Abstract
Background
There are limited information available related to neonatal characteristics of RASopathies, a group of autosomal dominant syndromes with considerable phenotypic overlap.
Methods
The retrospective review revealed 9 neonates born with congenital heart defects (CHDs) and diagnosed as RASopathies due to de novo mutations (DNMs) by trio-based exome sequencing (ES) between January 2017 and December 2020. We report in details of the neonatal course, molecular analysis and 180-days of age follow-up in affected individuals.
Results
The early clinical spectrum included various types of CHDs, less noticeable multiple extracardiac anomalies and unspecific symptoms like poor feeding. Of the 8 variants identified from 6 genes, 2 in RASA1 were novel: (NM_002890.2: c.2828 T > C (p.Leu943Pro)) and (NM_002890.2: c.2001del (p.Pro668Leufs*10)), which functionally impaired the protein structure. There was a relatively high mortality rate of 33.33% (3/9) for all the defects combined. A RAF1-deficient male and a RASA1-deficient male survived from severe heart failure by surgical interventions in early life.
Conclusions
Our results revealed that family-based ES was useful in identifying DNMs and causal genes for sporadic diseases and screening Rasopathies shortly after birth. We recommended a family-based ES and a full phenotypic evaluation including echocardiogram, magnetic resonance imaging, ultrasonography and coagulation screening in neonates with CHDs and a suspected genetic etiology.
Keywords: RASopathies, Neonatal, Congenital heart defect, Family-based exome sequencing, De novo mutation
Background
CHD accounts for nearly one-third of all congenital birth defects [1] and, therefore, the focus on CHDs is integral to eliminating preventable infant deaths. Recent preclinical [2] and clinical [3] have indicated a genetic etiology for CHDs. Another large genetic study of CHDs with next-generation-sequencing (NGS) [4] suggested that 8% and 2% of cases are attributable to de novo autosomal dominant and inherited autosomal recessive variation, respectively. These studies contributed to the understanding of the genetic etiology of CHDs and the underlying molecular mechanisms. This, in turn, will enable a multidisciplinary care to these individuals to extend lifespan and improve health [5].
The RASopathies are a group of autosomal dominant disorders with overlapping cardiac, facial, and neurodevelopmental features [6]. RASopathies include Noonan syndrome (NS) (MIM 163950), Noonan syndrome with multiple lentigines (NSML) (MIM 151100), Noonan-like syndrome with loose anagen hair (NS-LAH) (MIM 607721), cardio-facio-cutaneous syndrome (CFC) (MIM 115150), Costello syndrome (CS) (MIM 218040), Neurofibromatosis type 1 (MIM 162200), Legius syndrome (MIM 611431) and capillary malformation-arteriovenous malformation (CM-AVM) (MIM 608354) [7]. Taken together, the RASopathies represent one of the most prevalent groups of malformation syndromes affecting greater than 1 in 1,000 individuals equally distributed in females and males [6]. While most individuals with RASopathies share characteristic findings affecting multiple organ systems, the phenotypic spectrum is wide, ranging from a mild or attenuated phenotype to a severe phenotype with infantile lethal complications [8]. Furthermore, less major clinical features were recognized in neonatal or infantile period [9] than the manifestations in toddlerhood and beyond. This non-specificity of presentations means that clinically diagnosing RASopathies can be challenging in early life. Hence, this study aims to describe the neonatal features of RASopathies with de novo causative variants (the most extreme form of rare genetic variations) in order to facilitate prompt clinical diagnosis, enhance patient management and family counseling.
Methods
Patients and clinical data
This is a retrospective analysis of clinical records for neonates with various CHDs and a confirmed molecular diagnosis of RASopathies due to de novo pathogenic variants from two tertiary hospitals in China between January 2017 and December 2020. The institutions included Xinhua Hospital affiliated with Shanghai Jiao Tong University School of Medicine (XH) and Children's Hospital affiliated with Shanghai Jiao Tong University (CH). The institutional review board at Xinhua Hospital, Shanghai Jiao Tong University School of Medicine approved this study with a waiver of informed consent due to the nature of the study (Approval number: XHEC-D-2022-077).
All subjects were admitted to the corresponding neonatal intensive care units (NICUs) from two hospitals immediately after birth or shortly after. The sociodemographic, baseline characteristic, and phenotypic data were extracted from electronic medical records (EMRs) and the phenotypes of the affected neonates were further translated into Human Phenotype Ontology (HPO) terms [10]. All patients were followed up till 180 days of age. When a patient died, the investigator was asked to identify the primary causes of death and to assess the relationship of death to the primary disease.
Exome sequencing and molecular analysis
Those individuals that were performed with trio-based exome sequencing (ES) and analysis were included in our final study. Detailed sequencing and analysis information can be found in our previously published paper [11]. Only detected single-nucleotide variants (SNVs) and copy number variations (CNV) in RASA1, HRAS, BRAF1, CBL, KRAS, NRAS, PTPN11, RAF1, RRAS, RRAS2, RIT1, SHOC2, SOS1, SOS2, MAP2K1 (MEK1), MAP2K2 (MEK2), SPRED1 and NF1 were further interpreted and categorized according to the guidelines recommended by the American College of Medical Genetics [12, 13] for the corresponding SNV and CNV results. They were also addressed as novel or reported according to entries into the Human Gene Mutation Database (http://www.hgmd.org) [14] and the Genome Aggregation Database (gnomAD; http://gnomad.broadinstitute.org) [15]. The following available online analysis tools were also used to predict the effect of the novel missense variants. Polymorphism phenotyping (PolyPhen-2) [16] was used to predict their pathogenic effects. The multiple-sequence alignments were carried out by T-Coffee Multiple Sequence Alignment Program [17]. The protein structure analysis was performed by Project HOPE [18].
Results
Demographic feature
Data collection for this study began January 1, 2017 and ended on December 30, 2020, when 325 neonates with various CHDs had been clinically evaluated and ES tested. Patients with incomplete clinical EMRs or family-based ES data not yet fulfilled were excluded. Totally, 9 patients were identified with a de novo pathogenic or likely pathogenic variant that established a genetic diagnosis of RASopathies. Nine RASopathies were sporadic cases and they were categorized into NS and Noonan-related disorders in six patients (Patient (P)1, P2, P3, P4, P5 and P6), CM-AVM in two (P7 and P8) and CS in one (P9). Our cohort included 6 males and 3 females whose ages ranged from at birth and 23 days after birth (median: 1 day after birth) at the time of admission to NICUs. Two pregnancies (P8 and P9) were complicated by polyhydramnios. Nuchal cystic hygroma and renal anomalieswere prenatally detected in 2 patients (P2 and P3) by prenatal ultrasound. Two patients (P4 and P8) (22.22%, 2/9) were born before 37 weeks of gestation. Additional demographic characteristics of the individuals are summarized in Table 1.
Table 1.
Clinical and laboratory data of the nine neonates with RASopathies
| Patients | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 |
|---|---|---|---|---|---|---|---|---|---|
| Gender | M | M | M | M | M | F | M | F | F |
| Age at presentation(days) | 5 | 0 | 23 | 2 | 0 | 6 | 0 | 0 | 1 |
| Gestation Age (weeks) | 39.86 | 38.29 | 41 | 36.14 | 39.29 | 37 | 40.43 | 31.86 | 37.29 |
| Birth weight (g) | 3800 | 3050 | 4750 | 3350 | 3620 | 3760 | 4400 | 2735 | 3505 |
| Birth length (cm) | 51 | 48 | 56 | 49 | 48 | 49 | 56 | 46 | 48 |
| Paternal age (years) | 34 | 43 | 32 | Unknown | 28 | 30 | 35 | 37 | 35 |
| Prenatal assessment | Uneventful |
Nuchal cystic hygroma and renal abnormality |
Nuchal cystic hygroma and renal abnormality |
Uneventful | Uneventful | Uneventful | Uneventful | Polyhydramnios | Polyhydramnios |
| Routine pulse oximetry screening after birth | Negative | Positive | Positive | Positive | Negative | Negative | Negative | Negative | Negative |
| Cardiovascular malformations by Echo | |||||||||
| PVS (HP:0001642) | Positive | – | – | – | – | – | – | – | – |
| ASD (HP:0001684) | Positive | Positive | Positive | Positive | Positive | Positive | – | Positive | Positive |
| HCM (HP:0001639) | Positive | – | Positive | Positive | – | – | – | – | – |
| Other cardiac anomalies | Mitral valve defects (HP:0001633) | PDA (HP:0001643) | tricuspid valve defects (HP:0001702) | PDA (HP:0001643)、Mitral valve defects(HP:0001633) | VSD (HP:0001629) | PDA (HP:0001643) | |||
| Characteristic facies | |||||||||
| Hypertelorism (HP:0000316) | – | – | – | Positive | – | – | – | – | – |
| Low-set ears (HP:0000369) | – | – | – | Positive | – | – | – | – | – |
| Other facial anomalies |
High-arched palate (HP:0000218)、 Microphthalmia (HP:0000568)、Webbed neck (HP:0000465) |
Sparse and curly hair (HP:0002212)、Long eyelashes (HP:0000527) | Micrognathia (HP:0000347)、Webbed neck (HP:0000465) | ||||||
| Clinical features | |||||||||
| Tachypnea (HP:0002790) | Positive | Positive | Positive | Positive | – | – | – | Positive | Positive |
| Cyanosis (HP:0000961) | Positive | Positive | – | Positive | – | – | – | – | |
| Feeding difficulties (HP:0008872) | – | Positive | – | Positive | – | Positive | – | – | Positive |
| Pleural effusion (HP:0002202) | – | – | – | Positive | – | – | – | – | Positive |
| Bleeding tendency (HP:0001892) | Positive | – | – | Positive | – | – | – | Positive | – |
| Cryptorchidism(HP:0000028) | – | Positive | – | Positive | – | – | – | – | – |
| Pectus excavatum (HP:0000767) | – | Positive | – | – | – | – | – | – | – |
| Other extracardiac anomalies |
Abnormal renal pelvis morphology (HP:0010944) |
Laryngomalacia (HP:0001601)、 Abnormal renal pelvis morphology (HP:0010944) |
Abnormal renal pelvis morphology (HP:0010944) |
Erythema (HP:0031286) |
Arteriovenous malformation of the spine (HP:0002390) |
Arteriovenous malformation of the brain (HP:108010) |
Laryngomalacia (HP:0001601)、Abnormal renal pelvis morphology (HP:0010944) |
||
| Surgery or not | No | No | Yes | No | No | No | Yes | No | No |
| 180 days of age outcome | ID at 20 weeks, 6 days |
ID at 4 weeks, 3 days |
Survival |
ID at 10 weeks, 4 days |
Survival | Survival | Survival | Survival | Survival |
Abbreviations: ASD atrial septal defect, VSD ventricular septal defect, HCM hypertrophic cardiomyopathy, PDA patent ductus arteriosus, PVS pulmonary valve stenosis, ID infant death
Cardiovascular manifestations
Cardiovascular abnormalities constituted the main clinical phenotype of RASopathies while the type of CHDs is different among the different disorders (Table 1). Secundum type atrial septal defect (ASD II) (HP: 0001684) was prominent in all individuals with NS and Noonan-related disorders (100%, n = 6/6). The ASD II was associated with other cardiac anomalies in different patients: concomitant mitral (HP: 0001633) or tricuspid (HP: 0001702) valve defects in P1, P3 and P4 (50%, 3/6), pulmonic valve stenosis (PVS) (HP: 0001642) in P1 and ventricular septal defect (VSD) (HP: 0001629) in P6. Three patients (P1, P3 and P4) developed hypertrophic cardiomyopathy (HCM) (HP: 0001639) during their neonatal period. Isolated ASD occurred in a male (P5) with NS. The CM-AVM-associated CHDs included patent ductus arteriosus (PDA) (HP: 0001643) which was isolated in P7 and associated with an ASD II in P8. Isolated ASD was identified in the patient with CS (P9).
Other clinical features in RASopathies
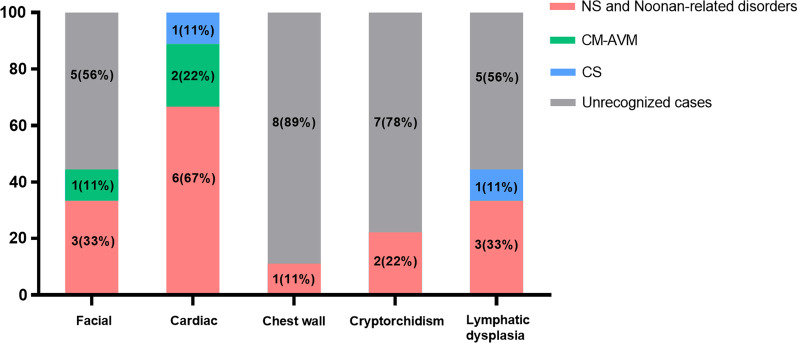
Since heterogeneity among neonates with RASopathies made early diagnosis with certainty difficult, we described their phenotypic spectrum, taking NS as the prototype, then focus on multiple extracardiac anomalies in addition to above CHDs that might distinguish the other syndromes (Fig. 1). Facial features identified in 2 (P3 and P4) out of 5 neonates with NS (40%, 2/5) included hypertelorism (HP: 0000316)/microphthalmia (HP: 0000568), high-arched palate (HP: 0000218) and low-set ears (HP: 0000369). The sparse curly hair (HP: 0002212) and long eyelashes (HP: 0000527) were observed in a female (P6) with NS-LAH. The patient (P9) with CS had a micrognathia (HP: 0000347) at birth. Other individuals (P1, P2, P5, P7 and P8) exhibited no distinctive facial features during the early neonatal period. Other extracardiac anomalies included abnormal renal pelvis morphology (HP: 0010944) in 3 patients (P2, P3 and P4) with NS and one (P9) with CS, cryptorchidism (HP: 0000028) in 2 patients (P2 and P4) with NS, laryngomalacia (HP: 0001601) in one (P3) with NS and another (P9) with CS, pleural effusion (HP: 0002202) in one (P4) with NS and another (P9) with CS, arteriovenous malformations (HP: 0100026) in 2 patients (P7 and P8) with CM-AVM, webbed neck (HP: 0000465) in one patient (P3) with NS and another (P9) with CS and pectus excavatum (HP: 0000767) in one patient (P2) with NS. Atypical skin rash (HP: 0000988) were observed in one patient (P5) with NS.
Fig. 1.
Distribution of major features in nine neonates with confirmed diagnosis of RASopathies in our cohort. As indicated in the legend, red, green and blue individually correspond to the number (percent) of the RASopathy categories and grey to those without the anomalies. Abbreviations: NS: Noonan syndrome; CM-AVM: capillary malformation-arteriovenous malformation; CS: Costello syndrome.
The major presentations for the initial visit to the NICUs in our cohort were summarized as followed. Six patients (P1, P2, P3, P4, P8 and P9) presented with tachypnea in the newborn period (range, 0–23 days) and three of them (P1, P2 and P4) had cyanosis concurrently that was attributable to their corresponding CHDs. Other common reasons were feeding difficulties in 3 patients (P2, P4 and P6) with NS and one (P9) with CS and bleeding tendency in 2 patients (P2 and P4) with NS and one (P8) with CM-AVM.
Molecular analysis of 9 neonates with RASopathies
As shown in Table 2, a definitive molecular diagnosis for 9 patients was provided. All pathogenic variants were uploaded to Leiden Open Variation Database (LOVD) (http://databases.LOVD.nl/shared). In our study, we identified six genes harboring a total of 8 de novo variants. The PTPN11, RAF1 and RASA1 were identified in 2 probands each and the remaining 3 genes per each. Six patients with NS and Noonan-related disorders and one with CS had reported gene mutations, including PTPN11 in 2 patients (P1 and P2), RAF1 in 2 patients (P3 and P4), BRAF in 1 patient (P5), SHOC2 in 1 patient (P6) and HRAS in 1 patient (P9).
Table 2.
Molecular profiles of nine neonates with RASopathies
| Patient number |
LOVD Individual ID |
Gene | Nucleotide change | Amino acid change | Molecular diagnosis | Inheritance pattern | Zygosity | Variant type | In silico prediction (SIFT/PolyPhen) |
ACMG classification | Novel/ published (PMID) |
De novo/ inherited |
Age of confirmed diagnosis (d) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | 00410414 |
PTPN11 |
c.1517A > C | p.Gln506Pro |
Noonan syndrome 1 (MIM:163,950) |
AD | Het | missense | – |
Pathogenic (PS2, PM2, PP3, PP4) |
Published (14961557) |
De novo | 61 |
| Patient 2 | 00410466 |
PTPN11 |
c.182A > G | p.Asp61Gly |
Noonan syndrome 1 (MIM:163,950) |
AD | Het | missense | – |
Pathogenic (PS2, PM2, PP3, PP4) |
Published (11704759) |
De novo | 49 |
| Patient 3 | 00410416 | RAF1 (NM_002880.3) | c.770C > T | p.Ser257Leu |
Noonan syndrome 5 (MIM:611,553) |
AD | Het | missense | – |
Pathogenic (PS2, PM2, PP3, PP4) |
Published (17603482) |
De novo | 84 |
| Patient 4 | 00410417 | RAF1 (NM_002880.3) | c.770C > T | p.Ser257Leu |
Noonan syndrome 5 (MIM:611,553) |
AD | Het | missense | – |
Pathogenic (PS2, PM2, PP3, PP4) |
Published (17603482) |
De novo | 78 |
| Patient 5 | 00410430 |
BRAF |
c.1405G > A | p.Gly469Arg |
Noonan syndrome 7 (MIM:613,706) |
AD | Het | missense | – |
Likely pathogenic (PS2, PM2, PP1,PP3, PP4) |
Published (25157968) |
De novo | 58 |
| Patient 6 | 00410431 |
SHOC2 |
c.4A > G | p.Ser2Gly |
Noonan syndrome -like with loose anagen hair 1 (MIM:607,721) |
AD | Het | missense | – |
Pathogenic (PS2, PM2, PP3, PP4) |
Published (19684605) |
De novo | 50 |
| Patient 7 | 00410432 | RASA1 (NM_002890.2) | c.2828 T > C | p.Leu943Pro |
Capillary malformation- arteriovenous malformation 1 (MIM:608,354) |
AD | Het | missense |
Deleterious/ Probably damaging |
Likely pathogenic (PS2, PM2, PM5, PP1, PP3, PP4) |
Novel | De novo | 64 |
| Patient 8 | 00410433 |
RASA1 |
c.2001del | p.Pro668Leufs*10 |
Capillary malformation- arteriovenous malformation 1 (MIM:608,354) |
AD | Het | frameshift | – |
Pathogenic (PVS1, PS2, PM2, PP3, PP4) |
Novel | De novo | 79 |
| Patient 9 | 00410435 |
HRAS |
c.37G > T | p.Gly13Cys |
Costello syndome (MIM:218,040) |
AD | Het | missense | – |
Pathogenic (PS2, PM2, PP3, PP4) |
Published (16835863) |
De novo | 55 |
Abbreviations AD Autosomal recessive inheritance disease, Het Heterozygous, WES Whole-exome sequencing, LOVD Leiden open variation database, ACMG American college of medical genetics
*Variants were classified according to the 2015 ACMG/AMP standards
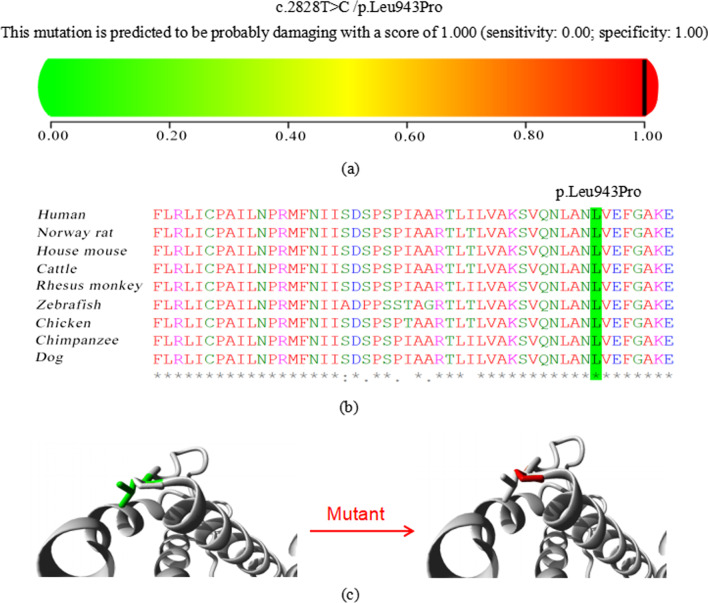
In 2 patients (P7 and P8) with CM-AVM, novel variants in RASA1 were detected. The novel missense variant, (NM_002890.2: c.2828 T > C, p.Leu943Pro), occurred with an amino acid change from a nonpolar amino acid of leucine (Leu) to another nonpolar amino acid of proline (Pro). It was found in a male (P7) presenting massive edema of the lower limbs (HP: 0010741) at birth and an initial chest X-ray demonstrated bilateral pleural effusions, pulmonary edema, and diffuse anasarca at admission. A bedside echocardiogram showed a cardiac overload. The neonate had magnetic resonance imaging (MRI) of his lower limbs on day of life (DOL) 9, which revealed extracranial arteriovenous fistulas (AVFs) (HP: 0,004,947) in spine. This variant was predicted to be “probably damaging” by PolyPhen-2 (Fig. 2a). The conservation analysis showed that the wild-type residue-Leu is conserved at this position (Fig. 2b). In addition, the RASA1 protein (the human RASA1 (UniProtKB/Swiss-Prot P20936) protein sequences were used as the reference sequence) was built based on a homologous structure using HOPE. The structure (Fig. 2c) revealed that the mutated residue-pro disrupted a α-helix leading to severe effects on the structure of the protein. ES identified another de novo in-frame deletion novel variant in RASA1 gene (NM_002890.2: c.2001del, p.Pro668Leufs*10) where no pathogenic variants have been previously reported to be responsible for CM-AVM. Since the typical phenotypes of both patients appropriately matched the disorder, 2 novel variants were considered to be disease-causing.
Fig. 2.
Detailed analysis of the novel missense variant (NM_002890.2: c.2828 T > C, p.Leu943Pro) in RASA1 gene. a Pathogenicity analyses by PolyPhen-2. b In silico analysis in different species by T-Coffee Multiple Sequence Alignment Program. c 3D structure models of wild type and mutant type in RASA1 by HOPE
180-Day outcomes in 9 neonates with RASopathies
During a 180-day of age follow-up interval, all patients in our cohort survived the neonatal period. At the end of the 6 months of age, 3 patients (P1, P2 and P4) diagnosed with NS died (Table 1). The 180-day mortality rate was 33.33% (3/9).
For the survival and severe manifestations/complications of RASopathies, we found among the 3 patients (P1, P2 and P4) who died, 2 patients (P1 and P4) with NS suffered severe coagulation defects and electrolyte disturbances which were difficult to correct during their hospital stay. HCM coexisted with structural malformations in both, evolving to congestive heart failure which may explain their worse survival. Another patient (P2) with NS presented persistent tachypnea and cyanosis because of the underlying heart failure from birth and developed feeding difficulties during early neonatal period. These life-threatening conditions made their families to give up treatments and discharge automatically.
We then evaluated the impact of expeditious treatments on the survival of this cohort. During follow-up, the clinical course of a patient (P3) with NS showed progressive aggravation of right ventricular hypertrophy compared with the initial echocardiographic evaluation. ASD device closure was performed at the age of 35 days after birth. A male (P7) with CM-AVM had life-threatening congestive heart failure from birth. The percutaneous cardiac catheterization and imaging were performed. A macrofistulas in spine was identified. This male patient was surgically treated after counseling with a multidisciplinary team including specialists in interventional radiology, surgery and cardiology. Both patients were in clinical remission until now.
Discussion
CHD is a large, rapidly emerging global problem in child health [19]. Without the ability to substantially alter the prevalence of CHD, early diagnoses and interventions must be used to improve survival. In this study, we presented a detailed clinical course of 9 neonates presenting various CHDs and having a confirmed genetic diagnosis of RASopathies due to de novo pathogenic variants. Our results emphasized the importance of family-based ES in neonates presenting with critical cardiac lesions, multiple extracardiac anomalies and other atypical symptoms with a suspected genetic etiology.
Cardiac symptoms were the most common initial presentation in children with RASopathies [20–22]. In this study, all individuals have cardiovascular abnormalities by initial echocardiographic evaluation. Previous studies indicated that the type of cardiac involvement was different among the different disorders [23, 24]. In our cohort, ASDII were detected in all individuals (P1, P2, P3, P4, P5 and P6) with NS and Noonan-related disorders. Three of them (P1, P3 and P4) developed HCM. A female with CS (P9) had isolated atrial septal defect. Furthermore, we found that the characteristic multiple extracardiac anomalies [7] were less noticeable during neonatal period and only two diagnose (P3 and P4) was included in the diagnostic criteria for NS proposed by van der Burgt [25]. Our results indicated that characteristic facies (P1, P2 and P5), typical chest wall (P1, P3, P4 and P5) and short stature (P1, P2, P3, P4 and P5) were clinically insignificant in most patients with NS. In spite of the unrecognized phenotypes in neonatal period, some reports have been made on the prenatal ultrasound findings in RASopathies [26, 27]. We detected four affected pregnancies (P2, P3, P8 and P9) in which prenatal ultrasonography demonstrated fetal abnormalities, providing clues for a suspected genetic etiology. Another five cases (P1, P4, P5, P6 and P7) had a normal ultrasound scan prenatally. This meant that prenatal diagnosis of RASopathies cannot be easily performed because only few of the phenotype features can be detected by ultrasound.
The germline pathogenic variants affect RAS-mitogen activated protein kinase (MAPK) pathway genes, resulting in comparable clinical phenotypes [28]. Nine patients in this study were sporadic cases and all harbored dominant de novo variants. Although trio-based Exome and genome sequencing have greatly facilitated the detection of DNMs in rare genetic disease, the current knowledge of its biogenesis mechanisms and functional effects remains limited. One of the prominent risk factors concerning germline de novo variants is the fact that their number increases steadily with the age of the father at conception. Our study cohort revealed that paternal ages at conception ranged from 28 to 43 y/o (median: 34.5 y/o), indicating mostly older fathers. Some studies were published in individuals with CHDs and also documented a paternal age effect on germline mutation rates in individuals with CHDs [4, 29, 30]. Collectively, these studies demonstrated that the dominant de novo variant rate among individuals might partially due to paternal age. With respect to the detection of novel de novo variants, interpreting their pathogenicity becomes more important. Our study assessed two novel de novo variants in two patients (P7 and P8) with CM-AVM and the results highlighted the relevance of combining bioinformatics analyses and clinical correlations for confidence that a particular de novo variant was disease-causing. For the frameshift one in P8 (NM_002890.2: c.2001del, p.Pro668Leufs*10), this type of mutation likely results in deleterious consequences. For the missense one in P7 (NM_002890.2: c.2828 T > C, p.Leu943Pro), we evaluated the evolutionary conservation of the affected nucleotide by T-Coffee, scored its functional impact using PolyPhen-2 and performed protein structure analysis by HOPE. Evidence of causal roles of two variant were additionally provided by their clinical correlations.
As with other rare diseases, the sole option for children with RASopathies has long been symptomatic therapies, notably for cardiopathies and growth delay [20–22, 31]. In this study, for both patients (P3 and P7) evolving to serious heart failure, beneficial effects of ASD device closure and intervention for AVFs in spine have been observed during our follow-up till 180-days of age. However, three patients with NS progressively aggravated and 2 of them (P1 and P4) had severe coagulation impairments and congestive heart failure caused by HCM. Another one (P2) required catheterization and had feeding difficulties. These might partially contribute to the clinical decision-making process and worse survival after the establishment of a molecular diagnosis.
Finally, our results demonstrated again that the power of family-based NGS was indisputable in the immediate detection of disorders that are clinically atypical and unrecognized in critically ill infants with CHDs. In a recent study [4], the trio-ES as first-line test in CHD probands revealed that DNMs accounted for 8% of cases. It is deemed that DNMs are more deleterious, on average, than inherited variation because they have been subjected to less stringent evolutionary selection [32, 33]. Consequently, an open question raised about the probable risks to existing or future siblings of the affected proband when parents who have had a child with a genetic disease caused by a DNM. Although the recurrence risk would be negligible if DNMs occurred exclusively in germ cells, new mutations can and do occur at any stage of gametogenesis and, indeed, of development, leading the indispensability of genetic counselling for DNMs [34]. We hope to find these answers in research yet to set up.
Conclusion
Our results highlighted the inherent limitations in phenotype-driven genetic testing in the neonatal population and support the application of family-based ES as a first-tier test in neonates with CHDs and a suspected genetic etiology.
Acknowledgements
We express our deep gratitude to the patients and the families for their willingness and cooperation in the study.
Abbreviations
- CHDs
Congenital heart defects
- DNMs
De novo mutations
- ES
Exome sequencing
- NGS
Next-generation-sequencing
- NS
Noonan syndrome
- NSML
Noonan syndrome with multiple lentigines
- NS-LAH
Noonan-like syndrome with loose anagen hair
- CFC
Cardio-facio-cutaneous syndrome
- CS
Costello syndrome
- CM-AVM
Capillary malformation-arteriovenous malformation
- XH
Xinhua Hospital affiliated with Shanghai Jiao Tong University School of Medicine
- CH
Children's Hospital affiliated with Shanghai Jiao Tong University
- NICUs
Neonatal intensive care units
- EMRs
Electronic medical records
- HPO
Human phenotype ontology
- CNVs
Copy number variations
- CMA
Chromosomal microarray analysis
- ASD II
Secundum type atrial septal defect
- VSD
Ventricular septal defect
- HCM
Hypertrophic cardiomyopathy
- PDA
Patent ductus arteriosus
- PVS
Pulmonary valve stenosis
- MRI
Magnetic resonance imaging
- DOL
Day of life
- AVFs
Arteriovenous fistulas
- MAPK
RAS-mitogen activated protein kinase
Author contributions
TZ conceived and designed the study. HH and SZ prepared an analytical plan, analyzed data, and drafted the initial manuscript. TZ collaborated in the revision and interpretation of the data and results, and revised the manuscript. TZ, HH SZ and LM involved in data collection, manuscript review and revision. All authors commented on the manuscript and approved the final manuscript as submitted.
Funding
None.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the Leiden Open Variation Database (LOVD) repository, [http://databases.LOVD.nl/shared/].
Declarations
Ethics approval and consent to participate
The Institutional Review Boards of Xinhua Hospital, Shanghai Jiao Tong University School of Medicine evaluated the design and implementation process and confirmed all methods performed were in accordance with the relevant guidelines and regulations. Ethics Committee of Xinhua Hospital Affiliated to Shanghai Jiaotong University School of Medicine approved this study with a waiver of informed consent due to the nature of the study (Approval number: XHEC-D-2022-077).
Consent for publication
Not applicable.
Competing interests
The authors declares that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Simin Zheng and Huanyang Huang contributed equally to this work
References
- 1.van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521(7553):520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaidi S, Brueckner M. Genetics and genomics of congenital heart disease. Circ Res. 2017;120(6):923–940. doi: 10.1161/CIRCRESAHA.116.309140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rauen KA. Defining RASopathy. Dis Model Mech. 2022 doi: 10.1242/dmm.049344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tajan M, Paccoud R, Branka S, Edouard T, Yart A. The RASopathy family: consequences of germline activation of the RAS/MAPK pathway. Endocr Rev. 2018;39(5):676–700. doi: 10.1210/er.2017-00232. [DOI] [PubMed] [Google Scholar]
- 7.Rauen KA. The RASopathies. Annu Rev Genomics Hum Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu KPT, Luk HM, Leung GKC, Mak CCY, Cheng SSW, Hau EWL, et al. Genetic landscape of RASopathies in Chinese: three decades' experience in Hong Kong. Am J Med Genet C Semin Med Genet. 2019;181(2):208–217. doi: 10.1002/ajmg.c.31692. [DOI] [PubMed] [Google Scholar]
- 9.Myers A, Bernstein JA, Brennan ML, Curry C, Esplin ED, Fisher J, et al. Perinatal features of the RASopathies: Noonan syndrome, cardiofaciocutaneous syndrome and Costello syndrome. Am J Med Genet A. 2014;164A(11):2814–2821. doi: 10.1002/ajmg.a.36737. [DOI] [PubMed] [Google Scholar]
- 10.Kohler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine JP, et al. Expansion of the human phenotype ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019;47(D1):D1018–D1027. doi: 10.1093/nar/gky1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu T, Gong X, Bei F, Ma L, Chen Y, Zhang Y, et al. Application of next-generation sequencing for genetic diagnosis in neonatal intensive care units: results of a multicenter study in China. Front Genet. 2020;11:565078. doi: 10.3389/fgene.2020.565078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandt T, Sack LM, Arjona D, Tan D, Mei H, Cui H, et al. Adapting ACMG/AMP sequence variant classification guidelines for single-gene copy number variants. Genet Med. 2020;22(2):336–344. doi: 10.1038/s41436-019-0655-2. [DOI] [PubMed] [Google Scholar]
- 13.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, et al. The human gene mutation database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 18.Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandalenakis Z, Giang KW, Eriksson P, Liden H, Synnergren M, Wåhlander H, et al. Survival in children with congenital heart disease: have we reached a peak at 97%? J Am Heart Assoc. 2020 doi: 10.1161/JAHA.120.017704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gripp KW, Morse LA, Axelrad M, Chatfield KC, Chidekel A, Dobyns W, et al. Costello syndrome: clinical phenotype, genotype, and management guidelines. Am J Med Genet A. 2019;179(9):1725–1744. doi: 10.1002/ajmg.a.61270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierpont ME, Magoulas PL, Adi S, Kavamura MI, Neri G, Noonan J, et al. Cardio-facio-cutaneous syndrome: clinical features, diagnosis, and management guidelines. Pediatrics. 2014;134(4):e1149–e1162. doi: 10.1542/peds.2013-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romano AA, Allanson JE, Dahlgren J, Gelb BD, Hall B, Pierpont ME, et al. Noonan syndrome: clinical features, diagnosis, and management guidelines. Pediatrics. 2010;126(4):746–759. doi: 10.1542/peds.2009-3207. [DOI] [PubMed] [Google Scholar]
- 23.Jhang WK, Choi JH, Lee BH, Kim GH, Yoo HW. Cardiac manifestations and associations with gene mutations in patients diagnosed with RASopathies. Pediatr Cardiol. 2016;37(8):1539–1547. doi: 10.1007/s00246-016-1468-6. [DOI] [PubMed] [Google Scholar]
- 24.Lee CL, Tan LTH, Lin HY, Hwu WL, Lee NC, Chien YH, et al. Cardiac manifestations and gene mutations of patients with RASopathies in Taiwan. Am J Med Genet A. 2020;182(2):357–364. doi: 10.1002/ajmg.a.61429. [DOI] [PubMed] [Google Scholar]
- 25.van der Burgt I, Berends E, Lommen E, Beersum SV, Hamel B, Mariman E. Clinical and molecular studies in a large Dutch family with Noonan syndrome. Am J Med Genet. 1994;53(2):187–91. doi: 10.1002/ajmg.1320530213. [DOI] [PubMed] [Google Scholar]
- 26.Scott A, Di Giosaffatte N, Pinna V, Daniele P, Corno S, D'Ambrosio V, et al. When to test fetuses for RASopathies? Proposition from a systematic analysis of 352 multicenter cases and a postnatal cohort. Genet Med. 2021;23(6):1116–1124. doi: 10.1038/s41436-020-01093-7. [DOI] [PubMed] [Google Scholar]
- 27.Sparks TN, Lianoglou BR, Adami RR, Pluym ID, Holliman K, Duffy J, et al. Exome sequencing for prenatal diagnosis in nonimmune hydrops fetalis. N Engl J Med. 2020;383(18):1746–1756. doi: 10.1056/NEJMoa2023643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170(1):17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewsk KJ, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science (New York, NY) 2015;350(6265):1262–6. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orme CM, Boyden LM, Choate KA, Antaya RJ, King BA. Capillary malformation–arteriovenous malformation syndrome: review of the literature, proposed diagnostic criteria, and recommendations for management. Pediatr Dermatol. 2013;30(4):409–415. doi: 10.1111/pde.12112. [DOI] [PubMed] [Google Scholar]
- 32.Goldmann JM, Veltman JA, Gilissen C. De novo mutations reflect development and aging of the human germline. Trends Genet. 2019;35(11):828–839. doi: 10.1016/j.tig.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13(8):565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 34.Breuss MW, Antaki D, George RD, Kleiber M, James KN, Ball LL, et al. Autism risk in offspring can be assessed through quantification of male sperm mosaicism. Nat Med. 2020;26(1):143–150. doi: 10.1038/s41591-019-0711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Leiden Open Variation Database (LOVD) repository, [http://databases.LOVD.nl/shared/].