ABSTRACT
Rift Valley fever virus (RVFV) is endemic in sub-Saharan Africa (SSA), with outbreaks reported in the Arabian Peninsula and throughout SSA. The natural reservoir for RVFV are ruminants, with livestock populations exceeding 50% exposure rates in some areas of SSA. Transmission to humans can occur through exposure to infected livestock products or multiple species of mosquito vectors. In 2013 and 2014, cross-sectional surveys occurred in two districts of Nacala-a-Velha and Mecubúri in northern Mozambique, and participants provided blood samples for later serological assays. IgG against the N protein of RVFV was detected through multiplex bead assay (MBA). Of the 2,278 persons enrolled between the two surveys and study sites, 181 (7.9%, 95% confidence interval (CI): 6.9%-9.1%) were found to be IgG seropositive with increasing seroprevalence with older age and significantly higher seroprevalence in Nacala-a-Velha (10.5%, 8.8%-12.5%) versus Mecubúri (5.7%, 4.5%-7.1%). Seroprevalence estimates were not significantly different between the 2013 and 2014 surveys. Significant spatial clustering of IgG positive persons were consistent among surveys and within the two districts, pointing toward the consistency of serology data for making population-level assumptions regarding RVFV seroprevalence. A subset of persons (n = 539) provided samples for both the 2013 and 2014 surveys, and a low percentage (0.81%) of these were found to seroconvert between these two surveys. Including the RVFV N protein in an MBA antigen panel could assist elucidate RVFV exposure in SSA.
IMPORTANCE Due to sporadic transmission, human contact with Rift Valley Fever Virus (RVFV) is difficult to ascertain at a population level. Detection of antibodies against RVFV antigens assist in estimating exposure as antibodies remain in the host long after the virus has been cleared. In this study, we show that antibodies against RVFV N protein can be detected from dried blood spot (DBS) samples being assayed by multiplex bead assay. DBS from two districts in northern Mozambique were tested for IgG against the N protein, and 7.9% of all enrolled persons were seropositive. Older persons, males, and persons residing closer to the coast had higher RVFV N protein seroprevalence. Spatial clustering of IgG positive persons was noted in both districts. These results show low exposure rates to RVFV in these two northern districts in Mozambique, and the ability to perform serology for the RVFV N protein from dried blood samples.
KEYWORDS: Rift Valley fever virus, seroprevalence, Mozambique, serology, IgG, risk factors, seroconversion
INTRODUCTION
Rift Valley fever virus (RVFV) is a Phlebovirus within the order Bunyavirales, and while most infections in humans are asymptomatic or present with mild symptoms, a small proportion of infections can progress to hemorrhagic fever syndrome (1). RVFV is endemic in sub-Saharan Africa (SSA), causing intermittent outbreaks of Rift valley fever (RVF) among ruminants and humans (2, 3). Transmission of RVFV can occur through mosquito vectors, primarily through Aedes spp. However, other genera have also been implicated in perpetuating transmission during the rainy season (4, 5). Additionally, physical contact with ruminants or consumption of ruminant products (such as meat and raw milk) can serve as a source of zoonotic transmission (6–8). Residing in proximity to ruminants has consistently been shown as a risk factor for human exposure to RVFV (7, 9, 10). Since the mid-20th century, multiple outbreaks have been identified in SSA and the Arabian peninsula, and, though typically confined to a few hundred human cases or less, large incidences such as the 1977 Egypt and 2000 Arabian outbreaks have involved thousands of human cases and hundreds of deaths (11).
Serological data estimating RVFV exposure have been collected in both humans and ruminants and have shown evidence for the presence of the virus in nearly all SSA countries as well as in the Arabian Peninsula (2, 12). As outbreaks are infrequent and infection often does not elicit treatment-seeking behavior, serological data provide an objective indicator of historic exposure in humans or other mammals and can be used to generate epidemiological estimates. Contemporary serological assays for RVFV have utilized different antigenic targets, including: nucleocapsid protein (N) (13, 14), glycoprotein (Gn) (15), non-structural proteins NSs and NSm (12), and complete virus particles (16, 17). Estimates within human populations are typically low for IgM or IgG antibody seropositivity and have rarely exceeded 10% (18, 19), except following an RVFV outbreak (16). Seroprevalence among ruminants is generally higher and can exceed 50%, but is highly variable among species, location, and immunoassay utilization (20–22).
This study investigated IgG seropositivity to RVFV N from persons residing in two districts in northern Mozambique from two cross-sectional surveys conducted in 2013 and 2014. The recombinant N was covalently bound to microspheres and included in a bead-based multiplex serological assay, and validation of this target was performed by comparison to inactivated whole-virus enzyme-linked immunosorbent assay (ELISA). Risk factors for seropositivity were assessed along with analysis of spatial clustering in the districts of Nacala-a-Velha and Mecubúri. For persons sampled in both surveys, the durability of IgG response to the N was evaluated, as well as seroconversion and seroreversion events.
RESULTS
Between the 2013 and 2014 cross-sectional surveys in Nacala-a-Velha and Mecubúri (Fig. 1A), a total of 2,278 DBS were collected with 539 (23.7% of all) persons providing a DBS sample for both surveys for RVFV serological testing (Table 1) (23). Of all participants, 46.7% were enrolled in Nacala-a-Velha and 53.3% in Mecubúri, and for the persons providing samples for both surveys, 51.0% were from Nacala-a-Velha and 49.0% were Mecubúri. Approximately half (50.8%) of persons were less than 15 years of age and 54.5% were female.
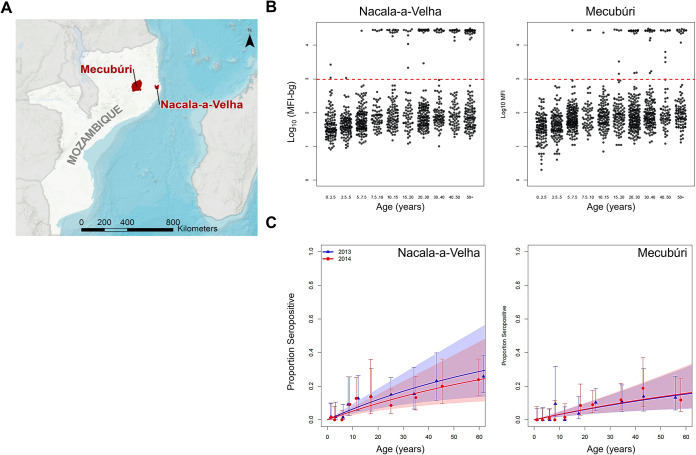
FIG 1.
Study sites and IgG levels and seropositivity to RVFV N by age. (A) Location of Nacala-a-Velha and Mecubúri districts in northern Mozambique. (B) Beeswarm plots for RVFV N IgG assay signal by age groups for the two districts with data for 2013 and 2014 surveys combined. Horizontal red hashed line denotes seropositivity threshold. (C) Seroprevalence curves for RVFV N by district and survey year. Modeling estimates for seroconversion and seroreversion rates displayed in Table 2.
TABLE 1.
Demographic characteristics of participants providing blood samples in Mozambique from 2013 and 2014
| Year |
|||
|---|---|---|---|
| Characteristic | 2013 | 2014 | Total (%) |
| Participants enrolled | 1118 | 1160 | 2278 (100%) |
| District | |||
| Nacala-a-Velha | 526 | 538 | 1064 (46.7%) |
| Mecubúri | 592 | 622 | 1214 (53.3%) |
| Age | |||
| 0–4 yrs | 249 | 243 | 492 (21.6%) |
| 5–9 yrs | 190 | 251 | 441 (19.4%) |
| 10–14 yrs | 105 | 119 | 224 (9.8%) |
| 15+ yrs | 574 | 547 | 1121 (49.2%) |
| Sex | |||
| Female | 612 | 630 | 1242 (54.5%) |
| Male | 506 | 530 | 1036 (45.5%) |
| Provided DBS in both 2013 and 2014 surveys | 539 (23.7%) | ||
IgG seroprevalence to RVFV N in the study population was low, with 181 (7.9%, 95% CI: 6.9%-9.1%) of persons seropositive between the two surveys, and no significant differences noted between the 2013 (8.5%, 6.9%-10.3%) and the 2014 (7.4%, 6.0%-9.1%) surveys. Seroprevalence was significantly higher in Nacala-a-Velha (10.5%, 8.8%-12.5%) versus Mecubúri (5.7%, 4.5%-7.1%). Increased seroprevalence by age were noted for both districts (Fig. 1B and C). Modeled seroconversion rates had overlapping confidence intervals between the two survey years within each district, but estimated rates were significantly higher for Nacala-a-Velha (0.0068 and 0.0054) versus Mecubúri (0.0033 versus 0.0034) (Table 2).
TABLE 2.
Estimates for Rift Valley fever virus N seroconversion by district and survey years in two districts of Mozambique from 2013 and 2014
| Modelled rate | Rate estimate | 95% confidence interval | |
|---|---|---|---|
| SCRa | |||
| Nacala-a-Velha 2013 | 0.0068 | 0.0042 | 0.0093 |
| Nacala-a-Velha 2014 | 0.0054 | 0.0033 | 0.0075 |
| Mecubúri 2013 | 0.0033 | 0.0019 | 0.0046 |
| Mecubúri 2014 | 0.0034 | 0.002 | 0.0048 |
| SRRb | 0.0055 | −0.0084 | 0.0194 |
Seroconversion rate.
Seroreversion rate.
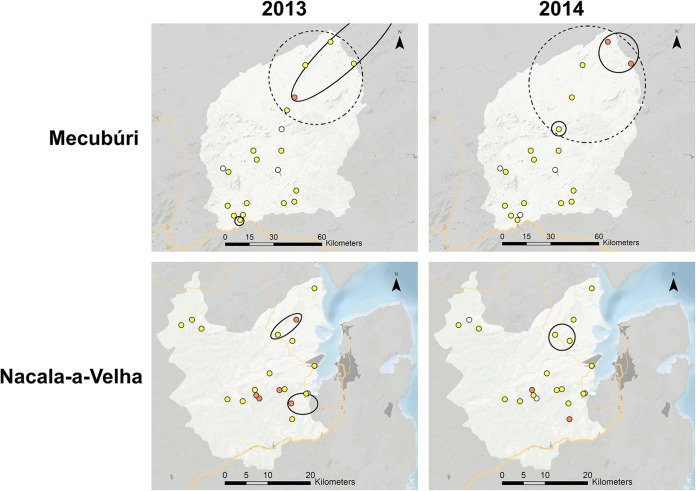
Assessing spatial clustering for seropositivity (binary data) or median mean fluorescence intensity (MFI)-bg assay signal (continuous data) found consistency in the two study sites between the two survey years. Fig. 2 shows the northern area of Mecubúri having statistically significant clusters identified for higher seroprevalence and MFI-bg signal in both the 2013 and 2014 surveys. Likewise, a statistically significant cluster for higher seroprevalence was observed in northeast Nacala-a-Velha for both surveys, though the 2013 survey alone also found a separate cluster in eastern Nacala-a-Velha. No spatially significant clusters were observed for increased MFI-bg signal in Nacala-a-Velha for either survey.
FIG 2.
Spatial patterns of IgG against RVFV N between two study sites for the two surveys. Spatially significant clusters of IgG positivity indicated by hashed ellipses and for IgG levels by solid lines. Marker intensity for seropositivity: 0%, white; 0-24%, yellow; 25-50%, orange.
Modeling for risk factors for IgG seropositivity to RVFV N found significantly increased odds of seropositivity with increasing age and for male sex, with males having an overall 60% increase in odds of being Np seropositive (Table 3). When assessing participants enrolled by sex, females were enrolled at higher percentages from ages approximately 15 to 40 (Fig. S4A), but the consistent higher proportion of males who were seropositive was not observed until approximate age 40 years and beyond (Fig. S4B). In the adjusted model, seroprevalence was not significantly different between the 2 districts, village elevation, household socioeconomic status (SES), or long-lasting insecticidal net (LLIN) use whether individual- or community-level. Correlation between village seroprevalence and elevation showed an overall negative trend, but with a high degree of variation and a non-significant negative slope (rho = 0.37)(Fig. S5).
TABLE 3.
Adjusted modeling of risk factors for seropositivity to RVFV N protein inMozambique from 2013 and 2014
| Variable | Seropositivity aOR (95% CI) |
|---|---|
| District | |
| Mecuburi | Ref |
| Nacala-a-Velha | 1.2 (0.62-2.3) |
| Elevation (m) | 1.0 (1.0-1.0) |
| Age | |
| <5 yrs | Ref |
| 5–10 yrs | 3.7 (0.41-33) |
| 10–15 yrs | 15 (1.9-123) |
| 15–20 yrs | 27 (3.4-223) |
| 20–30 yrs | 23 (3-174) |
| 30–40 yrs | 32 (4.3-244) |
| >40 yrs | 40 (5.4-289) |
| Sex | |
| Female | Ref |
| Male | 1.6 (1.0-2.5) |
| SES quintile | |
| 1st (Poorest) | Ref |
| 2nd | 0.52 (0.23-1.2) |
| 3rd | 0.67 (0.34-1.3) |
| 4th | 0.98 (0.54-1.8) |
| 5th (Richest) | 0.55 (0.27-1.1) |
| LLIN Use (individual) | 1.0 (0.55-1.9) |
| LLIN Use (community) | 0.5 (0.07-3.5) |
| SESa LLINb |
Socioeconomic status.
Long-lasting insecticidal net.
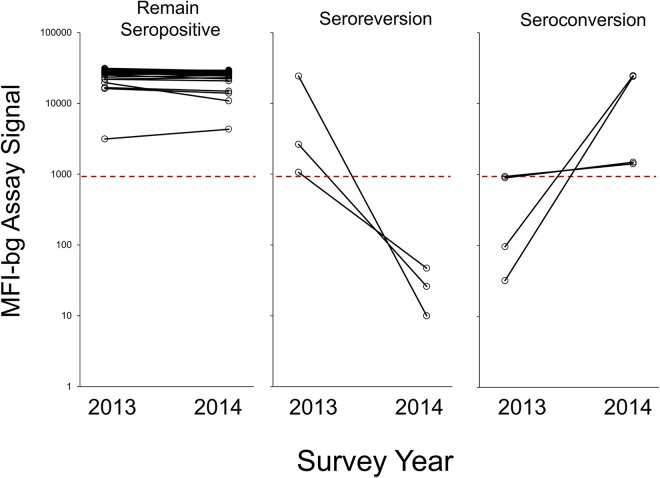
Among these 539 individuals sampled in both the 2013 and 201 surveys, the majority of persons (489, 90.7%) were seronegative at both time points. For the 50 persons with at least one sample showing IgG to N, most of these (43, 86.0%) were clearly seropositive at both time points with no substantial fluctuation in assay signal between the 2013 and 2014 surveys (Fig. 3). However, of the 46 persons seropositive during the 2013 survey, 3 (6.5%) had a decrease in IgG levels, causing them to become seronegative by 2014. Two of these persons were male children under 5 years of age and 1 person was a female in her mid-30s. Of the 494 persons seronegative in 2013, 4 (0.81%) had seroconverted by the 2014 survey, indicating an exposure to RVFV during this period of time. Two young males (both between 15 and 20 years old) had MFI-bg signals close to the seropositivity threshold in 2013, and slight increases in MFI-bg values in 2014 caused these persons to be classified as seropositive. However, 2 persons (both females over 40 years of age from Nacala-a-Velha) had log-folds increase in assay signal between the 2 surveys.
FIG 3.
Anti-RVFV N IgG level for persons sampled in 2013 and 2014 surveys. Plots shown for persons IgG seropositive to N remaining seropositive (n = 43), persons becoming IgG negative (seroreverting, n = 3), and persons becoming IgG positive (seroconverting, n = 4).
Comparison with RVFV ELISA results found moderate correlation in assay signal between the 2 platforms for the same sample (rho = 0.59, Fig. S2A). If the ELISA was considered as the gold standard immunoassay, receiver operating characteristic (ROC) analysis for MBA results found a high estimate for area under the curve (AUC = 0.923), but to maximize the specificity of binary classification for MBA results, a seropositivity threshold of >20,000 MFI-bg units would be required (Fig. S2B and C). No correlation was noted between an individual’s response to RVFV N and 2 other arboviral antigens included in the MBA panel: chikungunya virus E1, and dengue 3 virus virus-like particle (VLP) (Fig. S6).
DISCUSSION
These findings present serological evidence for human exposure to RVFV in the northern Mozambican districts of Nacala-a-Velha and Mecubúri. Though overall IgG seroprevalence to the RVFV N was low (7.9% among all participants), significant differences were noted between the Nacala-a-Velha (10.5%) and Mecubúri (5.7%) study sites. Enrollment numbers and participant characteristics were very similar between both study sites with no significant differences between distribution of participant ages or sex ratio (23). Notably, Mecubúri only saw 4 participants (0.33%) under 15 years of age seropositive for RVFV N, whereas 22 (2.1%) of participants of the same age range from Nacala-a-Velha were seropositive – a greater than 6-fold difference. The Nacala-a-Velha district is on the northeast coast of Mozambique whereas Mecubúri district is more geographically disperse and >100km inland. This observed difference in seroprevalence could potentially be due to differences in appropriate mosquito habitat between the 2 districts (4, 18, 24), or differences in close human interactions with ruminant livestock or consumption in ruminant products (6, 8, 9, 19). Reports for RVFV exposure among humans in Mozambique have consistently found low IgG seroprevalence estimates of less than 10% (18, 25, 26), though seroprevalence among Mozambican ruminants has been observed to be much higher with estimates routinely exceeding 10% (27–29), and even 50% in some studies (30, 31).
The IgG response against RVFV N appears to be largely durable over a time period of 1 year. Further evidence by modeling seroprevalence curves by age suggests that after seroconversion, persons would remain IgG positive for many years after exposure. After seropositive/seronegative binary classification, very few samples had assay signal near the seropositivity threshold, indicating that if an individual was IgG seropositive (previously exposed to RVFV), they typically harbored a high IgG titer against the N. Indeed, this was shown by the overall distribution of the assay signal for the entire study population with a clear unimodal sub-population of seronegative participants, and a unimodal subpopulation of seropositive individuals with very high assay signal. The finding of monotonic increase in population-level seropositivity with age to an infectious disease antigen mirrors other pathogen antigens known to induce long-lived memory B cells and circulating IgG antibodies (23, 32–34). As persons age in an endemic setting, the cumulative probability of lifetime exposure to a pathogen is continually increasing, so the oldest individuals have endured the highest odds of even a single infection event. For the 539 persons sampled in both 2013 and 2014, we found that 43 of the 46 (93.5%) individuals seropositive in 2013 not only retained their seropositivity status against N, but this level of IgG was largely unchanged 1 year later. Among the 3 persons (6.5% of 2013 IgG positives) who seroreverted between 2013 and 2014, 2 were under the age of 5 years, possibly indicating a less-durable B cell population in these younger persons (34). Though it was observed that 4 individuals (0.74%) seroconverted between 2013 and 2014, it is difficult to estimate if this 1-year span of time is a ‘typical’ rate of exposure for RVFV in northern Mozambique, though these findings from a 1-year follow-up do mirror the overall modeled estimates for seroconversion (0.47% probability per year of life) and seroreversion (0.55% probability). Low rates of observed and modeled seroconversion both point to low annual transmission rates in these districts, though it should be noted that RVFV infections during outbreak periods likely account for a large proportion of all exposures (35). At this same time period of 2013–2014, RVFV outbreaks were identified in human (18) and ruminant (30) populations in Mozambique, though these were all identified in southern districts of the country. The 60% increased odds for seropositivity in males differs from other serostudies which have generally found no differences in seroprevalence by sex (6, 18, 19), though a 2009 Kenya study did find over 2-fold increase in seroprevalence in males (16). Increased odds in males in this Mozambique study population may be due to behavioral differences leading to increased exposure to both ruminants and mosquitoes.
Both chikungunya virus and dengue virus serotype 3 are endemic in Mozambique and are common sources of acute febrile illness in the population (26, 36, 37). This study found no correlation between levels of IgG antibodies against these arboviruses and RVFV N. These data point to 2 findings: no evidence of IgG cross-binding among these 3 arboviral antigens, and different exposure patterns of the populace to these different arboviruses. The lack of cross-binding is not surprising as all 3 viruses are distantly related, with dengue and chikungunya viruses belonging to the phylum Kitrinoviricota, whereas RVFV belongs to the Negarnaviricota phylum. Chikungunya and dengue viruses are strictly transmitted through the Aedes vector (24), whereas RVFV has been found to be more liberal with its arthropod host (4, 5). Though non-significant, an overall negative trend observed by village elevation and RVFV N seroprevalence provides some evidence that residence in more optimal mosquito habitats (at lower elevations) may facilitate the mosquito/human transmission cycle in this population. RVFV transmission to humans through contact with livestock (or livestock products) has been reported, and studies sampling from Mozambican sheep, goats, and cows have consistently found levels of RVFV livestock exposure >10% and exceeding 60% in some districts (27–29, 31).
Limitations to this study include the inability to perform functional inhibition studies with this sample set. While RVFV exposure could be ascertained using the total anti-N IgG response, estimation of levels of protective antibodies was not possible with the MBA as run (38). Participants were not asked questions relating to livestock ownership or contact (and livestock from the communities were not sampled from), so associations with this important mode of viral transmission could not be investigated. Though the RVFV N is a common serological tool for immunoassays (13, 14, 20), other RVFV antigens were not included in our MBA panel, and IgG against the N is our only metric of exposure. As with some previous livestock studies (14, 39), the current study only provided a follow-up sampling of 1 year, and longitudinal assessment of populations (and individuals) could provide a more accurate illustration of rates of virus transmission and the associated risk factors.
In conclusion, the RVFV recombinant N provides an excellent serological marker to assess human exposure to RVFV. This study utilized DBS as a sample type to detect anti-N IgG, and being able to utilize DBS provides pragmatic sample collection in a variety of field settings. At a population level, though the Mozambican districts of Nacala-a-Velha and Mecubúri both saw overall low seroprevalence to this antigen, spatial and population estimates were consistent between household surveys conducted in 2013 and 2014. Future surveys in Mozambique may consider serological and other data to attempt to estimate human and ruminant exposure to RVFV in this area of Africa. For surveys from SSA employing a MBA technology, inclusion of this antigen target would provide a wealth of data regarding RVFV exposure in human populations.
MATERIALS AND METHODS
Ethics.
These surveys were performed to evaluate the impact of long-lasting insecticidal nets on Plasmodium falciparum prevalence (23). The National Bioethics Committee in Mozambique approved the original serosurvey study (#249/CNBS/13). Adult participants provided written consent before enrollment in the study and provided written consent on behalf of child participants, with agreement for future antibody testing against other infectious diseases. United States Centers for Disease Control and Prevention (CDC) investigators provided technical assistance without access to personally identifiable information and were not considered engaged in human subjects research (#2014-268).
Study design, participant enrollment, and blood sample collection.
Two cross-sectional household surveys occurred in September 2013 and October 2014 in coastal Nacala-a-Velha District and inland Mecubúri District in Nampula Province in northern Mozambique (23) (Fig. 1A). In each district, 20 enumeration areas were randomly selected with probability proportional to size; in each enumeration area, 20 households were randomly selected using simple random sampling after full enumeration of households in the area. In each household, all members present were invited to enroll in the survey which included questions on LLIN use, sociodemographic variables, and SES, and were asked to provide a dried blood sample on filter paper. In the second survey in 2014, teams revisited the same households from the 2013 surveys, with no replacement of households. In both surveys, participants provided a blood sample by finger prick, and 10 μL of whole blood was spotted onto each of six circular filter paper extensions configured in a wheel (Trop Bio Pty Ltd). Blood was dried overnight and the dried blood spots (DBS) were individually stored in plastic baggies with desiccant, temporarily stored at 4°C, and shipped to CDC in Atlanta, GA, USA at room temperature. Upon receipt at CDC, DBS were stored at −20°C until processed in the laboratory.
Antigen coupling to beads.
A recombinant RVFV N antigen (South African isolate M35/74, GenBank accession number JF784388, GenScript) fused to glutathione S-transferase (GST) was covalently bound to polystyrene microbeads (SeroMap Beads; Luminex Corporation) by the commonly used carbodiimide intermediate reaction with N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide (EDC, Calbiochem) cross-linker (23). A 1 mL bead coupling (12.5 × 106 beads) was performed at a concentration of 17 μg/mL protein in a buffer of 50 mM 2-(N-morpholino)-ethanesulfonic acid and 0.85% NaCl at pH 5.0. Chikungunya virus E1 antigen and dengue virus serotype 3 VLP were coupled as described previously (40, 41), and included to generate seroepidemiological estimates for these pathogens (as reported separately) and to assess potential cross-binding with RVFV N antigen. As a control for nonspecific binding, a bead conjugated to only GST was included in the multiplex panel, and this protein was coupled to beads at 15 μg/mL at pH 5.0. Beads were stored at 4°C until used for data collection by multiplex bead assay (MBA). The MBA panel also included microbeads coupled to other infectious disease and vaccine-preventable disease antigens (23).
Blood sample elution and MBA data acquisition.
Antibodies from one tab of the DBS filter paper extension were eluted overnight at 4°C using 200 μL PBS containing 0.3% Tween 20 and 0.05% sodium azide (an approximately 1:40 serum dilution based on an estimated hematocrit of 50%). A further 1:10 dilution of the serum was made with phosphate-buffered saline (PBS) containing 0.3% Tween 20, 0.02% sodium azide, 0.5% bovine serum albumin, 0.5% polyvinyl alcohol, 0.8% polyvinyl pyrrolidone, and 0.5% casein, and a final 3 μg/mL crude and unclarified Escherichia coli extract. This dilution was stored overnight at 4°C for absorption of E. coli antibodies and to eliminate nonspecific reactivity to polystyrene (42, 43).
The MBA was performed as described previously in 96-well filtered-bottom plates (Millipore) with wash steps (with PBS + 0.05% Tween 20) between incubation steps (23). The diluted sample (approximately 1:400 serum dilution) was incubated with antigen-coupled beads for 90 min protected from light at room temperature under gentle shaking. Bound antigen-specific IgG was detected with 45-minute incubation with biotinylated mouse monoclonal antibodies to human IgG (1:500 anti-human IgG, 1:625 anti-human IgG4, both from Southern Biotech) followed by a 30 min incubation with streptavidin-phycoerythrin (Invitrogen)(42, 44). Data were acquired with a BioPlex-200 instrument with BioPlex Manager 6.1 software (Bio-Rad) that calculated the median fluorescence intensity (MFI) from each bead region for each assay well. Background (bg) MFI from a buffer-only blank on each plate was subtracted from MFI readings for each bead region, providing an assay signal of MFI-bg. The mean MFI-bg value from duplicate wells was used for analysis. For the original study, samples with high CV% values (>15%) for >4 positive antibody responses were repeated (23). The average variability (measured as CV%) for the positive responses of control sera across the 64-plate study was <15% (Fig. S1).
RVFV ELISA.
RVF virus-specific IgG ELISA was performed as previously described to compare results for the novel MBA compared to the standard ELISA (45, 46). Briefly, following heat and detergent inactivation, DBS specimens were tested by anti- IgG ELISA using inactivated RVFV-infected VERO-E6 cell antigens. A cell slurry was prepared by sonicating gamma-irradiated lysate of VERO-E6 cells infected with virus. Four dilutions of each specimen (1:100, 1:400, 1:1600 and 1:6400) were tested. Titers and cumulative sum of optical densities of each dilution (SUMOD) minus background of absorbance of uninfected control VERO E6 cells (adjusted SUMOD) were recorded. Specimens were considered positive only if both the adjusted SUMOD and titer were above pre-established conservative cutoff values, which were set as >0.95 and ≥1:400 for IgG). Comparison between MBA and ELISA results for a subset of all study specimens (n = 98) is shown in Fig. S2 This subset was chosen based on a wide range of MBA IgG assay signal values to N protein.
Determination of threshold for RVFV N IgG seropositivity.
Three methods were employed to identify an MFI-bg assay signal cutoff to denote true IgG positivity to the N:
-
(i)
A panel of 86 serum specimens from U.S. residents who had no reported history of international travel was assayed by MBA, and the lognormal mean plus 3 standard deviations was calculated from the MFI-bg signals as the N cutoff. This threshold value was calculated to be 527 MFI-bg units.
-
(ii)
A ROC curve utilizing the ELISA results as a serological gold standard identified a threshold value of 26,878 MFI-bg units when maximizing sensitivity and specificity using a closest to 0,1 criterion (47).
-
(iii)
A two-component finite mixture model (FMM) approach attempted to find 2 sub-populations (putative seronegative and seropositive) within the sample set data (48). For N assay signal, the lognormal mean + 3 standard deviations (99.7% specificity) was calculated from the first (negative) component and found to be a 966 MFI-bg units.
In assessing the distribution of MFI-bg assay signals for the entire study population (Fig. S3) the threshold obtained by the two-component FMM approach appeared to be most appropriate by visual inspection, and this was the chosen threshold for dichotomizing assay results. A total of 24 samples (1.1% of entire sample set) were found to have an assay signal between 527 and 966, whereas 78 (3.4%) samples had an MFI-bg signal between 966 and 26,878.
Spatial analysis.
A centroid GPS of each village was used for analysis. Elevation above sea level in meters (m) were obtained by an ASTER Digital Elevation Model for the GPS coordinate for each village. The GPS coordinates and elevation for each village were averaged, and the percentage of individuals who were IgG positive for each antigen per village was plotted, using ArcGIS 10.3.1(Esri). SatScan software (v9.7) was used to identify spatial clusters of villages with elevated prevalence. In addition, SatScan was utilized for spatial clusters of villages with significantly elevated median IgG response (by MFI-bg assay signal values) (49, 50). An elliptical cluster shape was assumed and statistically significant clusters (P < 0.05 used for significance) are represented by the border of a convex hull of villages deemed to be inside the cluster.
Statistical analysis.
ROC analysis was performed in SAS software (v9.4) by the PROC LOGISTIC procedure and two-component finite mixture models by PROC FMM. A reversible catalytic model was fit to the seropositivity by age data for N in R version 3.3.2 (R Foundation for Statistical Computing). Estimates for the serological conversion rate (SCR) and serological reversion rate (SRR) per year were jointly estimated from the likelihood model, with a constant SRR across sites and years, but allowing SCR to vary by site and year (23). Poisson regression for seropositivity adjusting for adjusting for district of residence age, sex, village elevation, household socioeconomic status (SES), and long-lasting insecticidal net (LLIN) use was performed in R as described previously (23).
ACKNOWLEDGMENTS
The authors acknowledge Zach Matson for developing the Microsoft Excel template for the Shewart plot, Patrick J Lammie for support with coordinating laboratory objectives, and the United States Agency for International Development for survey support.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention or US President’s Malaria Initiative. Use of trade names is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention, the Public Health Service, or the U.S. Department of Health and Human Services.
Footnotes
Supplemental material is available online only.
Contributor Information
Eric Rogier, Email: erogier@cdc.gov.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Ikegami T, Makino S. 2011. The pathogenesis of Rift Valley fever. Viruses 3:493–519. 10.3390/v3050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nanyingi MO, Munyua P, Kiama SG, Muchemi GM, Thumbi SM, Bitek AO, Bett B, Muriithi RM, Njenga MK. 2015. A systematic review of Rift Valley fever epidemiology 1931–2014. Infect Ecol Epidemiol 5:28024. 10.3402/iee.v5.28024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshra H, Lorenzo G, Busquets N, Brun A. 2011. Rift Valley fever: recent insights into pathogenesis and prevention. J Virol 85:6098–6105. 10.1128/JVI.02641-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontenille D, Traore-Lamizana M, Diallo M, Thonnon J, Digoutte JP, Zeller HG. 1998. New vectors of Rift Valley fever in West Africa. Emerg Infect Dis 4:289–293. 10.3201/eid0402.980218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nepomichene TNJJ, Raharimalala FN, Andriamandimby SF, Ravalohery J-P, Failloux A-B, Heraud J-M, Boyer S. 2018. Vector competence of Culex antennatus and Anopheles coustani mosquitoes for Rift Valley fever virus in Madagascar. Med Vet Entomol 32:259–262. 10.1111/mve.12291. [DOI] [PubMed] [Google Scholar]
- 6.Grossi-Soyster EN, Lee J, King CH, LaBeaud AD. 2019. The influence of raw milk exposures on Rift Valley fever virus transmission. PLoS Negl Trop Dis 13:e0007258. 10.1371/journal.pntd.0007258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lancelot R, Béral M, Rakotoharinome VM, Andriamandimby S-F, Héraud J-M, Coste C, Apolloni A, Squarzoni-Diaw C, de La Rocque S, Formenty PBH, Bouyer J, Wint GRW, Cardinale E. 2017. Drivers of Rift Valley fever epidemics in Madagascar. Proc Natl Acad Sci USA 114:938–943. 10.1073/pnas.1607948114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholas DE, Jacobsen KH, Waters NM. 2014. Risk factors associated with human Rift Valley fever infection: systematic review and meta-analysis. Trop Med Int Health 19:1420–1429. 10.1111/tmi.12385. [DOI] [PubMed] [Google Scholar]
- 9.Tigoi C, Sang R, Chepkorir E, Orindi B, Arum SO, Mulwa F, Mosomtai G, Limbaso S, Hassan OA, Irura Z, Ahlm C, Evander M. 2020. High risk for human exposure to Rift Valley fever virus in communities living along livestock movement routes: A cross-sectional survey in Kenya. PLoS Negl Trop Dis 14:e0007979. 10.1371/journal.pntd.0007979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alhaji NB, Babalobi OO, Wungak Y, Ularamu HG. 2018. Participatory survey of Rift Valley fever in nomadic pastoral communities of North-central Nigeria: the associated risk pathways and factors. PLoS Negl Trop Dis 12:e0006858. 10.1371/journal.pntd.0006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bird BH, Ksiazek TG, Nichol ST, Maclachlan NJ. 2009. Rift Valley fever virus. J Am Vet Med Assoc 234:883–893. 10.2460/javma.234.7.883. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl JF, Ragan IK, Rowland RR, Wainaina M, Mbotha D, Wilson W. 2019. A multiplex fluorescence microsphere immunoassay for increased understanding of Rift Valley fever immune responses in ruminants in Kenya. J Virol Methods 269:70–76. 10.1016/j.jviromet.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Jansen van Vuren P, Potgieter AC, Paweska JT, van Dijk AA. 2007. Preparation and evaluation of a recombinant Rift Valley fever virus N protein for the detection of IgG and IgM antibodies in humans and animals by indirect ELISA. J Virol Methods 140:106–114. 10.1016/j.jviromet.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Fafetine JM, Tijhaar E, Paweska JT, Neves LCBG, Hendriks J, Swanepoel R, Coetzer JAW, Egberink HF, Rutten VPMG. 2007. Cloning and expression of Rift Valley fever virus nucleocapsid (N) protein and evaluation of a N-protein based indirect ELISA for the detection of specific IgG and IgM antibodies in domestic ruminants. Vet Microbiol 121:29–38. 10.1016/j.vetmic.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Ragan IK, Davis AS, McVey DS, Richt JA, Rowland RR, Wilson WC. 2018. Evaluation of fluorescence microsphere immunoassay for detection of antibodies to Rift Valley fever virus nucleocapsid protein and glycoproteins. J Clin Microbiol 56:e01626-17. 10.1128/JCM.01626-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaBeaud AD, Muiruri S, Sutherland LJ, Dahir S, Gildengorin G, Morrill J, Muchiri EM, Peters CJ, King CH. 2011. Postepidemic analysis of Rift Valley fever virus transmission in northeastern kenya: a village cohort study. PLoS Negl Trop Dis 5:e1265. 10.1371/journal.pntd.0001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subudhi S, Dakouo M, Sloan A, Stein DR, Grolla A, Jones S, Dibernardo A, Rosenke K, Sas M, Traore A, Lindsay R, Groschup MH, Misra V, Feldmann H, Sogoba N, Safronetz D, Niang M. 2018. Seroprevalence of Rift Valley fever virus antibodies in cattle in Mali, 2005–2014. Am J Trop Med Hyg 98:872–874. 10.4269/ajtmh.17-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudo ES, Pinto G, Weyer J, Le Roux C, Mandlaze A, José AF, Muianga A, Paweska JT. 2016. Serological evidence of rift valley fever virus among acute febrile patients in Southern Mozambique during and after the 2013 heavy rainfall and flooding: implication for the management of febrile illness. Virol J 13:96. 10.1186/s12985-016-0542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray GC, Anderson BD, LaBeaud AD, Heraud J-M, Fèvre EM, Andriamandimby SF, Cook EAJ, Dahir S, de Glanville WA, Heil GL, Khan SU, Muiruri S, Olive M-M, Thomas LF, Merrill HR, Merrill MLM, Richt JA. 2015. Seroepidemiological study of interepidemic rift valley fever virus infection among persons with intense ruminant exposure in Madagascar and Kenya. Am J Trop Med Hyg 93:1364–1370. 10.4269/ajtmh.15-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faburay B, Wilson WC, Secka A, Drolet B, McVey DS, Richt JA. 2019. Evaluation of an indirect enzyme-linked immunosorbent assay based on recombinant baculovirus-expressed rift valley fever virus nucleoprotein as the diagnostic antigen. J Clin Microbiol 57:e01058-19. 10.1128/JCM.01058-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jori F, Alexander KA, Mokopasetso M, Munstermann S, Moagabo K, Paweska JT. 2015. Serological evidence of Rift Valley fever virus circulation in domestic cattle and African buffalo in Northern Botswana (2010–2011). Front Vet Sci 2:63. 10.3389/fvets.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Britch SC, Binepal YS, Ruder MG, Kariithi HM, Linthicum KJ, Anyamba A, Small JL, Tucker CJ, Ateya LO, Oriko AA, Gacheru S, Wilson WC. 2013. Rift Valley fever risk map model and seroprevalence in selected wild ungulates and camels from Kenya. PLoS One 8:e66626. 10.1371/journal.pone.0066626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plucinski MM, Candrinho B, Chambe G, Muchanga J, Muguande O, Matsinhe G, Mathe G, Rogier E, Doyle T, Zulliger R, Colborn J, Saifodine A, Lammie P, Priest JW. 2018. Multiplex serology for impact evaluation of bed net distribution on burden of lymphatic filariasis and four species of human malaria in northern Mozambique. PLoS Negl Trop Dis 12:e0006278. 10.1371/journal.pntd.0006278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mordecai EA, Ryan SJ, Caldwell JM, Shah MM, LaBeaud AD. 2020. Climate change could shift disease burden from malaria to arboviruses in Africa. Lancet Planet Health 4:e416–e423. 10.1016/S2542-5196(20)30178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niklasson B, Liljestrand J, Bergstrom S, Peters CJ. 1987. Rift Valley fever: a sero-epidemiological survey among pregnant women in Mozambique. Epidemiol Infect 99:517–522. 10.1017/s0950268800068011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudo ES, Falk K, Cliff J. 2018. Historical perspective of arboviruses in Mozambique and its implication for current and future epidemics. Adv Exp Med Biol 1062:11–18. 10.1007/978-981-10-8727-1_2. [DOI] [PubMed] [Google Scholar]
- 27.Blomström A-L, Scharin I, Stenberg H, Figueiredo J, Nhambirre O, Abilio A, Berg M, Fafetine J. 2016. Seroprevalence of Rift Valley fever virus in sheep and goats in Zambezia, Mozambique. Infect Ecol Epidemiol 6:31343. 10.3402/iee.v6.31343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jäckel S, Eiden M, Balkema-Buschmann A, Ziller M, van Vuren PJ, Paweska JT, Groschup MH. 2013. A novel indirect ELISA based on glycoprotein Gn for the detection of IgG antibodies against Rift Valley fever virus in small ruminants. Res Vet Sci 95:725–730. 10.1016/j.rvsc.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Fafetine J, Neves L, Thompson PN, Paweska JT, Rutten VP, Coetzer JA. 2013. Serological evidence of Rift Valley fever virus circulation in sheep and goats in Zambezia Province, Mozambique. PLoS Negl Trop Dis 7:e2065. 10.1371/journal.pntd.0002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fafetine JM, Coetzee P, Mubemba B, Nhambirre O, Neves L, Coetzer JAW, Venter EH. 2016. Rift Valley fever outbreak in livestock, Mozambique, 2014. Emerg Infect Dis 22:2165–2167. 10.3201/eid2212.160310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagerqvist N, Moiane B, Mapaco L, Fafetine J, Vene S, Falk KI. 2013. Antibodies against Rift Valley fever virus in cattle, Mozambique. Emerg Infect Dis 19:1177–1179. 10.3201/eid1907.130332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold BF, van der Laan MJ, Hubbard AE, Steel C, Kubofcik J, Hamlin KL, Moss DM, Nutman TB, Priest JW, Lammie PJ. 2017. Measuring changes in transmission of neglected tropical diseases, malaria, and enteric pathogens from quantitative antibody levels. PLoS Negl Trop Dis 11:e0005616. 10.1371/journal.pntd.0005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogier E, Nace D, Dimbu PR, Wakeman B, Pohl J, Beeson JG, Drakeley C, Tetteh K, Plucinski M. 2021. Framework for characterizing longitudinal antibody response in children after plasmodium falciparum infection. Front Immunol 12:617951. 10.3389/fimmu.2021.617951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akkaya M, Kwak K, Pierce SK. 2020. B cell memory: building two walls of protection against pathogens. Nat Rev Immunol 20:229–238. 10.1038/s41577-019-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redding DW, Tiedt S, Lo Iacono G, Bett B, Jones KE. 2017. Spatial, seasonal and climatic predictive models of Rift Valley fever disease across Africa. Philos Trans R Soc Lond B Biol Sci 372. 10.6084/m9.figshare.c.3749951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muianga A, Pinto G, Massangaie M, Ali S, Oludele J, Tivane A, Falk KI, Lagerqvist N, Gudo ES. 2018. Antibodies against chikungunya in Northern Mozambique during dengue outbreak, 2014. Vector Borne Zoonotic Dis 18:445–449. 10.1089/vbz.2017.2261. [DOI] [PubMed] [Google Scholar]
- 37.Mugabe VA, Ali S, Chelene I, Monteiro VO, Guiliche O, Muianga AF, Mula F, António V, Chongo I, Oludele J, Falk K, Paploski IA, Reis MG, Kitron U, Kümmerer BM, Ribeiro GS, Gudo ES. 2018. Evidence for chikungunya and dengue transmission in Quelimane, Mozambique: results from an investigation of a potential outbreak of chikungunya virus. PLoS One 13:e0192110. 10.1371/journal.pone.0192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cartwright HN, Barbeau DJ, McElroy AK. 2021. Isotype-specific Fc effector functions enhance antibody-mediated Rift Valley fever virus protection in vivo. mSphere 6:e0055621. 10.1128/mSphere.00556-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olaleye OD, Tomori O, Schmitz H. 1996. Rift Valley fever in Nigeria: infections in domestic animals. Rev Sci Tech 15:937–946. 10.20506/rst.15.3.966. [DOI] [PubMed] [Google Scholar]
- 40.Rogier EW, Moss DM, Mace KE, Chang M, Jean SE, Bullard SM, Lammie PJ, Lemoine JF, Udhayakumar V. 2018. Use of bead-based serologic assay to evaluate Chikungunya virus epidemic, Haiti. Emerg Infect Dis 24:995–1001. 10.3201/eid2406.171447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poirier MJ, Moss DM, Feeser KR, Streit TG, Chang G-JJ, Whitney M, Russell BJ, Johnson BW, Basile AJ, Goodman CH, Barry AK, Lammie PJ. 2016. Measuring Haitian children's exposure to chikungunya, dengue and malaria. Bull World Health Organ 94:817–825A. 10.2471/BLT.16.173252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Priest JW, Moss DM. 2020. Measuring Cryptosporidium serologic responses by multiplex bead assay. Methods Mol Biol 2052:61–85. 10.1007/978-1-4939-9748-0_5. [DOI] [PubMed] [Google Scholar]
- 43.Waterboer T, Sehr P, Pawlita M. 2006. Suppression of non-specific binding in serological Luminex assays. J Immunol Methods 309:200–204. 10.1016/j.jim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Moss DM, Lammie PJ, Petri WA, Herbein J, Derado G, Hamlin K, Priest JW. Jr.. 2014. Longitudinal evaluation of enteric protozoa in Haitian children by stool exam and multiplex serologic assay. Am J Trop Med Hyg 90:653–660. 10.4269/ajtmh.13-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bird BH, Githinji JWK, Macharia JM, Kasiiti JL, Muriithi RM, Gacheru SG, Musaa JO, Towner JS, Reeder SA, Oliver JB, Stevens TL, Erickson BR, Morgan LT, Khristova ML, Hartman AL, Comer JA, Rollin PE, Ksiazek TG, Nichol ST. 2008. Multiple virus lineages sharing recent common ancestry were associated with a Large Rift Valley fever outbreak among livestock in Kenya during 2006–2007. J Virol 82:11152–11166. 10.1128/JVI.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shoemaker TR, Nyakarahuka L, Balinandi S, Ojwang J, Tumusiime A, Mulei S, Kyondo J, Lubwama B, Sekamatte M, Namutebi A, Tusiime P, Monje F, Mayanja M, Ssendagire S, Dahlke M, Kyazze S, Wetaka M, Makumbi I, Borchert J, Zufan S, Patel K, Whitmer S, Brown S, Davis WG, Klena JD, Nichol ST, Rollin PE, Lutwama J. 2019. First laboratory-confirmed outbreak of human and animal Rift Valley Fever virus in Uganda in 48 Years. Am J Trop Med Hyg 100:659–671. 10.4269/ajtmh.18-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zweig MH, Campbell G. 1993. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39:561–577. 10.1093/clinchem/39.4.561. [DOI] [PubMed] [Google Scholar]
- 48.Rogier E, Wiegand R, Moss D, Priest J, Angov E, Dutta S, Journel I, Jean SE, Mace K, Chang M, Lemoine JF, Udhayakumar V, Barnwell JW. 2015. Multiple comparisons analysis of serological data from an area of low Plasmodium falciparum transmission. Malar J 14:436. 10.1186/s12936-015-0955-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulldorf M. 1997. A spacial scan statistic. Commun Stat Theory 26:1481–1496. 10.1080/03610929708831995. [DOI] [Google Scholar]
- 50.Kulldorf M. 2015. Inc. SatScan TM v9.4: Software for the spatial and space-time scan statistics. https://www.satscan.org/. Accessed 8 August 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6. Download jvi.00672-22-s0001.pdf, PDF file, 0.5 MB (521KB, pdf)