Abstract
A novel gene, designated ohb1, which encodes the oxygen-sensitive and biotin-, ATP-, thiamin-, pyridoxal phosphate-, and metal-ion-independent, reversible 4-hydroxybenzoate decarboxylase (4-HOB-DC) from the obligate anaerobe Clostridium hydroxybenzoicum JW/Z-1T was sequenced (GenBank accession no. AF128880) and expressed. The 1,440-bp open reading frame (ORF) (ohb1) encodes 480 amino acids. Major properties of the heterologous enzyme (Ohb1) expressed in Escherichia coli DH5α were the same as those described for the native 4-HOB-DC (Z. He and J. Wiegel, J. Bacteriol. 178:3539–3543, 1996). The deduced amino acid sequence shows up to 57% identity and up to 74% similarity to hypothetical proteins deduced from ORFs in genomes from bacteria and archaea, suggesting a possible novel gene family.
Decarboxylation and carboxylation of aromatic compounds have been proposed to play important initial roles in the anaerobic degradation of (hydroxy)arylic acids (e.g., hydroxybenzoic acids) and phenolic compounds in methanogenic environments (2, 12, 19, 24, 33–35). Zhang and Wiegel (33) proposed a pathway comprised of the sequential actions of at least six bacteria for the mineralization of 2,4-dichlorophenol. The recently isolated Clostridium hydroxybenzoicum (34, 36) was proposed to transform the dechlorination product phenol to hydroxybenzoate. Subsequently, two reversible decarboxylases with a narrow substrate spectrum, a 4-hydroxybenzoate decarboxylase (4-HOB-DC) and a 3,4-dihydroxybenzoate decarboxylase, were purified and characterized (15, 16). Whereas many carboxylases and decarboxylases require the involvement of biotin, ATP, or thiamin pyrophosphate or the addition of a metal ion (27), the two enzymes from C. hydroxybenzoicum do not require such additions for either the decarboxylation or carboxylation reaction. Although 4-HOB-DC and 3,4-dihydroxybenzoate decarboxylase activities have been found in a few other organisms (references 11a–13, 19, and 25, and unpublished results), the distribution among microorganisms has not been well documented. To our knowledge, none of the enzymes has been purified nor the encoding genes cloned. We report here the characterization and expression (in Escherichia coli) of the gene encoding the oxygen-sensitive, cofactor-independent, reversible 4-HOB-DC from C. hydroxybenzoicum JW/Z-1T (ATCC 51151 and DSM 7310) (34). The host strain E. coli DH5α (obtained from S. Kushner, University of Georgia) was cultivated at 37°C in Luria broth (3, 28) and, when required, in the presence of ampicillin (50 μg/ml; Sigma, St. Louis, Mo.), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (30 μg/ml; Promega), and isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM; Inalco Pharmaceuticals, San Luis Obispo, Calif.).
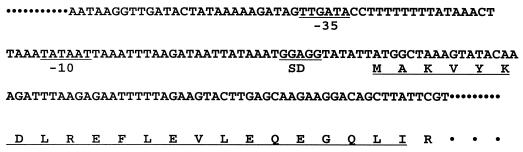
The 4-HOB-DC-encoding gene ohb1 from C. hydroxybenzoicum.
Standard molecular techniques and procedures (reference 28) were used to clone and sequence the gene. The N-terminal (M[A]KVYRDLREFLEVLXQXGXLI) (17) and three internal (SDLYDHLYVPAGSEVVLEGHIIPR, IVIVVDEFVDPFNLEQVMWALTTR, and YSVVTNVHGSWQNHALMLGLDK) (obtained from a pure enzyme as judged by sodium dodecyl sulfate gel electrophoresis; Wistar Protein Microsequencing Facility, Philadelphia, Pa.) sequences were used to design the degenerate PCR primers (underlined sections) Inter2 (5′-CKNGTNGTNARNGCCCACAT-3′) and Inter3 (5′-AAYGTNCAYGGNWSNTGGCA-3′) (K = G, T; N = A, C, G, T; R = G, A; Y = T, C; W = A, T; S = G, C), which were used in conjunction with a PCR DIG-labeling kit (Boehringer Mannheim) to amplify and label a 1-kb DNA fragment from genomic DNA. This fragment hybridized to a 2.3-kb fragment of HindIII-digested genomic DNA in Southern hybridization. Subsequently, a genomic mini-library was constructed in pUC18. Colony hybridization yielded two positive colonies from which two recombinant plasmids containing the same 2.3-kb DNA fragment (pJBH-1 and pJBH-2) were obtained and sequenced at the Molecular Genetics Instrumentation Facility (University of Georgia).
Using the Genetics Computer Group sequence analysis software package (University of Wisconsin, Madison), the computer analysis of the nucleotide sequence revealed one 1,440-bp open reading frame (ORF) with a presumptive promoter region (Fig. 1). As deduced from the DNA sequence data, the protein is composed of 480 amino acids. The ATG initiation codon is preceded directly by a putative ribosome-binding site (Shine-Dalgarno) GGAGG, with a spacer of 6 bp (Fig. 1). A putative −10 promoter region, TATAAT, and a −35 promoter region, TTGATA, were found 22 and 49 bp, respectively, upstream of the ribosomal binding site. These sequences are similar to that of a typical E. coli promoter. The G+C content of the gene is 39.2% compared with 35.6% of genomic DNA. AT-containing isocodons were preferentially used.
FIG. 1.
Nucleotide sequence of the promoter region of the gene ohb1 encoding 4-HOB-DC of C. hydroxybenzoicum JW/Z-1T. The underlined amino acids indicate the regions which had been previously sequenced from the purified enzyme. SD, Shine-Dalgarno sequence.
Expression of the gene ohb1 and characterization of the recombinant 4-HOB-DC Ohb1.
The heterologously expressed 4-HOB-DC reached specific activities of 1.12 and 0.09 μmol of 4-HOB min−1 · mg−1 decarboxylated in anaerobically prepared (34) crude extracts of anaerobically and aerobically grown E. coli DH5α, respectively, and 0.42 μmol of 3,4-dihydroxybenzoate decarboxylated min−1 · mg−1 in anaerobically grown cells. The ratio of these activities for the two substrates (measured as described before [34]) correlates well with the previously reported ratio (kcat of 3,300 versus 1,100) observed with the purified 4-HOB-DC (15). Strain DH5α, with or without harboring the parental plasmid pUC18, exhibited no 4-HOB-DC activity under either anaerobic or aerobic conditions after growth in the presence or absence of IPTG and/or 4-hydroxybenzoate. The orientation of the ohb1-containing chromosomal DNA fragment in the vector did not affect the level of the enzyme activity, suggesting that the gene was expressed in E. coli by its own promoter. In contrast to C. hydroxybenzoicum, in E. coli, the enzyme was expressed constitutively.
We concluded that the cloned gene encodes the previously purified 4-OHB-DC because all three internal sequences obtained from the protein were found to be identical with the corresponding regions in the deduced amino acid sequence (15), and the original N-terminal sequence shows only two minor differences (the start is M and A instead of M/A and the sixth amino acid changed from R to K). The recombinant enzyme in E. coli did not show properties different from those of the native enzyme. For both we observed that (i) they were oxygen sensitive; (ii) 2 mM metal ions, including 2 mM Na+ and K+, 2 mM NH4+, and cofactors such as 5 mM ATP, 0.5 mM biotin, and 0.1 mM thiamin diphosphate, had no effect on 4-HOB-DC activity; (iii) the addition of 2 mM NH4+ slightly decreased only the rate of decarboxylation of 3,4-dihydroxybenzoate; and (iv) 2 mM Zn2+ inactivated both decarboxylation activities. Furthermore, the calculated molecular mass of 53.9 kDa from the deduced amino acid sequence is similar to the sodium dodecyl sulfate-gel electrophoretic value of 57 kDa found for the parental subunit (15).
Homology to other proteins.
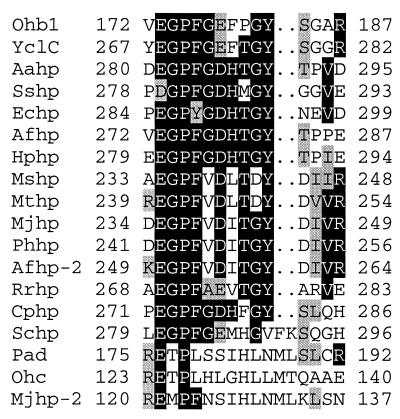
Using the deduced amino acid sequence of Ohb1 and the FASTA and BLAST search programs, a database search identified many homologous hypothetical protein sequences (Table 1), with the hypothetical protein YclC from Bacillus subtilis exhibiting the greatest identity (57.7%) and similarity (74.3%). Eighteen sequences exhibited similarities above 40%. The alignment revealed eight motifs of conserved regions, termed motif A (amino acid numbers in the Ohb1 sequence, 129 to 131, 138, and 177 with an arginine in position 168 in all but two proteins), B (201 to 222), C (274 to 284), D (383 to 397, 400), E (403 to 407), F (413 to 417), G(426 to 429), and H (437 to 443). The only proteins with a known enzymatic activity and some homology to Ohb1 were two arylic acid decarboxylases: Pad (phenyl acrylic acid decarboxylase) from Saccharomyces cerevisiae (8) and Ohd (3-octaprenyl-4-hydroxybenzoate carboxy-lyase) from E. coli (26). However, they exhibit the lowest similarity and identity (Table 1), contain only two of the eight identified motifs—motifs A (not shown) and C (Fig. 2)—and were significantly smaller (190 and 184 amino acids, respectively) than the 480 amino acids for Ohb1. A few arylic acid decarboxylases acting on other substrates have been sequenced (6–8, 26, 32). Although the sequences of the three decarboxylases from Lactobacillus and from Bacillus sp. showed high homologies among themselves, they exhibited only identities below 15% to the Ohb1 sequence. Two decarboxylases from S. cerevisiae (8), the diphosphomalonate decarboxylase and uroporphyrinogen decarboxylase, did not show any identity with the Ohb1 sequence. Because the sequence E-X-P in motif C (Fig. 2) is conserved in all of the listed protein sequences except in the two non-arylic acid decarboxylases, we speculate that this sequence E-X-P could play a role in the binding or catalysis of (hydroxy) arylic acids.
TABLE 1.
Similarity among amino acid sequences of 4-HOB-DC (Ohb1) from C. hydroxybenzoicum JW/Z-1 and deduced homologous proteins from other microorganisms
| Protein (microorganism)a | Accession no. | % Similarity | %Identity |
|---|---|---|---|
| YclC (hypothetical protein Bacillus subtilis) | D50453_66 | 74.3 | 57.7 |
| Sshp (Synechocystis sp PCC6803) | D90901_54 | 55.9 | 32.2 |
| Aahp (Aquifex aeolicus)A,T | AB000747_13 | 54.3 | 32.7 |
| Rrhp (Rhodospirillum rubrum) | U65510_2 | 53.5 | 31.0 |
| Phhp (Pyrococcus horikoshii)A,HT | AE000004_96 | 53.4 | 31.7 |
| Echp (Escherichia coli) | AE00459_13 | 53.0 | 29.7 |
| Afhp (Archaeoglobus fulgidus)A,T | AE001091_1 | 52.9 | 32.0 |
| Mjhp (Methanococcus jannaschii)A,T | D64441 | 52.8 | 27.5 |
| Mthp (Methanobacterium thermoautotrophicum)A,T | AE000902_5 | 52.6 | 27.8 |
| Afhp-2 (Archaeoglobus fulgidus)A,T | AE000989_12 | 52.1 | 25.1 |
| Mshp (Methanobrevibacter smithii)A | P22349 | 51.1 | 25.9 |
| Schp (Saccharomyces cerevisiae)E | S33751 | 50.3 | 25.2 |
| Hphp (Helicobacter pylori) | AE000555_16 | 48.4 | 25.2 |
| Mjhp-2 (Methanococcus jannaschii)A,T | Q57566 | 47.3 | 17.6 |
| Cphp (Chlamydia psittaci) | U88070_6 | 45.6 | 23.0 |
| Pad (Saccharomyces cerevisiae)E | P33751 | 41.9 | 17.2 |
| Ohc (Escherichia coli) | P09550 | 40.7 | 20.1 |
A = archaea; T = extreme thermophile (Topt ≥ 65°C); HT = hyperthermophile (Topt ≥ 85°C); E = eukaryote.
FIG. 2.
Motif C as an example of the regions with conserved amino acid sequences. White letters on a black background represent identical amino acids in more than 50% of the sequences, and black letters on a grey background represent conservative substitutions. Dots indicate gaps introduced to maximize the alignment (using values for gap weight of 5 and gap extension of 12). The numbers refer to the first and last positions of the motif in the corresponding sequence. Ohb1, 4-HOB-DC from C. hydroxybenzoium strain JW/Z-1T; Ohc, 3-octaprenyl-4-hydroxybenzoate carboxylase from E. coli (26); Pad, phenylacrylic acid decarboxylase from Saccharomyces cerevisiae (9). Designations at left are for hypothetical proteins from 12 different microorganisms as follows: Mjhp and Mjhp-2 from Methanococcus jannaschii (5), Mshp from Methanobrevibacter smithii (14), Mthp from Methanobacterium thermoautotrophicum (29), Phhp from Pyrococcus horikoshii (21), Afhp and Afhp-2 from Archaeoglobus fulgidus (23), Rrhp from Rhodospirillum rubrum (22), Yclc from Bacillus subtilis (31), Aahp from Aquifex aeolicus (10), Sshp from Synechocystis sp. strain PCC6803 (20); Echp from E. coli (1, 4), Hphp from Helicobacter pylori (30), Cphp from Chlamydia psittaci (18), Schp from Saccharomyces cerevisiae (11). Alignment was done with the PILEUP program, University of Wisconsin Genetics Computer Group.
So far, all characterized hydroxy arylic acid decarboxylases show very distinct, narrow substrate patterns, which may explain the lack of or low homology observed between the published sequences of arylic acid decarboxylases. It will be interesting to learn whether the identified homologous ORFs encode reversible decarboxylases independent of ATP, biotin, and thiamin and thus, as hypothesized, belong to a novel gene family of arylic acid decarboxylases. Such a gene could have been present before the separation of bacteria and archaea because the homologous gene sequences are observed in species of both. This includes hyperthermophiles, which are regarded by many as the closest relatives among the presently known microorganisms to the hypothetical common ancestor. Thus, these genes could represent an ancient type of carboxylase, which was substituted during later evolution by more efficient biotin- or thiamin-dependent and ATP-utilizing carboxylases (27).
Nucleotide sequence accession number.
The nucleotide sequence of the 1,480-bp ORF of ohb1 has been deposited in GenBank under accession no. AF128880.
Acknowledgments
We thank Cara Runsick-Mitchell and Delina Lyon for help in preparing the manuscript.
REFERENCES
- 1.Bailey M J, Koronakis V, Schmoll T, Hughes C. Escherichia coli HlyT protein, a transcriptional activator of haemolysin synthesis and secretion, is encoded by the rfaH(sfrB) locus required for expression of sex factor and lipopolysaccharide genes. Mol Microbiol. 1992;6:1003–1012. doi: 10.1111/j.1365-2958.1992.tb02166.x. [DOI] [PubMed] [Google Scholar]
- 2.Berry D F, Fransic A J, Bollag J M. Microbial metabolism of homocyclic and heterocyclic aromatic compounds under anaerobic conditions. Microbiol Rev. 1987;51:43–59. doi: 10.1128/mr.51.1.43-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bethesda Research Laboratories. BRL pUC host: E. coli DH5αTM competent cells. Bethesda Res Lab Focus. 1986;8:9–12. [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 6.Cavin J-F, Barthelmebs L, Diviès C. Molecular characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum: gene cloning, transcriptional analysis, overexpression in Escherichia coli, purification, and characterization. Appl Environ Microbiol. 1997;63:1939–1944. doi: 10.1128/aem.63.5.1939-1944.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavin J-F, Dartois V, Diviès C. Gene cloning, transcriptional analysis, purification, and characterization of phenolic acid decarboxylase from Bacillus subtilis. Appl Environ Microbiol. 1998;64:1466–1471. doi: 10.1128/aem.64.4.1466-1471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clausen M, Lamb C J, Megnet R, Doerner P W. PAD1 encodes phenylacrylic acid decarboxylase which confers resistance to cinnamic acid in Saccharomyces cerevisiae. Gene. 1994;142:107–112. doi: 10.1016/0378-1119(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 9.Daniels D L, Plunkett III G, Borland V, Blattner F R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992;257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 10.Dechert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich, F. S., J. Mulligan, E. Allen, R. Araujo, E. Aviles, A. Berno, J. Carpenter, E. Chen, J. M. Cherry, E. Chung, M. Duncan, S. Hunicke-Smith, R. Hyman, C. Komp, D. Kashkari, H. Lew, D. Lin, D. Mosedale, K. Nakahara, A. Namath, P. Oefner, C. Oh, F. X. Petel, D. Roberts, S. Schramm, M. Schroeder, T. Shogren, N. Shroff, A. Winant, M. Yelton, D. Botstein, and R. W. Davis. 1995. Direct submission to the EMBL Data Library. Accession no. P35751.
- 11a.Drake, H. Personal communication.
- 12.Gallert V, Winter J. Comparison of 4-hydroxybenzoate decarboxylase and phenol carboxylase activities in crude extracts in a defined, 4-hydroxybenzoate and phenol-degrading anaerobic consortium. Appl Microbiol Biotechnol. 1992;37:119–124. [Google Scholar]
- 13.Grant D J W, Patel J C. The non-oxidative decarboxylation of p-hydroxybenzoic acid, gentisic acid, protocatechuic acid and gallic acid by Klebsiella aerogenes (Aerobacter aerogenes) Antonie Leeuwenhoek. 1969;35:325–341. doi: 10.1007/BF02219153. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton P T, Reeve J N. Structure of genes and an insertion element in the methane producing archaebacterium Methanobrevibacter smithii. Mol Gen Genet. 1985;200:47–59. doi: 10.1007/BF00383311. [DOI] [PubMed] [Google Scholar]
- 15.He Z, Wiegel J. Purification and characterization of an oxygen-sensitive reversible 4-hydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. Eur J Biochem. 1995;229:77–82. doi: 10.1111/j.1432-1033.1995.tb20440.x. [DOI] [PubMed] [Google Scholar]
- 16.He Z, Wiegel J. Purification and characterization of an oxygen-sensitive, reversible 3,4-dihydroxybenzoate decarboxylase from Clostridium hydroxybenzoicum. J Bacteriol. 1996;178:3539–3543. doi: 10.1128/jb.178.12.3539-3543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Z. Reversible hydroxybenzoate decarboxylases from the anaerobic bacterium Clostridium hydroxybenzoicum. Ph.D. dissertation. Athens: University of Georgia; 1996. [Google Scholar]
- 18.Hsia R C, Pannekoek Y, Ingerowski R, Bavoil P M. Type III secretion genes identify a putative virulence locus of Chlamydia psittaci Mol. Microbiol. 1997;25:351–359. doi: 10.1046/j.1365-2958.1997.4701834.x. [DOI] [PubMed] [Google Scholar]
- 19.Hsu T, Lux M F, Drake H L. Expression of an aromatic-dependent decarboxylase which provides growth-essential CO2 equivalents for the acetogenic (Wood) pathway of Clostridium thermoaceticum. J Bacteriol. 1990;172:5091–5907. doi: 10.1128/jb.172.10.5901-5907.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain pcc6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 21.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, T. Yoshizawa, Y. Nakamura, F. T. Robb, K. Horikoshi, Y. Masuchi, H. Shizuya, and H. Kikuchi. Unpublished data.
- 22.Kerby R L, Hong S S, Ensign S A, Coppoc L J, Ludden D W, Roberts G P. Genetic and physiological characterization of the Rhodospirillum rubrum carbon monoxide dehydrogenase system. J Bacteriol. 1992;174:5284–5294. doi: 10.1128/jb.174.16.5284-5294.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn E P, Suflita J M, Rivera M D, Young L Y. Influence of alternate electron acceptors on the metabolic fate of hydroxybenzoate isomers in anoxic aquifer slurries. Appl Environ Microbiol. 1989;55:590–598. doi: 10.1128/aem.55.3.590-598.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin A K. The origin of urinary aromatic compounds excreted by ruminants. 3. The metabolism of phenolic compounds to simple phenols. Br J Nutr. 1982;48:487–507. doi: 10.1079/bjn19820135. [DOI] [PubMed] [Google Scholar]
- 26.Nonet M L, Marvel C C, Tolan D R. The hisT-purF region of the Escherichia coli K-12 chromosome. Identification of additional genes of the hisT and purF operons. J Biol Chem. 1987;262:12209–12217. [PubMed] [Google Scholar]
- 27.O’Leary M H. Catalytic strategies in enzymatic carboxylation and decarboxylation. Enzymes. 1992;20:235–269. [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H-M, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicare R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum delta H: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 31.Yamane K, Kumano M, Kurita K. The 25 degrees-36 degrees region of the Bacillus subtilis chromosome: determination of the sequence of a 146 kb segment and identification of 113 genes. Microbiology. 1996;142:3047–3056. doi: 10.1099/13500872-142-11-3047. [DOI] [PubMed] [Google Scholar]
- 32.Zago A, Degrassi G, Bruschi C V. Cloning, sequencing, and expression in Escherichia coli of the Bacillus pumilus gene for ferulic acid decarboxylase. Appl Environ Microbiol. 1995;61:4484–4486. doi: 10.1128/aem.61.12.4484-4486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Wiegel J. Sequential anaerobic degradation of 2,4-dichlorophenol in freshwater sediments. Appl Environ Microbiol. 1990;56:1119–1127. doi: 10.1128/aem.56.4.1119-1127.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Wiegel J. Isolation and partial characterization of a Clostridium species transforming p-hydroxybenzoate and 3,4-dihydroxybenzoate and producing phenols as the final transformation products. Microb Ecol. 1990;20:103–121. doi: 10.1007/BF02543871. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Wiegel J. Reversible conversion of 4-hydroxybenzoate and phenol by Clostridium hydroxybenzoicum. Appl Environ Microbiol. 1994;60:4182–4185. doi: 10.1128/aem.60.11.4182-4185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Mandelco L, Wiegel J. Clostridium hydroxybenzoicum sp. nov., an amino acid-utilizing, hydroxybenzoate-decarboxylating bacterium isolated from methanogenic freshwater pond sediment. Int J Syst Bacteriol. 1994;44:214–222. doi: 10.1099/00207713-44-2-214. [DOI] [PubMed] [Google Scholar]