. 1), a distance long enough to accommodate all of the fengycin synthetase genes.
FIG. 1.
Map of the B. subtilis F29-3 chromosome fragment containing the genes involved in fengycin synthesis. This fragment is arbitrarily divided into four regions (B0, B1, B2, and B3) according to the four BamHI sites on the map. The fragments used as probes for library screening are indicated. Plasmids pFC660, pFC3-1, and pFC6A5 are cosmid clones. B, BamHI; C, ClaI; D, DraIII; K, KpnI; N, NcoI; S, SacI.
Organization of fengycin synthetase genes.
The sequences of pFC6A5, pFC3-1, and pFC660 were determined. These three clones cover a 64-kb segment of the B. subtilis F29-3 chromosome that can be arbitrarily divided into four regions according to the BamHI sites on the map (Fig. 1). Computer analysis revealed that the B1 and the B2 regions contain five genes, fenC, fenD, fenE, fenA, and fenB (Fig. 1). Proteins encoded by these genes contain sequences highly conserved among peptide synthetases, suggesting that these proteins are peptide synthetases. Earlier studies demonstrated that inserting Tn917 or Tn917ac1 into these genes results in cells defective in fengycin synthesis (8, 9), suggesting that these five genes are involved in fengycin synthesis and that the enzymes encoded by these genes are fengycin synthetases.
Nucleotide sequence of fenC.
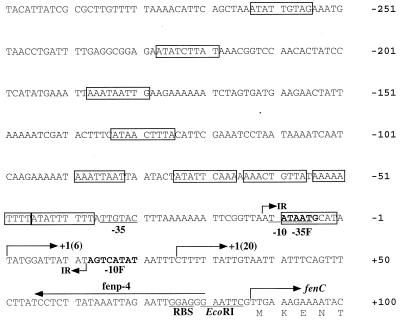
Sequence analysis revealed that fenC is the first gene in the cluster. This gene, 7,680 bp long, is present within the two NcoI fragments in the B1 region (Fig. 1). This gene starts from a TTG codon which is preceded by a ribosome-binding sequence, GGAGG (Fig. 2). The protein (FenC) encoded by this gene has an estimated molecular mass of 287,233 Da. FenC contains two sets of 10 core sequences that are highly conserved among peptide synthetases (Table 1) (32, 33), suggesting that FenC contains two amino acid activation modules, FenC1 and FenC2. In addition to these core sequences, FenC1 and FenC2 contain a condensation domain at N-terminal regions. FenC2 contains an epimerization domain with seven conserved motifs marking the end of the enzyme (Table 1).
FIG. 2.
Sequence of the region upstream from fenC. −10 and −35 denote a ςA-type promoter; −10F and −35F denote sequences homologous to the −10 and −35 sequences of ςF promoters. Sequences homologous to the 9-bp direct repeats in the gltA and gltC promoters are boxed. 1(6) and 1(20), transcription start sites identified with RNA prepared from cells cultured for 6 and 20 h, respectively; IR, 24-bp region of dyad symmetry; RBS, ribosome-binding site; fenp-4, primer used to identify the transcription start site.
TABLE 1.
Comparison of conserved sequences in peptide synthetases and FenC
| Domain | Core | Sequence conserved in peptide synthetasesa | Sequence in FenC | Module in FenC | Encoding region in fenC (nucleotides) |
|---|---|---|---|---|---|
| Adenylation | A1 | L(T/S)YxEL | LTYAQL | C1 | 1542–1559 |
| ISYRHL | C2 | 4650–4667 | |||
| A2 | LKAGxAYL(V/L)P(L/I)D | LKAGGAYVPLD | C1 | 1673–1705 | |
| MKAGGVYIPID | C2 | 4785–4817 | |||
| A3 | LAYxxYTSG(S/T)TGxPKG | LAYVIYTSGSTGQPKG | C1 | 1887–1934 | |
| SAYIIYTSGTTGAPKG | C2 | 5010–5057 | |||
| A4 | FDxS | FDAS | C1 | 2037–2048 | |
| FDVF | C2 | 5163–5174 | |||
| A5 | NxYGPTE | HGYGPTE | C1 | 2319–2339 | |
| NSYGVTE | C2 | 5454–5474 | |||
| A6 | GELxIxGxG(V/L)ARGYL | GELYIAGAGVARGYL | C1 | 2481–2525 | |
| GELCIAGAGVAKGYH | C2 | 5616–5660 | |||
| A7 | Y(R/K)TGDL | YKTGDL | C1 | 2589–2606 | |
| YRTGDL | C2 | 5724–5741 | |||
| A8 | GRxDxQVKIRGxRIELGEIE | GRLDDQVKIRGYRIEPGECE | C1 | 2643–2702 | |
| GRIDHQVKINGYRIETEEIE | C2 | 5778–5837 | |||
| A9 | LPxYM(I/V)P | LPGYMIP | C1 | 2835–2855 | |
| LPAYMVP | C2 | 5979–5999 | |||
| A10 | NGK(V/L)DR | SGKLDR | C1 | 2905–2922 | |
| NGKLDR | C2 | 6039–6056 | |||
| Thiolation | T | DxFFxxLGG(H/D)S(L/I) | DNFFDRGGNSL | C1 | 3039–3071 |
| DSFFELGGDSI | C2 | 6180–6212 | |||
| Epimerization | E1 | PIQxWF | PIQSRF | C2 | 6369–6386 |
| E2 | HHxISDG(W/V)S | HHLAVDGVS | C2 | 6768–6794 | |
| E3 | DxLLxAxG | ELLLTALG | C2 | 7113–7136 | |
| E4 | EGHGRE | EGHGGV | C2 | 7185–7202 | |
| E5 | RTVGWFTxxYP(Y/V)PFE | RTVGWFTSIYPILLD | C2 | 7230–7274 | |
| E6 | PxxGxGYG | PHKGTGYG | C2 | 7350–7373 | |
| E7 | FNYLG(Q/R) | FNYLGQL | C2 | 7422–7442 | |
| Condensation | C1 | SxAQxR(L/M)(W/Y)xL | THAQRRVWFT | C1 | 111–140 |
| SSAQKRMYVL | C2 | 3244–3273 | |||
| C2 | RHExLRTxF | RHESLRTSF | C2 | 3381–3408 | |
| C3 | MHHxISDG(W/V)S | FHHIIMDDIS | C1 | 492–521 | |
| MHHIISDGVS | C2 | 3606–3635 | |||
| C4 | YxD(F/Y)AVW | YKDYAVW | C2 | 3695–3715 | |
| C5 | (I/V)GxFVNT(Q/L)(C/A)xR | LGMFVSSLPIR | C1 | 949–981 | |
| LGMFVNTLALR | C2 | 4050–4082 | |||
| C6 | (H/N)QD(Y/V)PFE | HQDYPFE | C2 | 4157–4177 | |
| C7 | RDxSRNPL | RDMSRNPV | C2 | 4206–4229 |
From reference 25.
Expression and purification of the amino acid adenylation domains of FenC.
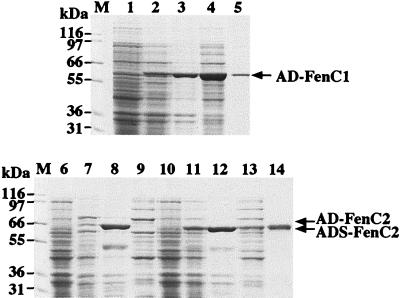
The N-terminal and C-terminal regions of a peptide synthetase can be deleted without significantly affecting the catalytic activity of the adenylation domain of the enzyme (32, 33). It is also known that the region required for amino acid adenylation in a peptide synthetase resides in the region from the 100th amino acid upstream from the core A2 sequence to the 100th amino acid downstream from the core A8 sequence of a peptide synthetase (Table 1) (27). Therefore, we cloned the DNA fragment (nucleotides 1389 to 2996 of fenC) encoding this region in FenC1 into pQE60 and overexpressed the adenylation domain in E. coli M15(pREP) (Fig. 3, lanes 1 and 2). The overexpressed recombinant protein (AD-FenC1) was present in both supernatant and pellet fractions of cell lysate (lanes 3 and 4). AD-FenC1 was purified from the supernatant fraction by affinity chromatography with a His-Bind column. This single purification step purified AD-FenC1 to near homogeneity (lane 5). A DNA fragment (nucleotides 4483 to 6134 of fenC) encoding the same region (AD-FenC2) in FenC2 was also cloned and overexpressed (lanes 6 to 9). However, the recombinant protein was insoluble and was present in the pellet fraction (lane 8) but not in the supernatant fraction (lane 9) of the cell lysate. AD-FenC2 was also shortened from the N-terminal end. A recombinant protein, ADS-FenC2 (lanes 10 to 14), which was three amino acids shorter than AD-FenC2, was present in both supernatant and pellet fractions of the cell homogenate (lanes 12 and 13). This allowed purification of the recombinant protein from the supernatant fraction by affinity chromatography with a His-Bind column (lane 14).
FIG. 3.
Overexpression and purification of recombinant FenC adenylation domains. Proteins were purified from cells overexpressing AD-FenC1 (lanes 1 to 5), AD-FenC2 (lanes 6 to 9), and ADS-FenC2 (lanes 10 to 14). Cell extracts were prepared before (lanes 1, 6, and 10) or after (lanes 2, 7, and 11) IPTG induction. Cell extracts were centrifuged and separated into pellet (lanes 3, 8, and 12) and supernatant (lanes 4, 9, and 13) fractions. Recombinant AD-FenC1 (lane 5) and ADS-FenC2 (lane 14) were further purified with a His-Bind column. Proteins were analyzed by SDS-PAGE and stained with Coomassie blue. Arrows indicate overexpressed recombinant proteins. The positions of molecular mass markers (M) are shown at the left.
Substrate specificity.
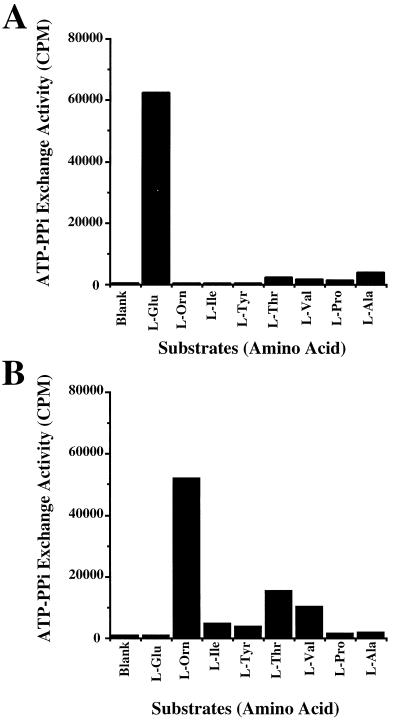
The enzymatic activities of recombinant AD-FenC1 and ADS-FenC2 proteins were determined by an ATP-PPi exchange assay (23). The amino acids used for the assay included the eight amino acids in the fengycin molecule (Fig. 4) as well as other common amino acids. Adding l-glutamic acid to the reaction mixture containing 6 μg of AD-FenC1 produced the highest ATP-PPi exchange activity (Fig. 4A), approximately 16- to 207-fold higher than the activities exhibited by the other amino acids. This finding suggests that FenC1 has a substrate specificity toward l-glutamic acid. The result also demonstrated that FenC2 specifically activates l-ornithine. Adding l-ornithine to the reaction mixture containing 20 μg of ADS-FenC2 also produced the highest ATP-PPi exchange activity (Fig. 4B), approximately 3- to 51-fold higher than the activities exhibited by the other amino acids. Adding the other 13 common amino acids to the reaction mixtures produced only background levels of ATP-PPi exchange activity (data not shown). Notably, ADS-FenC2 was less active than AD-FenC1. Approximately three times more ADS-FenC2 was necessary to achieve roughly the same level of ATP-PPi exchange activity exhibited by AD-FenC1.
FIG. 4.
ATP-PPi exchange activities of AD-FenC1 (A) and ADS-FenC2 (B).
Biochemical characteristics.
The catalytic activities of recombinant AD-FenC1 and ADS-FenC2 were examined under different reaction conditions. Similar to FenB (24), these two enzymes had an optimum activity at 25°C, at pH 4.5 to 5.0, and with an Mg2+ concentration at 10 mM in a buffer containing 2 mM EDTA.
Determination of the transcription start site of fenC mRNA.
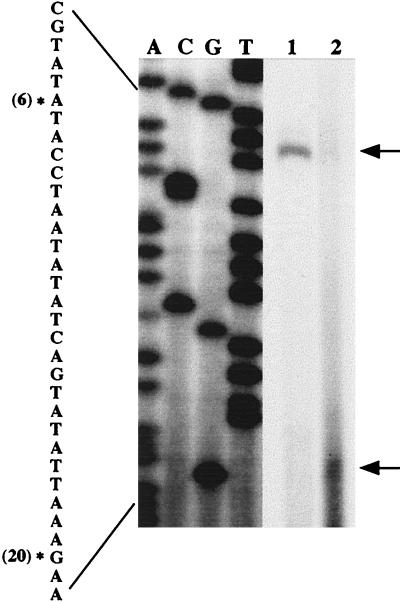
The presence of a promoter in the region immediately upstream from fenC could be predicted because fenC is the first gene of the gene cluster. Therefore, a primer complementary to the region between +56 and +80 (Fig. 2) was used to identify the transcription start site of the mRNA by primer extension. We use total RNA prepared from the cells cultured for 6 h and identified a start site that was located 86 nucleotides upstream from the initiation codon of fenC (Fig. 2 and 5). Sequence analysis identified −10 and −35 sequences typical for a ςA promoter. A 24-bp region of dyad symmetry was found between nucleotides −12 and +12. The function of this structure is unknown. We also performed a primer extension experiment with total mRNA prepared from cells cultured for 20 h and identified another transcription start site located 61 nucleotides upstream from the initiation codon of fenC (Fig. 2 and 5). Sequences that are homologous to the nine conserved 9-bp direct repeats (5′-ATATTGTTT) in gltA and gltC promoters (4) were also identified (Fig. 2).
FIG. 5.
Determination of the transcription start site. A, C, G, and T denote the dideoxynucleotides used to terminate the reactions. Asterisks indicate the 5′ terminus of RNA; arrows indicate the cDNA products of primer extension; (6) and (20) denote RNA prepared from cells cultured for 6 and 20 h, respectively.
Isolation of mutants containing Tn917lux insertion in fenC.
We have generated mutants defective in fengycin synthesis with Tn917lux. Among these mutants, five contained the Tn917lux insertion in fenC. Tn917lux in these mutants, including FX12, FX17, FX18, FS26, and FS41, was inserted at nucleotides 769, 1871, 5056, 5563, and 6989 in fenC, respectively. Among these five mutants, only the luxAB genes in mutant FS41 were oriented facing the promoter of fenC; this allowed the mutant to produce light. Tn917lux in the other mutants was inserted in the opposite orientation.
Transcription of fenC.
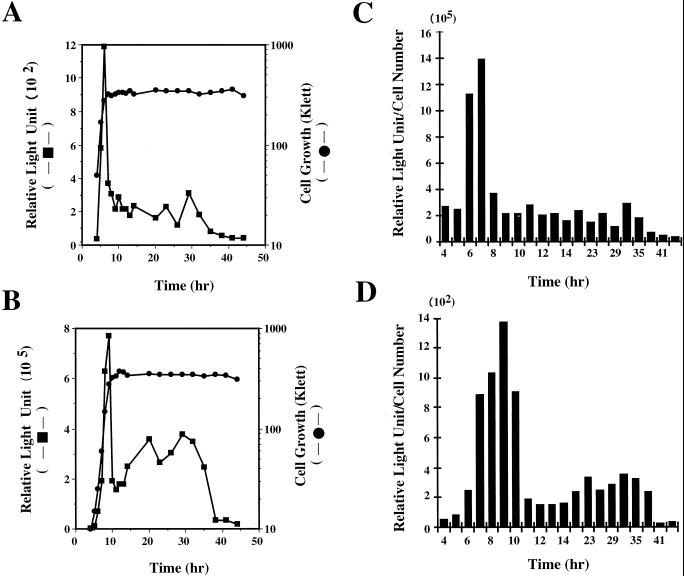
Mutant FS41 was cultured in nHA medium, and the transcription of fenC was examined by monitoring the luciferase activity exhibited by the cells. Cells were inoculated at a density of approximately 5 × 107 CFU/ml and cultured under constant shaking at 37°C for a total of 44 h. Under these conditions, cells reached late log phase approximately 7 h after inoculation. Cells exhibited low levels of luciferase activity during early log phase (Fig. 6A). The activity started to increase from h 6 and reached the highest level at h 7, when the cells were at late log phase (Fig. 6A). After cells entered early stationary phase, the activity immediately decreased by 75% (Fig. 6A). However, transcription of fenC was maintained at this level for approximately a day and then decreased to the basal level. A 2.1-kb fragment containing the promoter of fenC was also cloned into a fusion vector, pGHL6 (Fig. 2). In B. subtilis F29-3, this plasmid (pGFS) exhibited a level of luciferase activity higher than that displayed by mutant FS41 (Fig. 6B), presumably due to the copy numbers of luxAB genes in the cells. B. subtilis F29-3(pGFS) had a growth rate lower than that of mutant FS41. As in the experiment with mutant FS41, the cells were also inoculated at a concentration of 5 × 107 CFU/ml. The cells reached late log phase approximately 9 h after inoculation (Fig. 6A). B. subtilis F29-3(pGFS) exhibited the highest level of luciferase activity at late log phase; the activity decreased by 75% at the early stationary phase and then remained at the 50% level for approximately 24 h (Fig. 6B).
FIG. 6.
Transcription of luxAB genes by mutant FS41 (A) and by B. subtilis F29-3(pGFS) (B). Cells were cultured in nHA broth. Cell growth was monitored with a Klett-Summerson photoelectric colorimeter; luciferase activity was measured with a luminometer. Luciferase activities shown in panels A and B were also calculated on a per-cell basis (C and D) to demonstrate that increases in RLU during late log phase were unrelated to cell growth.
DISCUSSION
This study identified five fengycin synthetase genes clustered in a 37-kb region on the chromosome of B. subtilis F29-3. These genes, in the order fenC-fenD-fenE-fenA-fenB (Fig. 1), are separated by short intercistronic regions of 16 to 32 nucleotides. Two transcription start sites, situated 86 and 61 nucleotides upstream from the initiation codon of fenC, were also identified by primer extension (Fig. 2). Neither subcloning experiments with a promoter-probe vector (pGHL6) nor computer analysis revealed any additional promoter sequence within the gene cluster, implying that these five fengycin synthetase genes are organized in an operon and are transcribed from the identified promoter. An earlier study identified a sequence resembling a transcription stop signal located immediately downstream from fenB (24), suggesting that fenB is the last gene of the operon. This study demonstrated that the two amino acid activation modules encoded by fenC (the first gene of the operon) activate the first two amino acids of the fengycin molecule (Fig. 4). An earlier study also demonstrated that fenB, the last gene in the operon, activates the last amino acid of fengycin molecule (24). In addition to FenC and FenB, the functions of several other fengycin synthetase modules, including FenD2, FenE1, FenA1, FenA2, and FenA3, are now known (unpublished data). From these results, it seems likely that fengycin synthetase genes and the amino acid sequence in fengycin are colinear.
Tosato et al. have sequenced five pps genes of B. subtilis 168 (38). They reported that the sequences between FenB and PpsE, a protein encoded by ppsE, were 90% identical (38). Although this percentage was calculated incorrectly and the actual sequence identity is 73.9%, they postulated that PpsE may have the same enzymatic function as FenB. Owing to the sequence identity, that investigation postulated that the other four pps genes may also encode fengycin synthetases (38). From the sequence of the fengycin synthetase operon, this may indeed be the case because the extents of identity are as follows: between FenC and PpsA, 78.9%; between FenD and PpsB, 80.2%; between FenE and PpsC, 81.3%; between FenA and PpsE, 72.3%; and between FenB and PpsE, 73.9%. However, since B. subtilis 168 does not transcribe pps genes and does not synthesize fengycin (38), it is not clear whether these Pps proteins are still functional. Tosato et al. (38) did not demonstrate the functions of Pps proteins experimentally but did predict the functions of Pps proteins, although they predicted the functions of several enzymes incorrectly (24, 38). Interestingly, we have found a gene cluster in B. subtilis F29-3 which resembles the surfactin synthetase operon of B. subtilis 168 (unpublished data). The fact that strain F29-3 does not produce surfactin (39) raises the possibility that strains F29-3 and 168 evolved from a common ancestor that produced both fengycin and surfactin. The fengycin synthetase genes in strain 168 probably lost their functions during evolution and became pseudogenes. We also compared the sequences located upstream of fenC and ppsA. We found that the sequences in the fenC and the ppsA promoters differ by one nucleotide in the −10 region: TATAAT in the fenC promoter and TATAGT in the pps promoter. It is not known if this single-nucleotide change results in a lack of transcription of pps genes.
When the two amino acid adenylation domains of FenC were expressed in E. coli, it was found that recombinant AD-FenC1 was a soluble protein that could be purified from the cell homogenate (Fig. 4). However, AD-FenC2 was insoluble in E. coli and was difficult to refold. The solubility of the enzyme could be changed by deleting the three N-terminal amino acids of AD-FenC2 (Fig. 3). However, deleting these three amino acids may have influenced the catalytic activity of the protein since approximately three times more ADS-FenC2 was necessary to achieve the same level of PPi-exchange activity exhibited by AD-FenC1 (Fig. 4). This finding is unsurprising since changing the length of an adenylation domain has been demonstrated to influence enzymatic activity (13). This study found that like FenB (24), AD-FenC1 and ADS-FenC2 have a low pH optimum between 4.5 and 5.0 and a temperature optimum at 25°C.
A primer extension study, using total RNA prepared from log-phase cells, identified a transcription start site located 86 nucleotides upstream from the initiation codon of fenC. Also found was a −10 and a −35 sequence typical of a ςA promoter, explaining why fenC is transcribed during log phase (Fig. 6). Primer extension study also revealed that a second promoter which has a transcription start site located 61 nucleotides upstream from the initiation codon of fenC (Fig. 2 and 5) is involved in the transcription of fengycin synthetase genes during stationary phase. Upstream from this start site, we found a −10 sequence of AGTCATAT and a −35 sequence of ATAATG with a spacing of 16 nucleotides. These sequences show a low degree of homology with the −10 and −35 sequences of ςF promoters. Whether ςF regulates this promoter is not known. Fusion studies revealed that fenC transcription increases during log phase. We have calculated the RLU on a per-cell basis. The RLU-to-cell number ratios were relatively constant during early and mid log phase (Fig. 6C and D). However, the ratios increased significantly after mid log phase (Fig. 6C and D), indicating that the increase in RLU (Fig. 6A and B) is not simply due to cell growth. This increase also cannot be attributed to accumulation of luxAB mRNA in the cells, because the half-life of the mRNA at this stage is less than 3 min (data not shown). Therefore, it is likely that transcription of fenC is positively regulated during late log phase. When cells enter stationary phase, this positive regulatory mechanism is probably turned off, resulting in a 75% decrease in transcription activity (Fig. 6A and B). The ςA promoter, which is active during stationary phase (28), may continue to transcribe the fengycin synthetase operon at this stage, albeit at low levels (Fig. 5, lane 2), and the downstream ςF-like promoter may play an important role in transcribing the mRNA during stationary phase.
One of the fengycin synthesis mutants that we have isolated, FX28, contains a Tn917lux insertion in a gene highly homologous to the gltC of strain 168 (unpublished data). GltC is a transcription activator that regulates the transcription of gltA and gltC (4); this is the reason why we compared the sequence of the promoter of fenC with the sequences of gltA and gltC promoters. Although no GltC-binding sequence (2, 3) was found in the promoter of fenC, copies of 9-bp direct repeats, which were present in the promoters of gltA and gltC (4), were found. Because the functions of these repeats are unknown, it is not certain whether GltC regulates the fenC promoter. Furthermore, a nitrogen source in a culture medium can affect the timing and amount of fengycin synthesis (39). Therefore, investigation is under way to determine whether the genes involved in nitrogen metabolism, including gltC, regulate fengycin synthesis. Fengycin synthesis apparently involves large numbers of genes, and their expression probably involves complex regulatory mechanisms. Results presented herein provide valuable reference for future studies on the mechanisms involved in fengycin synthesis.
ACKNOWLEDGMENTS
This research was support by Medical Research Grant CMRP759 from the Change-Gung Memorial Hospital and by Biological Research Grant NSC-88-2314-B-182-004 from the National Science Council of the Republic of China.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Belitsky B R, Janssen P J, Sonenshein A L. Sites required for GltC-dependent regulation of Bacillus subtilis glutamate synthase expression. J Bacteriol. 1995;177:5686–5695. doi: 10.1128/jb.177.19.5686-5695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky B R, Sonenshein A L. Mutations in GltC that increase Bacillus subtilis gltA expression. J Bacteriol. 1995;177:5696–5700. doi: 10.1128/jb.177.19.5696-5700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohannon D E, Sonenshein A L. Positive regulation of glutamate biosynthesis in Bacillus subtilis. J Bacteriol. 1989;171:4718–4727. doi: 10.1128/jb.171.9.4718-4727.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 6.Bujard H, Gentz R, Lanzer M, Stueber D, Mueller M, Ibrahimi I, Haeuptle M T, Dobberstein B. A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Methods Enzymol. 1987;155:416–433. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- 7.Carmi O A, Stewart G S, Ulitzur S, Kuhn J. Use of bacterial luciferase to establish a promoter probe vehicle capable of nondestructive real-time analysis of gene expression in Bacillus spp. J Bacteriol. 1987;169:2165–2170. doi: 10.1128/jb.169.5.2165-2170.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang L K, Chen C L, Chang Y S, Tschen J S, Chen Y M, Liu S T. Construction of Tn917ac1, a transposon useful for mutagenesis and cloning of Bacillus subtilis genes. Gene. 1994;150:129–134. doi: 10.1016/0378-1119(94)90871-0. [DOI] [PubMed] [Google Scholar]
- 9.Chen C L, Chang L K, Chang Y S, Liu S T, Tschen J S. Transposon mutagenesis and cloning of the genes encoding the enzymes of fengycin biosynthesis in Bacillus subtilis. Mol Gen Genet. 1995;248:121–125. doi: 10.1007/BF02190792. [DOI] [PubMed] [Google Scholar]
- 10.Cohen S N, Chang A C. Recircularization and autonomous replication of a sheared R-factor DNA segment in Escherichia coli transformants. Proc Natl Acad Sci USA. 1973;70:1293–1297. doi: 10.1073/pnas.70.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosmina P, Rodriguez F, de Ferra F, Grandi G, Perego M, Venema G, van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993;8:821–831. doi: 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 12.de Crécy-Lagard V, Marliere P, Saurin W. Multienzymatic nonribosomal peptide biosynthesis: identification of the functional domains catalysing peptide elongation and epimerisation. Life Sci. 1995;318:927–936. [PubMed] [Google Scholar]
- 13.Elsner A, Engert H, Saenger W, Hamoen L, Venema G, Bernhard F. Substrate specificity of hybrid modules from peptide synthetases. J Biol Chem. 1997;272:4814–4819. doi: 10.1074/jbc.272.8.4814. [DOI] [PubMed] [Google Scholar]
- 14.Fuma S, Fujishima Y, Corbell N, D’Souza C, Nakano M M, Zuber P, Yamane K. Nucleotide sequence of 5′ portion of srfA that contains the region required for competence establishment in Bacillus subtilis. Nucleic Acids Res. 1993;21:93–97. doi: 10.1093/nar/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imanaka T, Fujii M, Aramori I, Aiba S. Transformation of Bacillus stearothermophilus with plasmid DNA and characterization of shuttle vector plasmids between Bacillus stearothermophilus and Bacillus subtilis. J Bacteriol. 1982;149:824–830. doi: 10.1128/jb.149.3.824-830.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleinkauf H, von Döhren H. Nonribosomal biosynthesis of peptide antibiotics. Eur J Biochem. 1990;192:1–15. doi: 10.1111/j.1432-1033.1990.tb19188.x. [DOI] [PubMed] [Google Scholar]
- 18.Kleinkauf H, von Döhren H. A nonribosomal system of peptide biosynthesis. Eur J Biochem. 1996;236:335–351. doi: 10.1111/j.1432-1033.1996.00335.x. [DOI] [PubMed] [Google Scholar]
- 19.Koch U. Fengycin: Strukuräufklarung eines mikroheterogene Lipopeptidolidantibiotikums. Dissertation, zur Erlangung des Grades eines Doktors, der Naturwissenschaften. Tübingen, Germany: Eberhard-Karls-Universität zu Tübingen; 1988. [Google Scholar]
- 20.Krätzschmar J, Krause M, Marahiel M A. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J Bacteriol. 1989;171:5422–5429. doi: 10.1128/jb.171.10.5422-5429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lee S G, Lipmann F. Tyrocidine synthetase system. Methods Enzymol. 1975;43:585–602. doi: 10.1016/0076-6879(75)43121-4. [DOI] [PubMed] [Google Scholar]
- 24.Lin G H, Chen C L, Tschen J S, Tsay S S, Chang Y S, Liu S T. Molecular cloning and characterization of fengycin synthetase gene fenB from Bacillus subtilis. J Bacteriol. 1998;180:1338–1341. doi: 10.1128/jb.180.5.1338-1341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marahiel M A, Stachelhaus T, Mootz H D. Molecular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 27.Mootz H D, Marahiel M A. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran C P, Jr, Lang N, LeGrice S F, Lee G, Stephens M, Sonenshein A L, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Schneider A, Marahiel M A. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis. Arch Microbiol. 1998;169:404–410. doi: 10.1007/s002030050590. [DOI] [PubMed] [Google Scholar]
- 31.Schneider A, Stachelhaus T, Marahiel M A. Targeted alteration of the substrate specificity of peptide synthetases by rational module swapping. Mol Gen Genet. 1998;257:308–318. doi: 10.1007/s004380050652. [DOI] [PubMed] [Google Scholar]
- 32.Stachelhaus T, Marahiel M A. Modular structure of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol Lett. 1995;125:3–14. doi: 10.1111/j.1574-6968.1995.tb07328.x. [DOI] [PubMed] [Google Scholar]
- 33.Stachelhaus T, Marahiel M A. Modular structure of peptide synthetases revealed by dissection of the multifunctional enzyme GrsA. J Biol Chem. 1995;270:6163–6169. doi: 10.1074/jbc.270.11.6163. [DOI] [PubMed] [Google Scholar]
- 34.Stachelhaus T, Mootz H D, Bergendahl V, Marahiel M A. Peptide bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain. J Biol Chem. 1998;273:22773–22781. doi: 10.1074/jbc.273.35.22773. [DOI] [PubMed] [Google Scholar]
- 35.Stein T, Vater J, Kruft V, Otto A, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris H R. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J Biol Chem. 1996;271:15428–15435. doi: 10.1074/jbc.271.26.15428. [DOI] [PubMed] [Google Scholar]
- 36.Steller S, Vollenbroich D, Leenders F, Stein T, Conrad B, Hofemeister J, Jacques P, Thonart P, Vater J. Structural and functional organization of the fengycin synthetase multienzyme system from Bacillus subtilis b213 and A1/3. Chem Biol. 1999;6:31–41. doi: 10.1016/S1074-5521(99)80018-0. [DOI] [PubMed] [Google Scholar]
- 37.Tognoni A, Franchi E, Magistrelli C, Colombo E, Cosmina P, Grandi G. A putative new peptide synthase operon in Bacillus subtilis: partial characterization. Microbiology. 1995;141:645–648. doi: 10.1099/13500872-141-3-645. [DOI] [PubMed] [Google Scholar]
- 38.Tosato V, Albertini A M, Zotti M, Sonda S, Bruschi C V. Sequence completion, identification and definition of the fengycin operon in Bacillus subtilis 168. Microbiology. 1997;143:3443–3450. doi: 10.1099/00221287-143-11-3443. [DOI] [PubMed] [Google Scholar]
- 39.Vanittanakom N, Loeffler W, Koch U, Jung G. Fengycin—a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J Antibiot. 1986;39:888–901. doi: 10.7164/antibiotics.39.888. [DOI] [PubMed] [Google Scholar]
- 40.Vater J, Stein T, Vollenbroich D, Kruft V, Wittmann-Liebold B, Franke P, Liu L, Zuber P. The modular organization of multifunctional peptide synthetases. J Protein Chem. 1997;16:557–564. doi: 10.1023/a:1026386100259. [DOI] [PubMed] [Google Scholar]
- 41.Weckermann R, Furbass R, Marahiel M A. Complete nucleotide sequence of the tycA gene coding the tyrocidine synthetase 1 from Bacillus brevis. Nucleic Acids Res. 1988;16:11841. doi: 10.1093/nar/16.24.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youngman P J, Perkins J B, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci USA. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zamenhof P J, Villarejo M. Construction and properties of Escherichia coli strains exhibiting α-complementation of β-galactosidase fragments in vivo. J Bacteriol. 1972;110:171–178. doi: 10.1128/jb.110.1.171-178.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]