Abstract
From December 2019, the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was started as a cluster of pneumonia cases in Wuhan, Hubei Province, China. The disturbing statistics of SARS-CoV-2 promoted scientists to develop an effective vaccine against this infection. NOM protein is a multi-epitope protein that designed based on Nucleocapsid, ORF3a, and Membrane proteins of SARS-CoV-2. Flagellin is a structural protein that binds to the Toll-like receptor 5 and can enhance the immune response to a particular antigen. In this study, NOM protein as vaccine candidate was linked to the carboxyl and amino terminals of flagellin adjuvant derived from Salmonella enterica subsp. enterica serovar Dublin. Then, informatics evaluations were performed for both NOM protein and NOM protein linked to flagellin (FNOM). The interaction between the NOM and FNOM proteins with the TLR5 were assessed using docking analysis. The FNOM protein, which compared to the NOM protein, had a more suitable 3D structure and a stronger interaction with TLR5, was selected for experimental study. The FNOM and Spike (S) proteins expressed and then purified by Ni-NTA column as vaccine candidates. For analysis of immune response, anti-FNOM and anti-S proteins total IgG and IFN-γ, TNF-α, IL-6, IL-10, IL-22 and IL-17 cytokines were evaluated after vaccination of mice with vaccine candidates. The results indicated that the specific antisera (Total IgG) raised in mice that received FNOM protein formulated with S protein were higher than mice that received FNOM and S proteins alone. Also, IFN-γ and TNF-α levels after the spleen cells stimulation were significantly increased in mice that received the FNOM protein formulated with S protein compared to other groups. Immunogenic evaluations showed that, the FNOM chimeric protein could simultaneously elicit humoral and cell-mediated immune responses. Finally, it could be concluded that the FNOM protein formulated with S protein could be considered as potential vaccine candidate for protection against SARS-CoV-2 in the near future.
Keywords: Docking, NOM protein, SARS-CoV-2, Vaccine, Immunoinformatics, Cytokine, ELISA
1. Introduction
Acute Respiratory Syndrome of Coronavirus 2 (SARS-CoV-2) appeared in December 2019 and is responsible for the epidemic of Coronavirus 2019 (COVID-19) [1]. Population vaccination is one of the most effective measures to respond to the epidemic caused by the new Coronavirus infection [2]. Spike protein (S) is the major protein for neutralizing SARS-CoV-2, and most vaccines on the market are developed based on protein S [3]. Membrane (M), Nucleocapsid (N), and ORF3a proteins are conserved between different strains of human Coronavirus [4]. ORF3a protein is a polymorphic, multifunctional viral protein that induces cellular innate immune and proinflammatory responses. This protein has significant potential in the SARS-CoV and SARS-CoV-2 pathogenesis. The SARS-CoV and SARS-CoV-2 ORF3a proteins are similar in their vital protein domains, all of which have three transmembrane regions [5]. The studies indicated that the M and N proteins of SARS-CoV and SARS-CoV-2 have strong antigenic characteristics. The abundance of M protein among common coronaviruses and the high antigenicity of N protein may help to expand the T cell response and improve cross-reactive immunity. Furthermore, the M protein is a highly conserved structural protein between SARS-CoV-2 and SARS-CoV that has been suggested a potential target for the development of cross-protective vaccines [[6], [7], [8], [9], [10]]. As a result, these proteins can be suitable candidate antigens for the design of vaccine against SARS-CoV-2. There were seven protein vaccines candidates in phase III clinical trial until April 13, 2021 [11]. Potent immune responses do not require the entire pathogen, but a single piece of the pathogen can be immunogenic and produce the appropriate immune response [12]. Many protein subunit candidates against SARS-CoV-2 are currently in human clinical trials. Each of these candidates uses different immunogens, mainly from various forms of the whole S protein or its receptor binding domain (RBD), a region of the S protein that mediates viral binding to the ACE2 receptor of target host cells [13].
Toll-like receptors (TLRs) play a crucial role in regulating innate and adaptive immune responses to antigens. Attempts are now being made to design subunit vaccines in a way that directly targets TLRs [14]. Flagellin is a major structural protein that binds to the Toll-like receptor 5 (TLR5) and can enhance the immune response to a particular antigen. The carboxyl and amino terminals of flagellin were derived from Salmonella enterica subsp. enterica serovar Dublin as an adjuvant. It is a potent immune stimulant and binds to the TLR5, which initiates an innate and acquired immune response [15]. In some studies, flagellin in vaccine formulations increased antibody-dependent protective responses, resulting in significantly higher antibody titers [16,17].
NOM protein is a designed chimeric protein based on epitope-rich domains N, M and ORF3a proteins [18]. In this research, we used the carboxyl and amino terminals of flagellin in the structure of NOM protein. In this way, the NOM protein with the (EAAAK)2 linker was placed between the carboxyl and amino terminals of flagellin. We hypothesized the structure of the NOM protein would become more appropriate with the addition of flagellin to it and may elicit a more robust immune response. Therefore, we compared the structure and interaction of the NOM and FNOM proteins with TLR5 using immunoinformatics tools. The FNOM protein, which, compared to the NOM protein, had a more suitable 3D structure and a stronger interaction with TLR5, was selected for experimental study. Clinical studies have suggested a protective role for humoral and cell-mediated immunity in improving SARS-CoV-2 infection. Given that no vaccine has developed 100% stable immunity, many studies are being conducted on the formulation and different types of vaccines simultaneously. In this study, we evaluated a new approach, which is the combination of other SARS-CoV-2 proteins with S protein to possibly provoke a more appropriate immune response against COVID-19 infection. We designed the FNOM protein using informatics tools. Subsequently, the FNOM protein and SARS-CoV-2 S protein were expressed in E. coli BL21 (DE3) and origami strains, respectively, and purified using affinity chromatography. Then, evaluated different formulations of FNOM and S proteins in the mouse model. Finally, we analyzed the humoral and cellular responses generated after vaccination.
2. Material and methods
2.1. Vaccine engineering
A previous study, we designed a protein vaccine candidate (NOM) against SARS-CoV-2, by exploiting the programs of reverse vaccinology [18]. In this study, we joined NOM protein to the carboxyl and amino terminals of the flagellin adjuvant that was derived from Salmonella enterica subsp. enterica serovar Dublin. The NOM protein domains are linked together by (EAAAK)2 linkers. The VaxiJen v2.0 server was used to predict the antigenicity of the FNOM protein (Doytchinova and Flower, 2007). The AllerTOP tool was also employed to predict protein allergenicity (http://www.ddg-pharmfac.net/AllerTOP/). The different physicochemical parameters were assessed by Expasy's ProtParam (http://us.expasy.org/tools/protparam.html) [19].
2.2. Secondary and tertiary structure prediction and validation
The secondary structure of the protein was predicted by the GOR V server [20]. The Galaxy WEB has been applied to predict the tertiary structure of the designed construct. The TBM approach is used in the Galaxy WEB [21]. The final evaluation of the 3D models was conducted utilizing MolProbity, ProSA web, and Ramachandran plot. The Ramachandran plot, which can be archived by RAMPAGE, estimates the torsional angles of the residues in the protein and shows if they are in allowed, favored or outliers regions [22]. Using the model's atomic coordinates, ProSA web can identify errors in the intended 3D structures. The ProSA web makes a Z-score and a diagram of protein residue energies [23]. MolProbity, a web service for structured validation, calculates structured validation metrics, including Poor Rotamers, Ramachandran plot, clash score, Protein-geometry, and MolProbity score. The validation of proteins relies significantly on clash analysis. The clash analysis dramatically enhances the validity of proteins calculated using MolProbity [24].
2.3. B cell epitope prediction based on conformation
The ElliPro server predict conformational epitopes of the produced protein using the final determined sequence. ElliPro is a web service that integrates Thornton's approach, the MODELLER tool, and the Jmol viewer to anticipate linear and conformational antibody epitopes in protein sequence structures [25].
2.4. Mapping of T-cell MHC class I epitopes of FNOM and NOM proteins
T-cell MHC class I-restricted epitopes of FNOM and NOM proteins were identified using the Immune Epitope Database Analysis Resource (IEDB-AR) programs that contain data sets of experimentally characterized B and T cell epitopes for humans (Parikesit et al., 2009; Rana et al., 2015). IEDB tool takes an amino acid sequence or set of sequences and determines the ability of each sequence to bind to a specific MHC class I molecule. The VaxiJen v2.0 server was used to predict the antigenicity of the predicted epitopes (Doytchinova and Flower, 2007). Also, the AllerTOP tool was employed to predict epitopes allergenicity (http://www.ddg-pharmfac.net/AllerTOP/).
2.5. Molecular docking analysis
T-cell MHC class I-restricted epitopes with the highest score were selected to evaluate their interaction with the HLA-A*11:01 receptor. The FASTA sequences of epitopes were subjected to PEP-FOLD server to obtain their 3D structure [26]. The selected epitopes and HLA-A*11:01 receptor were prepared for docking using the Autodock Tool version 1.5.644. Autodock Vina 1.1.245 was used for peptide docking with a grid space that covered the entire allele [27,28]. Also, ClusPro 2.0 has been utilized to predict the interaction patterns between the TRL5 (PDB ID: 3v47) and the NOM and FNOM proteins. Initially, various compounds and crystallographic water were eliminated from the receptor. ClusPro 2.0 uses PIPER, an FFT correlation method for docking proteins having pairwise interaction potential. The PROtein binDIng enerGY prediction (PRODIGY) webserver was used to estimate the protein-receptor complex's binding affinity and dissociation constant (Kd) [29]. Ultimately, LigPlot + v.4.5.3 [30] was used to examine and display the docked complexes. All structural images of the complexes were created using the PyMOL v2.3.4 program [31].
2.6. Plasmid construction, expression, and purification of FNOM and S proteins
The fusion gene of FNOM (∼60 kDa) was constructed in a pET28a vector (cloning site of NcoI at the 5′ end and XhoI at the 3′ end) and synthesized by Biomatik Corporation (Cambridge, Ont., Canada). The whole gene of S protein ((∼135 kDa) was synthesized by Generay Biotech (Shanghai, China) in pET22b vector with cloning site NdeI at the 5′ end and XhoI at the 3′ end. In addition, a hexahistidine tag (6xHis-tag) was added at the C-terminal of the S and FNOM proteins. The FNOM vector was transformed into the competent E. coli BL21 (DE3) cells, and the S protein vector was transformed into the competent E. coli origami strain by heat shock method. Expression of the recombinant proteins was induced by adding Lactose at the final concentrations of 10 g/L in LB Broth medium. FNOM and S proteins expression assessed by 12% SDS-PAGE. Expression of the FNOM and S proteins was confirmed by Western blot analysis using HRP conjugated His-specific antibody (Sigma, USA) at a dilution of 1:1000. To purify the FNOM and S proteins, Ni-NTA column according to the manufacturer's instruction (Qiagen, Hilden, Germany) was used by denaturation method. Endotoxin removed from FNOM and S proteins using ε-poly-l-lysine-agarose (Pierce High Capacity Endotoxin Removal Spin Column, 0.5 ml, #88274; Thermo Fisher Scientific, Inc., USA). The FNOM and S proteins were dialyzed overnight at 4 °C in PBS at pH 7.4, and finally, the amount of protein was measured by the Bradford method.
2.7. Animals and immunization
In this study, four groups of mice were used. 6–8 weeks old female BALB/c mice were purchased from the Royan Research Institute, Karaj, Iran. The animal studies were performed according to guidelines of the European Communities Council (86/609/EEC). Each group consisted of 18 mice that were divided as follows:
-
Group I
F-NOM protein (40 μg) formulated in alum adjuvant (n = 18).
-
Group II
S protein (40 μg) formulated in alum adjuvant (n = 18).
-
Group III
F-NOM protein (20 μg) + S protein (20 μg) formulated in alum adjuvant (n = 18).
-
Group IV
PBS (as a control group, n = 18).
Experimental mice were subcutaneously (s.c) immunized on days 0 and 14 with 100 μl containing 40 μg of the vaccine candidates (Groups I, II and III) formulated in alum adjuvant. The control group was injected with PBS buffer with the same condition. Two weeks after the last injection, the blood samples were collected, and sera were taken from all mice in each group by centrifugation and stored at −20 °C until immunoassay.
2.8. Assessment of humoral immune responses
Using optimized indirect ELISA to detect the availability of specific total IgG, the experimental sera underwent analysis. Summarily, after overnight coating of the 96 well ELISA plates (obtained from Greiner, Germany) with 100 μl of the FNOM and S proteins (1μg/well) at a temperature of 4 °C, they were blocked with a blocking buffer of 5% skimmed milk in PBS. After adding the diluted sera in blocking buffer (the ratio of 1:100 to 1:12800) into the plates, the 1:10000 dilutions of HRP-conjugated anti-mouse IgG obtained from Sigma, USA, was used as the secondary antibodies. The plates were incubated using a substrate of Tetramethylbenzidine (TMB) subsequent to rinsing in order to detect the reactivity of the antibody at 450 nm via an ELISA reader manufactured by Awareness Stat Fax 2100, USA.
2.9. Cytokine profile analysis by flow cytometry
To assess the pattern of cytokines induced by antigens put into separate groups of mice, two weeks following the second injection, they were sacrificed under sterile conditions, and their spleen was extracted and homogenized. Shortly, (∼2 × 106) spleen cells have cultured in 24-well microtiter plates (Greiner, Germany) with or without 10 μg/ml of filtered FNOM and S proteins. After 72 h of incubation, the supernatants were gathered and kept at −70 °C till the cytokines were tested. The supernatants' concentrations of IL-17F, IL-22, IL-6, IFN-γ, TNF-α, IL-17A, and IL-10 were measured by a mouse 7 Plex cytokine assay kit (BioLegend, USA) using a flow cytometer (BD FACSCaliburTM, USA). The cytokine pattern was compared between different groups of mice.
2.10. Assessment of IgG responses in human
Serum samples were collected from the whole blood of 10 SARS-CoV-2 RT-PCR positive female participants with clinical symptoms of COVID-19 who had been referred to the Infectious and Tropical Diseases Research Center of the Hormozgan University of Medical Sciences. Detection of IgG antibody against FNOM protein in human sera was done by ELISA method. As a control, pre-epidemic samples from ten female participants from Hormozgan province diabetes cohort (collected before the start of the COVID-19 pandemic) were used.
Briefly, the purified FNOM protein with a concentration of 1 μg/well was used to coat the surface of 96-well microtiter plates (Greiner, Germany). The wells were washed with PBST and the unoccupied sites were blocked with a blocking buffer of 5% skimmed milk in PBS. After washing the plates, serial dilutions of the human sera were added to the wells in triplicate and incubated at 37 °C for 1 h. Then, the plates were incubated with human conjugated secondary IgG antibody (Sigma, USA) diluted 1:10,000 in PBST for 1 h at 37 °C. Plates were incubated with TMB substrate and the reactions were stopped by 2 N H2SO4 to read at 450 nm using an ELISA plate reader.
2.11. Statistical analysis
Statistical analysis of humoral immune responses was performed using One Way Analysis of Variance (ANOVA) followed by Student t-test, and Tukey HSD tests. Kruskal–Wallis was used to analyzing cytokines results using the version 6 Prism (GraphPad) program. P values of 0.05 were considered statistically significant.
3. Results
3.1. Characteristics of designed protein
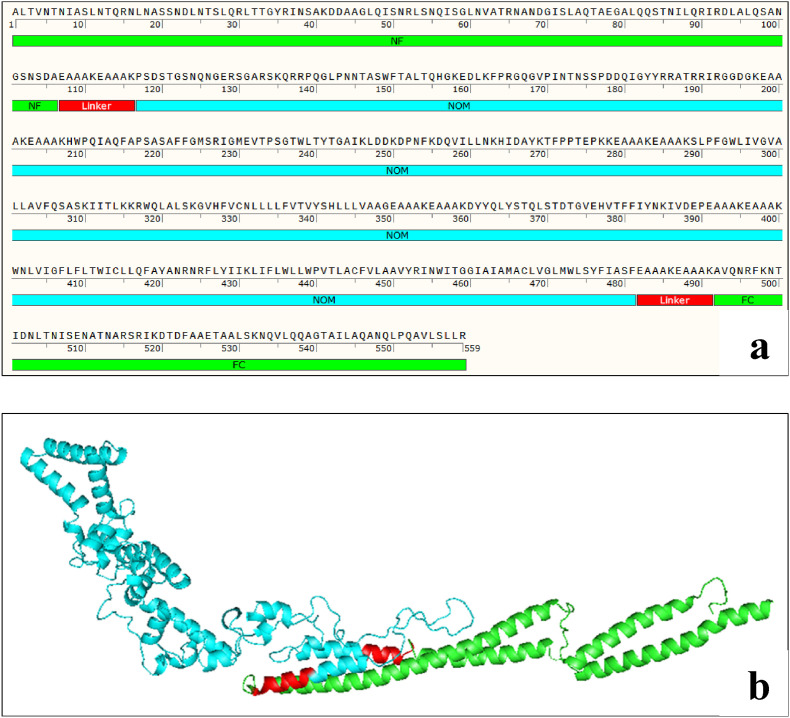
The carboxyl and amino domains of the flagellin bind to TLR5 and enhance the immune response to the antigen. For this reason, we added the carboxyl and amino terminals of the flagellin adjuvant of Salmonella enterica subsp. enterica serovar Dublin to the NOM vaccine structure. The vaccine pieces were linked to each other using (EAAAK)2 linker. The final FNOM multi-epitope vaccine had 559 amino acid residues (Fig. 1 a). Molecular weight of FNOM protein was 60 kDa. Antigenicity of the FNOM protein was estimated to be 0.5507 at a 0.4% threshold for the virus model, which indicates FNOM protein is probable antigen. The AllerTOP tool also predicted that the FNOM protein is not allergenic (Table 1 ). The primary structure analysis of NOM and FNOM showed that the FNOM protein has higher solubility, stability, and half-life in Ecoli compared to NOM protein (Table 1).
Fig. 1.
Schematic diagram of FNOM protein construct consists of NOM protein (blue) linked to the carboxyl and amino terminals of the flagellin adjuvant (green). The NOM protein domains are linked together by (EAAAK)2 linkers. a) FNOM protein sequence, b) FNOM protein 3D structure.
Table 1.
The physicochemical parameters, antigenicity and allergenicity of NOM and FNOM Proteins.
| Characteristics | FNOM | NOM |
|---|---|---|
| Number of amino acids | 559 | 337 |
| Molecular weight (dalton) | 60778.29 | 37513.55 |
| Total number of negatively charged residues (Asp + Glu) | 43 | 19 |
| Total number of positively charged residues (Arg + Lys) | 59 | 32 |
| Theoretical isoelectric point (pI) 5.14 | 9.53 | 9.62 |
| Estimated half-life | 4.4 h (mammalian reticulocytes, in vitro). >20 h (yeast, in vivo). >10 h (Escherichia coli, in vivo). | >20 h (mammalian reticulocytes, in vitro). >20 h (yeast, in vivo). ? (Escherichia coli, in vivo). |
| Instability index | 33.27 | 35.75 |
| Aliphatic index | 93.60 | 97.92 |
| Grand average hydropathy | −0.101 | 0.177 |
| Antigenicity | 0.5507 | 0.5999 |
| Allergenicity | Non-allergen | Non-allergen |
3.2. Protein secondary and tertiary structure prediction and validation
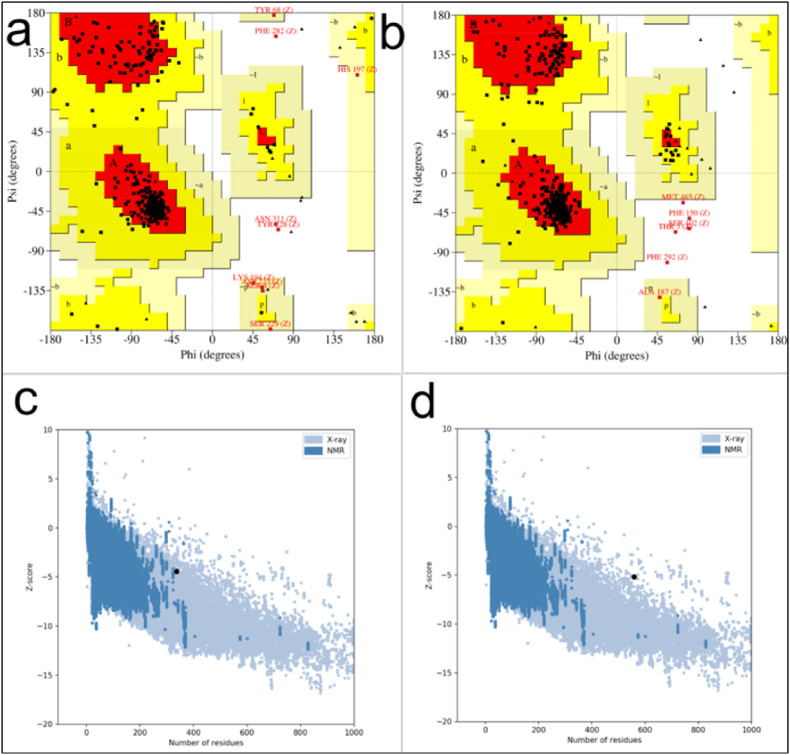
NOM protein secondary structure consisted of 43.92% alpha helix, 16.02% strand extension, and 40.06% random coil. In comparison, FNOM protein secondary structure consisted of 54.2% alpha helix (H), 11.63% extended strand (E), and 34.17% random coil (C). GalaxyWEB server created a 3D structure for FNOM designed protein (Fig. 1b). The best structural model was selected for subsequent evaluations. To compare and analyze the structure of NOM and FNOM proteins, ProSA web, Ramachandran plot and MolProbity were used to validate the models. In evaluating the third structure, the ProSA Z score indicated the quality of the structures based on the cα position. Z-scores acquired from ProSA web fell within the range of values generally observed for native proteins, suggesting that Z-scores of NOM and FNOM proteins are −4.42 and −5.18, respectively (Fig. 2 ). By analyzing Ramachandran plots, we determined that NOM and FNOM proteins possessed 85.8 and 92.7% of their residues in favored regions, respectively, when it comes to structural quality (F.g. 2). MolProbity calculates the protein structural clash score. The all-atom clash score of NOM protein was 9.37 (75% of the best among structures of comparable resolution) and FNOM protein was 6.61 (89% of the best among structures), Also, the Molprabity score of NOM protein was 2.21 (64% of the best among structures) and for FNOM protein was 1.61 (91% of the best among structures) (Table 2 ).
Fig. 2.
(a–d) Validation of tertiary structure of the NOM and FNOM proteins using (a) NOM Ramachandran plot, (b) FNOM Ramachandran plot (c) NOM ProSA web, (d) FNOM ProSA web.
Table 2.
The features of FNOM and NOM structure proteins 3D.
| Protein | Z-score | Clash score | Molprabity score | Residues in the favored regions of Ramachandran plots (%) |
|---|---|---|---|---|
| FNOM | −5.18 | 6.61 (89% of the best among structures) | 1.61 (91% of the best among structures) | 92.7% |
| NOM | −4.42 | 9.37 (75% of the best among structures) | 2.21 (64% of the best among structures) | 85.8% |
3.3. Defining discontinuous B cell epitopes
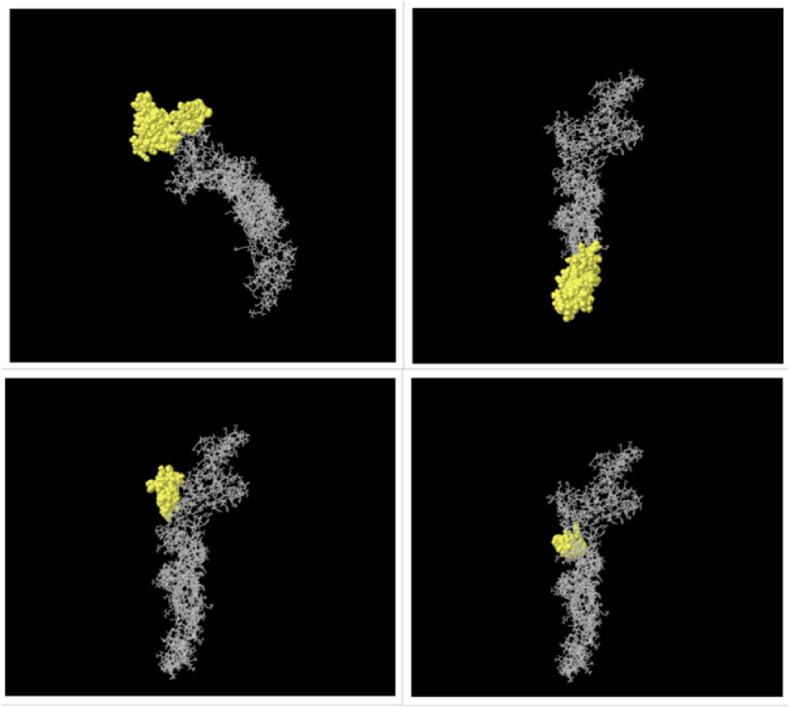
Analysis of the 3D structure of the FNOM by the ElliPro server indicated regions of conformational epitopes (Fig. 3 ). Conformational B cell epitopes were predicted, with scores ranging from 0.555 to 0.788 (Table 3 ).
Fig. 3.
Three-dimensional structure of the conformational epitopes of the FNOM protein. The yellow surface indicates the epitopes, and the bulk of the protein is represented by gray sticks.
Table 3.
Conformational B cell epitopes from the FNOM using the ElliPro server.
| Residues | Number of residues | Score |
|---|---|---|
| Q306, S309, K310, I311, I312, T313, L314, K315, K316, R317, W318, Q319, L320, A321, L322, S323, K324, G325, V326, H327, F328, V329, C330, N331, L332, L333, L334, L335, F336, V337, T338, V339, Y340, S341, H342, L343, L344, L345, V346, A347, A348, G349, E350, A351, A352, A353, K354, E355, A356, A357, A358, K359, D360, Y361, Y362, Q363, L364, Y365, S366, T367, Q368, L369, S370, D372, T373, G374, V375, E376, H377, V378, T379, F380, F381, I382, Y383, N384, K385, I386, V387, D388, E389, P390, E391, A392, A393, A394, K395, E396, A397, A398, A399, K400, N402, G406, F407 | 95 | 0.788 |
| A1, L2, T3, V4, N5, T6, N7, I8, A9, S10, L11, N12, T13, Q14, R15, N16, L17, N18, A19, S20, S21, N22, D23, L24, N25, T26, S27, L28, Q29, R30, L31, T32, T33, G34, Y35, R36, I37, N38, S39, A40, K41, D42, D43, A44, A45, G46, L47, Q48, I49, R52, N506, E509, N510, N513, A514, S516, R517, I518, D520, T521, D522, F523, A524, A525, E526, T527, A528, A529, L530, S531, K532, N533, Q534, V535, L536, Q537, Q538, A539, G540, T541, A542, I543, L544, A545, Q546, A547, N548, Q549, L550, P551, Q552, A553, V554, L555, S556, L557, L558, R559 | 98 | 0.778 |
| R227, I228, G229, M230, E231, V232, T233, P234, S235, G236, T237, W238, L239, T240, Y241, T242, I259, N262, K263, H264, I265, D266, A267, Y268, K269, T270, F271, P272, P273, T274, E275 | 31 | 0.68 |
| G193, G194, G196, A206, K207, W209, P210, Q211, I212, A213, Q214, F215, A216, P217, S218, A219, S220, A221, F222, F223, G224, M225, S226 | 23 | 0.629 |
| D93, L94, L96, Q97, S98, A99, N100, G101, S102, N103, S104, D105, A106, E107, A108, A109, A110, K111, A113, A114, P117, R185, R186, W436, M465, A466 | 26 | 0.585 |
| A187, T188, I191, R192 | 4 | 0.579 |
| L437, L438, W439 | 3 | 0.555 |
3.4. T-cell epitope mapping of NOM and FNOM constructs and prediction of antigenicity and allergenicity
To predict T-cell epitopes, we focused on all HLA-A allotypes, and also B*07:02, B*08:01, and B*15:01 allotypes because of having a much higher gene frequency. Peptides with the highest potential for NOM protein (Table 4 ) and FNOM protein (Table 5 ) were selected based on their high-affinity score, the antigenicity and allergenicity results, which indicated these epitopes are antigen and could not trigger an autoimmune response. The highest affinity score was related to the KTFPPTEPKK epitope (N1–F1), which overlapped between the FNOM and NOM proteins. In addition, the F4 (DAAGLQISNR) epitope with a high affinity score and antigenicity (Table 5) was specific to the FNOM protein, which was predicted from the amino domain of the flagellin. Epitopes N1–F1 and F4 were selected to evaluate their interaction with the HLA-A*11:01 receptor using molecular docking.
Table 4.
T cell-predicted epitopes of NOM protein with high affinity, Probable antigen and non allergen.
| Protein | Vaxigene Score | Antigenicity | Allergenicity | Epitopes | Name |
|---|---|---|---|---|---|
| HLA-A*03:01 | 0.7657 | Probable antigen | Non-allergen | KTFPPTEPKK | N1 |
| HLA-A*11:01 | 0.7657 | Probable antigen | Non-allergen | KTFPPTEPKK | N2 |
| HLA-A*01:01 | 0.4533 | Probable antigen | Non-allergen | SSPDDQIGYY | N3 |
| HLA-A*68:01 | 0.7633 | Probable antigen | Non-allergen | YKTFPPTEPK | N4 |
| HLA-A*30:01 | 0.7657 | Probable antigen | Non-allergen | KTFPPTEPKK | N5 |
| HLA-A*11:01 | 0.7633 | Probable antigen | Non-allergen | YKTFPPTEPK | N6 |
| HLA-A*68:01 | 0.8343 | Probable antigen | Probable allergen | TNIASLNTQR | N7 |
| HLA-A*26:01 | 0.5690 | Probable antigen | Non-allergen | DTGVEHVTFF | N8 |
| HLA-A*25:01 | 0.5690 | Probable antigen | Non-allergen | DTGVEHVTFF | N9 |
| HLA-A*03:01 | 0.7633 | Probable antigen | Non-allergen | YKTFPPTEPK | N10 |
| HLA-A*68:01 | 0.8238 | Probable antigen | Probable allergen | QISGLNVATR | N11 |
| HLA-A*68:01 | 0.5337 | Probable antigen | Probable allergen | NISENATNAR | N12 |
| HLA-A*01:01 | 0.6630 | Probable antigen | Non-allergen | NSSPDDQIGY | N13 |
| HLA-A*03:01 | 2.1298 | Probable antigen | Non-allergen | KLDDKDPNFK | N14 |
| HLA-A*68:01 | 0.7657 | Probable antigen | Non-allergen | KTFPPTEPKK | N15 |
| HLA-B*15:01 | 0.5986 | Probable antigen | Non-allergen | AQFAPSASAF | N16 |
| HLA-B*08:01 | 1.0185 | Probable antigen | Non-allergen | TLKKRWQLAL | N17 |
Table 5.
T cell-predicted epitopes of FNOM protein with high affinity, Probable antigen and non allergen.
| Protein | Vaxigene Score | Antigenicity | Allergenicity | Epitopes | Name |
|---|---|---|---|---|---|
| HLA-A*03:01 | 0.7657 | Probable antigen | Non-allergen | KTFPPTEPKK | F1 |
| HLA-A*11:01 | 0.7657 | Probable antigen | Non-allergen | KTFPPTEPKK | F2 |
| HLA-A*01:01 | 0.4533 | Probable antigen | Non-allergen | SSPDDQIGYY | F3 |
| HLA-A*68:01 | 1.3526 | Probable antigen | Non-allergen | DAAGLQISNR | F4 |
| HLA-A*68:01 | 0.7633 | Probable antigen | Non-allergen | YKTFPPTEPK | F5 |
| HLA-A*30:01 | 0.7657 | Probable antigen | Non-allergen | KTFPPTEPKK | F6 |
| HLA-A*11:01 | 0.7633 | Probable antigen | Non-allergen | YKTFPPTEPK | F7 |
| HLA-A*68:01 | 0.8343 | Probable antigen | Probable allergen | TNIASLNTQR | F8 |
| HLA-A*26:01 | 0.5690 | Probable antigen | Non-allergen | DTGVEHVTFF | F9 |
| HLA-A*25:01 | 0.5690 | Probable antigen | Non-allergen | DTGVEHVTFF | F10 |
| HLA-A*03:01 | 0.7633 | Probable antigen | Non-allergen | YKTFPPTEPK | F11 |
| HLA-A*68:01 | 0.8238 | Probable antigen | Probable allergen | QISGLNVATR | F12 |
| HLA-A*68:01 | 0.5337 | Probable antigen | Probable allergen | NISENATNAR | F13 |
| HLA-A*01:01 | 0.6630 | Probable antigen | Non-allergen | NSSPDDQIGY | F14 |
| HLA-A*03:01 | 2.1298 | Probable antigen | Non-allergen | KLDDKDPNFK | F15 |
| HLA-A*68:01 | 0.7657 | Probable antigen | Non-allergen | KTFPPTEPKK | F16 |
| HLA-B*15:01 | 0.5986 | Probable antigen | Non-allergen | AQFAPSASAF | F17 |
| HLA-B*08:01 | 1.0185 | Probable antigen | Non-allergen | TLKKRWQLAL | F18 |
3.5. Protein-protein and epitope-protein molecular docking
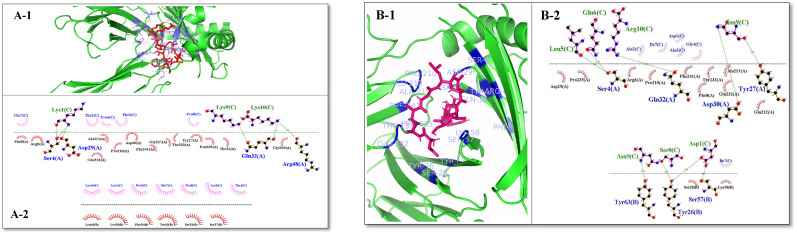
The results of evaluations indicated that, the binding affinity between the selected epitopes and the HLA-A*11:01 immune receptor are −7.8 kcal/mol and −8.1 kcal/mol, respectively. Analyzes showed that F1–N1 and F4 epitopes bind strongly to the HLA-A*11:01 receptor. In addition, We considered the best binding affinity and the number of residues involved in hydrogen and hydrophobic binding of intermolecular complexes as the main criteria for choosing the best complex. The results evaluation indicated the binding between the predicted epitope from the FNOM protein (F4) with HLA-A*11:01 receptor (Fig. 4 B-1, B-2) was stronger than binding between the predicted epitope from the NOM protein (F1–N1) with HLA-A*11:01 receptor (Figs. 4A–1, A-2).
Fig. 4.
The graphical illustration of the complex forms of the F1–N1 and F4 epitopes with the HLA-A*11:01 receptor. (A-1) HLA-A*11:01 is in green and the F1–N1 epitope is in red. In the close-up view, the residues involved in the binding site of HLA-A*11:01 that interact with the F1–N1 epitope are demonstrated (purple) (A-2) LigPlot representation of the interaction of HLA-A*11:01 and F1–N1 epitope. (B-1) HLA-A*11:01 is in green and the F4 epitope is in purple. In the close-up view, the residues involved in the binding site of HLA-A*11:01 that interact with the F4 epitope are demonstrated (blue). (B-2) LigPlot representation of the interaction of HLA-A*11:01 and F4 epitope.
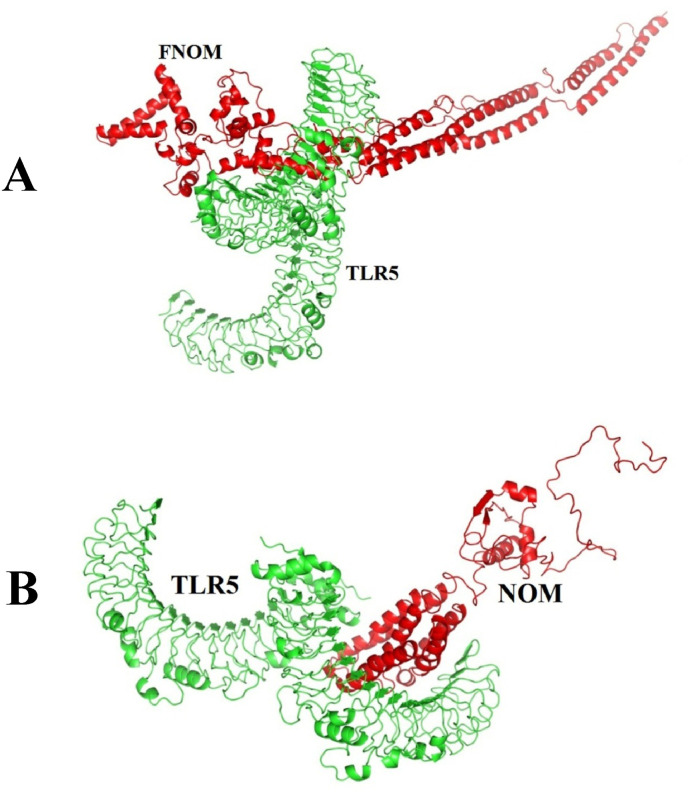
Molecular docking between the NOM and FNOM proteins with TLR5 was performed by ClusPro 2 (Fig. 5 ). The best docked complexes were selected based on the highest interaction tendency to TLR5, the most negative weighted score, the lowest PRODIGY (Kd) score, the binding affinity (ΔG), and the highest number of residues involved in hydrogen and hydrophobic bonds (Table 6 ). The results indicated strong interaction between the NOM and FNOM proteins with TLR5. But this bond between the FNOM protein and the TLR5 was stronger. Intermolecular interactions between the TLR5 with NOM and FNOM proteins were calculated using the program LIGPLOT which hydrogen bonds and hydrophobic contacts are defined in accordance with standard geometric parameters.
Fig. 5.
The graphical illustration of the complex forms of the FNOM (A) and NOM (B) proteins with the TLR5. TLR5 is in green and the NOM and FNOM proteins are in red.
Table 6.
Molecular docking results of the NOM-TLR5 and FNOM- TLR5 complexes.
| Complexes | Hydrogen bond | Hydrophobic bond | Weight Score | Binding affinity (ΔG) (Kcal/mol) | Kd (M) at 37.0 C |
|---|---|---|---|---|---|
| NOM-TLR5 | 13 | 22 | −1045.5 | −16.7 | 5.9E-13 |
| FNOM-TLR5 | 25 | 19 | −1172.1 | −23.3 | 8.3E-18 |
* LIGPLOT representation of the Hydrophobic and hydrogen interaction between the HLA-A*11:01 with F1–N1 and F4 epitopes. Hydrogen bonds are shown by green dashed lines between HLA-A*11:01 (blue) and epitopes (green) residues, and hydrophobic interactions are shown by spoked arcs representing residues of HLA-A*11:01 (black) and epitopes (blue).
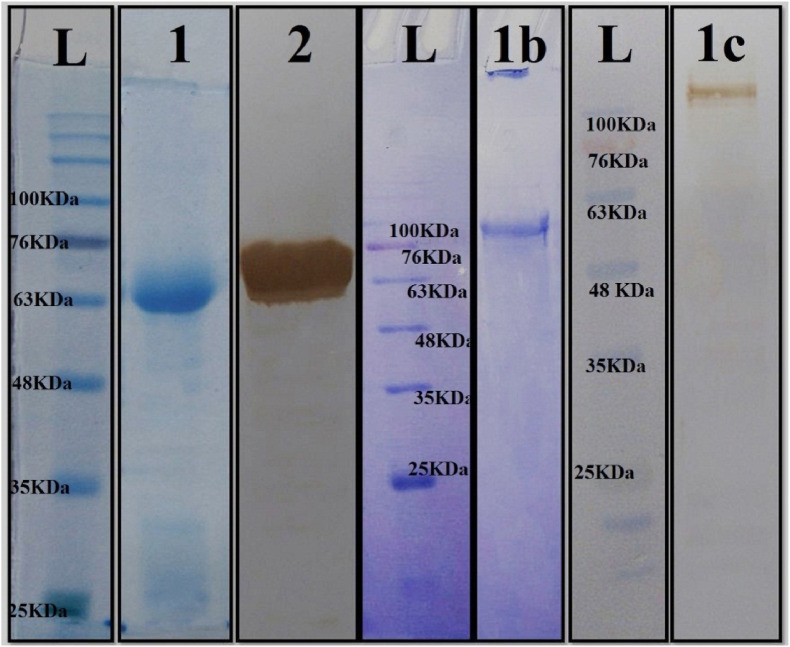
3.6. Expression and purification of FNOM and S proteins
After subcloning the gene of S protein into pET22b and FNOM protein into the pET-28a vector, S and FNOM proteins were expressed in E.coli origami strain and E.coli BL21 strain, respectively. According the analysis of 12% SDS-PAGE, 60 and 135 kDa FNOM and S proteins were present in the medium. Ni-NTA affinity chromatography was employed for the FNOM and S proteins purification (Fig. 6 ). The proteins detected by Western blot using anti-His tag antibody (Fig. 6).
Fig. 6.
Purification of FNOM and S proteins by Ni-NTA column. L: protein weight marker. lane 1: purified FNOM protein (∼60 kDa). 2: Western blot analysis of the purified FNOM protein, 1b: purified S protein (∼135 kDa), 1c: Western blot analysis of the purified S protein.
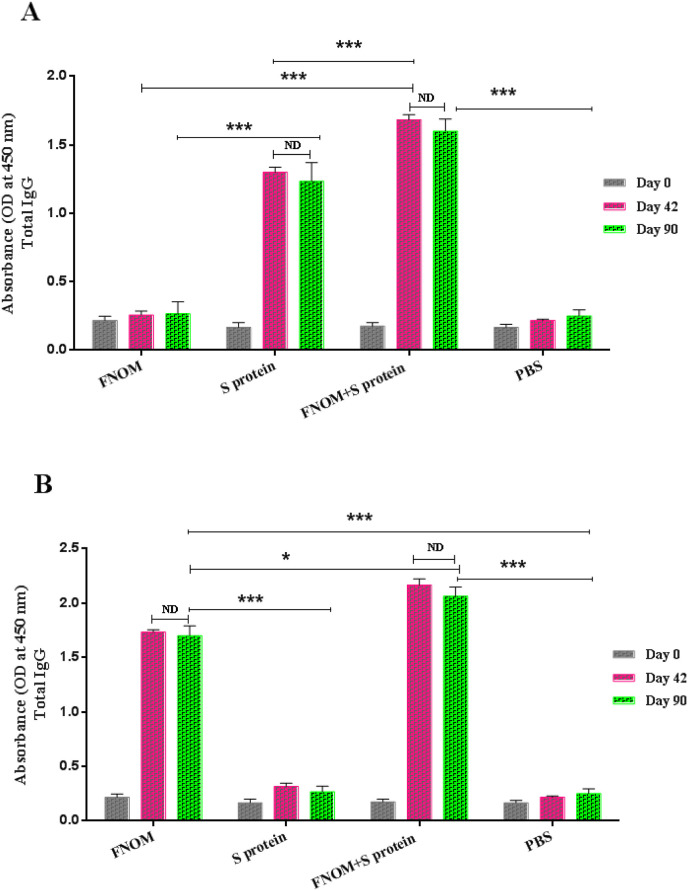
3.7. Specific total IgG responses against recombinant vaccine candidates
Specific total IgG levels against the FNOM and S proteins were assessed two weeks after the last immunization (Fig. 7 ). Results indicated higher FNOM-specific IgG antibodies in FNOM immunized mice compared to S protein-receiving mice, while the S + FNOM vaccine candidate induced stronger IgG antibody responses compared to FNOM and S proteins alone.
Fig. 7.
Evaluation of the longevity of total IgG responses against vaccine candidates. Mice were immunized and the longevity of FNOM and S proteins responses were measured. A Significant increase was observed in the mice immunized with the S, FNOM and S + FNOM proteins compared to the control group on day 90. a) The total IgG response of different immunized groups against the S protein, b) The total IgG response of different immunized groups against FNOM protein. ND represents no detectable difference, *** for p < 0.0001 and * for p < 0.05.
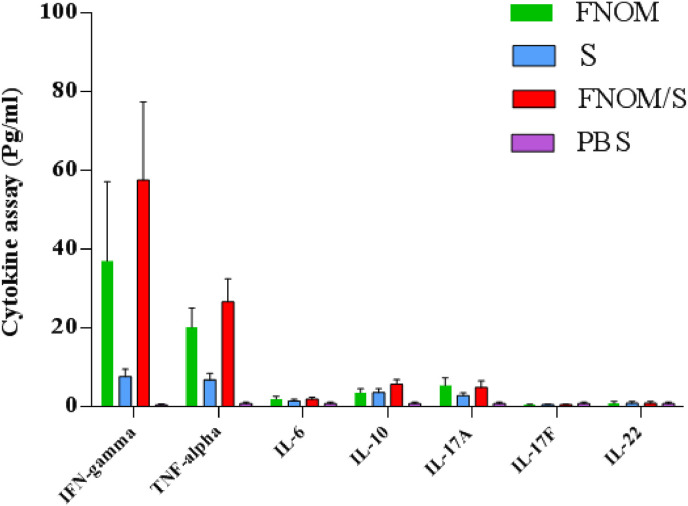
3.8. Cytokine assay
Multiplex cytokine analysis was carried out two weeks after vaccination subsequent to stimulation of the spleen cells with and without 10 μg/ml of the filtered SARS-CoV-2 S and FNOM proteins. Compared to the control group and S protein immunized group, in subjects receiving the FNOM and S + FNOM vaccine candidates, the levels of TNF-α, IFN-γ, IL-10 and IL-17A cytokines were increased after the spleen cells stimulation. This increase was significant for TNF-α and IFN-γ cytokines. However, the levels of TNF-α and IFN-γ cytokines in the FNOM + S receiving group were significantly higher than in the FNOM-receiving group. No increase was observed in IL-6, IL-17F, and IL-22 cytokines levels after the spleen cells stimulation in subjects receiving the vaccine candidates (P > 0.05 for all three cytokines). The measured cytokines secretion in the supernatant of the spleen cells in subjects receiving the FNOM and FNOM + S vaccine candidates were higher for TNF-α and IFN-γ cytokines (P < 0.001) compared to IL-10 and IL-17A cytokines (P > 0.05), which reflects a robust potential bias towards Th1 cytokines secretion as a result of stimulation via SARS-CoV-2 S and FNOM proteins (Fig. 8 ). The increase was not significant for IL-10 and IL-17A cytokines.
Fig. 8.
Cytokines profile following vaccination. Immunized mice were sacrificed 14 days after the last vaccination. Splenocytes were prepared and stimulated with the FNOM and S proteins for 72 h. The cytokines profile of the TNF-α, IFN-γ, IL-10, IL-17A, IL-17F, IL-6 and IL-22 were assessed. Compared to control and S protein groups, in subjects receiving the FNOM and S + FNOM vaccine candidates, the levels of TNF-α, IFN-γ, IL-10 and IL-17A cytokines were increased after the spleen cells stimulation. This increase was significant for TNF-α and IFN-γ cytokines (p < 0.001).
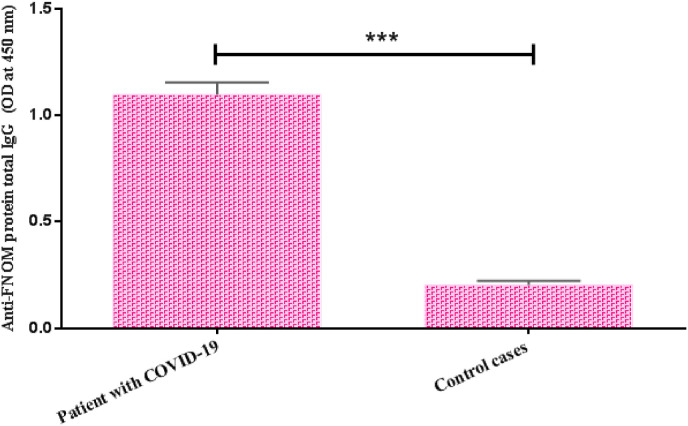
3.9. Determination of IgG responses in human cases
The level of IgG immune responses against purified FNOM protein in sera collected from COVID-19 patients was measured by ELISA. The results showed that in patients with COVID-19, the level of IgG is significantly higher than in people who had no history of infection with SARS-CoV-2 strains (p < 0.05) (Fig. 9 ).
Fig. 9.
Measurement of IgG anti-FNOM protein levels in the serum of COVID-19 patients by ELISA method. There was a significant difference between IgG levels of COVID-19 patients (1:100 dilution) and control cases. Bars represent mean ± S.D. of 10 human subjects in each group. Statistical analysis was performed using Student's t-test.
4. Discussion
The continued global spread of the COVID-19 pandemic necessitates urgent access to a protective and safe vaccine to stop the SARS-CoV-2 spread and effectuate population protection. Most of the certified vaccines have been conventionally concentrated on the robust protective antibodies induced against the target pathogens, as a result, they are aimed to confer sterilizing immunity in the vaccinated population [32]. The development of subunit vaccines relies on the observation that it is not necessary to administer the whole pathogen to elicit a strong immune response. Instead a mere immunogenic fragment suffices for this matter [13]. The polysaccharide, conjugated, virus-like particle and protein subunit vaccines are regarded as variants of subunit administration strategies with different chemical nature of the administered antigen, platform employed for the antigen administration, and the requirement of using an adjuvant for potently activation of the immune system [13]. The immune response to Flagellin that is a subunit protein of the flagellum can be boosted [33]. We employed it as an adjuvant within the vaccine structure to potentiate the immunogenicity of the antigens through binding to TLR5. TLR5 is predominantly present on monocytes, dendritic cells, epithelial cells, as well as on T cells [34]. The binding between TLR5 and flagellin leads to the induction of production of MyD88-dependent Th2 cytokine. At the same time, flagellin also signals through the NLRC4 inflammasome, which is a MyD88-independent approach for innate immune signaling [35].
The ORF3a protein has a number of highly conserved functional motifs that may be responsible for several of its functions, including ion channel activity, viral replication, and cytopathogenic effects associated with COVID-19 [5]. In COVID-19 patients, CD+4 and CD+8 T cell responses are involved in protective immunity against SARS-CoV-2, and a wide variety of T cell epitopes against structural and non-structural viral proteins have been reported in recovering individuals. Cellular epitopes against M protein are much higher among common coronaviruses and SARS-CoV-2 than S protein, indicating the potential of protein M as a target for cross-reactive immunity [10]. Also, N protein is the most abundant viral protein and is highly immunogenic during common coronavirus infections and may help to expand the T cell response and improve cross-reactive immunity. Dot blot analysis of M, N and S proteins to evaluate the humoral immune response to SARS-CoV-2 showed that N protein was the most immunized protein among all proteins analyzed [10,36]. Based on these findings, we have proposed a new approach to vaccine development against SARS-CoV-2. We used the SARS-CoV-2 ORF3a, M and N proteins plus the carboxyl and amino terminals of the flagellin that derived from Salmonella enterica subsp. enterica serovar Dublin to design a multi-epitope vaccine (FNOM). This study evaluated the immune responses of FNOM and S proteins in different formulations (FNOM protein, S protein, and S protein formulated with FNOM protein). We linked NOM protein [18] to the carboxyl and amino terminals of the flagellin as an adjuvant. The fusion gene of NOM contains the T cell and B cell epitopes of N, M and ORF3a proteins. The structures of the NOM and FNOM proteins were compared using informatic tools. The docking results indicated strong interaction between the NOM and FNOM proteins with TLR5. But this bond between the FNOM protein and the TLR5 was stronger, which may indicate the FNOM protein is more effective in eliciting an immune response. Also, T-cell MHC class I-restricted epitopes of FNOM protein showed stronger interaction with the HLA-A*11:01 receptor than NOM epitopes of class I-restricted T cells. Since other informatics analyses, such as the evaluation of tertiary structure and solubility indicated FNOM protein was more suitable, we selected FNOM protein in this study for in vivo investigation. To investigate the immune response, S and FNOM proteins were injected subcutaneously in different formulations (FNOM + S, FNOM and S) to BALB/c mice. The mice's collected sera were analyzed using ELISA to measure the specific antibodies against FNOM and S proteins. We observed an increase in cytokines related to cellular responses (TNF-α, IFN-γ, IL-10 and IL-17A) two weeks after the last injection in the immunized groups with FNOM and FNOM + S candidates compared to control groups. This increase was significant for TNF-α and IFN-γ cytokines. In a study, Ewer et al. evaluated the immune responses to the ChAdOx1 peptide vaccine against SARS-CoV-2. Analysis of cytokines secretion after peptide stimulation of PBMCs against this candidate showed that the secretion of IFN-γ and IL-2 cytokines increased in subjects receiving ChAdOx1 peptide vaccine compared to the control group. While IL-4 and IL-13 levels did not increase. Also, flow cytometric evaluations showed that CD4+ T cells mainly secreted Th1 cytokines (IFN-γ, IL-2 and TNF-α) [37]. The results of our investigations were in agreement with this study. In our evaluations, the levels of IFN-γ and TNF-α cytokines increased after the second injection in the immunized groups with FNOM and FNOM + S candidates [37]. In most studies evaluating vaccine candidates against SARS-CoV-2, increased levels of cytokines IFN-γ and TNF-α, and a potentially strong bias towards the secretion of Th1 cytokines have been observed [[37], [38], [39]]. Also, an increase in the titer of FNOM and S specific antibodies were observed from 14 days after the last injection until three months later. It was shown that FNOM protein formulated with S protein leads to dramatically increased IgG total antibody levels. Analysis of ELISA results showed that the serum of mice receiving FNOM + S vaccine candidate against S and FNOM proteins showed a higher titer of antibody compared to the groups receiving S and FNOM proteins alone. These results show that the two proteins have a synergistic effect and the simultaneous injection of these two proteins enhances the total IgG antibody response. Some studies have shown that predicted epitopes against SARS-CoV-2 antigen can increase antibody titers in mice [40]. SARS-CoV-2 specific T cell responses are induced by CD4+ and CD8+ T cells. The association of antiviral T cell responses during acute infection has been established for long-term immunity [41]. The results of our studies are consistent with other studies related to SARS-CoV-2 vaccine [[40], [41], [42]]. These results suggest that using FNOM protein could be considered to improve the immunogenicity of vaccines based on S protein against the SARS-CoV-2. Since the M, N and ORF3a structural proteins are highly conserved, the presence of this protein in the formulation of the vaccines against SARS-CoV-2 may be effective against mutations and increase the effectiveness of the vaccine.
5. Conclusion
This pilot study indicated that the presence of FNOM protein in the S protein vaccine formulation would increase the cellular and humoral immune response compared to the S protein vaccine candidate alone. FNOM recombinant protein could stimulate a higher level of Th1 cytokines compared to S protein. The coordination of these cytokines plays an important role in clearing the COVID-19 infection. Further research into the protective efficacy of the FNOM + S vaccine candidate against SARS-CoV-2 as well as the challenge with this virus will be welcomed.
Declaration of interest statment
The authors declare no competing financial interests.
CRediT authorship contribution statement
Narges Farshidi: Methodology, Conceptualization. Tayebeh Ghaedi: Methodology, Conceptualization. Mehdi Hassaniazad: Methodology, Investigation, Conceptualization. Ebrahim Eftekhar: Software, Methodology, Formal analysis. Hamed Gouklani: Methodology, Formal analysis, Software. Hossein Farshidi: Project administration, Conceptualization. Mohammad Reza Asadi Karam: Methodology. Behzad Shahbazi: Methodology, Formal analysis. Mehdi Kalani: Writing – original draft, Conceptualization. Khadijeh Ahmadi: Writing – original draft, Software, Methodology, Conceptualization.
Acknowledgments
This work was supported by a grant from the Hormozgan University of Medical Sciences, Iran (grant number: 990149) and Professor Alborzi Clinical Microbiology Research Center, Shiraz University of Medical Sciences, Iran.
References
- 1.Al-Qahtani A.A. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): emergence, history, basic and clinical aspects. Saudi J. Biol. Sci. 2020;27:2531–2538. doi: 10.1016/j.sjbs.2020.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Co-operation, O.f.E., and Development . OECD Publishing; 2021. Enhancing Public Trust in COVID-19 Vaccination: the Role of Governments. [Google Scholar]
- 3.Heinz F.X., Stiasny K. Distinguishing features of current COVID-19 vaccines: knowns and unknowns of antigen presentation and modes of action. npj Vaccines. 2021;6:1–13. doi: 10.1038/s41541-021-00369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmadi K., Zahedifard F., Mafakher L., Einakian M.A., Ghaedi T., Kavousipour S., Faezi S., Karmostaji A., Sharifi‐Sarasiabi K., Gouklani H. Chemical Biology & Drug Design; 2021. Active Site‐based Analysis of Structural Proteins for Drug Targets in Different Human Coronaviruses. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J., Ejikemeuwa A., Gerzanich V., Nasr M., Tang Q., Simard J.M., Zhao R.Y. Understanding the role of SARS-CoV-2 ORF3a in viral pathogenesis and COVID-19. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.854567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alharbi S.N., Alrefaei A.F. Comparison of the SARS-CoV-2 (2019-nCoV) M protein with its counterparts of SARS-CoV and MERS-CoV species. J. King Saud Univ. Sci. 2021;33 doi: 10.1016/j.jksus.2020.101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y., Wen J., Tang L., Zhang H., Zhang X., Li Y., Wang J., Han Y., Li G., Shi J. The M protein of SARS-CoV: basic structural and immunological properties. Dev. Reprod. Biol. 2003;1:118–130. doi: 10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lon J.R., Bai Y., Zhong B., Cai F., Du H. Prediction and evolution of B cell epitopes of surface protein in SARS-CoV-2. Virol. J. 2020;17:1–9. doi: 10.1186/s12985-020-01437-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopandić Z., Protić-Rosić I., Todorović A., Glamočlija S., Gnjatović M., Ćujic D., Gavrović-Jankulović M. IgM and IgG immunoreactivity of SARS-CoV-2 recombinant M protein. Int. J. Mol. Sci. 2021;22:4951. doi: 10.3390/ijms22094951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallejo A., Martín-Hondarza A., Gómez S., Velasco H., Vizcarra P., Haemmerle J., Casado J.L. Cellular responses to Membrane and Nucleocapsid viral proteins are also boosted after SARS-CoV-2 Spike mRNA vaccination in individuals with either past infection or cross-reactivity. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.812729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagy A., Alhatlani B. An overview of current COVID-19 vaccine platforms. Comput. Struct. Biotechnol. J. 2021;19:2508–2517. doi: 10.1016/j.csbj.2021.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasanzadeh S., Habibi M., Shokrgozar M.A., Cohan R.A., Ahmadi K., Karam M.R.A., Bouzari S. In silico analysis and in vivo assessment of a novel epitope-based vaccine candidate against uropathogenic Escherichia coli. Sci. Rep. 2020;10:1–16. doi: 10.1038/s41598-020-73179-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyriakidis N.C., López-Cortés A., González E.V., Grimaldos A.B., Prado E.O. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. npj Vaccines. 2021;6:1–17. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majewska M., Szczepanik M. The role of Toll-like receptors (TLR) in innate and adaptive immune responses and their function in immune response regulation. Postępy Higieny Medycyny Doświadczalnej. 2006;60:52–63. [PubMed] [Google Scholar]
- 15.Gupta S.K., Bajwa P., Deb R., Chellappa M.M., Dey S. Flagellin a toll-like receptor 5 agonist as an adjuvant in chicken vaccines. Clin. Vaccine Immunol. 2014;21:261–270. doi: 10.1128/CVI.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lockner J.W., Eubanks L.M., Choi J.L., Lively J.M., Schlosburg J.E., Collins K.C., Globisch D., Rosenfeld-Gunn R.J., Wilson I.A., Janda K.D. Flagellin as carrier and adjuvant in cocaine vaccine development. Mol. Pharm. 2015;12:653–662. doi: 10.1021/mp500520r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J., Bao C., Reinhardt R.L., Abraham S.N. Local induction of bladder Th1 responses to combat urinary tract infections. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2026461118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enayatkhani M., Hasaniazad M., Faezi S., Gouklani H., Davoodian P., Ahmadi N., Einakian M.A., Karmostaji A., Ahmadi K. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. J. Biomol. Struct. Dyn. 2021;39:2857–2872. doi: 10.1080/07391102.2020.1756411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. The Proteomics Protocols Handbook. Springer; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- 20.Kouza M., Faraggi E., Kolinski A., Kloczkowski A. Prediction of Protein Secondary Structure. Springer; 2017. The GOR method of protein secondary structure prediction and its application as a protein aggregation prediction tool; pp. 7–24. [DOI] [PubMed] [Google Scholar]
- 21.HeeShin W. Prediction of protein structure and interaction by GALAXY protein modeling programs. Biodes. 2014;2:1–11. [Google Scholar]
- 22.Oberholser K. Proteopedia entry: Ramachandran plots. Biochem. Mol. Biol. Educ. 2010;38 doi: 10.1002/bmb.20457. 430-430. [DOI] [PubMed] [Google Scholar]
- 23.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B. MolProbity: more and better reference data for improved all‐atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponomarenko J., Bui H.-H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Culshaw A., Ladell K., Gras S., McLaren J.E., Miners K.L., Farenc C., Van Den Heuvel H., Gostick E., Dejnirattisai W., Wangteeraprasert A. Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8+ T cell response. Nat. Immunol. 2017;18:1228–1237. doi: 10.1038/ni.3850. [DOI] [PubMed] [Google Scholar]
- 28.Yoon S.-i., Kurnasov O., Natarajan V., Hong M., Gudkov A.V., Osterman A.L., Wilson I.A. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 30.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 31.DeLano W.L. Pymol: an open-source molecular graphics tool. CCP4 Newsletter protein crystallography. 2002;40:82–92. [Google Scholar]
- 32.Pollard A.J., Bijker E.M. A guide to vaccinology: from basic principles to new developments. Nat. Rev. Immunol. 2021;21:83–100. doi: 10.1038/s41577-020-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hajam I.A., Dar P.A., Shahnawaz I., Jaume J.C., Lee J.H. Bacterial flagellin—a potent immunomodulatory agent. Exp. Mol. Med. 2017;49 doi: 10.1038/emm.2017.172. e373-e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen-Nissen E., Smith K.D., Strobe K.L., Barrett S.L.R., Cookson B.T., Logan S.M., Aderem A. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. USA. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Didierlaurent A., Ferrero I., Otten L.A., Dubois B., Reinhardt M., Carlsen H., Blomhoff R., Akira S., Kraehenbuhl J.-P., Sirard J.-C. Flagellin promotes myeloid differentiation factor 88-dependent development of Th2-type response. J. Immunol. 2004;172:6922–6930. doi: 10.4049/jimmunol.172.11.6922. [DOI] [PubMed] [Google Scholar]
- 36.Smits V.A., Hernández-Carralero E., Paz-Cabrera M.C., Cabrera E., Hernández-Reyes Y., Hernández-Fernaud J.R., Gillespie D.A., Salido E., Hernández-Porto M., Freire R. The Nucleocapsid protein triggers the main humoral immune response in COVID-19 patients. Biochem. Biophys. Res. Commun. 2021;543:45–49. doi: 10.1016/j.bbrc.2021.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ewer K.J., Barrett J.R., Belij-Rammerstorfer S., Sharpe H., Makinson R., Morter R., Flaxman A., Wright D., Bellamy D., Bittaye M. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2021;27:270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 38.DiPiazza A.T., Leist S.R., Abiona O.M., Moliva J.I., Werner A., Minai M., Nagata B.M., Bock K.W., Phung E., Schäfer A. COVID-19 vaccine mRNA-1273 elicits a protective immune profile in mice that is not associated with vaccine-enhanced disease upon SARS-CoV-2 challenge. Immunity. 2021;54:1869–1882. doi: 10.1016/j.immuni.2021.06.018. e1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I. MedRxiv; 2020. Safety and Immunogenicity of the Ad26. COV2. S COVID-19 Vaccine Candidate: Interim Results of a Phase 1/2a, Double-Blind, Randomized, Placebo-Controlled Trial. [Google Scholar]
- 40.Shehata M.M., Mahmoud S.H., Tarek M., Al-Karmalawy A.A., Mahmoud A., Mostafa A., Elhefnawi M.M., Ali M.A. In silico and in vivo evaluation of SARS-CoV-2 predicted epitopes-based candidate vaccine. Molecules. 2021;26:6182. doi: 10.3390/molecules26206182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heitmann J.S., Bilich T., Tandler C., Nelde A., Maringer Y., Marconato M., Reusch J., Jäger S., Denk M., Richter M. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature. 2022;601:617–622. doi: 10.1038/s41586-021-04232-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somogyi E., Csiszovszki Z., Molnár L., Lőrincz O., Tóth J., Pattijn S., Schockaert J., Mazy A., Miklós I., Pántya K. A peptide vaccine candidate tailored to individuals' genetics mimics the multi-targeted T cell immunity of COVID-19 convalescent subjects. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.684152. [DOI] [PMC free article] [PubMed] [Google Scholar]