ABSTRACT
The continuous emergence of novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants poses new challenges in the fight against the coronavirus disease 2019 (COVID-19) pandemic. The newly emerging Omicron strain caused serious immune escape and raised unprecedented concern all over the world. The development of an antibody targeting a conserved and universal epitope is urgently needed. A subset of neutralizing antibodies (NAbs) against COVID-19 from convalescent patients were isolated in our previous study. In this study, we investigated the accommodation of these NAbs to SARS-CoV-2 variants of concern (VOCs), revealing that IgG 553-49 neutralizes pseudovirus of the SARS-CoV-2 Omicron variant. In addition, we determined the cryo-electron microscopy (cryo-EM) structure of the SARS-CoV-2 spike (S) protein complexed with three monoclonal antibodies targeting different epitopes, including 553-49, 553-15, and 553-60. Notably, 553-49 targets a novel conserved epitope and neutralizes the virus by disassembling S trimers. IgG 553-15, an antibody that neutralizes all of the VOCs except Omicron, cross-links two S trimers to form a trimer dimer, demonstrating that 553-15 neutralizes the virus by steric hindrance and virion aggregation. These findings suggest the potential to develop 553-49 and other antibodies targeting this highly conserved epitope as promising therapeutic reagents for COVID-19.
IMPORTANCE The emergence of the Omicron strain of SARS-CoV-2 caused higher immune escape, raising unprecedented concerns about the effectiveness of antibody therapies and vaccines. In this study, we identified a SARS-CoV-2 neutralizing antibody, 553-49, which neutralizes all variants by targeting a completely conserved novel epitope. In addition, we revealed that IgG 553-15 neutralizes SARS-CoV-2 by cross-linking virions and that 553-60 functions by blocking receptor binding. Comparison of different receptor binding domain (RBD) epitopes revealed that the 553-49 epitope is hidden in the S trimer and keeps a high degree of conservation during SARS-CoV-2 evolution, making 553-49 a promising therapeutic reagent against the emerging Omicron and future variants of SARS-CoV-2.
KEYWORDS: COVID-19, SARS-CoV-2, neutralizing antibody, Omicron variant, NAb
INTRODUCTION
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a serious pandemic, with more than 150 million infections and over 5 million deaths to date (1–3). Coronaviruses are enveloped by a trimeric transmembrane spike (S) glycoprotein that has an essential role in viral entry into the host cell. The S1 subunit of S, consisting of a receptor binding domain (RBD) and an N-terminal domain (NTD), is responsible for receptor binding. The S2 subunit primes the fusion of viral and cellular membranes (4). SARS-CoV-2 S recognizes angiotensin-converting enzyme 2 (ACE2) of the host cell through the receptor binding motif (RBM) on the RBD (5). During infection, the S protein undergoes dynamic conformational change, with the RBD adopting an “up” (or “open”) or “down” (or “closed”) state. Only up RBD can recognize the receptor and initiate the viral entry (4, 6). The S protein has been used as a major target for the development of vaccines and therapeutic neutralizing monoclonal antibodies (MAbs) against COVID-19 (7).
As SARS-CoV-2 escalated to circulate in the human population, many SARS-CoV-2 variants appeared, and five of them have been defined as variants of concern (VOCs), including Alpha (B.1.1.7), Beta (P.1), Gamma (B.1.351), Delta (B.1.617.2), and Omicron (B.1.1.529) (8). The S protein of the Alpha strain bears one substitution (a substitution of Y for N at position 501 [N501Y]) in the RBM, which enhances its transmissibility (9). The Gamma strain contains three substitutions (K417N, E484K, and N501Y) in the RBM, resulting in immune escape (10). The Delta strain bears L452R and E484Q in the RBD domain and a P681R mutation in the furin cleavage site that enhances the fusogenic activity of the S protein and the pathogenicity of the virus (11). The Omicron strain, other than K417N, E484A, N501Y, and P681H, contains another 30 mutations in the spike and escapes almost 85% of existing SARS-CoV-2 neutralizing antibodies (NAbs) (12).
Considering the ongoing SARS-CoV-2 pandemic and constantly emerging variants, antibodies and vaccines targeting conserved and universal epitopes are urgently needed. Antibodies neutralize virus infection through different mechanisms, such as aggregation of virus particles (13), prevention of viral attachment, inhibition of S fusion, or disassembly of the virion. The combination of antibodies with different epitopes and various neutralization mechanisms might be an effective way to fight against the SARS-CoV-2 variants. Our previous study identified a set of antibodies from convalescent patients targeting different epitopes of S, including four RBD-targeting IgGs, 553-15, 553-60, 553-63, and 553-49, and one non-RBD-targeting IgG, 413-2 (14). In this study, we investigated the neutralization activities of these NAbs against SARS-CoV-2 VOCs. Binding assays and pseudovirus-based neutralization assays revealed that IgG 553-60 lost its neutralizing ability against all of the VOCs except Alpha. IgG 553-15 neutralized all of the VOCs except Omicron. Only 553-49 neutralized the SARS-CoV-2 Omicron variant. In addition, cryo-electron microscopy (cryo-EM) structural studies of Omicron S in complex with 553-49 and wild-type (WT) S in complex with 553-15 and 553-60 revealed their specific epitopes and neutralization mechanisms. Notably, IgG 553-49 disassembled Omicron S trimers by targeting a completely conserved novel epitope that is buried inside the trimer and exposed only when the trimers are disassembled. The highly conserved epitope makes 553-49 and other antibodies targeting this epitope ideal therapeutic reagents to fight against the emerging and future variants of SARS-CoV-2.
RESULTS
Broad neutralizing activity of IgG 553-49.
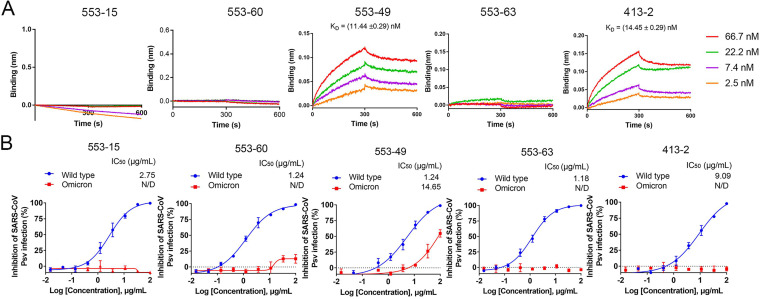
We first used biolayer interferometry (BLI) to investigate the affinities between the S proteins of different SARS-CoV-2 variants (WT, Alpha, Beta, Gamma, Delta, and Omicron) and the five IgGs, 553-15, 553-60, 553-63, 553-49, and 413-2, to determine whether these monoclonal antibodies accommodated the emerged variants. The Alpha S could bind to all five antibodies, while the Beta and Gamma S proteins were not able to bind with 553-60 and 553-63 (Fig. S1 in the supplemental material). IgG 553-15 retained high binding ability to the S proteins of Alpha, Beta, Gamma, and Delta, at 5.12, 4.03, 10.14, and 1.65 nM, respectively, but could not bind Omicron S (Fig. S1). Fortunately, 553-49 maintained its binding affinity to the S proteins of all five VOCs, at 1.89, 2.58, 5.87, 40.05, and 3.43 nM, respectively (Fig. 1 and Fig. S1).
FIG 1.
Accommodation of monoclonal antibodies to SARS-CoV-2 Omicron variant. (A) BLI sensorgrams and kinetics of monoclonal antibodies binding to SARS-CoV-2 Omicron spike protein. (B) Neutralizing activities of IgGs (553-15, 553-60, 553-49, 553-63, and 413-2) against SARS-CoV-2 WT and Omicron pseudoviruses (Psv). Data are shown as mean values ± standard deviations. N/D, not detected.
We next assayed the neutralizing activities of these five antibodies against SARS-CoV-2 variants and SARS-CoV pseudoviruses. They neutralized WT SARS-CoV-2 pseudoviruses with 50% inhibition concentration (IC50) values of 2.94, 0.79, 0.11, 13.91, and 10.82 μg/mL, respectively (Fig. S1). The neutralization activities of 553-60 and 553-63 against SARS-CoV-2 Beta and Gamma strains were completely abolished, which was consistent with the results of BLI (Fig. S1). The Delta strain evaded the neutralization activity mediated by 553-60, 553-49, 553-63, and 413-2, although its S bound tightly to these antibodies (Fig. S1), which was consistent with the finding that Delta pseudovirus was relatively resistant to neutralizing antibodies (15) because the P681R mutation enhanced the fusogenic activity of the S protein. As for the Omicron strain, 553-49 and 413-2 maintained their binding activities; however, only 553-49 was able to neutralize the pseudovirus, with an IC50 of 11.44 ng/mL (Fig. 1).
Thus, among the 5 antibodies targeting different epitopes of S, only IgG 553-49 retained affinity with Omicron S and neutralization activity against the Omicron strain.
IgG 553-49 targets conserved epitopes and induces the S trimer to disassemble.
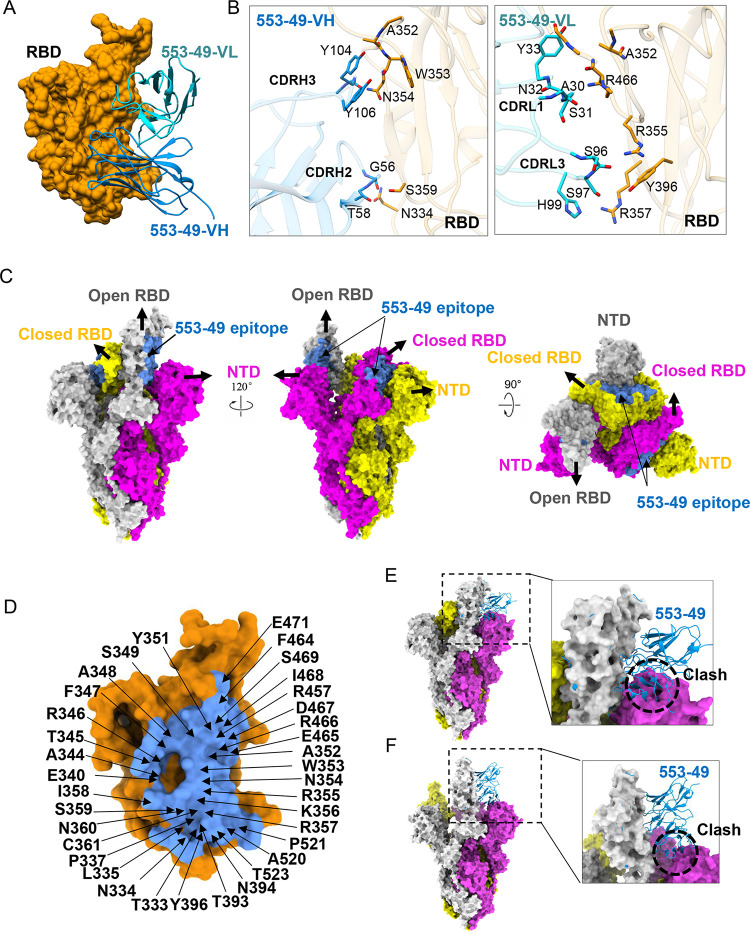
To understand the broad neutralization mechanism of 553-49, we used cryo-EM to determine the structure of IgG 553-49 complexed with the stabilized prefusion ectodomain of SARS-CoV2 Omicron S. The purified Omicron S trimer was mixed with 553-49 at a 1:1.5 molar ratio, incubated at 4°C for 2 h, and further purified by gel filtration. The peak fraction of the gel filtration was used for cryo-EM data collection. Two states of particles were observed, the Omicron S trimer in the apo state (apo-OS) and the Omicron S monomer complexed with 553-49 (OS–553-49) (Fig. S2, S3, and S4). Thus, the Omicron S trimers were disassembled into monomers upon 553-49 binding. The cryo-EM structure of the Omicron S trimer was determined to a resolution of 3.40 Å (Fig. 2C and Fig. S3 and S4). For the OS–553-49 complex, due to the flexibility of the S monomer, only the S RBD and 553-49 Fab region was locally refined to 4.06-Å resolution (Fig. 2A and Fig. S3 and S4), sufficient for model building of RBD and 553-49 Fab.
FIG 2.
Cryo-EM structure of SARS-CoV-2 Omicron S in complex with IgG 553-49. (A) Structure of Omicron S RBD–553-49. The RBD is displayed in orange surface mode. The heavy chain and light chain of 553-49 are shown as ribbons colored in cornflower blue and cyan, respectively. (B) The interfaces between the RBD and 553-49. (C) Molecular-surface representation of the apo-Omicron S model displayed in side and top views. The 553-49 epitope (colored in cornflower blue) on the RBD was covered by the NTD of the adjacent protomer. (D) Close-up view of the 553-49 epitope on the RBD. The residues involved in the interaction are labeled. (E, F) Binding of 553-49 to a spike trimer with one up RBD (E) or a wide-open RBD (PDB 7WHK) (F) would clash with the NTD of the adjacent protomer (indicated by the black dashed circles).
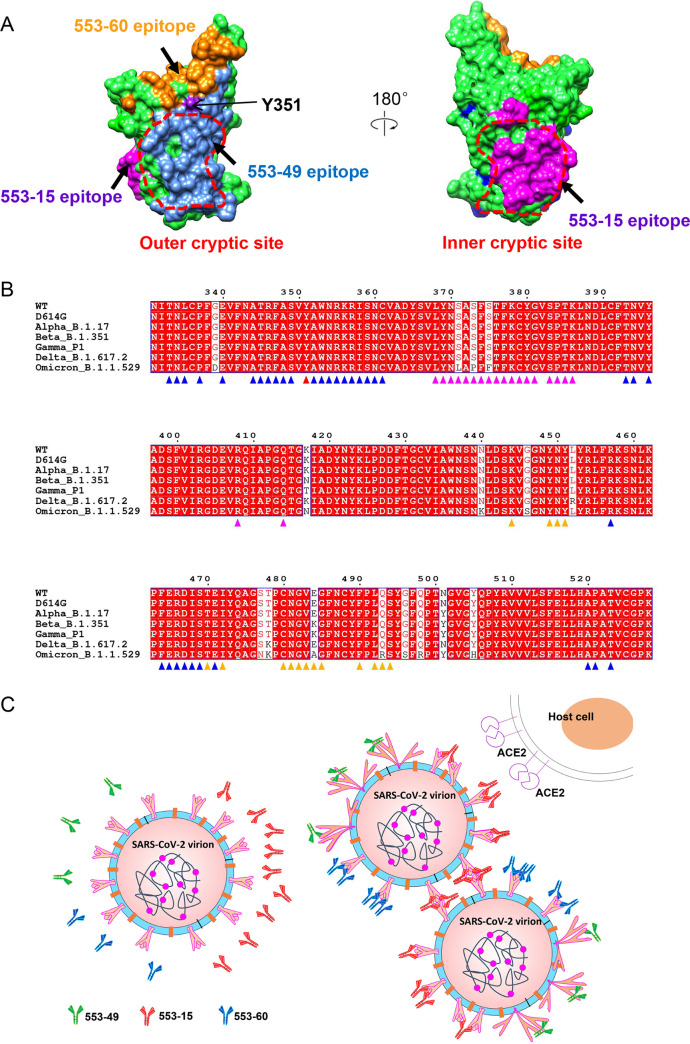
IgG 553-49 buried a surface area of 1,076 Å2 on the RBD (Fig. 2D). The binding of 553-49 with RBD was mainly mediated by complementarity-determining regions (CDRs) CDRH2, CDRH3, CDRL1, and CDRL3 (Fig. 2A and B). Residues N334, A352, W353, N354, R355, R357, S359, Y396, and R466 of the RBD showed direct contacts with 553-49 (Fig. 2B and Fig. S3E, S3F). In total, 36 residues of the RBD might be involved in the interaction, and they are completely conserved among all VOCs (Fig. 2D, and see Fig. 6A and B), revealing an extremely conservative epitope. In the S trimer, this epitope was covered by the NTD of the neighboring protomer no matter whether RBD adopted an up or down conformation (Fig. 2C). Structural alignments of the OS–553-49 complex with apo-Omicron S and with the wide-open RBD of Omicron S induced by a nanobody binding (16) both indicated that 553-49 would clash with the NTD of the adjacent protomer (Fig. 2E and F). The binding of 553-49 triggered dramatic movement of the RBD and NTD, resulting in disassembly of the S trimer.
FIG 6.
Epitopes and neutralization mechanisms of three NAbs. (A) Comparison of 553-49, 553-15, and 553-60 epitopes on the S RBD. The RBD is displayed in green surface representation. The 553-49, 553-15, and 553-60 epitopes are colored cornflower blue, magenta, and orange, respectively. The inner and outer cryptic sites are indicated by red dotted lines. (B) Sequence alignment of S RBDs of SARS-CoV-2 WT and all VOCs, showing that IgG 553-49 targets to a completely conserved epitope. Conserved amino acids are highlighted in red. Residues involved in 553-49, 553-15, and 553-60 interactions are marked with triangles in blue, magenta, and orange, respectively. Residues involved in both 553-49 and 553-60 binding are marked with triangles in red. (C) Schematic model of the neutralization mechanisms of 553-49, 553-15, and 553-60.
IgG 553-15 induces the formation of dimers of trimers.
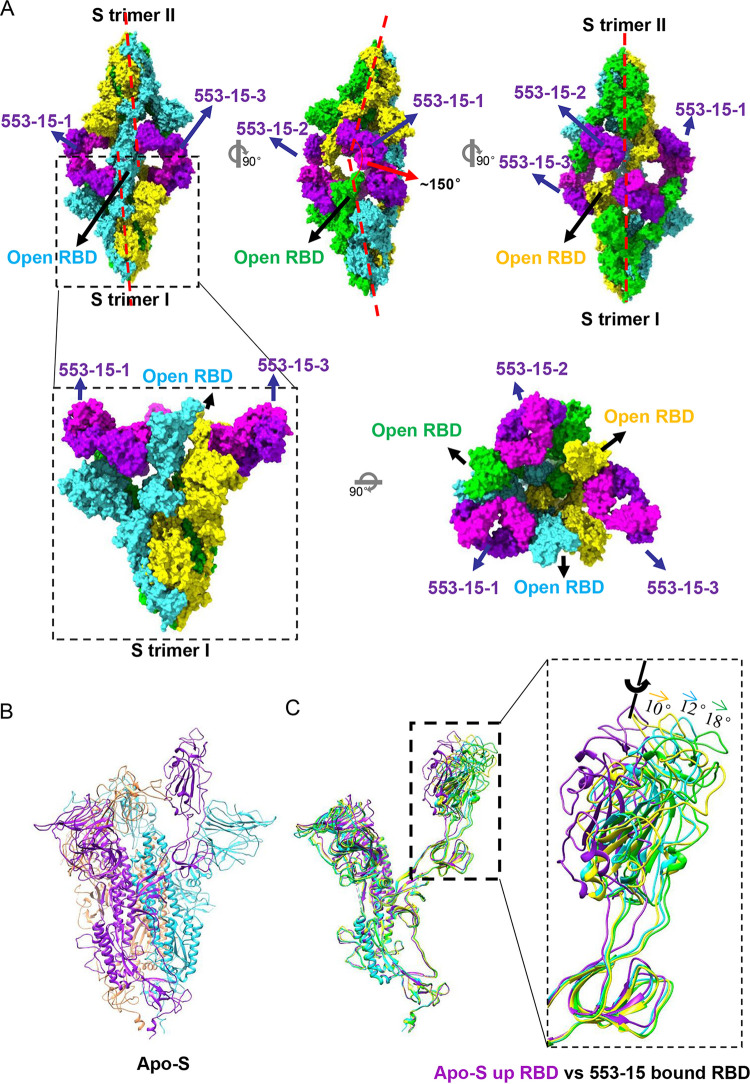
To characterize the cross-neutralization mechanism of 553-15, we determined the cryo-EM structures of the stabilized prefusion SARS-CoV-2 S(D614G) ectodomain trimer (2.7 Å) and its complex with 553-15. Negative-stain micrographs showed that incubation of IgG 553-15 with S cross-linked two S trimers (Fig. S7). Further cryo-EM study unveiled a previously uncharacterized structure of S complexed with 553-15 in which three IgG molecules cross-linked two S trimers, forming an unsymmetrical head-to-head dimer of trimers (Fig. 3 and Fig. S8 and S9). The cryo-EM structure of the whole complex was determined to a resolution of 4.47 Å (Fig. S8 and S9 and Movie S1). To improve the resolution, a single trimer was locally refined to 3.45-Å resolution and three RBD Fab regions were locally refined to resolutions of 3.80, 3.89, and 4.11 Å, respectively (Fig. S8 and S9). The Fc region is missing in the final reconstruction because of its flexibility, although weak density can be observed in the cryo-EM map (Fig. S10).
FIG 3.
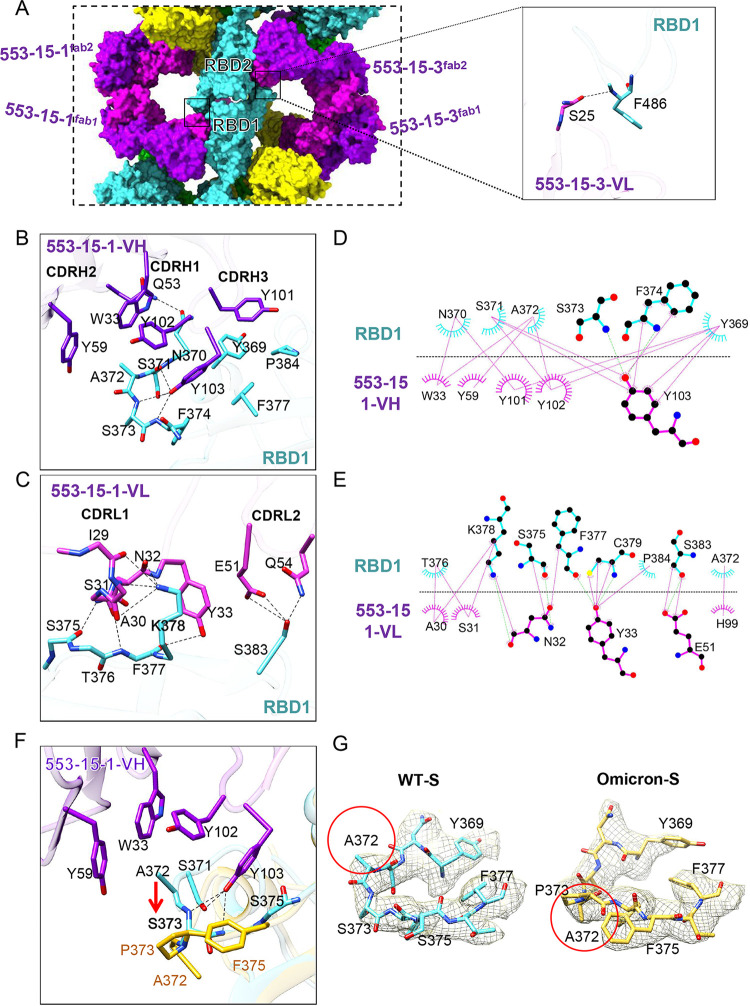
Cryo-EM structure of SARS-CoV-2(D614G) S in complex with IgG 553-15. (A) Molecular-surface representation of S–553-15 complex. The dimer was formed by two S trimers with different pseudo-3-fold rotational symmetry axes (red dashed lines). All RBDs are open. (B) Model of the apo S(D614G) trimer (with one up RBD) displayed in ribbon mode. (C) Comparison of 553-15-bound S protomers with the up S protomer of the apo-S trimer (purple). IgG 553-15 induced all RBDs to the open state with different angles.
Binding of IgG 553-15 induced all RBDs of S trimers to the open state (Fig. 3), although the apo-S trimer alone used in this study adopted one up RBD and two down RBDs (Fig. 3B and Fig. S5 and S6). Compared with the up RBD in the unliganded S trimers, the three RBDs were rotated ~18°, ~12°, and ~10°, respectively (Fig. 3C and Fig. S8E and F). Thus, the three up RBDs of the S trimer did not follow 3-fold symmetry (Fig. 3A). Each trimer had its own pseudo-3-fold axis with two axes forming an angle of ~150°. Despite the different RBD conformations, all three 553-15 Fabs interacted with RBDs in similar conformations (Fig. S8F). The interaction was focused on the inner surface of the RBD, without much overlap with the RBM region (Fig. S11A and E). The binding of the 553-15 Fab with RBD buried 787 Å2 of surface area. A total of 21 residues from the RBD were involved in the interaction, with 9 residues binding to the heavy chain and 17 residues to the light chain (Fig. 4B, 4C, 4D and 4E). All three CDRs of the heavy chain participated in the binding through hydrophobic and hydrophilic interactions. Extensive hydrophobic contacts were mainly formed between the CDRH3 loop (101-YYY-103) and residues Y369, N370, S371, A372, F374, F377, and P384 of the RBD (Fig. 4B, 4D and Fig. S8F). Residue A372 inserted into the hydrophobic cavity consisting of Y102 of CDRH3, W33 of CDRH1, and Y59 of CDRH2, forming intensive hydrophobic contacts (Fig. 4B, 4D, and Fig. S8F). Four pairs of hydrogen bonds were formed between Y103 of CDRH3, Q53 of CDRH2, and the loop (370-NSAS-373) of the RBD (Fig. 4B, 4D and Fig. S8F).
FIG 4.
The interaction between 553-15 and the SARS-CoV-2(D614G) S RBD. (A) Close-up-view of two opposite RBD regions (left). The main chain of RBD1 F486 forms a hydrogen bond (dashed line) with S25 from the opposite 553-15-3 CDRL1 (right), introducing an extra contact. (B) Three-dimensional display showing the key interactions between the 553-15 heavy chain (purple) and Omicron S RBD (cyan). Hydrogen bonds are represented by dashed lines. (C) Three-dimensional display showing the key interactions between the 553-15 light chain (pink) and Omicron S RBD (cyan). (D, E) Two-dimensional representations showing the interactions between RBD1 and the 553-15-1 heavy chain (D) and the 553-15-1 light chain (E). Hydrogen bonds are shown as light green dashed lines. The interactions were analyzed by LigPlot (38). (F) Superimposition of the Omicron S RBD (yellow) with the 553-15 (purple)-bound WT S RBD (cyan), showing the conformational change of residues 372 to 375 caused by the S373P mutation (red arrow). (G) Density maps of residues 369 to 377 of the WT and Omicron S RBDs (PDB 7WOP) shown in the same view. The directions of residue A372 are different (red circles).
The binding of the 553-15 light chain to the RBD was mainly mediated by hydrogen bonds between CDRL1 (29-IASNY-33) and the β sheet (375-STFK-379) of the RBD, as well as E51, Q54 of CDRL2, and S383 of the RBD (Fig. 4C, 4E). Interestingly, the two Fabs of IgG 553-15-1 (553-15-1Fab1 and 553-15-1Fab2) interacted simultaneously with two RBDs from opposite trimers, introducing additional interactions between 553-15-1 and the opposite RBD by forming a hydrogen bond between S25 and the main chain of F486 (Fig. 4A). Thus, these two extra contacts contributed to the stabilization of the dimer.
Based on the structure of the complex, the failure of 553-15 to neutralize the Omicron variant was mainly due to the S373P mutation, which destroyed the hydrophobic contacts mediated by A372 of the RBD and the three pairs of hydrogen bonds mediated by Y103 of the VH region (Fig. 4F and G).
IgG 553-15 neutralizes the virus by aggregating virions and blocking receptor attachment.
Although 553-15 targets the non-RBM region, the previous study showed that 553-15 was able to compete with ACE2 for binding with membrane-bound S protein (14). Structure superimposition of RBD-ACE2 and the S–553-15 complex indicated that 553-15 would clash with an ACE2 glycosylated residue (N322) (Fig. S11A, S11B, S11C). It has been reported that a nanobody that cross-links SARS-CoV-2 trimers into dimers of trimers could induce the aggregation of virions (17). Thus, IgG cross-linking might contribute to the neutralization by 553-15, as well as the antibody targeting to this epitope, which is absent of ACE2 blocking ability.
IgG 553-60 epitope overlaps the RBM region.
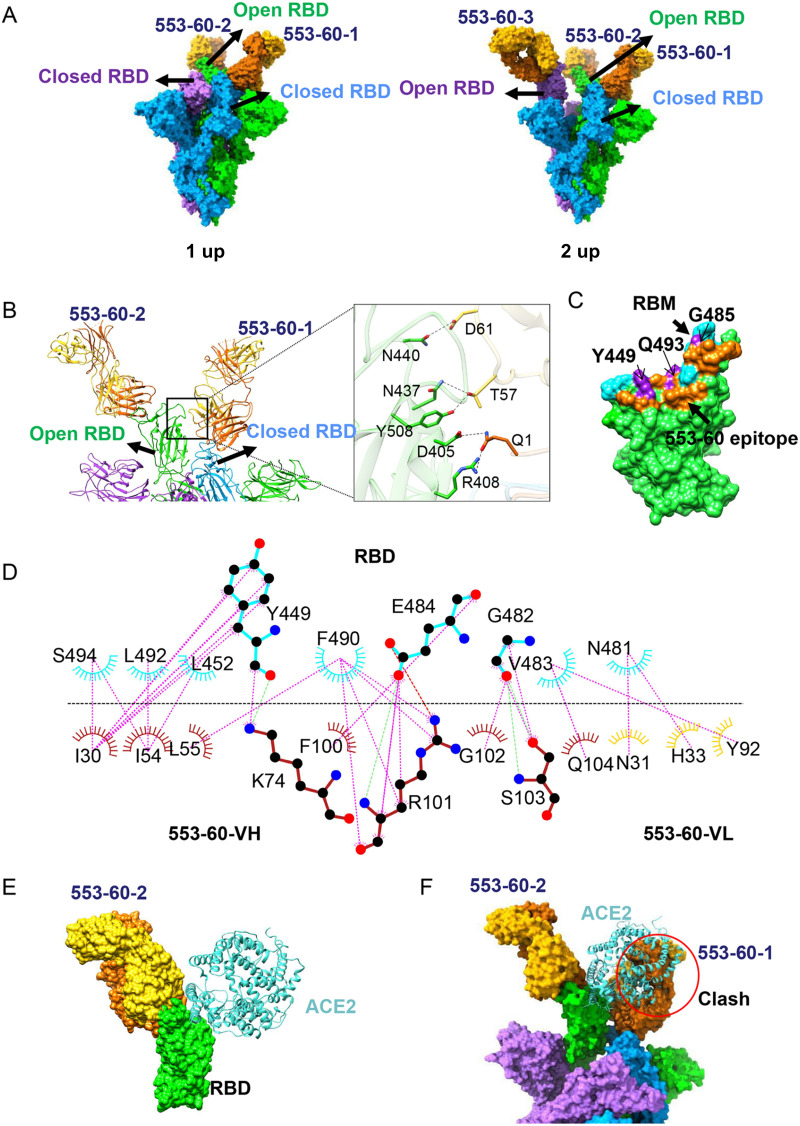
The previous study showed that 553-60 targeted two epitopes of S. To identify its precise epitopes, we studied the cryo-EM structure of IgG 553-60 complexed with the stabilized SARS-CoV-2 S ectodomain. Two distinct conformational states were observed (Fig. 5A), of which one had one RBD in the open state (1-up) and the other had two RBDs in the open state (2-up). Both structures were determined to 3.25 Å (Fig. S12 and S13). In 1-up state, two 553-60 molecules were bound to the S trimer, one on the up RBD (named 60-up) and the other on the down RBD (named 60-down), which is next to the up RBD in the clockwise direction. In the 2-up state, each RBD bound to a 553-60 molecule, regardless of up or down conformation (Fig. 5A).
FIG 5.
Cryo-EM structure of the D614G S in complex with IgG 553-60. (A) Molecular-surface representation of S–553-60 structures in two states, with 1 up RBD and 2 up RBDs, respectively. (B) Ribbon and stick representations of two neighboring up and down RBDs, each interacting with one 553-60 Fab. Fab 553-60-1 interacts with both up and down RBDs, introducing additional contacts. (C) Surface representation of RBD showing the buried binding site, including 553-60 (orange), RBM (cyan), and 553-60 epitope overlaps with the RBM region (purple). (D) Two-dimensional representations showing the interaction between the RBD (cyan) and 553-60 (heavy chain, orange; light chain, gold). Hydrogen bonds are shown as light dashed lines. The interactions were analyzed by LigPlot (38). (E, F) Superimposition of RBD-553-60-2 with RBD-ACE2 (PDB 6LZG) complex structures revealed a minor clash between ACE2 and 553-60-2 (E) but a major clash between ACE2 and 553-60-1 Fab which binds to the neighboring down RBD (F).
The epitope of 60-up was near the RBM region and overlapped residues G485, Q493, and Y449, which are involved in ACE2 binding (Fig. 5C). In contrast to 553-15, the 553-60 Fab molecule had a smaller footprint, with the heavy and light chains burying surface areas of 557 Å2 and 101 Å2, respectively. The interaction between 553-60 and the RBD was primarily driven by CDRH1, CDRH2, CDRH3, and CDRL1 through hydrophilic and hydrophobic interactions (Fig. 5D and Fig. S12E). Residues G482, E484, Y449, and N481 of the RBD were involved in the hydrophilic interaction by forming one salt bridge and hydrogen bonds. Hydrophobic contacts that formed between V483, F490, L452, and Y449 from the RBD and Y92, L55, L54, and I30 of 553-60 further enhanced the interaction (Fig. 5D and Fig. S12E).
In addition, the binding of 553-60 to the down RBD introduced additional contacts through interacting with the neighboring up RBD (Fig. 5B and Fig. S12E). Salt bridges and hydrogen bonds were formed between Q1, T57, and D61 from the VH and VL regions and R408, D405, N437, Y508, and N440 of the open RBD (Fig. 5C and Fig. S13E). This additional contact locked two S protomers together and hindered the RBD’s further opening. Superimposition of the up RBD with the ACE2-RBD complex structure revealed a minor clash between ACE2 and 553-60-2 (Fig. 5B). Interestingly, ACE2 clashed with the neighboring 553-60-1 Fab. Thus, the binding of 553-60 with the down RBD not only limited the opening of the RBD but also prevented the up RBD from recognizing and attaching to ACE2. This explains the mechanism of 553-60-mediated neutralization. In addition, the cross-linking of the S trimer could also be observed from the negative staining of the S–553-60 complex (Fig. S7), even though stable dimers of trimers were not observed in the cryo-EM data. Similarly to 553-15, IgG cross-linking might partially contribute to antibody neutralization.
DISCUSSION
The continuous emergence of novel SARS-CoV-2 variants reduced the potency of vaccines and therapeutic antibodies induced by first-generation virus. The development of antibodies targeting conserved epitopes is urgently needed. Here, we identified an outer cryptic conserved epitope of the RBD that has never been reported.
Antibodies neutralize virus infection through different mechanisms (18). Many antibodies occupy the RBM to block receptor attachment (19, 20). Some antibodies lock the S protein in all-RBD-down conformation to prevent receptor attachment and conformational change that is essential for viral infection (18, 21). In addition, antibodies may induce S trimer cross-linking, resulting in steric hindrance or virion aggregation, to neutralize virions (13, 22, 23). The neutralization mechanism of antibody disassembling the trimeric S proteins was rarely reported, due to the instability of the complex. In this study, we observed that IgG 553-49 disassembled recombinant Omicron S trimers into monomers upon binding (Fig. 6C). It has been reported that the bispecific nanobody bn03 induces the RBD to a wide-open state and eventually disassembles the S trimer to neutralize SARS-CoV-2 (16). Although occupying different epitopes, both 553-49 and bn03 would clash with the NTD when aligned to the up RBD of apo-Omicron S. Thus, significant movement of the RBD and NTD might trigger the disassembling of the S trimers on virions.
In addition, we determined the structures of SARS-CoV-2 WT S complexed with IgGs 553-60 and 553-15. IgG 553-60 targets the RBM region of S, which explains why it lost its neutralization ability against all of the VOCs except Alpha, since the RBM is the most variable region in all variants. IgG 553-15 targets the inner cryptic surface of the RBD, partly overlapping with the CR3022 epitope (24). Similar as 553-15, CR3022 exhibits highly resistant activity to Alpha, Beta, Gamma, and Delta but not Omicron. The neutralizing mechanism of antibodies targeting this inner cryptic epitope has remained unclear. For example, the IgG COVA1-16 can neutralize SARS-CoV and SARS-CoV-2 pseudovirus, but the Fab alone cannot (25). Here, we observed that IgG 553-15 cross-linked two S trimers to form dimers, demonstrating its neutralization mechanism of inducing virion aggregation, which may contribute to understanding the neutralization mechanism of COVA1-16 antibody in the absence of ACE2 blocking.
Thus, antibodies 553-49, 553-15, and 553-60 neutralize SARS-CoV-2 by different mechanisms. The epitope of 553-60 partly overlaps the RBM, which is easily exposed on the surface of the virus and easily mutated during viral adaptations. IgG 553-15 targets a relatively conserved inner site of the RBD that is exposed when at least two RBDs are open (24), which enables it to neutralize all of the VOCs except Omicron. The epitope of 553-49 is not exposed on the S trimer surface and keeps a high degree of conservation during the evolution of SARS-CoV-2. Our results provide a new potential therapeutic against SARS-CoV-2 Omicron and highlight a novel conserved epitope on the S RBD that may be used to achieve broad protection against the emerging variants.
MATERIALS AND METHODS
Production of S ectodomains of recombinant SARS-CoV-2(D614G) and variants.
The gene encoding the SARS-CoV-2 S ectodomain (residues 1 to 1208, GenBank accession number MN908947) was synthesized (GenScript) and cloned into the mammalian expression construct pcDNA-3.1, with proline substitutions at residues 986 and 987, a GSAS substitution at the furin cleavage site (residues 682 to 685), a T4 fibritin trimerization motif, an HRV3C protease cleavage site, a Twin-Strep-Tag, and an 8×His tag at the C-terminal end being introduced simultaneously using the multiS one-step cloning kit (Vazyme). Using the SARS-CoV-2 WT plasmid as the template, mutations including D614G, B.1.17 mutations (Del 69H70V, N501Y, P681H, S982A, Del 145Y, A570D, T716I, and D1118H), B.1.351 mutations (K417N, E484K, and N501Y), and P1 mutations (K417T, E484K, and N501Y) were introduced using a multi-site-directed mutagenesis kit (Yeasen).
The SARS-CoV-2 Omicron S ectodomain (human codon) was purchased from GenScript (catalog number: MC_0101274). An expression plasmid of Omicron S with HexaPro mutations (26), a GSAS substitution at the furin cleavage site (residues 682 to 285), a T4 fibritin trimerization motif, a Twin-Strep-Tag, and a C-terminal 8×His tag was constructed in the pcDNA3.1 vector using the multiS one-step cloning kit (Vazyme).
Transient transfection of the expression plasmid in an HEK293F cell suspension was conducted by using polyethyleneimine. After 72 h, the supernatants were harvested and filtered for further purification in 20 mM Tris, pH 8.0, 200 mM NaCl using the HisTrap HP (GE) and the Superose 6 increase 10/300 column (GE Healthcare).
Expression and purification of recombinant antibodies.
A pair of plasmids separately expressing the heavy and light chains of antibodies were transiently cotransfected into HEK293F cells. After 72 h, the supernatants with secreted antibodies were collected and purified by using protein A/G Sepharose (Smart Lifesciences). The bound antibodies on the Sepharose were eluted with 100 mM sodium acetate (pH 3.0), and then the pH was adjusted to 6.0 by adding 1 M Tris (pH 8.5). The purified antibodies were used in the following binding and neutralization analyses.
BLI binding assays.
Biolayer interferometry (BLI) was carried out on an Octet red96 device (Pall FortéBio) to detect the binding kinetics of S proteins with antibodies. First, the recombinant S proteins at 20 μg/mL in sodium acetate buffer (pH 5.0) were immobilized for 300 s onto activated AR2G biosensors (Pall FortéBio) and then immersed in 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride/N-hydroxysuccinimide for 300 s. Next, the biosensors loaded with saturated S proteins were incubated with 3-fold serial dilutions of antibodies with an initial concentration of 300 nM at 37°C for 300 s and then immersed into wells containing the kinetics buffer of 0.02% phosphate-buffered saline supplemented with 0.02% Tween 20 (PBST) for another 300 s. The baseline was established in the kinetics buffer. For data analysis, all the curves were fitted by a 1:1 binding model using the Data Analysis software (FortéBio). Mean kon (association constant), koff (dissociation constant), and KD (equilibrium dissociation constant) values were determined by averaging binding curves within a dilution series having R2 values greater than the 95% confidence level.
Pseudotyped-virus neutralization.
A pseudotyped-virus neutralization assay was performed to determine the neutralization activities of antibodies involved in this study. Briefly, whole S glycoprotein sequences of SARS-CoV and the wild type or variants of SARS-CoV-2 were inserted into the pcDNA3.1+ vector and severally cotransfected into 293T cells (ATCC, Manassas, VA, USA) with a defective HIV-1 genome that encodes a luciferase reporter. Supernatants containing S pseudoviruses were collected 48 h posttransfection. Serial 1/3 dilutions of antibodies were incubated with pseudoviruses at 37°C for 1 h, and then the mixtures were added to ACE2-expressing Huh-7 cells (104 per well in 96-well plates). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). The culture medium was refreshed 12 h postinfection, followed by an additional 48-h incubation. The Huh-7 cells were subsequently lysed with 50 μL lysis reagent (Promega), and 30-μL amounts of the lysates were transferred to 96-well Costar flat-bottom luminometer plates (Corning Costar) for the detection of relative light units using a firefly luciferase assay kit (Promega) on an ultra luminometer (Tecan). A nonlinear regression analysis was performed on the resulting curves, using Prism (GraphPad), to calculate the half-maximal inhibitory concentration (IC50) values.
The formation of SARS-CoV-2 S–553-15 and S–553-60 complexes.
The S trimer at 1.032 mg/mL was mixed with 553-60 or 553-15 at 1 mg/mL in a 1:1.4 molar ratio (S trimer/IgG), incubated at 4°C for 1 h, and further purified by using the Superose 6 increase 10/300 column (GE Healthcare). The peak tube was concentrated to 0.4 to 0.5 mg/mL in 20 mM Tris, pH 8.0, 200 mM NaCl.
The formation of the SARS-CoV-2 Omicron S–553-49 complex.
The Omicron S trimer at 1.928 mg/mL was mixed with 553-49 at 1.549 mg/mL in a 1:1.5 molar ratio (Omicron S trimer/IgG), incubated at 4°C for 2 h, and further purified by using the Superose 6 increase 10/300 column (GE Healthcare). The peak tube was concentrated to 0.4 to 0.5 mg/mL in 20 mM Tris, pH 8.0, 200 mM NaCl.
Cryo-EM sample preparation.
A 3-μL complex sample was added to a freshly glow-discharged holey amorphous nickel-titanium alloy film supported by 400-mesh gold grids. The sample was plunge-frozen in liquid ethane using the Vitrobot IV (FEI/Thermo Fisher Scientific), with a 2-s blot time, −3 blot force, and 10-s wait time.
Cryo-EM data collection.
Cryo-EM data were collected on a Titan Krios microscope (FEI/Thermo Fisher Scientific) operated at 300 kV and equipped with a K3 Summit direct detector (Gatan) and a GIF quantum energy filter (Gatan) set to a slit width of 20 eV. Automated data acquisition was carried out with SerialEM software (27).
For the S–553-15 complex, movies were taken in the superresolution mode at a nominal magnification of ×81,000, corresponding to a physical pixel size of 1.07 Å, and a defocus range from −1.2 to −2.5 μm. Each movie stack was dose fractionated to 41 frames, with a total exposure dose of about 61 e−/Å2.
For the S–553-60 complex, movies were taken in the superresolution mode at a nominal magnification of ×105,000, corresponding to a physical pixel size of 0.82 Å, and a defocus range from −1.2 to −2.5 μm. Each movie stack was dose fractionated to 40 frames, with a total exposure dose of about 60 e−/Å2.
For the Omicron S–553-49 complex and apo-S(D614G), movies were taken in the superresolution mode at a nominal magnification of ×81,000, corresponding to a physical pixel size of 1.064 Å, and a defocus range from −1.2 to −2.5 μm. Each movie stack was dose fractionated to 40 frames, with a total exposure dose of about 58 e−/Å2.
Cryo-EM image processing.
All the data processing was carried out using RELION 3.0 and cryoSPARC (28).
For the S–553-15 complex (Fig. S10), a total of 6,943 movie stacks were binned 2 by 2, dose weighted, and motion corrected using MotionCor2 (29). The parameters of the contrast transfer function (CTF) were estimated by using Gctf (30, 31). All micrographs were then manually selected based on ice conditions, defocus rangge, and estimated resolution. Particles were initially autopicked by using the Laplacian of Gaussian method and then subjected to two-dimensional (2-D) classification. The top classes were used as the 2-D reference for template picking, yielding 403,685 particles. Particles were extracted and subjected to a routine process of 2-D classification, three-dimensional (3-D) initial model, 3-D classification. Finally, 188,942 particles were grouped and subjected to auto-refinement and postprocessing, yielding a trimer dimer density map at 4.47-Å overall resolution with C1 symmetry.
To get a higher-resolution, asymmetric, single-trimer map, particles were subjected to symmetry expansion with C2 symmetry. All 377,884 expanded-symmetry particles were autorefined, CTF refined, and polished, yielding a trimer map at 3.45-Å overall resolution with C1 symmetry. To improve resolution at the three RBD–553-15 interfaces of a trimer, we performed local refinement focused on the NTD/RBD domains with Fab (NRAb). Three copies of particles targeted on different NRAb regions (NRAb1, NRAb2, and NRAb3) were subjected to focused 3-D classification without alignment (tau_fudge = 40) by using the local mask separately. A total of 169,450 NRAb1 particles were selected and then subtracted for focused refinement, yielding a 3.89-Å overall-resolution density map. A total of 199,441 NRAb2 particles were selected and subtracted for focused refinement, yielding a 3.80-Å overall-resolution density map. A total of 249,722 NRAb3 particles were selected and then subtracted for focused refinement, yielding a 4.11-Å overall-resolution density map. The focused, refined maps of NRAbs were fitted into the trimer map and merged with it using the “vop maximum” command in UCSF Chimera. The composite map for the S–553-15 asymmetric dimer of trimers was formed by using “vop maximum” with 2× NRAb maps and 2× trimer maps, which were all fitted in the original 4.47-Å trimer dimer map.
For S–553-60 (Fig. S13), the same procedure was performed. After 3-D classification, two dominant classes were selected separately. Then, 88,701 particles of state 1 (one-up RBD, two Fabs) were reconstructed into a 3.25-Å overall-resolution density map, and 99,762 particles of state 2 (two-up RBDs, three Fabs) were reconstructed into a 3.25-Å overall-resolution density map. To improve resolution at the RBD-MAb interfaces of the S–553-60 protein complex, the same procedure of local refinement was carried out. Three clusters of particles targeted on different NRAb regions (NRAb1 from state 1, NRAb2 from state 1, and NRAb3 from state 2) were subjected to focused 3-D classification without alignment (tau_fudge = 40) by using the local mask separately. A total of 88,701 NRAb1 particles were selected and then subtracted for focused refinement, yielding a 3.35-Å overall-resolution density map. 57,260 NRAb2 particles were selected for focused refinement, yielding a 3.79-Å overall-resolution density map. A total of 29,291 NRAb3 particles were selected and then subtracted for focused refinement, yielding a 3.94-Å overall-resolution density map. The focused, refined maps of NRAb1/NRAb2 were fitted into the map of state 1 particles and merged with it using the “vop maximum” command in UCSF Chimera. Similarly, the focused refined maps of NRAb1/NRAb2/NRAb3 were fitted into the map of state 2 particles and merged with it using the “vop maximum” command in UCSF Chimera.
For apo-S(D614G) (Fig. S6), the routine procedure was carried out. At last, 2,001,157 particles yielded a 2.70-Å map of apo-S.
For Omicron S–553-49 (Fig. S4), 7,294 movies were motion corrected and CTF estimated. Among these, 5,896 were selected for further processing. After blob picking in cryoSPARC and 2-D classification, trimer and monomer particles were observed. Separate heterogenous refinements resulted in two major structures: the Omicron S trimer in the apo state and the S monomer complexed with 553-49. Finally, 28,589 trimer particles yielded a 3.40-Å map of apo-Omicron S after nonuniform (NU) refinement, and 597,461 monomer particles yielded a 4.06-Å map of the RBD–553-49 interface region.
The reported resolutions above are based on the gold-standard Fourier shell correlation (FSC) 0.143 criterion. All the visualization and evaluation of 3-D density maps were performed with UCSF Chimera (32), and the local resolution variations were calculated using RELION (33). These composite maps were then subjected to “vop zflip” in UCSF Chimera to get the correct handedness and used for subsequent model building and analysis.
Model building and structure refinement.
Apo-OS trimer models were generated by SWISS-MODEL (34) and fitted into the map using UCSF Chimera (32). The model was manually adjusted, glycans at N-linked glycosylation sites were added in COOT (35), and then several iterative rounds of real-space refinement were carried out in PHENIX (36).
For Omicron S complexed with 553-49, the S-RBD model obtained from the apo-OS trimer model and the antibody model generated by Alpha fold2 were fitted into the map using UCSF Chimera (32). The model was further refined in COOT (35) and PHENIX (36) iteratively.
For model building of the S(D614G) (apo-S) trimer model, the WT S model (PDB accession number 6VYB) was fitted into the map using UCSF Chimera (32) and further refined in COOT (35) and PHENIX (36) iteratively.
For model building of S–553-15 and S–553-60, the apo-S trimer model and the antibody (553-15 and 553-60) models generated by SWISS-MODEL were fitted into the map using UCSF Chimera (32), followed by manual adjustment in COOT (35) and real-space refinement in Phenix. The RBD–553-15 Fab (553-15–S NR15) region was built and refined against the locally refined map (NRAb2) and then docked back into the global-refinement trimer maps. The RBD–553-60 Fab (553-60–S NR60) region was built and refined against the locally refined map (NRAb1) and then docked back into the global-refinement trimer maps. Model validation was performed using MolProbity. Details of the refinement statistics of the complexes are summarized in Table S1. Figures were prepared using UCSF Chimera and UCSF ChimeraX (37).
Data availability.
Coordinates and maps associated with data reported in the manuscript have been deposited to the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) with accession numbers EMD-32638 and PDB 7WO4 (S–553-15 dimer of trimers), EMD-32639 and PDB 7WO5 (S–553-15 trimer), EMD-32641 and PDB 7WO7 (S–553-15 NRAb2 local refinement), EMD-32646 and PDB 7WOA (S–553-60 trimer with 1 up-state RBDs binding 2 Fabs), EMD-32647 and PDB 7WOB (S–553-60 trimer with 2 up-state RBDs binding 3 Fabs), EMD-32648 and PDB 7WOC (S–553-60 NRAb1 local refinement), EMD-32651 and PDB 7WOG (Omicron RBD–553-49 Fab), EMD-32901 and PDB 7WZ1 (apo-Omicron S), and EMD-32902 and PDB 7WZ2 [apo-S(D614G)].
ACKNOWLEDGMENTS
We thank the Center of Cryo-Electron Microscopy, Fudan University, and the Center for Biological Imaging of the Institute of Biophysics (IBP) for their support in cryo-EM data collection.
This work was supported by grants from the Ministry of Science and Technology of China (grants number 2021YFC2302500 to L.S. and 2021YFE0201800 to F.L.), the National Natural Science Foundation of China (grants number 81900729 to L.S., 31925010 to F.L. and 31970146 to Z.C.), and the National Key R&D Program of China (grant number 2019YFA0904400 to T.Y.).
L.S., F.L., Z.C., and T.Y. conceived and supervised the study. W.Z., S.X., Q.L., A.H., and Y.H. purified the spike proteins and IgG antibodies. X.Z. and Z.C. performed the cryo-EM study. X.T. and W.S. performed BLI assays and the pseudovirus neutralization assay. W.Z., X.Z., Z.C., and L.S. analyzed the data. The manuscript was written by W.Z. and L.S. and reviewed, commented on, and approved by all the authors.
Fei Lan is listed as one of the inventors on two patents (WO2021207962A1 and O2021248279A1) related to the antibodies used in this study. The other authors declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
[This article was published on 4 August 2022 with inaccuracies in the authors’ affiliations. The affiliations were corrected in the current version, posted on 24 August 2022.]
Contributor Information
Tianlei Ying, Email: tlying@fudan.edu.cn.
Zhenguo Chen, Email: ZhenguoChen@fudan.edu.cn.
Fei Lan, Email: fei_lan@fudan.edu.cn.
Lei Sun, Email: LLSun@fudan.edu.cn.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kissler SM, Tedijanto C, Goldstein E, Grad YH, Lipsitch M. 2020. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science 368:860–868. 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Wan Y, Liu P, Zhao J, Lu G, Qi J, Wang Q, Lu X, Wu Y, Liu W, Zhang B, Yuen KY, Perlman S, Gao GF, Yan J. 2015. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res 25:1237–1249. 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO. 2021. Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 26 November 2021.
- 9.Zhu X, Mannar D, Srivastava SS, Berezuk AM, Demers JP, Saville JW, Leopold K, Li W, Dimitrov DS, Tuttle KS, Zhou S, Chittori S, Subramaniam S. 2021. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol 19:e3001237. 10.1371/journal.pbio.3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD. 2021. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593:130–135. 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 11.Takeda M. 2022. Proteolytic activation of SARS-CoV-2 spike protein. Microbiol Immunol 66:15–23. 10.1111/1348-0421.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Huang W, Li Q, Wang P, An R, Wang J, Wang Y, Niu X, Yang S, Liang H, Sun H, Li T, Yu Y, Cui Q, Liu S, Yang X, Du S, Zhang Z, Hao X, Shao F, Jin R, Wang X, Xiao J, Wang Y, Xie XS. 2022. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602:657–663. 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klasse PJ, Sattentau QJ. 2002. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J Gen Virol 83:2091–2108. 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- 14.Wan J, Xing S, Ding L, Wang Y, Gu C, Wu Y, Rong B, Li C, Wang S, Chen K, He C, Zhu D, Yuan S, Qiu C, Zhao C, Nie L, Gao Z, Jiao J, Zhang X, Wang X, Ying T, Wang H, Xie Y, Lu Y, Xu J, Lan F. 2020. Human-IgG-neutralizing monoclonal antibodies block the SARS-CoV-2 infection. Cell Rep 32:107918. 10.1016/j.celrep.2020.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, Kosugi Y, Shirakawa K, Sadamasu K, Kimura I, Ito J, Wu J, Iwatsuki-Horimoto K, Ito M, Yamayoshi S, Loeber S, Tsuda M, Wang L, Ozono S, Butlertanaka EP, Tanaka YL, Shimizu R, Shimizu K, Yoshimatsu K, Kawabata R, Sakaguchi T, Tokunaga K, Yoshida I, Asakura H, Nagashima M, Kazuma Y, Nomura R, Horisawa Y, Yoshimura K, Takaori-Kondo A, Imai M, Tanaka S, Nakagawa S, Ikeda T, Fukuhara T, Kawaoka Y, Sato K, Genotype to Phenotype Japan (G2P-Japan) Consortium . 2022. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 602:300–306. 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, Zhan W, Yang Z, Tu C, Hu G, Zhang X, Song W, Du S, Zhu Y, Huang K, Kong Y, Zhang M, Mao Q, Gu X, Zhang Y, Xie Y, Deng Q, Song Y, Chen Z, Lu L, Jiang S, Wu Y, Sun L, Ying T. 2022. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell 185:1389–1401.e18. 10.1016/j.cell.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanke L, Das H, Sheward DJ, Perez Vidakovics L, Urgard E, Moliner-Morro A, Kim C, Karl V, Pankow A, Smith NL, Porebski B, Fernandez-Capetillo O, Sezgin E, Pedersen GK, Coquet JM, Hallberg BM, Murrell B, McInerney GM. 2022. A bispecific monomeric nanobody induces spike trimer dimers and neutralizes SARS-CoV-2 in vivo. Nat Commun 13:155. 10.1038/s41467-021-27610-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD. 2020. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe 28:867–879.e5. 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv Z, Deng YQ, Ye Q, Cao L, Sun CY, Fan C, Huang W, Sun S, Sun Y, Zhu L, Chen Q, Wang N, Nie J, Cui Z, Zhu D, Shaw N, Li XF, Li Q, Xie L, Wang Y, Rao Z, Qin CF, Wang X. 2020. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 369:1505–1509. 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou D, Duyvesteyn HME, Chen CP, Huang CG, Chen TH, Shih SR, Lin YC, Cheng CY, Cheng SH, Huang YC, Lin TY, Ma C, Huo J, Carrique L, Malinauskas T, Ruza RR, Shah PNM, Tan TK, Rijal P, Donat RF, Godwin K, Buttigieg KR, Tree JA, Radecke J, Paterson NG, Supasa P, Mongkolsapaya J, Screaton GR, Carroll MW, Gilbert-Jaramillo J, Knight ML, James W, Owens RJ, Naismith JH, Townsend AR, Fry EE, Zhao Y, Ren J, Stuart DI, Huang KA. 2020. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat Struct Mol Biol 27:950–958. 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Xu W, Chen Z, Fu W, Zhan W, Gao Y, Zhou J, Zhou Y, Wu J, Wang Q, Zhang X, Hao A, Wu W, Zhang Q, Li Y, Fan K, Chen R, Jiang Q, Mayer CT, Schoofs T, Xie Y, Jiang S, Wen Y, Yuan Z, Wang K, Lu L, Sun L, Wang Q. 2022. An ultrapotent pan-beta-coronavirus lineage B (beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope. Protein Cell 13:655–675. 10.1007/s13238-021-00871-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastie KM, Li H, Bedinger D, Schendel SL, Dennison SM, Li K, Rayaprolu V, Yu X, Mann C, Zandonatti M, Diaz Avalos R, Zyla D, Buck T, Hui S, Shaffer K, Hariharan C, Yin J, Olmedillas E, Enriquez A, Parekh D, Abraha M, Feeney E, Horn GQ, Aldon Y, Ali H, Aracic S, Cobb RR, Federman RS, Fernandez JM, Glanville J, Green R, Grigoryan G, Lujan Hernandez AG, Ho DD, Huang K-YA, Ingraham J, Jiang W, Kellam P, Kim C, Kim M, Kim HM, Kong C, Krebs SJ, Lan F, Lang G, Lee S, Leung CL, Liu J, Lu Y, MacCamy A, et al. 2021. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science 374:472–478. 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583:290–295. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 24.Yuan M, Wu NC, Zhu X, Lee C-CD, So RTY, Lv H, Mok CKP, Wilson IA. 2020. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368:630–633. 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Wu NC, Yuan M, Bangaru S, Torres JL, Caniels TG, van Schooten J, Zhu X, Lee CD, Brouwer PJM, van Gils MJ, Sanders RW, Ward AB, Wilson IA. 2020. Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity 53:1272–1280.e5. 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh CL, Goldsmith JA, Schaub JM, DiVenere AM, Kuo HC, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, Chou CW, Byrne PO, Hjorth CK, Johnson NV, Ludes-Meyers J, Nguyen AW, Park J, Wang N, Amengor D, Lavinder JJ, Ippolito GC, Maynard JA, Finkelstein IJ, McLellan JS. 2020. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369:1501–1505. 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. 2017. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14:290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 29.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Turer E, Li X, Zhan X, Choi M, Tang M, Press A, Smith SR, Divoux A, Moresco EM, Beutler B. 2016. Insulin resistance and diabetes caused by genetic or diet-induced KBTBD2 deficiency in mice. Proc Natl Acad Sci USA 113:E6418–E6426. 10.1073/pnas.1614467113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 33.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7:e42166. 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD. 2018. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74:531–544. 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Download jvi.00480-22-s0001.mp4, MP4 file, 11.5 MB (11.8MB, mp4)
Table S1, Fig. S1-S13, and Movie S1. Download jvi.00480-22-s0002.pdf, PDF file, 8.6 MB (8.1MB, pdf)
Data Availability Statement
Coordinates and maps associated with data reported in the manuscript have been deposited to the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) with accession numbers EMD-32638 and PDB 7WO4 (S–553-15 dimer of trimers), EMD-32639 and PDB 7WO5 (S–553-15 trimer), EMD-32641 and PDB 7WO7 (S–553-15 NRAb2 local refinement), EMD-32646 and PDB 7WOA (S–553-60 trimer with 1 up-state RBDs binding 2 Fabs), EMD-32647 and PDB 7WOB (S–553-60 trimer with 2 up-state RBDs binding 3 Fabs), EMD-32648 and PDB 7WOC (S–553-60 NRAb1 local refinement), EMD-32651 and PDB 7WOG (Omicron RBD–553-49 Fab), EMD-32901 and PDB 7WZ1 (apo-Omicron S), and EMD-32902 and PDB 7WZ2 [apo-S(D614G)].