ABSTRACT
Emerging severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) variants, especially the Omicron variant, have impaired the efficacy of existing vaccines and most therapeutic antibodies, highlighting the need for additional antibody-based tools that can efficiently neutralize emerging SARS-CoV-2 variants. The use of a “single” agent to simultaneously target multiple distinct epitopes on the spike is desirable in overcoming the neutralizing escape of SARS-CoV-2 variants. Herein, we generated a human-derived IgG-like bispecific antibody (bsAb), Bi-Nab35B5-47D10, which successfully retained parental specificity and simultaneously bound to the two distinct epitopes on receptor-binding domain (RBD) and S2. Bi-Nab35B5-47D10 showed improved spike binding breadth among wild-type (WT) SARS-CoV-2, variants of concern (VOCs), and variants being monitored (VBMs) compared with its parental monoclonal antibodies (MAbs). Furthermore, pseudotyped virus neutralization demonstrated that Bi-Nab35B5-47D10 can efficiently neutralize VBMs, including Alpha (B.1.1.7), Beta (B.1.351), and Kappa (B.1.617.1), as well as VOCs, including Delta (B.1.617.2), Omicron BA.1, and Omicron BA.2. Crucially, Bi-Nab35B5-47D10 substantially improved neutralizing activity against Omicron BA.1 (IC50 = 0.15 nM) and Omicron BA.2 (IC50 = 0.67 nM) compared with its parental MAbs. Therefore, Bi-Nab35B5-47D10 represents a potential effective countermeasure against SARS-CoV-2 Omicron and other variants of concern.
IMPORTANCE The new, highly contagious SARS-CoV-2 Omicron variant caused substantial breakthrough infections and has become the dominant strain in countries across the world. Omicron variants usually bear high mutations in the spike protein and exhibit considerable escape of most potent neutralization monoclonal antibodies and reduced efficacy of current COVID-19 vaccines. The development of neutralizing antibodies with potent efficacy against the Omicron variant is still an urgent priority. Here, we generated a bsAb, Bi-Nab35B5-47D10, which simultaneously targets SARS-CoV-2 RBD and S2 and improves the neutralizing potency and breadth against SARS-CoV-2 WT and the tested variants compared with their parental antibodies. Notably, Bi-Nab35B5-47D10 has more potent neutralizing activity against the VOC Omicron pseudotyped virus. Therefore, Bi-Nab35B5-47D10 is a feasible and potentially effective strategy by which to treat and prevent COVID-19.
KEYWORDS: COVID-19, SARS-CoV-2, bispecific antibodies, Omicron variants, neutralization, neutralizing antibodies
INTRODUCTION
Owing to the continuous SARS-CoV-2 evolution caused by mutations and recombination, numerous genetically distinct SARS-CoV-2 lineages have emerged, and five major variants, including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and the newly identified Omicron (B.1.529 and BA), have been designed sequentially, based on criteria such as various transmissibility and the ability to escape immunity (1–6). Most mutations of SARS-CoV-2 concern variants primarily clustered on the N-terminal domain (NTD) and RBD, two regions of spike S1 which are the two major domains being targeted by neutralizing antibodies (5, 7–10). Many of these mutations in S1 have been previously reported to undermine the effectiveness of COVID-19 vaccines and therapeutic neutralizing antibodies (11, 12). The recently identified SARS-CoV-2 Omicron variant of concern worsened the situation (13). The Omicron BA.1 variant harbors an unusually high number of mutations, a rapid spread capacity, and over 30 mutations in the viral spike protein, including 15 mutations in RBD (14). The Omicron variant has rapidly replaced the previously dominant Delta variant and has become the dominant circulating strain in many countries across the world since it was first reported in November 2021 in Botswana and South Africa (15, 16). The substantial mutations of the Omicron variant make it completely or partially resistant to neutralization by most potent MAbs against other VOCs and also result in significantly reduced efficacy of existing COVID-19 vaccines (4, 17–19). Therefore, it is still a top priority to develop highly potent and broadly monospecific or multispecific neutralizing MAbs targeting SARS-CoV-2 heavily mutated variants, such as the Omicron variant (20).
Monoclonal antibody cocktails and bispecific antibodies are effective strategies by which to counter the escape of highly mutated SARS-CoV-2 variants (21). The bsAb strategy has been successfully applied to the treatment of cancer, inflammatory disorders, and viral infectious diseases (22–25). bsAbs are advantageous over antibody cocktails, given the complicated formulations and costs of antibody cocktail strategies (26–29). The SARS-CoV-2 spike ectodomain is segregated into two units, termed S1 and S2. The S1 subunit of SARS-CoV-2, containing NTD and C-terminal RBD, is responsible for cellular binding and is targeted by most neutralizing antibodies, whereas the S2 subunit is relatively more conserved and mediates membrane fusion (30, 31). The S2 region, harboring neutralizing epitopes, is an alternative conserved target on the spike (32). To avail the cellular binding and fusion of two key steps of SARS-CoV-2 entry, we generated a human-derived, IgG1-like bispecific antibody, Bi-Nab35B5-47D10, based on two human neutralizing antibodies, 35B5 and 47D10, which target the spike RBD and S2 region, respectively. Human MAb 35B5 is a potent human MAbs pan-neutralizing against WT SARS-CoV-2 and VOCs, whereas 47D10 is an anti-S2 human neutralizing antibody with crossing activity against several beta coronaviruses (CoVs). Our results show that Bi-Nab35B5-47D10 neutralizing activities successfully maintained parental specificity and simultaneously targeted two epitopes of the RBD and S2 region and that the two arms of this bsAb potently neutralized various circulating SARS-CoV-2 variants. Notably, it has improved neutralization activity against the relatively resistant SARS-CoV-2 Delta and Omicron BA.1 variants. These results indicate the potential development of therapeutic strategies of Bi-Nab35B5-47D10 against SARS-CoV-2 Omicron and other variants.
RESULTS
Design and expression of bispecific antibodies.
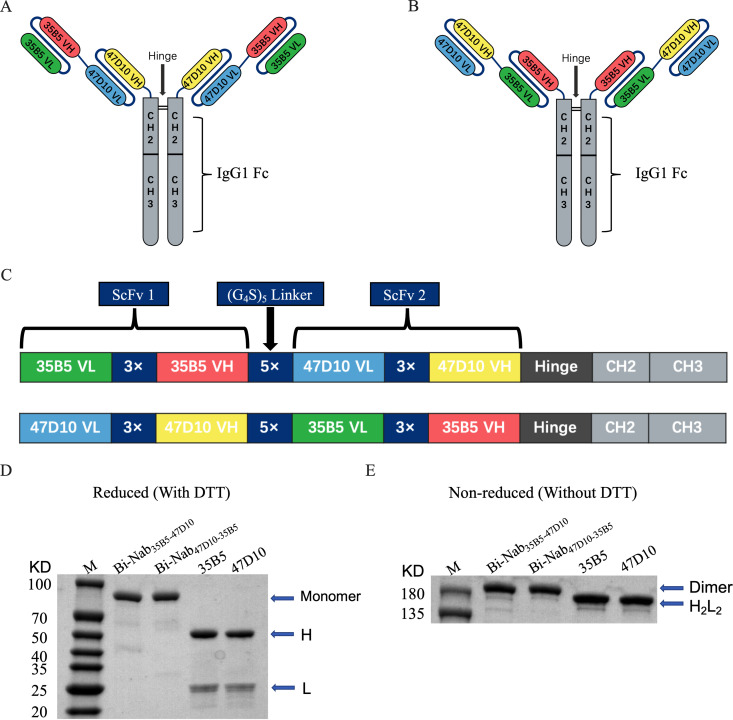
To obtain functional bsAbs, we designed and generated bsAbs containing tandem single-chain variable fragment (ScFv) domains of two potent neutralizing MAbs (35B5 and 47D10) with glycine-serine (G4S) linker separation based on structural information and computational simulations of spike trimers as previously described (33). We selected 35B5 and 47D10 as the two neutralizing antibodies, given that their epitopes are located in different regions of the spike and lack an unfavorable steric clash. The 35B5 MAb is a new class, RBD-targeting, potent neutralizing human antibody through a distinctive spike glycan displacement mechanism, and its epitope is invariant among WT SARS-CoV-2 and circulating variants (18, 34). The 47D10 MAb, isolated from WT SARS-CoV-2-infected patients’ memory B cells, was identified as a SARS-CoV-2 S2-specific MAb (Fig. S1A and B). Based on 35B5 and 47D10, we designed two molecular topologies of bsAbs: 35B5VL-VH → G4S linker → 47D10VL-VH (Fig. 1A) and 47D10VL-VH → G4S linker → 35B5VL-VH (Fig. 1B). To prevent possible steric resistance caused by the SARS-CoV-2 spike trimer, the empirical choice of 5× G4S linkers was made. To maintain the antibody Fc-mediated activities, the individual 35B5 and 47D10 ScFvs were then connected using 5× G4S linkers and fused to the IgG1 Fc to generate the IgG-like bsAbs molecule (Fig. 1C). Two bsAbs, Bi-Nab47D10-35B5 and Bi-Nab35B5-47D10, were produced by using the ExpiCHO Expression System and purified by affinity chromatography.
FIG 1.
Construction and generation of bsAbs. (A and B) Overview of the strategy for designing bsAbs. Schematic diagram of the molecular configurations of Bi-Nab35B5-47D10 (A) and Bi-Nab47D10-35B5 (B). (C) Schematic presentation of the bsAbs. Antibody domains are color-coded according to their architecture (green, variable light chain of 35B5; red, variable heavy chain of 35B5; blue, variable light chain of 47D10; yellow, variable heavy chain of 47D10; gray, human IgG1 Fc). (D) Reduced SDS-PAGE analysis of two bsAbs and two parental MAbs. The proteins were analyzed under reducing conditions (+DTT). H and L denote the heavy and light chains, respectively. The molecular weight of the bsAbs monomer was 90 kDa. M, molecular weight standard. (E) Nonreduced SDS-PAGE analysis of affinity purified bsAbs and parental MAbs. All four antibodies were expressed in ExpiCHO-S cells and captured on protein A affinity resin. The proteins were analyzed under direct affinity elution conditions (without DTT). Three independent experiments were performed at this scale and yielded the same results. Additional independent experiments yielded similar results at larger culture volumes.
To evaluate the biological activity of Bi-Nab47D10-35B5 and Bi-Nab35B5-47D10, we initially verified whether they were correctly folded soluble proteins by reducing and nonreducing SDS-PAGE (Fig. 1D and E). As expected, both Bi-Nab47D10-35B5 and Bi-Nab35B5-47D10 ran as homogeneous species at the expected molecular weights: monomer 90 kDa in the reducing SDS-PAGE gel and dimer 180 kDa in the nonreducing SDS-PAGE gel, respectively (Fig. 1D and E). These results indicate that eukaryotically expressed bsAbs fold correctly.
Binding and inhibiting properties of bsAbs.
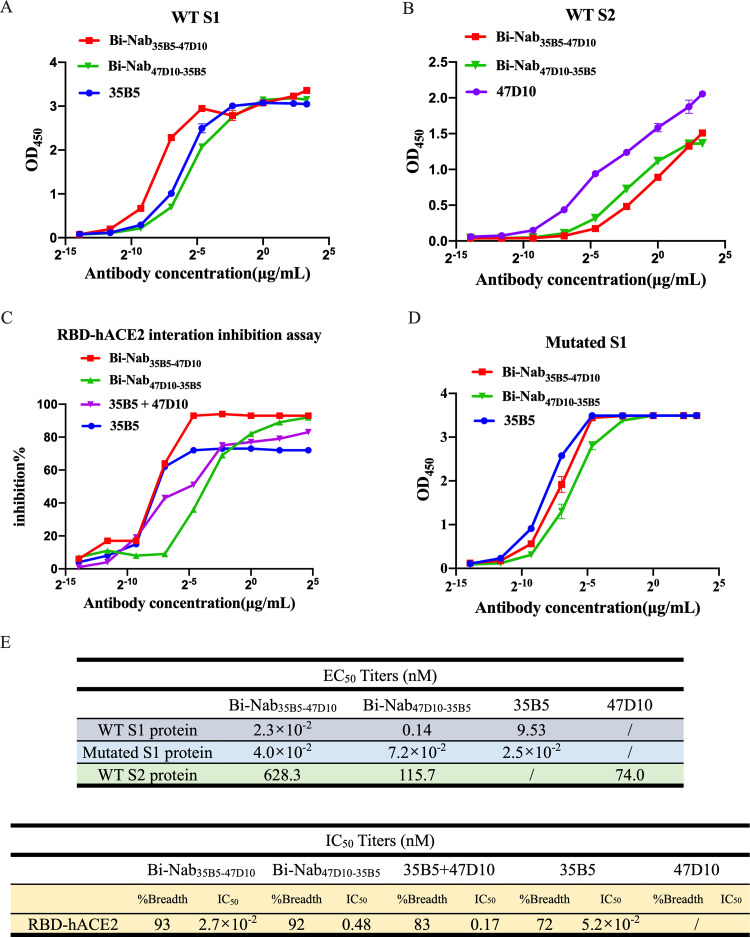
Next, to validate the simultaneous spike engagement of the two arms of the bsAbs, we performed an indirect enzyme-linked immunosorbent assay (ELISA) binding assay. Equal concentrations of the S1 protein and the S2 protein of the WT SARS-CoV-2 strain were used as the coating antigens (Fig. 2A and B). Bi-Nab35B5-47D10 showed the best binding ability to the WT SARS-CoV-2 S1 with the lowest concentration for the 50% of maximal effect (EC50) value, 2.3 × 10−2 nM (Fig. 2A and E). Both Bi-Nab35B5-47D10 and Bi-Nab47D10-35B5 display similar affinity to the S2 protein, although a reduced affinity of bsAbs to the S2 protein compared with the parental MAb 47D10 was observed (Fig. 2B). These findings confirm that Bi-Nab47D10-35B5 and Bi-Nab35B5-47D10 can simultaneously bind both the RBD and the S2 epitopes, demonstrating that both arms of the bsAbs are functional. Simultaneously, we assessed the potential of bsAbs to block the binding of angiotensin-converting enzyme 2 (ACE2) to the WT SARS-CoV-2 RBD using ELISA-based binding inhibition assays as previously described (35). Consistent with the binding results of bsAbs to WT S1, the Bi-Nab35B5-47D10 exhibited a potent blocking interaction between RBD and ACE2, similar to that of parental 35B5, with a half maximum inhibitory concentration (IC50) value of 2.7 × 10−2 nM, and was superior to both Bi-Nab47D10-35D5 and the cocktail (Fig. 2C and E). Furthermore, Bi-Nab35B5-47D10 also substantially bound to the mutated S1 proteins of SARS-CoV-2 VOCs, which included HV69-70 deletion and N501Y and D614G mutations, and harbored a binding capacity to the VOC S1 similar to that of parental 35B5 (Fig. 2D). Indeed, Bi-Nab35B5-47D10 demonstrated activity equal to, or in some cases better than, that of parental MAbs (Fig. 2E). These results indicate that Bi-Nab35B5-47D10 retains the binding capacity and breadth of its parental MAbs.
FIG 2.
Binding and inhibition properties of bsAbs. (A and B) ELISA binding assay of bsAbs and parental MAbs to the S1 protein (A) or the S2 protein (B) of WT SARS-CoV-2. EC50, concentration for 50% of maximal effect. (C) ELISA analysis of bsAbs or a cocktail or parental MAb-mediated inhibition of WT RBD protein binding to ACE2. For the antibody cocktail, the value on the x axis refers to the final antibody concentration. IC50, half maximal inhibitory concentration. (D) ELISA analysis of bsAbs or parental MAbs binding to the mutated S1 protein of SARS-CoV-2, including HV69-70 deletion, N501Y, and D614G. (E) Representative EC50 and IC50 titers of bsAbs and parental MAbs showing the effective binding and inhibiting activity of Bi-Nab35B5-47D10.
Crossing activity of bsAbs on SARS-CoV-2 VOCs and VBM S proteins.
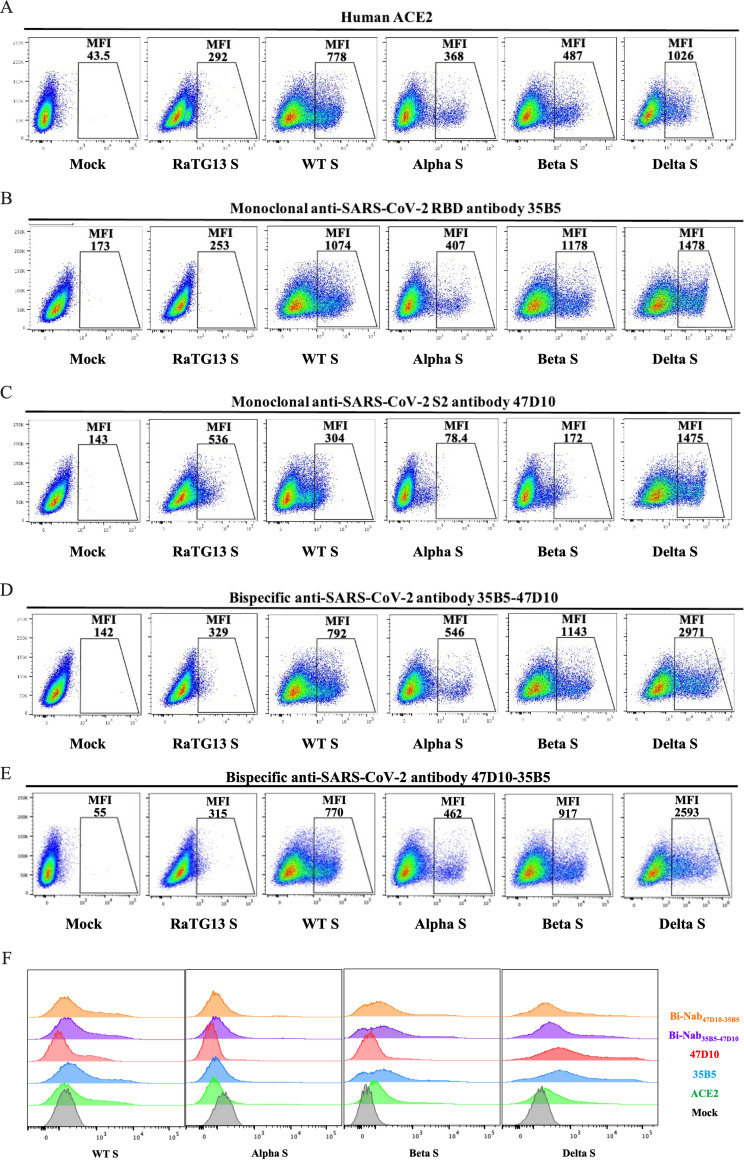
Previous work has identified that several CoVs, such as SARS-CoV and bat CoV RaTG13, utilize a similar binding mechanism with ACE2 to achieve cross-species infection and transmission (36, 37). Next, we further investigated whether bsAbs could induce cross-reactivity by recognizing different CoV S proteins in the native conformation. Human embryonic kidney (HEK) 293T cells transiently expressing RaTG13 S protein, SARS-CoV S protein, WT SARS-CoV-2 S protein, Alpha variant (B.1.1.7, N501Y in RBD) S protein (38), Beta variant (B.1.351, K417N, E484K, and N501Y in RBD) S protein (39, 40), or Delta variant (B.1.617.2, L452R, and T478K in RBD) S protein (41) were incubated with Bi-Nab35B5-47D10, Bi-Nab47D10-35B5, MAb 35B5. or MAb 47D10, respectively, followed by a flow cytometry analysis. ACE2 was used as a positive-control, and all CoV S proteins bound to ACE2 well, except RaTG13 (Fig. 3A). Previous work has reported that RaTG13 binds to hACE2 with a much lower affinity than to SARS-CoV-2 (36, 42). Consistent with the previous results that MAb 35B5 is a SARS-CoV-2-specific neutralizing antibody targeting a conserved epitope on RBD, 35B5 easily detected the expression of four different SARS-CoV-2 S proteins on the HEK293T cell surface with no cross-reactivity of the RaTG13 S protein (Fig. 3B). Neither the two parental MAbs nor the two bsAbs showed a binding signal for the SARS-CoV S protein (data not shown). We found that MAb 47D10 cross-reacted with the RaTG13 S protein (Fig. 3C), which expands the minor cross-reaction of Bi-Nab35B5-47D10 and Bi-Nab47D10-35B5 to the RaTG13 S protein (Fig. 3D and E).
FIG 3.
Binding kinetics of bsAbs to cell surface-associated coronavirus S protein. (A to E) Binding of bsAbs or parental MAbs to RaTG13 S, WT SARS-CoV-2 S, Alpha S, Beta S, or Delta S proteins. HEK293T cells were transfected to transiently express RaTG13 S, WT SARS-CoV-2, Alpha S, Beta S, or Delta S proteins and were incubated with MAb 35B5 (B), MAb 47D10 (C), Bi-Nab35B5-47D10 (D), and Bi-Nab47D10-35B5 (E), respectively, for 1 h on ice. Soluble hACE2 with a His tag was used as a positive-control (A), followed by a FITC-conjugated anti-human IgG Fc antibody or a FITC-conjugated anti-His antibody. Then, the cells were analyzed by flow cytometry. Mean fluorescence intensity (MFI) was normalized to the empty vector (mock) group (F). The experiments were performed three times, and one representative is shown.
The surface spike protein of Alpha variant, Beta variant, and Delta variant was readily detected by both Bi-Nab35B5-47D10 and Bi-Nab47D10-35B5, similar to the parental 35B5 MAb. Notably, both Bi-Nab35B5-47D10 and Bi-Nab47D10-35B5 increased the affinity of the Delta variant S protein, displaying mean fluorescent intensity (MFI) values of 2,971 and 2,593, respectively (Fig. 3D and E). Taken together, both bsAbs can specifically bind to the native S proteins of SARS-CoV-2 VOCs and VBMs (Fig. 3F).
Elevated neutralization sensitivity of Bi-Nab35B5-47D10 to SARS-CoV-2 VBM variants.
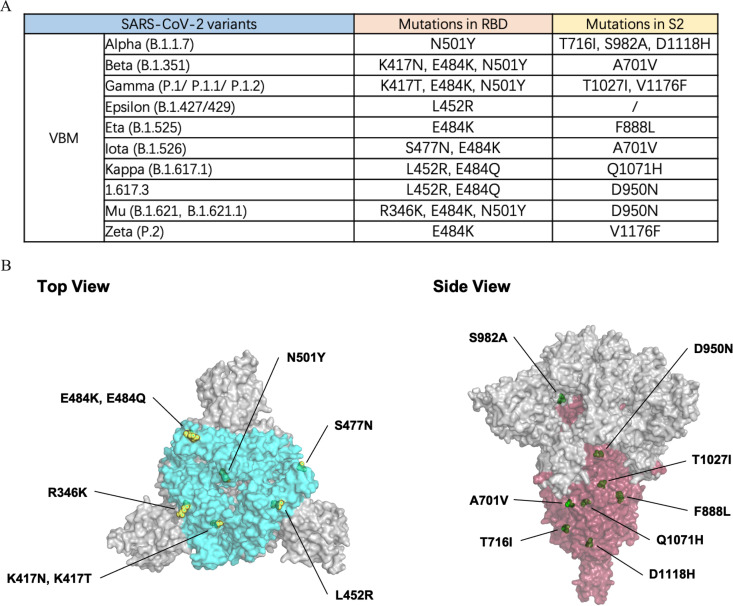
The emergence of SARS-CoV-2 VBMs with various mutations, including Alpha (with N501Y in RBD, T716I, S982A, and D1118H in S2), Beta (with K417N, E484K, and N501Y in RBD, A701V in S2), Gamma (with K417T, E484K and N501Y in RBD, T1027I and V1176F in S2), Epsilon (with L452R in RBD), Eta (with E484K in RBD, F888L in S2), Iota (with S477N and E484K in RBD, A701V in S2), Mu (with R346K, E484K, and N501Y in RBD, D950N in S2), Zeta (with E484K in RBD, V1176F in S2), 1.617.3 (with L452R and E484K in RBD, D950N in S2), and Kappa (with L452R and E484Q in RBD, Q1071H in S2), results in the escape of neutralization and threatens efforts to contain the COVID-19 pandemic (Fig. 4A).
FIG 4.
RBD and S2 mutations of VBM variants. (A) Statistics on VBM RBD and S2 mutations are displayed. (B) The crystal structure of the SARS-CoV-2 spike trimer (PDB ID: 7KRQ), highlighting the mutational landscape of VBM variants relative to WT SARS-CoV-2. The epitopes of the RBD (bright blue) and S2 (dark red) regions are shown. The mutations are indicated by yellow (RBD mutations) and green (S2 mutations) spheres on the surface of the S trimer, using the PyMOL software suite.
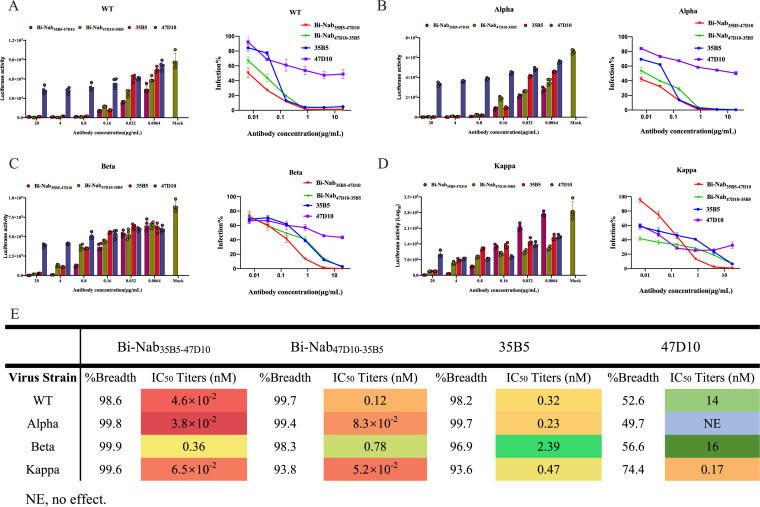
The rational design of bsAb is to avoid escape mutants. To initially assess the virus neutralization capacity of the two bsAbs in comparison to their parental MAbs 35B5 and 47D10, we performed neutralization assays with pseudovirus. Because the two parental MAbs target the RBD and S2, respectively, we focused on mutations of VBMs that occur in the RBD and S2 regions (Fig. 4B). Alpha, Beta, and Kappa variants were chosen to represent VBMs because these variants contain most of the VBM mutations (Fig. 4A). WT SARS-CoV-2, Alpha, Beta, and Kappa variants S protein pseudovirus were incubated with 35B5, 47D10, Bi-Nab35B5-47D10, or Bi-Nab47D10-35B5, respectively, and their transduction was measured according to luciferase activities. As expected, Bi-Nab35B5-47D10 neutralized WT SARS-CoV-2 S pseudovirus effectively, with an IC50 value of 4.6 × 10−2 nM (Fig. 5A), and displayed substantially improved neutralization breadth among the three VBM variants, especially for the Alpha and Kappa variants (Fig. 5B to D). Among the four antibodies, Bi-Nab35B5-47D10 and Bi-Nab47D10-35B5 potently neutralized the Alpha and Kappa variants and exhibited comparable neutralization potencies with much lower IC50 values than those displayed by the parental MAbs (Fig. 5B, D, and E). Among the three VBM variants, the Beta variant exhibited the least sensitivity to the 35B5 MAb (Fig. 5C). Notably, Bi-Nab35B5-47D10 increased the neutralization activity against the Beta pseudovirus approximately 6-fold, with the lowest IC50 value being 0.36 nM compared with 35B5 (Fig. 5C and E). Therefore, these results suggest an enhanced cross-reactivity of Bi-Nab35B5-47D10, as indicated by the elevated potency against the WT SARS-CoV-2, Alpha, Beta and Kappa variants (Fig. 5E).
FIG 5.
Neutralization of bsAbs against WT and VBM pseudoviruses. (A to D) Two parental MAbs and two bsAbs mediated neutralization of the indicated pseudovirus. WT SARS-CoV-2 S (A), Alpha S (B), Beta S (C), or Kappa S (D) pseudovirus were preincubated with 5-fold serially diluted Bi-Nab47D10-35B5, Bi-Nab35B5-47D10, 47D10, 35B5. Then, the mixture was added to HEK293 cells transiently expressing hACE2 and lysed 48 h later, and their transduction was measured according to luciferase activities. Potencies were calculated against sensitive viruses, and heatmaps of IC50 titers were generated in Excel. Warmer colors indicate more potent neutralization. Breadths based on IC50s are also summarized (E). Representative IC50 titers and neutralization breadths of bsAbs and the parental MAbs showing the improved neutralization activity of Bi-Nab35B5-47D10. NE, no effect. The experiment was performed twice, and one representative is shown. Error bars represent the standard error of the mean of technical triplicates.
Bi-Nab35B5-47D10 potently neutralizes SARS-CoV-2 Delta and Omicron variants.
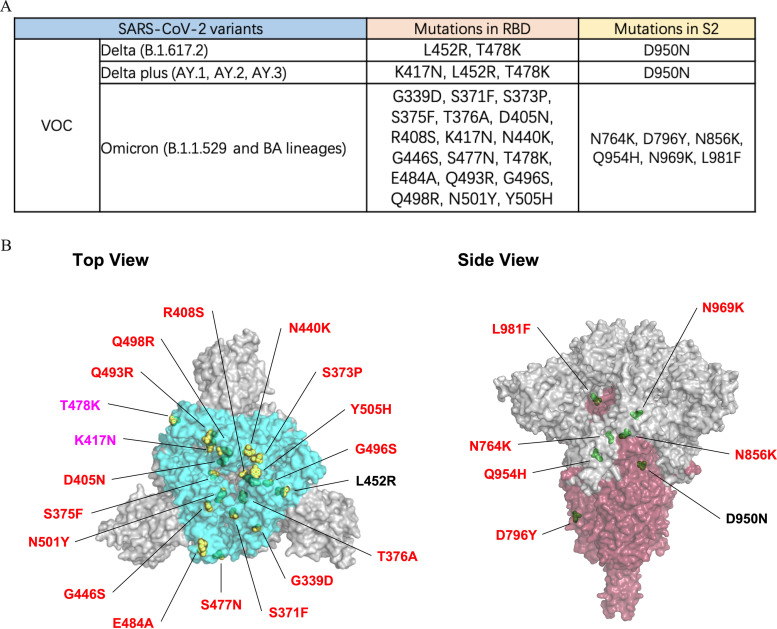
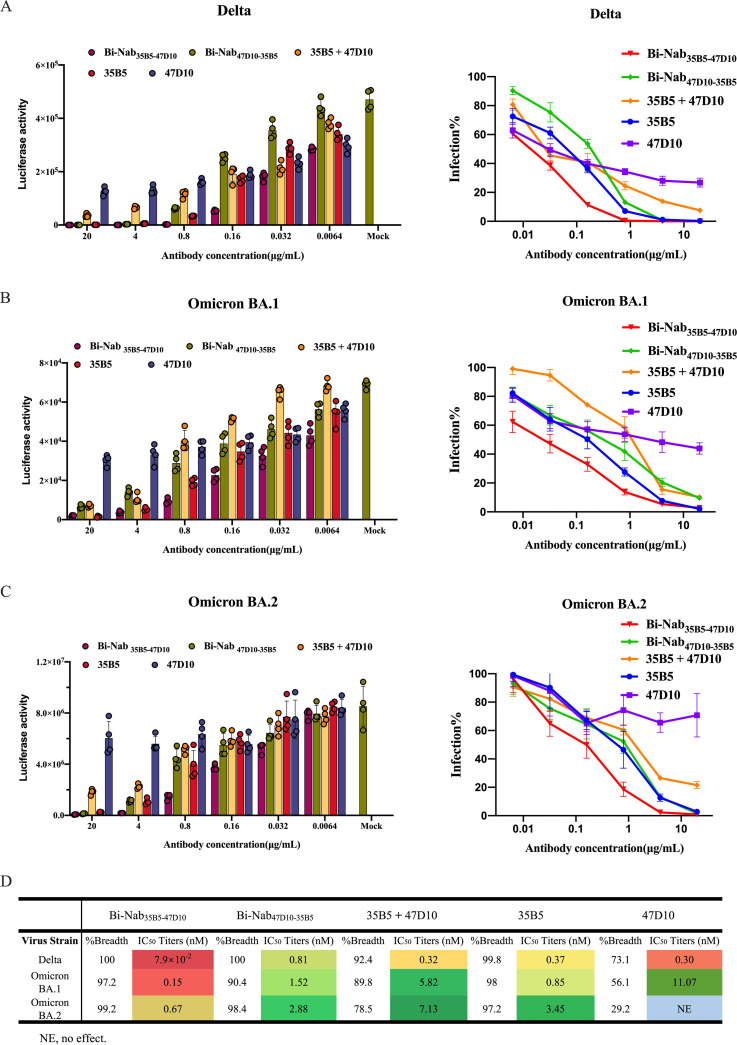
The Delta variant and Omicron variant are more transmissible than other variants and are linked to a resurgence of COVID-19 in many countries across the world. Finally, we set out to identify the neutralization properties of bsAbs on SARS-CoV-2 VOCs, namely, the Delta and Omicron variants. Compared to the mutations found in Delta, the Omicron lineage harbors more than 30 amino acid mutations in the spike protein, and most of the mutations are structurally focused in RBD (Fig. 6A) in regions accessible to antibodies at the top of the spike, increasing the likelihood of immune evasion (Fig. 6B) (43). Parental MAbs 35B5 and 47D10 exhibited comparable neutralization potencies against the Delta variant, with IC50 values of 0.37 nM and 0.30 nM (Fig. 7A), respectively, whereas the neutralizing activity of MAbs 35B5 and 47D10 against Omicron variants was reduced substantially, possibly due to the increased number of mutations in both the S1 and the S2 regions compared to that of Delta variants (Fig. 6B). 47D10 only slightly neutralized Omicron BA.1, with an IC50 value of 11.07 nM (Fig. 7B). At the same time, 47D10 did not effectively neutralize Omicron BA.2 (Fig. 7C). These results are consistent with published studies which show that Omicron displays enhanced neutralization escape compared with other SARS-CoV-2 variants. Interestingly, Bi-Nab47D10-35B5 exhibited substantially lower, but detectable, neutralization (0.81 nM for Delta, 1.52 nM for Omicron BA.1, 2.88 nM for Omicron BA.2). Among these antibodies, Bi-Nab35B5-47D10 showed substantially higher neutralization titers against the Delta, Omicron BA.1, and Omicron BA.2 variants, with IC50 values of 7.9 × 10−2 nM, 0.15 nM, and 0.67 nM, respectively, which were substantially lower than those of Bi-Nab47D10-35B5 or the cocktail (Fig. 7D). Specifically, Bi-Nab35B5-47D10 neutralizes Omicron BA.1 with 39-fold higher potency than a cocktail. This indicates that the molecular topology of bsAbs substantially impacts the neutralizing activity against VOCs. Notably, the neutralizing activity of Bi-Nab35B5-47D10 against Omicron was largely enhanced when linking the ScFv of 35B5 and 47D10, although MAb 47D10 only slightly neutralized Omicron (Fig. 7D). Collectively, Bi-Nab35B5-47D10 potently neutralized both the Delta and Omicron variants and substantially increased the neutralization activity in comparison with the parental MAbs.
FIG 6.
RBD and S2 mutations of VOC variants. (A) Schematic of VOC RBD and S2 mutations is illustrated. (B) Top view (left panel) and side view (right panel) of spike trimer are shown with mutations in RBD and S2 and highlighted with residue atoms as colored spheres, indicated in yellow (RBD mutations, the red font indicates the mutations unique to the Omicron variant, and the pink refers to the mutations common to VOCs) and green (S2 mutations) on the surface of the S trimer (PDB ID: 7KRQ).
FIG 7.
Cross-reactive neutralization of bsAbs against Delta and Omicron pseudoviruses. Using a lentiviral-based pseudovirus system, the neutralization potency of two parental MAbs and two bsAbs against Delta (A), Omicron BA.1 (B), and Omicron BA.2 (C) pseudoviruses were analyzed. The IC50 was determined by the log (inhibitor) response of a nonlinear regression analysis, and bars and error bars depict the mean and the standard error of the mean, respectively. (D) Bi-Nab35B5-47D10 exhibited significantly improved neutralization activity compared to Bi-Nab47D10-35B5 and parental MAbs. IC50 titers, breadth, and potency of two bsAbs and two parental MAbs against Delta and Omicron pseudoviruses are presented with heat maps. NE, no effect.
DISCUSSION
The ongoing evolution of SARS-CoV-2 and the emergence of new SARS-CoV-2 variants compromise the efficacy of current SARS-CoV-2 vaccines and licensed MAb therapies (44, 45). The unusually high mutations of the Omicron variant with high spreads have resulted in breakthrough infections in the world since its first report in November 2021 (46, 47). It is still urgent to develop highly potent and broadly neutralizing MAbs targeting multiple SARS-CoV-2 variants (9, 48). In this study, we generated a potent bispecific MAb Bi-Nab35B5-47D10 targeting spike RBD and S2 in two distinct regions. Our results demonstrate that Bi-Nab35B5-47D10 retained the specificity of their parental MAbs and displayed increased potency and breadth compared with their parental MAbs. Bi-Nab35B5-47D10 exhibited pan-neutralizing activities against WHO-stated SARS-CoV-2 VBMs and VOCs, including the B.1.617.2 (Delta), Omicron BA.1, and Omicron BA.2 variants, highlighting its potential application in the prevention and treatment of SARS-CoV-2 VOCs.
Bispecific or multiple specific antibodies targeting different regions of the spike protein are a favorable strategy by which to treat COVID-19 caused by the ongoing emergence of new SARS-CoV-2 VOCs (49, 50). Except for the increased threshold to produce neutralizing escape mutants, bispecific antibodies have practical and cost advantages over the MAb cocktail strategy since the complicated formulation of MAb cocktails routinely increases manufacturing costs and volumes. Here, we explored two SARS-CoV-2 neutralizing MAbs, 35B5 and 47D10, which target divergent spike regions and block SARS-CoV-2 infection by distinct mechanisms. 47D10 is an S2-targeting MAb, and 35B5 binds to an invariant epitope in the RBD and causes dissociation of the spike trimer by a glycan displacement action (34). The epitope of 35B5 in the RBD is invariant in SARS-CoV-2 WT, four VBMs, and two Delta and Omicron VOCs (18, 34). The rationale for bsAbs targeting both RBD and S2 is that RBD and S2 are responsible for two different critical steps of viral entry: viral binding, and viral fusion, respectively. Furthermore, the S2 region is relatively conserved and has fewer mutations than does the RBD. As expected, Bi-Nab35B5-47D10 shows improved binding neutralization activity compared to their parent MAbs. The superior neutralization of Bi-Nab35B5-47D10 relative to Bi-Nab47D10-35B5 against Delta and Omicron variants may be related to the position order of the neutralization variable region and spike structure. Although the combination of 35B5 and 47D10 through the bispecific IgG-like design enhanced the binding and neutralization activity, the cocktail of 35B5 and 47D10 did not show a synergistic effect compared with 35B5 alone (Fig. 7). Structural data of Bi-Nab35B10-47D10 and spike complexes will provide mechanistic clues to the enhanced function of Bi-Nab35B5-47D10.
SARS-CoV-2 evolution is ongoing, and continuously emerging SARS-CoV-2 variants have led to successive waves of infection. In addition to the earlier Omicron variants BA.1 and BA.2, more Omicron subvariants have emerged, including BA.3, BA.4, and BA.5 (51, 52). Omicron BA.4 and BA.5, the two most recent Omicron sublineages, exhibit further neutralization escape from COVID-19-vaccinated serum and are fueling a new wave of infection in South Africa and in other areas in the world (53, 54). Fortunately, the epitope of 35B5 on RBD does not contain the mutations of Omicron BA.1/2 and BA.4 (Fig. 6) (34, 51, 53). Crucially, Bi-Nab35B5-47D10 further improved the potency to neutralize the Omicron VOC in comparison with parental 35B5 (Fig. 7). Therefore, it is rational that Bi-Nab35B10-47D10 will potently be of interest for further clinical development for COVID-19 treatment.
MATERIALS AND METHODS
Design and expression of bispecific antibodies.
Plasmids containing the heavy and light chain genes for the production of the monoclonal antibodies 35B5 and 47D10 were prepared as previously described (55). Single-chain Fv format bispecific antibodies were designed from the sequences of the variable regions of monoclonal antibodies 35B5 and 47D10 (ScFv 35B5-47D10) or 47D10 and 35B5 (ScFv 47D10-35B5), utilizing tandem glycine-serine (G4S) peptide linkers. Codon-optimized ScFvs DNA sequences were synthesized and cloned into the pUC57 cloning vector (GenScript, Piscataway, NJ) and subcloned into the eukaryotic cell expression vector AbVec-hIgG1 between the AgeI and HindIII sites.
The ExpiCHO Expression System (Thermo Fisher) was used to produce bsAbs. Briefly, following the manufacturer’s max titer protocol, 25 mL (6 × 106 cells/mL) ExpiCHO-S cells in a 125 mL flask were transfected with a master mixture containing 25 μg bispecific antibody plasmid and 80 μL ExpiFectamine CHO reagent diluted in 1 mL cold OptiPRO SFM complexation medium. The ExpiFectamine CHO/DNA complexes were added to the cells immediately or after up to 5 min of incubation at room temperature without any loss of performance. The cells were incubated on an orbital shaker at 37°C with a humidified atmosphere of 8% CO2 humidified in air, without the need to change or add media. On the day after transfection, 150 μL ExpiCHO enhancer and 6 mL ExpiCHO feed were added to the flask. A second volume of ExpiCHO feed was added to cultured cells on day 5 posttransfection, and the flask was immediately returned to the shaking incubator. Supernatants were harvested at 4,000 to 5,000 × g for 30 min in a refrigerated centrifuge, and the supernatant was filtered through a 0.22 μm filter on day 12 post transfection.
Bispecific antibody isolation and purification.
Briefly, bsAbs were efficiently purified by using protein A-Sepharose affinity chromatography medium (GenScript, L00210-10). The purified antibodies were separated on a 7.5 to 12% polyacrylamide gel and revealed with Coomassie blue under reducing or nonreducing conditions. To ensure functionality, stability, and batch-to-batch consistency, all antibodies were subjected to quality control and biophysical characterization.
Cells and plasmids.
HEK293T cells producing pseudovirus and HEK293 cells overexpressing recombinant human ACE2 (293/hACE2) were preserved in our laboratory and maintained in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher) containing 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin and were incubated at 37°C with 5% CO2 and 95% humidity. ExpiCHO-S cells (Thermo Fisher) were cultured in ExpiCHO expression medium in 125 mL shaker flasks in a 37°C incubator with ≥80% relative humidity and 8% CO2 on an orbital shaker with a 50 mm shaking diameter rotating at 95 rpm. For routine maintenance, ExpiCHO-S cells were typically passaged every 3 days at a ratio of 1:20 when they were grown to 3 to 5 × 106 cells/mL. The pcDNA3.1-hACE2 plasmid with human codon optimization, plasmids encoding WT SARS-CoV-2 S glycoprotein, SARS-CoV-2 VBM spike protein, SARS-CoV-2 VOC S glycoprotein, RaTG13 S glycoprotein, lentiviral packaging plasmid psPAX2, and pLenti-GFP lentiviral reporter plasmid, were generously gifted by Zhaohui Qian.
Enzyme-linked immunosorbent assay.
To evaluate antibody characterization in vitro, the ELISA method with modifications was used as reported previously (56). In brief, 50 ng of the SARS-CoV-2 S1 protein of the WT strain (Sino Biological, 40591-V08H) or B.1.1.7 (Sino Biological, 40591-V08H7) or SARS-CoV-2 S2 protein (Sino Biological, 40590-V08B) or SARS-CoV-2 RBD protein (Sino Biological, 40592-V08B) was coated on ELISA plates in 100 μL per well at 37°C for 2 h or at 4°C overnight. After washing 3 times with PBST, blocking buffer with 5% FBS and 0.05% Tween 20 was added to the ELISA plates and incubated for 1 h at 37°C. Next, 100 mL serially diluted MAbs or bsAbs was added to each well in 100 μL blocking buffer for 1 h at 37°C. Following washing 3 times, mouse anti-human IgG Fc secondary antibody with HRP (Abcam) was added and incubated at 37°C for 1 h, followed by washing with PBST. The ELISA plates were reacted with 3,30,5,50-tetramethylbenzidine (TMB, Sigma) substrate at 25°C for 5 min and then stopped by 0.2 M H2SO4 stop buffer. The optical density (OD) at 450 nm was measured using an iMark microplate absorbance reader (BIO-RAD). Nonlinear regression was used to calculate the EC50.
ELISA-based receptor-binding inhibition of hACE2.
The ability of antibodies to inhibit the binding of the SARS-CoV-2 RBD to hACE2 was investigated by ELISA. The 96-well ELISA plates were coated with 200 ng of hACE2 protein (Sino Biological, 10108-H08H) per well overnight at 4°C and washed with PBST and blocked for 1 h with blocking buffer as above. Meanwhile, 5-fold serial dilutions of MAbs or bispecific antibodies or a cocktail were incubated with 4 ng/mL SARS-CoV-2 RBD with mouse IgG Fc tag protein (Sino Biological, 40592-V05H) for 1 h at 25°C. Then, the mixtures were added to ELISA plates and incubated for 1 h at 37°C. After further washing, bound SARS-CoV-2 RBD protein was detected with anti-mouse Fc HRP antibody (Abcam) diluted 1:10000 in blocking solution, followed by washing with PBST. The ELISA plates were reacted with TMB substrate at 25°C for 5 min and then stopped by 0.2 M H2SO4 stop buffer and determined at OD 450 nm. The IC50 was determined by using 4-parameter logistic regression.
SARS-CoV-2 pseudotyped reporter virus and pseudotyped virus neutralization assay.
The generation of SARS-CoV-2 or SARS-CoV-2 VOC-type pseudoviruses was performed as previously described (57). In brief, pseudoviruses were produced by using PEI to cotransfect 293T cells with psPAX2, pLenti-GFP, and plasmids encoding SARS-CoV-2 S, SARS-CoV-2 VOCs S, SARS-CoV S, RaTG13 S, or empty vector at a ratio of 1:1:1. The medium was replaced with fresh media containing 10% fetal bovine serum and 1% penicillin-streptomycin 4 h post transfection. The supernatants were harvested 48 h post transfection and centrifuged at 800 × g for 5 min to remove cell debris before passing through a 0.45 μm filter.
For the pseudovirus neutralization assay, HEK293 (hACE2/293) cells 80 to 90% confluent in T75 cell culture flasks were transfected with 20 μg of plasmid encoding hACE2 for 36 h and seeded into 24-well plates the day before transduction with pseudovirus. The initial concentration of 35B5, 47D10, cocktail, Bi-Nab35B5-47D10, and Bi-Nab47D10-35B5 to pseudovirus was 20 μg/mL. Fivefold serially diluted bsAbs or MAbs or a cocktail were incubated with SARS-CoV-2 pseudotyped virus for 1 h. The 500 μL per well mixture was subsequently incubated with hACE2/293 cells overnight, and then the mixture was changed to fresh media. Approximately 48 h post incubation, the luciferase activity of SARS-CoV-2-type pseudovirus-infected hACE2/293 cells was detected by the Dual-Luciferase Reporter Assay System (Promega), and cells were lysed with 120 μL medium containing 50% Steady-Glo and 50% complete cell growth medium at room temperature for 5 min. The percentage of infection was normalized to those derived from cells infected with SARS-CoV-2 pseudotyped virus in the absence of antibodies. The neutralization breadth of an antibody against SARS-CoV-2 VBMs and VOCs was based on the maximum percentage of pseudovirus that could have been neutralized by that antibody. The IC50 was determined by using 4-parameter logistic regression.
Flow cytometry-based bsAb binding assay.
HEK293T cells 80 to 90% confluent in 6-well cell culture plates were transfected with 2 μg of plasmids encoding either WT SARS-CoV-2 S or SARS-CoV-2 mutated S protein or SARS-CoV S protein or RaTG13 S protein using linear polyethylenimine (PEI, Sigma–Aldrich, 408727). A total amount of 2 μg DNA diluted in 1 mL per well of DMEM was mixed with PEI in a 1:2 ratio. The transfection mixture was added to cell culture plates with DMEM in a dropwise manner after 15 min of incubation at room temperature. After 4 to 6 h of incubation at 37°C under 5% CO2, the medium was changed to fresh complete cell growth medium (DMEM supplemented with 10% FBS and 1% penicillin-streptomycin). Cells were detached by using PBS with 1 mM EDTA 40 h post transfection and washed with cold PBS containing 2% FBS. After washing, the cells were incubated with bsAbs, monoclonal human anti-SARS-CoV-2 RBD antibody 35B5, or monoclonal human anti-SARS-CoV-2 S2 antibody 47D10 (5 μg per well) for 1 h on ice, followed by FITC-conjugated anti-human IgG (1:100) (ZSGB-Bio, ZF-0308) for 1 h on ice away from light. Cells were acquired by flow cytometry (BD Biosciences) and analyzed using FlowJo. Normalized MFI represents data for each sample with the mean MFI of the control cells subtracted.
Statistical analysis.
All statistical analyses were performed using GraphPad Prism 9.0 software. In the ELISA, three independent experiments were performed, and the mean values ± standard error of the mean and the EC50 values were calculated by using sigmoidal dose-response nonlinear regression. In the ELISA-based inhibition assay and pseudovirus neutralization assay, three or two independent experiments were performed, and the mean values ± standard error of the mean and the IC50 values were plotted by fitting a nonlinear 4-parameter dose-response curve. The figure legends show all of the statistical details of the experiments. PyMol was used to prepare all of the structure figures (Schrodinger: https://www.schrodinger.com/pymol).
ACKNOWLEDGMENTS
The work was supported by the National Natural Science Foundation of China (32192453) and the Chinese Universities Scientific Fund (2022RC019 and 2022TC163). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding resources.
Footnotes
Supplemental material is available online only.
Contributor Information
Zhaohui Qian, Email: zqian2013@sina.com.
Lilin Ye, Email: yelilinlcmv@tmmu.edu.cn.
Pinghuang Liu, Email: liupinghuang@cau.edu.cn.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Arora P, Kempf A, Nehlmeier I, Graichen L, Sidarovich A, Winkler MS, Schulz S, Jäck H-M, Stankov MV, Behrens GMN, Pöhlmann S, Hoffmann M. 2021. Delta variant (B.1.617.2) sublineages do not show increased neutralization resistance. Cell Mol Immunol 18:2557–2559. 10.1038/s41423-021-00772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruel T, Hadjadj J, Maes P, Planas D, Seve A, Staropoli I, Guivel-Benhassine F, Porrot F, Bolland W-H, Nguyen Y, Casadevall M, Charre C, Péré H, Veyer D, Prot M, Baidaliuk A, Cuypers L, Planchais C, Mouquet H, Baele G, Mouthon L, Hocqueloux L, Simon-Loriere E, André E, Terrier B, Prazuck T, Schwartz O. 2022. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat Med 28:1297–1302. 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- 3.Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IATM, Datir R, Collier DA, Albecka A, Singh S, Pandey R, Brown J, Zhou J, Goonawardane N, Mishra S, Whittaker C, Mellan T, Marwal R, Datta M, Sengupta S, Ponnusamy K, Radhakrishnan VS, Abdullahi A, Charles O, Chattopadhyay P, Devi P, Caputo D, Peacock T, Wattal C, Goel N, Satwik A, Vaishya R, Agarwal M, Mavousian A, Lee JH, Bassi J, Silacci-Fegni C, Saliba C, Pinto D, Irie T, Yoshida I, Hamilton WL, Sato K, Bhatt S, Flaxman S, James LC, Corti D, Piccoli L, Barclay WS, Rakshit P, CITIID-NIHR BioResource COVID-19 Collaboration , et al. 2021. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599:114–119. 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Iketani S, Guo Y, Chan JF-W, Wang M, Liu L, Luo Y, Chu H, Huang Y, Nair MS, Yu J, Chik KK-H, Yuen TT-T, Yoon C, To KK-W, Chen H, Yin MT, Sobieszczyk ME, Huang Y, Wang HH, Sheng Z, Yuen K-Y, Ho DD. 2022. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602:676–681. 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 5.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. 2021. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596:276–280. 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 6.Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, Wang B, Paesen GC, Lopez-Camacho C, Slon-Campos J, Hallis B, Coombes N, Bewley K, Charlton S, Walter TS, Skelly D, Lumley SF, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert S, James W, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. 2021. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 184:2348–2361.e6. 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan M, Liu H, Wu NC, Lee C-CD, Zhu X, Zhao F, Huang D, Yu W, Hua Y, Tien H, Rogers TF, Landais E, Sok D, Jardine JG, Burton DR, Wilson IA. 2020. Structural basis of a shared antibody response to SARS-CoV-2. Science 369:1119–1123. 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Zhang B, Chuang G-Y, Ho DD, Sheng Z, Kwong PD, Shapiro L. 2021. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe 29:819–833.e7. 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes CO, Jette CA, Abernathy ME, Dam K-MA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, Robbiani DF, Nussenzweig MC, West AP, Bjorkman PJ. 2020. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588:682–687. 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, Zepeda S, di Iulio J, Bowen JE, Montiel-Ruiz M, Zhou J, Rosen LE, Bianchi S, Guarino B, Fregni CS, Abdelnabi R, Foo S-YC, Rothlauf PW, Bloyet L-M, Benigni F, Cameroni E, Neyts J, Riva A, Snell G, Telenti A, Whelan SPJ, Virgin HW, Corti D, Pizzuto MS, Veesler D. 2021. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184:2332–2347.e16. 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, Lambson BE, de Oliveira T, Vermeulen M, van der Berg K, Rossouw T, Boswell M, Ueckermann V, Meiring S, von Gottberg A, Cohen C, Morris L, Bhiman JN, Moore PL. 2021. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 27:622–625. 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. 2021. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184:2372–2383.e9. 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odak I, Schultze-Florey CR, Hammerschmidt SI, Ritter C, Willenzon S, Friedrichsen M, Ravens I, Sikora R, Bayir LM, Gutierrez Jauregui R, Bernhardt G, Stankov MV, Cossmann A, Hansen G, Krey T, Cornberg M, Koenecke C, Behrens GMN, Bošnjak B, Förster R. 2022. Longitudinal tracking of immune responses in COVID-19 convalescents reveals absence of neutralization activity against Omicron and staggered impairment to other SARS-CoV-2 variants of concern. Front Immunol 13:863039. 10.3389/fimmu.2022.863039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye G, Liu B, Li F. 2022. Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain. Nat Commun 13:1214. 10.1038/s41467-022-28882-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torjesen I. 2021. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ 375:n2943. 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- 16.Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, Dushoff J, Mlisana K, Moultrie H. 2022. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science 376:eabn4947. 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carreno JM, et al. 2022. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 602:682–688. 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 18.Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Huang W, Li Q, Wang P, An R, Wang J, Wang Y, Niu X, Yang S, Liang H, Sun H, Li T, Yu Y, Cui Q, Liu S, Yang X, Du S, Zhang Z, Hao X, Shao F, Jin R, Wang X, Xiao J, Wang Y, Xie XS. 2022. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602:657–663. 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer A-S, Winkler MS, Lier M, Dopfer-Jablonka A, Jäck H-M, Behrens GMN, Pöhlmann S. 2022. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell 185:447–456.e11. 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter JD, Scherer M, Hutter CAJ, Garaeva AA, Zimmermann I, Wyss M, Rheinberger J, Ruedin Y, Earp JC, Egloff P, Sorgenfrei M, Hürlimann LM, Gonda I, Meier G, Remm S, Thavarasah S, van Geest G, Bruggmann R, Zimmer G, Slotboom DJ, Paulino C, Plattet P, Seeger MA. 2022. Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance. EMBO Rep 23:e54199. 10.15252/embr.202154199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Gasparo R, Pedotti M, Simonelli L, Nickl P, Muecksch F, Cassaniti I, Percivalle E, Lorenzi JCC, Mazzola F, Magrì D, Michalcikova T, Haviernik J, Honig V, Mrazkova B, Polakova N, Fortova A, Tureckova J, Iatsiuk V, Di Girolamo S, Palus M, Zudova D, Bednar P, Bukova I, Bianchini F, Mehn D, Nencka R, Strakova P, Pavlis O, Rozman J, Gioria S, Sammartino JC, Giardina F, Gaiarsa S, Pan-Hammarström Q, Barnes CO, Bjorkman PJ, Calzolai L, Piralla A, Baldanti F, Nussenzweig MC, Bieniasz PD, Hatziioannou T, Prochazka J, Sedlacek R, Robbiani DF, Ruzek D, Varani L. 2021. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature 593:424–428. 10.1038/s41586-021-03461-y. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Yu J, Lanzi A, Yao X, Andrews CD, Tsai L, Gajjar MR, Sun M, Seaman MS, Padte NN, Ho DD. 2016. Engineered bispecific antibodies with exquisite HIV-1-neutralizing activity. Cell 165:1621–1631. 10.1016/j.cell.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Q. 2020. Bispecific antibodies for autoimmune and inflammatory diseases: clinical progress to date. BioDrugs 34:111–119. 10.1007/s40259-019-00400-2. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Bardelli M, Espinosa DA, Pedotti M, Ng T-S, Bianchi S, Simonelli L, Lim EXY, Foglierini M, Zatta F, Jaconi S, Beltramello M, Cameroni E, Fibriansah G, Shi J, Barca T, Pagani I, Rubio A, Broccoli V, Vicenzi E, Graham V, Pullan S, Dowall S, Hewson R, Jurt S, Zerbe O, Stettler K, Lanzavecchia A, Sallusto F, Cavalli A, Harris E, Lok S-M, Varani L, Corti D. 2017. A human bi-specific antibody against Zika virus with high therapeutic potential. Cell 171:229–241.e15. 10.1016/j.cell.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bournazos S, Gazumyan A, Seaman MS, Nussenzweig MC, Ravetch JV. 2016. Bispecific anti-HIV-1 antibodies with enhanced breadth and potency. Cell 165:1609–1620. 10.1016/j.cell.2016.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao H, Sun Y, Deng Y-Q, Wang N, Tan Y, Zhang N-N, Li X-F, Kong C, Xu Y-P, Chen Q, Cao T-S, Zhao H, Yan X, Cao L, Lv Z, Zhu D, Feng R, Wu N, Zhang W, Hu Y, Chen K, Zhang R-R, Lv Q, Sun S, Zhou Y, Yan R, Yang G, Sun X, Liu C, Lu X, Cheng L, Qiu H, Huang X-Y, Weng T, Shi D, Jiang W, Shao J, Wang L, Zhang J, Jiang T, Lang G, Qin C-F, Li L, Wang X. 2021. Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection. Cell Res 31:25–36. 10.1038/s41422-020-00444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Trial Investigators . 2021. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med 384:238–251. 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shima M, Hanabusa H, Taki M, Matsushita T, Sato T, Fukutake K, Fukazawa N, Yoneyama K, Yoshida H, Nogami K. 2016. Factor VIII-mimetic function of humanized bispecific antibody in hemophilia A. N Engl J Med 374:2044–2053. 10.1056/NEJMoa1511769. [DOI] [PubMed] [Google Scholar]
- 29.Suurs FV, Lub-de Hooge MN, de Vries EGE, de Groot DJA. 2019. A review of bispecific antibodies and antibody constructs in oncology and clinical challenges. Pharmacol Ther 201:103–119. 10.1016/j.pharmthera.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann M, Kleine-Weber H, Pöhlmann S. 2020. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78:779–784.e5. 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou P, et al. 2022. Broadly neutralizing anti-S2 antibodies protect against all three human betacoronaviruses that cause severe disease. bioRxiv. 10.1101/2022.03.04.479488. [DOI] [PMC free article] [PubMed]
- 33.Steinhardt JJ, Guenaga J, Turner HL, McKee K, Louder MK, O'Dell S, Chiang C-I, Lei L, Galkin A, Andrianov AK, A Doria-Rose N, Bailer RT, Ward AB, Mascola JR, Li Y. 2018. Rational design of a trispecific antibody targeting the HIV-1 Env with elevated anti-viral activity. Nat Commun 9:877. 10.1038/s41467-018-03335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, et al. 2022. 35B5 antibody potently neutralizes SARS-CoV-2 Omicron by disrupting the N-glycan switch via a conserved spike epitope. Cell Host Microbe 30:887–895.e4. 10.1016/j.chom.2022.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Pan Z, Yue S, Yu F, Zhang J, Yang Y, Li R, Liu B, Yang X, Gao L, Li Z, Lin Y, Huang Q, Xu L, Tang J, Hu L, Zhao J, Liu P, Zhang G, Chen Y, Deng K, Ye L. 2020. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct Target Ther 5:180. 10.1038/s41392-020-00301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu K, Pan X, Li L, Yu F, Zheng A, Du P, Han P, Meng Y, Zhang Y, Wu L, Chen Q, Song C, Jia Y, Niu S, Lu D, Qiao C, Chen Z, Ma D, Ma X, Tan S, Zhao X, Qi J, Gao GF, Wang Q. 2021. Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell 184:3438–3451.e10. 10.1016/j.cell.2021.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown KA, Gubbay J, Hopkins J, Patel S, Buchan SA, Daneman N, Goneau LW. 2021. S-gene target failure as a marker of variant B.1.1.7 among SARS-CoV-2 isolates in the Greater Toronto Area, December 2020 to March 2021. JAMA 325:2115–2116. 10.1001/jama.2021.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, Krüger N, Graichen L, Hofmann-Winkler H, Kempf A, Winkler MS, Schulz S, Jäck H-M, Jahrsdörfer B, Schrezenmeier H, Müller M, Kleger A, Münch J, Pöhlmann S. 2021. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184:2384–2393.e12. 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao N-Y, Korsman S, Davies M-A, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Kosakovsky Pond SL, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. 2021. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592:438–443. 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 41.Edara VV, et al. 2021. Infection and vaccine-induced neutralizing antibody responses to the SARS-CoV-2 B.1.617.1 variant. bioRxiv. 10.1101/2021.05.09.443299. [DOI] [PMC free article] [PubMed]
- 42.Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. 2020. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol 27:763–767. 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, Berrios C, Ofoman O, Chang CC, Hauser BM, Feldman J, Roederer AL, Gregory DJ, Poznansky MC, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. 2022. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 185:457–466.e4. 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mileto D, Micheli V, Fenizia C, Cutrera M, Gagliardi G, Mancon A, Bracchitta F, De Silvestri A, Rizzardini G, Lombardi A, Biasin M, Gismondo MR. 2022. Reduced neutralization of SARS-CoV-2 Omicron variant by BNT162b2 vaccinees' sera: a preliminary evaluation. Emerg Microbes Infect 11:790–792. 10.1080/22221751.2022.2045878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, Chawansuntati K, Malasit P, Mongkolsapaya J, Screaton G. 2010. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328:745–748. 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He X, Hong W, Pan X, Lu G, Wei X. 2021. SARS-CoV-2 Omicron variant: characteristics and prevention. MedComm (2020) 2:838–845. 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gobeil SM, et al. 2022. Structural diversity of the SARS-CoV-2 Omicron spike. Mol Cell 82:2050–2068.e6. 10.1016/j.molcel.2022.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H, et al. 2022. Neutralization of SARS-CoV-2 Omicron BA.2 by therapeutic monoclonal antibodies. bioRxiv. 10.1101/2022.02.15.480166. [DOI]
- 49.Cho H, Gonzales-Wartz KK, Huang D, Yuan M, Peterson M, Liang J, Beutler N, Torres JL, Cong Y, Postnikova E, Bangaru S, Talana CA, Shi W, Yang ES, Zhang Y, Leung K, Wang L, Peng L, Skinner J, Li S, Wu NC, Liu H, Dacon C, Moyer T, Cohen M, Zhao M, Lee FE-H, Weinberg RS, Douagi I, Gross R, Schmaljohn C, Pegu A, Mascola JR, Holbrook M, Nemazee D, Rogers TF, Ward AB, Wilson IA, Crompton PD, Tan J. 2021. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci Transl Med 13:eabj5413. 10.1126/scitranslmed.abj5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z, Li S, Zhang G, Peng W, Chang Z, Zhang X, Fan Z, Chai Y, Wang F, Zhao X, Li D, Zhang R, He Z, Zou W, Xu K, Lei W, Liu P, Hao J, Zhang J, Sun L, Wu G, Tan S, Gao GF, Gao F, Wu Y. 2022. An engineered bispecific human monoclonal antibody against SARS-CoV-2. Nat Immunol 23:423–430. 10.1038/s41590-022-01138-w. [DOI] [PubMed] [Google Scholar]
- 51.Arora P, et al. 2022. Comparable neutralisation evasion of SARS-CoV-2 omicron subvariants BA.1, BA.2, and BA.3. Lancet Infect Dis 22:766–767. 10.1016/S1473-3099(22)00224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan K, et al. 2022. Omicron sub-lineages BA.4/BA.5 escape BA.1 infection elicited neutralizing immunity. medRxiv. 10.1101/2022.04.29.22274477. [DOI] [Google Scholar]
- 53.Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, Zhou D, Ginn HM, Selvaraj M, Liu C, Mentzer AJ, Supasa P, Duyvesteyn HME, Das R, Skelly D, Ritter TG, Amini A, Bibi S, Adele S, Johnson SA, Constantinides B, Webster H, Temperton N, Klenerman P, Barnes E, Dunachie SJ, Crook D, Pollard AJ, Lambe T, Goulder P, Paterson NG, Williams MA, Hall DR, Fry EE, Huo J, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR, Conlon C, Deeks A, Frater J, Frending L, Gardiner S, Jämsén A, Jeffery K, Malone T, Phillips E, Rothwell L, Stafford L. 2022. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 185:2422–2433.e13. 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desingu PA, Nagarajan K. 2022. The emergence of Omicron lineages BA.4 and BA.5, and the global spreading trend. J Med Virol 10.1002/jmv.27967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Li R, Pan Z, Qian C, Yang Y, You R, Zhao J, Liu P, Gao L, Li Z, Huang Q, Xu L, Tang J, Tian Q, Yao W, Hu L, Yan X, Zhou X, Wu Y, Deng K, Zhang Z, Qian Z, Chen Y, Ye L. 2020. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol 17:647–649. 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yue S, et al. 2021. Sensitivity of SARS-CoV-2 variants to neutralization by convalescent sera and a VH3-30 monoclonal antibody. Front Immunol 12:751584. 10.3389/fimmu.2021.751584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11:1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download jvi.00775-22-s0001.pdf, PDF file, 0.2 MB (159KB, pdf)