Abstract
The tumor microenvironment in pancreatic ductal adenocarcinoma (PDAC) involves a significant accumulation of fibroblasts as part of the host response to cancer. Employing single-cell RNA-sequencing, multiplex immunostaining, and several genetic mouse models, we identify carcinoma-associated fibroblasts (CAFs) with opposing functions in PDAC progression. Depletion of fibroblast activation protein (FAP)+ CAFs results in increased survival, in contrast to depletion of alpha smooth muscle actin (αSMA)+ CAFs that leads to decreased survival. Tumor-promoting FAP+ CAFs (TP-CAFs) and tumor-restraining αSMA+ CAFs (TR-CAFs) differentially regulate cancer-associated pathways and accumulation of Tregs. Improved efficacy of gemcitabine is observed when IL-6 is deleted from αSMA+ CAFs but not from FAP+ CAFs employing dual-recombinase genetic PDAC models. Improved gemcitabine efficacy due to lack of IL-6 synergizes with anti-PD1 immunotherapy to significantly improve survival of PDAC mice. Our study identifies functional heterogeneity of CAFs in PDAC progression and their different roles in therapy response.
Introduction
Fibroblasts accumulate in tumors with a putative capacity to regulate pancreatic ductal adenocarcinoma (PDAC) progression (1–3). Collectively, these cells are referred to as carcinoma-associated fibroblasts (CAFs). CAFs may contribute to the emergence and progression of PDAC, and response to treatment (3–7). The focus on CAFs in pancreatic cancer has been driven by the fact that, in some patients, stromal cells (including fibroblasts) can out-number the cancer cells (8–11). Pancreatic stellate cells likely become ‘activated’ by TGFβ1 and other mediators to become CAFs in pancreatic cancer (3,7,10). Such activated pancreatic stellate cells (aPSCs) are also referred to in the literature as myofibroblasts due to their expression of αSMA (12,13). The biology of CAFs in PDAC is still evolving, with increased recognition of their role in shaping the tumor immune microenvironment (1,3,14,15). Moreover, it is likely that CAFs in mouse and human PDAC are a heterogeneous population and have been tentatively classified into inflammatory, myofibroblastic/extracellular matrix (ECM)-producing, and antigen-presenting CAFs by several different research groups (16–21). However, their precise function in PDAC progression remains unknown.
A subtype of CAFs has been identified in PDAC, defined by expression of FAP, with an ability to promote PDAC tumor growth, possibly via CXCL12 (SDF1) (22) and CCL2 signaling (23). Recent studies have suggested that targeting FAP+ CAFs may lead to suppression of tumor growth (24–26). Loss of FAP protein in stromal cells delays PDAC disease progression in mice (24). Alternatively, αSMA+ CAFs have been shown to function in restraining tumors in genetically engineered mouse models (GEMMs) of PDAC and help polarize tumor infiltrating T cells (27). A debate continues whether FAP+ CAFs and αSMA+ CAFs are the same population with model-specific data interpretations from different laboratories (15,22,25,28–30). Therefore, the question that remains unanswered to date is whether FAP and αSMA identify the same CAF population or subsets of fibroblasts with distinct function(s) in PDAC biology and therapy. We conducted an unbiased study to identify different CAFs, with a specific focus to determine the function of FAP+ CAFs and αSMA+ CAFs in PDAC, employing multiparametric analysis coupled with new genetic mouse models. Our results unravel functional heterogeneity of CAFs within the tumor microenvironment with distinct roles in PDAC progression and therapy response.
Results
scRNA-seq analysis identifies FAP+ CAFs and αSMA+ CAFs as distinct populations
To explore whether FAP+ CAFs and αSMA+ CAFs are distinct subsets of CAFs, we performed scRNA-seq on Pdx1Cre/+; LSL-KrasG12D/+; Trp53R172H/+; LSL-YFP (KPCY, see nomenclature of GEMMs in Supplementary Table 1) tumors and identified different cell types present in the tumor microenvironment, including cancer cells, immune cells, endothelial cells, pericytes, and CAFs (Figure 1A, Supplementary Figure 1A, Supplementary Tables 2–3). The CAF cluster was characterized by expression of mesenchymal genes (Col1a1, Col1a2, Dcn) and lack of expression of epithelial genes (Krt8, Krt18, Krt19), and correlated with previously identified subsets based on similar transcriptome analysis (Supplementary Figure 1A–B). To further appreciate the precise association of FAP+ CAFs and αSMA+ CAFs within the CAF clusters, a deeper computational analysis of the CAF clusters was performed to reveal 6 subsets with distinct transcriptional profiles, termed aCAF, bCAF, cCAF, dCAF, eCAF, and fCAF (Figure 1A, Supplementary Figure 1C, Supplementary Table 4). To further investigate potential changes in CAF clusters during PDAC progression, we compared early-stage PDAC with late-stage PDAC in KPC mice. Early-stage PDAC presented with primarily normal and PanIN histology, whereas late-stage PDAC presented with more advanced disease histology (Supplementary Figure 2A). The distribution of CAF subsets shifted during disease progression, with aCAFs as the most abundant population in early-stage PDAC and cCAFs as the most abundant population in late-stage PDAC, respectively (Figure 1A).
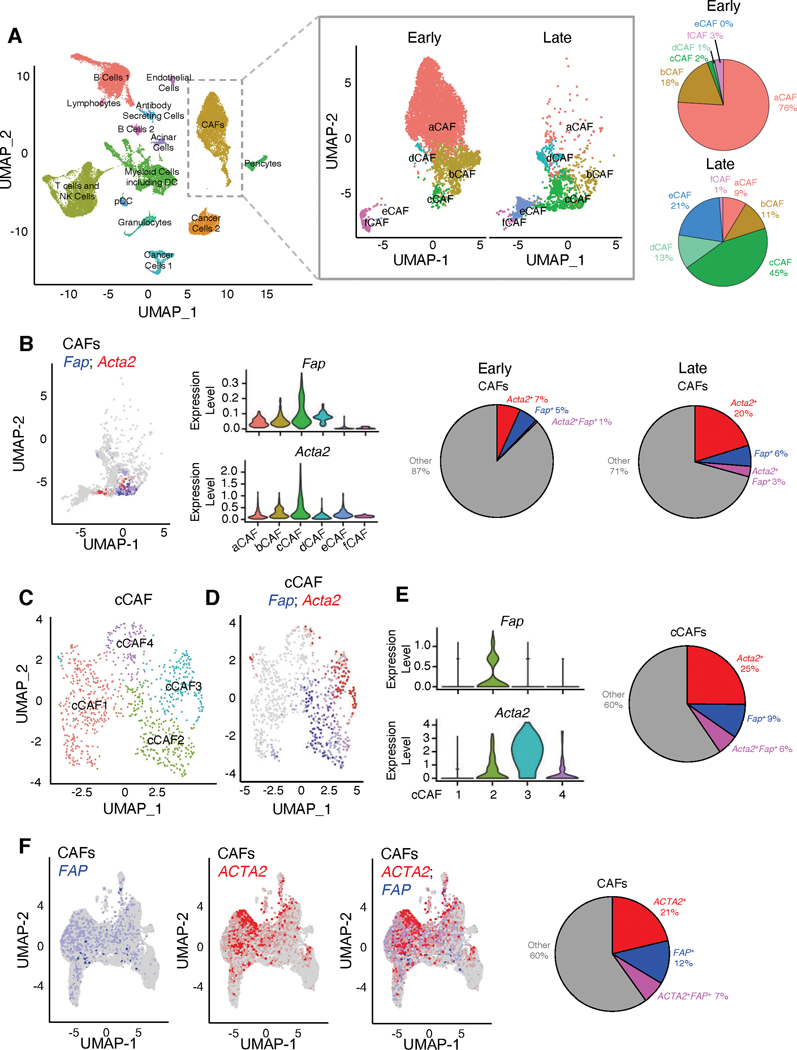
Figure 1. Distinct CAFs populations are identified by single-cell RNA-sequencing (scRNA-seq) in pancreatic tumors.
(A) UMAP projection of cell populations in KPC pancreatic tumors as determined by scRNA-seq (n=8 mice; 31,861 cells; left panel). UMAP projection of the digitally selected CAF cluster in early-stage and late-stage KPC mice (early-stage, n=5 mice, 6,018 cells; late-stage, n=3 mice, 1,606 cells; center panel). Percentages of CAF subsets in early-stage and late-stage KPC mice (right panel).
(B) UMAP projection (left panel) and violin plots (center panels) of Fap and Acta2 transcripts in late-stage KPC tumors (n=3 mice; 1,606 cells). Relative percentages of Fap+, Acta2+, and Acta2+Fap+ CAFs in early-stage (n=5 mice; 6,018 cells) and late-stage KPC tumors (right panel).
(C) UMAP projection of subpopulations of cCAF cluster identified in (B).
(D) UMAP projection (left panel) of Fap and Acta2 transcripts in cCAF cluster.
(E) Violin plots of Fap and Acta2 expression levels in cCAF subsets (left panels). Relative percentages of Fap+, Acta2+, and Acta2+Fap+ cCAFs (right panel).
(F) UMAP projections of FAP-expressing and ACTA2-expressing CAFs (left panel) and relative percentages of ACTA2+, FAP+, and ACTA2+FAP+ CAFs from human PDAC samples (right panel). scRNA-seq data from Peng et al. (31) was reanalyzed to overlay ACTA2-expressing and FAP-expressing CAFs. Fibroblast clusters were identified based on the expression of mesenchymal genes COL1A1, COL1A2, DCN and PDPN.
We evaluated the expression of well-established fibroblast-associated genes within the 6 CAF subsets. Several genes (Col1a1, Col1a2, Pdgfra, Vim, and Pdpn) were expressed in all 6 subsets of CAFs, whereas other genes (Tagln, Fap, S100a4, Acta2, and Pdgfrb) were restricted to specific subsets of CAFs (Supplementary Figure 2B). To ensure that our analysis was not overtly influenced by the presence of pericytes, we assessed the CAF cluster for common pericyte markers (Mcam and Cspg4), and found minimal expression of pericyte marker genes, as also reported by others (16,18,20) (Supplementary Figure 2C). The CAF cluster was also separated into three subpopulations previously termed as inflammatory CAF (iCAF), myofibroblastic CAF (myCAF) and antigen-presenting CAF (apCAF) (16,18,20) (Supplementary Figure 3A–B). Clustering of CAFs into iCAF, myCAF, and apCAF was associated with differences in subpopulation composition based on PDAC stage, with iCAF being the most abundant subset in the early-stage PDAC and myCAF as the most abundant subset in the late-stage PDAC (Supplementary Figure 3A–B). This analysis further confirms that clustering CAFs into 6 sub-clusters provides more in-depth appreciation of the changing landscape of CAF subsets with PDAC progression (Figure 1A).
Next, we specifically determined the localization of Fap (encoding FAP) and Acta2 (encoding αSMA) within the CAF subsets and found that while Fap and Acta2 expressing cells largely localized to the cCAF subset (Figure 1B), more detailed analysis of this subset revealed minimal overlap of FAP+ CAFs and αSMA+ CAFs within cCAFs (Figure 1C–E). The cCAF cluster was further subdivided into cCAF1 (defined by expression of Sfrp1, Serpina3n, Chl1, Mmp2, Igfbp4), cCAF2 (defined by expression of Igfbp7, Serpine2, Col8a1, Id3, Cdkn2a), cCAF3 (defined by expression of Acta2, Tagln, Lgals1, Tmsb4x, Rpl41), and cCAF4 (defined by expression of Ero1l, Egln3, Slc2a1, Pgk1, Bnip3) (Figure 1C, Supplementary Table 5). Fap was predominantly identified in cCAF2 sub-cluster and displayed minimal overlap with Acta2 within the cCAF cluster (Figure 1D–E). Acta2 was predominantly associated with cCAF3 with some association also observed with cCAF2 and cCAF4 (Figure 1D–E). The existence of Fap+ CAFs and Acta2+ CAFs as distinct subsets of cells was further confirmed using a murine PDAC scRNA-seq dataset (CAFs enriched by sorting for CD45−/EpCAM−/CD31− cells) (16) (Supplementary Figure 3C–D). This provides the confidence that our findings identifying such separation of CAFs subsets can be validated in multiple datasets. Next, we analyzed scRNA-seq database with 23 human PDAC tumor CAF samples (31), wherein CAFs were identified based on the expression of mesenchymal genes Col1a1, Col1a2, Dcn and Pdpn. Human CAFs from 23 human PDAC tumors also present with FAP+ CAFs and ACTA2+ CAFs as distinct subsets of cells (Figure 1F).
Identification of FAP+ and αSMA+ CAFs as distinct subpopulations with different prognostic values
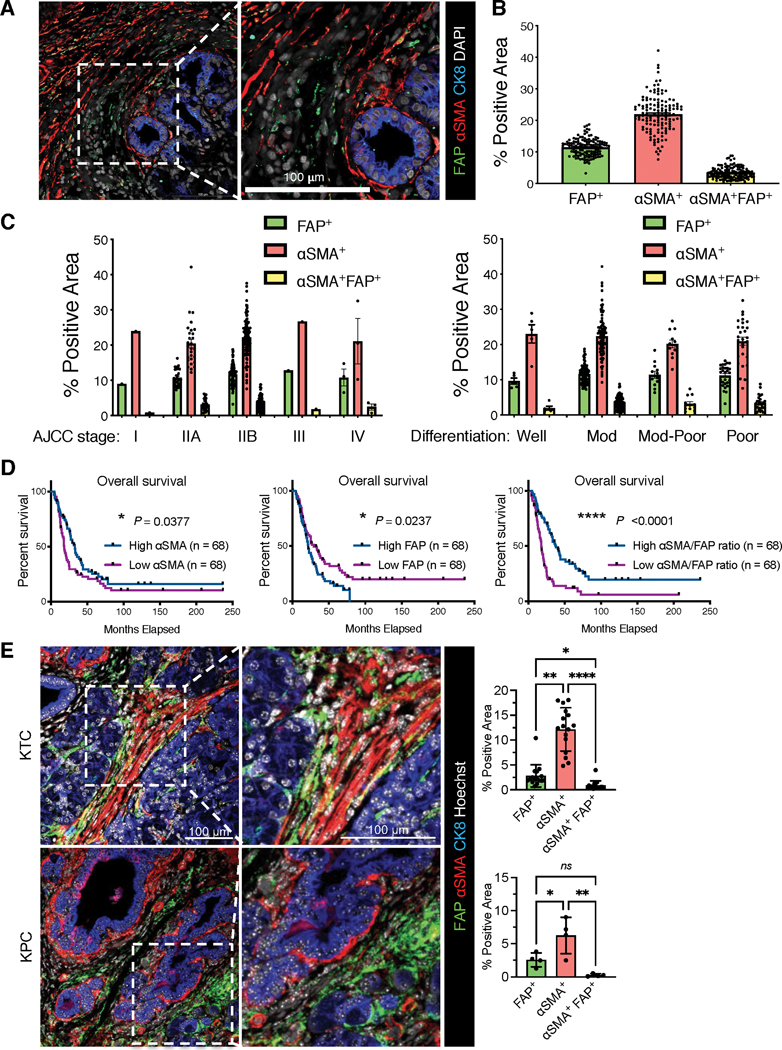
Based on our scRNA-seq analyses showing the minimal overlap between FAP and αSMA expression in CAFs of human and mouse PDAC, we next examined treatment naïve human PDAC tissue microarray (TMA) samples (n = 136) for FAP and αSMA protein expression. The results validated a minimal overlap between FAP and αSMA expression, independent of tumor stage or differentiation status (Fig. 2A–C, Supplementary Table 6). Moreover, αSMA positivity correlated with significantly increased overall survival of patients, in contrast to FAP positivity which was associated with significantly decreased overall survival (Fig. 2D). Furthermore, the ratio of αSMA/FAP demonstrated a significant prognostic value, showing that patients with high αSMA expression and low FAP expression exhibited significantly better overall survival (Fig. 2D). Consistent results were observed on pancreatic tumors from PDAC mouse models (Fig. 2E). These results further suggest that FAP+ CAFs and αSMA+ CAFs are distinct CAF subsets with potentially distinct functions in PDAC.
Figure 2. Distinct CAF subpopulations are identified by immunostaining in human pancreatic tumors.
(A-D) Human PDAC tissue microarray (TMA) of MDACC cohort with 136 cases were examined by FAP, αSMA, and cytokeratin-8 (CK8) immunofluorescence staining. (A) Representative image of FAP, αSMA, and cytokeratin-8 stained PDAC tissue (left panel). Scale bar: 100 μm. (B) The percentage of FAP+αSMA−, αSMA+FAP−, and αSMA+FAP+ positive area for all cases. (C) The percentages of FAP+αSMA−, αSMA+FAP−, and αSMA+FAP+ positive area in the subgroups with indicated AJCC stage (left panel). The percentages of FAP+αSMA−, αSMA+FAP−, and αSMA+FAP+ positive area in the subgroups with indicated tumor histology identities (right panel). (D) The comparison of overall survival between patient subgroups stratified by the values of FAP level, αSMA level, or αSMA/FAP ratio. Log rank Mantel-Cox test performed.
(E) The immunofluorescent staining of FAP and αSMA on pancreatic tumor tissue sections from KTC and KPC transgenic mouse models. Scale bar, 100 μm. KTC, n=16 mice; KPC, n=4 mice. Kruskall-Wallis with Dunn’s multiple comparison test performed for KTC tumors, one-way ANOVA with Tukey’s multiple comparison performed for KPC tumors. * p <0.05, ** p <0.01, **** p <0.0001, ns: not significant.
FAP+ and αSMA+ CAFs exhibit opposing functions in PDAC progression in identical autochthonous models of PDAC
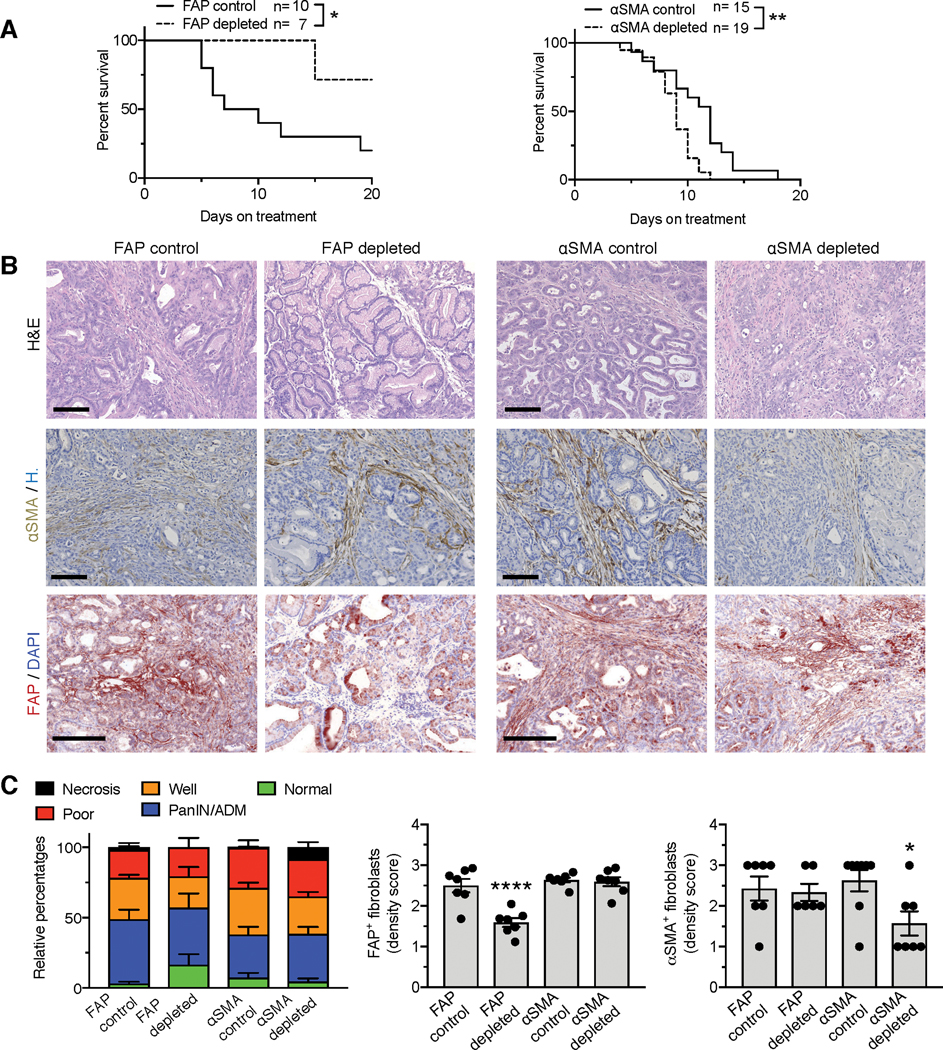
Guided by the non-overlapping expression and the clinical data associated with FAP and αSMA in PDAC CAFs, we generated FAP-thymidine kinase (TK) transgenic mice and crossed them with KTC PDAC GEMMs to disable the accumulation of FAP+ CAFs. These mice were studied alongside KTC;αSMA-TK mice (27), which prevent the accumulation of αSMA+ CAFs in PDAC (Supplementary Table 1). Specific depletion of FAP+ CAFs resulted in significant suppression of PDAC tumor progression and a significant increase in overall survival of mice (Figure 3A–C, Supplementary Figure 4A–B). In contrast, depletion of αSMA+ CAFs resulted in a more aggressive PDAC tumor with decreased overall survival of mice (27) (Figure 3A–C, Supplementary Figure 4A–B). Successful depletion of FAP+ CAFs or αSMA+ CAFs was confirmed by immunohistochemistry analyses of PDAC tumors (Figure 3B–C). The FAP-TK and αSMA-TK transgenes disable the accumulation of proliferating FAP-expressing and αSMA-expressing cells with similar efficiency upon ganciclovir administration, respectively (Figure 3B–C). Importantly, depletion of FAP+ CAFs did not change the number of αSMA+ CAFs, and depletion of αSMA+ CAFs did not alter the number of FAP+ CAFs the tumors (Figure 3C). This further affirms our previous (vide supra) assessment that FAP+ CAFs and αSMA+ CAFs are distinct cell subsets in the TME with opposing functions.
Figure 3. FAP+ CAFs and αSMA+ CAFs have opposing roles in PDAC progression.
(A) Survival curves of FAP-TK KTC mice and αSMA-TK KTC mice after start of treatment with GCV or PBS. FAP control, n=10; FAP depleted, n=7; αSMA control, n=15; αSMA depleted, n=19. Log rank Mantel-Cox test performed comparing the indicated groups.
(B) Representative micrographs of H&E (top panel), αSMA (middle panel) or FAP (bottom panel) immunostaining on control, FAP+ CAF-depleted, or αSMA+ CAF-depleted KTC pancreatic tumor sections. Scale bar: 100 μm.
(C) Relative percentages of each tumor histological phenotype (left panel). FAP control, n=7; FAP depleted, n=7; αSMA control, n=14; αSMA depleted, n=11. Quantification of FAP (middle panel) and αSMA (right panel) density scores in indicated groups of tumors. FAP scoring: n=7 mice per group. αSMA scoring: FAP control, n=7; FAP depleted, n=6; αSMA control, n=8; αSMA depleted, n=7. One-way ANOVA with Tukey’s multiple comparisons test was performed comparing control to depleted mice. * p < 0.05, ** p < 0.01, **** p < 0.0001.
Moreover, scRNA-seq analysis revealed that Pcna expression levels were consistent across the CAF subsets (Supplementary Figure 4C). The percentage of Fap+ and Acta2+ CAFs expressing Pcna was also similar (Supplementary Figure 4C), indicating that these cell types exhibit comparable proliferative indexes. The percentage of Fap+ and Acta2+ cancer cells was 2.3% and 0.9%, respectively, suggesting that FAP-TK and αSMA-TK transgenes have minimal impact on cancer cells (Supplementary Figure 4D). Desmin expression (a dominant pericyte marker) did not significantly change in FAP+ CAF-depleted tumors or αSMA+ CAF-depleted tumors when compared to their respective controls (Supplementary Figure 4E). Altogether, these data indicate that pericytes are not depleted in these TK model systems, and the observed phenotypes are due to the function(s) of CAFs and not due to perivascular or cancer cells.
Histopathological analysis of tumors from FAP+ CAF-depleted mice were associated with increased prevalence of normal tissue, whereas tumors from αSMA+ CAF-depleted mice revealed a more aggressive tumor phenotype (Figure 3C, Supplementary Figure 4A–B). The phenotypes observed upon depletion of FAP+ CAFs and αSMA+ CAFs were confirmed using alternative models of PDAC. A similar phenotype, together with decreased overall survival of mice, was observed when αSMA+ CAFs are depleted in Ptf1acre/+; LSL-KrasG12D/+; Trp53R172H/+ (KPCp48) mice, a different genetic mouse model of PDAC (Supplementary Figure 4F–H). Improved histological scoring was also observed when FAP+ CAFs were depleted in the context of orthotopically implanted KPC-derived cell lines, when compared to control mice (Supplementary Figure 4I). These findings further support the specific targeting of FAP+ CAFs without any significant impact on cancer cells. Previous studies implicated FAP depletion in the development of a cachexic phenotype in mice (32,33), albeit with a distinct genetic strategy. Our genetic targeting strategy, limited to only actively proliferating cells, was not associated with body weight loss or muscle wasting over time in non-tumor bearing adult mice (Supplementary Figure 5A–B). Loss in FAP+ cells in the spleen of healthy (tumor-free) FAP-TK mice was also not observed (Supplementary Figure 5C).
FAP+ CAFs and αSMA+ CAFs distinctly influence the PDAC immune microenvironment
To decipher the mechanistic underpinning for the opposing functions of FAP+ CAFs and αSMA+ CAFs in PDAC progression, we performed global transcriptomic analyses of control, FAP+ CAF-depleted KTC tumors, and αSMA+ CAF-depleted KTC tumors. Comparative analyses of gene expression following the depletion of FAP+ CAFs or αSMA+ CAFs revealed minimal overlap of both downregulated and upregulated genes (Figure 4A–B). To ascertain whether such distinct transcriptomic profiles were associated with specific biological processes, we evaluated the enriched pathways in FAP+ CAF-depleted tumors and αSMA+ CAF-depleted tumors. Interestingly, FAP+ CAF depletion was associated with upregulation of largely distinct pathways related to protein processing, proteolysis, fibrinogen and blood coagulation, cell junctions, endopeptidase inhibitor activity, and pancreatic secretion, potentially reflecting the improved histology in FAP+ CAF-depleted tumors (Figure 4C). In αSMA+ CAF-depleted tumors, gene expression changes were associated with pathways related to epithelial migration, cell proliferation, cytokine production, inflammatory responses, as well as T and B cell-related immunity (Figure 4D).
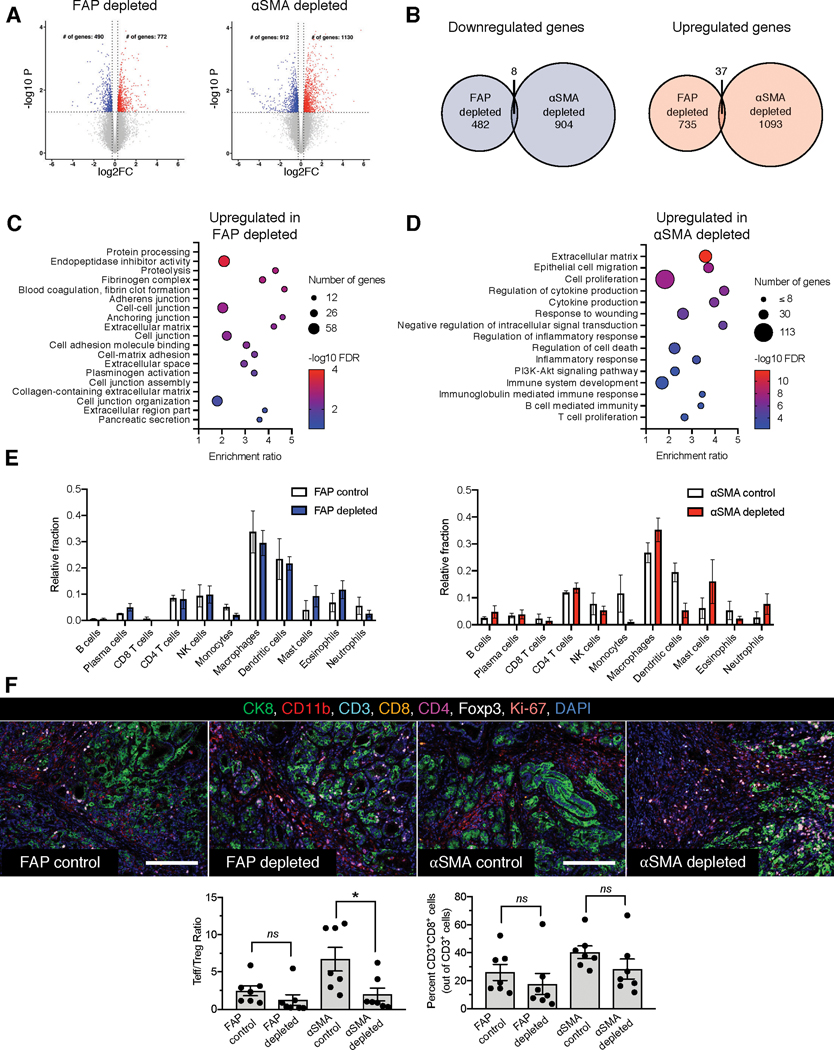
Figure 4. FAP+ and αSMA+ CAFs distinctly polarize the PDAC tumor immune microenvironment.
(A-B) Differentially regulated genes in endpoint FAP+ CAF-depleted and αSMA+ CAF-depleted KTC tumors. (A) Volcano plots of downregulated and upregulated genes in FAP+ CAF-depleted and αSMA+ CAF-depleted KTC tumors. n=3 mice per group. (B) Unique and common downregulated and upregulated genes in FAP+ CAF-depleted and αSMA+ CAF-depleted KTC tumors.
(C-D) GSEA pathways identified after depletion of FAP+ (C) or αSMA+ cells (D) from KTC tumors.
(E) CIBERSORT analysis of abundance of immune cell subsets in FAP+ CAF-depleted and αSMA+ CAF-depleted KTC tumors. n=3 mice per group.
(F) Multiplex immunohistochemical analysis of tumor immune infiltrate of endpoint FAP+ CAF-depleted and αSMA+ CAF-depleted KTC mice (top panel). Scale bar: 100μm. Quantification of Teff/Treg ratio (bottom left panel) and percent CD3+CD8+ cells (bottom right panel). n=7 mice per group. One-way ANOVA and unpaired two-tailed t-test performed comparing control to depleted mice. * p < 0.05, ns = not significant.
As several pathways related to immune cells and inflammatory signaling were upregulated in αSMA+ CAF-depleted tumors (Figure 4C), we performed CIBERSORT analysis to evaluate the abundance of immune cell subsets. Macrophages and B cells were decreased in FAP+ CAF-depleted tumors, whereas an increase in macrophages and a decrease in dendritic cells were observed in αSMA+ CAF-depleted tumors (Figure 4E). Gene Set Enrichment Analysis (GSEA) of differentially expressed pathways between the control and depleted groups also revealed that certain pathways (such as immune response pathway and adaptive immune system pathway) were oppositely regulated by FAP+ CAF depletion and αSMA+ CAF depletion, with downregulation in FAP+ CAF-depleted tumors and upregulation in αSMA+ CAF-depleted tumors (Supplementary Figure 5D).
To further probe the tumor immune microenvironment of FAP+ CAF-depleted or αSMA+ CAF-depleted KTC tumors, multiplex immunohistochemistry analysis for immune markers was conducted (Figure 4F). Depletion of αSMA+ CAFs reduced the T-effector cell (Teff) to T-regulatory cell (Treg) ratio (Figure 4F). FAP+ CAFs depletion had no significant influence on T cells (Figure 4F). FAP+ CAFs depletion, in contrast to αSMA+ CAFs depletion, was associated with decreased CD11b+ (myeloid) cell infiltration (Supplementary Figure 5E). Depletion of FAP+ or αSMA+ CAFs was not associated with changes in CD8+ cells (Figure 4F). These results support the notion that opposing functions of FAP+ CAFs or αSMA+ CAFs are, at least in part, likely due to differential polarization of tumor immune microenvironment.
FAP+ CAF and αSMA+ CAF secretomes distinctly impact PDAC response to therapy
To further identify the role of αSMA+ CAFs and FAP+ CAFs in PDAC progression and response to therapy, the secretome of the CAFs was evaluated, keeping in mind the sum functional contribution of TP-CAFs and TR-CAFs in the PDAC immune microenvironment (Figure 4E–F). The immune modulatory cytokine IL-6 is a critical mediator of polarization of immune cells (34) and has been implicated in PDAC cancer progression and response to chemotherapy (35–38).
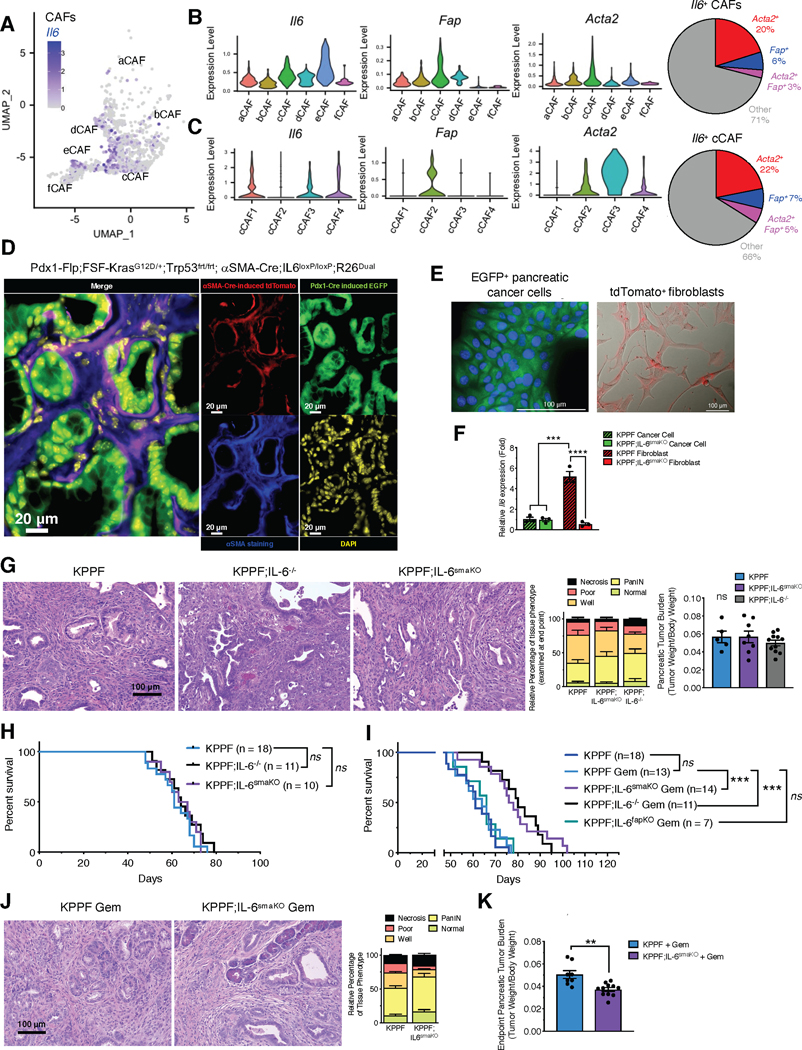
Previous studies identified Il6 as a putative marker gene of iCAFs (16,18); however, we observed that Il6 was also expressed in myCAFs, at both early-stage PDAC and late-stage PDAC in the KPC mice (Supplementary Figure 3A–B). Il6 transcripts are enriched in both cCAFs and eCAFs of early and late stage PDAC tumors (Figure 5A–B and Supplementary Figure 5F), the former composed of both Fap+ and Acta2+ CAFs. Further analysis of the cCAF cluster revealed Il6 is enriched in cCAF1, 3, and 4, albeit some expression also detected in cCAF2 (Figure 5C). Within total CAFs and the cCAFs sub-cluster, both Acta2+ and Fap+ CAFs express Il6 (Figure 5B–C and Supplementary Figure 5F). This observation was confirmed in a published dataset of murine PDAC (Supplementary Figure 5G) (16).
Figure 5. IL-6 produced by distinct CAFs populations in pancreatic cancer.
(A-C) The expression profile of Il6, Fap, and Acta2 of CAF populations from late-stage KPC pancreatic tumors, presented in UMAP projection (A), violin plots (B-C, left panels), and percentages (B-C, right panels) as determined by scRNA-seq originally shown in Figure 1.
(D) Representative fluorescence microscopic images of Pdx1-Flp-induced EGFP (green) expression, αSMA-Cre-induced tdTomato (red) expression, and αSMA immunofluorescence staining (blue) in the PDAC tissues of KPPF;IL-6smaKO;R26Dual mice. Scale bars, 20 μm.
(E) Representative fluorescence images of FACS isolated EGFP-expressing cancer cells and tdTomato-expressing αSMA+ CAFs (depleted for IL-6) from PDAC tissues of KPPF;IL-6smaKO;R26Dual mice shown in (D). Scale bars: 100 μm.
(F) qRT-PCR examination of IL-6 (Il6) in primary cancer cells and αSMA+ CAFs isolated from KPPF;IL-6smaKO;R26Dual mice (shown in E) and KPPF;αSMA-Cre;R26Dual mice (without IL-6 deletion in αSMA+ CAFs). Expression relative to Gapdh and KPPF cancer cells reported. n=3 biological replicates per group. One-way ANOVA with Sidak’s multiple comparisons test based on ΔCT values performed.
(G) Representative pancreatic sections with H&E staining of KPPF, KPPF;IL-6smaKO, and KPPF;IL-6−/− mice examined at endpoint (8–11 weeks of age, left panel), relative percentage of each histological tissue phenotype (center panel), and tumor burden (right panel). Scale bar: 100 μm. Histological phenotypes: KPPF, n=14; KPPF;IL-6smaKO, n=11; KPPF;IL-6−/−, n=11. Tumor burden: KPPF, n=5; KPPF;IL-6smaKO, n=8; KPPF;IL-6−/−, n=11. Two-way ANOVA (histological phenotypes) and one-way ANOVA (tumor burden) with Tukey’s multiple comparison test performed.
(H) Overall survival of KPPF (n=18), KPPF;IL-6smaKO (n=10), and KPPF;IL-6−/− (n=11) mice. Log rank Mantel-Cox test performed comparing the indicated groups.
(I) Overall survival of untreated KPPF (n=18, from Figure 5H) and KPPF (n=13), KPPF;IL-6smaKO (n=14), KPPF;IL-6−/− (n=11), and KPPF;IL-6fapKO (n=7) mice treated with gemcitabine (Gem). Log rank Mantel-Cox test performed comparing the indicated groups.
(J-K) Representative H&E sections of pancreatic tissues and relative percentage of each histological tissue phenotype (J), and pancreatic tumor burden (K) of gemcitabine-treated KPPF and KPPF;IL-6smaKO mice at endpoint. For (J): KPPF, n=6; KPPF;IL-6smaKO Gem, n=5. For (K): KPPF, n=8; KPPF;IL-6smaKO Gem, n=11. Scale bar: 100 μm. Two-way ANOVA with Sidak’s multiple comparison test performed for (J). Unpaired two-tailed t-test performed for (K). * p < 0.05, ** p < 0.01, *** p < 0.001, ns = not significant.
To investigate the functional contribution of CAF-derived IL-6 to PDAC progression and therapy response, we generated GEMMs in which two distinct gene recombination systems (Flippase- and Cre-mediated recombination, Supplementary Table 1) independently drive cancer formation (Pdx1-Flp;FSF-KrasG12D/+;Trp53frt/frt; KPPF; Supplementary Figure 6A–B) and allow conditional gene recombination in αSMA+ CAFs (αSMA-Cre; floxed-gene of interest) or FAP+ CAFs (FAP-Cre; floxed-gene of interest) (39). KPPF mice presented with a similar disease progression as the comparable Cre-driven KPPC model (Pdx1-Cre;LSL-KrasG12D/+;Trp53loxP/loxP; Supplementary Figure 6C–D). Potential for recombination events in the CAFs in KPPF;αSMA-Cre;R26Confetti reporter mice was visualized by capture of GFP, RFP, YFP and CFP fluorescent cells in the desmoplastic stroma associated with PDAC (Supplementary Figure 6E).
First, KPPF;αSMA-Cre;R26Dual reporter mice were bred to a conditional IL-6 (Il6) gene knockout allele (KPPF;IL-6smaKO;R26Dual), effectively abrogating IL-6 transcription in αSMA+ CAFs (Figure 5D–F, Supplementary Table 1). Using KPPF;αSMA-Cre;R26Dual reporter mice, we confirmed enrichment of IL-6 transcripts in purified tdTomato+ fibroblasts and noted elevated IL-6 expression in αSMA+ CAFs than cancer cells, which was largely ablated in the αSMA+ CAFs isolated from the KPPF;IL-6smaKO;R26Dual mice (Figure 5F). Genomic recombination events captured by PCR reactions in tumors and control cells and organs also confirmed the specificity of the genetic strategy employed to ensure conditional deletion of IL-6 in αSMA+ cells from the KPPF;IL-6smaKO;R26Dual mice (Supplementary Figure 6F).
To examine the production of IL-6 by cells other than αSMA+ CAFs, the KPPF mice were also bred with systemic (whole body) IL-6 deleted mice (KPPF; IL-6−/−). PDAC progression, overall survival and PDAC histology was similar in both the KPPF;IL-6smaKO and KPPF; IL-6−/− mice when compared to the KPPF control (Figure 5G–H, Supplementary Figure 7A–C). Il6 expression was significantly decreased in tumors of KPPF; IL-6smaKO mice compared to KPPF control tumors, and absent in KPPF;IL-6−/− tumors (Supplementary Figure 7D). Il1b transcript (encoding IL-1β) abundance was unchanged (Supplementary Figure 7D). Collectively these findings support that IL-6 is likely produced by multiple cell types in PDAC, including αSMA+ CAFs and FAP+ CAFs, but its impact on cancer progression (treatment naïve mice harboring p53 loss) is not rate-limiting (36).
IL-6 deletion in αSMA+ CAFs improves gemcitabine efficacy and synergizes with checkpoint blockade therapy
IL-6 signaling is reported to confer pro-survival signals to cancer cells via the JAK/STAT signaling pathway in the context of chemotherapy (35,36). We next investigated whether CAF-derived IL-6 might have a functional contribution in the context of chemotherapy response. IL-6 was genetically deleted in FAP+ CAFs also for this assessment (Supplementary Table 1 and Supplementary Figure 7E). In contrast with loss of IL-6 in FAP+ CAFs (KPPF;IL-6fapKO mice), KPPF;IL-6smaKO mice (with loss of IL-6 in αSMA+ CAFs) were associated with a significant increase in the overall survival of mice upon treatment with gemcitabine (Figure 5I) and identical to KPPF; IL-6−/− mice (Figure 5I). Loss of IL-6 from αSMA+ CAFs resulted in improved histopathology and reduced tumor burden in the context of gemcitabine treatment (Figure 5J–K). Analysis of TCGA human pancreatic cancer cohort further revealed that the ACTA2/IL6 low expression subgroup exhibited less progressive disease in response to primary therapy than the ACTA2/IL6 high expression subgroup (Supplementary Figure 7F). Minimal impact on lung and liver metastasis was observed in KPPF;IL-6smaKO mice compared to KPPF mice treated with gemcitabine (Supplementary Figure 8A). Gemcitabine treatment increased the number of putative IL-6 producing αSMA+ CAFs in KPPF mice (Figure 6A), further supporting their role in therapy resistance. Gemcitabine-enhanced αSMA+ CAFs abundance was also reversed in KPPF;IL-6smaKO mice (Figure 6A).
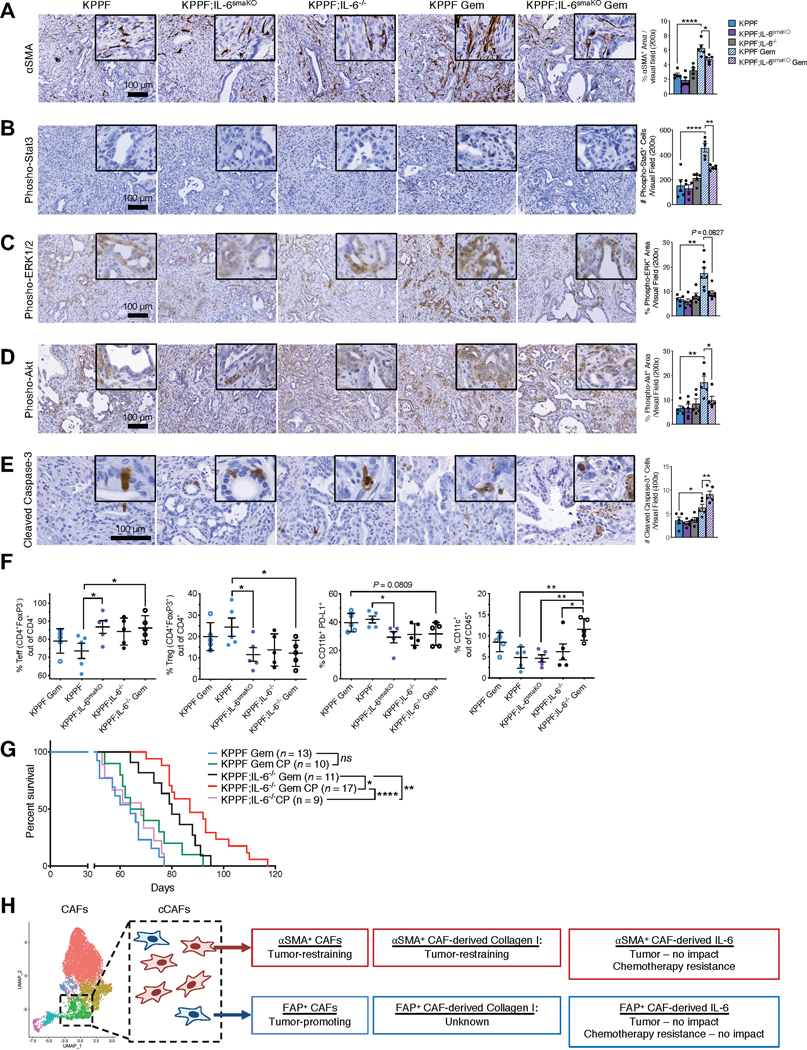
Figure 6. IL-6 produced by CAFs differentially regulates pancreatic tumor immune profile and therapeutic response.
(A-E) Representative images of PDAC tissues from KPPF, KPPF;IL-6smaKO, or KPPF;IL-6−/− mice with or without gemcitabine treatment, stained with αSMA (A), phospho-Stat3 (B), phospho-ERK1/2 (C), phospho-Akt (D), and cleaved caspase-3 (E). n = 5 mice per group for (A), (B), (D), and (E). For (C), KPPF, n=5; KPPF;IL-6smaKO, n=5; KPPF;IL-6−/−, n=5; KPPF Gem, n=7; KPPF;IL-6smaKO Gem, n=7. Scale bars: 100 μm. One-way ANOVA with Sidak’s multiple comparison test performed for A-B, D-E. Kruskal-Wallis with Dunn’s multiple comparison test performed for C.
(F) Percentages of CD4+FoxP3− effector T cells (Teff), CD4+FoxP3+ regulatory T cells (Treg), Teff/Treg ratio, CD11b+PD-L1+ cells, and CD11c+ cells in PDAC tissues of the indicated experimental groups (n = 5 per group, examined at 2.5-months age after saline or gemcitabine treatment for 2 weeks). One-way ANOVA with Sidak’s multiple comparison test performed.
(G) Overall survival of KPPF and KPPF;IL-6−/− mice, treated with gemcitabine (Gem) and/or anti-CTLA4/anti-PD1 (CP) as either single-agent treatment or combined treatment. KPPF Gem, n=13, from Figure 5I; KPPF Gem CP, n=10; KPPF;IL-6−/− Gem, n=11, from Figure 5I; KPPF;IL-6−/− Gem CP, n=17; KPPF;IL-6−/− CP, n=9. Log rank Mantel-Cox test performed comparing the indicated groups. See Supplementary Table 1 for the complete nomenclature list of all mouse strains.
(H) Schematic of the functional roles of CAF subsets and their secretome in PDAC progression and response to therapy.
* p < 0.05, ** p < 0.01, **** p < 0.0001, ns: not significant.
Cancer cells in the tumors of KPPF mice treated with gemcitabine showed elevated levels of phosphorylated Stat3, ERK1/2 and Akt, which was attenuated in KPPF;IL-6smaKO mice treated with gemcitabine (Figure 6B–D). Gemcitabine treatment in KPPF;IL-6smaKO mice did not significantly impact tumor collagen deposition or vasculature, when compared to controls (Supplementary Figure 8B–C). Gemcitabine treatment or IL-6 genetic depletion did not significantly alter the presence of FSP1+ or FAP+ CAFs (Supplementary Figure 8D–E). In contrast, cleaved caspase-3, indicative of apoptosis, increased upon gemcitabine treatment in KPPF;IL-6smaKO mice treated with gemcitabine (Figure 6E). Collectively, these results suggest that αSMA+ CAF derived IL-6 confers tumor resistance to gemcitabine by promoting cancer cell survival, whereas FAP+ CAF derived IL-6 exerts an insignificant impact in the response to gemcitabine (Figure 5I).
Given the well documented role of IL-6 on the tumor immune microenvironment (34), we evaluated the immune composition of tumor, spleen, and peripheral blood of KPPF, KPPF;IL-6−/−, and KPPF;IL-6smaKO mice, with and without gemcitabine therapy (Supplementary Figure 9A–B). Intra-tumoral immune cell frequencies were impacted by loss of IL-6, whereas immune frequencies in the spleen and peripheral blood were largely unchanged (Figure 6F, Supplementary Figure 10A–C). The number of regulatory T cells (Treg) and effector T cells (Teff) significantly changed in the tumors of both KPPF;IL-6−/− and KPPF;IL-6smaKO mice (Figure 6F). Further, the frequency of CD11b+PD-L1+ cells were significantly reduced in KPPF;IL-6−/− and KPPF;IL-6smaKO mice compared to control KPPF mice (Figure 6F). Gemcitabine treatment with IL-6 loss resulted in increased frequency of CD11c+ cells (Figure 6F). These observed alterations in the immune microenvironment suggested that tumors with loss of IL-6 may be sensitive to immune checkpoint blockade in combination with gemcitabine. Therefore, we tested the therapeutic benefit of dual immune checkpoint blockade using anti-CTLA4 and anti-PD-1 antibodies (anti-CTLA4 and anti-PD-1; αCP). While αCP did not reveal efficacy in KPPF or KPPF;IL-6−/− mice, it synergized with gemcitabine to reveal efficacy with significant increase in overall survival of KPPF;IL-6−/− mice (Figure 6G). The immune profiling of the tumor suggested that gemcitabine treatment results in an increase in putative CD11c+ dendritic cells (Figure 6F) with the likely potential for enhanced tumor antigen presentation for an improved effector T cell response upon αCP and depletion of IL-6 (Figure 6G). These results further emphasize the contribution of IL-6 in PDAC therapy response.
Discussion
Several elegant studies have identified potential diversity of CAFs in pancreatic cancer (15–17,19,20,31). While specific markers for different classes of CAFs have not been identified, multiple studies, employing scRNA-seq, have shown that one can classify CAFs into different, transcriptionally defined clusters (16–19,31). Immunohistochemistry studies and mouse models fluorescently labeling mesenchymal cell populations have also suggested that CAFs can express diverse markers that do not always overlap (40–43). In this regard, FAP+ CAFs and αSMA+ CAFs have been identified as being part of the same scRNA-seq cluster and purported to exhibit similar functions; however, other studies showed that an additional subset of CAFs (Meflin+ CAFs) give rise to increasing numbers of αSMA+ CAFs but not FAP+ CAFs during tumor progression (44).
Although recent studies in mice and clinical trial results have demonstrated that suppression of CAFs can lead to acceleration of pancreatic cancer in some contexts (27,44–47), the question remains whether all CAFs are targeted by such strategies or a sub-set of CAFs are manipulated, resulting in more aggressive PDAC. This become an important unaddressed question because other studies have suggested that targeting FAP+ CAFs results in control of PDAC (24), although therapeutic efficacy data from an ongoing PDAC clinical trial is currently unavailable (ClinicalTrials.gov identifier: NCT03932565). Therefore, employing scRNA-seq, multiplex immunostaining, and new genetic mouse models, our studies focused on identifying whether FAP+ CAFs and αSMA+ CAFs possess similar biological and functional relevance in PDAC or display diversity with respect to their biology and actions.
We identify that FAP+ CAFs and αSMA+ CAFs are distinct populations of fibroblasts in both human PDAC samples and transgenic mouse models. The αSMA+ CAFs predominantly act to restrain PDAC (TR-CAFs) and the net function of FAP+ CAFs is to promote PDAC (TP-CAFs, Figure 6H). This data was supported by computational analyses of scRNA-seq data. FAP+ CAFs and αSMA+ CAFs present in the same scRNA-seq sub-clusters of CAFs (cCAFs) due to some commonalities in their transcriptomes; however, they are distinct populations of cells with largely distinct, non-overlapping transcriptomes. We also show that FAP+ CAFs and αSMA+ CAFs regulate different cancer-associated transcriptomic networks and define the tumor immune composition in distinct manners. Suppression of αSMA+ CAFs leads to decreased Teff/Treg ratio, potentially contributing to the accelerated tumor growth. Deletion of type I collagen in αSMA+ CAFs was associated with decreased abundance as well as impaired activation of T cells (21), suggesting that the observed changes in T cells upon depletion of αSMA+ CAFs is in part mediated by reduced collagen I. In contrast, the FAP+ CAFs have an impact on inflammation and their suppression leads to a decrease in CD11b+ myeloid cells and an inhibition of PDAC with increased overall survival of mice. Macrophages have been reported to be tumor promoting in PDAC (48–50), suggesting that alterations in macrophages may contribute to increased survival of FAP+ CAF-depleted mice.
Previous studies showed that the overall functional contribution of αSMA+ CAFs is tumor-restraining (20,27,47). The tumor-restraining contribution of αSMA+ CAFs is in part mediated by their production of type I collagen (21). Our studies identify that IL-6 produced by αSMA+ CAFs does not contribute to progression of PDAC, despite depletion of about 50% of total IL-6 present in the TME. To address the impact of total IL-6 in the TME on PDAC progression, we also crossed IL-6 whole body knockout mice (IL-6−/−) with KPPF mice. Complete lack of IL-6 in the TME of PDAC also has no impact on PDAC progression. These data clearly indicate that IL-6 does not play a role PDAC progression; therefore, it is not surprising that IL-6 deleted from subset of cells in the TME also does not reveal a functional importance. The total loss of IL-6 from the TME or just from the αSMA+ CAFs only has an impact when gemcitabine is provided to suppress cancer cell proliferation and tumor growth. This shows that the optimal response to chemotherapy is compromised by the presence of IL-6 in the TME likely by providing cancer cell survival benefit. This aspect of drug resistance is an applied role of IL-6 but not its natural role in the context of PDAC progression. Therefore, the sum totality of the actions of αSMA+ CAFs is ‘tumor restraining’. When one begins to audit one by one the specific proteins produced by αSMA+ CAFs using dual recombinase genetic mouse models, we begin to unravel their specific role in PDAC progression. Specific deletion of type I collagen from αSMA+ CAFs revealed its tumor restraining action (21) but without an impact on chemotherapy resistance (27). Driven by many published reports and scRNA-seq data, we show that deletion of IL-6 from αSMA+ CAFs does not alter the progression of PDAC, and therefore does not contribute to the ‘tumor restraining’ properties of αSMA+ CAFs. In contrast, IL-6 produced by αSMA+ CAFs has an ‘applied’ function in providing protection against chemotherapy induced inhibition of tumor growth, resulting in perceived therapy resistance. In addition, the abundance of αSMA+ CAFs was significantly increased by gemcitabine treatment, presumably leading to further increased IL-6 production by αSMA+ CAFs. Our study reveals the complexity of CAF biology in PDAC, specifically the different CAF roles in natural progression of the disease and how their function(s) impacts therapeutic intervention.
Collectively our studies show that targeting FAP+ CAFs emerges as a viable strategy to achieve inhibition of PDAC. Such strategy must contemplate sparing the αSMA+ CAFs, and this study suggests that this may be feasible since they are minimally overlapping in their respective protein biomarker presentation. We show that depletion of FAP+ CAFs does not impact αSMA+ CAFs content in PDAC. In this regard, FAP directed-CAR T cell therapy might be an effective strategy as previously demonstrated (51). Targeting chemokines that are selectively secreted by FAP+ CAFs might be a viable strategy also (22). Importantly, the strategy to inhibit FAP+ CAFs must consider prevention of bone marrow toxicities and cachexia (32,33,51). Strategies to increase the number of αSMA+ CAFs can represent another strategy to control PDAC (52). In this regard, activating vitamin D receptor on αSMA+ CAFs could program them to further control PDAC (53).
Our studies indicate that αSMA+ CAF-derived IL-6 confers chemoresistance and negatively regulates T cells in the tumor microenvironment. In this regard, organoid-based studies, wherein a subset of CAFs were highlighted to represent an immunomodulatory phenotype that included IL-6 production (18). While αSMA+ CAFs as a whole emerged as tumor-restraining, in the context of gemcitabine therapy stress, cancer cells likely utilize the IL-6 produced by αSMA+ CAFs to promote their survival through activation of STAT3 signaling. Previous studies reported that high CAF abundance was associated with increased MAPK and STAT3 co-signaling in proliferative and invasive cancer cells (54). In mice treated with gemcitabine, deletion of IL-6 in αSMA+ CAFs led to a reduction in phospho-STAT3 and phospho-ERK1/2 levels in cancer cells, indicating potential paracrine signaling between αSMA+ CAFs and cancer cells to promote cancer cell survival. Alternatively, Ly6Chi monocytes have been reported to express IL6R (55); thus, it is possible αSMA+ CAF derived IL-6 acts indirectly on cancer cells to promote chemoresistance.
It is conceivable that the IL-6 production by the αSMA+ CAFs is for self-preservation purposes in non-treatment conditions but is also utilized by cancer cells for induction of survival signaling pathways in the context of gemcitabine resistance. Previous studies have reported that IL-6 signaling confers pro-survival signals to cancer cells via the JAK/STAT signaling pathway in the context of chemotherapy (35,36). We also observed a significant polarization of the PDAC immune microenvironment upon deletion of αSMA+ CAF produced IL-6, however such changes in Teff and Treg frequencies did not impact PDAC progression. The immune checkpoint blockade therapy was ineffective when combined with gemcitabine in KPPF mice, however a combinatorial benefit was observed with inhibition of IL-6 signaling, indicating that αSMA+ CAF produced IL-6 may be a critical suppressor of immune checkpoint blockade therapy in PDAC. Suppression of IL-6 likely favors the emergence of effector T cells that when combined with the cell death and generation of neoantigens induced by gemcitabine associated with increased CD11c+ cells, augmenting the efficacy of immune checkpoint blockade. A trial targeting IL-6R with tocilizumab in conjunction with nab-paclitaxel and gemcitabine is ongoing (NCT02767557); however, our data suggests that addition of checkpoint blockade to this treatment regimen has the potential to improve therapeutic response.
In summary, this study demonstrates that CAFs are not uniform in their biology and exhibit functional diversity with therapeutic implications for pancreatic cancer.
Methods
Mice
All acronyms designating specific genetically engineered mice (GEMM) are listed in Supplementary Table 1. LSL-KrasG12D/+ (56), Pdx1-Cre (56), Ptf1a-Cre (56), αSMA-TK (57), αSMA-RFP (58), FAP-TK (59), Rosa26-loxP-Stop-loxP-YFP (60), Rosa26-CAG-Brainbow 2.1 (61), LSL-Trp53R172H/+ (56), FSF-KrasG12D/+ (62), Pdx1-Flp (62), Trp53frt/+ (63), αSMA-Cre (57), CMV-Cre (64), IL-6loxP/loxP (65), Trp53loxP/loxP (66), and Tgfbr2loxP/loxP (67) mouse strains were previously documented. The FAP-Cre transgenic strain was newly generated. A 5 kb sequence flanking the FAP promoter and partial Exon 1 (Ex1) was cloned into pORF-HSV1-TK vector (Invivogen) using Not I and Age I. The sequence-confirmed FAP-TK construct was released from the vector using Not I and Swa I before purification and injection into fertilized eggs. FAP-Cre was generated similarly, where the Cre sequence was inserted into the pORF-FAP-TK plasmid with Not I and BstE II digestion. The FAP-Cre plasmid was digested with Not I and Swa I, excised, and injected into fertilized eggs as described for FAP-TK. The transgenic mice were generated in the MD Anderson Genetically Engineered Mouse Facility on the C57Bl/6 genetic background. FAP-TK have been deposited at Jackson Laboratory (stock 034655). These mice were bred onto PDAC GEMMs or implanted orthotopically with 689KPC cancer cells, as previously described (68). The KPPF and R26Dual mice of dual-recombinase system were kindly provided by D. Saur. Mice were maintained on a mixed genetic background and both male and female mice were evaluated.
Ganciclovir (GCV; sud-gcv, Invivogen) was administered i.p. daily at 50 mg/kg of body weight (approximately 1.5 mg per 25g mouse). GCV was administered to KTC mice at 21 to 37 days of age, and to KPCp48 mice at 50–51 days of age. Control groups were TK negative mice that received GCV, phosphate buffer saline (PBS), or were not injected. In the orthotopic tumor model (689KPC), GCV was administered 15 days following tumor implantation and mice were euthanized at 40 days following tumor implantation. All mice were housed under standard housing conditions at MD Anderson Cancer Center (MDACC) animal facility, and all animal procedures were approved by the MDACC Institutional Animal Care and Use Committee. Investigators were not blinded to group allocation but were blinded for the histological assessment of phenotypic outcome with no randomization method used. The experimental endpoint was defined when the animals developed significant signs of illness leading to their death or requiring euthanasia. Survival curves for KTC mice are plotted based on the number of days after start of GCV or PBS treatment and KPCp48 survival curves are plotted based on the entire lifespan of the mice with GCV or PBS treatment start time indicated. For therapeutic treatments, mice were given gemcitabine (G-4177, LC Laboratories) intraperitoneally (i.p.) twice per week at 40 mg/kg of body weight. Anti-CTLA4 (9H10; BioXCell) and anti-PD-1 antibodies (RMP1–14; BioXCell) were i.p. administered at 200 μg/mouse twice per week.
Flow cytometry
For analysis of immune cell populations in spontaneous tumors of indicated transgenic mouse strains, the staining and flow cytometry procedures were conducted as previously described (21). For analysis of cells from KPC tumors, the tumors were minced and digested in collagenase IV (4 mg/mL) and dispase (4 mg/mL) in DMEM media for 1 hour at 37°C. Digestion was stopped by the addition of FBS to neutralize the protease activity, followed by washing with FACS buffer 3 times. Digested tissues were then filtered through a 70 μm mesh followed by a 40 μm mesh, centrifuged, and incubated in ACK lysis buffer for 3 minutes at room temperature. Prior to staining, spleen was minced and filtered through a 40 μm mesh and was incubated in ACK lysis buffed for 3 minutes at room temperature. FAP and its corresponding isotype antibody were conjugated with Zenon Alexa Fluor 647 Rabbit IgG labeling kit according to manufacturer’s instructions. Samples were stained with antibody and fixable viability dye eFluor 780 in FACS buffer for 30 minutes on ice followed by washing prior to analysis on a BD LSR Fortessa X20. For sorting experiments, samples were analyzed and sorted on a BD FACS Aria. Unstained and single-stained samples were used for compensation controls. Data analysis was performed in FlowJo software (TreeStar, Inc). Details on the antibodies, source, and dilution are listed in Supplementary Table 7.
Single-cell RNA-sequencing (scRNA-seq)
PDAC samples with less than 10% pancreatic adenocarcinoma areas were defined as early-stage PDAC, while PDAC samples with greater than 50% pancreatic adenocarcinoma areas were defined as late-stage PDAC. The tumors of KPC mice were processed to obtain single cell suspensions (see flow cytometry method section). scRNA-seq on these samples was conducted at the MDACC Advanced Technology Genomics Core. Single cell Gel Bead-In-Emulsions (GEMs) generation and barcoding, post GEM-RT cleanup and cDNA amplification, library construction and Illumina-ready sequencing library generation were prepared by following the manufacturer’s guidelines. High Sensitivity dsDNA Qubit kit was used to estimate the cDNA and library concentration. HS DNA Bioanalyzer was used for the quantification of cDNA. DNA 1000 Bioanalyzer was used for the quantification of libraries. The “c-loupe” files were generated by using Cell Ranger software pipelines following manufacturer’s guidelines. Cells from unfractionated tumor were encapsulated using 10X Genomics’ Chromium controller and Single Cell 3’ Reagent Kits v2. Following capture and lysis, cDNA was synthesized and amplified to construct Illumina sequencing libraries. The libraries from about 1,000 cells per sample were sequenced with Illumina Nextseq 500. The run format was 26 cycles for read 1, 8 cycles index 1, and 124 cycles for read 2. scRNA-seq data was processed by the Advanced Technology Genomics Core at MDACC.
Cell clustering was performed using the Seurat R software as previously described (69,70). Specifically, cell clustering was conducted using the non-linear dimensional reduction technique by the Uniform Manifold Approximation and Projection (UMAP) algorithm. To identify marker genes of cell clusters, we compared each of the cell clusters using pairwise differential expression analysis with settings recommended for data with batch effect, with average FC expression compared to other included clusters was >2. The clustering of CAFs was performed using an analogous gene signature sets used in a recently published dataset (16).
Library Seurat (version 3.6.1), dplyr and cowplot were loaded into R to explore QC metrics, filter cells, normalize data, cluster cells, and identify cluster biomarkers. To filter out low-quality cells, a threshold with a minimum of 200 and a maximum of 4000–7000 genes per cell was used. Cells with more than 10% of the mitochondrial genome were also removed for further analysis. To remove the influence of technical characteristics from downstream analyses, “sctransform” package was used for normalization. “RunUMAP” function was used for clustering the cells. “FindAllMarkers” function was used to identify the specific markers for each cell cluster. “DoHeatmap” function was used to show the top genes in each cluster. “VlnPlot” function was used to show expression probability distributions across cell clusters of the genes we selected to assign the cell type identity, and the genes that we were interested in. The Markov affinity-based graph imputation of cells (MAGIC) algorithm, which utilizes the nearest neighbor graphing and a diffusion operator to restore or “smooth” missing transcripts from the single-cell expression data based on the expression of similar cells, was employed (71). MAGIC smoothing of the cancer cell cluster was performed using the library Matrix and library Rmagic (71) based on a pooled gene signature of CAFs.
Histopathological scoring
Mouse tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned at 5 μm thickness. Sections were processed for hematoxylin and eosin (H&E) staining, Masson’s trichrome staining (MTS) using Gomori’s Trichome Stain Kit (38016SS2, Leica Biosystems), or picrosirius red (Direct Red 80, Sigma-Aldrich) according to manufacturer’s instructions. Histopathological assessments were conducted in a blinded fashion by scoring H&E-stained sections for relative percentages of the listed histopathological phenotypes. A weighted histology score was then applied to the percentages as follows: for tumors with less than 5% normal tissue two points, else zero points; greater than 30% PanIN or ADM tissue two points, else zero points; cancer area greater than 30% 4 points, else zero points; poor differentiated PDAC area greater than 30% 5 points, else zero points; and necrosis area greater than 5% 6 points, else zero points. The weighted scores were then summed for each animal to be interpreted as a higher value meaning worse histopathology. Images were obtained with a Leica DM 1000 LED microscope using a 20x objective and an MC120 HD Microscope Camera with Las V4.4 Software (Leica). Tumor scores for orthotopic tumors were evaluated based on H&E sections of the entire pancreas and attributed a score on a scale from 1 (minor involvement) to 4 (extensive involvement).
Immunofluorescent labeling and immunohistochemistry
All antibodies, source, and dilutions are listed in Supplementary Table 7. Formalin-fixed, paraffin-embedded (FFPE) sections were processed for immunohistochemical (IHC) staining as previously described (72), after citrate-based antigen retrieval (pH=6). Staining for αSMA was performed with M.O.M. kit (Vector Laboratories) following the manufacturer’s instructions. For all stainings, counterstaining with hematoxylin was performed and DAB positivity was examined in ten visual fields at 200× magnification. For αSMA IHC on mouse tumor sections, immunoreactive score (IRS) were obtained from the sum of distribution and intensity scores for each section, established on a scale of 1 to 4 (73). FAP immunohistochemistry was performed and imaged identically as the FAP stain in the multiplex fibroblast panel (see below). Pseudocolored images with FAP-520 channel being colored brown and the DAPI channel colored blue on a white background were used for the scoring. The stromal region was scored on a scale of 0–3 for the density of FAP+ fibroblasts in each image. The scores were averaged for each mouse and presented.
Patient cohort for PDAC tissue microarray
All human pancreatic tumor sections (n = 136) were fixed on tissue microarray slides that contain three representative 1 mm cores for each patient (two representative cores of tumor and one core of matched benign pancreatic tissue). The tissue microarrays were constructed from FFPE blocks of archived PDAC specimens as previously described (74). This study was approved by the Institutional Review Board of MDACC (IRB LAB05–0854) and in accordance with the U.S. Common Rule. Written informed consent was obtained from all patients. The patients included in this cohort received no neoadjuvant therapy. Cases and clinical information were retrieved from the surgical pathology files of the Department of Pathology, MDACC (Supplementary Table 6). Immunofluorescence images were obtained using the LSM800 confocal laser scanning microscope under 100x magnification and analyzed by ZEN software (Zeiss). The quantification of indicated staining was based on the average reading of the two representative tumor cores for each patient.
Multispectral imaging of multiplex stained tissue sections
The multiplex staining procedures, spectral unmixing and cell segmentation using the Nuance and inForm imaging softwares were previously published (75). Antibody concentrations used for the multiplex staining can be found in Supplementary Table 7. Multiplex stained slides were imaged with the Vectra Multispectral Imaging System, using Vectra software version 3.0.3 (Perkin Elmer). Each tissue section was scanned in its entirety using a 4x objective, and up to 50 cancer regions (at 20x) were selected for multispectral imaging using the Phenochart software (Perkin Elmer). Each multiplex field was scanned every 20 nm of the emission light spectrum across the range of each emission filter cube. Filter cubes used for multispectral imaging were DAPI (440–600 nm), FITC (520 nm-680 nm), Cy3 (570–690 nm), Texas Red (580–700 nm) and Cy5 (680–720 nm). Multispectral images from single marker stained slides with the corresponding fluorophores were used to generate a spectral library using the Nuance Image Analysis software (Perkin Elmer). The library contained the emitting spectral peaks of all fluorophores and was used to unmix each multispectral image (spectral unmixing) to its individual 8 components by using the inForm 2.4 image analysis software. All spectrally unmixed image cubes were subsequently segmented into individual cells based on the nuclear DAPI counterstain. For the immune cell population analysis, all spectrally unmixed and segmented images were analyzed using the inForm phenotyping algorithm. This allows for the individual identification of each DAPI-stained cell according to their pattern of fluorophore expression and nuclear/cellular morphological features. Cells were phenotyped into eight different classes according to the markers they expressed: CD3+ T cells (CD3+), Teff cells (CD3+CD4+Foxp3−), cytotoxic T cells (CD3+CD8+), Treg cells (CD3+CD4+Foxp3+), myeloid cells (CD11b+), cancer cells (CK8+), and other cells (negative for all markers). For fibroblast cell population analysis, FAP staining was performed after antigen retrieval with TE buffer (pH=9), and αSMA and CK8 staining performed after antigen retrieval with citrate buffer (pH=6.0). Slides were imaged with a Zeiss Axio Scan.Z1 and LSM800 confocal microscope. Percent positive area for a given fibroblast marker and negative for CK8 was quantified in ImageJ.
EGFP/tdTomato visualization and immunofluorescence.
Tissues from the R26Dual lineage tracing mouse strain, expressing Pdx1-Flp-driven intrinsic EGFP and αSMA-Cre-driven tdTomato, were fixed in 4% paraformaldehyde overnight at 4°C and equilibrated in 30% sucrose overnight at 4°C. Tissues were then embedded in O.C.T. compound (TissueTek) and processed for 5-μm-thick frozen sections. Sections were blocked for 1 h with 4% cold water fish gelatin (Aurion) and immunostained overnight at 4°C with anti-αSMA antibody (followed by goat anti-mouse Alexa Fluor 647 secondary antibody). Slides were then mounted with Vectashield Mounting Medium (Vector Laboratories) to a glass coverslip and visualized under the LSM800 confocal laser scanning microscope and ZEN software (Zeiss).
Isolation of primary pancreatic adenocarcinoma cells and myofibroblasts from PDAC tissues
Establishment of primary PDAC cancer cell and myofibroblast lines was conducted as previously described with minor modifications (72,76). Fresh PDAC tissues from KPPF;IL-6smaKO;R26Dual mice and KPPF;αSMA-Cre;R26Dual mice were minced, digested with collagenase IV (17104019, Gibco, 4 mg/mL) and dispase II (17105041, Gibco, 4 mg/mL) in RPMI at 37°C for 1 h, filtered by 70 μm cell strainers, resuspended in RPMI with 20% FBS, and then seeded on type-I collagen-coated dishes (354401, Corning). Cells were cultured in RPMI medium containing 20% FBS and 1% penicillin, streptomycin, and amphotericin B (PSA) antibiotic mixture. Pdx1-lineage traced cancer cells and αSMA-lineage traced myofibroblasts were further purified by FACS (BD FACSAria™ II) based on EGFP and tdTomato signals, respectively. The sorted cells were subsequently maintained in vitro. All studies were performed on cells cultivated less than 25 passages. DNA from these primary cell lines was extracted using the DNA Mini Kit (51304 QIAGEN). Total RNA was extracted from indicated cells using the Direct-zol RNA Kit (Zymo Research), processed for cDNA synthesis using the Reverse Transcription Kit (Applied Biosystems), and subjected to the qRT-PCR using SYBR Green Master Mix (Applied Biosystems). Primer sequences are as follows: Il6 Forward: 5’-GCTTAATTACACATGTTCTCTGGGAAA-3’; Il6 Reverse: 5’-CAAGTGCATCATCGTTGTTCATAC-3’; Il1b Forward: 5’-GGGCTGCTTCCAAACCTTTG-3’; Il1b Reverse: 5’-TGATACTGCCTGCCTGAAGCTC-3’; Gapdh Forward: 5’-AGGTCGGTGTGAACGGATTTG-3’; Gapdh Reverse: 5’-TGTAGACCATGTAGTTGAGGTCA-3’.
Global gene expression profiling
Total RNA was also isolated from tumors of age-matched KTC;αSMA-TK, and KTC;FAP-TK mice (n = 3 mice per in each group), that were administrated with GCV or PBS. RNA extraction was carried out using the QIAGEN RNeasy Mini Kit and submitted to the Microarray Core Facility at MD Anderson Cancer Center. Gene expression analysis was performed using Affymetrix MTA 1.0 Genechip. The Limma package (77) from R Bioconductor was used for quantile normalization of expression arrays and to analyze differential gene expression between the TK groups (KTC;αSMA-TK and the KTC-FAP-TK groups) and their respective control (KTC-αSMA-TKcontrol and KTC;FAP-TKcontrol) groups (p ≤ 0.05 and fold change ≥ 1.2). Analyses of differentially expressed pathways between the TK and control groups were performed using Gene Set Enrichment Analysis (GSEA) (78). For CIBERSORT analysis, R package biomaRt (version 2.50.3) was used to convert mouse genes into human gene symbols and CIBERSORT with built-in LM22 gene signatures used for deconvolution analysis (79).
Statistical analyses
The statistical tests used for the comparative analyses presented are listed in the figure legends. Statistical analyses were carried out using GraphPad Prism (GraphPad Software version 8). Kaplan-Meier plots were used for survival analysis and the log rank Mantel-Cox test was used to evaluate statistical differences with GraphPad Prism. Error bars represent standard error of the mean unless specified in the figure legends. Statistical significance was defined as P < 0.05.
Data availability
Source data for each figure are included. Microarray data from KTC;αSMA-TK GEMMs were previously deposited at Gene Expression Omnibus under accession number GSE52812 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52812) (27). Gene expression microarray data from KTC;FAP-TK GEMMs was deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120577).
Supplementary Material
Statement of Significance.
Pancreatic adenocarcinoma (PDAC) is associated with accumulation of dense stroma consisting of fibroblasts and extracellular matrix that regulate tumor progression. Here, we identify two distinct populations of fibroblasts with opposing roles in the progression and immune landscape of PDAC. Our findings demonstrate that fibroblasts are functionally diverse with therapeutic implications.
Acknowledgements
We wish to thank Erika J. Thompson, David P. Pollock and Yunxin Chen for their assistance with scRNA-seq, Tapsi Kumar and Nicholas Navin for their assistance with analysis of human scRNA-seq data, Judith Kaye for mouse husbandry support, and Ehsan Ehsanipour for assistance with generation and validation of the FAP-Cre mouse. This work was primarily supported by the Cancer Prevention and Research Institute of Texas (CPRIT). Research in the Kalluri laboratory is also supported by NCI P01CA117969 and CPRIT Award RP150231. RK is a Distinguished University Chair supported by the Sid W. Richardson Foundation. JKD was supported by the American Legion Auxiliary Fellowship and Center for Clinical and Translational Sciences TL1 training grant (NIH UL1TR000371). KMM and YC were supported by Ergon Foundation Postdoctoral Fellowships. Other support includes: UT MDACC Flow Cytometry Core (NIH P30CA016672), UT MDACC Advanced Technology Genomics Core (NIH P30CA016672), and UT MDACC Genetically Engineered Mouse Facility (NIH P30CA016672).
Footnotes
Conflict of interest: KMM and YC received speaker honorarium from Stellanova Therapeutics. VSL is a Scientific Advisory Board member and stockholder of Stellanova Therapeutics.
References
- 1.Chen Y, McAndrews KM, Kalluri R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nature Reviews Clinical Oncology 2021;18:792–804 doi 10.1038/s41571-021-00546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LeBleu VS, Kalluri R. A peek into cancer-associated fibroblasts: origins, functions and translational impact. Dis Model Mech 2018;11(4) doi 10.1242/dmm.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016;16(9):582–98 doi 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 2012;21(3):309–22 doi 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 5.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19(11):1423–37 doi 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosein AN, Brekken RA, Maitra A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat Rev Gastroenterol Hepatol 2020;17(8):487–505 doi 10.1038/s41575-020-0300-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20(3):174–86 doi 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pure E. The road to integrative cancer therapies: emergence of a tumor-associated fibroblast protease as a potential therapeutic target in cancer. Expert Opin Ther Targets 2009;13(8):967–73 doi 10.1517/14728220903103841. [DOI] [PubMed] [Google Scholar]
- 9.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, et al. Pancreatic cancer. Nat Rev Dis Primers 2016;2:16022 doi 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 10.Sherman MH. Stellate Cells in Tissue Repair, Inflammation, and Cancer. Annu Rev Cell Dev Biol 2018;34:333–55 doi 10.1146/annurev-cellbio-100617-062855. [DOI] [PubMed] [Google Scholar]
- 11.Sherman MH, Downes M, Evans RM. Nuclear receptors as modulators of the tumor microenvironment. Cancer Prev Res (Phila) 2012;5(1):3–10 doi 10.1158/1940-6207.CAPR-11-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apte MV, Wilson JS. Dangerous liaisons: pancreatic stellate cells and pancreatic cancer cells. J Gastroenterol Hepatol 2012;27 Suppl 2:69–74 doi 10.1111/j.1440-1746.2011.07000.x. [DOI] [PubMed] [Google Scholar]
- 13.Rockey DC, Weymouth N, Shi Z. Smooth muscle alpha actin (Acta2) and myofibroblast function during hepatic wound healing. PLoS One 2013;8(10):e77166 doi 10.1371/journal.pone.0077166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neesse A, Algul H, Tuveson DA, Gress TM. Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut 2015;64(9):1476–84 doi 10.1136/gutjnl-2015-309304. [DOI] [PubMed] [Google Scholar]
- 15.Ohlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med 2014;211(8):1503–23 doi 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov 2019;9(8):1102–23 doi 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosein AN, Huang H, Wang Z, Parmar K, Du W, Huang J, et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight 2019;4(16):e129212 doi 10.1172/jci.insight.129212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017;214(3):579–96 doi 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernard V, Semaan A, Huang J, San Lucas FA, Mulu FC, Stephens BM, et al. Single-Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. Clinical Cancer Research 2019;25(7):2194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. IL1-Induced JAK/STAT Signaling Is Antagonized by TGFbeta to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancer Discovery 2019;9(2):282–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Kim J, Yang S, Wang H, Wu CJ, Sugimoto H, et al. Type I collagen deletion in alphaSMA(+) myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer. Cancer Cell 2021;39(4):548–65 e6 doi 10.1016/j.ccell.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A 2013;110(50):20212–7 doi 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Lin Y, Shi Y, Li B, Liu W, Yin W, et al. FAP Promotes Immunosuppression by Cancer-Associated Fibroblasts in the Tumor Microenvironment via STAT3-CCL2 Signaling. Cancer Res 2016;76(14):4124–35 doi 10.1158/0008-5472.CAN-15-2973. [DOI] [PubMed] [Google Scholar]
- 24.Lo A, Li CP, Buza EL, Blomberg R, Govindaraju P, Avery D, et al. Fibroblast activation protein augments progression and metastasis of pancreatic ductal adenocarcinoma. JCI Insight 2017;2(19) doi 10.1172/jci.insight.92232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 2010;330(6005):827–30 doi 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 26.Santos AM, Jung J, Aziz N, Kissil JL, Pure E. Targeting fibroblast activation protein inhibits tumor stromagenesis and growth in mice. J Clin Invest 2009;119(12):3613–25 doi 10.1172/JCI38988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell 2014;25(6):719–34 doi 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017;171(7):1611–24 e24 doi 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018;33(3):463–79 e10 doi 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Kieffer Y, Hocine HR, Gentric G, Pelon F, Bernard C, Bourachot B, et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov 2020;10(9):1330–51 doi 10.1158/2159-8290.CD-19-1384. [DOI] [PubMed] [Google Scholar]
- 31.Peng J, Sun B-F, Chen C-Y, Zhou J-Y, Chen Y-S, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Research 2019;29(9):725–38 doi 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts EW, Deonarine A, Jones JO, Denton AE, Feig C, Lyons SK, et al. Depletion of stromal cells expressing fibroblast activation protein-alpha from skeletal muscle and bone marrow results in cachexia and anemia. J Exp Med 2013;210(6):1137–51 doi 10.1084/jem.20122344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran E, Chinnasamy D, Yu Z, Morgan RA, Lee CC, Restifo NP, et al. Immune targeting of fibroblast activation protein triggers recognition of multipotent bone marrow stromal cells and cachexia. J Exp Med 2013;210(6):1125–35 doi 10.1084/jem.20130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesina M, Wormann SM, Neuhofer P, Song L, Algul H. Interleukin-6 in inflammatory and malignant diseases of the pancreas. Seminars in immunology 2014;26(1):80–7 doi 10.1016/j.smim.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Nagathihalli NS, Castellanos JA, VanSaun MN, Dai X, Ambrose M, Guo Q, et al. Pancreatic stellate cell secreted IL-6 stimulates STAT3 dependent invasiveness of pancreatic intraepithelial neoplasia and cancer cells. Oncotarget 2016;7(40):65982–92 doi 10.18632/oncotarget.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wormann SM, Song L, Ai J, Diakopoulos KN, Kurkowski MU, Gorgulu K, et al. Loss of P53 Function Activates JAK2-STAT3 Signaling to Promote Pancreatic Tumor Growth, Stroma Modification, and Gemcitabine Resistance in Mice and Is Associated With Patient Survival. Gastroenterology 2016;151(1):180–93 e12 doi 10.1053/j.gastro.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 37.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 2011;19(4):456–69 doi 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Yan W, Collins MA, Bednar F, Rakshit S, Zetter BR, et al. Interleukin-6 is required for pancreatic cancer progression by promoting MAPK signaling activation and oxidative stress resistance. Cancer Res 2013;73(20):6359–74 doi 10.1158/0008-5472.CAN-13-1558-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, LeBleu VS, Carstens JL, Sugimoto H, Zheng X, Malasi S, et al. Dual reporter genetic mouse models of pancreatic cancer identify an epithelial-to-mesenchymal transition-independent metastasis program. EMBO Mol Med 2018;10:e9085 doi 10.15252/emmm.201809085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugimoto H, Mundel TM, Kieran MW, Kalluri R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol Ther 2006;5(12):1640–6 doi 10.4161/cbt.5.12.3354. [DOI] [PubMed] [Google Scholar]
- 41.Neuzillet C, Tijeras-Raballand A, Ragulan C, Cros J, Patil Y, Martinet M, et al. Inter- and intra-tumoural heterogeneity in cancer-associated fibroblasts of human pancreatic ductal adenocarcinoma. J Pathol 2019;248(1):51–65 doi 10.1002/path.5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raz Y, Cohen N, Shani O, Bell RE, Novitskiy SV, Abramovitz L, et al. Bone marrow-derived fibroblasts are a functionally distinct stromal cell population in breast cancer. J Exp Med 2018;215(12):3075–93 doi 10.1084/jem.20180818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell 2011;19(2):257–72 doi 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizutani Y, Kobayashi H, Iida T, Asai N, Masamune A, Hara A, et al. Meflin-Positive Cancer-Associated Fibroblasts Inhibit Pancreatic Carcinogenesis. Cancer Res 2019;79(20):5367–81 doi 10.1158/0008-5472.CAN-19-0454. [DOI] [PubMed] [Google Scholar]
- 45.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med 2013;19(11):1410–22 doi 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 46.Ramanathan RK, McDonough SL, Philip PA, Hingorani SR, Lacy J, Kortmansky JS, et al. Phase IB/II Randomized Study of FOLFIRINOX Plus Pegylated Recombinant Human Hyaluronidase Versus FOLFIRINOX Alone in Patients With Metastatic Pancreatic Adenocarcinoma: SWOG S1313. J Clin Oncol 2019;37(13):1062–9 doi 10.1200/JCO.18.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 2014;25(6):735–47 doi 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steele CW, Karim SA, Leach JDG, Bailey P, Upstill-Goddard R, Rishi L, et al. CXCR2 Inhibition Profoundly Suppresses Metastases and Augments Immunotherapy in Pancreatic Ductal Adenocarcinoma. Cancer Cell 2016;29(6):832–45 doi 10.1016/j.ccell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Y, Herndon JM, Sojka DK, Kim KW, Knolhoff BL, Zuo C, et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity 2017;47(2):323–38 e6 doi 10.1016/j.immuni.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Candido JB, Morton JP, Bailey P, Campbell AD, Karim SA, Jamieson T, et al. CSF1R(+) Macrophages Sustain Pancreatic Tumor Growth through T Cell Suppression and Maintenance of Key Gene Programs that Define the Squamous Subtype. Cell Rep 2018;23(5):1448–60 doi 10.1016/j.celrep.2018.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lo A, Wang LS, Scholler J, Monslow J, Avery D, Newick K, et al. Tumor-Promoting Desmoplasia Is Disrupted by Depleting FAP-Expressing Stromal Cells. Cancer Res 2015;75(14):2800–10 doi 10.1158/0008-5472.CAN-14-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. IL1-Induced JAK/STAT Signaling Is Antagonized by TGFbeta to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancer Discov 2019;9(2):282–301 doi 10.1158/2159-8290.CD-18-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 2014;159(1):80–93 doi 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ligorio M, Sil S, Malagon-Lopez J, Nieman LT, Misale S, Di Pilato M, et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. Cell 2019;178(1):160–75 e27 doi 10.1016/j.cell.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long KB, Tooker G, Tooker E, Luque SL, Lee JW, Pan X, et al. IL6 Receptor Blockade Enhances Chemotherapy Efficacy in Pancreatic Ductal Adenocarcinoma. Mol Cancer Ther 2017;16(9):1898–908 doi 10.1158/1535-7163.MCT-16-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7(5):469–83 doi 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 57.LeBleu VS, Taduri G, O’Connell J, Teng Y, Cooke VG, Woda C, et al. Origin and function of myofibroblasts in kidney fibrosis. Nat Med 2013;19(8):1047–53 doi 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.LeBleu VS, Teng Y, O’Connell JT, Charytan D, Muller GA, Muller CA, et al. Identification of human epididymis protein-4 as a fibroblast-derived mediator of fibrosis. Nat Med 2013;19(2):227–31 doi 10.1038/nm.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McAndrews KM, Miyake T, Ehsanipour EA, Kelly PJ, Becker LM, McGrail DJ, et al. Dermal alphaSMA(+) myofibroblasts orchestrate skin wound repair via beta1 integrin and independent of type I collagen production. EMBO J 2022:e109470 doi 10.15252/embj.2021109470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Developmental Biology 2001;1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 2010;143(1):134–44 doi 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 62.Schonhuber N, Seidler B, Schuck K, Veltkamp C, Schachtler C, Zukowska M, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med 2014;20(11):1340–7 doi 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee CL, Moding EJ, Huang X, Li Y, Woodlief LZ, Rodrigues RC, et al. Generation of primary tumors with Flp recombinase in FRT-flanked p53 mice. Dis Model Mech 2012;5(3):397–402 doi 10.1242/dmm.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 1995;23(24):5080–1 doi 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quintana A, Erta M, Ferrer B, Comes G, Giralt M, Hidalgo J. Astrocyte-specific deficiency of interleukin-6 and its receptor reveal specific roles in survival, body weight and behavior. Brain, behavior, and immunity 2013;27(1):162–73 doi 10.1016/j.bbi.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 66.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005;436(7051):725–30 doi 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev 2006;20(22):3147–60 doi 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017;546(7659):498–503 doi 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015;33(5):495–502 doi 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci 2017;20(3):484–96 doi 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 2018;174(3):716–29 e27 doi 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015;527(7579):525–30 doi 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meyerholz DK, Beck AP. Principles and approaches for reproducible scoring of tissue stains in research. Lab Invest 2018;98(7):844–55 doi 10.1038/s41374-018-0057-0. [DOI] [PubMed] [Google Scholar]
- 74.Wang H, Wang H, Zhang W, Fuller GN. Tissue microarrays: applications in neuropathology research, diagnosis, and education. Brain Pathol 2002;12(1):95–107 doi 10.1111/j.1750-3639.2002.tb00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun 2017;8:15095 doi 10.1038/ncomms15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Y, LeBleu VS, Carstens JL, Sugimoto H, Zheng X, Malasi S, et al. Dual reporter genetic mouse models of pancreatic cancer identify an epithelial-to-mesenchymal transition-independent metastasis program. EMBO Mol Med 2018;10(10) doi 10.15252/emmm.201809085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smyth GK. limma: Linear Models for Microarray Data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S. (eds) Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Statistics for Biology and Health Springer, New York, NY: 2005. [Google Scholar]
- 78.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12(5):453–7 doi 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for each figure are included. Microarray data from KTC;αSMA-TK GEMMs were previously deposited at Gene Expression Omnibus under accession number GSE52812 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52812) (27). Gene expression microarray data from KTC;FAP-TK GEMMs was deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120577).