Abstract
The Pseudomonas fluorescens YT101 gene narG, which encodes the catalytic α subunit of the respiratory nitrate reductase, was disrupted by insertion of a gentamicin resistance cassette. In the Nar− mutants, nitrate reductase activity was not detectable under all the conditions tested, suggesting that P. fluorescens YT101 contains only one membrane-bound nitrate reductase and no periplasmic nitrate reductase. Whereas N2O respiration was not affected, anaerobic growth with NO2 as the sole electron acceptor was delayed for all of the Nar− mutants following a transfer from oxic to anoxic conditions. These results provide the first demonstration of a regulatory link between nitrate and nitrite respiration in the denitrifying pathway.
Dissimilatory denitrification refers to the respiratory reduction of nitrate and/or nitrite to dinitrogen via nitric oxide and nitrous oxide. This alternative anaerobic process of energy conservation is phylogenetically widespread in bacteria. The first step, reduction of nitrate to nitrite, can be performed not only by most denitrifiers but also by nitrate reducers, such as Escherichia coli, that are unable to reduce the nitrite produced into gas. It has been shown that the reaction sequence NO2−→NO→N2O→N2 forms a functional unit since several steps of the reduction chain are interdependent in various bacteria (1, 6, 9). For example, mutation in the nir genes of Pseudomonas stutzeri resulted in the simultaneous loss of nitrite and nitric oxide reduction (10). Similarly, a significant decrease in nitric oxide reductase activity was observed after Tn5 insertion in the nirS gene of Pseudomonas fluorescens YT101 (21). Finally, inactivation of the norQ gene of Paracoccus denitrificans encoding the nitric oxide reductase eliminated both nitrite reduction and nitric oxide reduction (20). To date, regulatory links between nitrate reductases and the other reductases of the denitrifying pathway have never been demonstrated, and it should therefore be interesting to determine if nitrate respiration is a part of this complex regulatory network. To address this possibility, we constructed and characterized isogenic mutants of P. fluorescens unable to synthesize the membrane-bound nitrate reductase A (NRA). Of the different structural genes encoding this enzyme, narG was chosen as the target for mutagenesis since it encodes the α catalytic subunit, and the chromosomal copy of narG was replaced with an in vitro-inactivated copy of this gene. To our knowledge, this is the first time that mutants deficient in the synthesis of the NRA have been obtained by allelic exchange of the narG gene in a denitrifying bacterium.
Construction of Nar− mutants of P. fluorescens.
P. fluorescens YT101 is a Rifr derivative of the strain AK15 (21). To obtain the Nar− mutants, the wild-type chromosomal copy of narG of P. fluorescens YT101 was replaced after homologous recombination by a copy of the deleted gene with an insertion of the apra3 gentamicin resistance gene. DNA restriction, agarose gel electrophoresis, ligation, and transformation were carried out by standard methods (16). Plasmid pNR25 (11) carrying 3.3 kb of narG was digested with NarI, and ends were made blunt by the action of Klenow and T4 DNA polymerase. The SmaI fragment containing the 1.8-kb apra3 gene from plasmid pHP45Ω (12) was blunt ended and ligated into the NarI-digested pNR25. The presence of the apra3 gene was confirmed by restriction enzyme analysis. The ΔnarG::apra3 construct was then excised from pSDNG3 by using the ClaI and SpeI enzymes and cloned into the EcoRI site of pLAFR3 plasmid (19), yielding pLDNR5. The pLDNR5 plasmid was then mobilized from E. coli to P. fluorescens YT101 by triparental mating by using the conjugative plasmid helper pRK2013 (11). Transconjugants were then selected on Luria-Bertani (LB) medium containing rifampin (50 μg/ml), gentamicin (10 μg/ml), and tetracycline (10 μg/ml). Recombinants showing double crossover were identified after several rounds of growth in LB medium containing gentamicin at 4°C to cure the pLDNR5 and scoring for Tcs Gmr Rifr colonies. Three mutants, namely LP59JG, LP37JG, and LP15JG, were obtained from independent experiments. Replacement of the wild-type chromosomal copy of the narG gene by the deleted copy containing the apra3 gentamicin resistance gene was verified by Southern blot analysis of chromosomal DNA of the wild-type strain YT101 and of the Nar− mutants digested with BglII. The 3.3-kb fragment of the narG gene, the deleted fragment of the narG gene, and the 1.8-kb apra3 gene were used as probes and labeled with digoxigenin-11-dUTP by using the PCR DIG Probe Synthesis kit (Boehringer, Mannheim, Germany) as described by the manufacturer, and the results were in complete agreement with the predicted patterns. Southern blot analysis of DNA of Nar− mutants using labeled pLAFR3 as a probe allowed us to verify that no additional insertion of the vector occurred (data not shown).
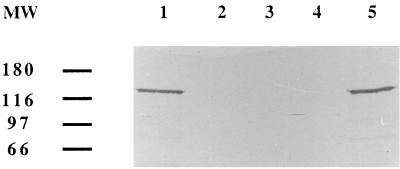
Expression of NRA was compared in wild-type and mutant strains by immunoblotting techniques. For this purpose, the NRA α catalytic subunit of P. fluorescens YT101 was purified to homogeneity as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by silver staining according to the procedure described previously (11). The identification of the α catalytic subunit was confirmed by determining the amino-terminal sequence by automated gas phase Edman degradation (Centre d’Analyse du CNRS, Solaize, France). An antiserum was raised in rabbits (Valbex, Villeurbanne, France) and was used to detect NRA expression by conventional immunoblotting techniques. As shown in Fig. 1, the antiserum recognized a 118-kDa protein in total protein extracts from the wild-type strain incubated for 4 h under anaerobiosis in the presence of 20 mM nitrate (lane 1). As expected, this peptide was not detected in Nar− mutants incubated for 4 h or more under inducing conditions (lanes 2, 3, and 4).
FIG. 1.
Western blot of total proteins from the wild-type strain YT101 (lane 1) and LP59JG (lane 2), LP37JG (lane 3), and LP15JG mutants (lane 4), and LPCJG21 (lane 5) complemented strain. The blot was developed with polyclonal antibodies raised against the α subunit of nitrate reductase purified from P. fluorescens YT101. Molecular sizes are indicated on the left in kilodaltons.
Furthermore, nitrate reductase activities were compared in dense cultures and in cell extracts of the wild-type strain and Nar− mutants by using methyl viologen or benzyl viologen as an electron donor (8). In the case of the wild-type strain, no nitrate reductase activity could be detected with either electron donor when cells were grown aerobically with or without nitrate. A nitrate reductase activity was observed with benzyl viologen as the electron donor for wild-type cells incubated anaerobically with nitrate (10 mM), but no activity could be detected under the same conditions when methyl viologen was the electron donor. For the mutants strains, no activity was detected under all the conditions tested with either electron donor (results not shown). The existence of an additional periplasmic nitrate reductase has been reported in a wide variety of phylogenetically unrelated bacteria (2–5, 7, 13–15, 17, 18). Since all of our Nar− mutants presented a total lack of nitrate reductase activity whatever the conditions tested, this strongly suggests that P. fluorescens YT101 contains only one nitrate reductase. Therefore, the existence of more than one nitrate reductase in nitrate-reducing or denitrifying bacteria cannot be generalized.
Effects of narG disruption on growth characteristics of Nar− mutants.
The ability of Nar− mutants to use nitrate, nitrite, or nitrous oxide as the sole electron acceptor to sustain growth was compared to that of the wild-type strain YT101. For this, 150-ml plasma flasks containing 38 ml of LB medium supplemented with either 20 mM KNO3 or 10 mM KNO2 were made anaerobic by evacuation and flushing three times with helium. Flasks containing 20 mM N2O were prepared by replacing (after flushing) 18 ml of the atmosphere of 150-ml plasma flasks containing 38 ml of LB medium with 18 ml of gaseous N2O. After inoculation with either the wild-type strain YT101 or the Nar− mutant strains, flasks were incubated at 28°C on an orbital shaker. Each treatment was tested in triplicate, and bacterial growth was monitored at 580 nm. Nitrate and nitrite concentrations were determined by ionic chromatography (Centre d’Analyse du CNRS). Nitrous oxide concentration was determined by using a GIRDEL 30 gas chromatograph equipped with a thermal conductivity detector.
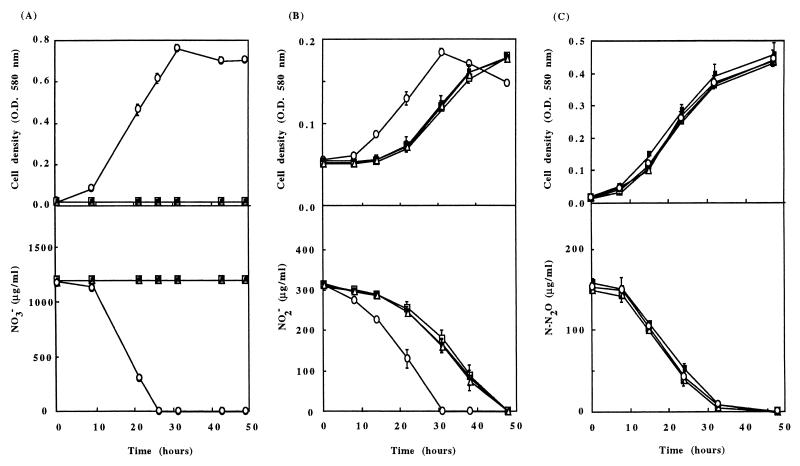
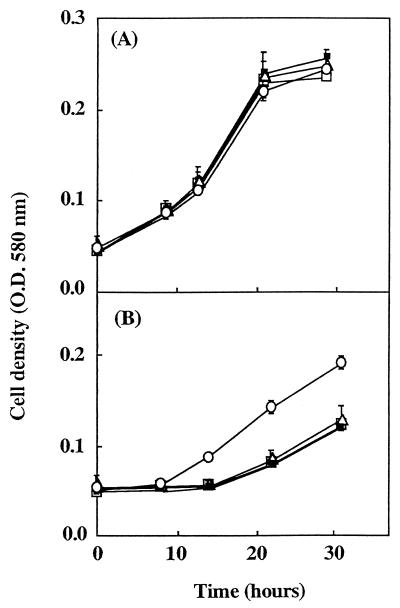
Under aerobic conditions, the wild-type strain and the Nar− mutants had similar growth rates (result not shown). The growth characteristics of the wild-type strain and the Nar− mutants under anaerobic conditions with various nitrogen oxides as electron acceptors are given in Fig. 2. When nitrate was the sole electron acceptor, Nar− mutants were unable to grow while the wild-type strain reached optical density at 580 nm of 0.75 within 30 h (Fig. 2A). Moreover, no significant reduction of nitrate was observed for the Nar− mutants after 50 h while nitrate decreased below the detection threshold within 25 h for the wild-type strain. When nitrous oxide was the sole electron acceptor, the wild-type strain and Nar− mutants had similar growth curves and nitrous oxide was similarly reduced within 40 h in all cultures (Fig. 2C). When nitrite was the sole electron acceptor, the wild-type strain reached an optical density at 580 nm of 0.18 within 30 h with a concomitant decrease of nitrite concentration in the culture medium (Fig. 2B). A 14-h lag was observed for both growth resumption and nitrite consumption for all Nar− mutants. The generation times of the wild type and mutants were similar, i.e., 16 h 30 min and 17 h, respectively. At the end of this experiment, mutant cells were reinoculated into fresh medium containing nitrite and anaerobic growth was monitored. In this case, the lag period was no longer observed, and both the wild-type strain and mutant strains reinitiated growth immediately with the same generation time, 7 h 50 min (Fig. 3A). This could be due to the selection of variants or spontaneous mutants in which the characteristics of anaerobic growth with nitrite have been restored. This possibility was ruled out by showing that the delay reappeared when cells grown twice under anaerobiosis with nitrite were then grown aerobically before being recultivated under anoxic conditions with nitrite (Fig. 3B). One of the Nar− mutants (strain LP59JG) containing the ΔnarG::apra3 gene was restored to the wild-type phenotype by complementing the narG gene in cis by using plasmid pLNR4. After introduction of this plasmid into the LP59JG Nar− mutant, transconjugants were then screened for their abilities to grow anaerobically with nitrate as the sole electron acceptor. NarG synthesis by the complemented strain LPCJG21 was confirmed by Western blotting (Fig. 1, lane 5) and by measuring nitrate reductase activity with benzyl viologen as an electron donor. When oxygen, nitrate, nitrite, and nitrous oxide were used as electron acceptors the complemented strain exhibited growth patterns that were identical to those of the wild-type strain (data not shown). In particular, the delay before growth resumption observed under anaerobiosis with nitrite as the sole electron acceptor was no longer observed. Therefore, the phenotypic changes observed for Nar− mutants can be clearly attributed to the insertional inactivation of narG.
FIG. 2.
Anaerobic growth (top) and electron acceptor consumption (bottom) of the wild-type strain YT101 (○) and the LP59JG (□), LP37JG (▵), and LP15JG (■) mutants in LB medium supplemented with KNO3 (20 mM) (A), KNO2 (10 mM) (B), and N2O (20 mM) (C). When not visible, the standard deviation (vertical bar) is within symbol dimensions. O. D., optical density.
FIG. 3.
Anaerobic growth in LB medium with 10 mM KNO2 as the sole electron acceptor of wild-type YT101 strain (○) and the LP57JG (□), LP37JG (▵), and LP15JG (■) mutants. Cells were precultured anaerobically with 10 mM KNO2 (A), or cells were precultured anaerobically with 10 mM KNO2 to late exponential phase and then grown aerobically before being recultivated under anaerobiosis with 10 mM KNO2 (B). When not visible, the standard deviation (vertical bar) is within symbol dimensions. O. D., optical density.
The effects of narG mutation on the denitrifying pathway were limited to nitrite respiration. Since the DNA fragment that was used to disrupt narG contains a transcription termination sequence (hairpin) which is oriented so as to block transcription coming out from the apra3 fragment (12), it can be expected that transcription of downstream narG sequences was inhibited or dramatically reduced. Therefore, the phenotype of the Nar− mutants can be attributed either to the lack of the product of narG itself or to the lack of the product(s) of the gene(s) located downstream of narG in the narGHJI operon.
The growth resumption delay observed in all the Nar− mutants of P. fluorescens after the transfer from aerobic to anaerobic conditions with nitrite as the sole electron acceptor suggests that there is a genetic and/or functional relationship between the dissimilatory reduction of nitrate and that of nitrite. Since nitrite respiration was not inhibited but only delayed in the Nar− mutants, the reasons for this phenotypic change are likely to be complex and may include the following. (i) Some specific gene(s) may exist in the nar operon of P. fluorescens, whose product(s) is involved in the regulation of nir gene expression. Therefore, either the synthesis of nitrite reductase and/or a specific nitrite transport system could be delayed or could proceed at a slower rate in Nar− mutants. In the latter case, growth on nitrite should occur only when a sufficient cellular concentration of nitrite reductase and/or nitrite carrier is reached within the cells. (ii) The genes coding for nitrate and nitrite reduction in P. fluorescens YT101 may be localized on the same cluster, and disruption of a single transcriptional unit could affect all the regulatory circuits of this denitrification gene cluster. In support of this hypothesis, Ye et al. (22) have recently identified a DNA region involved in reduction of nitrate, nitrite, and nitric oxide by the denitrifying bacterium Pseudomonas sp. strain G-179. (iii) Besides this putative transcriptional control of nir genes by the products of the nar operon, some enzyme regulation at the activity level may also exist. Given that the membrane-bound nitrate reductase faces the cytoplasm whereas nitrite reductase is periplasmic, a protein-protein interaction seems unlikely. (iv) Finally, tolerance to nitrite may have been decreased in Nar− mutants. It is therefore possible that following a shift from oxic to anoxic conditions, anaerobic growth with nitrite may require a physiological acclimation, a process in which a functional nitrate reductase could be involved. In mutants lacking nitrate reductase, this acclimation process would be hindered, resulting in a longer lag phase before growth resumption. This hypothesis is supported by the fact that when wild-type and mutant cells were cultivated twice under anaerobiosis with nitrite, growth occurred at a rate which was half that of cells precultured under oxic conditions. The data, however, do not allow us to distinguish among the various explanations.
Our results have highlighted intriguing questions, not the least being the nature of the linkage between the respiratory nitrate reductase and nitrite reduction. The Nar− mutants constructed in this study may facilitate further investigation. The relationship between nitrate and nitrite reduction observed in P. fluorescens YT101 raises the question of whether it is similar in other denitrifying bacteria.
REFERENCES
- 1.Arai H, Igarashi Y, Kodama T. Structure and ANR-dependent transcription of the nir genes for denitrification from Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1994;58:1286–1291. doi: 10.1271/bbb.58.1286. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Carter J P, Hsiao Y H, Spiro S, Richardson D J. Soil and sediment bacteria capable of aerobic nitrate respiration. Appl Environ Microbiol. 1995;61:2852–2858. doi: 10.1128/aem.61.8.2852-2858.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choe M, Reznikoff W S. Anaerobically expressed Escherichia coli genes identified by operon fusion techniques. J Bacteriol. 1991;173:6139–6146. doi: 10.1128/jb.173.19.6139-6146.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe M, Reznikoff W S. Identification of the regulatory sequence of anaerobically expressed locus aeg-46,5. J Bacteriol. 1993;175:1165–1172. doi: 10.1128/jb.175.4.1165-1172.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.deBoer A P N, Reijnders W N M, Kuenen J G, Stouthamer A H, van Spanning R J M. Isolation, sequencing and mutational analysis of a gene cluster involved in nitrite reduction in Paracoccus denitrificans. Antonie Leeuwenhoek. 1994;66:111–127. doi: 10.1007/BF00871635. [DOI] [PubMed] [Google Scholar]
- 7.Grove J, Tanapongpipat S, Thomas G, Griffiths L, Crooke H, Cole J. Escherichia coli K-12 genes essential for the synthesis of c-type cytochromes and a third nitrate reductase located in the periplasm. Mol Microbiol. 1996;19:467–481. doi: 10.1046/j.1365-2958.1996.383914.x. [DOI] [PubMed] [Google Scholar]
- 8.Jones R W, Garland P B. Sites and specificity for the reduction of bipyridilium compounds with anaerobic respiratory systems of Escherichia coli. Effects of permeability barriers imposed by the cytoplasmic membrane. Biochem J. 1997;190:79–94. doi: 10.1042/bj1640199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jüngst A, Wakabayashi S, Matsubara H, Zumft W G. The nirSTBM region coding for cytochrome cdl-dependent nitrite respiration of Pseudomonas stutzeri consist of a cluster of mono-, di- and tetraheme proteins. FEBS Lett. 1991;279:205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- 10.Jüngst A, Zumft W G. Interdependence of respiratory NO reduction and nitrite reduction revealed by mutagenesis of nirQ, a novel gene in the denitrification gene cluster of Pseudomonas stutzeri. FEBS Lett. 1992;314:308–314. doi: 10.1016/0014-5793(92)81495-8. [DOI] [PubMed] [Google Scholar]
- 11.Philippot L, Clays-Josserand A, Lensi R, Trinsoutrot I, Normand P, Potier P. Purification of the dissimilative nitrate reductase of Pseudomonas fluorescens and cloning and sequencing of its corresponding genes. Biochim Biophys Acta. 1997;1350:272–276. doi: 10.1016/s0167-4781(97)00007-9. [DOI] [PubMed] [Google Scholar]
- 12.Prentki P, Krish H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 13.Reyes F, Rodan M D, Klipp W, Castillo F, Moreno-Vivian C. Isolation of periplasmic nitrate reductase genes from Rhodobacter sphaeroides DSM 158: structural and functional differences among prokaryotic nitrate reductases. Mol Microbiol. 1996;19:1307–1318. doi: 10.1111/j.1365-2958.1996.tb02475.x. [DOI] [PubMed] [Google Scholar]
- 14.Reyes F, Gavira M, Castillo F, Moreno-Vivian C. Periplasmic nitrate-reducing system of the phototrophic bacterium Rhodobacter sphaeroides DSM 158: transcriptional and mutational analysis of the napKEFDABC gene cluster. Biochem J. 1998;331:897–904. doi: 10.1042/bj3310897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richterich, P., N. Lakey, G. Gryan, L. Jaehn, L. Mintz, K. Robinson, and G. M. Church. 1994. Automated multiplex sequencing of the E. coli genome. Unpublished DNA sequence. GenBank accession no. U00008.
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Sears H J, Ferguson S J, Richardson D J, Spiro S. The identification of a periplasmic nitrate reductase in Paracoccus denitrificans. FEMS Microbiol Lett. 1993;113:107–112. [Google Scholar]
- 18.Siddiqui R A, Warnecke-Eberz U, Hengsberger A, Schneider B, Kostka S, Friedrich B. Structure and function of a periplasmic nitrate reductase in Alcaligenes eutrophus H16. J Bacteriol. 1993;175:5867–5876. doi: 10.1128/jb.175.18.5867-5876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stouthamer A H, de Boer A P N, van der Oost J, van Spanning R J M. Emerging principles of inorganic nitrogen metabolism in Paracoccus denitrificans and related bacteria. Antonie Leeuwenhoek. 1997;71:33–41. doi: 10.1023/a:1000113824961. [DOI] [PubMed] [Google Scholar]
- 21.Ye R W, Arunakumari A, Averill B A, Tiedje J M. Mutants of Pseudomonas fluorescens deficient in dissimilatory nitrite reductase are also altered in nitric oxide reduction. J Bacteriol. 1992;174:2560–2564. doi: 10.1128/jb.174.8.2560-2564.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye, R. W., L. Bedzyk, and T. Wang. 1998. Identification and characterization of a DNA region involved in reduction of nitrate, nitrite, and nitric oxide by the denitrifying bacteria Pseudomonas sp. G-179. Unpublished DNA sequence. GenBank accession no. AF083948.