Abstract
The goal of an efficacious cancer therapy is to specifically target diseased cells at high accuracy while sparing normal, healthy cells. Over the past three decades, immunotherapy, based on the use of monoclonal antibodies (mAbs) directed against tumor-associated antigens, to inhibit their oncogenic function, or against immune checkpoints, to modulate specific T cell responses against cancer, has proven to be an important strategy for cancer therapy. Nevertheless, the number of mAbs approved for clinical use is still limited because of significant drawbacks to their applicability. Oligonucleotide aptamers, similarly to antibodies, form high-affinity bonds with their specific protein targets, thus representing an effective tool for active cancer targeting. Compared to antibodies, aptamers’ use as therapeutic agents benefits from their low size, low/no immunogenicity, simple synthesis and design flexibility for improving efficacy and stability. This review intends to highlight recently emerged applications of aptamers as recognition elements, from biomarker discovery to targeted drug delivery and targeted treatment, showing aptamers’ potential to work in conjunction with antibodies for attacking cancer from multiple flanks.
Keywords: Aptamers, monoclonal antibodies, targeted therapy, theranostics, SELEX
Introduction
Cancer, often referred to as the “Pathology of the Century”, represents the second leading cause of death worldwide, affecting millions of people each year [1, 2]. Cytotoxic chemotherapy and radiotherapy are the main treatment options for many patients as neoadjuvant or adjuvant to surgery, however their success is often limited by several drawbacks, in part due to the lack of selectivity for tumor cells.
Typically, conventional chemotherapeutics target highly proliferating cells, resulting unable to discriminate cancer cells from rapidly dividing healthy cells and thus producing severe side effects to normal tissues. This strongly reduces the amount of drug that can be systemically administered and, together with pharmacokinetic effects, hampers its effectiveness to the tumor site. Moreover, the development of multiple chemoresistance mechanisms, including decreased uptake or increased efflux of the drug, low activation of prodrug or higher drug inactivation, DNA damage repair, mutations of drug targets, occurrence of compensatory signaling pathways and evasion of cell death, represents the major cause of tumor relapse and metastasis, hampering the clinical outcome [3–5]. Also, a paradoxical effect of chemotherapy has been evidenced by which its therapeutic efficacy on the primary tumor can be balanced by the stimulation of responses capable of inducing epithelial-mesenchymal transition, stem phenotype and other protective mechanisms, which in turn modify tumor microenvironment (TME), causing detrimental effects on tumor relapse and promotion of metastases [6]. Healthy secretome is composed by a great variety of factors and contributes to maintain homeostasis in the microenvironment. Chemotherapy can induce alterations in the tumor cell secretome composition, shaping it in a collection of protumorigenic factors that promote the formation of an immunosuppressive TME [7]. Therefore, TME can become a shield for cancer cells, protecting them from drugs insults and contributing to the development of chemoresistance.
Radiotherapy is equally subject to mechanisms of resistance and has profound effects on TME. Interestingly, radiotherapy, in addition to attacking cancer cells by inducing DNA damage, modulates diverse components of the TME, producing anti-tumor immunomodulatory effects [8]. However, radiotherapy, as chemotherapy, encounters the bottlenecks caused by increased resistance to treatment by multiple mechanisms including genes mutation or their aberrant expression, epigenetic alterations, cancer stem cells existence, the over activation of same signaling pathways and TME alteration [9]. To overcome the limitations of conventional chemotherapy and radiotherapy, a huge effort is devoted to identify cancer-specific biomarkers, usually proteins found mutated or abnormally expressed in cancer cells, which can enable effective targeted therapies that target cancer cells specifically, killing them or stopping their growth or spread, while sparing healthy cells [10]. Two main classes of targeting cancer agents are currently available for use in clinical practice, monoclonal antibodies (mAbs) and small-molecule inhibitors [11]. So far, Food and Drug Administration (FDA) has approved several agents for specific types of cancer for which established therapeutic targets have been identified [12–14]. As example, trastuzumab and pertuzumab (mAbs) on the one hand, and lapatinib and neratinib (small molecules) on the other, all targeting and interfering with the human epidermal growth factor receptor 2 (HER2), have shown to be beneficial for treating patients affected by a subtype of breast cancer overexpressing HER2 [15]. Furthermore, the opportunity to use drugs in combination is a viable strategy to overcome resistance to single-agent targeted therapy, which often occurs. Indeed, crosstalk between pathways and feedback loops, which are essential aspects of cell physiology, could modify therapeutic vulnerability in cancer cells. Combinations of two or more therapeutic agents can be more effective than monotherapy approach, because they work by different mechanisms in a characteristically synergistic or additive manner [16, 17].
Unfortunately, targetable proteins have not yet been identified for several human cancers that still lack targeted therapy, including heterogeneous triple-negative breast cancer (TNBC) and glioblastoma (GBM) [18, 19]. In search for efficacious strategies to both identify new actionable biomarkers and specifically target cancer cells, alternatively and/or in combination with existing drugs, oligonucleotide aptamers have sparked intense interest [20, 21].
Aptamers: the chemical antibodies
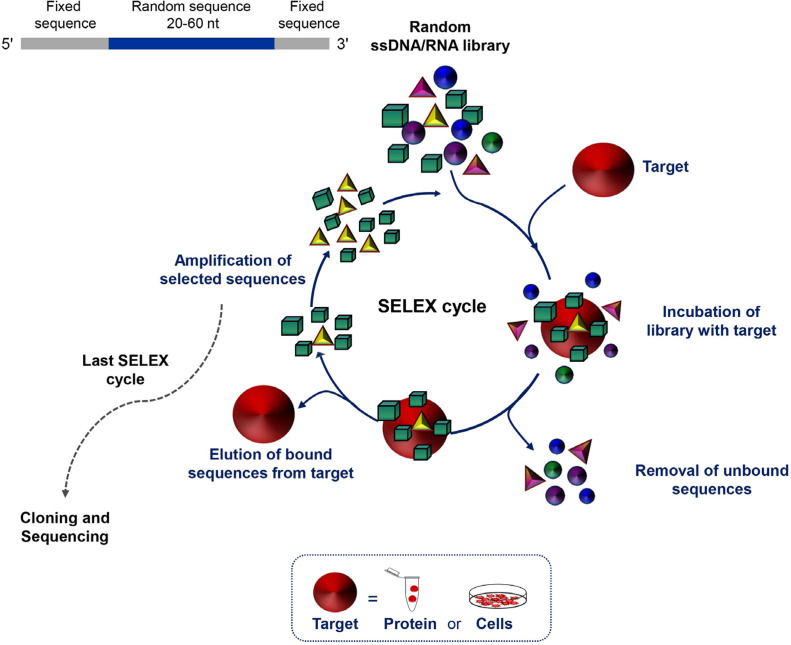
Aptamers, often termed “chemical antibodies”, are short, single-stranded DNA or RNA that assume three-dimensional structures to bind at high affinity to a specific target molecule. The targets of aptamers range from small chemicals to large cell-surface and transmembrane proteins even embedded in their natural environment, i.e. living cells, tissues or animals. Their easy and reproducible manufacture, combined with the nucleic acid composition that supports a plethora of chemical modifications, which in turn allow for different conjugation chemistries, makes aptamers suitable for molecular recognition-based applications, including cancer diagnosis and targeted therapy [22]. Aptamers are selected in vitro from large libraries of random sequence oligonucleotides using systematic evolution of ligands by exponential enrichment (SELEX) technology (Figure 1). SELEX consists of a multi-step process involving repeated cycles of: i) incubation of the library with the target; ii) partition of target-bound oligonucleotides from unbound sequences; iii) elution of the bound sequences frSom the target; and iv) amplification of the selected oligonucleotides. Individual sequences are identified by classical cloning of the final selected pool and/ or high-throughput sequencing and bioinformatics analyses, which allow to monitor the selection of aptamers during all SELEX rounds [23, 24]. Finally, the selected sequences are tested for binding affinity and specificity.
Figure 1.
Schematic representation of the SELEX technology. SELEX is a multistep process starting with random libraries of single-stranded DNA (ssDNA) or RNA flanked by two fixed primer-binding sequences on both ends necessary for enzymatic amplification and in vitro transcription (in the case of a RNA library). It includes reiterated rounds (usually 8-20) of binding, partitioning and amplification (see text for details). nt: nucleotide
Introduced for the first time about thirty years ago [25–27], SELEX has aroused growing interest and has been successfully applied, to date, for the isolation of aptamers against proteins of biomedical interest for use as research agents, diagnostic agents, therapeutics, biosensor components and tools for biomarker and drug discovery [28]. In recent years, sophisticated SELEX protocols have been developed, optimizing the starting library and the separation techniques, in order to reduce the process times and the number of cycles for the selection of high efficacious aptamers endowed with the proper features required for their final application [29–31].
It is well known that cell surface protein biomarkers are of particular interest for cancer diagnosis and targeted therapy, and mAbs against antigens expressed on the surface of cancer cells represent a revolutionary therapeutic approach. Aptamers can be selected to target specific cell surface proteins, so they may have the same potential as mAbs immunotherapy.
To generate aptamers against a cell surface protein, a purified protein can be used as target for the selection, usually the peptide corresponding to the extracellular region (protein-SELEX approach) or, otherwise, live cells that overexpress the pre-identified target of interest (cell-SELEX approach). The latter approach allows selecting aptamers against protein targets as close as possible to their natural distributions and conformations. Protein-SELEX has the advantage of easy control of the enrichment conditions and takes usually shorter time than cell-SELEX. However, it often happens that aptamers generated against a recombinant protein do not recognize the protein in the endogenous environment [32]. For this reason, selection on whole cells [33] or strategies that alternate protein- and cell-SELEX [34, 35], even if trickier, are better suited for generating aptamers targeting cell-surface proteins especially for in vivo applications. Furthermore, an important application of the cell-SELEX consists in generating aptamers targeting cancer cells without prior knowledge of biomarkers present on the cell surface by alternating positive selection on target cells and counter-selection steps on off-target cells. At the end of the selection, a panel of aptamers will be identified that recognizes a surface molecular signature specific of the tumor cell type used for the positive selection. Importantly, at the end of cell-SELEX, the best binding aptamers are used as bait to identify their targets, thus proving to be an amazing tool for biomarker discovery. Using this approach, aptamers with cell-type discriminating properties were selected, which identify the surface molecular signatures of cancer cells associated with important behaviors, including resistance to treatment, tumorigenicity, stemness and metastatic potential [36–43]. Recently, by applying a differential cell-SELEX approach to TNBC cells, our group generated six novel nuclease-resistant RNA aptamers able to distinguish TNBC cells from both non-malignant and non-TNBC breast cancer cells, as well as to differentiate TNBC histological specimens in aptamer-based histochemical approaches [44]. Importantly, they are endowed with some biological activities, such as internalization within the target cells and anti-proliferative and anti-mammosphere activity [44]. The availability of a set of aptamers acting as high efficacious recognition probes for functional surface signatures of TNBC cells fulfills the great challenge of simultaneously targeting multiple protein targets.
Importantly, SELEX can also be realized in more complex cellular environments as cellular spheroids in three-dimensional cell culture systems [45], or even tissues [46] and whole-organisms in vivo [47–49]
Unique properties of aptamers for targeted cancer therapy
Aptamers possess several unique features as recognition elements that rend them a valid alternative or complement to conventional antibodies in targeted cancer therapy (Table 1).
Table 1.
Strengths and weaknesses of aptamers in comparison with antibodies
| Criteria | Aptamers | Antibodies |
|---|---|---|
| Size | 5–15 kDa | 150–180 kDa |
| • Target accessibility | High | Low |
| • Minimal target size | 60 Da | 600 Da |
| • Tissue/tumor penetration | High | Low |
| • Clearance rate | Rapid | Slow |
| Basic composition | Nucleotides | Amino acids |
| • Resistance to harsh environment conditions (pH and temperature) | High | Low |
| • Shelf-life | Long | Limited |
| • Versatility to chemical | High | Limited |
| • Nuclease degradation | Sensitive; limited half-life in vivo (unmodified) | Resistant; long half-life in vivo |
| Therapeutic efficacy | ||
| • Affinity and specificity | KD, nano/pico | KD, nano/pico |
| • Immunogenicity | Low/none | High |
| • Modulation of target activity | Yes | Yes |
| • Fc-mediated effector | No | Yes |
| Discovery | ||
| • Time | In vitro SELEX, 2–8 weeks | In vivo biological process, 6 |
| Production | ||
| • Scale up | Easy | Hard |
| • Batch to batch variation | None | High |
KD values: dissociation constants
First, because of their production by chemical synthesis, the reproducibility of aptamers is very accurate and manufacture does not require the use of cell cultures and animals, which are instead required for the production of antibodies. In addition, with the increasing development of sophisticated technologies, the production of aptamers is becoming even more cost- and time-effective.
As a result of their complex tridimensional structures, aptamers like antibodies have high affinity and specificity for the target with KD values typically in the low nanomolar-picomolar range. Like antibodies, aptamers can recognize with high specificity proteins of the same family, or even the same protein present in different conformational states [22, 50]. However, unlike antibodies, aptamers have a low size, usually from 5 to 15 kDa, which allows them to bind low molecular weight targets or some hidden binding domains that are inaccessible to larger antibodies. Further, their smaller size, 20- to 25-fold as compared with full-sized mAbs, is a crucial property that allows high tissue penetration, as needed for cancer treatment. Most importantly, aptamers support a huge variety of chemical modifications that are crucial to overcome undoubted limitations to their clinical applications, such as uncertain stability and rapid renal clearance. For instance, the susceptibility of aptamers to nuclease degradation is easily overcome by applying analogues of phosphate, ribose and bases [51], thus increasing their stability and half-life in vivo especially in the complex microenvironment that surrounds the tumor. Further, to increase their permanence in the circulation, bulky molecules are appended to the aptamer including polyethylene glycol (PEG) [52] or even antibodies [53], without affecting the high tumor penetration and target accessibility. More importantly, chemical modification allows reducing the potential in vivo toxicity and immunogenicity of aptamers, thus allowing for the safe administration of repeated doses that are precluded for antibodies, unless they are humanized or produced completely in humans (Table 1). An elegant example of an ad hoc chemical modification of aptamers was recently reported by Zhou et al. [54], which generated aptamers able to work only in hypoxic TMEs (conditional aptamers). Indeed, the addition of a PEG5000-azobenzene-NHS to the aptamer prevents binding to its target protein. When the conjugate reaches the tumor site, the hypoxic environment causes the release of the PEG-group from the aptamer that assumes a conformation competent for the recognition of the target. This strategy allows limiting the action of the aptamer exclusively to cancer cells, thus reducing toxicity towards healthy cells, in the case in which the target of the aptamer is not expressed only in tumors.
Finally, aptamers modifications allow to set up sophisticated combination therapies by chemically conjugating aptamers with other type of therapeutic molecules or drugs [22, 55], thus improving their effectiveness for clinical application.
Applications of aptamers in cancer targeted therapy
For all the properties described above, cell-targeting aptamers have proven to be a valid alternative to mAbs in various cancer applications (Figure 2).
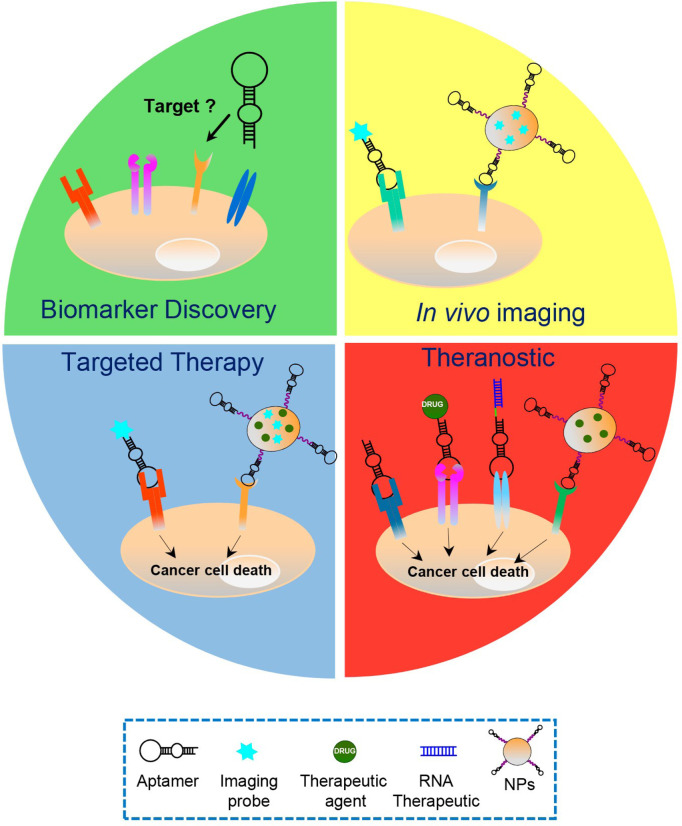
Figure 2.
Aptamers’ applications in cancer treatment. Schematic representation of the various cancer applications of aptamers, including biomarker discovery, in vivo imaging, targeted therapy and theranostic approaches, which play key roles in the personalized cancer therapy. Aptamers have been used as: i) recognition agents for biomarkers discovery; ii) imaging agents upon conjugation to fluorescent probes, radionuclides or nanoparticles (NPs) functionalized with biomedical imaging agents; iii) stand-alone therapeutics or delivery agents for chemotherapeutics, RNA therapeutics and drug-loaded NPs; and iv) theranostic agents, upon conjugation to imaging agents (in case the aptamer itself has anti-tumor activities) or NPs loaded with both drug and imaging agents
First, aptamers are foreseen as promising stand-alone therapeutic agents; indeed, by binding to their proper cancer-related target, they can interfere with its biological function, thus ultimately inhibiting tumor development and progression. They can act either as inhibitors of the target, competing with small molecules and protein ligands, as in the case of aptamers that inhibit human receptor tyrosine kinases [37, 56–61], or as activators, promoting the function of target receptors [62, 63]. In the latter case, usually the generation of a dimeric aptamer is able to bind to two receptor monomers, thus inducing dimerization and activation of the receptor. As an example of aptamers’ ability to equalize antibodies as targeted therapeutics, Mahlknecht et al. [64], compared the efficacy of the trimeric anti-HER2 DNA aptamer with a monoclonal antibody against the same receptor in reducing tumor growth in gastric cancer xenograft mice, showing the superior efficacy of the aptamer over the antibody. Recently, as demonstration of the striking therapeutic potential of aptamers, Yang et al. [65], has developed a new approach that harnesses the power of aptamer technology in cancer immunotherapy. They decorated the surface of natural killer (NK) cells with a CD30 aptamer by conjugating the aptamer to different lipophilic anchors intercalated into the cell membrane. Importantly, under the guidance of aptamers, NK cells specifically targeted CD30-expressing lymphoma cells, triggering cell apoptosis and death due to their innate killing capacity. This aptamer-based strategy, which conforms NK cells for cell-specific adaptive immunotherapy without genetic manipulation, opens a powerful new avenue for cancer-targeted immunotherapy.
The development of aptamer-based cancer therapy includes not only aptamers used as stand-alone agents, but also conjugated with other drugs. Indeed, many aptamers internalize upon binding to their protein target on the cell-surface through receptor-mediated endocytosis or, as in the case of the nucleolin-aptamer, macropinocytosis [66]. Thus, they can function as selective delivery vehicles for secondary therapeutic cargos into cancer cells that exclusively express and/or overexpress the aptamer target on their surface. Overall, the development of actively targeted aptamer-based systems will improve methods for delivery of drugs whose therapeutic benefit is limited by instability, lack of specificity or poor bioavailability. So far, different types of aptamer-based drug delivery systems have been devised where, depending on the drug, it is usually intercalated into the aptamer structure or appended to the aptamer sequence through a flexible linker or, otherwise, loaded into diverse formulations of nanocarriers conjugated with aptamers. In such ways, successful delivery of chemotherapeutics [67], toxins [68], therapeutic small interfering RNAs (siRNAs), microRNAs (miRNAs), anti-miRNAs [22, 69] and even large functional RNAs [70] specifically to cancer cells has been achieved by using aptamers as cancer cell-targeting ligands, thus resulting in highly effective therapies. Furthermore, with the aim of overcoming the biological barriers that hinder the efficient and safe delivery of drugs specifically to tumor sites, sophisticated aptamer-based constructs have been designed, such as those allowing therapeutics to reach tumor cells protected by the blood-brain barrier (BBB) [71]. Recently, Macdonald et al. [72], conceived a bifunctional doxorubicin (DOX)-intercalated aptamer, able to bind both to transferrin, highly expressed on the endothelial BBB cell membranes, and to the epithelial cell adhesion molecule (EpCAM) on metastatic TNBC cells. The resulting construct was capable of transcytosing the BBB, upon binding to transferrin and delivering DOX specifically into EpCAM-positive tumor cells, both in an in vitro model of BBB and in mouse models of brain metastasis of TNBC.
Using a different approach, our group proved the ability of the anti-platelet-derived growth factor receptor beta (PDGFRβ) Gint4.T aptamer, which we previously generated by GBM cell-SELEX and validated as a high affinity ligand/inhibitor of the receptor in GBM [37, 58] and bone marrow-derived human mesenchymal stem cells [73], to efficiently deliver a drug (Bez 235, PI3K-mTOR inhibitor) to orthotopic (intracranial) GBM xenografts across the BBB. We indeed generated multifunctional nanosystems made up of polymeric NPs, loaded with drug and fluorescent dye, decorated on their surface with the aptamer for both BBB crossing and cancer targeting, exploiting the high expression of PDGFRβ both on BBB endothelial cells and GBM cells [74].
Encapsulating therapeutic agents, in aptamer-decorated nanocarriers offers numerous advantages, including: i) several specific ligands can be linked to the surface of the NPs, increasing the specificity of targeting; ii) multiple drugs can be loaded, allowing for combined therapy; and iii) NPs are very stable, inert and have no toxicity [74, 75].
A recent demonstration of the efficacy of aptamer-based nanoplatforms for hydrophobic drugs’ delivery is given by the elegant study by the Guo et al. [76]. The authors constructed a four-way RNA junction NP for delivering of paclitaxel (PTX), one of the most poorly bioavailable drugs because of its water insolubility and toxicity, with the purpose of maximizing the thermodynamic stability of the construct and the solubility of the PTX cargo. Upon intravenous injections, the construct, equipped with the anti-epidermal growth factor receptor (EGFR) CL4 aptamer [41, 58, 59] as a specific ligand for tumor-targeting, resulted able to drastically inhibit breast cancer growth, with nearly undetectable toxicity and immune responses in mice.
In addition to this, the applications of aptamer-functionalized NPs for cancer therapy appear varied, including their use to remove tumors “surgically”. Indeed, a dynamic magneto nano-therapy using the AS-14 DNA aptamer targeted to mouse Ehrlich’s carcinoma fibronectin was developed to eradicate a tumor in mice by applying an aptamer-guided alternating magnetic field at the single cell level [77].
Moreover, the efficacy of cancer immunotherapy can be improved by nanotechnology and several examples show the potential of nanoparticle-based delivery systems assembled with Abs as active disease-targeting agents to regulate TME and modulate immune response [78, 79]. Similarly, in the last years, aptamers have been proven to act as immunoagents by themselves [80] or upon integration into immunomodulatory nanosystems [81]. Notably, the first CAR-like multivalent aptamer NPs were assembled that could activate T cells and inhibit melanoma growth, opening the way to use aptamer-based constructs for overcoming the safety issues of CAR-T cell therapy [82].
The possibility to directly conjugate aptamers to fluorescent probes and radionuclides makes them exquisite agents for in vivo tumor imaging modalities and even theranostic approaches in case the aptamer itself has anti-tumor activities [83]. As we have recently shown, PDGFRβ identifies a subgroup of mesenchymal TNBCs with invasive and stem-like phenotype, and has a crucial role in driving TNBC cell invasiveness and metastases formation [84]. Importantly, due to its inhibitory effect on PDGFRβ activation and downstream signaling pathways, the Gint4.T aptamer was found to be highly effective for both imaging of TNBC xenografts and TNBC cell-derived lung metastases, upon labeling with a near-infrared dye, and for the suppression of metastases formation when administrated intravenously in a mouse model [84]. Aside from direct conjugation of the aptamers to imaging probes, again, aptamer-conjugated NPs can be functionalized with biomedical imaging agents or can be used as containers of imaging and therapeutic agents [85, 86]. Importantly, in both cases the aptamers can achieve comparable or even superior performance than their antibody counterparts [87, 88].
Aptamers for complementing the use of antibodies
Some recent studies clearly show that the combined use of aptamers and antibodies can be more effective than the monotherapy approach by working in a synergistic or an additive manner. Most of these studies provide support for the clinical evaluation of novel synergistic combinations of immune checkpoint inhibitors and aptamer-based agents for cancer treatment. Ajona et al. [89], tested the anti-tumor efficacy of the combination of an anti-programmed cell death protein 1 (PD-1) mAb and an anti-C5a spiegelmer (a L-stereoisomer aptamer, highly stable because of its non-natural conformation) in preclinical lung cancer models. Interestingly, the aptamer, used to inhibit the signaling of complement C5a with its receptors, revealed only modest antitumor effects as monotherapy but in combination with the antibody, strongly influenced the immune response causing a significant reduction of tumor growth and metastases. Furthermore, Gefen et al. [90], constructed a trimeric version of the T cell immunoglobulin-3 (TIM-3) aptamer, selected by combining a protein- and cell-SELEX protocol, able to delay colon carcinoma cells-derived tumor growth, upon systemic administration to mice as single agent. The blockade of TIM-3 by the currently available TIM-3 antibodies had only little antitumor effect. Importantly, the aptamer was found to be more efficacious than the TIM-3 antibody even when injected at a dose 4.5-fold lower than the antibody. However, as demonstration of the great potential of combining aptamers and antibodies for dual blocking, the aptamer strongly synergized with the PD-1 antibody in prolonging the survival of tumor-bearing mice.
Programmed death-ligand 1 (PD-L1) expression is higher in TNBC than in other breast cancers, providing the rational for recently approved immunotherapy with anti-PD-L1 antibodies, in combination with PTX, for advanced PD-L1-positive TNBC [91]. Currently, enormous effort is devoted to identifying novel actionable biomarkers that may allow combination therapies with immune checkpoint blockade in TNBC. Recently, our group demonstrated the efficacy of a novel combination treatment with the previously validated PDGFRβ aptamer [37, 73, 84] and anti-PD-L1 mAbs in both human and murine in vitro settings, consisting of TNBC cells cultured as monolayer or co-cultured with lymphocytes [92]. Importantly, the PDGFRβ aptamer (intravenous administration) potentiated the efficacy of the anti-PD-L1 antibody in inhibiting tumor growth and lung metastasis formation in a syngeneic 4T1 TNBC mouse model by acting on both tumor cells and immune populations. Indeed, by analyzing TME, after the sacrifice of mice, we demonstrated that the combined blockade caused the depletion of FOXP3+ Treg cells and the increase of CD8+ T cells more consistently than single monotherapies.
Also, we proved that the combined treatment of cancer cells with the highly efficacious anti-EGFR CL4 2’fluoro-pyrimidines RNA aptamer and either an anti-HER2 (Erb-hcAb) antibody [93] or the immunomodulators anti-PD-L1 (10_12 mAb) [93] or anti-CTL4 (ipilimumab) mAbs [94] resulted in more efficient killing of cancer cells than single treatments. These findings provided the rational for generating new bispecific antibody-aptamer conjugates that combine the biological functions of both aptamer and antibody in a single molecule, resulting in increased anti-cancer activity [93, 94]. Specifically, the 5’ amino-terminated CL4 aptamer, conjugated with formylbenzamide formed a stable covalent hydrazone bond with the antibody conjugated with an aromatic hydrazine. The resulting new conjugates acquired increased specificity for double antigen-positive tumor cells, accordingly to the presence of two binding sites in the chimeric construct. When tested on tumor cells, immune cells and tumor and immune cell co-cultures, they inhibited the growth of tumor target cells more efficiently than the parental compounds and, in the case of the EGFR aptamer conjugation with immune-checkpoint mAbs modulators, induced the efficient activation of T cells against cancer cells. Although not yet tested in vivo due to the low purification yields of the final constructs, we believe the combination of aptamers and antibodies is a promising approach to potentiate each other’s strengths, overcoming the current therapeutic limitations of both aptamers and antibodies as single agents (Table 1). Indeed, aptamer-based immunoconjugates could have optimal biological features for therapeutic applications such as increased specificity for tumor cells, crosslinking of immune and cancer cells and improved pharmacokinetic and pharmacodynamic properties due to the combined advantages of the small-size aptamer, for increasing tumor penetration, with those of the antibody, which allows a longer half-life in circulation.
Accordingly, a previous study proved that the conjugation of a mAb to an aptamer allows overcoming the therapeutic limitations of aptamers, thus representing a novel approach for rapid, low-cost and high-throughput cancer therapy [53]. Indeed, linking an antibody to the anti-vascular endothelial growth factor (VEGF) pegaptanib aptamer extended the in vivo pharmacokinetic of the aptamer without affecting its binding affinity and, on the other hand, the aptamer part of the monospecific immune conjugate penetrated deep into the tumor while anti-VEGF antibody did not.
Conclusions
An area of intense research is dedicated to finding next generation strategies to efficiently target cancer cells without affecting healthy cells. Cell-SELEX-generated aptamers have many applications (i.e. research reagents, diagnostics, therapeutics and theranostics), thus giving this technology great potential in cancer medicine.
Targeted anti-cancer therapies have recently gained increasing attention in the anti-cancer drug development industry, as these therapies constitute the main branch of precision medicine. The target market for targeted cancer therapies includes drugs used as precision medicine for the treatment of malignant and benign tumors. Numerous cancer targeted therapies have been approved by the FDA to treat various types of cancer and have been commercialized, while many others are still being studied in both clinical trials and preclinical tests.
This market is highly competitive with continuous launches, approvals, and a robust pipeline of new biopharmaceutical products. Nevertheless, the majority of these products show high costs that risk limiting their development. In this view perspective, aptamers may represent a cost-effective alternative, but it is clear that, despite the promising properties, their commercial applications are still limited. Even though thirty years have passed since the first SELEX description and several aptamers have been selected and successful applied in both diagnostic and therapeutic modalities, no aptamers have so far been approved for cancer treatment. However, two aptamers are in clinical trials for cancer therapy: i) AS1411, a DNA quadruplex aptamer against nucleolin, an ubiquitous intranuclear and cytoplasmic phosphoprotein involved in the regulation of cell proliferation, often over-expressed on the surface of tumour cells; and ii) NOX-A12, a Spiegelmer that binds and neutralizes CXCL12 chemokine, which has a crucial role in cell growth and migration [95].
While it is true that there is some reticence about the introduction in the market of new molecules, and that moving from laboratory to clinic typically takes a long time and requires significant financial investment, it is also true that many limitations have kept aptamers from realizing the practical potential that they seemed to have when they were first developed. Major challenges for aptamers practical use cover their discovery process and the strategies for improving their binding affinities for diagnostic applications, and therapeutic efficacy, half-life and stability for patients’ treatment. Nevertheless, researchers worldwide are working hard to overcome those obstacles. Specialized partitioning methodology, e.g., capillary electrophoresis-based techniques make possible to select aptamers even by a single-round selection [96]. In addition, by integrating the SELEX technology with high-throughput sequencing technology allows identifying the best candidate aptamers in early selection cycles thus strongly reducing times and costs of the process [23]. Further, the types of chemical modifications for adapting aptamers to any desired applications is growing fast [97]. Efficacious approaches have been developed for engineering aptamers toward their use as diagnostics [98], in vivo imaging agents [99], therapeutics [100] and delivery agents [67, 97]. Notably, the global aptamers market was estimated at 32 million of dollars in 2018 and is expected to reach 100 million of dollars in 2025, with a compound annual growth rate (CAGR) of 15.5% [101].
Thus, it is possible to envisage that in the near future, in the era of combination therapy, we will fight cancer by attacking it from multiple flanks by complementing existing drugs with novel oligonucleotide aptamer-based therapeutics, overcoming drug resistance and increasing therapeutic efficacy.
Abbreviations
- BBB:
blood-brain barrier
- DOX:
doxorubicin
- EGFR:
epidermal growth factor receptor
- EpCAM:
epithelial cell adhesion molecule
- GBM:
glioblastoma
- HER2:
human epidermal growth factor receptor 2
- mAbs:
monoclonal antibodies
- NK:
natural killer
- NPs:
nanoparticles
- PD-1:
programmed cell death protein 1
- PDGFRβ:
platelet-derived growth factor receptor beta
- PD-L1:
programmed death-ligand 1
- PEG:
polyethylene glycol
- PTX:
paclitaxel
- SELEX:
systematic evolution of ligands by exponential enrichment
- TIM-3:
T cell immunoglobulin-3
- TME:
tumor microenvironment
- TNBC:
triple-negative breast cancer
- VEGF:
vascular endothelial growth factor
Declarations
Author contributions
Conceptualization: LC; writing-original draft preparation: LC; writing-review and editing: LA, SC, MF, LC. All the authors gave final approval of the version to be published.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
This work was supported by Fondazione AIRC per la Ricerca sul Cancro (IG 23052) to LC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Copyright
© The Author(s) 2021.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Falzone L, Salomone S, Libra M. Evolution of cancer pharmacological treatments at the turn of the third millennium. Front Pharmacol. 2018;9:1300. 10.3389/fphar.2018.01300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–26. 10.1038/nrc3599 [DOI] [PubMed] [Google Scholar]
- 4.Xu G, McLeod HL. Strategies for enzyme/prodrug cancer therapy. Clin Cancer Res. 2001;7:3314–24. [PubMed] [Google Scholar]
- 5.Cree IA, Charlton P. Molecular chess? Hallmarks of anti-cancer drug resistance. BMC Cancer. 2017;17:10. 10.1186/s12885-016-2999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Alterio C, Scala S, Sozzi G, Roz L, Bertolini G. Paradoxical effects of chemotherapy on tumor relapse and metastasis promotion. Semin Cancer Biol. 2020;60:351–61. 10.1016/j.semcancer.2019.08.019 [DOI] [PubMed] [Google Scholar]
- 7.Madden EC, Gorman AM, Logue SE, Samali A. Tumour cell secretome in chemoresistance and tumour recurrence. Trends Cancer. 2020;6:489–505. 10.1016/j.trecan.2020.02.020 [DOI] [PubMed] [Google Scholar]
- 8.Colton M, Cheadle EJ, Honeychurch J, Illidge TM. Reprogramming the tumour microenvironment by radiotherapy: implications for radiotherapy and immunotherapy combinations. Radiat Oncol. 2020;15:254. 10.1186/s13014-020-01678-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirata E, Sahai E. Tumor microenvironment and differential responses to therapy. Cold Spring Harb Perspect Med. 2017;7:a026781. 10.1101/cshperspect.a026781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudino TA. Targeted cancer therapy: the next generation of cancer treatment. Curr Drug Discov Technol. 2015;12:3–20. 10.2174/1570163812666150602144310 [DOI] [PubMed] [Google Scholar]
- 11.Imai K, Takaoka A. Comparing antibody and small-molecule therapies for cancer. Nat Rev Cancer. 2006;6:714–27. 10.1038/nrc1913 [DOI] [PubMed] [Google Scholar]
- 12.Aggarwal S. Targeted cancer therapies. Nat Rev Drug Discov. 2010;9:427–8. 10.1038/nrd3186 [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Sun Q. Novel targeted drugs approved by the NMPA and FDA in 2019. Signal Transduct Target Ther. 2020;5:65. 10.1038/s41392-020-0164-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seebacher NA, Stacy AE, Porter GM, Merlot AM. Clinical development of targeted and immune based anti-cancer therapies. J Exp Clin Cancer Res. 2019. 11;38:156. 10.1186/s13046-019-1094-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh DY, Bang YJ. HER2-targeted therapies-a role beyond breast cancer. Nat Rev Clin Oncol. 2020;17:33–48. 10.1038/s41571-019-0268-3 [DOI] [PubMed] [Google Scholar]
- 16.Doroshow JH, Simon RM. On the design of combination cancer therapy. Cell. 2017;171:1476–8. 10.1016/j.cell.2017.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayat Mokhtari R, Homayouni TS, Baluch N, Morgatskaya E, Kumar S, Das B, et al. Combination therapy in combating cancer. Oncotarget. 2017;8:38022–43. 10.18632/oncotarget.16723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016;3:674–90. 10.1038/nrclinonc.2016.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qazi MA, Vora P, Venugopal C, Sidhu SS, Moffat J, Swanton C, et al. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol. 2017;28:1448–56. 10.1093/annonc/mdx169 [DOI] [PubMed] [Google Scholar]
- 20.Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol Ther Nucleic Acids. 2014;3:e182. 10.1038/mtna.2014.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerchia L. Aptamers: promising tools for cancer diagnosis and therapy. Cancers (Basel). 2018;10:132. 10.3390/cancers10050132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16:181–202. 10.1038/nrd.2016.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blind M, Blank M. Aptamer selection technology and recent advances. Mol Ther Nucleic Acids. 2015;4:e223. 10.1038/mtna.2014.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinghorn AB, Fraser LA, Lang S, Shiu SCC, Tanner JA. Aptamer bioinformatics. Int J Mol Sci. 2017;18:2516. 10.3390/ijms18122516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- 26.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- 27.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–8. 10.1038/344467a0 [DOI] [PubMed] [Google Scholar]
- 28.Li L, Xu S, Yan H, Li X, Yazd HS, Li X, et al. Nucleic acid aptamers for molecular diagnostics and therapeutics: advances and perspectives. Angew Chem Int Ed Engl. 2020;60:2221–31. 10.1002/anie.202003563 [DOI] [PubMed] [Google Scholar]
- 29.Wang B. A new design for the fixed primer regions in an oligonucleotide library for SELEX aptamer screening. Front Chem. 2020;8:475. 10.3389/fchem.2020.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol. 2013;31:453–7. 10.1038/nbt.2556 [DOI] [PubMed] [Google Scholar]
- 31.Ozer A, Pagano JM, Lis JT. New technologies provide quantum changes in the scale, speed, and success of SELEX methods and aptamer characterization. Mol Ther Nucleic Acids. 2014;3:e183. 10.1038/mtna.2014.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Kuan CT, Mi J, Zhang X, Clary BM, Bigner DD, et al. Aptamers selected against the unglycosylated EGFRvIII ectodomain and delivered intracellularly reduce membrane-bound EGFRvIII and induce apoptosis. Biol Chem. 2009;390:137–44. 10.1515/BC.2009.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shangguan D, Bing T, Zhang N. Cell-SELEX: aptamer selection against whole cells. In: Tan W, Fang X, editors. Aptamers selected by cell-SELEX for theranostics. Berlin: Springer; 20. pp. 13–33. [Google Scholar]
- 34.Fechter P, Cruz Da Silva E, Mercier MC, Noulet F, Etienne-Seloum N, Guenot D, et al. RNA aptamers targeting integrin α5β1 as probes for cyto- and histo-fluorescence in glioblastoma. Mol Ther Nucleic Acids. 2019;17:63–77. 10.1016/j.omtn.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, et al. Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem. 2001;276:48644–54. 10.1074/jbc.M104651200 [DOI] [PubMed] [Google Scholar]
- 36.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem. 2007;79:4900–7. 10.1021/ac070189y [DOI] [PubMed] [Google Scholar]
- 37.Camorani S, Esposito CL, Rienzo A, Catuogno S, Iaboni M, Condorelli G, et al. Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRβ aptamer. Mol Ther. 2014;22:828–41. 10.1038/mt.2013.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sefah K, Tang ZW, Shangguan DH, Chen H, Lopez-Colon D, Li Y, et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia. 2009;23:235–44. 10.1038/leu.2008.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, et al. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3:991–1001. 10.1002/cmdc.200800030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103:11838–43. 10.1073/pnas.0602615103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esposito CL, Passaro D, Longobardo I, Condorelli G, Marotta P, Affuso A, et al. A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS One. 2011;6:e24071. 10.1371/journal.pone.0024071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Zhang W, Liu L, Zhu Z, Ouyang G, An Y, et al. In vitro selection of DNA aptamers for metastatic breast cancer cell recognition and tissue imaging. Anal Chem. 2014;86:6596–603. 10.1021/ac501205q [DOI] [PubMed] [Google Scholar]
- 43.Liu M, Wang Z, Tan T, Chen Z, Mou X, Yu X, et al. An aptamer-based probe for molecular subtyping of breast cancer. Theranostics. 2018;8:5772–83. 10.7150/thno.28949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Camorani S, Granata I, Collina F, Leonetti F, Cantile M, Botti G, et al. Novel aptamers selected on living cells for specific recognition of triple-negative breast cancer. iScience. 2020;23:100979. 10.1016/j.isci.2020.100979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Souza AG, Marangoni K, Fujimura PT, Alves PT, Silva MJ, Bastos VA, et al. 3D cell-SELEX: development of RNA aptamers as molecular probes for PC-3 tumor cell line. Exp Cell Res. 2016;341:147–56. 10.1016/j.yexcr.2016.01.015 [DOI] [PubMed] [Google Scholar]
- 46.Zhong W, Pu Y, Tan W, Liu J, Liao J, Liu B, et al. Identification and application of an aptamer targeting papillary thyroid carcinoma using tissue-SELEX. Anal Chem. 2019;91:8289–97. 10.1021/acs.analchem.9b01000 [DOI] [PubMed] [Google Scholar]
- 47.Sola M, Menon AP, Moreno B, Meraviglia-Crivelli D, Soldevilla MM, Cartón-García F, et al. Aptamers against live targets: is in vivo SELEX finally coming to the edge? Mol Ther Nucleic Acids. 2020;21:192–204. 10.1016/j.omtn.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, et al. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol. 2010;6:22–4. 10.1038/nchembio.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng C, Chen YH, Lennox KA, Behlke MA, Davidson BL. In vivo SELEX for identification of brain-penetrating aptamers. Mol Ther Nucleic Acids. 2013;2:e67. 10.1038/mtna.2012.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gelinas AD, Davies DR, Janjic N. Embracing proteins: structural themes in aptamer-protein complexes. Curr Opin Struct Biol. 2016;36:122–32. 10.1016/j.sbi.2016.01.009 [DOI] [PubMed] [Google Scholar]
- 51.Diafa S, Hollenstein M. Generation of aptamers with an expanded chemical repertoire. Molecules. 2015;20:16643–71. 10.3390/molecules200916643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Da Pieve C, Blackshaw E, Missailidis S, Perkins AC. PEGylation and biodistribution of an anti-MUC1 aptamer in MCF-7 tumor-bearing mice. Bioconjug Chem. 2012;23:1377–81. 10.1021/bc300128r [DOI] [PubMed] [Google Scholar]
- 53.Heo K, Min SW, Sung HJ, Kim HG, Kim HJ, Kim YH, et al. An aptamer-antibody complex (oligobody) as a novel delivery platform for targeted cancer therapies. J Control Release. 2016;229:1–9. 10.1016/j.jconrel.2016.03.006 [DOI] [PubMed] [Google Scholar]
- 54.Zhou F, Fu T, Huang Q, Kuai H, Mo L, Liu H, et al. Hypoxia-activated PEGylated conditional aptamer/ antibody for cancer imaging with improved specificity. J Am Chem Soc. 2019;141:18421–7. 10.1021/jacs.9b05063 [DOI] [PubMed] [Google Scholar]
- 55.Dassie JP, Giangrande PH. Current progress on aptamer-targeted oligonucleotide therapeutics. Ther Deliv. 2013;4:1527–46. 10.4155/tde.13.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Camorani S, Crescenzi E, Fedele M, Cerchia L. Oligonucleotide aptamers against tyrosine kinase receptors: prospect for anticancer applications. Biochim Biophys Acta Rev Cancer. 2018;1869:263–77. 10.1016/j.bbcan.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 57.Cerchia L, Esposito CL, Camorani S, Rienzo A, Stasio L, Insabato L, et al. Targeting Axl with an high-affinity inhibitory aptamer. Mol Ther. 2012;20:2291–303. 10.1038/mt.2012.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camorani S, Crescenzi E, Colecchia D, Carpentieri A, Amoresano A, Fedele M, et al. Aptamer targeting EGFRvIII mutant hampers its constitutive autophosphorylation and affects migration, invasion and proliferation of glioblastoma cells. Oncotarget. 2015;6:37570–87. 10.18632/oncotarget.6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Camorani S, Crescenzi E, Gramanzini M, Fedele M, Zannetti A, Cerchia L. Aptamer-mediated impairment of EGFR-integrin αvβ3 complex inhibits vasculogenic mimicry and growth of triple-negative breast cancers. Sci Rep. 2017;7:46659. 10.1038/srep46659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueki R, Sando S. A DNA aptamer to c-Met inhibits cancer cell migration. Chem Commun (Camb). 2014;50:13131–4. 10.1039/c4cc06016d [DOI] [PubMed] [Google Scholar]
- 61.Camorani S, Fedele M, Zannetti A, Cerchia L. TNBC challenge: oligonucleotide aptamers for new imaging and therapy modalities. Pharmaceuticals (Basel). 2018;11:123. 10.3390/ph11040123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, et al. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest. 2008;118:376–86. 10.1172/JCI33365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueki R, Ueki A, Kanda N, Sando S. Oligonucleotide-based mimetics of hepatocyte growth factor. Angew Chem Int Ed Engl. 2016;55:579–82. 10.1002/anie.201508572 [DOI] [PubMed] [Google Scholar]
- 64.Mahlknecht G, Maron R, Mancini M, Schechter B, Sela M, Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc Natl Acad Sci U S A. 2013;110:8170–5. 10.1073/pnas.1302594110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang S, Wen J, Li H, Xu L, Liu Y, Zhao N, et al. Aptamer-engineered natural killer cells for cell-specific adaptive immunotherapy. Small. 2019;15:e1900903. 10.1002/smll.201900903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoon S, Rossi JJ. Aptamers: uptake mechanisms and intracellular applications. Adv Drug Deliv Rev. 2018;134:22–35. 10.1016/j.addr.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Powell Gray B, Kelly L, Ahrens DP, Barry AP, Kratschmer C, Levy M, et al. Tunable cytotoxic aptamer-drug conjugates for the treatment of prostate cancer. Proc Natl Acad Sci U S A. 2018;115:4761–6. 10.1073/pnas.1717705115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chu TC, Marks JW, 3rd, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, et al. Aptamer: toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–92. 10.1158/0008-5472.CAN-05-4583 [DOI] [PubMed] [Google Scholar]
- 69.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–33. 10.1038/nature07758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Porciani D, Cardwell LN, Tawiah KD, Alam KK, Lange MJ, Daniels MA, et al. Modular cell-internalizing aptamer nanostructure enables targeted delivery of large functional RNAs in cancer cell lines. Nat Commun. 2018;9:2283. 10.1038/s41467-018-04691-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deeken JF, Löscher W. The blood-brain barrier and cancer: transporters, treatment, and Trojan horses. Clin Cancer Res. 2007;13:1663–74. 10.1158/1078-0432.CCR-06-2854 [DOI] [PubMed] [Google Scholar]
- 72.Macdonald J, Denoyer D, Henri J, Jamieson A, Burvenich IJG, Pouliot N, et al. Bifunctional aptamer-doxorubicin conjugate crosses the blood-brain barrier and selectively delivers its payload to EpCAM-positive tumor cells. Nucleic Acid Ther. 2020;30:117–28. 10.1089/nat.2019.0807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Camorani S, Hill BS, Fontanella R, Greco A, Gramanzini M, Auletta L, et al. Inhibition of bone marrow-derived mesenchymal stem cells homing towards triple-negative breast cancer microenvironment using an anti-PDGFRβ aptamer. Theranostics. 2017;7:3595–607. 10.7150/thno.18974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Monaco I, Camorani S, Colecchia D, Locatelli E, Calandro P, Oudin A, et al. Aptamer functionalization of nanosystems for glioblastoma targeting through the blood-brain barrier. J Med Chem. 2017;60:4510–6. 10.1021/acs.jmedchem.7b00527 [DOI] [PubMed] [Google Scholar]
- 75.Prusty DK, Adam V, Zadegan RM, Irsen S, Famulok M. Supramolecular aptamer nano-constructs for receptor-mediated targeting and light-triggered release of chemotherapeutics into cancer cells. Nat Commun. 2018;9:535. 10.1038/s41467-018-02929-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo S, Vieweger M, Zhang K, Yin H, Wang H, Li X, et al. Ultra-thermostable RNA nanoparticles for solubilizing and high-yield loading of paclitaxel for breast cancer therapy. Nat Commun. 2020;11:972. 10.1038/s41467-020-14780-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belyanina IV, Zamay TN, Zamay GS, Zamay SS, Kolovskaya OS, Ivanchenko TI, et al. In vivo cancer cells elimination guided by aptamer-functionalized gold-coated magnetic nanoparticles and controlled with low frequency alternating magnetic field. Theranostics. 2017;7:3326–37. 10.7150/thno.17089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feng X, Liu J, Xu W, Li G, Ding J. Tackling autoimmunity with nanomedicines. Nanomedicine (Lond). 2020;15:1585–97. 10.2217/nnm-2020-0102 [DOI] [PubMed] [Google Scholar]
- 79.Feng X, Xu W, Li Z, Song W, Ding J, Chen X. Immunomodulatory nanosystems. Adv Sci (Weinh). 2019;6:1900101. 10.1002/advs.201900101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pastor F. Aptamers: A new technological platform in cancer immunotherapy. Pharmaceuticals (Basel). 2016;9:64. 10.3390/ph9040064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ravichandran G, Rengan AK. Aptamer-mediated nanotheranostics for cancer treatment: a review. ACS Appl Nano Mater. 2020;3:9542–59. 10.1021/acsanm.0c01785 [DOI] [Google Scholar]
- 82.Bai C, Gao S, Hu S, Liu X, Li H, Dong J, et al. Self-assembled multivalent aptamer nanoparticles with potential CAR-like characteristics could activate T cells and inhibit melanoma growth. Mol Ther Oncolytics. 2020;17:9–20. 10.1016/j.omto.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bouvier-Müller A, Ducongé F. Application of aptamers for in vivo molecular imaging and theranostics. Adv Drug Deliv Rev. 2018;134:94–106. 10.1016/j.addr.2018.08.004 [DOI] [PubMed] [Google Scholar]
- 84.Camorani S, Hill BS, Collina F, Gargiulo S, Napolitano M, Cantile M, et al. Targeted imaging and inhibition of triple-negative breast cancer metastases by a PDGFRβ aptamer. Theranostics. 2018;8:5178–99. 10.7150/thno.27798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim MW, Jeong HY, Kang SJ, Jeong IH, Choi MJ, You YM, et al. Anti-EGF receptor aptamer-guided co-delivery of anti-cancer siRNAs and quantum dots for theranostics of triple-negative breast cancer. Theranostics. 2019;9:837–52. 10.7150/thno.30228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tan J, Yang N, Zhong L, Tan J, Hu Z, Zhao Q, et al. A new theranostic system based on endoglin aptamer conjugated fluorescent silica nanoparticles. Theranostics. 2017;7:4862–76. 10.7150/thno.19101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiang D, Zheng C, Zhou SF, Qiao S, Tran PH, Pu C, et al. Superior performance of aptamer in tumor penetration over antibody: implication of aptamer-based theranostics in solid tumors. Theranostics. 2015;5:1083–97. 10.7150/thno.11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Melancon MP, Zhou M, Zhang R, Xiong C, Allen P, Wen X, et al. Selective uptake and imaging of aptamer- and antibody-conjugated hollow nanospheres targeted to epidermal growth factor receptors overexpressed in head and neck cancer. ACS Nano. 2014;8:4530–8. 10.1021/nn406632u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ajona D, Ortiz-Espinosa S, Moreno H, Lozano T, Pajares MJ, Agorreta J, et al. A combined PD-1/ C5a blockade synergistically protects against lung cancer growth and metastasis. Cancer Discov. 2017;7:694–703. 10.1158/2159-8290.CD-16-1184 [DOI] [PubMed] [Google Scholar]
- 90.Gefen T, Castro I, Muharemagic D, Puplampu-Dove Y, Patel S, Gilboa E. A TIM-3 oligonucleotide aptamer enhances T cell functions and potentiates tumor immunity in mice. Mol Ther. 2017;25:2280–8. 10.1016/j.ymthe.2017.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cortés J, André F, Gonçalves A, Kümmel S, Martín M, Schmid P, et al. IMpassion132 phase III trial: atezolizumab and chemotherapy in early relapsing metastatic triple-negative breast cancer. Future Oncol. 2019;15:1951–61. 10.2217/fon-2019-0059 [DOI] [PubMed] [Google Scholar]
- 92.Camorani S, Passariello M, Agnello L, Esposito S, Collina F, Cantile M, et al. Aptamer targeted therapy potentiates immune checkpoint blockade in triple-negative breast cancer. J Exp Clin Cancer Res. 2020;39:180. 10.1186/s13046-020-01694-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Passariello M, Camorani S, Vetrei C, Cerchia L, De Lorenzo C. Novel human bispecific aptamer-antibody conjugates for efficient cancer cell killing. Cancers (Basel). 2019;11:1268. 10.3390/cancers11091268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Passariello M, Camorani S, Vetrei C, Ricci S, Cerchia L, De Lorenzo C. Ipilimumab and its derived EGFR aptamer-based conjugate induce efficient NK cell activation against cancer cells. Cancers (Basel). 2020;12:331. 10.3390/cancers12020331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hori SI, Herrera A, Rossi JJ, Zhou J. Current advances in aptamers for cancer diagnosis and therapy. Cancers (Basel). 2018;10:9. 10.3390/cancers10010009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hirose K, Tsuchida M, Asakura H, Wakui K, Yoshimoto K, Iida K, et al. A single-round selection of selective DNA aptamers for mammalian cells by polymer-enhanced capillary transient isotachophoresis. Analyst. 2017;142:4030–8. 10.1039/c7an00909g [DOI] [PubMed] [Google Scholar]
- 97.Odeh F, Nsairat H, Alshaer W, Ismail MA, Esawi E, Qaqish B, et al. Aptamers chemistry: chemical modifications and conjugation strategies. Molecules. 2019;25:3. 10.3390/molecules25010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Elskens JP, Elskens JM, Madder A. Chemical modification of aptamers for increased binding affinity in diagnostic applications: current status and future prospects. Int J Mol Sci. 2020;21:4522. 10.3390/ijms21124522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dougherty CA, Cai W, Hong H. Applications of aptamers in targeted imaging: state of the art. Curr Top Med Chem. 2015;15:1138–52. 10.2174/1568026615666150413153400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ni S, Zhuo Z, Pan Y, Yu Y, Li F, Liu J, et al. Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl Mater Interfaces. 2020;[Epub ahead of print]. [DOI] [PubMed]
- 101.Global aptamer market research report 2019 [Internet]. Selbyville: Market Study Report; c2019. [cited 2020 Dec 09]. Available from: https://www.marketstudyreport.com/reports/global-aptamer-market-research-report-2019 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.