Abstract
Malaria and other apicomplexan-caused diseases affect millions of humans, agricultural animals, and pets. Cell traversal is a common feature used by multiple apicomplexan parasites to migrate through host cells and can be exploited to develop therapeutics against these deadly parasites. Here, we provide insights into the mechanism of the Cell-traversal protein for ookinetes and sporozoites (CelTOS), a conserved cell-traversal protein in apicomplexan parasites and malaria vaccine candidate. CelTOS has previously been shown to form pores in cell membranes to enable traversal of parasites through cells. We establish roles for the distinct protein regions of Plasmodium vivax CelTOS and examine the mechanism of pore formation. We further demonstrate that CelTOS dimer dissociation is required for pore formation, as disulfide bridging between monomers inhibits pore formation, and this inhibition is rescued by disulfide-bridge reduction. We also show that a helix-destabilizing amino acid, Pro127, allows CelTOS to undergo significant conformational changes to assemble into pores. The flexible C terminus of CelTOS is a negative regulator that limits pore formation. Finally, we highlight that lipid binding is a prerequisite for pore assembly as mutation of a phospholipids-binding site in CelTOS resulted in loss of lipid binding and abrogated pore formation. These findings identify critical regions in CelTOS and will aid in understanding the egress mechanism of malaria and other apicomplexan parasites as well as have implications for studying the function of other essential pore-forming proteins.
Keywords: CelTOS, malaria, Plasmodium, liposome, phosphatidic acid, pore-forming protein, pore-formation assay, cell migration, protein crosslinking, mass spectrometry
Abbreviations: AMBER, assisted model building with energy refinement; aMD, accelerated molecular dynamics; CelTOS, Cell-traversal protein for ookinetes and sporozoites; cMD, conventional molecular dynamics; FA, fatty acid; MS, mass spectrometry; PA, phosphatidic acid; PDB, Protein Data Bank; PFP, pore-forming protein; POPA, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate; RMSF, root mean square fluctuation; RT, room temperature; SEC, size-exclusion chromatography
Apicomplexan parasites infect a wide range of hosts and affect millions of humans and agricultural animals (1). Apicomplexan parasites include Plasmodia spp., the most severe parasite in the category that causes malaria (2, 3); Toxoplasma, an opportunistic parasite that can cause severe toxoplasmosis in immunocompromised individuals; Babesia, the causative agent of babesiosis (4); Theileria (5), a tick-born parasite that causes theileriosis in cattle; and Cytauxzoon felis, which causes cytauxzoonosis, an emerging tick-born disease in cats (6). Among these apicomplexa-caused diseases, malaria results in the most significant mortality and morbidity, causing over 600,000 deaths and 200 million cases worldwide annually (7).
Apicomplexan parasites exhibit diverse biology and infect diverse hosts but share common features such as the presence of a definitive cell structure (the apical complex), a nonphotosynthetic plastid (the apicoplast), and actin-based motility. Cell traversal is another important common feature shared by most but not all apicomplexan parasites. Parasites traverse several cell types to complete essential segments of their life cycle inside the host and the vector. Cell-traversal protein for ookinetes and sporozoites (CelTOS), Sporozoite Protein Essential for Cell Traversal 1 (SPECT1), and SPECT2 have established roles in cell traversal (8). Among these, CelTOS is relatively unique because it is required for cell traversal in both the human host and the arthropod vector. In addition, CelTOS is highly conserved among arthropod-borne apicomplexan parasites (9).
Disruption of the CelTOS gene in Plasmodium berghei causes a 200-fold reduction in infectivity during mosquito stage development, and the resultant salivary gland sporozoites show defects in their ability to migrate through different cell types (10). Furthermore, CelTOS is a promising malaria antigen (11), and immunization with recombinant CelTOS shows humoral and cellular responses against the parasite (12, 13, 14). Finally, Amazonian populations elicit natural immune responses against Plasmodium vivax CelTOS (15). These findings clearly highlight the importance of CelTOS in malaria parasites and in the human host response and confirm the potential of CelTOS as a malaria vaccine candidate and malaria therapeutic target.
The structure of P. vivax CelTOS revealed that the helical organization in CelTOS resembled other membrane-disrupting proteins (9). Soluble CelTOS exists as a dimer in which the two monomers protect the hydrophobic core and expose their hydrophilic residues on the surface (9). CelTOS specifically binds to phosphatidic acid (PA) and forms pores in the inner leaflet of the host membrane (9). However, how the CelTOS dimer rearranges and incorporates into the cell membrane is not clearly understood. This lack of information on the functional regions of CelTOS hinders structure-based design of effective CelTOS immunogens.
In this study, we report how the CelTOS dimer in solution transitions into a CelTOS pore in membranes and identify primary, secondary, tertiary, and quaternary changes in CelTOS structure that regulate its fundamental function in pore formation. We tested the hypothesis that the soluble CelTOS dimer is in an inactive state and undergoes mandatory conformational rearrangements to penetrate cell membranes and form pores. We generated distinct CelTOS mutants and studied their dynamics and functional characteristics using 500 ns accelerated molecular dynamics (aMD) simulations and pore-forming assays. We developed a dimer-locked CelTOS using structure-based engineering and introduced cysteines at specific locations in CelTOS that prevent dimer dissociation. The disulfide-stabilized CelTOS dimer was inactive in the pore-forming assay. We identified a kink in helix 4 of CelTOS caused by Pro127, a residue conserved in other apicomplexans. Mutation of Pro127 to alanine stabilizes CelTOS and reduces its pore-forming activity, suggesting Pro127 may provide inherent instability to the CelTOS dimer. We further truncated the flexible N-terminal and C-terminal segments individually and observed enhanced pore-forming activity upon deleting the C-terminal segment. On the contrary, deletion of the N-terminal region has no significant difference from the WT protein. Finally, we showed that Lys122 and Leu164 that comprise two conserved lipid-binding residues that likely interact with a phospholipid were essential for pore-forming activity. These data suggest that the soluble CelTOS dimer undergoes a functionally essential conformation change upon contact with the lipid membrane. These insights will inform understanding the egress mechanism of Plasmodium and apicomplexan parasites.
Results
Engineered Cys residues stabilize CelTOS in an inactive dimer state
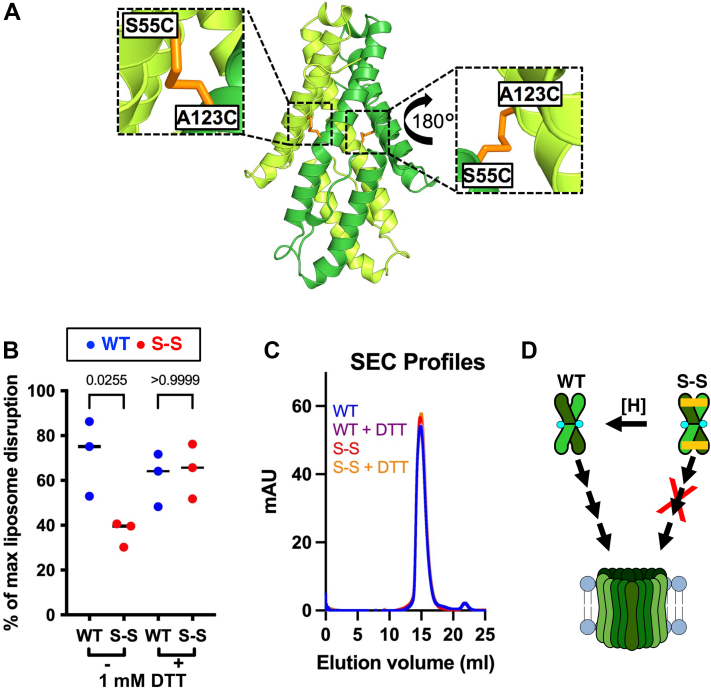
We proposed that CelTOS is inactive in the dimer state observed in the crystal structure (Protein Data Bank [PDB] ID: 5TSZ) and undergoes significant conformational changes to form pores and incorporate into the inner leaflet of host cell membranes (9). To test this hypothesis, we incorporated disulfide bonds in CelTOS that lock the dimer conformation and prevent dimer dissociation. The CelTOS structure (PDB ID: 5TSZ) was analyzed for potential disulfide bridging residues, and Ser55 and Ala123 were identified; when mutated to cysteine (Ser55Cys and Ala123Cys), these residues formed a stable disulfide bridge (Fig. 1A) (16).
Figure 1.
Disulfide-locked CelTOS dimer is inactive in pore formation.A, CelTOS dimer (PDB ID: 5TSZ) is shown in the cartoon highlighting the location of Ala123 and Ser55. Note that the shown modeled structure highlights residues that were computationally mutated to cysteines to lock CelTOS in the dimer state. B, pore-forming assay: disulfide-locked CelTOS dimer (S–S) is significantly less active than the WT protein. Note that this loss of activity was rescued in the presence of 1 mM DTT that reduces the disulfide bridge, enables dimer dissociation, and facilitates pore formation. The graph represents the median value and three independent biological replicates each with three to four technical replicates (please note that the graph shows the combined data from three individual biological replicates demonstrating the reproducibility of the data. Please see Fig. S3 for these three individual biological replicates). Significance was determined using Kruskal–Wallis analysis and Dunn’s multiple comparison. C, size-exclusion chromatography (SEC) profiles of WT and S–S in the presence or the absence of 1 mM DTT. D, a model depicting the plausible mechanism of conversion of soluble CelTOS dimer to the multimeric membrane pore (left); S–S inactivation (right) and its rescue by disulfide reduction (shown as [H]). CelTOS, Cell-traversal protein for ookinetes and sporozoites; PDB, Protein Data Bank.
The disulfide mutant CelTOS (referred to as S–S) was readily expressed using a similar procedure as for WT CelTOS, and the size-exclusion chromatography (SEC) profiles of S–S and WT were indistinguishable (Fig. S1). A nonreduced SDS-PAGE analysis that retains disulfide bridging in the analysis but denatures the protein structure was conducted to evaluate the disulfide bridging of the various proteins (Fig. S1). This analysis revealed a band of ∼37 kDa comprising a disulfide-locked dimer for the S–S mutant and a band of ∼18 kDa corresponding to the uncrosslinked protein for WT. Together, the size-exclusion profile and the SDS-PAGE analysis suggest that the incorporated disulfide linkages create a cross-linked dimer but do not affect the native oligomeric state or structure of CelTOS dimer.
One concern of the introduction of the disulfide bridges is that CelTOS may form the expected disulfide bridges (C55–C123) of the parallel dimer but could also form the alternate disulfide bridges (C55–C55 or C123–C123) of an antiparallel dimer. We investigated which disulfide bridging patterns were observed using mass spectrometry (MS)–based bottom–up proteomic analysis. In nonreduced conditions, only the interchain disulfide linkage between Cys55 and Cys123 (SGSTASSSLEGGSEF55CER-123CALEPTEK) was detected with a precursor abundance of 70 ± 7% cross-linked species and 30 ± 7% noncrosslinked species (Figs. 1A and S2). A large fraction of noncrosslinked species (30 ± 7%) suggests the heterogeneity in disulfide-locked mutant because of doubly cross-linked and singly cross-linked CelTOS dimer, and/or all-reduced CelTOS dimer (WT). Singly cross-linked CelTOS dimer arise by (a) Cys55 (chain A) crosslinked with Cys123 (chain B); or (b) Cys55 (chain B) crosslinked with Cys123 (chain A). In three independent biological replicate experiments with independent purifications of the proteins, no disulfide linkage between Cys55–Cys55 or Cys123–Cys123 of the potential antiparallel dimer was detected, thus ruling out the possibility of alternate dimer organization in the S–S mutant (Fig. S2). Together, with the SEC profile, the MS analysis demonstrates the presence of the WT-like parallel dimer state in the S–S mutant.
We examined the pore-forming activity of the S–S mutant in nonreducing conditions and observed significantly less activity than the WT protein (Figs. 1B and S3). Interestingly, this loss of pore-forming activity of the S–S mutant could be rescued in reducing conditions (1 mM DTT) that eliminate the disulfide linkage (Figs. 1B and S3). We also examined if added DTT has any detrimental effect on CelTOS dimerization. The elution profile cannot distinguish between WT- and disulfide-locked CelTOS variant in the presence or the absence of 1 mM DTT suggesting a similar dimeric conformation of WT and S–S variants in all conditions tested (Fig. 1C). This in turn indicates that the dimer is stable in solution but must dissociate upon interaction with lipids. Together, these results indicate that the addition of the cysteine residues has no effect on CelTOS dimeric state and function unless the cysteines are crosslinked to create a stabilized dimer.
Despite engineered disulfide linkages in the S–S mutant, the purified protein was a mixture of locked dimer (S–S) and noncrosslinked dimer (WT) (Fig. S1), and therefore, the S–S mutant showed residual membrane disruption activity. Indeed, MS analysis of the nonreduced S–S dimer when the protein sample was alkylated with iodoacetamide to modify free sulfides revealed over 25 carbamidomethylated peptide spectral matches as free sulfides suggesting ∼30% of all S–S dimer variants have a single disulfide bond or remained entirely unlocked (Fig. S2). These findings clearly indicate heterogeneity in the disulfide-locked CelTOS mutant. Taken together, these results suggest that, for parasite cell traversal, the native CelTOS dimer must undergo dimer dissociation prior to significant conformational rearrangement to form pores in the lipid bilayer (Fig. 1D).
P127 causes a kink in helix 4 of CelTOS and is important for pore-forming activity
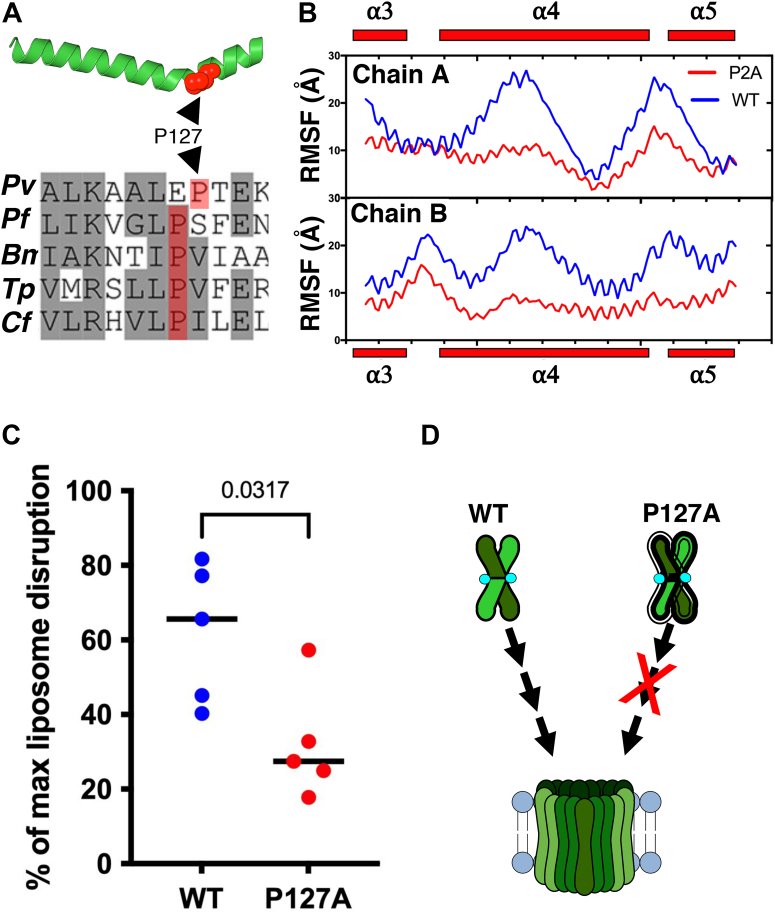
CelTOS is always observed as a dimer in solution (9). However, we demonstrated that covalently stabilizing the dimer state by disulfide bridging inactivates CelTOS, and activity can be restored only by eliminating this artificial disulfide bridge. We hypothesized that CelTOS contains inherently unstable region(s) that enable conformational rearrangements required for pore assembly and incorporation into the cell membrane. We found that the two monomers of the CelTOS dimer are not identical and deviate structurally from helix 4 to the C terminus. The conformation of helix 4 differs between the two CelTOS monomers when the N termini of the CelTOS monomers are aligned (Fig. S4). A deviation of 5.7 Å is observed between the C⍺ atoms of Thr135 of the two monomers (Fig. S4). The structural mobility is caused by Pro127 in helix 4 of CelTOS, which causes a bend in the helix. Pro127 is also conserved among different CelTOS orthologs in other Plasmodium species (Figs. 2A and S5) suggesting a critical role in its physiological function. Therefore, we suspected that Pro127 may provide internal flexibility to CelTOS structure.
Figure 2.
Pro127 is required for pore-forming activity of CelTOS.A, CelTOS helix 4 (PDB ID: 5TSZ) is shown. Pro127 (shown in red sphere) causes a bend in the helix 4 (top panel). Multiple sequence alignment (MSA) of CelTOS orthologs in apicomplexan parasites showing conserved Pro127 (highlighted in red). Residues in gray background represent conserved residues, and residues in white background represent nonconserved residues. Bm, Babesia microti; Cf, Cytauxzoon felis; Pf, Plasmodium falciparum; Pv, Plasmodium vivax; Tp, Theileria parva (bottom panel). B, root mean square fluctuation (RMSF) plot of two monomers indicates that the Pro127Ala (P127A) mutant is less flexible in different regions including helix 4 (⍺4). C, pore-formation assay of CelTOS-WT and Pro127Ala (P127A) mutant. The graph represents the median value and five independent biological replicates each with eight technical replicates (see Fig. S6 for five independent biological replicates demonstrating the reproducibility of the data). Significance was determined using a Mann–Whitney U test. D, a model showing the enhanced rigidity in P127A mutant causes loss of pore-forming activity. CelTOS, Cell-traversal protein for ookinetes and sporozoites; PDB, Protein Data Bank.
We mutated Pro127 to Ala (P127A mutant), which was readily expressed with a SEC profile similar to the WT protein, suggesting P127A exhibits a similar architecture to WT (Fig. S1). In 500 ns aMD simulations, the P127A mutant was less flexible than the WT protein as evident from the lower root mean square fluctuation (RMSF) values (Fig. 2B). The P127A mutant showed a significant reduction in pore-forming activity compared with the WT protein, consistent with flexibility as a requirement for activity (Figs. 2C and S6). The aMD simulation and pore-forming assay results imply that Pro127 provides flexibility to the CelTOS structure, which is functionally essential to provide conformational changes for CelTOS to incorporate into the inner leaflet of the host cell membrane (Fig. 2D).
Truncation of the flexible C-terminal residues increases CelTOS activity
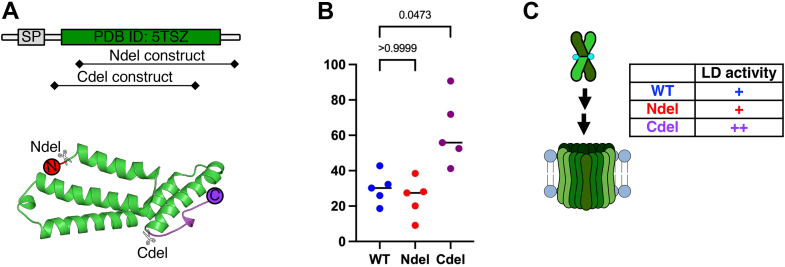
The C-terminal disordered region lacks a transmembrane domain or sequence similarity to any known membrane-interacting domains (Fig. 3A). When the two CelTOS monomers were aligned with respect to the N-terminal region, we observed a shift of 11.1 Å between Cα atoms of Glu143, suggesting that the C-terminal region is the most flexible region in the CelTOS structure (Fig. S7). The structural resemblance of C-terminal helices to membrane-disrupting proteins (9) and its flexibility in the CelTOS structure suggest that the C terminus may play a role in membrane penetration. We designed a C-terminal truncation mutant (C-del) comprising Leu36–Leu164 that omits residues C terminal to helix 5 (His165–onward) (Figs. 3A and S5). The purified C-del mutant eluted later than WT protein during SEC (Fig. S1), suggesting either an alternate conformation or a dissociation of the dimer into monomers. Strikingly, C-del showed a ∼50% increase in pore-forming activity compared with the WT protein (Figs. 3B and S8). Taken together, these results establish that the flexible C-terminal region is a regulator that constrains the major conformation rearrangements in CelTOS required for the pore-forming activity (Fig. 3C).
Figure 3.
Truncation of flexible ends affect pore-forming activity.A, a schematic of Plasmodium vivax CelTOS protein sequence. Predicted domains are shown. SP, signal peptide (top). Chain A of CelTOS dimer structure (PDB ID: 5TSZ) is extracted to show the regions of truncations highlighting flexible N (red) and C (purple) regions: N-del = Gly51–Asp196; C-del = Leu36–Leu164 (bottom). B, pore-forming activity of N-del and C-del mutants and their comparison to the WT protein. The graph represents the median value and five independent biological replicates each with eight technical replicates (see Fig. S8 for five independent biological replicates demonstrating the reproducibility of the data). Significance was determined using Kruskal–Wallis analysis and Dunn’s multiple comparisons test. C, a model comparing the pore-forming activity of the two mutants to the WT protein. While the N-del mutant (+) is similar to WT (+), the C-del mutant (++) shows enhanced pore-forming activity. CelTOS, Cell-traversal protein for ookinetes and sporozoites; PDB, Protein Data Bank.
To study the role of N-terminal residues, we designed an N-terminal truncation mutant that comprises residues Gly51–Asp196 (N-del) (Figs. 3A and S5). N-del was readily expressed in Escherichia coli and eluted as a dimer as confirmed by SEC profile (Fig. S1). Among five different biological replicates, a modest but nonsignificant reduction in activity of the N-del mutant compared with WT was observed. Collectively, the pore-forming activity of N-del was statistically nonsignificant from the WT protein (Figs. 3, B and C and S8).
The two lipid-interacting residues Lys122 and Leu164 are required for pore formation
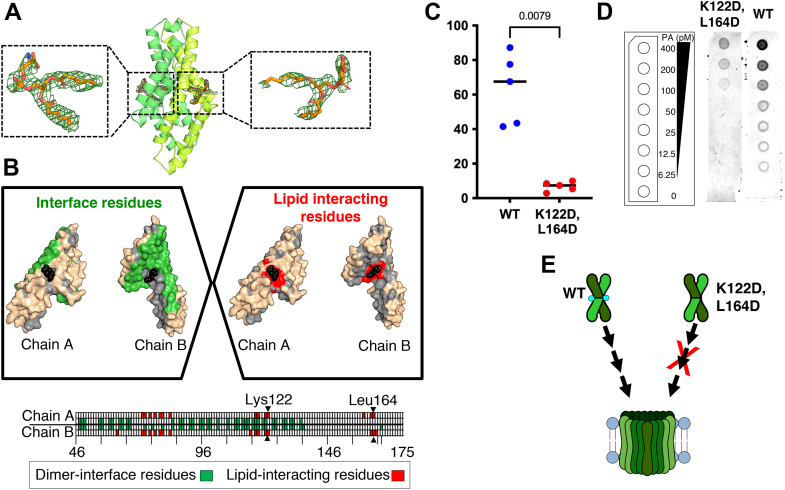
CelTOS specifically binds to PA, and the CelTOS structure (PDB ID: 5TSZ) contains a buried surface area of 3003 Å2 that protects a hydrophobic core (9). We observed two regions of unmodeled electron density in this hydrophobic core that could accommodate bidentate lipid molecules (Fig. 4A). We hypothesized that these two unmodeled densities may correspond to two lipid molecules, and therefore, the CelTOS dimer may contain two lipid-binding sites.
Figure 4.
Lys122 and Leu164 are important for pore-forming activity of CelTOS.A, cartoon view of CelTOS structure (PDB ID: 5TSZ). The two monomers are shown in green and lime colors. Two POPA molecules (blue sticks) modeled in the electron densities are shown in the enlarged views. 2fo–fc map is contoured at 1.0 sigma. B, top, the two monomer structures of CelTOS showing the interface residues in green (left). CelTOS monomers showing the plausible lipid-interacting residues in red (right). Bottom, an illustration of the CelTOS dimer showing the residues that are involved at the monomer–monomer interactions (green in color). Residues within 5 Å radius of bound lipid molecules are highlighted in red. Lys122 and Leu164 are shown. C, pore-formation assay of Lys122Asp,Leu164Asp(K122D,L164D) mutant and its comparison to the WT protein. The graph represents the median value and five independent biological replicates each with eight technical replicates (see Fig. S9 for five independent biological replicates demonstrating the reproducibility of the data). Significance was determined using a Mann–Whitney U test. D, evaluation of binding to phosphatidic acid of WT and K122D,L164D mutant. An equimolar protein concentration of 1 μM was used for WT and K122D,L164D mutant. Left, a schematic layout of the phosphatidic acid (PA) lipid strips containing a concentration gradient of PA from 400 to 0 pmol. Middle, the double mutant shows poor binding to PA. Right, the WT protein shows binding to PA. One representative biological replicate of three is shown. Two additional biological replicates are shown in Fig. S10. E, a model showing the loss of pore-forming activity of the double mutant (K122D,L164D). CelTOS, Cell-traversal protein for ookinetes and sporozoites; PDB, Protein Data Bank; POPA, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate.
Two 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA) molecules were readily modeled into the electron density (Fig. 4, A and B). Residues within a radius of 5 Å around the bound lipid molecules were then examined in detail. Among these, Glu73, Asn77, and Lys122 showed favorable polar interactions with the hydrophilic head group of POPA. Leu72, Ile75, Leu79, Ala80, Ile83, Leu117, Val118, and Leu121 showed nonpolar interactions with the hydrophobic tails. These residues are conserved in CelTOS orthologs in different Plasmodium species (Fig. S5), suggesting CelTOS orthologs likely contain similar lipid-binding regions.
Most of these lipid-interacting residues are buried in the CelTOS dimer interface, and we suspected that attempts to manipulate these residues may affect the integrity and stability of the CelTOS dimer (Fig. 4B). Therefore, we limited our approach to mutating lipid-binding residues that were located outside the CelTOS dimer interface. We identified Lys122, which makes hydrophilic interactions with the polar head of POPA and Leu164, which makes hydrophobic interactions with the nonpolar tail of POPA.
The double mutant, Lys122Asp, Leu164Asp (referred to as K122D,L164D), was readily expressed in E. coli. The SEC profile of this mutant was similar to WT, suggesting the double mutations did not affect the dimeric state or the overall shape of the protein (Fig. S1). Next, we tested the pore-forming activity of K122D,L164D and found that this double mutant was significantly less active than the WT protein (Figs. 4C and S10). We have previously established a lipid-binding assay that revealed CelTOS can bind PA (9, 17). We leveraged this lipid-binding assay to evaluate if the mutations in the lipid-binding region affect binding to PA. The K122D,L164D mutations showed a significant loss of lipid binding in comparison to the WT CelTOS (Figs. 4D and S10). These results suggest that Lys122 and Leu164 are functionally conserved lipid-binding residues essential for pore-forming activity of CelTOS (Fig. 4E).
Discussion
Malaria causes the highest burden of mortality and morbidity of all apicomplexan-caused diseases and affects millions of humans annually. Plasmodium spp., the causative agent of malaria, undergoes a complex life cycle within the mosquito vector and host. Plasmodium parasites must traverse several cell types within the vector and host before they find a suitable cell type to commit to development. Attesting to the crucial role of this cellular traversal, at least two classes of pore-forming proteins (PFPs) are vital for Plasmodium growth and propagation (18). Parasites lacking functional CelTOS expression are defective in mosquito and liver stage development (10), suggesting CelTOS is essential and a promising therapeutic or vaccine target that can both block transmission to mosquitoes and prevent infections in humans.
PFPs exist in a vast diversity of organisms but consistently require interaction with a target cell membrane, often in a receptor-specific manner, and undergo significant conformational rearrangement to penetrate into the membrane (19). This conformational rearrangement is required for the PFP to transform from a soluble to membrane-embedded protein (20). In at least one case, the PFP is maintained in an inactive state by a bound chaperone, and membrane activation occurs upon chaperone release (21). We have previously established that CelTOS specifically binds to PA and disrupts membranes through pore formation (9). This suggests CelTOS needs neither a membrane-bound receptor for its activity nor a chaperone to maintain its inactive state. Therefore, we hypothesized that the dimer state of CelTOS is an inactive state that must undergo significant conformational rearrangements to interact with and form pores in the lipid membrane. To test this hypothesis, we structurally designed a disulfide-stabilized dimer of CelTOS through incorporation of site-targeted cysteine substitutions. Valle et al. (22) used a similar disulfide bridge approach to lock sticholysin I in the homodimer state. The disulfide-locked sticholysin I homodimer could not form pores and was 193 times less hemolytic than the WT protein (22). Similarly, Nguyen et al. (23) created double-cysteine LukF (Leukocidin fast fraction) mutants and showed that the disulfide-trapped mutants were inactive because of the transition arrest from prepore to pore. Similarly, the locked CelTOS dimer was inactive in pore formation. This loss of pore-forming activity was completely restored when the disulfide bond was reduced. These results clearly indicate that the soluble CelTOS dimer is indeed in an inactive conformation and establishes dimerization as a general method to regulate diverse PFPs.
Two monomers in the CelTOS dimer structure (PDB ID: 5TSZ) protect a hydrophobic core from the aqueous environment. We observed density in CelTOS structure that can represent two PA-containing lipid molecules in this position. The modeled PA molecules made polar and hydrophobic contacts with surface and core-region residues, respectively. Such lipid-binding pockets have been reported to have a role in pore formation in other PFPs (24). For example, the monomeric lipid-bound conformation of fragaceatoxin C, a pore-forming toxin protein, contains two conserved lipid-binding pockets, which are primary sites of this molecule’s interaction with the cell membrane (24).
Purified CelTOS is always isolated as a dimer (9), suggesting the dimer is the stable state in the aqueous solution. This raises immediate questions: why and how does this stable dimer undergo significant conformational rearrangement in contact with the lipid bilayer? To answer these questions, we analyzed the available CelTOS structure (PDB ID: 5TSZ) and identified a proline, Pro127, in the middle of helix 4. Prolines are well-known helix breakers (25) and, as expected, Pro127 causes a bend in the CelTOS helix 4. A similar proline-guided helix bend has been reported previously to confer the structural flexibility seen in importin-β (26). When these proline residues are mutated to alanine in importin-β, nuclear transportation is drastically reduced because the mutant’s structural flexibility is lost and cannot undergo rapid functional conformational changes (26). Membrane alignment of alpha helices of toroidal pores demonstrates that proline-induced bends play a critical functional role in stabilizing the pores (27, 28). Similarly, in our current work, the CelTOS Pro127Ala mutant showed loss of pore-forming activity. The aMD simulation of Pro127Ala mutant showed less RMSF values compared with WT, suggesting a loss of structural flexibility in this mutant. The conservation of Pro127 in CelTOS orthologs in other Plasmodium spp., as well as in highly divergent apicomplexan parasites, is strongly indicative that this residue has a functionally conserved mechanism for conferring inherent flexibility. In the current work, we have studied the effect of Pro127Ala on the pore-forming activity of CelTOS. While a clear effect has been observed for the Pro127Ala mutant, it is plausible that mutations of other nearby residues may cause similar loss of pore formation activity in CelTOS. Future studies may expand on additional residues in this region to identify if the phenomenon is exclusive to the proline or generalizable to other residues.
We examined the role of flexible N-terminal and C-terminal regions in CelTOS. These flexible regions flank segments with structural similarity to membrane-disrupting proteins (9). To test our hypothesis if the N-terminal region assists CelTOS to undergo rearrangement, we truncated CelTOS before the start of helix 2 (between Gly51 and Gly52). This mutant was readily expressed in bacteria but showed similar pore-forming activity to the WT protein. These results suggest the noncritical role of N-terminal residues in pore formation.
We further analyzed the potential functional importance of the C-terminal region of CelTOS by creating a C-terminal truncation by deleting His165–Asp196. Unexpectedly, this mutant showed a dramatic increase in pore-forming activity, suggesting the C-terminal region is a negative regulator of CelTOS activity. This finding is similar to our understanding of the functional importance of C-terminal regulatory domains defined in the BCl-2 family of PFPs (29). The deletion of functionally homologous regulatory regions in a membrane fusion protein, Snap receptor (SNARE), promotes fusion of phospholipid endosomal vesicles (30). Therefore, it is plausible that the malarial parasites may use the CelTOS C-terminal regulatory region to control the precise timing of pore formation and parasite egress from the host or vector cell (Fig. 5).
Figure 5.
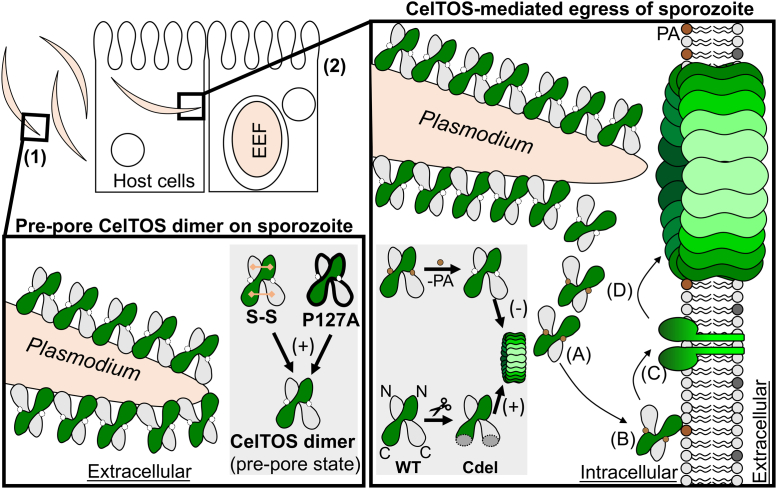
Proposed model for CelTOS-mediated pore formation (1). Prepore CelTOS exists as dimer on parasite surface. The engineered disulfide linkages (S–S) and single proline to alanine mutation (P117A) stabilize the dimer state (shown in blue panel) (2). Inside the host cell, interaction between phosphatidic acid (PA)–bound CelTOS (A) and cell membrane leads to conformational rearrangement in CelTOS and penetration into the membrane (B). C, membrane-inserted dimer dissociates into two monomers and forms prepore in the cell membrane. D, prepore states lead to generation of the mature pore. Small-molecule inhibitors may target CelTOS at all stages, whereas antibodies may be effective at extracellular stages of CelTOS. CelTOS is localized on the Plasmodium surface of the extracellular parasite and forms pores at the inner leaflet of the host cell to disrupt the host cell membrane to enable parasites to exit the host cell (9). Protective CelTOS-specific antibodies may function by binding to the extracellular stages of the prepore CelTOS states facilitating parasite recognition and clearance or by binding to the extracellular face of the mature CelTOS pore facilitating infected-cell recognition and clearance. In contrast, small-molecule inhibitors that target any state of CelTOS for inhibition of activity may also prove effect in preventing malarial infection or transmission. CelTOS, Cell-traversal protein for ookinetes and sporozoites.
In conclusion, our findings clearly demonstrate that CelTOS must undergo conformational rearrangements, including dimer dissociation, to form pores. We have shown that CelTOS contains lipid-binding pockets, and these lipid-binding residues are conserved across apicomplexan parasites. We identified Pro127 as a conserved key residue that provides inherent flexibility to CelTOS, assisting the required conformational rearrangement. We have also shown that the C-terminal tail is a negative regulatory region in CelTOS. These studies may inform the design of future generations of CelTOS-based small-molecule inhibitors and vaccines and have widespread implications for the mechanisms of regulation and activation of PFPs.
Experimental procedures
Bioinformatics
Multiple sequence alignment of CelTOS sequences from diverse apicomplexan parasites confirm that CelTOS is a cell-traversal protein conserved in diverse apicomplexan parasites. To compare different CelTOS sequences, the following CelTOS sequences were retrieved from the UniProt database (https://www.uniprot.org/): P. vivax CelTOS (UniProt ID: A5JZX5), Plasmodium falciparum CelTOS (UniProt ID: Q8I5P1), Babesia microti (UniProt ID: I7J9F8), Theileria parva CelTOS (UniProt ID: Q4N982), and C. felis (PiroplasmaDB ID: CF003135). These CelTOS sequences were submitted to the online Clustal Omega server (https://www.ebi.ac.uk/Tools/msa/clustalo/) in the FASTA format to perform multiple sequence alignment. Clustal Omega uses seeded guide trees and hidden Markov model profile–profile techniques to perform a global alignment of sequences. The input protein sequences were submitted with the following default parameters: dealign input sequences: no, mbed-like clustering guide-tree: yes, mbed-like clustering iteration: yes, number of combined iterations: 0, max guide tree iterations: −1, max hmm iterations: −1, order: input. The percentage identity matrices and globally aligned sequences were downloaded.
To color the alignment files based on the conserved residues, the multiple sequence alignment file was submitted to the boxshade server (https://embnet.vital-it.ch/software/BOX_form.html). Boxshade is an algorithm that converts the input alignment files into the user-desired colored formats. The following parameters were used: output-format: RTF_new, Font-size: 10, consensus-line: none, and Fraction-of-sequences: 0.5. The output gray-colored files were downloaded from the server.
Next, the secondary structure information was added to these aligned CelTOS sequences using P. vivax CelTOS structure (PDB ID: 5TSZ; www.rcsb.org/) (9).
Cloning, expression, and purification of different CelTOS mutants
Structure-guided design
P. vivax CelTOS coordinates (PDB ID: 5TSZ) were retrieved from the RCSB-PDB database and analyzed in PyMol (https://pymol.org/2/). CelTOS dimer (chains A and B) was extracted from the ternary complex and analyzed different protein regions in this dimer structure. The following mutants were shortlisted based on our structure-guided designs: Pro127Ala (to remove the bend in the helix 4 region); Lys122Asp and Leu164Asp (to confirm the interaction of Lys122 and Leu164 interaction with the bound lipid); Ser55Cys and Ala123Cys (to lock the dimer through disulfide linkage); N-terminal truncation (Gly51–Asp196; to study the effect of N-terminal residues); and C-terminal truncation (Leu36–Leu164; to study the effect of C-terminal residues).
Cloning
Previously cloned pET28+ vector that contains codon-optimized PvCelTOS (Gene ID: PVX_123510) sequence was used (L36-D196) (9). This vector contains the T7 promoter followed by PvCelTOS sequence and a 6×-His tag at the C terminus of PvCelTOS. Inserts were cloned using SalI and XhoI restriction sites.
Expression and purification
The cloned mutants were transformed in BL21(DE3)-competent E. coli cells, grown in the lysogeny broth media containing kanamycin (10 μg/ml) at 37 °C. The culture was induced with 1 mM IPTG at an absorbance of 0.6 to 0.8 for 3 h at 37 °C.
To purify the CelTOS mutants, cell pellets were resuspended in lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM KCl, 5 mM imidazole, 0.25 mg/ml lysozyme, and protease inhibitor tablets) and lysed by sonication at 70% power for 3 min (0.5 s: ON; 0.5 s: OFF). The lysate was centrifuged at 10,000g for 20 min, and the His6-tagged proteins were purified by nickel–nitrilotriacetic acid chromatography. Thus, eluted protein was concentrated using a 10 kDa AMICON centrifugal concentrator. The concentrated protein was filtered through 0.22 μm filter for further purification by SEC using ÄKTA pure (GE Healthcare). Approximately 500 μl of filtered protein was loaded on Superdex 200 10/300 GL column (GE Healthcare Life Sciences). Protein sample was eluted isocratically using AKTA buffer (filtered, degassed solution of 10 mM Hepes and 150 mM KCl in millipore water) at a maximum flow rate of 0.7 ml/min. Protein peaks were analyzed by monitoring their UV absorbance at 280 nm. Sample fractions of 400 μl each were collected. SDS-PAGE gel of different fractions was run to pool fractions of interest. Finally, the pooled protein fractions were concentrated using 10 kDa centrifugal concentrator.
To study effect of DTT on CelTOS architecture, we compared the SEC profiles of PvCelTOS WT and PvCelTOS S–S mutant as follows: PvCelTOS WT and PvCelTOS S–S were purified by nickel resin and SEC as described previously. The resulting peak from each size-exclusion purification was pooled, concentrated, and split into two equal aliquots. The aliquots were then incubated in 10 mM Hepes (pH 7.4), 150 mM KCl ± 1 mM DTT for an hour before further purification by size exclusion (Superdex 200 Increase 10/300 GL column) using either 10 mM Hepes (pH 7.4), 150 mM KCl or 10 mM Hepes (pH 7.4), 150 mM KCl, and 1 mM DTT as the running buffer. The chromatograms were then overlayed for comparison.
Pore-forming assay
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and POPA lipids (Avanti Polar Lipids) were purchased as chloroform dissolved lipid mixture (4 mM POPA + 16 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and stored at −80 °C. For the pore-formation assay, one vial of this lipid mixture was retrieved and kept on ice. The vial was gently broken, and chloroform-dissolved lipid mixture was transferred to a glass tube. The glass tube containing the lipid mixture was dried under N2 gas (20–30 min) to remove chloroform. Dried lipids were dissolved in diethyl ether and hydrated in the buffer (10 mM Hepes [pH 7.4] and 150 mM KCl). Prior to making liposomes, 20 mM carboxyfluorescein dye was added to the hydration buffer to detect pore formation by fluorescence dequenching. Liposomes were prepared by serial cycles of vigorous mixing and ultrasonic bath treatment. Thus prepared, quenched 6-carboxyfluorescein-filled liposomes were extruded through Whatman Nuclepore Track-Etched Membrane (pore size = 200 nm) (Sigma–Aldrich) as previously described (17). Finally, 6-carboxyfluorescein-filled liposomes were separated from the unincorporated 6-carboxyfluorescein by SEC using Sephadex G-25–300 resin (Sigma–Aldrich).
To study the effect of different mutations on CelTOS structure, we added varying concentrations (25, 100, and 250 μM) of each mutant to the 250 nM liposome solution. The solution was incubated for 5 min at room temperature (RT), and the carboxyfluorescein release from liposome was observed using a fluorescent Biotek Synergy H1 plate reader at 512 nm upon excitation at 492 nm. Finally, Triton X-100 was added to complete dequenching of liposome in each experiment. Three replicates of each experiment were performed for all the mutants. The percent liposome disruption (LD) was calculated as:
Lipid blot assay
CelTOS binding to lipid strips was performed as previously described (9, 17) and per manufacturer’s recommended protocol. Customized PA membrane strips (Echelon Biosciences, Inc) containing a concentration gradient of PA from 400 to 0 pmol (400, 200, 100, 50, 25, 12.5, 6.25, and 0 pmol) were incubated in blocking solution (50 mM Tris [pH 8.0], 150 mM NaCl, 0.1% Tween-20, and 3% bovine serum albumin) for 1 h at RT. Purified CelTOS variant was added to a membrane at a final concentration of 1 μM in blocking solution for 1 h at RT. Membranes were next treated with 1:1000 dilution of primary antibody (6-His tag mouse antitag; Invitrogen) for 1 h at RT. The membrane was subjected to 1:10,000 dilution of secondary antibody (IRDye 800CW Goat antimouse IgG Secondary Antibody; LI-COR Biosciences) in blocking solution for 1 h at RT. The membrane was washed three times between each step with wash solution (50 mM Tris [pH 8.0], 100 mM NaCl, 0.1% Tween-20). Finally, the fluorescence signal was detected using Odyssey CLx imaging system.
Molecular dynamics simulations
To study the effect of different mutants on CelTOS structure, we performed aMD simulation as described later.
In silico mutagenesis
Prior to mutagenesis, structure was prepared using Protein Preparation Wizard (Impact 6.3; Schrodinger 2014-2, Maestro 9.8) (31) as previously described (32). In brief, the structure was corrected for atoms and bonds, and energy was minimized. Maestro visualizer was used to mutate the residues (in both the chains) and generate the following CelTOS mutants: Pro127Ala (Pro127 to Ala); Lys122Asp and Leu164Asp (K122D,L164D); S–S (Ser55/Ser123 to Cys); N-del (Leu36–Phe54 fragment was deleted); and C-del (His165–Asp196 fragment was deleted).
System preparations
All the designed mutants were individually prepared using tleap module of AMBER14 (Assisted Model Building with Energy Refinement). FF14SB force field parameters were set for the protein using the AMBER14 LEaP module (33). The AM1-BCC method was used to assign partial atomic charges for bound inhibitor (POPA), and general amber force field was used to create its topology (34). Na+ ions were treated according to the “nonbonded” model method (35). The prepared systems were solvated with TIP3P water model by creating a cubic water box, where distance of the box was set to 10 Å from periphery of protein (36). Molecular systems were neutralized through the AMBER LEaP module by the addition of a necessary amount of counter ions (Na+) to construct the system in an electrostatically preferred position. Na+ and Cl− ions were added to maintain the ionic strength of 150 mM. The whole assembly was saved as the prepared topology and coordinates files to use as input for the PMEMD module of the AMBER (37).
System minimization, heating, and equilibration
Prepared systems were energy minimized in a two-step process: initial 1000 steps of steepest descent, during which each complex was fixed to allow water and ion movement, followed by 500 steps of conjugate gradient minimization of the whole system (complex, water, and ions). The minimized systems were gradually heated from 0 to 298 K using an NVT ensemble for 100 ps where the protein–ligand complex was restrained with a large force constant of 5 kcal/mol/Å2.
Following heating, the systems were equilibrated under constant pressure at 298 K, and the restrain was gradually removed at NPT ensemble as follows: 5 kcal/mol/Å2 (40 ps), 2 kcal/mol/Å2 (20 ps), 1 kcal/mol/Å2 (20 ps), and 0.5 k cal/mol/Å2 (10 ps).
Initial conventional molecular dynamics
Prior to an aMD simulation, a 50 ns long conventional molecular dynamics (cMD) simulation was performed to calculate the system-specific parameters. Each cMD simulation was performed on NPT ensemble at 298 K temperature and 1 atm pressure. The step size of 2 fs was kept for whole simulation study. Langev in thermostat and barostat were used for temperature and pressure coupling. The SHAKE algorithm was applied to constrain all bonds containing hydrogen atoms (38). Nonbonded cutoff was kept on 10 Å, and long-range electrostatic interactions were treated by particle mesh Ewald method with fast Fourier transform grid spacing of approximately 0.1 nm (39).
aMD
aMD is an all-atom enhanced sampling method to identify metastable conformational states in the protein structure. The following parameters were extracted from the initial cMD simulation to start a 500 ns aMD simulation for each system:
-
(1)
Average total potential energy threshold (EthreshP; kcal/mol) = total potential energy (kcal/mol) + 0.16 kcal/mol/atom × (number of total atoms)
-
(2)
Inverse strength boost factor for the total potential energy (alphaP) = 0.16 kcal/mol/atom × (number of total atoms)
-
(3)
Average dihedral energy threshold (Edih; kcal/mol) = 4 kcal/mol/residue × (number of solute residues)
-
(4)
Inverse strength boost factor for the dihedral energy (alphaD) = (1/5) × 4 kcal/mol/residue × (number of solute residues)
The last 200 ns production run was analyzed using the cpptraj module of the AMBER14 and VMD (40). Trajectory snapshots were taken at each 100 ps, which were used for final analysis. The minimization and equilibration were performed by PMEMD module of AMBER14. The production simulations were performed using PMEMD program of AMBER running on NVIDIA Tesla C2050 GPU workstation (41).
Analysis
The stability of each system during the simulation was studied by calculating the RMSD of the backbone atoms of different frames to the initial conformation, and therefore, RMSD is the measure of the average distance between the atoms (usually the backbone atoms) of superimposed protein structures. The stable trajectories were further processed for RMSF analysis.
RMSF
To study the effect of different mutations on the overall protein flexibility, average fluctuation values were followed for individual amino acid residues during the simulations. The RMSFs were calculated with the backbone atoms of amino acid residues for each structure using the following formula:
T = number of frames
RMSF values were calculated using the CPPTRAJ module of AMBER14.
Abundance measurement of the disulfide-linked and nonlinked peptides of S–S dimer by MS
Sample preparation and LC–MS/MS
Purified protein was buffer exchanged to 25 mM ammonium bicarbonate (pH 7.8), and the protein concentration was measured by nanodrop at an absorbance at 280 nm. Approximately 10 μg proteins were alkylated with 5 mM iodoacetamide in dark for 30 min. Trypsin/Lys-C mix (Mass Spec Grade; Promega) was added to protein solution at 1:50 ratio (trypsin:protein) and incubated for 16 h at 37 °C. The enzymatic digestion was terminated by adding 0.1% fatty acid (FA). The tryptic digest was further purified by C18 Zip Tips (MilliporeSigma) and reconstituted in 0.1% FA. The peptide solution was centrifuged at 15,000 RPM before loading on a reversed-phase C18 column. Approximately, 800 ng of peptide solution was loaded onto the EASY-Spray PepMap RSLC C18 (15 cm × 150 μm) column in mobile phase A (0.1% FA in water [LCMS grade]) using UltiMate 3000 UHPLC System (Thermo Fisher Scientific). The peptides were separated using a 60-min gradient from 2 to 30% mobile phase B (0.1% FA in acetonitrile) from 5 to 40 min and ramped up the mobile phase B up to 90% for another 8 min and hold at 90% B for 2 min at a flow rate of 300 nl/min. The peptide eluent from the C18 column was electrosprayed in Thermo Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) in positive ion mode using 2.2 kV voltage. Peptides with the precursor mass in the m/z range 375 to 1800 with two to eight charges were selected for higher energy collisional dissociation activation (normalized collision energy = 35%) with 60 s of dynamic exclusion. The resolution for MS1 was set at 120,000 (full width at half maximum at m/z 200) and higher energy collisional dissociation fragment ions were analyzed with a resolution of 30,000 (full width at half maximum at m/z 200). Samples were run in three independent biological triplicates.
LC–MS/MS data analysis
The raw data files were analyzed on Thermo Proteome Discoverer, version 2.5 (Thermo Fisher Scientific) using SEQUEST HT and XlinkX/PD search nodes for the peptide and disulfide linkage search, respectively. Enzyme digestion was set as full trypsin with a maximum of three missed cleavages. Mass tolerances for precursor and fragment ions were set to 5 ppm and 0.02 Da, respectively. Variable modifications included methionine oxidation (+15.995 Da) and carbamidomethyl cysteine (+57.021 Da). A disulfide (−2.016 Da) linkage on cysteine residue was set as a crosslink modification in XlinkX/PD node. Peptides and crosslinks with a 1% false discovery rate were selected for further analysis. Disulfide-linked peptides with a minimum XlinkX score of 40 were considered as confident identifications. The precursor abundance of peptides and disulfide-linked peptides were used for quantification. The sum of log2-transformed precursor abundance of each peptide and crosslinked species was plotted, and the mean of three was used to calculate the percent abundance of disulfide-linked and nonlinked cysteine residue containing peptides.
Statistical analyses and software
All analyses were performed using Prism 9 (GraphPad Software, Inc).
Data availability
All data generated or analyzed during this study are included in this article and its supporting information files.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the Intramural Research Program and the Extramural Research Program (grant no.: R01AI137162) of National Institute of Allergy and Infectious Diseases, National Institutes of Health and the Burroughs Wellcome Fund. We thank J. Patrick Gorres for editing the article.
Author contributions
N. H. T., H. K., and J. R. J. conceptualization; N. H. T., H. K., J. R. J., S. A. M., N. D. S., M. F., and P. H. S. methodology; N. H. T., H. K., and S. A. M. validation; N. H. T., H. K., and S. A. M. formal analysis; N. H. T., H. K., J. R. J., S. A. M., and N. D. S. investigation; N. H. T. and M. F. resources; N. H. T. and H. K. data curation; N. H. T., H. K., and J. R. J. writing–original draft; N. H. T., H. K., J. R. J., S. A. M., N. D. S., M. F., and P. H. S. writing–review & editing; N. H. T., H. K., J. R. J., S. A. M., M. F., and P. H. S. visualization; N. H. T., M. F., and P. H. S. supervision; N. H. T. and M. F. project administration; N. H. T. and M. F. funding acquisition.
Funding and additional information
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Karen Fleming
Supporting information
References
- 1.Levine N.D., Corliss J.O., Cox F.E., Deroux G., Grain J., Honigberg B.M., et al. A newly revised classification of the protozoa. J. Protozool. 1980;27:37–58. doi: 10.1111/j.1550-7408.1980.tb04228.x. [DOI] [PubMed] [Google Scholar]
- 2.Black R.E., Cousens S., Johnson H.L., Lawn J.E., Rudan I., Bassani D.G., et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet (London, England) 2010;375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 3.Price R.N., Tjitra E., Guerra C.A., Yeung S., White N.J., Anstey N.M. Vivax malaria: neglected and not benign. Am. J. Trop. Med. Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- 4.Homer M.J., Aguilar-Delfin I., Telford S.R., 3rd, Krause P.J., Persing D.H. Babesiosis. Clin. Microbiol. Rev. 2000;13:451–469. doi: 10.1128/cmr.13.3.451-469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw M.K. Cell invasion by theileria sporozoites. Trends Parasitol. 2003;19:2–6. doi: 10.1016/s1471-4922(02)00015-6. [DOI] [PubMed] [Google Scholar]
- 6.Sherrill M.K., Cohn L.A. Cytauxzoonosis: diagnosis and treatment of an emerging disease. J. Feline Med. Surg. 2015;17:940–948. doi: 10.1177/1098612X15610681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO . World Health Organization; Geneva: 2019. World Malaria Report 2019. [Google Scholar]
- 8.Kumar H., Tolia N.H. Getting in: the structural biology of malaria invasion. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jimah J.R., Salinas N.D., Sala-Rabanal M., Jones N.G., Sibley L.D., Nichols C.G., et al. Malaria parasite CelTOS targets the inner leaflet of cell membranes for pore-dependent disruption. eLife. 2016;5 doi: 10.7554/eLife.20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kariu T., Ishino T., Yano K., Chinzei Y., Yuda M. CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol. Microbiol. 2006;59:1369–1379. doi: 10.1111/j.1365-2958.2005.05024.x. [DOI] [PubMed] [Google Scholar]
- 11.Doolan D.L., Southwood S., Freilich D.A., Sidney J., Graber N.L., Shatney L., et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergmann-Leitner E.S., Hosie H., Trichilo J., Deriso E., Ranallo R.T., Alefantis T., et al. Self-adjuvanting bacterial vectors expressing pre-erythrocytic antigens induce sterile protection against malaria. Front. Immunol. 2013;4:176. doi: 10.3389/fimmu.2013.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergmann-Leitner E.S., Legler P.M., Savranskaya T., Ockenhouse C.F., Angov E. Cellular and humoral immune effector mechanisms required for sterile protection against sporozoite challenge induced with the novel malaria vaccine candidate CelTOS. Vaccine. 2011;29:5940–5949. doi: 10.1016/j.vaccine.2011.06.053. [DOI] [PubMed] [Google Scholar]
- 14.Bergmann-Leitner E.S., Mease R.M., De La Vega P., Savranskaya T., Polhemus M., Ockenhouse C., et al. Immunization with pre-erythrocytic antigen CelTOS from Plasmodium falciparum elicits cross-species protection against heterologous challenge with Plasmodium berghei. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues-da-Silva R.N., Soares I.F., Lopez-Camacho C., Martins da Silva J.H., Perce-da-Silva D.S., Teva A., et al. Plasmodium vivax cell-traversal protein for ookinetes and sporozoites: naturally acquired humoral immune response and B-cell epitope mapping in Brazilian amazon inhabitants. Front. Immunol. 2017;8:77. doi: 10.3389/fimmu.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig D.B., Dombkowski A.A. Disulfide by design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinform. 2013;14:346. doi: 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimah J.R., Schlesinger P.H., Tolia N.H. Liposome disruption assay to examine lytic properties of biomolecules. Bio-Protoc. 2017;7 doi: 10.21769/BioProtoc.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerra A.J., Carruthers V.B. Structural features of apicomplexan pore-forming proteins and their roles in parasite cell traversal and egress. Toxins (Basel) 2017;9:265. doi: 10.3390/toxins9090265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dal Peraro M., van der Goot F.G. Pore-forming toxins: ancient, but never really out of fashion. Nat. Rev. Microbiol. 2016;14:77–92. doi: 10.1038/nrmicro.2015.3. [DOI] [PubMed] [Google Scholar]
- 20.Anderluh G., Lakey J.H. Disparate proteins use similar architectures to damage membranes. Trends Biochem. Sci. 2008;33:482–490. doi: 10.1016/j.tibs.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Fraser S.A., Karimi R., Michalak M., Hudig D. Perforin lytic activity is controlled by calreticulin. J. Immunol. 2000;164:4150–4155. doi: 10.4049/jimmunol.164.8.4150. [DOI] [PubMed] [Google Scholar]
- 22.Valle A., Perez-Socas L.B., Canet L., Hervis Y.P., de Armas-Guitart G., Martins-de-Sa D., et al. Self-homodimerization of an actinoporin by disulfide bridging reveals implications for their structure and pore formation. Sci. Rep. 2018;8:6614. doi: 10.1038/s41598-018-24688-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen V.T., Higuchi H., Kamio Y. Controlling pore assembly of staphylococcal gamma-haemolysin by low temperature and by disulphide bond formation in double-cysteine LukF mutants. Mol. Microbiol. 2002;45:1485–1498. doi: 10.1046/j.1365-2958.2002.03125.x. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka K., Caaveiro J.M., Morante K., González-Mañas J.M., Tsumoto K. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat. Commun. 2015;6:6337. doi: 10.1038/ncomms7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cingolani G., Petosa C., Weis K., Muller C.W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 26.Kumeta M., Konishi H.A., Zhang W., Sakagami S., Yoshimura S.H. Prolines in the alpha-helix confer the structural flexibility and functional integrity of importin-beta. J. Cell Sci. 2018;131:jcs206326. doi: 10.1242/jcs.206326. [DOI] [PubMed] [Google Scholar]
- 27.Tuerkova A., Kabelka I., Králová T., Sukeník L., Pokorná Š., Hof M., et al. Effect of helical kink in antimicrobial peptides on membrane pore formation. eLife. 2020;9 doi: 10.7554/eLife.47946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlesinger P.H., Ferdani R., Liu J., Pajewska J., Pajewski R., Saito M., et al. Scmtr: a chloride-selective, membrane-anchored peptide channel that exhibits voltage gating. J. Am. Chem. Soc. 2002;124:1848–1849. doi: 10.1021/ja016784d. [DOI] [PubMed] [Google Scholar]
- 29.Cosentino K., García-Sáez A.J. Bax and bak pores: are we closing the circle? Trends Cell Biol. 2017;27:266–275. doi: 10.1016/j.tcb.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parlati F., Weber T., McNew J.A., Westermann B., Sollner T.H., Rothman J.E. Rapid and efficient fusion of phospholipid vesicles by the alpha-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12565–12570. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Computer-Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 32.Sharma V., Wakode S. Pharmacophore generation and atom based 3D-QSAR of quinoline derivatives as selective phosphodiesterase 4B inhibitors. RSC Adv. 2016;6:75805–75819. [Google Scholar]
- 33.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 35.Stote R.H., Karplus M. Zinc binding in proteins and solution: a simple but accurate nonbonded representation. Proteins. 1995;23:12–31. doi: 10.1002/prot.340230104. [DOI] [PubMed] [Google Scholar]
- 36.Kiss P.T., Baranyai A. Sources of the deficiencies in the popular SPC/E and TIP3P models of water. J. Chem. Phys. 2011;134:054106. doi: 10.1063/1.3548869. [DOI] [PubMed] [Google Scholar]
- 37.Case D.A., Cheatham T.E., 3rd, Darden T., Gohlke H., Luo R., Merz K.M., Jr., et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunsteren W.F.v., Berendsen H.J.C. Algorithms for macromolecular dynamics and constraint dynamics. Mol. Phy. 2006;34:1311–1327. [Google Scholar]
- 39.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98 [Google Scholar]
- 40.Humphrey W., Dalke A., Schulten K. Vmd: visual molecular dynamics. J. Mol. Graphics. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 41.Gotz A.W., Williamson M.J., Xu D., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theor. Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supporting information files.