ABSTRACT
The zinc finger antiviral protein (ZAP) is an interferon-stimulated gene (ISG) with potent intrinsic antiviral activity. ZAP inhibits the replication of retroviruses, including murine leukemia virus (MLV) and HIV-1, as well as alphaviruses, filoviruses, and hepatitis B virus, and also the retrotransposition of LINE-1 and Alu retroelements. ZAP operates posttranscriptionally to reduce the levels of viral transcripts available for translation in the cytoplasm, although additional functions might be involved. Recent studies have shown that ZAP preferentially binds viral mRNAs containing clusters of CpG dinucleotides via its four CCCH-type zinc fingers. ZAP lacks enzymatic activity and utilizes other cellular proteins to suppress viral replication. Tripartite motif 25 (TRIM25) and the nuclease KHNYN have been identified as ZAP cofactors. In this study, we identify Riplet, a protein known to play a central role in the activation of the retinoic acid-inducible gene I (RIG-I), as a novel ZAP cofactor. Overexpression of Riplet acts to strongly augment ZAP’s antiviral activity. Riplet is an E3 ubiquitin ligase containing three domains, an N-terminal RING finger domain, a central coiled-coil domain, and a C-terminal P/SPRY domain. We show that Riplet interacts with ZAP via its P/SPRY domain and that the ubiquitin ligase activity of Riplet is not required to stimulate ZAP-mediated virus inhibition. Moreover, we show that Riplet interacts with TRIM25, suggesting that both Riplet and TRIM25 may operate as a complex to augment ZAP activity.
IMPORTANCE The ZAP is a potent restriction factor inhibiting replication of many RNA viruses by binding directly to viral RNAs and targeting them for degradation. We here identify RIPLET as a cofactor that stimulates ZAP activity. The finding connects ZAP to other innate immunity pathways and suggests oligomerization as a common theme in sensing pathogenic RNAs.
KEYWORDS: retrovirus restriction, innate immunity, zinc finger antiviral protein, host cofactors, protein-protein interactions
INTRODUCTION
Pattern recognition receptors (PRRs) mediate detection of pathogen-associated molecular patterns (PAMPs) present on many pathogens, including viruses, in the cytoplasm of an infected cell. The PRRs include RNA sensors such as RIG-I and MDA-5 (1) and DNA sensors such as cyclic GMP-AMP synthase (2). Activation of PRRs initiates various signal cascades that result in the induction of interferons (IFNs) and subsequent release of cytokines and chemokines. The induction of IFN is a hallmark of the cell’s innate immune defense. When interferons exit the infected cell and bind to IFN receptors on the surface of neighboring cells, they trigger pathways that lead to an upregulation of hundreds of host proteins known as interferon-stimulated genes (ISGs) (3). While the functions of many ISGs remain unknown, many of these genes contribute to the establishment of an antiviral state of the cell and represent the crucial first line of defense again viral infection (4, 5). One such gene is the zinc finger antiviral protein (ZAP).
ZAP was first identified in a screen for host factors restricting Moloney murine leukemia virus (MLV) infection (6) and was subsequently shown to inhibit other retroviruses as well as alphaviruses, filoviruses, hepatitis B virus, vaccinia virus, and, more recently, the coronavirus SARS-CoV-2 (7–16). ZAP has potent restriction activity that operates at the posttranscriptional stage to reduce the number of viral transcripts available for translation in the cytosol (6, 15, 17). Recent studies have shown that ZAP preferably binds viral mRNAs containing clustered CpG dinucleotides via its four CCCH-type zinc fingers (18, 19). ZAP has been suggested to have played a major role in the evolution of the low CpG content of many eukaryotic genomes, allowing for discrimination between host mRNAs (low CpG) and pathogen RNAs (high CpG) (19–21).
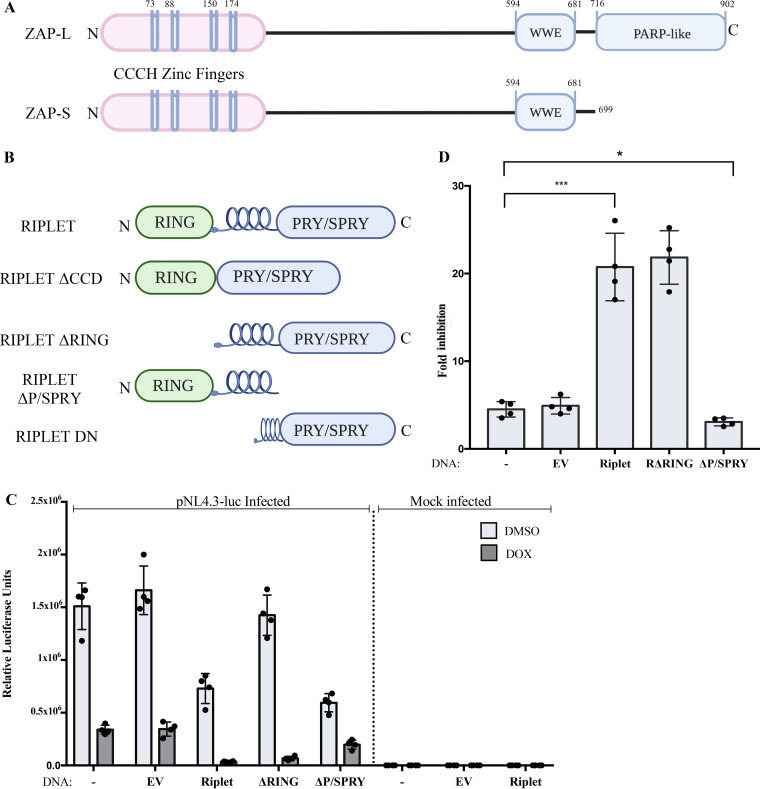
ZAP derives from an ancestral gene widely present in eukaryotic genomes, including mammals, birds, reptiles, and fish (22). The human orthologue, ZC3HAV1, gives rise to multiple isoforms of the ZAP protein (23), including long and short forms designated ZAP-L and ZAP-S, respectively, both having antiviral activity (15, 24, 25). Some other short forms of the ZAP protein can serve as dominant negative regulators of the active isoforms and thereby inhibit ZAP activity (26). All forms of ZAP contain four zinc fingers located at the N terminus and a central domain that contains an additional zinc finger of the CCCH-type as well as two WWE motifs, and it has recently been shown to form a binding site for poly(ADP-ribose) (27). The long isoform contains a poly(ADP-ribose) polymerase (PARP)-like domain at the C terminus, which is catalytically inactive by virtue of alterations at the active site but nevertheless can potentiate ZAP activity, perhaps by altering intracellular localization (Fig. 1A).
FIG 1.
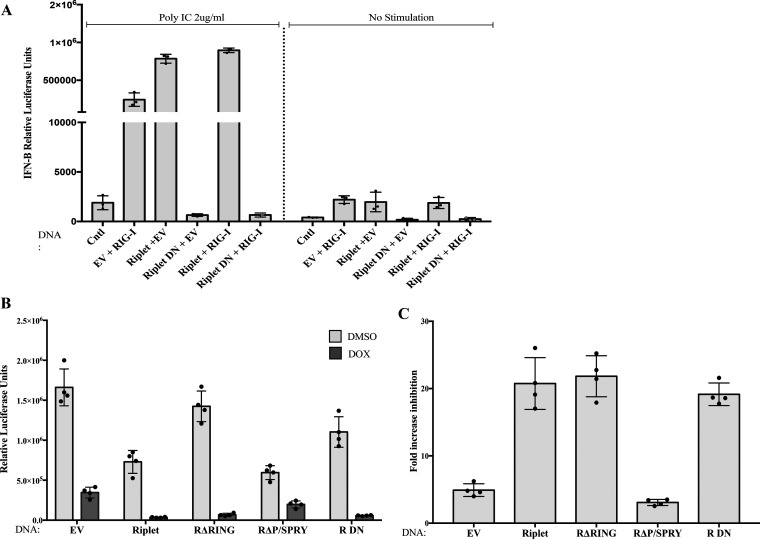
Riplet augments ZAP-mediated restriction of HIV-1 reporter virus. (A) Schematic representation of two major isoforms of ZAP. ZAP-L, the long form; ZAP-S, the short form. Zinc fingers, WWE domain, and PARP-like domain are indicated. (B) Schematic representation of the domains of Riplet and mutant constructs. RING, coil domain, and PRY/SPRY domain are indicated. (C) Riplet overexpression augments ZAP-mediated restriction of retroviral infection. 293TrexhZAP cell lines expressing the indicated DNAs were infected with VSVG-pseudotyped HIV-luc reporter virus followed by ZAP induction with doxycycline or DMSO as a control at 6 h postinfection. Firefly luciferase reporter activity was measured at 24 h postinfection and normalized to total protein content measured by a Bradford assay for each sample. Data points presented are the mean RLU/mg ± SD values of four independent experiments done in triplicate. (D) Riplet-mediated increase in ZAP-mediated HIV-1 inhibition, derived from data in panel C. 293TrexhZAP cell lines expressing the indicated DNAs were infected with VSVG-pseudotyped HIV-luc reporter virus followed by ZAP induction with doxycycline or DMSO as a control. Fold inhibition of virus was calculated as the ratio of luciferase expression levels in DMSO-treated cells to those in doxycycline-treated cells. Data points represent the mean ±SD values of four independent experiments. Student's t test was used for statistical analysis. ***, P < 0.0002; *, P < 0.02.
ZAP binds directly to target RNAs through its CCCH zinc finger motifs (18), and mutations altering its second and fourth zinc fingers have been shown to obliterate its antiviral action (28). ZAP can also disrupt the formation of the translational initiation complex on the RNA by directly interacting with the translation initiation factor eIF4A, and this shutoff of translation may be required for subsequent degradation of the RNA (29). Furthermore, ZAP interacts with Ago2 to modulate microRNA (miRNA)-mediated gene silencing upon activation of the cell’s stress response (30, 31). ZAP may have other functions (32), including the regulation of host gene expression. A small set of cellular mRNAs, including those encoding CCL5 and TRAILR4, a regulator of the apoptotic cytokine TRAIL mRNA, have been shown to be modestly elevated (up to 4-fold) in ZAP knockout (KO) cell lines (33).
The ability of ZAP to inactivate and degrade its target RNAs is promoted or regulated by a number of other factors. ZAP recruits components of the 5′-to-3′ and 3′-to-5′ degradation pathways to target viral RNAs for degradation by the exosome machinery (17, 28, 29). Other studies have identified the nuclease KHNYN as a ZAP cofactor, and overexpression of KHNYN has been shown to enhance ZAP-mediated restriction of CpG-enriched HIV-1 replication (34). The relative contribution of these various nucleases to ZAP activity is unclear and may vary between cell types. Recently, one of the tripartite motif (TRIM) family of proteins, TRIM25, has been reported to act as a ZAP cofactor that stimulates its antiviral activity (35, 36). The E3 ligase activity of TRIM25 has been shown to enhance ZAP-mediated virus restriction, though mutation of all of the consensus sites for ubiquitinylation of ZAP did not impact its antiviral activity (35, 36). TRIM25 has been reported to play an important role in innate immune defense as the main cofactor of another antiviral protein, RIG-I (37), though its involvement in RIG-I activation has been recently questioned (38, 39). ZAP has been shown to regulate RIG-I signaling in human cells (40), but ZAP is not required for the RIG-I-mediated induction of IFN in primary mouse cells (41). Finally, the nuclear protein Matrin 3 has been shown to negatively regulate ZAP-mediated restriction of retroviral infection (42). Given the complexity of ZAP activity, additional positive and negative regulators are likely to be found.
In the present study, we identify Riplet, a protein known to play a central role in innate immune detection of PAMPS, as a bona fide ZAP cofactor that enhances ZAP’s antiviral activity. Riplet is an E3 ubiquitin ligase encoded by the RNF135 gene in humans, and it contains three domains, a RING finger domain at the N terminus, a central coiled-coil domain, and a PRY/SPRY domain at the C terminus (here called P/SPRY) (Fig. 1B). Although Riplet resembles proteins in the tripartite motif (TRIM) family, it lacks a B-Box domain that is characteristic of TRIM proteins. The P/SPRY domain of Riplet has very high sequence similarity to the P/SPRY domain of TRIM25 out of all the TRIM family members, suggesting that Riplet might overlap or share TRIM25 functions. Riplet, widely expressed in human cells, serves to modulate the innate immune activation of the retinoic-acid inducible gene I (RIG-I) pathway involved in the sensing of viral RNAs. Riplet was initially shown to interact with RIG-I and deliver K63-linked polyubiquitin chains to both the N-terminal CARD domain, as well as the C-terminal domain (CTD), of RIG-I (43, 44). Ubiquitinylation of RIG-I by Riplet leads to its activation and further amplification of downstream signaling to induce the expression of IFNs and thereby trigger upregulation of ISGs (43, 44). The E3 ligase activity of Riplet is required to catalyze the final reaction of this ubiquitin transfer cascade. Additionally, it has been demonstrated that Riplet can stimulate RIG-I activation in a ubiquitin-independent manner by cross-bridging filaments of RIG-I bound to RNAs (38). This mechanism leads to a concentration of RIG-I assemblies that induces receptor clustering and ultimately amplifies antiviral signaling to initiate IFN production and, thereby, expression of ISGs. Although the role of Riplet in the RIG-I pathway has been well characterized, its involvement in the ZAP antiviral pathway has not been previously investigated. Here, we demonstrate that Riplet enhances ZAP restriction of viral RNA reporters. We further show that Riplet interacts with TRIM25, suggesting that Riplet and TRIM25 may operate cooperatively to augment ZAP activity.
RESULTS
Riplet augments ZAP-mediated inhibition of HIV-1 reporter gene expression.
Previous studies of the ZAP restriction factor ZAP have identified several cofactors that enhance its activity, and we expect that more remain to be found. One cofactor, TRIM25, is a member of the large TRIM family with E3 ubiquitin ligase activity and is potentially involved with the RIG-I RNA sensing pathway (37). An isoform of human ZAP has been reported to associate with RIG-I, promote its oligomerization, and activate the IRF3- and NF-κB-mediated induction of interferon production (40). These observations led us to examine whether Riplet, a protein with similarity to the TRIM proteins and also involved in the RIG-I pathway (43–45), might stimulate ZAP functions. To monitor the antiviral activity of ZAP, we utilized 293TrexhZAP cells, a derivative of the 293A cell line in which doxycycline induces the expression of myc-tagged full-length ZAP (15). We transduced these cells and various derivative cell lines with single-round HIV reporter genomes and monitored expression of the reporter gene with and without induction of ZAP expression. To test for the potential involvement of Riplet in ZAP-mediated inhibition of HIV-1, we generated pools of cells overexpressing Riplet by transfection of 293TrexhZAP cells with a DNA construct expressing full-length Riplet followed by antibiotic selection. While the endogenous levels of expression in the parental 293TrexhZAP cells are low, and the protein was not detectable by Western blotting with commercially available antibodies, the protein was readily detected in the pools of transfected cells (Fig. 2). These pooled cell populations were then tested for ZAP-mediated virus restriction by infecting them with a vesicular stomatitis virus envelope glycoprotein (VSVG)-pseudotyped HIV-1 luciferase reporter vector (pNL4.3env-luc+). At 6 h postinfection, fresh medium containing doxycycline to induce ZAP expression, or dimethyl sulfoxide (DMSO) as a control, was added, and luciferase reporter virus expression was measured at 24 h postinfection (Fig. 1C). We note that the luciferase sequence in the reporter genome is relatively rich in CpG dinucleotides (with 120 copies in the 2,154-nucleotide [nt] sequence) and thus provides a highly responsive RNA substrate for ZAP restriction. We found that overexpression of Riplet alone in these cells, without addition of doxycycline to induce ZAP expression, led to a modest reduction (2-fold or less) in virus reporter expression relative to the parental line (Fig. 1C), likely due to its known activation of interferon expression. ZAP induction in the parent line, and in cells expressing an empty vector control, induced an approximately 5-fold restriction of HIV-1-luc expression, in accordance with previous reports. ZAP induction in the lines with overexpression of Riplet led to a dramatic increase in restriction of the reporter gene expression to about 20-fold over the DMSO control (Fig. 1D). This result demonstrates that Riplet overexpression substantially augments ZAP-mediated inhibition of viral gene expression.
FIG 2.
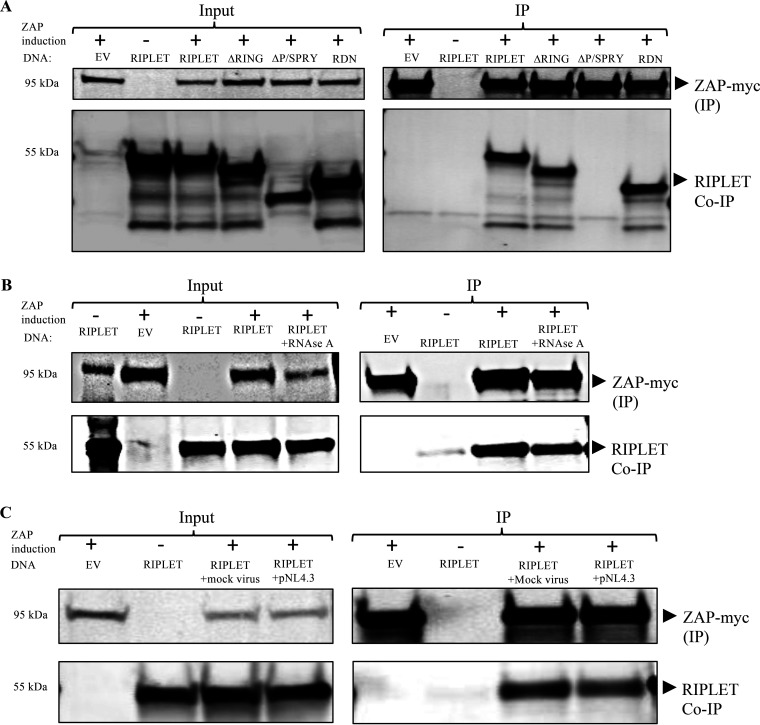
Riplet binds to ZAP to augment restriction of HIV-1 reporter virus. (A) Riplet interacts with ZAP via its PRY/SPRY domain. 293TrexhZAP cells were transiently transfected with DNAs overexpressing Riplet, Riplet mutants, or an empty vector control (EV), treated with doxycycline to induce ZAP expression as indicated (+/−), and lysed 48 h later. Riplet mutants included deletion of the RING domain (ΔRING), deletion of the P/SPRY domain (ΔP/SPRY), and dominant negative Riplet (RDN). (A, Left) In the input data, total proteins were analyzed by gel electrophoresis, blotted, and probed for myc-tagged ZAP (top) or Riplet (bottom). (A, Right) In the IP data, myc-tagged ZAP was recovered by immunoprecipitation using a mouse α-myc antibody, and bound proteins were analyzed by electrophoresis and probed for myc-tagged ZAP (top) or Riplet (bottom). (B) Riplet interaction with ZAP is RNase A-resistant. 293TrexhZAP cells were transfected with DNAs overexpressing Riplet or an EV control, treated with doxycycline or control DMSO to induce ZAP expression as indicated (+/−), and lysed 48 h later. Lysates were treated with RNase A (50 μg/mL) as indicated. (B, Left) In the input data, total proteins were analyzed by gel electrophoresis, blotted, and probed for myc-tagged ZAP with mouse α-myc antibody (top) or Riplet antibodies (bottom). (B, Right) In the IP data, myc-tagged ZAP was recovered by immunoprecipitation with mouse α-myc antibody, and bound proteins were analyzed by electrophoresis and probed for myc-tagged ZAP (top) or Riplet (bottom). Approximate molecular weights of major proteins estimated from size markers are indicated on left. (C) Riplet interacts with ZAP in pNL4.3-luc-infected cells. 293TrexhZAP cells transfected with an EV control or DNA overexpressing Riplet were either left uninfected or infected with a VSVG-pseudotyped pNL4.3-luc reporter virus or with a mock virus preparation lacking the VSVG envelope (mock virus). Cultures were treated with doxycycline or control DMSO to induce ZAP expression as indicated (+/−) and lysed 48 h later. (C, Left) In the input data, total proteins were analyzed by gel electrophoresis, blotted, and probed for myc-tagged ZAP with mouse α-myc antibody (top) or Riplet antibodies (bottom). (C, Right) In the IP data, myc-tagged ZAP was recovered by immunoprecipitation with mouse α-myc antibody, and bound proteins were analyzed by electrophoresis and probed for myc-tagged ZAP (top) or Riplet (bottom). Approximate molecular weights of major proteins estimated from size markers are indicated on left.
One possible explanation for the suppression of viral expression by Riplet would be an effect on viral DNA integration. To test for this possibility, we analyzed the extent of viral integration after infection of cells overexpressing Riplet or the empty vector control, or the parental 293TrexhZAP cells. Cells were infected with the reporter and propagated for 10 days after infection to allow decay of unintegrated DNAs. Total genomic DNA was then extracted, and viral DNA was measured by quantitative real-time PCR using specific primers targeting the HIV-1 genome. Proviral DNA was detectable in all cells tested, and cells overexpressing Riplet displayed only a slight reduction in the extent of viral integration compared to control cells (see Fig. S1 in the supplemental material). The minor reduction could not account for the substantial decrease of reporter gene expression measured in cells overexpressing Riplet. This finding indicates that Riplet stimulates ZAP inhibition of viral RNA without affecting DNA integration. Global transcription levels were not affected by Riplet overexpression as measured by reverse transcription-quantitative PCR (RT-qPCR) assays of mRNAs of the housekeeping genes Tert or GAPDH (glyceraldehyde-3-phosphate dehydrogenase), with or without ZAP induction (Fig. S2).
The P/SPRY domain of Riplet, but not its E3 ligase activity, is important for stimulation of ZAP-mediated inhibition.
Riplet consists of three domains, an N-terminal RING domain required for its E3 ligase activity, a central coiled-coil domain (CCD), and a P/SPRY domain at the C terminus. The P/SPRY domain of a number of cellular proteins, including Riplet, has been shown to be essential for RNA binding, mediating protein-protein interactions, and substrate specificity (46–48). To analyze the functional importance of the different domains of Riplet in the ZAP antiviral pathway, three deletion mutants were generated (Fig. 1B). One mutant contains a deletion of the RING domain (ΔRING), a second mutant has a deletion of the central CCD (ΔCCD), and a third mutant has a deletion of the C-terminal P/SPRY domain (ΔP/SPRY). 293TrexhZAP cells were transfected with DNAs encoding each of these proteins, and cell populations expressing the full-length and the three deletion mutants of Riplet were generated as before. Two of the mutants, ΔRING and ΔP/SPRY, as well as the full-length protein, were expressed at comparable levels (Fig. 2A), while the mutant lacking the central CCD was not expressed and was not used for further experiments. The cell populations were challenged by infection with HIV-luc reporter virus, with and without induction of ZAP expression, and the level of restriction was determined by measuring luciferase activity as before.
Expression of the Riplet RING domain deletion mutant without ZAP induction did not impact virus expression, as levels of luciferase reporter were comparable to those in the parental cell line and in cells expressing an empty vector control without ZAP induction. Induction of ZAP in the ΔRING-expressing cells demonstrated an enhancement of viral restriction comparable to that measured in cells expressing full-length Riplet and was far greater than the empty vector (EV) control (Fig. 1D). These results indicate that the RING domain, its encoded E3 ligase activity, and Riplet-mediated ubiquitination are not necessary for Riplet to enhance ZAP’s antiviral activity.
Expression of the Riplet mutant lacking the P/SPRY domain alone without ZAP induction led to a modest decrease of virus reporter expression, roughly 2-fold (Fig. 1C). However, expression of the P/SPRY domain deletion mutant with induction of ZAP did not enhance ZAP activity but, rather, led to a significant reduction of viral inhibition by ZAP compared to the inhibition mediated by full-length Riplet (Fig. 1D). These results suggest that overexpression of Riplet with a P/SPRY domain deletion not only fails to amplify ZAP-mediated viral restriction but even modestly inhibits it.
Riplet P/SPRY domain interacts with ZAP.
To test for interactions between Riplet and ZAP, we performed coimmunoprecipitation experiments. 293TrexhZAP cells were induced to overexpress Riplet by transient transfection with a cDNA expression construct or an empty vector control. The cells were then treated with doxycycline to induce ZAP expression or with DMSO as a control, and lysates were prepared 48 h later. The ZAP protein was immunoprecipitated with a monoclonal antibody against the myc tag; the associated proteins were eluted, separated by gel electrophoresis, and blotted; and the blots were probed with antibodies directed against Riplet. These Western blots showed a specific interaction between Riplet and ZAP. No background signal was detected in cells transfected with an empty vector and treated with doxycycline to induce ZAP expression. Similarly, Riplet-expressing cells treated with DMSO as a control without ZAP induction failed to show the Riplet-ZAP association (Fig. 2A).
To test whether Riplet-ZAP interaction might require RNA, the coimmunoprecipitations were carried out in the presence of RNase A. The association was resistant to treatment with RNase A (Fig. 2B). To test whether virus infection had any impact on the association of Riplet and ZAP, coimmunoprecipitation (co-IP) experiments were performed after infection with a VSVG-pseudotyped pNL4.3env-luc+ reporter virus or with a mock virus preparation lacking the VSVG envelope. At 6 h postinfection, cells were treated with doxycycline to induce ZAP or with control DMSO, and lysates were prepared 48 h later. The interaction was not affected by HIV-luc infection, as similar levels of Riplet protein were recovered in the immunoprecipitates from both infected and uninfected cells (Fig. 2C). Together, these results demonstrate that Riplet interacts with ZAP, the interaction is not affected by virus infection, and the interaction can occur even after harsh RNase treatment.
To determine which domain of Riplet is important for its interaction with ZAP, we carried out coimmunoprecipitation experiments with our two Riplet mutants. 293TrexhZAP cells were transiently transfected with plasmids encoding ΔRING or ΔP/SPRY Riplet as before. At 6 h posttransfection, ZAP expression was induced with doxycycline, and lysates were prepared after 48 h. ZAP was immunoprecipitated with anti-myc antibodies as before, and associated proteins were detected by Western blotting. ZAP bound to the ΔRING domain deletion mutant but not to the ΔP/SPRY mutant (Fig. 2A). These results show that the P/SPRY domain of Riplet is required both for its interaction with ZAP and for augmenting ZAP-mediated inhibition of HIV-1 in HEK293 cells.
We verified the interaction of Riplet and ZAP in the course of an innate immune response by adding treatment with “universal” IFN-α to cells expressing both proteins and subjecting them to coimmunoprecipitation experiments. We found that Riplet and ZAP interact, with and without IFN treatment, suggesting that the interaction is not impaired by the innate immune response (Fig. S3).
Riplet enhances degradation of unspliced, partially spliced, and multiply spliced viral mRNAs.
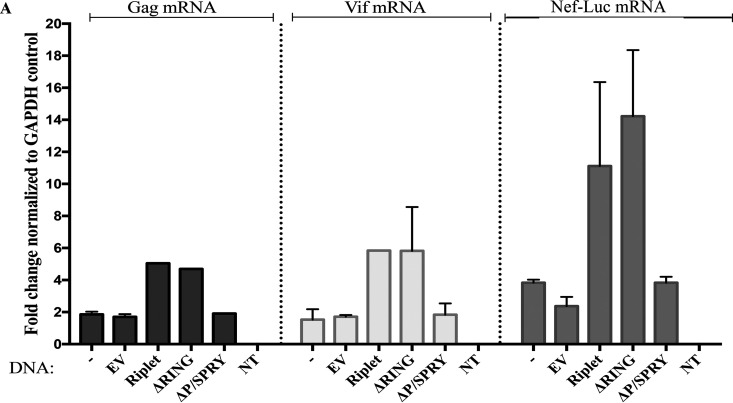
The HIV-1 genome produces multiple species of messenger RNAs that differ by alternative splicing of the common initial transcript (49). ZAP has been reported to selectively prevent the accumulation of the multiply spliced forms of viral mRNA in infected cells (15). We used qRT-PCR with specific primer pairs to analyze the abundance of unspliced, partially spliced, and multiply spliced viral mRNAs encoded by the reporter virus after ZAP induction and Riplet overexpression. We note that all three classes of mRNAs contain the luciferase sequences that may serve as a major ZAP target. 293TrexhZAP cells overexpressing Riplet or an empty vector control were infected with the HIV-1 reporter virus (pNL4.3env-luc+), and 6 h later, doxycycline was added to induce ZAP expression. Total cytoplasmic RNAs were extracted at 24 h postinfection, and mRNA abundance was analyzed by qRT-PCR. We calculated fold inhibition of the viral mRNAs as the ratio of mRNA levels in DMSO-treated cells to those in doxycycline-treated cells after normalization to mRNA levels of the housekeeping gene GAPDH. Our data showed that the fold inhibition of unspliced gag-pol mRNA and partially spliced vif mRNAs in the parental 293TrexhZAP cell line was approximately 2-fold, while inhibition of multiply spliced nef-luc mRNA was about 4-fold (Fig. 3A). A similar fold restriction of HIV-1 mRNAs was measured in cells expressing an empty vector control, with approximately 2-fold inhibition of gag and vif mRNAs and approximately 3-fold reduction of nef-luc mRNAs (Fig. 3A). These reductions were smaller than the reduction in reporter gene activity but may reflect the level of impact on viral mRNAs without including any effects on subsequent steps of gene expression. ZAP-mediated restriction of all species of viral mRNAs was further amplified in cells overexpressing Riplet after ZAP induction compared to control cells expressing an empty vector control. Fold inhibition of gag mRNA was increased to about 5-fold, and fold inhibition of vif mRNAs was increased to about 6-fold in cells overexpressing Riplet. The levels of the multiply spliced nef-luc mRNA were most heavily impacted by Riplet and ZAP expression compared to ZAP-only control, with the fold inhibition amplified to approximately 12-fold (Fig. 3A). Finally, we observed a similar reduction of viral mRNAs in cells overexpressing the ΔRING domain deletion mutant to that observed in cells overexpressing a full-length Riplet. This enhancement of viral inhibition was not observed in cells expressing the ΔP/SPRY mutant, suggesting that the P/SPRY domain of Riplet is important for the reduction of unspliced, partially spliced, and multiply spliced viral mRNAs by ZAP.
FIG 3.
Riplet enhances degradation of unspliced, partially spliced, and multiply spliced viral mRNAs. (A) 293TrexhZAP cells were left unmanipulated (−) or transfected with either an empty vector control (EV) or DNAs overexpressing the indicated Riplet constructs. Cells were infected with HIV-1 reporter virus (pNL4.3env-luc+), and 6 h later, doxycycline or DMSO was added. At 24 h postinfection, total cytoplasmic RNAs were extracted, and the abundance of three viral mRNA species was analyzed by qRT-PCR, using PCR primer pairs targeting gag, vif, nef (in this vector, nef-luc), and GAPDH as standard. Fold inhibition of the viral mRNAs was calculated as the ratio of mRNA levels in DMSO-treated cells to those in doxycycline-treated cells after normalization to mRNA levels of the housekeeping gene GAPDH. An additional sample containing no template (NT) for PCR amplification was included to control for DNA contamination.
Riplet self-associates and localizes to the cytoplasm.
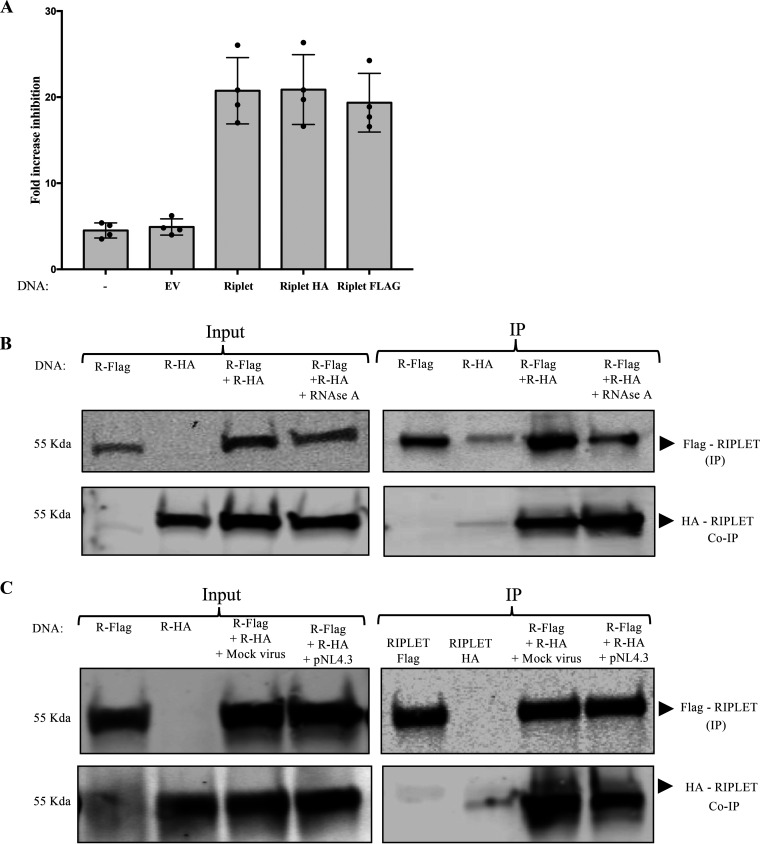
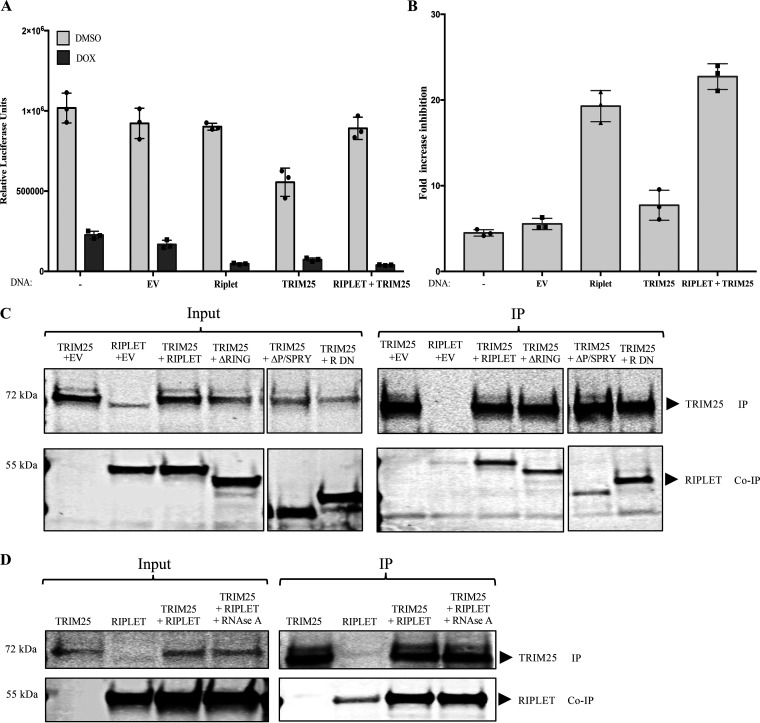
Previous studies have shown that TRIM and TRIM-like proteins self-associate into dimers through their coiled-coil domain and that these dimers can form higher-order oligomers that promote dimerization of the RING domain and efficient E3 ligase activity (38, 47, 48, 50, 51). The central coiled-coil domain of Riplet might also mediate the formation of multimers, and although the Riplet-mediated activation of ZAP did not seem to require the RING domain or the associated ubiquitin transferase activity, its activity could still require multimerization. To test this, we explored the potential of Riplet to self-associate in infected and uninfected cells. We generated plasmids expressing N-terminal Flag-tagged Riplet and a C-terminal HA-tagged Riplet and first verified their abilities to augment ZAP-mediated virus inhibition in single-round reporter virus infection experiments as before. Reporter virus expression assays confirmed that Riplet’s effect on ZAP’s antiviral function was not impacted by the presence of the N-terminal Flag tag or by the C-terminal hemagglutinin (HA) tag (Fig. 4A). To test for Riplet’s potential self-association, we expressed both tagged constructs in 293T cells, prepared lysates, and assayed for the coimmunoprecipitation of the two proteins. We recovered the Flag-tagged Riplet with a monoclonal anti-Flag antibody and monitored the levels of associated HA-tagged Riplet by Western blotting with anti-HA antibody. The analysis showed a specific interaction between HA-tagged Riplet and Flag-tagged Riplet (Fig. 4B). The interaction was resistant to RNase treatment, suggesting that Riplet multimerization can occur in the absence of RNAs (Fig. 4B). Furthermore, the interaction was not disrupted or increased by HIV-1 reporter virus infection (Fig. 4C).
FIG 4.
Riplet self-associates. (A) Fold increase of ZAP-mediated HIV-1 inhibition in cells expressing Riplet, Riplet-HA, and Riplet-Flag. 293TrexhZAP cells transfected with the indicated DNAs were infected with HIV-1 reporter virus followed by ZAP induction with doxycycline or DMSO as a control. Cells were lysed 4 h postinfection and assayed for luciferase. The fold virus inhibition was calculated as the ratio of luciferase expression levels in DMSO-treated cells to those in doxycycline-treated cells after normalization to total protein content by Bradford assay for each sample. Data points represent the mean ± SD values of four independent experiments. (B) Coimmunoprecipitation of tagged Riplet proteins. 293T cells were transfected with DNAs overexpressing Riplet-Flag (R-Flag) or Riplet-HA (R-HA) or both, as indicated, and lysed at 48 h posttransfection. Lysates were treated with RNase A (50 μg/mL). (B, Left) In the input data, total proteins were analyzed by gel electrophoresis, blotted, and probed for Flag-tagged Riplet with mouse α-Flag antibody (top) or HA-tagged Riplet with anti-HA antibodies (bottom). (B, Right) In the IP data, Flag-tagged Riplet was recovered by immunoprecipitation with α-Flag antibody, and bound proteins were analyzed by electrophoresis, blotted, and probed for Flag-tagged Riplet (top) or HA-tagged Riplet (bottom). Approximate molecular weights of major proteins estimated from size markers are indicated on left. (C) Coimmunoprecipitation of tagged Riplet proteins in 293TrexhZAP cells after various treatments. 293TrexhZAP cells were transfected with DNAs overexpressing Riplet-Flag (R-Flag) or Riplet-HA (R-HA) or both and infected with HIV-luc reporter virus (pNL4.3) or with a mock virus preparation lacking the VSVG envelope (mock virus). Lysates were prepared 48 h postinfection. (C, Left) In the input data, total proteins were analyzed by gel electrophoresis, blotted, and probed for Flag-tagged Riplet with mouse α-Flag antibody (top) or HA-tagged Riplet with anti-HA antibodies (bottom). (C, Right) In the IP data, Flag-tagged Riplet was recovered by immunoprecipitation with α-Flag antibody, and bound proteins were analyzed by electrophoresis, blotted, and probed for Flag-tagged Riplet (top) or HA-tagged Riplet (bottom). Approximate molecular weights of major proteins estimated from size markers are indicated on left.
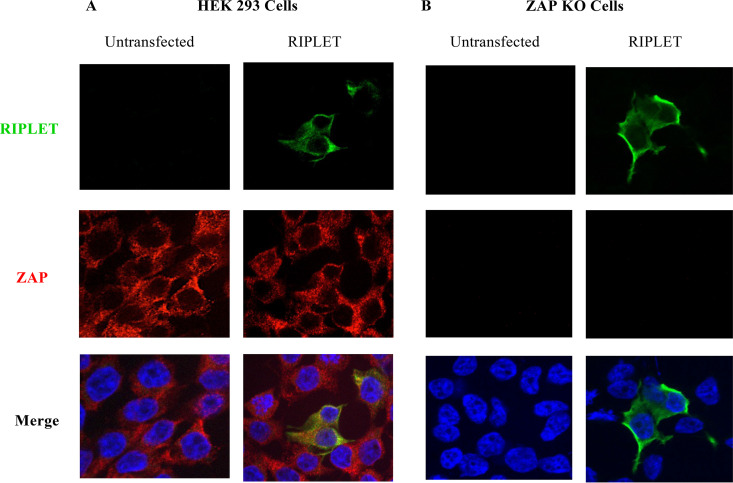
The intracellular localization of Riplet was examined by immunofluorescence microscopy. We tested both HEK293 cells expressing ZAP and ZAP knockout (KO) cells previously described (40). These cells were transfected with a Riplet-Flag expression vector, and 24 h later, they were fixed, permeabilized, and stained for Riplet with an anti-Flag tag antibody and for ZAP with a ZAP-specific antibody. After incubation with secondary antibodies, cells were examined by confocal imaging. We observed that Riplet was localized to the cytoplasm and overlapped with the localization of ZAP. Moreover, the absence of ZAP expression in the KO cell line did not affect its cytoplasmic localization (Fig. 5A and B).
FIG 5.
Riplet and ZAP are both cytoplasmic proteins. Cells were either untreated or transfected with a Riplet-Flag expression vector (Riplet) and, 24 h later, were fixed, permeabilized, and stained for Riplet with an anti-Flag tag antibody (green), for ZAP with a ZAP-specific antibody (red), and for nuclei with DAPI (blue). (A) HEK 293 cells. (B) ZAP knockout (KO) cells (40).
Riplet exhibits distinctive requirements for activation of the ZAP pathway compared to the RIG-I pathway.
A dominant negative form of Riplet, which abolishes RIG-I activation and subsequent induction of IFN-β, has been previously described (43, 44). This dominant negative Riplet mutant (here Riplet DN) lacks the RING domain as well as part of the central coiled-coil domain. Overexpression of Riplet DN in HEK293 cells strongly suppresses RIG-I signaling and IFN-β induction after stimulation with poly(I·C), a synthetic RIG-I ligand (38, 39, 43, 44). To determine the effect of the Riplet DN mutant in the ZAP antiviral pathway, we first verified its function in the RIG-I pathway by assessing whether the Riplet DN mutant blocked IFN-β induction in our experimental system. A reporter plasmid expressing firefly luciferase under IFN-β promoter control was introduced by transient transfection along with a plasmid encoding RIG-I into 293TrexhZAP cell pools overexpressing either wild-type Riplet, Riplet DN, or an empty vector control. Twenty-four hours later, we transfected poly(I·C) into the cells and measured reporter expression at 6 h posttransfection. Wild-type Riplet expression greatly stimulated IFN-β induction after poly(I·C) stimulation compared to the empty vector control, and as predicted, expression of Riplet DN potently suppressed this induction of IFN-β (Fig. 6A).
FIG 6.
Riplet DN suppresses RIG-I signaling but augments ZAP-mediated restriction of HIV-1 reporter virus. (A) Function of Riplet DN in the RIG-I pathway measured by a surrogate reporter of IFN-β induction upon poly(I·C) stimulation. HEK293 cells were untreated (Cntl) or transiently transfected with the indicated DNAs along with a construct expressing luciferase reporter under IFN-β promoter control. Twenty-four hours later, cells were stimulated with poly(I·C) (left) or were unstimulated (right), and reporter expression was measured 6 h later. Data points represent the mean ± SD values of three independent experiments. (B) Riplet DN overexpression augments ZAP-mediated restriction of retroviral infection. 293TrexhZAP cells transfected with empty vector (EV) or with DNAs expressing the indicated constructs were infected with HIV-luc reporter virus. Cells were treated with doxycycline to induce ZAP expression or DMSO as control at 6 h postinfection. Firefly luciferase reporter activity was measured at 24 h postinfection and normalized to total protein content measured by a Bradford assay for each sample. Data points presented are the mean RLU/mg ± SD values of four independent experiments done in triplicate. (C) Fold ZAP-mediated inhibition of HIV-1 reporter expression derived from data in panel B. Fold virus inhibition by ZAP was calculated as the ratio of luciferase expression levels in DMSO-treated cells to those in doxycycline-treated cells. Data points represent the mean ± SD values of four independent experiments.
We then tested the effect of Riplet DN on the antiviral activity of ZAP. 293TrexhZAP cell populations stably overexpressing wild-type Riplet or Riplet DN were established as before and then challenged by infection with the HIV-1 luciferase reporter virus. Riplet DN, without ZAP induction, did not impact the levels of reporter expression compared to cells expressing an empty vector control (Fig. 6B). Addition of doxycycline to induce ZAP in Riplet DN-expressing cells led to enhanced virus inhibition by ZAP at the same levels to those observed with the full-length Riplet, approximately 20-fold (Fig. 6C). These data show that the effect of the dominant negative Riplet applies only to the RIG-I pathway and that ZAP activity is not antagonized by this mutant but, instead, is stimulated to the same extent as by the wild-type Riplet. Thus, the absence of the RING domain in the DN mutant did not impact Riplet’s function in the ZAP pathway. Conversely, this result confirms that the presence of the P/SPRY domain, as well as a truncated coiled-coil domain in the Riplet DN mutant, suffices to act as a cofactor of ZAP and amplify its antiviral activity.
Riplet and TRIM25 both stimulate ZAP activity.
Previous studies have identified TRIM25 as a ZAP cofactor which acts to enhance ZAP-mediated inhibition of Sindbis virus (SINV) and MLV (35, 36). To examine the function of TRIM25 in our system, we overexpressed TRIM25 and/or Riplet in 293TrexhZAP cells as before and measured luciferase reporter expression after infection with HIV-1-luc reporter virus. As before, ZAP-mediated virus inhibition was calculated as the ratio of luciferase levels in mock-treated cells to those in doxycycline-treated cells. TRIM25 expression without ZAP induction resulted in slightly lower levels of luciferase reporter expression than those measured in empty vector control cells, and Riplet expression without ZAP induction in these experiments had almost no impact on the reporter (Fig. 7A), in this case, even less than seen in some experiments (Fig. 1C and Fig. 6B). The ZAP induction alone yielded an approximately 5-fold inhibition of luciferase expression. Under our conditions, we observed only a very modest increase of reporter virus inhibition with TRIM25 overexpression after ZAP induction, to approximately 7-fold (Fig. 7B). Riplet overexpression followed by ZAP induction, however, consistently led to about 20-fold virus inhibition. Overexpression of both TRIM25 and Riplet and subsequent ZAP induction resulted in only slightly more virus inhibition than cells with overexpression of Riplet alone and ZAP induction (Fig. 7B). These results suggest that both TRIM25 and Riplet augment ZAP-mediated inhibition of HIV-1 but that Riplet is a more potent ZAP cofactor than TRIM25.
FIG 7.
Riplet and TRIM25 both enhance ZAP activity and coimmunoprecipitate. (A) Effect of Riplet and TRIM25 on ZAP-mediated restriction of HIV-1 reporter expression. 293TrexhZAP cells were left untreated (−) or transfected with empty vector (EV) or DNAs expressing the indicated proteins and then infected with HIV-luc reporter virus. At 6 h postinfection, cells were treated with doxycycline to induce ZAP expression (dark bars) or DMSO as a control (light bars). Firefly luciferase reporter activity was measured at 24 h postinfection and normalized to total protein content measured by a Bradford assay for each sample. Data points presented are the mean RLU/mg ± SD values of three independent experiments done in triplicate. (B) Fold ZAP-mediated inhibition of HIV-1 expression in Riplet- and TRIM25-expressing cells. The fold inhibition was calculated from data in panel A. (C) Coimmunoprecipitation of TRIM25 and Riplet. 293TrexhZAP cells were transiently transfected with DNAs expressing the indicated proteins. Lysates were prepared 48 h later and analyzed directly or subjected to immunoprecipitation. (C, Left) In the input data, total proteins were analyzed by gel electrophoresis, blotted, and probed with α-TRIM25 antibody (top) or anti-Riplet antibodies (bottom). (C, Right) In the IP data, TRIM25 was recovered by immunoprecipitation with α-TRIM25 antibody, and bound proteins were analyzed by electrophoresis and probed for TRIM25 (top) or Riplet (bottom). Molecular weights of major protein estimated from size markers are indicated on left. (D) Coimmunoprecipitation of Riplet and TRIM25 is RNase resistant. 293TrexhZAP cells were transiently transfected by the indicated cDNAs or an empty vector control. Lysates were prepared 48 h later and treated with RNase A (50 μg/mL). Lysates were then analyzed directly or subjected to immunoprecipitation. (D, Left) In the input data, total proteins were analyzed by gel electrophoresis, blotted, and probed with α-TRIM antibody (top) or Riplet antibodies (bottom). Molecular weight of major protein estimated from size markers are indicated on left. (D, Right) In the IP data, TRIM25 was recovered by immunoprecipitation with α-TRIM25 antibody, and bound proteins were analyzed by electrophoresis and probed for TRIM25 (top) or Riplet (bottom). Approximate molecular weights of major proteins estimated from size markers are indicated on left.
ZAP and Riplet interact with TRIM25.
We investigated the potential interaction of Riplet with TRIM25 by performing coimmunoprecipitation experiments in 293TrexhZAP cells engineered to overexpress the proteins by transient transfection. Immunoprecipitation of Flag-tagged TRIM25 resulted in the recovery of high levels of Riplet without induction of ZAP (Fig. 7C), and similar results were obtained with induction of ZAP. Moreover, the interaction was resistant to RNase A treatment (Fig. 7D). To examine which domains of Riplet are important for its interaction with TRIM25, we performed coimmunoprecipitation experiments of TRIM25 and our Riplet mutants. We found that TRIM25 bound to both deletion Riplet mutants ΔRING and ΔP/SPRY, as well as Riplet DN (Fig. 7C), although the ΔP/SPRY Riplet mutant showed diminished binding to TRIM25 compared to full-length Riplet. Together, these results indicate that Riplet and TRIM25 interact with each other and that the interaction is independent of ZAP.
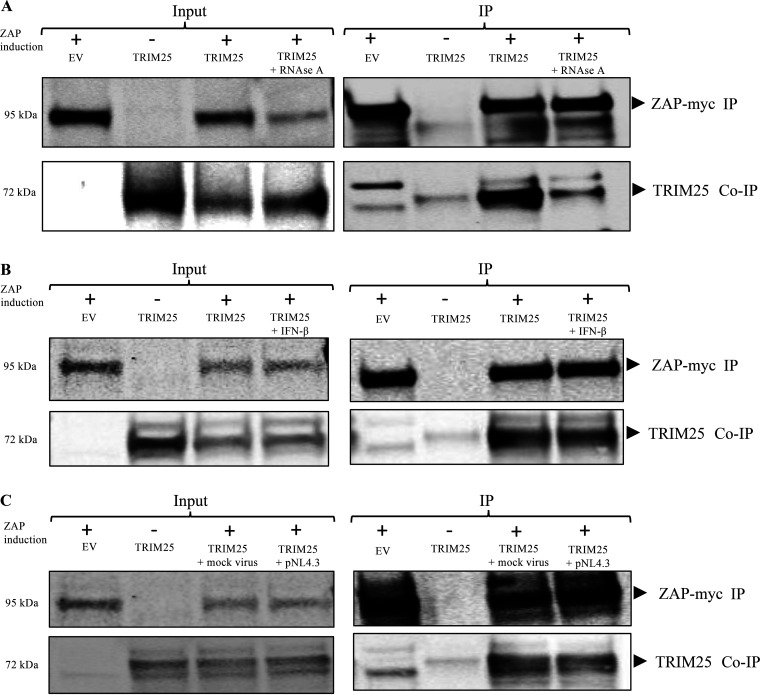
Finally, we carried out immunoprecipitation experiments to test for the interaction of TRIM25 and ZAP in our system. 293TrexhZAP cells were transfected to induce transient expression of TRIM25, and at 6 h posttransfection, ZAP was induced by doxycycline. Lysates were harvested 24 h later and subjected to coimmunoprecipitation with an antibody against the myc tag on ZAP. We observed that TRIM25 was associated with ZAP as previously reported (35, 36) (Fig. 8A). Importantly, we found that this interaction is RNase A sensitive, suggesting that the interaction of TRIM25 and ZAP depends on RNA binding (Fig. 8A). We performed coimmunoprecipitations after universal IFN-α treatment and observed no change in their binding (Fig. 8B). We repeated the tests after HIV-1 infection and again saw no changes (Fig. 8C).
FIG 8.
ZAP interaction with TRIM25. (A) ZAP and Trim25 coimmunoprecipitate, and the interaction is RNase sensitive. 293TrexhZAP cells were transfected with an empty vector control (EV) or DNAs expressing the indicated proteins and treated with doxycycline to induce ZAP expression (+) or with DMSO control (−) as indicated. Lysates were prepared 48 h later and treated with RNase as indicated. Lysates were either analyzed directly or subjected to immunoprecipitation for ZAP using a mouse α-myc antibody. (A, Left) In the input data, total proteins in lysate were analyzed by gel electrophoresis, blotted, and probed for myc-tagged ZAP (top) or TRIM25 (bottom). (A, Right) In the IP data, myc-tagged ZAP was recovered by immunoprecipitation using a mouse α-myc antibody, and bound proteins were analyzed by electrophoresis and probed for myc-tagged ZAP (top) or for co-IP of TRIM25 (bottom). Approximate molecular weights of major proteins estimated from size markers are indicated on left. (B) ZAP and TRIM25 co-IP after universal type I interferon treatment. 293TrexhZAP cells were transfected with an EV control or DNAs expressing the indicated proteins and treated with doxycycline to induce ZAP expression (+) or with DMSO control (−) as indicated. Cells were then treated with universal type I interferon at 1,000 U/mL as indicated. Lysates were prepared at 24 h posttransfection and either analyzed directly or subjected to immunoprecipitation for myc-tagged ZAP using a mouse α-myc antibody. (B, Left) In the input data, total proteins in lysate were analyzed by gel electrophoresis, blotted, and probed for myc-tagged ZAP (top) or TRIM25 (bottom). (B, Right) In the IP data, myc-tagged ZAP was recovered by immunoprecipitation using a mouse α-myc antibody, and bound proteins were analyzed by electrophoresis and probed for myc-tagged ZAP (top) or for co-IP of TRIM25 (bottom). Approximate molecular weights of major proteins estimated from size markers are indicated on left. (C) TRIM25 interacts with ZAP in pNL4.3-luc-infected cells. 293TrexhZAP cells were transfected with an EV control or DNAs expressing the indicated proteins and infected with a VSVG-pseudotyped HIV-1luc reporter virus (pNL4.3) or with a mock virus preparation lacking the VSVG envelope (mock virus). Six hours later, cells were treated with dox to induce ZAP expression (+) or with DMSO control (−) as indicated. Lysates were prepared 48 h later and either analyzed directly or subjected to immunoprecipitation for myc-tagged ZAP using a mouse α-myc antibody. (C, Left) In the input data, total proteins in lysate were analyzed by gel electrophoresis, blotted, and probed for myc-tagged ZAP (top) or TRIM25 (bottom). (C, Right) In the IP data, myc-tagged ZAP was recovered by immunoprecipitation using a mouse α-myc antibody, and bound proteins were analyzed by electrophoresis and probed for myc-tagged ZAP (top) or for co-IP of TRIM25 (bottom). Approximate molecular weights of major proteins estimated from size markers are indicated on left.
DISCUSSION
Host proteins with intrinsic antiviral activity, such as ZAP, are critical to control virus replication in the infected cell. The antiviral activity of ZAP accounts for an extensive reduction in the production of viral progeny in many settings, most often by mediating degradation of viral RNAs. How ZAP induces this degradation of target mRNAs remains to be elucidated, but several other host proteins are involved. We here demonstrate a role for Riplet in ZAP activity. Riplet is an E3 ligase involved in innate immune sensing of PAMPs. Riplet was initially described to be essential for modulating a RIG-I-dependent innate immune response against virus infection in vivo (43, 44). Since then, its role in the RIG-I pathway has been extensively characterized (38, 39, 43, 44, 50, 52). Riplet induces RIG-I activation by delivering polyubiquitin chains to RIG-I’s CARDs and its C-terminal domain (CTD) (38, 39, 43, 44, 50, 52). Additionally, it can induce the concentration of RIG-I filaments, thereby amplifying RIG-I signaling (38, 39, 50). Independent of the RIG-I pathway, Riplet has been shown to activate the transcription factor IRF3 to induce IFN production (53). ZAP has also been linked to the RIG-I pathway as a cytosolic RNA sensor capable of inducing the degradation of viral transcripts by the exosome (41). Our data presented here indicate that the functions of Riplet extend into the ZAP antiviral pathway where Riplet binds to ZAP and augments ZAP-mediated restriction of HIV-1 reporter virus.
In the present study, we show that overexpression of Riplet enhances ZAP inhibition of virus expression. It should be emphasized that the effects we document are all in the setting of Riplet overexpression. We note that Riplet overexpression per se has little impact on reporter genome expression and only has strong effects in the context of ZAP induction, giving some confidence that Riplet is truly a player in the ZAP pathway. The basal levels of Riplet expression in all cell lines that we have tested are very low, below our level of detection by Western blotting with available antibodies. Thus, the assays of ZAP that we and others routinely measure likely reflect its activity in the absence of detectable Riplet. We have not been able to knock down the already low levels of Riplet expression to monitor any potential loss of ZAP activity, and knockdowns performed after infection were not tolerated well by our cells. We thus do not know whether ZAP activity would be lost, or even diminished, upon complete elimination of Riplet by knockout. Nevertheless, the activation of ZAP by Riplet overexpression is strong, and it is possible that in some cell types, or in the course of infection, Riplet is induced to higher levels to enhance ZAP function. It is not known what cell types express the highest levels of Riplet, nor which cells might induce Riplet most strongly upon infection. Such cells would be predicted to exhibit the highest levels of ZAP-mediated restriction.
We find that Riplet interacts with ZAP via its P/SPRY domain and that this region is essential for enhancement of ZAP-mediated virus inhibition (Fig. 1C and D; Fig. 2A). A conserved mechanism by which Riplet and other TRIM/TRIM-like proteins engage their cognate RLRs via their P/SPRY domains has recently come to light (50), underscoring the importance of this domain for Riplet function. RIG-I binding also occurs via the PRY/SPRY domain of Riplet (38, 39, 50). Moreover, the PRY/SPRY domain of many TRIM and TRIM-like proteins has been deemed important for RNA binding (46–48, 54). While it is possible that Riplet’s PRY/SPRY domain binds to RNAs, treatment with RNase A did not disrupt the Riplet-ZAP association, suggesting that the interaction does not necessitate RNA binding (Fig. 2B), though we cannot rule out the possibility of an RNA bridge that is resistant to RNase treatment. It also remains possible that the interaction is mediated by a protein bridge.
It is noteworthy that the E3 ligase activity of Riplet, as well as the entire RING domain, were dispensable for its effect on ZAP’s antiviral activity: deletion of the RING domain did not affect Riplet’s ability to augment ZAP activity (Fig. 1C and D). In contrast, the E3 ligase activity of TRIM25 has been deemed essential for its function as a ZAP cofactor (35, 36), indicating that the mechanism by which each ligase enhances ZAP activity is markedly different. Riplet’s ubiquitin-independent ability to augment ZAP activity supports the idea that Riplet could function to induce higher-order oligomerization of ZAP. An analogous role of Riplet has been shown to take place in the RIG-I pathway, where Riplet bridges filaments of RIG-I bound to viral RNAs (38). TRIM25 is necessary for optimal binding of ZAP to RNAs (36), and it is plausible that Riplet also assists ZAP binding to target mRNAs. Given the importance of the PRY/SPRY domain of Riplet, it was not surprising that the Riplet DN mutant, which lacks the RING domain but retains the PRY/SPRY domain, continued to augment ZAP-mediated virus inhibition (Fig. 6B and C). Expression of Riplet DN in our system led to a profound decrease of IFN-β induction by poly(I·C), while ZAP activity was significantly enhanced (Fig. 6A to C). This finding indicates that the underlying mechanism by which Riplet augments ZAP activity is likely different from its ubiquitin-dependent function in RIG-I signaling. It may be similar to Riplet’s ubiquitin-independent role in inducing the formation of higher-order RIG-I multimers.
It is known that the central coiled-coil domain of TRIM/TRIM-like proteins can mediate dimerization (38, 47, 48, 50, 51). Here, we provide evidence that Riplet self-associates in ZAP-expressing cells (Fig. 4B and C). Riplet’s dimeric architecture is essential for recognition and ubiquitination of filamentous RIG-I (38). This finding is consistent with the notion that Riplet might be amplifying ZAP’s antiviral activity by promoting ZAP oligomerization. Given the structural similarity of Riplet and TRIM25, we examined the potential binding between these two proteins. Indeed, we detected a specific interaction between Riplet and TRIM25 (Fig. 7C and D). This observation is in line with the idea that Riplet and TRIM25 might form heterodimers. Our data showed a small yet consistent enhancement of ZAP-mediated virus inhibition when both proteins were expressed in the presence of ZAP compared to when either ligase was expressed alone in the presence of ZAP (Fig. 7A and B). It is plausible that Riplet or TRIM25 can act as homodimers, or heterodimers, and that they might operate cooperatively to enhance virus inhibition by ZAP.
In our system, TRIM25 bound to ZAP as previously reported (35, 36). We observed that the interaction was sensitive to treatment with RNase, suggesting that binding of RNAs is required. Our data suggest that the ZAP antiviral system requires the formation of larger complexes for effective reduction of viral mRNA transcripts. Because ZAP forms extensive interactions with target mRNAs (55), it is likely that multiple ZAP monomers engage different regions along a single mRNA molecule. Under this model, Riplet would function to bridge ZAP monomers bound to mRNAs, providing stability for larger complexes and inducing the formation of polymers. Further research to uncover additional ZAP cofactors will be critical to understanding the underlying mechanism of ZAP antiviral activity.
MATERIALS AND METHODS
Cell culture.
293TrexhZAP cells have been previously described (15). Briefly, these cells were generated by the transfection of 293Trex cells (Invitrogen) with the pcDNA4TO/myc-hZAP-v2 plasmid encoding a tetracycline-inducible myc-tagged hZAP-v2, one of the isoforms of human ZAP derived by alternative splicing. ZAP-knockout 293T cells (clone 89) were obtained from Akinori Takaoka at Hokkaido University, Japan (40). Cell populations with inducible ZAP and overexpressing Riplet or Riplet mutants (here denoted hZAP-R and hZAP-R mutant, respectively) were generated by transfecting 293TrexhZAP cells with various Riplet constructs by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol, followed by puromycin selection, and they were propagated for approximately 5 to 7 days. Extended culture resulted in loss of Riplet expression. All cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Atlas Bio), 2 mM glutamine, 1,000 U/mL of penicillin, and 100 mg/mL of streptomycin (Thermo Fisher) and maintained in a 37°C incubator with 5% CO2.
Plasmids and viral constructs.
Full-length Riplet was recovered from a plasmid containing a cDNA copy of the human RNF135 gene (pCMV SPORT 6; Invitrogen) by PCR amplification using KOD Hot Start DNA polymerase (Novagen) with the following oligonucleotides: Riplet forward (5′-GCGCCCCGGCGGTACCTTGGCCATGGCGGGCCTGGGCCTGGGCTCCG-3′) and Riplet reverse (5′-TAATCCCTTAGCTCGAGTTACACCTTTACTTGCTTTATTATC-3′). The PCR product was digested with KpnI and XhoI restriction enzymes and inserted into three different vectors, including a pCDNA 3.1(+) vector (Invitrogen), a pcDNA3.1+/C-DYK vector (Invitrogen), and the pcDNA3.1+/N-HA vector (Invitrogen). Riplet derivatives, including domain deletion mutants ΔRING, ΔP/SPRY, and dominant negative Riplet (R DN), were generated from full-length Riplet constructs according to standard cloning techniques. Newly generated DNA plasmids were verified by DNA sequencing (Genewiz). Plasmid pcDNA4/TO/myc-ZAP expressing a tagged ZAP has been described previously (15). A plasmid encoding Flag-tagged TRIM25 (36) was a gift of Guangxia Gao at the Chinese Academy of Sciences, Beijing, China. HIV-1-based reporter virus construct pNL4.3.E–R– Luc+ was obtained from AIDS Reagent Repository (catalog no. 3418). The pMD.G vector expresses the vesicular stomatitis virus (VSV) envelope glycoprotein, here denoted VSVG (Addgene; plasmid no. 12259). All plasmids were expanded in Escherichia coli DH5α cells, and DNA was purified using the Macherey-Nagel maxiprep kit according to the manufacturer’s instructions. Plasmids were transfected into HEK293T cells and 293TrexhZAP cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
Single-round infections.
293TrexhZAP cells and derivative cell lines stably expressing Riplet, Riplet mutants, or TRIM25 were infected with VSVG-pseudotyped HIV-luc reporter virus, and fresh media containing doxycycline (100 ng/mL) or DMSO were added at 6 h postinfection. Firefly luciferase reporter activity was measured at 24 h postinfection using the POLARstar Omega multimode microplate reader (BMG Labtech) after adding the luciferase assay reagent (luciferase assay system; Promega) to lysates. Virus inhibition was then calculated as the ratio of luciferase expression levels in DMSO-treated cells to those in doxycycline-treated cells. Protein levels were normalized to total protein content measured by a Bradford assay for each sample. Data points presented are the mean relative light units (RLU)/mg ± standard deviation (SD) values of four independent experiments done in triplicate.
Virus production.
HEK293T cells were seeded at a density of 8 × 106 cells per 100-mm plate (Nalgene)/mL 24 h before transfection. The next day, cells were cotransfected with 13 μg of the pNL4.3.E–R– Luc+ vector and 4 μg of VSVG expression vector by using Lipofectamine 2000 (Sigma-Aldrich) according to the manufacturer’s instructions. For mock controls, an empty vector plasmid instead of one expressing VSVG was used. Culture media containing reporter virus were harvested at 48 h posttransfection, filtered through a 0.45-μm filter, and used directly for transduction experiments.
Antibodies.
All of the antibodies were obtained commercially, including anti-Myc monoclonal antibody (Santa Cruz Biotechnology; 9E19; catalog no. sc-40), anti-TRIM25 antibody (dubbed “EFP” [for estrogen-responsive finger protein] antibody; Santa Cruz Biotechnology; catalog no. sc-135893), anti-ZAP mouse monoclonal antibody (Thermo Fisher Scientific, Proteintech anti-ZC3HAV1 monoclonal antibody; clone no. 1G10B9; catalog no. 66413-1-IG), RNF135 anti-Riplet polyclonal antibody (Thermo Fisher Scientific; catalog no. PA5-54457), anti-Flag monoclonal antibody (Sigma-Aldrich; catalog no. F1804), and anti-HA monoclonal antibody (Cell Signaling Technology [C29F4] MAb; catalog no. 3724). For microscopy, we utilized mouse anti-Flag (Sigma-Aldrich; catalog no. F1804) and rabbit anti-ZAP polyclonal antibody (Abcam; catalog no. ab154680).
RNA extraction and quantitative RT-PCR.
Levels of viral and cellular RNA were determined by quantitative reverse transcription-PCR (qRT-PCR). Total cytoplasmic RNA was extracted from 293TrexhZAP cells 24 h postinfection with VSVG-pseudotyped HIV-1 virus using the Qiagen RNeasy kit accordingly to the manufacturer’s instructions. The amount of isolated RNA for each sample was quantified by measuring the A260. Samples were treated with RNase-free DNase I (Roche) at 2 U/mL for 30 min at 37°C to digest any potential DNA plasmid contamination. Reverse transcription (RT) reactions were performed with 100 ng of purified cellular RNA using Applied Biosystems RT kit with random hexamer primers. Real-time quantitative PCRs were performed in 96-well plates with approximately 2 μL of RT product using a LightCycler 480 II real-time PCR system (Roche). The SYBR green PCR master mix (Roche) containing 15 pmol of indicated primers was used in 25-μL reaction mixtures with the following conditions: 10 min at 95°C, followed by 40 cycles of 30 s at 95°C and 1 min at 60°C. The genomic primers were unspliced HIV-1 genome (Pol mRNA) forward/reverse, GAATTTGCCAGGAAGATGGA/GCAGCCAATCTGAGTCAACA; partially spliced HIV-1 genome (vif mRNA) forward/reverse, GGCGACTGGGACAGC/CACACAATCATCACCTGCC; multiply spliced HIV-1 genome (nef-luc mRNA) forward/reverse, ACAGTCAGACTCATCAAGCTTCTCT/CGGGTCCCCTCGGGATT; and human GAPDH gene forward/reverse, TCGGAGTCAACGGATTTG/GCATCGCCCCACTTGATT. Relative mRNA abundance was calculated using the threshold cycle (2−ΔΔCT) method by first normalizing resulting viral mRNA levels to human GAPDH gene levels. Fold inhibition was determined as the ratio of mRNA levels in cells without ZAP induction (DMSO treated) to those in ZAP-expressing (doxycycline-treated) cells after normalization to viral mRNA levels to the housekeeping gene GAPDH. The results shown are means ± standard errors of the mean (SEMs) from three independent experiments performed in triplicates. Student’s t test was used for statistical analysis, and significance was attributed to P values of <0.05.
Genomic DNA extraction and quantitative RT-PCR.
To determine the extent of viral integration in cells overexpressing Riplet and empty vector control cells, infections were carried out with virus preparations of VSVG-pseudotyped HIV-1 or a mock virus lacking the VSVG envelope in the presence of 8 μg/mL polybrene (Sigma-Aldrich). Prior to infection, viral supernatants were incubated with RNase-free DNase I (Roche) at 2 U/mL for 30 min at 37°C to digest any potential viral plasmid DNA carryover. A control sample containing VSVG-pseudotyped HIV-1 virus was subsequently incubated at 90°C for 30 min to inactivate the virus and thus serve as control for potential plasmid carryover. Six hours postinfection, fresh medium containing doxycycline (100 ng/mL) or DMSO was added. Cells were passaged for 10 days in culture, after which cells were harvested and washed with phosphatase-buffered saline (PBS), and total DNA was isolated using a Qiagen DNeasy kit according to the manufacturer’s instructions. Approximately 80 ng of total DNA per sample was combined with SYBR green PCR master mix (Roche) containing 15 pmol of both forward (5′-GAATTTGCCAGGAAGATGGA-3′) and reverse (5′-GCAGCCAATCTGAGTCAACA-3′) primers targeting the HIV-1 gag-pol gene. A second set of reactions was run in parallel with primer pairs targeting the human GAPDH gene, forward (5′-GAAGGTGAAGGTCGGAGTC-3′) and reverse (5′-GAAGATGGTGATGGGATTTC-3′). An additional sample containing no template (NT) for PCR amplification was included to control for DNA contamination. All qPCRs were performed in 96-well plates using a 7900 Fast real-time PCR system (Applied Biosystems) with the following conditions: 10 min at 95°C, followed by 40 cycles of 30 s at 95°C and 1 min at 60°C. Levels of viral DNA were calculated by first normalizing resulting values with the 2−ΔΔCT method to the value for the human GAPDH gene. Values obtained were then normalized to the values for 293TrexhZAP cells without the addition of doxycycline (DMSO control). Results shown are means ± SEMs from three independent experiments done in triplicate. Student’s t test was used for statistical analysis.
Microscopy.
Coverslips on 6-well plates were pretreated with 0.2% poly-lysine (Thermo Fisher) and then seeded with ZAP knockout 293T cells (clone 89) and control cells. The next day, 200 ng of Riplet or an empty vector were transfected using Lipofectamine 2000 according to the manufacturer’s instructions. Twenty-four hours posttransfection, cells were fixed with 4% paraformaldehyde for 15 min at room temperature and then washed with PBS, and 10 mM glycine solution in PBS was added. Cells were permeabilized for 15 min with 1% bovine serum albumin (BSA) and 0.1% Triton-X in PBS. To stain cells, the following antibodies were used for 1 h at room temperature: mouse anti-Flag (1:500) and rabbit anti-ZAP (1:500) diluted in PBS-0.01% Triton-X. Cells were then washed three times with PBS-0.01% Triton-X and incubated in the dark for 1 h with secondary antibodies Alexa Fluor 594 anti-mouse and Alexa Fluor 488 anti-rabbit at 1:500 dilution (Molecular Probes). Cells were washed again three times with PBS-0.01% Triton-X, and DAPI (4′,6-diamidino-2-phenylindole) stain (Sigma-Aldrich) was added at 1:10,000 dilution and incubated for 20 min at room temperature. Slides were then mounted with Prolong Diamond antifade mountant (Invitrogen). Images were visualized on a motorized spinning disk confocal microscope (Yokogawa CSU-X1 A1 confocal head and Zeiss Axio Observer Z1 microscope), and SlideBook software (Intelligent Imaging Innovations) was used for image processing and colocalization analysis.
Coimmunoprecipitation experiments and immunoblot analysis.
Cells were engineered to express various cDNAs with or without ZAP induction. In some experiments, cells were treated with universal type I interferon alpha (1,000 U/mL; Pestka Biomedical Laboratories; catalog no. 11200-1). Cells were washed with cold PBS twice and then harvested with IP lysis buffer (Pierce) and incubated in ice for 30 min in the presence of protease and phosphatase inhibitor tablets (Roche). To test for RNase sensitivity, RNase A was added at 50 μg/mL to lysates and incubated at room temperature for 1 h. Samples were then centrifuged at 4°C for 30 min at 13,000 rpm. The soluble fraction was collected, and 10% of the sample was stored to serve as input control, while the other 90% of the lysate was subjected to immunoprecipitation. Anti-c-Myc magnetic beads (Pierce) or anti-Flag M2 magnetic beads (Sigma) were washed with Tris-buffered saline with 0.1% Tween 20 detergent four times, and cell extracts were then added. Beads and cell lysates were incubated with lysates overnight at 4°C. The next day, samples were washed four times with lysis buffer for a total of 35 min. The pellet was resuspended in SDS sample buffer with 10% β-mercaptoethanol, and samples were then boiled, separated from the beads, and resolved on a 4 to 20% precast polyacrylamide gel (Bio-Rad). After transfer to a nitrocellulose membrane (GE Healthcare), blots were incubated in blocking buffer (Rockland) for 1 h at room temperature. Primary antibodies were incubated overnight at 4°C, and then blots were washed 4 times and incubated for 1 h at room temperature with the appropriate secondary antibody. Bound antibodies were visualized on the LI-COR Odyssey CLx.
ACKNOWLEDGMENTS
This work was supported by NCI grant R01 CA 30488 to S.P.G. and National Science Foundation GRFP grant no. 1644869 to M.V.B.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Stephen P. Goff, Email: spg1@cumc.columbia.edu.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.Chow KT, Gale M, Jr., Loo YM. 2018. RIG-I and other RNA sensors in antiviral immunity. Annu Rev Immunol 36:667–694. 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- 2.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat Rev Immunol 14:36–49. 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoggins JW. 2019. Interferon-stimulated genes: what do they all do? Annu Rev Virol 6:567–584. 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 6.Gao G, Guo X, Goff SP. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703–1706. 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 7.Bick MJ, Carroll J-WN, Gao G, Goff SP, Rice CM, MacDonald MR. 2003. Expression of the zinc finger antiviral protein inhibits alphavirus replication. J Virol 77:11555–11562. 10.1128/jvi.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH. 2015. The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet 11:e1005252. 10.1371/journal.pgen.1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, Cuconati A, Block TM, Guo J-T, Guo H. 2013. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog 9:e1003494. 10.1371/journal.ppat.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moldovan JB, Moran JV. 2015. The zinc finger antiviral protein ZAP inhibits LINE and Alu retrotransposition. PLoS Genet 11:e1005121. 10.1371/journal.pgen.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller S, Möller P, Bick MJ, Wurr S, Becker S, Günther S, Kümmerer BM. 2007. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol 81:2391–2400. 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nchioua R, Kmiec D, Müller JA, Conzelmann C, Groß R, Swanson CM, Neil SJD, Stenger S, Sauter D, Münch J, Sparrer KMJ, Kirchhoff F. 2020. SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans. mBio 11:e01930-20. 10.1128/mBio.01930-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng C, Wyatt LS, Glushakow-Smith SG, Lal-Nag M, Weisberg AS, Moss B. 2020. Zinc-finger antiviral protein (ZAP) is a restriction factor for replication of modified vaccinia virus Ankara (MVA) in human cells. PLoS Pathog 16:e1008845. 10.1371/journal.ppat.1008845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Tu F, Zhu Y, Gao G. 2012. Zinc finger antiviral protein inhibits XMRV infection. PLoS One 7:e39159. 10.1371/journal.pone.0039159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y, Chen G, Lv F, Wang X, Ji X, Xu Y, Sun J, Wu L, Zheng YT, Gao G. 2011. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci USA 108:15834–15839. 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ficarelli M, Neil SJD, Swanson CM. 2021. Targeted restriction of viral gene expression and replication by the ZAP antiviral system. Annu Rev Virol 8:265–283. 10.1146/annurev-virology-091919-104213. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Y, Gao G. 2008. ZAP-mediated mRNA degradation. RNA Biol 5:65–67. 10.4161/rna.5.2.6044. [DOI] [PubMed] [Google Scholar]
- 18.Meagher JL, Takata M, Gonçalves-Carneiro D, Keane SC, Rebendenne A, Ong H, Orr VK, MacDonald MR, Stuckey JA, Bieniasz PD, Smith JL. 2019. Structure of the zinc finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences. Proc Natl Acad Sci USA 116:24303–24309. 10.1073/pnas.1913232116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takata MA, Gonçalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550:124–127. 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff SP. 2017. Evolution: zapping viral RNAs. Nature 550:46–47. 10.1038/nature24140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia X. 2020. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol Biol Evol 37:2699–2705. 10.1093/molbev/msaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonçalves-Carneiro D, Takata MA, Ong H, Shilton A, Bieniasz PD. 2021. Origin and evolution of the zinc finger antiviral protein. PLoS Pathog 17:e1009545. 10.1371/journal.ppat.1009545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li MMH, Aguilar EG, Michailidis E, Pabon J, Park P, Wu X, de Jong YP, Schneider WM, Molina H, Rice CM, MacDonald MR. 2019. Characterization of novel splice variants of zinc finger antiviral protein (ZAP). J Virol 93:e00715-19. 10.1128/JVI.00715-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Xu Y, Zhang K, Wang X, Sun J, Gao G, Liu Y. 2012. Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat Struct Mol Biol 19:430–435. 10.1038/nsmb.2243. [DOI] [PubMed] [Google Scholar]
- 25.Kerns JA, Emerman M, Malik HS. 2008. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc finger antiviral protein. PLoS Genet 4:e21. 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law LMJ, Albin OR, Carroll J-WN, Jones CT, Rice CM, Macdonald MR. 2010. Identification of a dominant negative inhibitor of human zinc finger antiviral protein reveals a functional endogenous pool and critical homotypic interactions. J Virol 84:4504–4512. 10.1128/JVI.02018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue G, Braczyk K, Goncalves-Carneiro D, Dawidziak DM, Sanchez K, Ong H, Wan Y, Zadrozny KK, Ganser-Pornillos BK, Bieniasz PD, Pornillos O. 2022. Poly(ADP-ribose) potentiates ZAP antiviral activity. PLoS Pathog 18:e1009202. 10.1371/journal.ppat.1009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X, Carroll J-WN, Macdonald MR, Goff SP, Gao G. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78:12781–12787. 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y, Wang X, Goff SP, Gao G. 2012. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J 31:4236–4246. 10.1038/emboj.2012.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung AKL, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. 2011. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42:489–499. 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang T-Y, Krug RM, Sullivan CS. 2013. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 14:435–445. 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmer MM, Kibe A, Rand U, Pekarek L, Ye L, Buck S, Smyth RP, Cicin-Sain L, Caliskan N. 2021. The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting. Nat Commun 12:7193. 10.1038/s41467-021-27431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Todorova T, Bock FJ, Chang P. 2014. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat Commun 5:5362. 10.1038/ncomms6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ficarelli M, Wilson H, Pedro Galão R, Mazzon M, Antzin-Anduetza I, Marsh M, Neil SJD, Swanson CM. 2019. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. Elife 8:e46767. 10.7554/eLife.46767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li MMH, Lau Z, Cheung P, Aguilar EG, Schneider WM, Bozzacco L, Molina H, Buehler E, Takaoka A, Rice CM, Felsenfeld DP, MacDonald MR. 2017. TRIM25 enhances the antiviral action of zinc finger antiviral protein (ZAP). PLoS Pathog 13:e1006145. 10.1371/journal.ppat.1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng X, Wang X, Tu F, Wang Q, Fan Z, Gao G. 2017. TRIM25 is required for the antiviral activity of zinc finger antiviral protein. J Virol 91:e00088-17. 10.1128/JVI.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gack MU, Shin YC, Joo C-H, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 38.Cadena C, Ahmad S, Xavier A, Willemsen J, Park S, Park JW, Oh S-W, Fujita T, Hou F, Binder M, Hur S. 2019. Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell 177:1187–1200.e16. 10.1016/j.cell.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayman TJ, Hsu AC, Kolesnik TB, Dagley LF, Willemsen J, Tate MD, Baker PJ, Kershaw NJ, Kedzierski L, Webb AI, Wark PA, Kedzierska K, Masters SL, Belz GT, Binder M, Hansbro PM, Nicola NA, Nicholson SE. 2019. RIPLET, and not TRIM25, is required for endogenous RIG-I-dependent antiviral responses. Immunol Cell Biol 97:840–852. 10.1111/imcb.12284. [DOI] [PubMed] [Google Scholar]
- 40.Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, Goto S, Kameoka S, Fujikura D, Yamada T, Mizutani T, Kazumata M, Sato M, Tanaka J, Asaka M, Ohba Y, Miyazaki T, Imamura M, Takaoka A. 2011. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol 12:37–44. 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- 41.Lee H, Komano J, Saitoh Y, Yamaoka S, Kozaki T, Misawa T, Takahama M, Satoh T, Takeuchi O, Yamamoto N, Matsuura Y, Saitoh T, Akira S. 2013. Zinc finger antiviral protein mediates retinoic acid inducible gene I–like receptor-independent antiviral response to murine leukemia virus. Proc Natl Acad Sci USA 110:12379–12384. 10.1073/pnas.1310604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erazo A, Goff SP. 2015. Nuclear matrix protein Matrin 3 is a regulator of ZAP-mediated retroviral restriction. Retrovirology 12:57. 10.1186/s12977-015-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-β induction during the early phase of viral infection. J Biol Chem 284:807–817. 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 44.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. 2010. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8:496–509. 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Oshiumi H, Miyashita M, Matsumoto M, Seya T. 2013. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog 9:e1003533. 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choudhury NR, Heikel G, Trubitsyna M, Kubik P, Nowak JS, Webb S, Granneman S, Spanos C, Rappsilber J, Castello A, Michlewski G. 2017. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol 15:105. 10.1186/s12915-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez JG, Sparrer KMJ, Chiang C, Reis RA, Chiang JJ, Zurenski MA, Wan Y, Gack MU, Pornillos O. 2018. TRIM25 binds RNA to modulate cellular anti-viral defense. J Mol Biol 430:5280–5293. 10.1016/j.jmb.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H-T, Hur S. 2021. Substrate recognition by TRIM and TRIM-like proteins in innate immunity. Semin Cell Dev Biol 111:76–85. 10.1016/j.semcdb.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mary C, Telles JN, Cheynet V, Oriol G, Mallet F, Mandrand B, Verrier B. 1994. Quantitative and discriminative detection of individual HIV-1 mRNA subspecies by an RNAse mapping assay. J Virol Methods 49:9–23. 10.1016/0166-0934(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 50.Kato K, Ahmad S, Zhu Z, Young JM, Mu X, Park S, Malik HS, Hur S. 2021. Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. Mol Cell 81:599–613.e8. 10.1016/j.molcel.2020.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, Aiken C, Olsen JC, Kar AK, Sodroski JG, Sundquist WI. 2008. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol 82:11682–11694. 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao D, Yang Y-K, Wang R-P, Zhou X, Diao F-C, Li M-D, Zhai Z-H, Jiang Z-F, Chen D-Y. 2009. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One 4:e5760. 10.1371/journal.pone.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vazquez C, Tan CY, Horner SM. 2019. Hepatitis C virus infection is inhibited by a noncanonical antiviral signaling pathway targeted by NS3-NS4A. J Virol 93:e00725-19. 10.1128/JVI.00725-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choudhury NR, Heikel G, Michlewski G. 2020. TRIM25 and its emerging RNA-binding roles in antiviral defense. Wiley Interdiscip Rev Rna 11:e1588. 10.1002/wrna.1588. [DOI] [PubMed] [Google Scholar]
- 55.Luo X, Wang X, Gao Y, Zhu J, Liu S, Gao G, Gao P. 2020. Molecular mechanism of RNA recognition by zinc finger antiviral protein. Cell Rep 30:46–52.e4. 10.1016/j.celrep.2019.11.116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download jvi.00526-22-s0001.pdf, PDF file, 0.5 MB (509.5KB, pdf)