Graphical abstract
Keywords: Allostery, Kinase, Chronic myeloid leukemia, Bcr-Abl, Allosteric drug, Molecular dynamics
Abstract
c-Abl, a non-receptor tyrosine kinase, regulates cell growth and survival in healthy cells and causes chronic myeloid leukemia (CML) when fused by Bcr. Its activity is blocked in the assembled inactive state, where the SH3 and SH2 domains dock into the kinase domain, reducing its conformational flexibility, resulting in the autoinhibited state. It is active in an extended ‘open’ conformation. Allostery governs the transitions between the autoinhibited and active states. Even though experiments revealed the structural hallmarks of the two states, a detailed grasp of the determinants of c-Abl autoinhibition and activation at the atomic level, which may help innovative drug discovery, is still lacking. Here, using extensive molecular dynamics simulations, we decipher exactly how these determinants regulate it. Our simulations confirm and extend experimental data that the myristoyl group serves as the switch for c-Abl inhibition/activation. Its dissociation from the kinase domain promotes the SH2-SH3 release, initiating c-Abl activation. We show that the precise SH2/N-lobe interaction is required for full activation of c-Abl. It stabilizes a catalysis-favored conformation, priming it for catalytic action. Bcr-Abl allosteric drugs elegantly mimic the endogenous myristoyl-mediated autoinhibition state of c-Abl 1b. Allosteric activating mutations shift the ensemble to the active state, blocking ATP-competitive drugs. Allosteric drugs alter the active-site conformation, shifting the ensemble to re-favor ATP-competitive drugs. Our work provides a complete mechanism of c-Abl activation and insights into critical parameters controlling at the atomic level c-Abl inactivation, leading us to propose possible strategies to counter reemergence of drug resistance.
1. Introduction
The core of the Abelson tyrosine kinase (c-Abl, hereafter referred to as Abl), encoded by the human ABL1 gene in chromosome 9, is a non-receptor tyrosine kinase [1], [2]. Abl is localized at subcellular sites, such as the mitochondria, cytoplasm, nucleus, the cell cortex, and the endoplasmic reticulum, interacting with other signaling proteins, including kinases, signaling adaptors, cell-cycle regulators, phosphatases, transcription factors, and cytoskeletal proteins [3], [4], [5]. It transduces normal cell signals for cytoskeleton remodeling for cell motility, regulation, autophagy, DNA damage response, and apoptosis [6], [7], [8], [9], [10]. Human cells express two alternatively spliced variants of Abl, giving rise to 1a and 1b isoforms. The 1b isoform is 19-residue longer than 1a in the N-terminal region [11]. At the N-terminus of isoform 1b, a myristoyl moiety is covalently linked to the Gly2 residue. Myristoyl is established as an essential element in autoinhibition, but not in membrane binding [11]. In contrast, the 1a isoform lacks the N-terminal myristoyl moiety. Following the N-terminal region, the two isoforms share the same sequence and structural components, the SH3, SH2, and kinase domains.
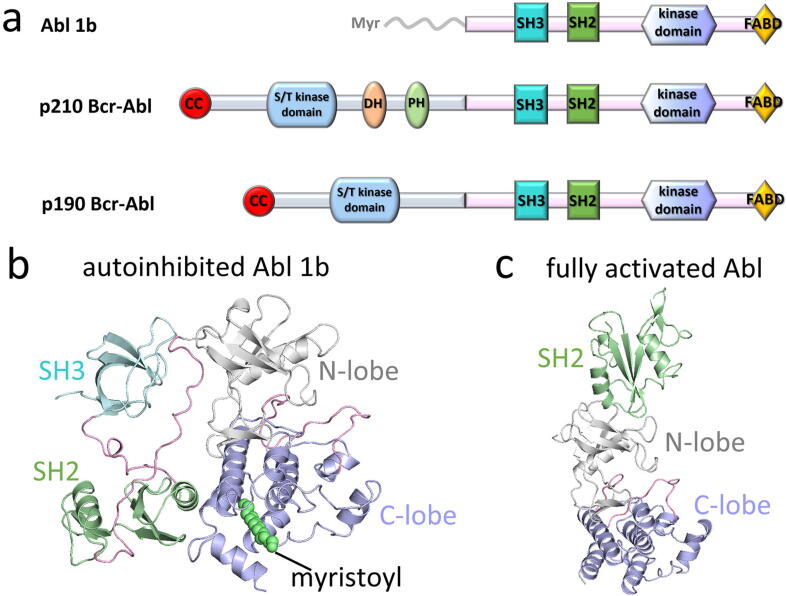
A mutation translocating the breakpoint cluster region (BCR) gene in chromosome 22 to the ABL1 gene yields the fusion BCR-ABL1 gene, which expresses the chimeric Bcr-Abl protein [12]. Bcr-Abl exhibits constitutive phosphorylated tyrosine activity that may induce aberrant cell signals and chronic myeloid leukemia (CML). Depending on the translocation breakpoint in the Bcr protein, two major isoforms of p190 and p210 Bcr-Abl proteins can be expressed [13], [14], [15], [16]. The p210 isoform contains a Dbl-homology (DH)–Pleckstrin-homology (PH) tandem module which does not exist in the p190 isoform (Fig. 1a). The PH domain is responsible for membrane binding by interacting with phosphoinositides [14]. The two isoforms present significant differences in their interactome and tyrosine phosphoproteome [15], [17], suggesting that the activation of the two Bcr-Abl isoforms is induced by different signaling pathways. They result in different leukemia phenotypes. p210 Bcr-Abl is the hallmark of chronic myelogenous leukemia (CML), with the Bcr-Abl protein observed in approximately 1/4 of adult B-cell acute lymphoblastic leukemias (B-ALL), whereas p190 Bcr-Abl accounts for 3/4 [14], [18]. In addition to CML and B-ALL, in recent years, the Abl protein has been implicated in neurodegenerative diseases. Increasing evidence suggests that Abl regulates the degradation of parkin and α-synuclein in Parkinson’s diseases [19], [20].
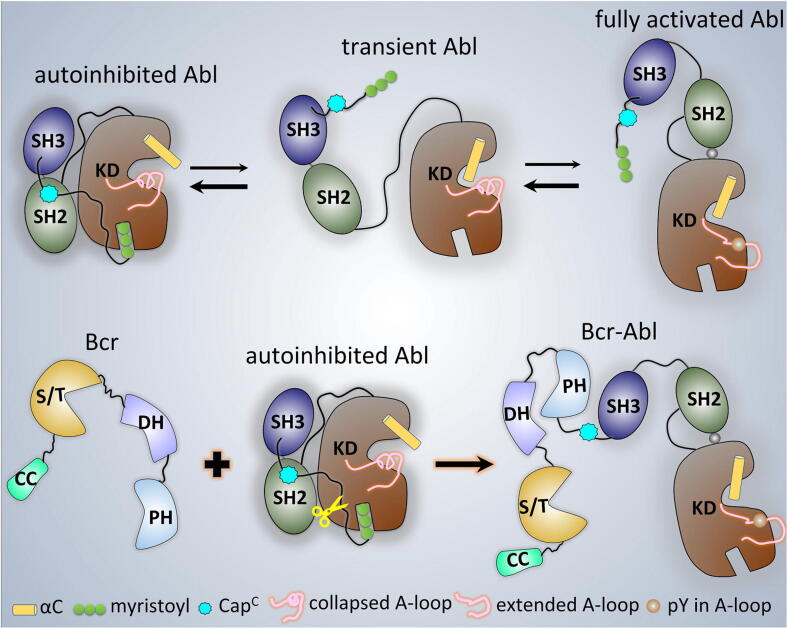
Fig. 1.
Domain structures of Abl 1b and Bcr-Abl. (a) Domain structures of Abl 1b and two Bcr-Abl isoforms, p210 and p190. Replacing the N-terminal region of Abl with Bcr generates the fusion Bcr-Abl protein. Two Bcr-Abl isoforms are different in the Bcr region. In the p190 isoform, the Bcr region consists of the coiled-coil (CC) and the serine/threonine (S/T) kinase domains. In the p210 isoform, in addition the Dbl homology (DH) and pleckstrin homology (PH) domains are fused to Abl. Abl is composed of the SH3, SH2, and kinase domains, followed by the F-actin binding domain (FABD). Structures of (b) autoinhibited (PDB: 1OPL) and (c) fully activated (PDB: 1OPL) Abl.
Many orthosteric tyrosine kinase inhibitors (TKIs) have been approved for the treatment of CML or Bcr-Abl-dependent B-ALL, such as imatinib, dasatinib, nilotinib, bosutinib, ponatinib, and bosutinib [21], [22], [23]. They compete with the nucleotide, adenosine triphosphate (ATP), at the ATP-binding pocket of the kinase domain. Unfortunately, their efficacies are affected in patients harboring point mutations, especially the gatekeeper T315I (numbered in Abl 1b) mutation [24], [25], [26]. The disease can relapse or be refractory in some patients who have already received the TKIs. To date, ponatinib appears to be the only orthosteric TKI that is able to overcome Bcr-Abl T315I-based resistance [27], [28]. However, resistance emerges by mutations at the binding site in the same BCR-ABL1 allele [25]. Phase 2 clinical trials was stopped due to its adverse effect on the patients’ arteries [27]. A temporary drug switching with imatinib was proposed [26]. Recently, an allosteric inhibitor of Bcr-Abl, asciminib (ABL001), was clinically demonstrated to have high inhibition potency, including of Bcr-Abl mutants with the T315I alteration, and was approved by the U.S. Food and Drug Administration (FDA) for the treatment of CML [29]. This allosteric drug avoids side effects resulting from the orthosteric TKIs. In combination with orthosteric inhibitors (nilotinib or ponatinib), asciminib shows a considerably enhanced efficacy. It appears to control the disease, eradicating CML in xenograft tumors, and to suppress drug resistance by point mutations [30], [31]. Distinguished from orthosteric inhibitors binding to the ATP-binding pocket, asciminib binds to the C-lobe allosteric pocket of the Abl kinase domain, which is occupied by the myristoyl group in Abl 1b, distantly regulating the Abl in Bcr-Abl. Asciminib, like the earlier GNF-2 and GNF-5 allosteric inhibitors, exquisitely mimics the endogenous biological myristoyl group action [32], [33], [34]. The emergence of drug resistance to the allosteric drugs led to an innovative drug regimen combining asciminib with ATP-competitive drugs [35]. Allosteric drugs can re-sensitize the ATP-binding pocket to the orthosteric drugs.
Insight into Abl structures and dynamics is vastly important for understanding the mechanisms of Abl activation and inhibition. Like other kinases, Abl populates two conformations, the compact conformation of the autoinhibited state and the extended conformation in the fully active state [36], [37], [38]. Domains and modules in Abl are the regulators of its activation and inactivation. The crystal structure of autoinhibited Abl 1b is mainly featured by (i) the juxtaposition between the SH3 domain and the N-lobe kinase domain mediated by the linkerSH2-KD, where KD denotes the kinase domain, (ii) the direct contact of the SH2 domain with the C-lobe kinase domain, and (iii) the binding of the myristoyl group at the hydrophobic pocket of the C-lobe of the kinase domain, conferring a “kinked” αI’-helix for facilitating the SH2/C-lobe interaction (Fig. 1b). The extended active conformation of Abl displays the SH2 domain docking onto the top of the N-lobe kinase domain (Fig. 1c). In solution, the two states can interconvert rapidly and achieve an equilibrium when the myristoyl motif is absent [38]. In the fully active state, despite the ligand binding, the αC-helix and the activation loop (A-loop) in the kinase domain adopt the “IN” and “extended” conformations, respectively. However, as captured by different orthosteric inhibitors, the kinase domain inhibitory states of Abl are diverse [39]. Type-Ⅰ inhibitors (e.g., dasatinib and bosutinib) prefer the catalytically active conformation, while type-Ⅱ inhibitors (e.g., imatinib, nilotinib, and ponatinib) favor the inactive form [40]. NMR data described two different inactive states (I1 and I2) of the unliganded kinase domain of Abl [37]. The I1 state adopts an αC-in, extended A-loop, and DFG-out conformation, similar to the PD173955-bound state [36]. The I2 state is identical to the imatinib-binding state, displaying the αC-out, collapsed A-loop, and DFG-out conformation. A crystal structure of the Src-like kinase domain of the inactive Abl conjugated with the ATP-peptide presents an αC-out, collapsed A-loop, and DFG-out conformation [41].
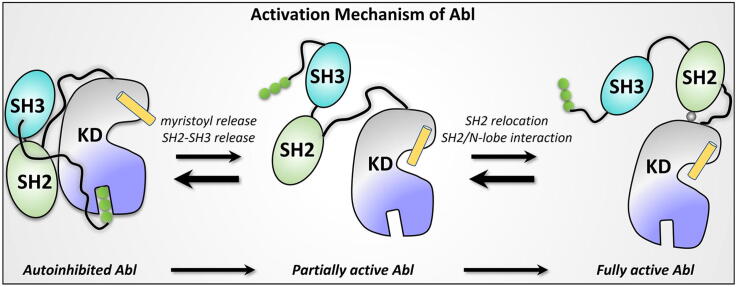
Despite these structural details of Abl [11], [33], [37], [38], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], some puzzles persist, and understanding its activation and inhibition at the atomistic level is still lacking. The exact role of individual modules in Abl’s allosteric regulation has also been unclear. Using molecular modeling and extensive molecular dynamics (MD) simulations, we aim to determine the component function in Abl regulation and decipher the full mechanism of Abl activation. Cells usually contain abundant ATP molecules, the main biochemical energy source for life activities [53]. Kinases are believed to be mostly bound with ATP. Different from most of the previously presented structures and models of Abl, which are in the unliganded or inhibitor-bound states, the models constructed in this work are liganded with ATP (Tables S1 & S2). Our results are consistent with experimental observations [11], [37], [38], [46], [47], [54], [55] and provide mechanistic details. Biophysical and biological data have shown that the release of the myristoyl group can activate Abl, and removal of the SH2-SH3 module can activate Abl too. But it has been unclear which one is the direct factor activating Abl. Our work clarifies the activation process. Abl activation involves multiple conformational changes. The release of the myristoyl switches the activation status of Abl but cannot instantly activate it. An additional step is required for Abl activation: the SH2-SH3 release. This is a key step in the initiation of Abl activation. However, the SH2-SH3 release cannot fully activate Abl either. After the SH2-SH3 release, the SH2 domain is free, allowing its translocation and relocation to interact with the N-lobe of the kinase domain. The interaction between the SH2 domain and the N-lobe can sample multiple states. Only one SH2/N-lobe interaction can fully activate c-Abl, as well as stabilize its fully active conformation. Here we propose the regulation mechanism of the precise SH2/N-lobe interaction for the activation of Abl. The scenarios described in this work ascertain the exact determinants of Abl activation and expand the understanding of the autoinhibition of its two isoforms, as well as the activation of Bcr-Abl.
2. Materials and methods
2.1. Modeling of inactive c-Abl systems
There are two available conformations of the kinase domain of inactive c-Abl (Fig. S1). One is bound to ATP linked to a peptide, displaying an OUT αC-helix, DFG-in, and collapsed A-loop (model 1) [41]. The other is bound to an inhibitor at the ATP pocket, adopting an IN αC-helix, DFG-out, and extended A-loop (model 2) [36]. To determine which model of the kinase domain of inactive Abl is suitable as the initial conformation, we first modelled the isolated kinase domain and the kinase domain with the SH2 and SH3 domains of Abl in the absence (configurations KC and CC) and presence (configurations KC-M and CC-M) of the myristoyl motif based on two models of the kinase domain (Fig. S2). KC/KC-M and CC/CC-M denote the isolated kinase domain and complex systems, respectively. In this work, we developed the topology and parameters of the myristoyl group. The ‘-M’ in KC-M and CC-M indicates the myristoyl group is involved in the systems. The systems of model 2 were constructed completely based on one crystal structure (chain A in PDB: 1OPL). Since only the isolated kinase domain of model 1 is available in the protein data bank, two crystal structures (PDB: 2G1T & chain A in PDB: 1OPL) were used to construct the systems of model 2. The four systems of model 1 were modelled via replacing the region from the N-lobe to the αI-helix in the C-lobe of the corresponding systems of model 2. The ATP-binding pocket of these systems were bound with the ATP molecule and Mg2+ (Table S1). The resulting eight systems were subjected to relaxation: (1) 500-step steepest descent minimizations by constraining the protein backbones with harmonic force constant of 5 kcal/mol/Å2; (2) 200 steps of steepest descent minimizations by constraining the backbones of the protein with harmonic force constant of 5 kcal/mol/Å2; and (3) 200-step minimizations using Adopted Basis Newton-Raphson (ABNR) without constraining atoms. Then, the models were solvated using explicit TIP3P water. Counterions of Na+ and Cl- were added into the solvated systems to neutralize the systems and mimic 150 mM ionic strength. 2 µs MD simulations were performed for the 8 resulting systems to determine the model (model 1 or model 2) for study. The conformation of the kinase domain in the systems of model 1 is more stable than that of model 2. In the systems of model 2, the αC-helix significantly moves outwards with the disruption of the K290-E305 salt bridge (Fig. S3), and the DFG motif shifts from OUT conformation (DFG-out) to the intermediate state (DFG-inter) (Fig. S4). These phenomena indicate that for inactive Abl in the presence of ATP, the conformation of model 1 is more favorable. Herein, the systems of model 1 were used as the studied systems.
2.2. Modeling of active Abl systems
Two crystal structures (chain B in PDB: 1OPL and PDB: 2G2I) were used to construct the active Abl systems. Three active systems of Abl were constructed: the wild-type complex containing the SH2 domain and the kinase domain (activeWT), the isolated kinase domain activeΔSH2, and the mutant by substitution of the I164 residue in the SH2 domain with glutamic acid (activeI164E) (Fig. S5). The activeΔSH2 model was constructed based on the crystal structure of pure kinase domain (PDB: 2G2I) by substituting the P396 residue with histidine, activeWT was modelled by replacing the kinase domain in the complex crystal structure (chain B in PDB: 1OPL) with the kinase domain of the activeΔSH2 system, and the activeI164E system was created by mutating the I164 residue in the SH2 domain to glutamic. In the three active models, the ATP molecule and Mg2+ occupy the ATP-binding pocket and the Y412 residue in the A-loop was phosphorylated (Table S2). Then, the three active models were subjected to relaxation, solvation, and neutralization, same to the inactive systems.
2.3. Molecular dynamics simulation protocols
Our simulations were carried out based on the protocol published in our previous works [56], [57], [58], [59], [60]. Prior to MD simulations, 10,000-step energetical minimizations were conducted to the constructed systems by using the conjugate gradient minimization method to remove bad contacts between atoms in the systems. Then, a total of 2 μs all-atom explicit-solvent MD simulations were performed for each system under the NPT ensemble (constant number of atoms, pressure, and temperature) and 3D periodic boundary conditions, using NAMD 2.13 package [61] with CHARMM [62] all-atom force field (version 36m) [63], [64]. The pressure and temperature were maintained at 1 atm and 310 K using the Langevin piston control algorithm and the Langevin thermostat method with a damping coefficient of 1 ps−1, respectively. The canonical phosphorylation of tyrosine generates negative charge of −2e. The standard CHARMM program provides the parameter and topology for the post-translational modification on the sidechain of the phosphotyrosine (pY412). All covalent bonds including hydrogen atoms were constrained using the RATTLE method so that the velocity Verlet algorithm was used to integrate the Newton motion equation with a larger time step of 2 fs. The interaction potentials between atoms were calculated by the long-range electrostatic interactions using particle mesh Ewald (PME) method with a grading space of 1.0 Å and the short-range van der Waals (vdW) interactions using switching functions with the twin-range cutoff at 12 and 14 Å. Trajectories were saved each 2 ps for data analysis. The result analysis was performed in the CHARMM [62] and VMD packages using the FORTRAN and TCL scripts. The root-mean-square deviation (RMSD) profiles show that all the studied systems can be converged after 500 ns (Fig. S6). Herein, averages were also taken over from the last 1 µs trajectories.
3. Results
3.1. Myristoyl is the switch for Abl autoinhibition and activation
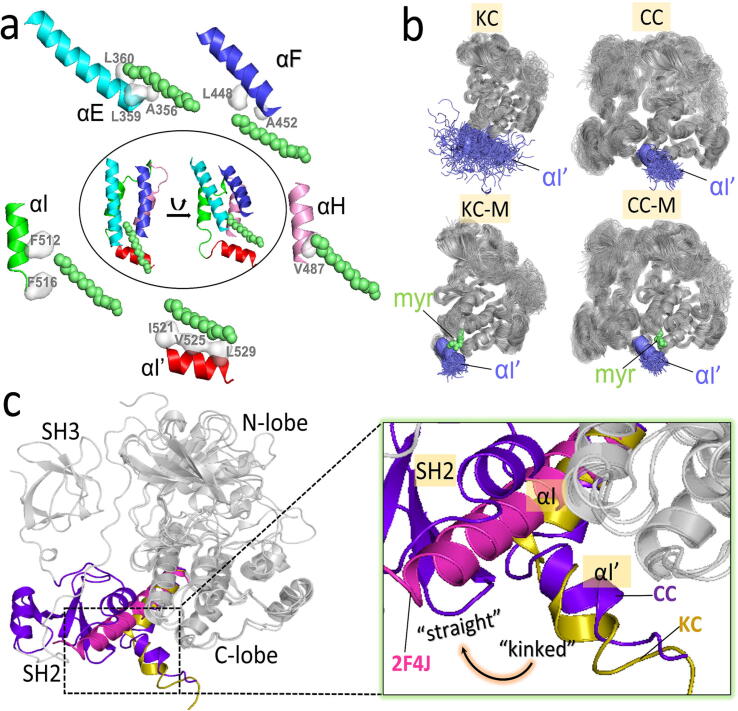
In autoinhibited Abl (Abl 1b isoform), the binding of the myristoyl motif to the C-lobe pocket (myristoyl pocket) is believed to maintain the compact inactive conformation [36]. The substitution of Gly with Ala at position 2 (G2A) results in an un-myristoylated Abl, dramatically increasing its activity [11]. Enhanced activity was also tested in assays that mutate residues in the myristoyl-binding pocket of Abl, which is expected to destabilize the binding environment. These support the premise that myristoyl binding is required for the autoinhibition of Abl. To gain insight into how myristoyl binding influences Abl function at the atomistic level, we modeled and simulated the isolated kinase domain (configuration KC) and the complex containing the SH2, SH3, and kinase domains (configuration CC) of Abl in the absence of myristoyl motif. In the presence of the myristoyl motif, we generated two additional configurations, KC-M and CC-M, where “M” denotes the myristoyl binding (Fig. S2). In the myristoyl-binding pocket of the kinase domain, the myristoyl moiety forms hydrophobic interactions with L512 and F516 in the αI-helix, A356, L359, and L360 in the αE-helix, L448 and A452 in the αF-helix, V487 in the αH-helix, and I521, V525, and L529 in the αI’-helix (Fig. 2a). Within the simulation timescale, prominent differences in the active site, αC-helix, and A-loop of the kinase domain caused by the myristoyl motif were not observed between the compared systems (KC vs KC-M and CC vs CC-M), leading us to conclude that the myristoyl binding or release is unlikely to directly and instantly affect the Abl kinase domain. Instead, upon myristoyl release, the αI’-helix in the C-lobe kinase domain was highly dynamic and underwent large structural changes from the ordered helix to random conformations (Fig. 2b). We speculate that this structural transition would lead to a straight αI’-helix, which can extend the αI-helix’s length and yield a longer αI-αI’-helix [34] (Fig. 2c). A straight αI-αI’-helix produces steric hinderance, preventing the SH2 domain from binding to the kinase domain. In contrast, the αI’-helix in the myristoyl-binding systems (KC-M and CC-M) can maintain the stable and ordered helix conformation (Fig. 2b). The N-terminal disordered region has been implied as a contributor for Abl autoinhibition [3], [11], [55], [65]. It carries two important motifs, the proline-rich (capPxxP) and C-terminal caps (CapC) [38] (Fig. S7). CapC interacts with the SH2 domain, which fastens the SH2-SH3 module to the kinase domain [43], [55]. The released myristoyl group likely disturbs the CapC/SH2 interaction, further promoting the SH2-SH3 release. The myristoyl axis is parallel to the αI’-helix which displays a kinked conformation (Fig. 2a). The kinked αI’-helix is probably an indispensable contributor to the myristoyl binding. Its deletion likely causes the myristoyl release, thus the CapC release. This can be supported by αI’-helix deletion leading to considerably enhanced activity of Abl, comparable to the un-myristoylated Abl with the G2A substitution [11]. Taken together, these suggest that the myristoyl group serves as the switch for Abl autoinhibition and activation. Its release may induce significant conformational changes in the αI’-helix of the kinase domain, which, in turn, promote the release of SH2 and SH3 domains for Abl activation. As we discuss below, additional steps are required for Abl activation.
Fig. 2.
Interaction of the myristoyl motif with the kinase domain in Abl autoinhibition. (a) Interaction between the myristoyl moiety and the αI’-, αI-, αE-, αF-, and αH-helix of the C-lobe in the autoinhibited Abl. (b) Superimposed snapshots showing the dynamic behaviors of helix αI’ (blue) of the kinase domain in the four inactive systems (KC, KC-M, CC, and CC-M). The aligned structures of the representative clusters are extracted from last 1-μs trajectories. The myristoyl group is displayed as the lime spheres. (c) Structural alignment of Abl from the systems of KC and CC to the Abl kinase domain from a crystal structure (PDB ID: 2F4J) in the absence of the myristoyl group. The aligned structures for KC and CC are the average conformations over the last 1-μs trajectories. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. SH2-SH3 release initiates Abl activation
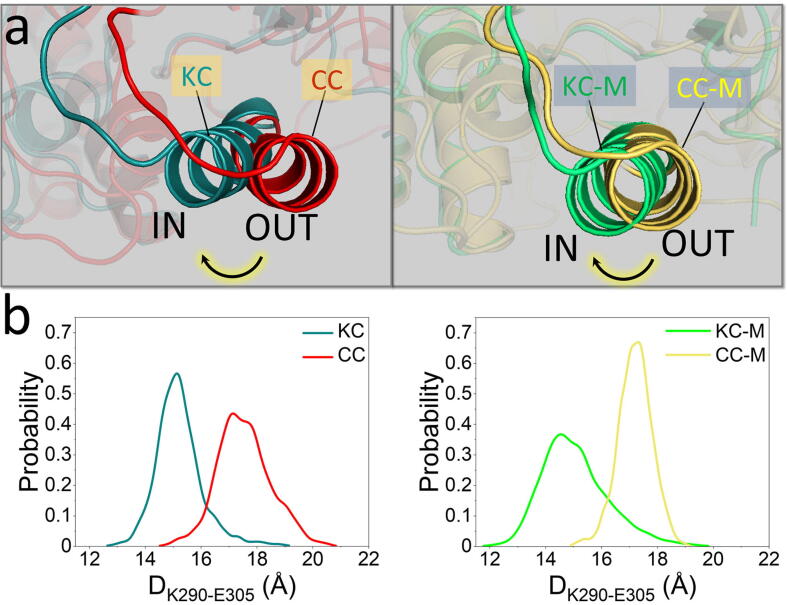
Like the Src family kinases [66], [67], [68], Abl contains SH2 and SH3 domains docking onto the kinase domain, which is critical for the autoinhibition [69]. Pioneering simulation works have implied that the SH2-SH3 module is rigid, maintained by the SH2-SH3 connector [43], [70]. The SH2 release, thereby, allows the release of the whole SH2-SH3 module. The structural features, their changes, and dynamics of some motifs (e.g., αC-helix and A-loop) in the kinase domain are usually used to determine the inhibiting/activating state or trend for kinases [71], [72]. Their conformational transitions clearly demonstrate the regulation of Abl activity. To determine how the SH2-SH3 module exactly regulates Abl, we compared the structural features of the kinase domains in various systems of the isolated kinase domain with that in the corresponding complex systems of Abl with and without SH2-SH3 domains (KC vs CC, and KC-M vs CC-M). To activate Abl, the A-loop and αC-helix usually shift the equilibrium towards the collapsed and IN conformations, respectively. Within the simulation timescale, we did not observe significant structural transitions of the A-loop in the four inactive systems. However, we were able to observe that the αC-helix tends to orient inwards (IN) in the two systems of the isolated kinase domain (KC and KC-M) (Fig. 3a). This observation is consistent with NMR data that Abl encompassing the SH2-SH3 module (34 %) has much higher population of the I2 state in which the αC-helix adopts an OUT conformation, than the isolated kinase domain (6 %) [37]. As a result of the IN αC-helix motion, E305 in the αC-helix tends to approach K290 in the β3-strand to form a salt bridge, a structural hallmark of active Abl. Within the simulation timescale, although E305 in the αC-helix did not completely move inwards to form the salt bridge, it shows this tendency. To quantify the αC-helix action, the distances between E305 in the αC-helix and K290 in the β3-strand (DK290-E305) were measured for the four systems (Fig. 3b). The distribution profiles show the highly populated DK290-E305 for KC and KC-M without the SH2-SH3 module peaking at ∼14.5–15 Å, which is shorter than those distances with the SH2-SH3 module (CC and CC-M, ∼17–17.5 Å). This indicates that removal of the SH2-SH3 module induces the activation characteristics of the Abl kinase domain. Remarkably, the fully inactive model (CC-M) displays the narrow distribution and highest peak, in contrast to the wide distribution and lower peaks for the other three systems. This suggests that simultaneous binding of the SH2-SH3 module and the myristoyl group is capable of preserving the inactive Abl. Together, all the above results suggest that the release of the SH2-SH3 module is a key step in the initiation of Abl activation. This activation step is independent of the myristoyl release or removal, in agreement with experiments [44], [54], [73], [74]. The SH2-kinase domain linker (linkerSH2-KD) mediates the conjunction between the SH3 domain and the N-lobe kinase domain. Unstable or weak SH3-linkerSH2-KD binding may lead to SH2-SH3 release. Mutations or deletion of this linker disrupt the binding of SH3 to the linker, elevating Abl kinase activity [11], [54]. Mutations in the linker which can enhance the SH3-linkerSH2-KD interaction favor Abl’s inhibitory state, countering its activation [54]. Strong SH2/C-lobe interaction also prevents SH2-SH3 release. The intramolecular π-π stacking formed between Y158 and Y361 contributes considerably to the SH2/C-lobe interaction. The Y158D substitution hinders this interaction, significantly enhancing Abl kinase activity [11].
Fig. 3.
Release of the SH3-SH2 module promotes the IN αC-helix conformation. (a) Representative snapshots and (b) probability distribution functions of the distance between K290 in the β3-strand and E305 in the αC-helix for the compared systems of KC and CC (left panel), and KC-M and CC-M (right panel). DK290-E305 is the distance between the nitrogen atom in the sidechain of the K290 residue and the carbon atom in the carboxy group of the E305 residue. Representative conformations in (a) were averaged and data of the K290-E305 distance in (b) were extracted from the last 1-μs trajectories.
3.3. Precise SH2/N-lobe interaction is required for full activation of Abl
Different from other kinases, to fully activate Abl, the SH2 domain undergoes reorientation and translation to dock onto the N-lobe of the kinase domain, resulting in an extended conformation [36], [38], [42], [43], [75]. This unique arrangement indicates that the SH2/N-lobe interaction acts as a necessary determinant for maximizing Abl activity. The intrinsic mechanism of allosteric regulation of this activation are not fully understood. To explore how the SH2/N-lobe interaction impacts Abl activity from the structural point of view, we constructed two active models of Abl in the presence (activeWT) and absence of the SH2 domain (activeΔSH2) (Fig. S5). In the initial construction, we adopted an active conformation of the kinase domain with the IN αC-helix and the extended A-loop. In the A-loop, Y412 was phosphorylated. Recent experiments indicated that the SH2 domain needs to interact precisely with the N-lobe of the kinase domain for full Abl activation and identified key residues contributing to the interaction at the SH2/N-lobe interface; I164, H233, T231, Y234 in the SH2 domain and E294 in the N-lobe kinase domain [37], [47]. Among these interface residues, they further identified I164 as a tether, linking between the SH2 domain and the N-lobe kinase domain, a linkage which is important for the active conformation of Bcr-Abl [47]. Mutations of I164 into polar (e.g., Gln or Thr) and charged (e.g., Asp, Glu, or Lys) amino acids resulted in significant decrease of Abl autophosphorylation and in vitro kinase activity. However, the hydrophobic substitutions, I164V and I164L led to weaker effects. The I164E substitution is expected to efficiently disrupt the SH2/kinase domain interface, consistent with the significant reduction of Abl and Bcr-Abl activity observed in a range of experiments [43], [47], [75]. We modelled another active state of Abl by replacing Ile with Glu at position 164 (activeI164E) and evaluated the structural changes of Abl’s kinase domain that the mutation caused (Fig. S5). During the simulations, we observed that the SH2 domain harboring the I164E mutation quickly separated from the kinase domain, but came back to interact with the kinase domain, as indicated by the increased and decreased distances between the SH2 and kinase domains (Fig. S8a). However, the activeI164E system changed the SH2/N-lobe interface, as evidenced by E164 pointing away from the interface (Fig. S8b). This suggests that in addition to the interface defined in the crystal structure, the SH2 domain may use other surfaces to interact with the kinase domain. However, we anticipate that the new interface is less populated and hard to capture by experimental structural analysis.
3.3.1. P-loop and αC-helix conformations
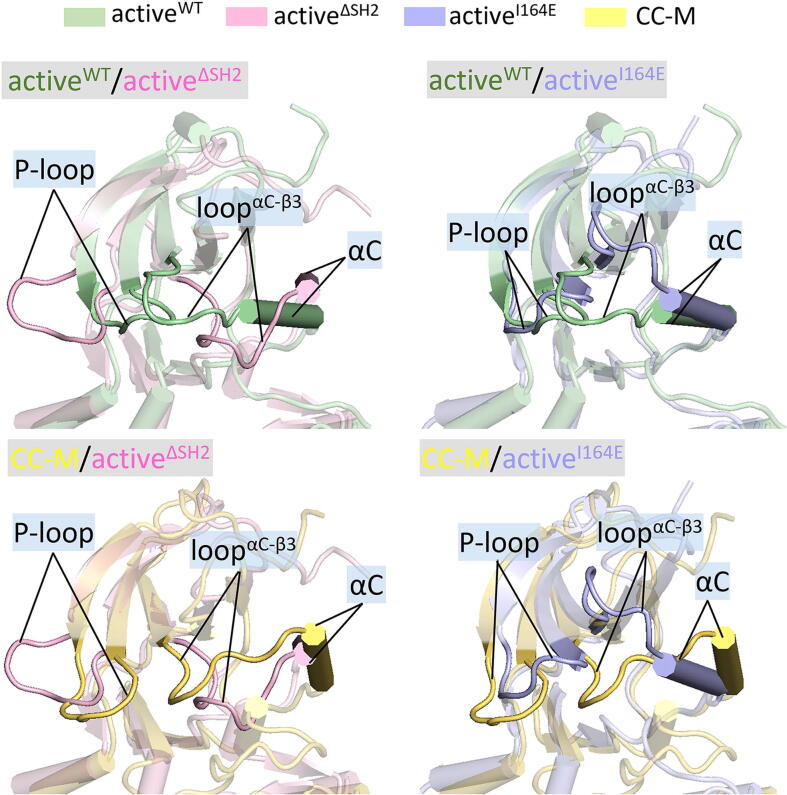
The P-loop and αC-helix in the N-lobe present different conformations in the fully active and inactive Abl (Fig. 4). It is regulated by the loop linking the αC-helix to the β3-strand (loopαC-β3). For the fully active Abl system (activeWT), the loopαC-β3 floats above the P-loop and the αC-helix, allowing the P-loop and the αC-helix to interact, resulting in a favored inward orientation of the αC-helix. On the other hand, loopαC-β3 is positioned between the αC-helix and the P-loop, predisposed to block the P-loop/αC-helix interaction, and an outwards conformation of the αC-helix. During the simulations, the interaction of the P-loop with the αC-helix was maintained in the kinase domain of activeWT. However, for the Abl system in the absence of SH2 (activeΔSH2), the P-loop retracts and clashes with the αC-helix. In this process, the P-loop shifts its position toward the inactive Abl, becoming increasingly closed, in line with NMR observations [37]. For activeI164E, rather than loopαC-β3 retraction, it is lifted from the P-loop and the αC-helix. The lifting motion of loopαC-β3 is able to push the αC-helix outwards, locating near the P-loop, as observed in the kinase domain of activeWT. It is interesting to note that for activeI164E, the P-loop/αC-helix separation leads to a suitable space for the sidechain of Y272 in the P-loop pointing towards the αC-helix (Fig. S9a). For activeWT, however, Y272 retreats its sidechain away from the αC-helix due to the absence of gap between the P-loop and the αC-helix (Fig. S9b).
Fig. 4.
Superimposed conformations of the N-lobe kinase domain to compare the P-loop and αC-helix conformations for activeWT vs activeΔSH2, activeWT vs activeI164E, CC-M vs activeΔSH2, and CC-M vs activeI164E. In cartoon representations, the N-lobe structures in the activeWT, activeΔSH2, activeI164E, and CC-M systems are colored in light green, light pink, light blue, and yellow, respectively. The P-loop, αC-helix, and the loopαC-β3 are highlighted. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3.2. αC-helix motion
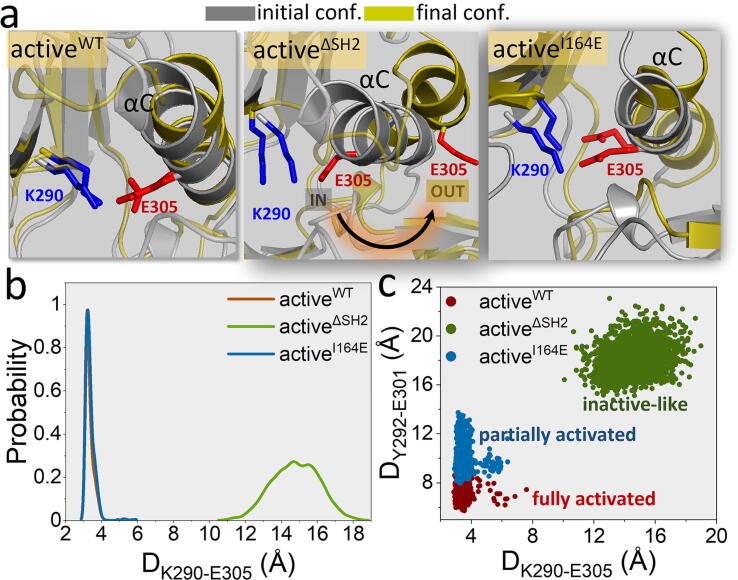
The IN and OUT conformations of the αC-helix are the structural signatures for active and inactive Abl, respectively. A salt bridge formed between K290 in the β3-strand and E305 in the αC-helix can be used to evaluate the fully or partially IN αC-helix. To understand the impact of the precise SH2/N-lobe interaction on the αC-helix orientation, we monitored the αC-helix motion by calculating the distance between K290 and E305, DK290-E305, for the three active models. In the simulations, we observed that the αC-helix in the isolated kinase domain (activeΔSH2) evidently shifts outwards, accompanied by the disruption of the K290-E305 salt bridge (Fig. 5a). The disruption of the K290-E305 salt bridge implies an inactive conformation with the αC-helix OUT orientation. When the SH2 domain is present, the K290-E305 salt bridge is consistently preserved in the activeWT and activeI164E systems. These observations are supported by the distribution profile of DK290-E305 peaked at ∼3.2, ∼15.9, and ∼3.2 Å for activeWT, activeΔSH2, and activeI164E, respectively (Fig. 5b). However, for activeI164E, a portion of the αC-helix is flexible and shifts between the IN and OUT conformations, which is not strong enough to completely push the whole αC-helix OUT, in contrast to the stable IN conformation observed in the activeWT system. The partial movement of the αC-helix towards the OUT conformation cannot completely disrupt the K290-E305 salt bridge, which we term ‘a partially active state’. We suspect that the kinase activity of the partially active Abl is not as strong as that of the fully active species. The αC-helix OUT motion occurs at its N-terminal portion driven by the loopαC-β3 dynamics, resulting in increased distance between the P-loop and the αC-helix. We measured the distance between Y272 in the P-loop and E301 in the αC-helix, DY272-E301, to observe its distribution on a two-dimensional plane along with DK290-E305 (Fig. 5c). Distance distributions can quantify activation levels of the kinase domain in the wild-type, isolated kinase domain, and I164E mutant systems. The distributions at both shorter DK290-E305 and DY272-E301 denote the fully active state, and at the shorter DK290-E305 with the longer DY272-E301 imply the partially active state. In contrast, the distributions at both longer DK290-E305 and DY272-E301 indicate the inactive-like state. The kinase domains in the activeWT and activeI164E systems mostly adopt fully active and partially active conformations, respectively, while that in activeΔSH2 has high population of inactive-like states.
Fig. 5.
Precise SH2/N-lobe kinase domain interaction favors the IN αC-helix conformation in the active Abl. (a) Initial and final snapshots, (b) probability distribution functions of DK290-E305, and (c) DK290-E305–DY292-E301 matrix for the three active systems (activeWT, activeΔSH2, and activeI164E) to evaluate the αC-helix motion. DY292-E301 is the distance between the Cα atom of the Y292 residue and the Cα atom of the E301 residue. The data in (b) and (c) were extracted from the last 1-μs trajectories.
3.3.3. Hydrophobic spines
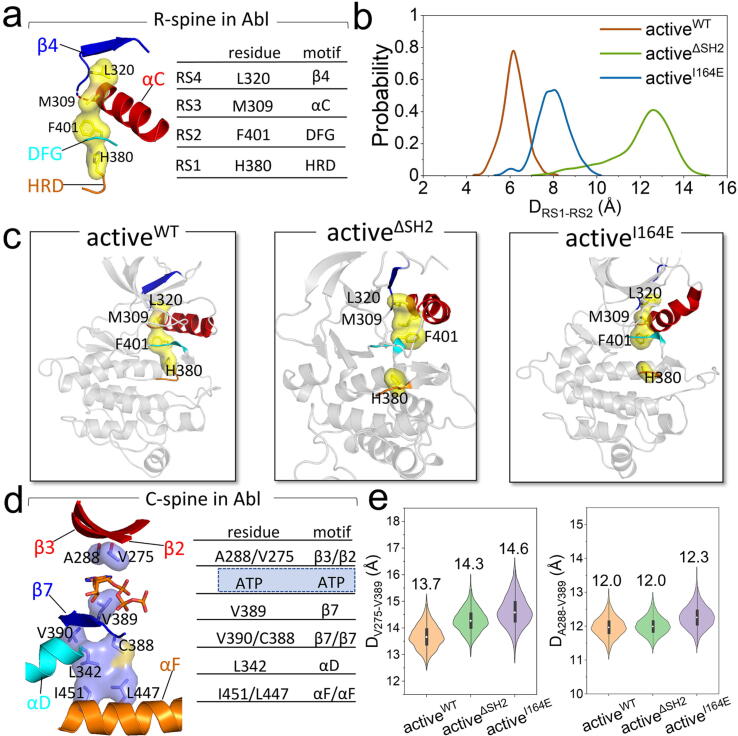
The regulatory (R-spine) and catalytic (C-spine) spines connecting the N-lobe and the C-lobe are essential for orchestrating allostery in active kinases [76], [77]. In Abl, the R-spine is formed by four residues of RS1 (H380 in the HRD motif), RS2 (F401 in the DFG motif), RS3 (M309 in the αC-helix), and RS4 (L320 in β4-strand) (Fig. 6a). The functional relevance of the R-spine has been evidenced by experiments [78], [79]. The spatial assembly of these residues via a chain of hydrophobic interactions from the N-lobe to the C-lobe indicates active Abl. This assembly is disrupted in the inactive state [79]. In our studies, we observed the considerably different behaviors of the R-spine in the activeWT and activeΔSH2 systems. In activeWT, the R-spine assembly is well maintained (Fig. 6c). Whereas the R-spine is dismantled in the activeΔSH2 and activeI164E systems, displaying the RS1-RS2 dissociation as H380 in the HRD motif separates from F401 in the DFG motif. The distances between RS1 and RS2 for these three systems display an increasing order of activeWT (∼6.2 Å) < activeI164E (∼8.0 Å) < activeΔSH2 (∼12.5 Å) (Fig. 6b), implying that it is more difficult to assemble the R-spine when the SH2 domain is absent. The C-spine assembles transiently upon ATP binding [77]. The assembly is required to orient the substrates for phosphorylation. In Abl, the C-spine constitutes residues A288/V275 in the β3-/β2-strands, V389 in the β7-strand, V390/C388 in the β7-strand, L342 in the αD-helix, and I451/L447 in the αF-helix (Fig. 6d). ATP is positioned in-between residues A288/V275 in the β3-/β2-strands and V389 in the β7-strand, forming a sandwich-like posture. This conformation implies that A288, V275, and V389 can modulate the ATP-binding space, thus affecting the stability of ATP binding. To characterize the ATP-binding space, the distances between A288/V275 in the β3-/β2-strands and V389 in the β7-strand (DA288-V389 and DV275-V389) were measured (Fig. 6e). Averaged DA288-V389 and DV275-V389 for activeWT are ∼12.0 and ∼13.7 Å, respectively, slightly shorter than those for activeΔSH2 (∼12.0 and ∼14.3 Å) and activeI164E (∼12.3 and ∼14.6 Å). This indicates that the precise docking of the SH2 domain onto the top of the N-lobe leads to a more compact space favorable for the ATP binding.
Fig. 6.
Precise SH2/N-lobe interaction stabilizes the hydrophobic spines in active Abl. (a) Abl’s R-spine is formed by L320 in the β4-strand, M309 in the αC-helix, F401 in the DFG, and H380 in the HRD. (b) Probability distribution functions of RS1-RS2 distances and (c) representative snapshots of the R-spine for the activeWT, activeΔSH2, and activeI164E systems. The DRS1-RS2 is the distance between the center of mass of the imidazole ring in H380 and the benzene ring in F401. (d) C-spine of Abl is composed of A288/V275 in the β3-/β2-strands, V389 in the β7-strand, V390/C388 in the β7-strand, L342 in the αD-helix, and I451/L477 in the αF-helix. (e) Statistics of the distances between V275/A288 and V389, DV275-V389 and DA288-V389, for the three active systems to characterize the ATP-binding space.
3.3.4. The mechanism of precise SH2/N-lobe interaction regulating Abl activation
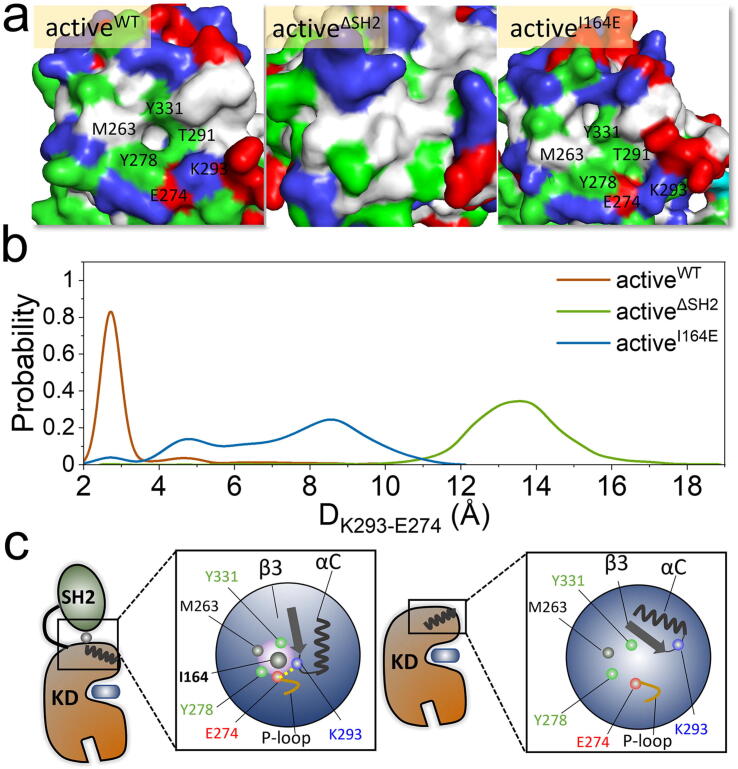
The above results show that precise SH2/N-lobe interactions are required for maintaining the active Abl conformation. The active kinase domain is characterized by the P-loop/αC-helix interaction, the IN αC-helix conformation, a well-assembled R-spine, and a compact ATP-binding space favorable for catalysis. In addition to these observations, we further examine the kinase domain surface at the SH2/N-lobe interface. Interestingly, we observed a deep pocket formed by M263, E274, Y278, T291, K293, and Y331 on the surface of N-lobe kinase domain, which exists in the activeWT system, but not in the activeΔSH2 or activeI164E systems (Fig. 7a). I164 in the SH2 domain occupied this pocket to coordinate kinase domain residues (M263, E274, Y278, T291, K293, and Y331). The coordination may affect the local conformation of the N-lobe of the kinase domain. In the pocket, a salt bridge forms between the positively charged K293 in the loopαC-β3 and the negatively charged E274 in the β2-strand. The salt bridge distance, DK293-E274, peaks at ∼2.8 Å for activeWT and ∼13.5 Å for activeΔSH2 (Fig. 7b). For the mutant system, the distance profile exhibits a wide distribution in the range of ∼4–10 Å without a considerable peak. Taken together, the K293-E274 salt bridge is stably maintained in the activeWT system due to the I164 coordination but disrupted in the activeΔSH2 and activeI164E systems due to its absence (Fig. 7c). The K293-E274 disruption confers a more mobile loopαC-β3, which breaks the P-loop/αC-helix interaction. The αC-helix consequently tends to move outward, shifting the equilibrium of Abl towards the inactive state.
Fig. 7.
Mechanism of the precise SH2/N-lobe interaction in active Abl. (a) Morphology of the N-lobe surfaces exposed to the SH2 domain and (b) probability distribution functions of K293-E274 distances for the activeWT, activeΔSH2, and activeI164E systems. DK293-E274 is the distance between the nitrogen atom in K293 sidechain and the carbon atom in the carboxy group of E274. (c) Schematic diagrams representing the precise SH2/N-lobe interaction for regulating the P-loop/αC-helix interaction and IN or OUT αC-helix motion in active Abl. In (a), the positively, negatively, hydrophobic, and polar residues are colored in blue, red, white, and green, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Allosteric and competitive active-site inhibitors
Autoinhibition is a common inactivating switch mechanism in nature [80], [81], [82], [83], [84], [85]. The principle is straightforward and since the inactivating segment is covalently linked, it is highly efficient. In autoinhibition, the inactivation segment obstructs the access of the ligand to the active site and its coordination for catalysis. It can do so by covering the active site, as in the case of PI3K [58], or allosterically, as in our case here. Either way, to be usefully regulated during cell life and respond swiftly to the changing conditions, its binding should be fairly loose, permitting its ON/OFF transition without needing to overcome high kinetic barriers. On the downside, this is also likely to result in basal activation, as observed in Abl 1b. The transition between the ‘closed’, autoinhibited state and the active, extended ‘open’ state is coupled with conformational changes, which reflect the shift in the population between the states. An allosteric oncogenic driver mutation can weaken the interactions of the inactive state, or (and) strengthen those of the active state, resulting in a shift in the equilibrium toward the active state. In kinases, allosteric drivers often work in this way, with the mutations mimicking the native activation scenario [86], reversing the autoinhibition switch. In the case of the Bcr-Abl, it is by removing Abl’s N-terminal segment to which the myristoyl group is linked. Allosteric drug resistance mutations away from the active site can confer resistance, for example, by shifting the ensemble toward conformations that introduce steric hindrance to drug binding at the ATP-binding pocket. They do this through long range conformational changes. On the other hand, allosteric inhibitors work by shifting the ensemble to preorganize a drug binding-favored active site, which mimics that of the wild-type [87]. Thus, from the translational standpoint, allosteric drug action, including by mimicking the endogenous autoinhibition by the myristoyl in Abl, work by blocking the mechanism of activation.
T315 substitutions resist imatinib, an ATP-competitive drug that binds to the inactive state. The Gray team identified GNF-2, and subsequently GNF-5 allosteric drugs [33], that bound Bcr-Abl at the myristoyl pocket through hydrophobic and vdW interactions and water-mediated H-bonds, and shifted the ensemble, sensitizing it to imatinib and nilotinib. Asciminib resembles the GNF drugs [30]. Thus, its combination with ATP-competitive drugs can successfully block tumor growth. Even though mutations at the myristate pocket can confer resistance to asciminib, they are sensitive to the ATP-competitive drugs, rendering the orthosteric-allosteric combination an attractive strategy [40]. Recently, asciminib with ponatinib were shown to also strongly suppress the Y253H and E255V resistance mutations and to restore efficacy [31], making Nathaniel Gray's Team strategy a promising CML treatment.
Nevertheless, resistance mutations may still arise. The most straightforward strategy appears selection of variant ATP-competitive drugs and allosteric drugs at the myristoyl pocket. Alternatively, if these fails, we suggest three possible strategies. (i) Allosteric drugs binding at sites other than the myristoyl pocket, mimicking ‘rescue’ mutations. Anaplastic lymphoma kinase (ALK) allosteric mutation L1198F is one example. While it led to resistance to lorlatinib through steric interference, it also resensitized ALK to crizotinib [88]. Like allosteric drugs, the allosteric mutation redistributed the ensemble, to populate conformations favorable for crizotinib binding. If, however, the location of a potential favorable allosteric site is unknown, a search for substitutions that can confer allosterically a bent αI′-helix could be one way, or inspection of the cross-correlated motions between the sites could provide clues [89]. (ii) A second possible option is blocking the SH2/N-lobe interface, as done in protein–protein interactions. That however could be energetically costly. At the SH2/N-lobe interface in the active Abl (or Bcr-Abl), a cystine residue (Cys324) is found in a pocket of the N-lobe, which may be a potential site covalently linking a drug to block the SH2/N-lobe interface, thus disturbing the active conformation. (iii) Another possible strategy is enhancing the SH2/C-lobe interface. As observed in the simulations, the SH2-SH3 release initiates Abl activation. Strong SH2/C-lobe interface avoids this initiation. Molecular glue is a promising and potential candidate, rendering a strong SH2/C-lobe interface to allosterically restrict Abl (or restore Bcr-Abl) inactive ensemble. This is analogous to the role of the myristoyl group or allosteric inhibitors (asciminib, GNF-5, and GNF-2). The combination of the molecular glue with the orthosteric drug is worth better to be adopted. An excellent inhibitory efficacy of molecular glue has been successfully achieved in SHP2 phosphatase [90], [91], [92], [93].
Other possible methods employed to identify allosteric residues or cryptic allosteric sites include artificial intelligence (AI) learned on allosteric residues, genetic algorithms and neural network, Markov State Models combined with simulations, or community network analysis, or deep scanning [94], [95], [96], [97]. Deep mutational scanning is a yet another option [98], [99].
4. Discussion
Abl is a non-receptor kinase regulating physiological cell signals. Its inhibition and activation are allosterically regulated via intramolecular interactions between modules [36], [38]. In this work, we depict the allosteric regulation mechanisms of Abl, the key kinase in leukemia, its autoinhibition and activation, by examining their exact roles in regulation at the atomic level. The scenarios observed in the simulations are consistent with, and clarify, available experimental data and provide a comprehensive and detailed description of the autoinhibited and active states, and the events taking place during the transition between the states [11], [37], [38], [46], [47], [54], [55]. The autoinhibited state of Abl presents a compact conformation with the SH2 and SH3 domains docking onto the back side of the kinase domain (Fig. 1b). Upon their release, Abl undergoes dissociation, transformation, and reorientation, with its ensemble shifting towards the active conformation (Fig. 8). The strongly active conformation of Abl adopts an extended conformation, in which the SH2 domain docks onto the top of the N-lobe kinase domain (Fig. 1c).
Fig. 8.
Schematic diagrams of the mechanism of Abl and Bcr-Abl activation.
The myristoyl motif and the SH2-SH3 module are essential contributors for inhibiting Abl (Abl 1b isoform). The myristoyl group is the switch between the inhibition and activation of Abl. The binding of the myristoyl group to the C-lobe pocket stabilizes Abl’s compact, autoinhibited conformation. Its release triggers the release of SH2-SH3 via steric repulsion produced by the conformational transition of the αI’-helix in the C-lobe kinase domain from the “kinked” to the “straight” state. The N-terminal disordered region is a contributor to Abl autoinhibition [3], [11], [55], [65]. It carries two important motifs, the proline-rich (capPxxP) and C-terminal caps (CapC) [38] (Fig. S7). CapC interacts with the SH2 domain, which fastens the SH2-SH3 module to the kinase domain [43], [55]. Since Abl 1a isoform contains the CapC region, CapC’s function likely applies to it too. With a different sequence (residues 1–26), Abl 1a N-terminal region can assist the docking of the SH2-SH3 to the kinase domain, with a mechanism similar to the myristoyl in the Abl 1b, and the pY-peptide at the C-terminus in c-Src kinase [100], [101]. CapC is retained in the Bcr-Abl oncoprotein (Fig. 8 and Fig. S7) [102]. The Abl region in the Bcr-Abl protein populates the active conformation. This further supports the requirement of myristoyl binding in Abl 1b isoform for the fastening of the SH2-SH3 module to the kinase domain. The released myristoyl group likely disturbs the CapC/SH2 interaction, further promoting the SH2-SH3 release.
Following myristoyl release, the SH2-SH3 module dissociates from the kinase domain. As observed in our simulation, SH2-SH3 release induces a structural transition of the αC-helix from the OUT to the IN conformation, in agreement with NMR data [38]. This indicates that the SH2-SH3 release initiates Abl activation. That activation is independent of the myristoyl release or removal, which can be supported by experimental data that optimize mutations at the SH3-linkerSH2-KD, or by the SH2/C-lobe interfaces that eliminate Abl activity, while mutations that disrupt the interfaces pronouncedly increase Abl activity [11], [44], [54], [73], [74]. Occasionally, protein dynamics, or competitive interactions in cells, e.g., the interaction between the SH2 domain and proteins containing the phosphotyrosine (pY) motif [103], and between the SH3 domain and proline-rich motifs [104], may release the SH2-SH3 module, allosterically activating Abl.
Upon SH2-SH3 release, the Abl’s kinase domain conformation will be rearranged. The SH2 domain migrates and interacts with the N-lobe, leading to the extended Abl conformation. The most stable conformation of active Abl is unique and precise, which is required for maximal catalytic activity. This conformation dominates Abl in Bcr-Abl. Importantly, the SH2 I164 residue is key for the precise SH2/N-lobe interactions. It can coordinate residues (M263, E274, Y278, T291, K293, and Y331) in the N-lobe (Fig. 7a), in which K293 in loopαC-β3 forms a salt bridge with E274 in β2-strand, thus restraining the loopαC-β3 to keep the P-loop/αC-helix interaction and the stable IN αC-helix conformation. Experimental data have shown that polar or charged substitutions of the SH2 I164 residue dramatically eliminated Bcr-Abl kinase activity [47]. Hydrophobic substitutions resulted in weaker effects. Results from our simulation support and clarify these experimental observations. We mutated the SH2 I164 to glutamic acid (I164E) in the extended Abl conformation and observed that the SH2 domain still interacted with the N-lobe, but the SH2/N-lobe interface changed. This implies diverse interactions between the SH2 domain and the N-lobe. However, the N-lobe coordination by I164 is absent in this interaction. The K293-E274 salt bridge is disrupted, resulting in a dynamic loopαC-β3 breaking the P-loop/αC-helix interaction and rendering the αC-helix OUT motion. This leads to a partially active Abl. When the SH2/N-lobe interaction is absent, the K293-E274 salt bridge is also disrupted, allowing loopαC-β3 retraction. The P-loop and αC-helix are completely separated and can more readily push the αC-helix OUT, inducing an inactive-like conformation. However, the precise SH2/N-lobe interaction is able to maintain a compact ATP-binding pocket for catalysis, as well as the assembled R-spine. The binding of orthosteric inhibitors to the ATP-site pocket of Abl can induce considerable disruption of the R-spine assembly in the RS1-RS2-RS3 due to the F401 in DFG motif flipping from its favorable IN to the OUT state [79]. We speculate that since steric hindrance is relieved, RS1-RS2 dissociation can provide a more accessible pocket for inhibitors. This speculation can be strongly supported by an assay performed by Lorenz et al., showing that the presence of the SH2 domain significantly decreased the binding rate of imatinib to the kinase domain of Abl from 0.82 ± 0.01 to 0.69 ± 0.02 µM−1s−1 [42]. The Abl I164E mutant restored somewhat the rate of imatinib binding (0.78 ± 0.02 µM−1s−1), nearly that of the isolated kinase domain.
5. Conclusions
In this work, we detailed how domains or modules of Abl allosterically regulate its inhibition and activation at the atomic level. The docking of the SH3 and SH2 domains to the back of the kinase domain, and the binding of the myristoyl group to the C-lobe’s pocket provide the structural basis for the autoinhibition of Abl. Our simulations show that release of the myristoyl group turns on the switch for Abl activation, triggering the SH2-SH3 release for Abl’s full activation. The SH2-SH3 release initiates the Abl activation, featured by the inwards motion of the αC-helix. The released SH2 domain undergoes reorientation and translocation, permitting it to interact with the N-lobe, to promote Abl activation. To fully activate Abl, precise SH2/N-lobe interaction, with SH2 I164 coordinating some N-lobe residues, is required. These interactions can stabilize the interacting P-loop/αC, the IN αC-helix, the well-assembled R-spine, and the compact ATP-binding space, in the specific catalysis-favored conformation. Conformations lacking the SH2/N-lobe interactions weaken or disable the kinase activity. Collectively, we outlined a complete mechanism of Abl activation. The structural events within the mechanism that we observed are verified by their ability to explain experimental observations. The newly gained insights broaden the knowledge of the elements controlling the inhibited and active population of Abl, as well as the activation of both Abl and Bcr-Abl.
Allosteric inhibitors, like the GNF series and asciminib, mimic the endogenous action of myristoyl in Abl 1b and work by inducing a bend in αI′-helix. A ligand that docks into the myristate pocket but prevents the bent conformation likely works as an activator [24]. Here we propose three new strategies for Abl (or Bcr-Abl) inhibition: (i) screening rescue mutations conferring allosterically a bent αI′-helix, (ii) blocking the SH2/N-lobe interface by a covalent inhibitor, and (iii) strengthening the SH2/C-lobe interface by molecular glue.
CRediT authorship contribution statement
Yonglan Liu: Methodology, Software, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Project administration. Mingzhen Zhang: Validation, Resources, Data curation, Writing – review & editing. Chung-Jung Tsai: Validation, Resources. Hyunbum Jang: Conceptualization, Validation, Resources, Data curation, Writing – review & editing. Ruth Nussinov: Conceptualization, Supervision, Project administration, Writing – review & editing, Visualization, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261201500003I. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, CCR. The calculations had been performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov/).
Footnotes
Supplementary data (the parameterization of the myristoyl group is provided in appendices (topo in appendix_A.rtf and para in appendix_B.prm)) to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.014.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Schindler T., Bornmann W., Pellicena P., Miller W.T., Clarkson B., Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289(5486):1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y., Jang H., Zhang M., Tsai C.J., Maloney R., Nussinov R. The structural basis of BCR-ABL recruitment of GRB2 in chronic myelogenous leukemia. Biophys J. 2022;121(12):2251–2265. doi: 10.1016/j.bpj.2022.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hantschel O., Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5(1):33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- 4.Pendergast A.M. The Abl family kinases: mechanisms of regulation and signaling. Adv Cancer Res. 2002;85:51–100. doi: 10.1016/s0065-230x(02)85003-5. [DOI] [PubMed] [Google Scholar]
- 5.Smith J.M., Mayer B.J. Abl: mechanisms of regulation and activation. Front Biosci. 2002;7:d31–d42. doi: 10.2741/a767. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida K., Komatsu K., Wang H.G., Kufe D. c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol Cell Biol. 2002;22(10):3292–3300. doi: 10.1128/MCB.22.10.3292-3300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raina D., Pandey P., Ahmad R., Bharti A., Ren J., Kharbanda S., et al. c-Abl tyrosine kinase regulates caspase-9 autocleavage in the apoptotic response to DNA damage. J Biol Chem. 2005;280(12):11147–11151. doi: 10.1074/jbc.M413787200. [DOI] [PubMed] [Google Scholar]
- 8.Tanos B., Pendergast A.M. Abl tyrosine kinase regulates endocytosis of the epidermal growth factor receptor. J Biol Chem. 2006;281(43):32714–32723. doi: 10.1074/jbc.M603126200. [DOI] [PubMed] [Google Scholar]
- 9.Michael M., Vehlow A., Navarro C., Krause M. c-Abl, Lamellipodin, and Ena/VASP proteins cooperate in dorsal ruffling of fibroblasts and axonal morphogenesis. Curr Biol. 2010;20(9):783–791. doi: 10.1016/j.cub.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobbaut M., Derua R., Doppler H., Lou H.J., Vandoninck S., Storz P., et al. Differential regulation of PKD isoforms in oxidative stress conditions through phosphorylation of a conserved Tyr in the P+1 loop. Sci Rep. 2017;7(1):887. doi: 10.1038/s41598-017-00800-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hantschel O., Nagar B., Guettler S., Kretzschmar J., Dorey K., Kuriyan J., et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112(6):845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 12.Hantschel O., Grebien F., Superti-Furga G. The growing arsenal of ATP-competitive and allosteric inhibitors of BCR-ABL. Cancer Res. 2012;72(19):4890–4895. doi: 10.1158/0008-5472.CAN-12-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deininger M.W., Goldman J.M., Melo J.V. The molecular biology of chronic myeloid leukemia. Blood. 2000;96(10):3343–3356. [PubMed] [Google Scholar]
- 14.Reckel S., Gehin C., Tardivon D., Georgeon S., Kukenshoner T., Lohr F., et al. Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase. Nat Commun. 2017;8(1):2101. doi: 10.1038/s41467-017-02313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reckel S., Hamelin R., Georgeon S., Armand F., Jolliet Q., Chiappe D., et al. Differential signaling networks of Bcr-Abl p210 and p190 kinases in leukemia cells defined by functional proteomics. Leukemia. 2017;31(7):1502–1512. doi: 10.1038/leu.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reckel S., Hantschel O. Bcr-Abl: one kinase, two isoforms, two diseases. Oncotarget. 2017;8(45):78257–78258. doi: 10.18632/oncotarget.20874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cutler J.A., Tahir R., Sreenivasamurthy S.K., Mitchell C., Renuse S., Nirujogi R.S., et al. Differential signaling through p190 and p210 BCR-ABL fusion proteins revealed by interactome and phosphoproteome analysis. Leukemia. 2017;31(7):1513–1524. doi: 10.1038/leu.2017.61. [DOI] [PubMed] [Google Scholar]
- 18.Luong-Gardiol N., Siddiqui I., Pizzitola I., Jeevan-Raj B., Charmoy M., Huang Y., et al. gamma-catenin-dependent signals maintain BCR-ABL1(+) B cell acute lymphoblastic leukemia. Cancer Cell. 2019;35(4):649–63 e10. doi: 10.1016/j.ccell.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Mahul-Mellier A.L., Fauvet B., Gysbers A., Dikiy I., Oueslati A., Georgeon S., et al. c-Abl phosphorylates alpha-synuclein and regulates its degradation: implication for alpha-synuclein clearance and contribution to the pathogenesis of Parkinson's disease. Hum Mol Genet. 2014;23(11):2858–2879. doi: 10.1093/hmg/ddt674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imam S.Z., Zhou Q., Yamamoto A., Valente A.J., Ali S.F., Bains M., et al. Novel regulation of parkin function through c-Abl-mediated tyrosine phosphorylation: implications for Parkinson's disease. J Neurosci. 2011;31(1):157–163. doi: 10.1523/JNEUROSCI.1833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M., Turhan A.G., Ding H., Lin Q., Meng K., Jiang X. Targeting BCR-ABL+ stem/progenitor cells and BCR-ABL-T315I mutant cells by effective inhibition of the BCR-ABL-Tyr177-GRB2 complex. Oncotarget. 2017;8(27):43662–43677. doi: 10.18632/oncotarget.18216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah N.P., Sawyers C.L. Mechanisms of resistance to STI571 in Philadelphia chromosome-associated leukemias. Oncogene. 2003;22(47):7389–7395. doi: 10.1038/sj.onc.1206942. [DOI] [PubMed] [Google Scholar]
- 23.Nussinov R., Zhang M., Maloney R., Liu Y., Tsai C.J., Jang H. Allostery: allosteric cancer drivers and innovative allosteric drugs. J Mol Biol. 2022:167569. doi: 10.1016/j.jmb.2022.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teng M., Luskin M.R., Cowan-Jacob S.W., Ding Q., Fabbro D., Gray N.S. The dawn of allosteric BCR-ABL1 drugs: from a phenotypic screening hit to an approved drug. J Med Chem. 2022;65(11):7581–7594. doi: 10.1021/acs.jmedchem.2c00373. [DOI] [PubMed] [Google Scholar]
- 25.Zabriskie M.S., Eide C.A., Tantravahi S.K., Vellore N.A., Estrada J., Nicolini F.E., et al. BCR-ABL1 compound mutations combining key kinase domain positions confer clinical resistance to ponatinib in Ph chromosome-positive leukemia. Cancer Cell. 2014;26(3):428–442. doi: 10.1016/j.ccr.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindstrom H.J.G., Friedman R. Rotating between ponatinib and imatinib temporarily increases the efficacy of imatinib as shown in a chronic myeloid leukaemia model. Sci Rep. 2022;12(1):5164. doi: 10.1038/s41598-022-09048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortes J.E., Kim D.W., Pinilla-Ibarz J., le Coutre P., Paquette R., Chuah C., et al. A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N Engl J Med. 2013;369(19):1783–1796. doi: 10.1056/NEJMoa1306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauro M.J., Hughes T.P. Asciminib in relapsed chronic myeloid leukemia. Reply. N Engl J Med. 2020;382(14):1379. doi: 10.1056/NEJMc2000116. [DOI] [PubMed] [Google Scholar]
- 29.Hughes T.P., Mauro M.J., Cortes J.E., Minami H., Rea D., DeAngelo D.J., et al. Asciminib in chronic myeloid leukemia after ABL kinase inhibitor failure. N Engl J Med. 2019;381(24):2315–2326. doi: 10.1056/NEJMoa1902328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wylie A.A., Schoepfer J., Jahnke W., Cowan-Jacob S.W., Loo A., Furet P., et al. The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature. 2017;543(7647):733–737. doi: 10.1038/nature21702. [DOI] [PubMed] [Google Scholar]
- 31.Eide C.A., Zabriskie M.S., Savage Stevens S.L., Antelope O., Vellore N.A., Than H., et al. Combining the allosteric inhibitor asciminib with ponatinib suppresses emergence of and restores efficacy against highly resistant BCR-ABL1 mutants. Cancer Cell. 2019;36(4):431–43 e5. doi: 10.1016/j.ccell.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee B.J., Shah N.P. Identification of TKI-sensitive point mutations that activate c-ABL kinase activity and transformation potential and confer in vitro resistance to the allosteric ABL inhibitor GNF-5. Blood. 2015;126(23) [Google Scholar]
- 33.Zhang J., Adrian F.J., Jahnke W., Cowan-Jacob S.W., Li A.G., Iacob R.E., et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463(7280):501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young M.A., Shah N.P., Chao L.H., Seeliger M., Milanov Z.V., Biggs W.H., 3rd, et al. Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680. Cancer Res. 2006;66(2):1007–1014. doi: 10.1158/0008-5472.CAN-05-2788. [DOI] [PubMed] [Google Scholar]
- 35.Jahnke W., Grotzfeld R.M., Pelle X., Strauss A., Fendrich G., Cowan-Jacob S.W., et al. Binding or bending: distinction of allosteric Abl kinase agonists from antagonists by an NMR-based conformational assay. J Am Chem Soc. 2010;132(20):7043–7048. doi: 10.1021/ja101837n. [DOI] [PubMed] [Google Scholar]
- 36.Nagar B., Hantschel O., Young M.A., Scheffzek K., Veach D., Bornmann W., et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112(6):859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 37.Xie T., Saleh T., Rossi P., Kalodimos C.G. Conformational states dynamically populated by a kinase determine its function. Science. 2020;370(6513) doi: 10.1126/science.abc2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saleh T., Rossi P., Kalodimos C.G. Atomic view of the energy landscape in the allosteric regulation of Abl kinase. Nat Struct Mol Biol. 2017;24(11):893–901. doi: 10.1038/nsmb.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y., Portelli S., Pat M., Rodrigues C.H.M., Nguyen T.B., Pires D.E.V., et al. Structure-guided machine learning prediction of drug resistance mutations in Abelson 1 kinase. Comput Struct Biotechnol J. 2021;19:5381–5391. doi: 10.1016/j.csbj.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoepfer J., Jahnke W., Berellini G., Buonamici S., Cotesta S., Cowan-Jacob S.W., et al. Discovery of asciminib (ABL001), an allosteric inhibitor of the tyrosine kinase activity of BCR-ABL1. J Med Chem. 2018;61(18):8120–8135. doi: 10.1021/acs.jmedchem.8b01040. [DOI] [PubMed] [Google Scholar]
- 41.Levinson N.M., Kuchment O., Shen K., Young M.A., Koldobskiy M., Karplus M., et al. A Src-like inactive conformation in the abl tyrosine kinase domain. Plos Biol. 2006;4(5):e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lorenz S., Deng P., Hantschel O., Superti-Furga G., Kuriyan J. Crystal structure of an SH2-kinase construct of c-Abl and effect of the SH2 domain on kinase activity. Biochem J. 2015;468(2):283–291. doi: 10.1042/BJ20141492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagar B., Hantschel O., Seeliger M., Davies J.M., Weis W.I., Superti-Furga G., et al. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol Cell. 2006;21(6):787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 44.Brasher B.B., Van Etten R.A. c-Abl has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. J Biol Chem. 2000;275(45):35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 45.Skora L., Mestan J., Fabbro D., Jahnke W., Grzesiek S. NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc Natl Acad Sci U S A. 2013;110(47):E4437–E4445. doi: 10.1073/pnas.1314712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith K.M., Yacobi R., Van Etten R.A. Autoinhibition of Bcr-Abl through its SH3 domain. Mol Cell. 2003;12(1):27–37. doi: 10.1016/S1097-2765(03)00274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grebien F., Hantschel O., Wojcik J., Kaupe I., Kovacic B., Wyrzucki A.M., et al. Targeting the SH2-kinase interface in Bcr-Abl inhibits leukemogenesis. Cell. 2011;147(2):306–319. doi: 10.1016/j.cell.2011.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corbi-Verge C., Marinelli F., Zafra-Ruano A., Ruiz-Sanz J., Luque I., Faraldo-Gomez J.D. Two-state dynamics of the SH3-SH2 tandem of Abl kinase and the allosteric role of the N-cap. Proc Natl Acad Sci U S A. 2013;110(36):E3372–E3380. doi: 10.1073/pnas.1303966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolker N., Gorna M.W., Sutto L., Torralba A.S., Superti-Furga G., Gervasio F.L. The SH2 domain regulates c-Abl kinase activation by a cyclin-like mechanism and remodulation of the hinge motion. Plos Comput Biol. 2014;10(10):e1003863. doi: 10.1371/journal.pcbi.1003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tse A., Verkhivker G.M. Molecular dynamics simulations and structural network analysis of c-Abl and c-Src kinase core proteins: capturing allosteric mechanisms and communication pathways from residue centrality. J Chem Inf Model. 2015;55(8):1645–1662. doi: 10.1021/acs.jcim.5b00240. [DOI] [PubMed] [Google Scholar]
- 51.Georgoulia P.S., Todde G., Bjelic S., Friedman R. The catalytic activity of Abl1 single and compound mutations: Implications for the mechanism of drug resistance mutations in chronic myeloid leukaemia. Biochim Biophys Acta Gen Subj. 2019;1863(4):732–741. doi: 10.1016/j.bbagen.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Oruganti B., Friedman R. Activation of Abl1 kinase explored using well-tempered metadynamics simulations on an essential dynamics sampled path. J Chem Theory Comput. 2021;17(11):7260–7270. doi: 10.1021/acs.jctc.1c00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greiner J.V., Glonek T. Intracellular ATP concentration and implication for cellular evolution. Biology (Basel) 2021;10(11) doi: 10.3390/biology10111166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panjarian S., Iacob R.E., Chen S., Wales T.E., Engen J.R., Smithgall T.E. Enhanced SH3/linker interaction overcomes Abl kinase activation by gatekeeper and myristic acid binding pocket mutations and increases sensitivity to small molecule inhibitors. J Biol Chem. 2013;288(9):6116–6129. doi: 10.1074/jbc.M112.431312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azam M., Latek R.R., Daley G.Q. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112(6):831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhang M., Jang H., Li Z., Sacks D.B., Nussinov R. B-Raf autoinhibition in the presence and absence of 14-3-3. Structure. 2021;29(7):768–77 e2. doi: 10.1016/j.str.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maloney R.C., Zhang M., Jang H., Nussinov R. The mechanism of activation of monomeric B-Raf V600E. Comput Struct Biotechnol J. 2021;19:3349–3363. doi: 10.1016/j.csbj.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang M., Jang H., Nussinov R. The mechanism of PI3Kalpha activation at the atomic level. Chem Sci. 2019;10(12):3671–3680. doi: 10.1039/c8sc04498h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jang H., Smith I.N., Eng C., Nussinov R. The mechanism of full activation of tumor suppressor PTEN at the phosphoinositide-enriched membrane. Iscience. 2021;24(5) doi: 10.1016/j.isci.2021.102438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jang H., Zhang M., Nussinov R. The quaternary assembly of KRas4B with Raf-1 at the membrane. Comput Struct Biotechnol J. 2020;18:737–748. doi: 10.1016/j.csbj.2020.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooks B.R., Brooks C.L., 3rd, Mackerell A.D., Jr., Nilsson L., Petrella R.J., Roux B., et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14(1):71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klauda J.B., Venable R.M., Freites J.A., O'Connor J.W., Tobias D.J., Mondragon-Ramirez C., et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou H.-X. How often does the myristoylated N-terminal latch of c-Abl come off? Febs Lett. 2003;552(2–3):160–162. doi: 10.1016/s0014-5793(03)00911-6. [DOI] [PubMed] [Google Scholar]
- 66.Daday C., de Buhr S., Mercadante D., Grater F. Mechanical force can enhance c-Src kinase activity by impairing autoinhibition. Biophys J. 2022;121(5):684–691. doi: 10.1016/j.bpj.2022.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng Y., Lin Y.L., Roux B. Computational study of the “DFG-flip” conformational transition in c-Abl and c-Src tyrosine kinases. J Phys Chem B. 2015;119(4):1443–1456. doi: 10.1021/jp511792a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meng Y., Shukla D., Pande V.S., Roux B. Transition path theory analysis of c-Src kinase activation. Proc Natl Acad Sci U S A. 2016;113(33):9193–9198. doi: 10.1073/pnas.1602790113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu W., Doshi A., Lei M., Eck M.J., Harrison S.C. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3(5):629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 70.Brasher B.B., Roumiantsev S., Van Etten R.A. Mutational analysis of the regulatory function of the c-Abl Src homology 3 domain. Oncogene. 2001;20(53):7744–7752. doi: 10.1038/sj.onc.1204978. [DOI] [PubMed] [Google Scholar]
- 71.Galdadas I., Carlino L., Ward R.A., Hughes S.J., Haider S., Gervasio F.L. Structural basis of the effect of activating mutations on the EGF receptor. Elife. 2021;10 doi: 10.7554/eLife.65824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin-Fernandez M.L., Clarke D.T., Roberts S.K., Zanetti-Domingues L.C., Gervasio F.L. Structure and dynamics of the EGF receptor as revealed by experiments and simulations and its relevance to non-small cell lung cancer. Cells. 2019;8(4):316. doi: 10.3390/cells8040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S., Brier S., Smithgall T.E., Engen J.R. The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Sci. 2007;16(4):572–581. doi: 10.1110/ps.062631007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Panjarian S., Iacob R.E., Chen S., Engen J.R., Smithgall T.E. Structure and dynamic regulation of Abl kinases. J Biol Chem. 2013;288(8):5443–5450. doi: 10.1074/jbc.R112.438382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Filippakopoulos P., Kofler M., Hantschel O., Gish G.D., Grebien F., Salah E., et al. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell. 2008;134(5):793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kornev A.P., Taylor S.S. Dynamics-driven allostery in protein kinases. Trends Biochem Sci. 2015;40(11):628–647. doi: 10.1016/j.tibs.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim J., Ahuja L.G., Chao F.A., Xia Y., McClendon C.L., Kornev A.P., et al. A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci Adv. 2017;3(4):e1600663. doi: 10.1126/sciadv.1600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu J., Ahuja L.G., Meharena H.S., Kannan N., Kornev A.P., Taylor S.S., et al. Kinase regulation by hydrophobic spine assembly in cancer. Mol Cell Biol. 2015;35(1):264–276. doi: 10.1128/MCB.00943-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Azam M., Seeliger M.A., Gray N.S., Kuriyan J., Daley G.Q. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15(10):1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nussinov R., Tsai C.J., Jang H. Autoinhibition can identify rare driver mutations and advise pharmacology. FASEB J. 2020;34(1):16–29. doi: 10.1096/fj.201901341R. [DOI] [PubMed] [Google Scholar]
- 81.Ibrahim M.T., Trozzi F., Tao P. Dynamics of hydrogen bonds in the secondary structures of allosteric protein Avena Sativa phototropin 1. Comput Struct Biotechnol J. 2022;20:50–64. doi: 10.1016/j.csbj.2021.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mu J., Zhou J., Gong Q., Xu Q. An allosteric regulation mechanism of Arabidopsis Serine/Threonine kinase 1 (SIK1) through phosphorylation. Comput Struct Biotechnol J. 2022;20:368–379. doi: 10.1016/j.csbj.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Byun J.A., VanSchouwen B., Akimoto M., Melacini G. Allosteric inhibition explained through conformational ensembles sampling distinct “mixed” states. Comput Struct Biotechnol J. 2020;18:3803–3818. doi: 10.1016/j.csbj.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Selvaratnam R., Chowdhury S., VanSchouwen B., Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc Natl Acad Sci USA. 2011;108(15):6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akimoto M., Martinez Pomier K., VanSchouwen B., Byun J.A., Khamina M., Melacini G. Allosteric pluripotency: challenges and opportunities. Biochem J. 2022;479(7):825–838. doi: 10.1042/BCJ20210528. [DOI] [PubMed] [Google Scholar]
- 86.Nussinov R., Zhang M., Maloney R., Tsai C.J., Yavuz B.R., Tuncbag N., et al. Mechanism of activation and the rewired network: New drug design concepts. Med Res Rev. 2022;42(2):770–799. doi: 10.1002/med.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nussinov R., Jang H., Gursoy A., Keskin O., Gaponenko V. Inhibition of nonfunctional Ras. Cell Chem Biol. 2021;28(2):121–133. doi: 10.1016/j.chembiol.2020.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shaw A.T., Friboulet L., Leshchiner I., Gainor J.F., Bergqvist S., Brooun A., et al. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med. 2016;374(1):54–61. doi: 10.1056/NEJMoa1508887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu J., Nussinov R. Allosteric effects in the marginally stable von Hippel-Lindau tumor suppressor protein and allostery-based rescue mutant design. Proc Natl Acad Sci U S A. 2008;105(3):901–906. doi: 10.1073/pnas.0707401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.LaMarche M.J., Acker M., Argintaru A., Bauer D., Boisclair J., Chan H., et al. Identification of TNO155, an allosteric SHP2 inhibitor for the treatment of cancer. J Med Chem. 2020;63(22):13578–13594. doi: 10.1021/acs.jmedchem.0c01170. [DOI] [PubMed] [Google Scholar]
- 91.Garcia Fortanet J., Chen C.H., Chen Y.N., Chen Z., Deng Z., Firestone B., et al. Allosteric inhibition of SHP2: identification of a potent, selective, and orally efficacious phosphatase inhibitor. J Med Chem. 2016;59(17):7773–7782. doi: 10.1021/acs.jmedchem.6b00680. [DOI] [PubMed] [Google Scholar]
- 92.Calligari P., Santucci V., Stella L., Bocchinfuso G. Discriminating between competing models for the allosteric regulation of oncogenic phosphatase SHP2 by characterizing its active state. Comput Struct Biotechnol J. 2021;19:6125–6139. doi: 10.1016/j.csbj.2021.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marasco M., Kirkpatrick J., Nanna V., Sikorska J., Carlomagno T. Phosphotyrosine couples peptide binding and SHP2 activation via a dynamic allosteric network. Comput Struct Biotechnol J. 2021;19:2398–2415. doi: 10.1016/j.csbj.2021.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang J., Jain A., McDonald L.R., Gambogi C., Lee A.L., Dokholyan N.V. Mapping allosteric communications within individual proteins. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-17618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Papaleo E., Saladino G., Lambrughi M., Lindorff-Larsen K., Gervasio F.L., Nussinov R. The role of protein loops and linkers in conformational dynamics and allostery. Chem Rev. 2016;116(11):6391–6423. doi: 10.1021/acs.chemrev.5b00623. [DOI] [PubMed] [Google Scholar]
- 96.Rivalta I., Batista V.S. In: Allostery: methods and protocols. Di Paola L., Giuliani A., editors. New York, NY; Springer, US: 2021. Community network analysis of allosteric proteins; pp. 137–151. [Google Scholar]
- 97.Kuzmanic A., Bowman G.R., Juarez-Jimenez J., Michel J., Gervasio F.L. Investigating cryptic binding sites by molecular dynamics simulations. Acc Chem Res. 2020;53(3):654–661. doi: 10.1021/acs.accounts.9b00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huss P., Meger A., Leander M., Nishikawa K., Raman S. Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by deep mutational scanning. Elife. 2021;10 doi: 10.7554/eLife.63775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fajer M., Meng Y., Roux B. The activation of c-Src tyrosine kinase: conformational transition pathway and free energy landscape. J Phys Chem B. 2017;121(15):3352–3363. doi: 10.1021/acs.jpcb.6b08409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shukla D., Meng Y., Roux B., Pande V.S. Activation pathway of Src kinase reveals intermediate states as targets for drug design. Nat Commun. 2014;5(1):3397. doi: 10.1038/ncomms4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Breitkopf S.B., Yuan M., Pihan G.A., Asara J.M. Detection of a rare BCR-ABL tyrosine kinase fusion protein in H929 multiple myeloma cells using immunoprecipitation (IP)-tandem mass spectrometry (MS/MS) Proc Natl Acad Sci U S A. 2012;109(40):16190–16195. doi: 10.1073/pnas.1212759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu B.A., Engelmann B.W., Nash P.D. The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction. Febs Lett. 2012;586(17):2597–2605. doi: 10.1016/j.febslet.2012.04.054. [DOI] [PubMed] [Google Scholar]
- 104.Liao T.J., Jang H., Nussinov R., Fushman D. High-affinity interactions of the nSH3/cSH3 domains of Grb2 with the C-terminal proline-rich domain of SOS1. J Am Chem Soc. 2020;142(7):3401–3411. doi: 10.1021/jacs.9b10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.