Abstract
Four proteins, PomA, PomB, MotX, and MotY, appear to be involved in force generation of the sodium-driven polar flagella of Vibrio alginolyticus. Among these, PomA and PomB seem to be associated and to form a sodium channel. By using antipeptide antibodies against PomA or PomB, we carried out immunoprecipitation to verify whether these proteins form a complex and examined the in vivo stabilities of PomA and PomB. As a result, we could demonstrate that PomA and PomB functionally interact with each other.
Many bacteria swim by rotating flagellar filaments, which are powered by rotary motors. The flagellar filament is attached via a flexible coupling to the basal body. Multiple torque-generating units exist in the cytoplasmic membrane surrounding the basal body and function independently (1). Energy to drive the motor comes from the transmembrane electrochemical potential of specific ions. For some bacteria, such as Escherichia coli and Bacillus subtilis, the flagellar motor is driven by the proton motive force, whereas alkaliphilic Bacillus and marine Vibrio species are driven by the sodium motive force (11, 16).
E. coli has two motor proteins, MotA and MotB, which are essential for rotating the proton-driven flagellar motor. MotA and MotB are localized in the cytoplasmic membrane and have four and one transmembrane segment(s), respectively (8, 22). From genetic and biochemical studies, it has been suggested that MotA and MotB form a complex via transmembrane regions and function as a proton channel (6, 21, 23, 24). The C terminus of MotB, which has a peptidoglycan binding motif, is thought to bind to peptidoglycan to make MotA–MotB complexes function as the stator (7, 9). Vibrio alginolyticus has two types of flagellar motor in the same cell: sodium driven (polar flagella) and proton driven (lateral flagella) (4, 12). Four genes, pomA, pomB, motX, and motY, have been shown to be essential for rotation of the sodium-driven polar flagella (3, 17, 18, 20). PomA and PomB exhibit about 20 to 30% similarity to the proton-driven motor proteins, MotA and MotB, respectively. Thus, it is plausible that PomA and PomB have roles similar to those of MotA and MotB (3). In this study, we prepared antipeptide antibodies against PomA and PomB and investigated the functional interaction between expressed PomA and PomB proteins.
Strains and plasmids.
Strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| V. alginolyticus strains | ||
| VIO5 | laf (Pof+ Laf−) | 20 |
| NMB155 | laf (Pofm Laf−) | S. Kojima |
| NMB191b | ΔpomAB mutant of VIO5 | This work |
| Plasmids | ||
| pSU41 | kan (Kmr) Plac lacZα | 5 |
| pYA301 | pSU41 Plac pomA | 14 |
| pYA303 | pSU41 Plac pomA pomB | 14 |
| pSK603 | pSU41 Plac pomB | 14 |
Kmr, kanamycin resistant; Plac, lac promoter; Pof, polar flagella; Laf, lateral flagella; Pofm, multipolar flagella.
The pomAB deletion mutant was constructed by a procedure similar to that described elsewhere (2). pYA303 was digested with DraI (inside the pomA gene) and HpaI (inside the pomB gene) and ligated. The resulting plasmid (pYA303 ΔpomAB) contains codons 1 to 68 of pomA fused to codons 126 to 301 of pomB. An XbaI-SacI fragment of pYA303 ΔpomAB was moved to a suicide vector, pKY704. The deletion was introduced into strain VIO5.
Detection of PomA and PomB by antipeptide antibody.
Antipeptide antibodies against PomA or PomB, which were called PomA91 or PomB93, respectively, were produced by Biologica (Nagoya, Japan). Another antipeptide antibody against PomA, called PomA1312, was produced by Sawady Technology (Tokyo, Japan) and was affinity purified. Peptide fragments were synthesized to correspond to the amino acid sequence predicted from the DNA sequence of each gene. Those for antibody PomA91 were raised against mixtures of fragment 1 (K126 to P151), fragment 2 (T185 to T212), and fragment 3 (Q226 to K246). Those for antibody PomA1312 were raised against fragment 4 (P231 to E253). Those for anti-PomB antibody were raised against mixtures of fragments 5 (T104 to G128), 6 (K219 to S243), and 7 (S266 to R290).
The reactivities of antibodies PomA91 and PomB93 against cellular proteins were assessed by immunoblotting (Fig. 1). An overnight culture of NMB191 cells in VC medium (3) was diluted 1:50 into VPG medium. At mid-log phase, cells were harvested by centrifugation and suspended at an optical density at 660 nm of 10 in sodium dodecyl sulfate (SDS) loading buffer (50 mM Tris-HCl [pH 6.8], 10% glycerol, 1% SDS, 0.1% bromophenol blue) containing β-mercaptoethanol. The suspension was boiled for 5 min, and then proteins in the samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred to a polyvinylidene difluoride membrane (Millipore Japan, Tokyo) by using a semidry blotting apparatus (Biocraft, Tokyo, Japan). PomA91 or PomB93 antisera were used for the primary antibody, and an alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) was used as the secondary antibody. Detection was performed as described previously (19). The plasmids were introduced into cells by electroporation as described previously (13).
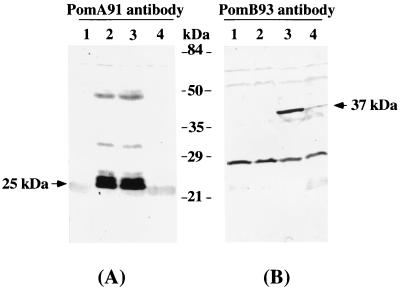
FIG. 1.
Detection of proteins by immunoblotting with antipeptide antibodies against PomA (A) or PomB (B). Lanes 1, pSU41; lanes 2, pYA301; lanes 3, pYA303; lanes 4, pSK603. Molecular mass markers are indicated.
With antibody PomA91, a protein with a molecular mass of 25 kDa was specifically detected in cells expressing the pomA gene from a plasmid. Antibody PomB93 specifically recognized a protein of 37 kDa in cells harboring a plasmid carrying pomB. The molecular masses of 25 kDa and 37 kDa correspond to the predicted molecular masses of PomA (27,224 Da) and PomB (35,461 Da), respectively. The appearance of those bands was correlated to the presence of the pomA or pomB gene on the plasmid. Both proteins were detected in a membrane fraction (data not shown).
In this detection system, neither PomA nor PomB was detected in cells expressing at wild-type levels. However, two proteins with masses of 25 and 37 kDa could be immunoprecipitated from lysates of 35S-labeled wild-type cells (Fig. 2). For the immunoprecipitation assay, cells of strains VIO5 (wild-type polar flagella and NMB155 multipolar flagella) were cultured overnight in VC medium and inoculated 1:50 in synthetic medium (25). At mid-log phase, Tran35S-label (ICN Biomedicals Inc., Costa Mesa, Calif.) was added to 100 μCi/ml; then the mixture was incubated at 30°C for 30 min. The radioactively labeled cells were harvested by centrifugation and lysed at 4°C for 30 min with 1 ml of TNET buffer (50 mM Tris-HCl [pH 7.8], 0.15 M NaCl, 5 mM EDTA, 1% Triton X-100); then the lysate was centrifuged at 10,000 × g for 30 min. The labeled proteins were immunoprecipitated with either antibody PomA1312 or antibody PomB93 by a method described previously (10). The resulting precipitates were subjected to SDS-PAGE followed by fluorography.
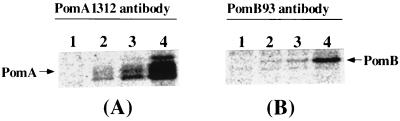
FIG. 2.
Immunoprecipitation assays with PomA1312 (A) and PomB93 (B). Lanes 1, NMB191 transformed with pSU41; lanes 2, VIO5; lanes 3, NMB155; lanes 4, NMB191 transformed with pYA303.
In both VIO5 and NMB155 cells, the 25-kDa PomA protein or the 37-kDa PomB protein was immunoprecipitated. The 25- and 37-kDa bands were not detected in a ΔpomAB mutant strain. These results indicate that PomA and PomB can be specifically detected by the antibodies prepared in this study. Other bands in addition to the PomA and PomB bands were also detected by the antibodies. A 45-kDa band was always present in cells overexpressing PomA. This band might represent the different conformations of PomA or an SDS-resistant complex of PomA with itself or another protein.
Stabilities of PomA and PomB proteins in vivo.
MotA and MotB proteins of E. coli are thought to be associated in a complex (21, 23, 24). It has been shown that MotB is unstable unless excess MotA is expressed together in the same cell (26). The PomA and PomB proteins might show features similar to those of MotA and MotB. In fact, the amount of PomB was much smaller when it was expressed alone than when PomA and PomB were expressed simultaneously (Fig. 2, lanes 3 and 4).
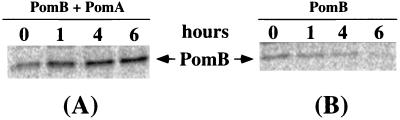
To examine the stabilities of PomA and PomB, NMB191 cells harboring the pomA gene and/or the pomB gene were cultured for 3, 6, 12, or 24 h and the amount of expressed PomA or PomB in the cells was analyzed by immunoblotting (Fig. 3). The intensity of the PomA band did not change during the entire period of the experiment, whether PomB was present or not (Fig. 3A). In contrast, PomB was stable when coexpressed with PomA but decreased in amount, and disappeared completely after 12 h, when expressed by itself (Fig. 3B). Pulse-chase analysis (Fig. 4) supported the PomA-dependent stabilization of PomB, and the half-time for disappearance of PomB was calculated to be about 4.5 h. These results suggest that PomA and PomB functionally interact with each other. They show that PomA is quite stable whether expressed by itself or together with PomB. PomB is slowly degraded in the absence of PomA, however, which indicates that simultaneous synthesis of PomA may facilitate the overproduction of PomB. This result is similar to that observed with the MotA and MotB proteins of E. coli (26). These facts support the idea that PomA and PomB may interact with each other in the sodium-driven motor, as MotA and MotB do in the proton-driven motor.
FIG. 3.
Stabilities of PomA and PomB. Overnight cultures of NMB191 cells with the pomA and/or pomB gene on a plasmid were inoculated at 1:50, and cells were harvested after 3, 6, 12, and 24 h. PomA or PomB was detected by immunoblotting with antibody PomA91 (A) or antibody PomB93 (B), respectively, as described in the text. Numbers below the lanes correspond to the times when cells were harvested.
FIG. 4.
Pulse-chase analysis of PomB. NMB191 cells harboring plasmids carrying pomA plus pomB (pYA303) (A) or pomB alone (pSK603) (B) were cultured in synthetic medium. At mid-log phase, Tran35S-label was added to 100 μCi/ml, and the mixture was incubated at 30°C for 30 min (pulse). Excess unlabeled methionine and cysteine were added to stop the labeling (chase at time zero). At the times indicated above the lanes, the cells were harvested by centrifugation, suspended in 1 ml of TNET buffer, and then immunoprecipitated with antibody PomB93, followed by SDS-PAGE and fluorography, as described in the text.
Interaction of PomA and PomB.
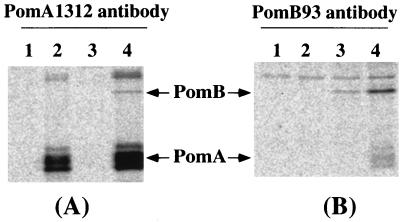
To test for physical interaction between PomA and PomB, we carried out immunoprecipitation assays (Fig. 5). Radiolabeled whole-cell lysate was incubated with antibody PomA1312, and PomA was immunoprecipitated; then the protein associated with PomA was analyzed by SDS-PAGE followed by fluorography. In addition to the 25-kDa band due to PomA, a 37-kDa band corresponding to PomB was also observed. The 37-kDa band was seen only in cells harboring pomA and pomB genes (Fig. 5A), which suggests that PomA forms a complex with PomB. Similar results were obtained with antibody PomB93. A 25-kDa protein was specifically coprecipitated along with PomB when the pomA gene was present (Fig. 5B). These results indicate that PomA and PomB physically interact with each other.
FIG. 5.
Coprecipitation of PomA and PomB. NMB191 cells transformed with the plasmid were labeled with Tran35S-label for 30 min and lysed, and the samples were immunoprecipitated either with antibody PomA1312 (A) or with antibody PomB93 (B) as described in the text. Samples were analyzed by SDS-PAGE followed by fluorography. The transformed plasmids were as follows: lanes 1, pSU41; lanes 2, pYA301; lanes 3, pSK603; lanes 4, pYA303.
Amiloride and its analogs, which are inhibitors of Na+ channels, Na+/H+ exchangers, and Na+/Ca+ exchangers, specifically inhibit the Na+-driven flagellar motor. Phenamil is the most potent inhibitor of the Na+ motor. Previously, we isolated mutants whose motility is resistant to phenamil (Mpar) and mapped the Mpar mutations to specific sites in pomA and pomB (14, 15). Further mutational analysis suggested that a certain structural change around these sites affects the sensitivity of the motor to phenamil. When mutations were present in both PomA and PomB, motility was less sensitive to phenamil than with either mutation alone. These findings also support the proposal that PomA and PomB interact with each other.
MotA and MotB are required for the rotation of proton-driven flagella, while sodium-driven flagella require four components: PomA, PomB, MotX, and MotY (3, 17, 18, 20). The motX and motY genes, whose homologs are not found in the E. coli genome, are located apart from each other on the Vibrio chromosome. On the other hand, pomA and pomB are in the same operon. It has been proposed that MotX is a sodium channel component and MotY functions to connect the stator to peptidoglycan (17, 18). Thus, we might speculate that the sodium-driven motor contains a complex containing four proteins rather than only two as in the proton-driven motor. Further analysis will be required to determine the functions of MotX and MotY.
Acknowledgments
We thank R. Elizabeth Sockett and David F. Blair for critically reading the manuscript and S. Kojima of our laboratory for providing us with the Pofm strain.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan (to I.K. and M.H.) and from the Japan Society for the Promotion of Science (to Y.A.).
REFERENCES
- 1.Aizawa S-I. Flagellar assembly in Salmonella typhimurium. Mol Microbiol. 1996;19:1–5. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- 2.Asai, Y., I. Kawagishi, E. Sockett, and M. Homma. Hybrid motor with the H+- and Na+-driven components can rotate Vibrio polar flagella using sodium ions. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 3.Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I, Homma M. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature (London) 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 5.Bartolomé B, Jubete Y, Martínez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 6.Blair D F, Berg H C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 7.Chun S Y, Parkinson J S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- 8.Dean G D, Macnab R M, Stader J, Matsumura P, Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984;159:991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 10.Homma M, Kutsukake K, Iino T. Structural genes for flagellar hook-associated proteins in Salmonella typhimurium. J Bacteriol. 1985;163:464–471. doi: 10.1128/jb.163.2.464-471.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imae Y, Atsumi T. Na+-driven bacterial flagellar motors. J Bioenerg Biomembr. 1989;21:705–716. doi: 10.1007/BF00762688. [DOI] [PubMed] [Google Scholar]
- 12.Kawagishi I, Maekawa Y, Atsumi T, Homma M, Imae Y. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J Bacteriol. 1995;177:5158–5160. doi: 10.1128/jb.177.17.5158-5160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawagishi I, Okunishi I, Homma M, Imae Y. Removal of the periplasmic DNase before electroporation enhances efficiency of transformation in a marine bacterium, Vibrio alginolyticus. Microbiology. 1994;140:2355–2361. [Google Scholar]
- 14.Kojima S, Asai Y, Atsumi T, Kawagishi I, Homma M. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations of putative channel components. J Mol Biol. 1999;285:1537–1547. doi: 10.1006/jmbi.1998.2377. [DOI] [PubMed] [Google Scholar]
- 15.Kojima S, Atsumi T, Muramoto K, Kudo S, Kawagishi I, Homma M. Vibrio alginolyticus mutants resistant to phenamil, a specific inhibitor of the sodium-driven flagellar motor. J Mol Biol. 1997;265:310–318. doi: 10.1006/jmbi.1996.0732. [DOI] [PubMed] [Google Scholar]
- 16.Manson M D, Tedesco P, Berg H C, Harold F M, van der Drift C. A proton motive force drives bacterial flagella. Proc Natl Acad Sci USA. 1977;74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarter L L. MotX, the channel component of the sodium-type flagellar motor. J Bacteriol. 1994;176:5988–5998. doi: 10.1128/jb.176.19.5988-5998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarter L L. MotY, a component of the sodium-type flagellar motor. J Bacteriol. 1994;176:4219–4225. doi: 10.1128/jb.176.14.4219-4225.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okumura H, Nishiyama S-I, Sasaki A, Homma M, Kawagishi I. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the methyl-accepting chemoreceptors. J Bacteriol. 1998;180:1862–1868. doi: 10.1128/jb.180.7.1862-1868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp L L, Zhou J D, Blair D F. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry. 1995;34:9166–9171. doi: 10.1021/bi00028a028. [DOI] [PubMed] [Google Scholar]
- 22.Stader J, Matsumura P, Vacante D, Dean G E, Macnab R M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986;166:244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stolz B, Berg H C. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J Bacteriol. 1991;173:7033–7037. doi: 10.1128/jb.173.21.7033-7037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang H, Braun T F, Blair D F. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 25.Tokuda H, Nakamura T, Unemoto T. Potassium ion is required for the generation of pH-dependent membrane potential and delta pH by the marine bacterium Vibrio alginolyticus. Biochemistry. 1981;20:4198–4203. doi: 10.1021/bi00517a038. [DOI] [PubMed] [Google Scholar]
- 26.Wilson M L, Macnab R M. Co-overproduction and localization of the Escherichia coli motility proteins MotA and MotB. J Bacteriol. 1990;172:3932–3939. doi: 10.1128/jb.172.7.3932-3939.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]