Abstract
Formaldehyde is a toxic metabolite that is formed in large quantities during bacterial utilization of the methoxy sugar 6‐O‐methyl‐d‐galactose, an abundant monosaccharide in the red algal polysaccharide porphyran. Marine bacteria capable of metabolizing porphyran must therefore possess suitable detoxification systems for formaldehyde. We demonstrate here that detoxification of formaldehyde in the marine Flavobacterium Zobellia galactanivorans proceeds via the ribulose monophosphate pathway. Simultaneously, we show that the genes encoding the key enzymes of this pathway are important for maintaining high formaldehyde resistance. Additionally, these genes are upregulated in the presence of porphyran, allowing us to connect porphyran degradation to the detoxification of formed formaldehyde.
Keywords: Bacteroidetes, carbohydrates, CAZymes formaldehyde detoxification, RuMP pathway
The oxidative demethylation of 6‐O‐methyl‐d‐galactose produces formaldehyde. Marine bacteria that utilize this sugar in the degradation of porphyran must therefore possess suitable formaldehyde detoxification pathways. The connection between the degradation of marine carbohydrates and the detoxification of formaldehyde has been demonstrated for the marine model organism Zobellia galactanivorans.
Marine algae are considered to be one of the most important primary producers in the marine ecosystem and one of the largest sources of marine carbohydrates.[ 1 , 2 ] Serving as energy storage and structural cell wall components, carbohydrates constitute up to 70 % of algae dry mass. [3] Compared to their terrestrial counterparts, marine polysaccharides differ in the backbone structure and side‐group modifications. [4] One bacterial phylum considered to be specialist in the degradation of high molecular weight organic matter such as marine carbohydrates are the Bacteroidetes.[ 5 , 6 , 7 ] Marine Bacteroidetes harbor gene clusters which are referred to as polysaccharide utilization loci (PULs) encoding carbohydrate‐active enzymes (CAZymes) as well as specific proteins for the binding and uptake of sugar units.[ 5 , 8 ] Their tremendous repertoire of CAZymes allows them to depolymerize complex marine carbohydrates and utilize the imported monosaccharides as a carbon and energy source.[ 4 , 8 ] Observations that Bacteriodetes are among the first responders after micro‐ and macroalgal blooms are related to their abilities of rapid growth on colonizable surfaces such as macroalgae as well as their CAZymes production.[ 9 , 10 ]
A model bacterium for the bioconversion of algal biomass is the marine Flavobacterium Zobellia galactanivorans DsijT, which was originally isolated from the red alga Delesseria sanguinea near the coast of Roscoff (Brittany, France). [11] In‐depth analysis of its complete genome and growth studies revealed that this microorganism possesses 50 PULs, is able to grow on numerous marine polysaccharides and utilizes them as a carbon source.[ 12 , 13 ] Extensive biochemical studies have elucidated essential CAZymes from Z. galactanivorans and their roles in the complex degradation pathways for alginate and laminarin from brown algae[ 14 , 15 , 16 , 17 , 18 , 19 ] as well as for carrageenan, agar and porphyran from red algae.[ 20 , 21 , 22 , 23 ] Porphyran is the common name of the agar from red algae of the genus Porphyra and is their main cell wall polysaccharide. [24] The porphyran backbone consists mainly of the alternating monosaccharide units 4‐linked‐α‐l‐galactose‐6‐sulfate (L6S) and 3‐linked‐β‐d‐galactose (Gal) or 3,6‐anhydro‐α‐l‐galactose (LA) (Figure S1, Supporting Information).[ 25 , 26 ] In addition, O‐methylation of d‐galactose is a frequent modification that results in the presence of up to 28 % of the methoxy sugar 6‐O‐methyl‐d‐galactose (G6Me) within the porphyran chain.[ 24 , 25 , 27 ]
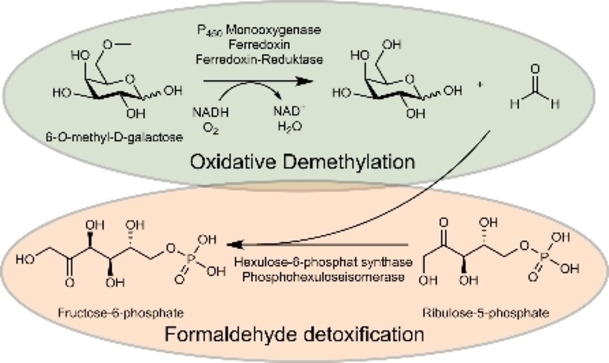
Considering the stability of methyl ethers, it is reasonable to assume that G6Me must first be demethylated before it can enter the cellular metabolism. We recently have demonstrated that the oxidative demethylation of G6Me is catalyzed by a cytochrome P450 monooxygenase with the appropriate redox partners ferredoxin and ferredoxin reductase. [28] The crystal structure of the cytochrome P450 monooxygenase from Z. galactanivorans informed on the binding of G6Me as well as other mechanistic insights. [29] The products of this demethylation are d‐galactose and formaldehyde in equimolar amounts. [28] However, formaldehyde formation leads to a problem for the organism, since formaldehyde is a toxic metabolite in cells due to its high reactivity as an electrophile. [30] The polarized carbonyl group of formaldehyde can be attacked by nucleophiles such as free amine or thiol groups of amino acids [31] and proteins [32] or nucleic acids, [33] resulting in protein and DNA damages and covalent cross‐links. [34]
Marine bacteria capable of degrading porphyran and utilizing G6Me should therefore possess suitable detoxification pathways for formaldehyde. Focusing on the discovery of possible pathways of formaldehyde detoxification, we first searched through the genomes of the Flavobacteria Z. galactanivorans DsijT and Formosa agariphila KMM 3901T, [35] which possess the cytochrome P450 monooxygenases and thus catalyze the oxidative demethylation of G6Me, [28] for genes encoding enzymes from well‐known detoxification pathways. Both organisms harbor annotated genes for enzymes found in the serine and tetrahydrofolate pathways (Table S1, Supporting Information). However, unlike F. agariphila, Z. galactanivorans additionally carries the genes for the putative key enzymes of the ribulose monophosphate (RuMP) pathway. This putative RuMP pathway was proposed to be an advantageous adaptive trait for Z. galactanivorans to cope with the release of formaldehyde when degrading red algal cell walls. [12] Altogether, F. agariphila and Z. galactanivorans should provide different responses to the accumulation of formaldehyde. The RuMP pathway is the most efficient pathway of formaldehyde assimilation in terms of ATP consumption and biomass yield.[ 36 , 37 , 38 ] It can be divided into three parts: fixation, cleavage, and regeneration. [39] While the cleavage and regeneration part can take place via different routes and are catalyzed by common enzymes of the central carbon cycle, the fixation of formaldehyde takes place via two unique key enzymes: a 3‐hexulose‐6‐phosphate synthase (HPS) and a 6‐phospho‐3‐hexuloisomerase (PHI).[ 39 , 40 ] HPS, a member of the class 2 aldolases, [41] catalyzes the Mg2+‐dependent aldol reaction between formaldehyde and d‐ribulose‐5‐phosphate (R5P) to give the intermediate d‐arabino‐3‐hexulose‐6‐phosphate (AH6P), which is then isomerized by PHI to d‐fructose‐6‐phosphate (F6P) (Figure 2, top). [40] F6P is consumed in the cleavage part to generate triose phosphates such as glyceraldehyde‐3‐phosphate or dihydroxyacetone phosphate which then can be metabolized in the glycolysis or the Entner‐Doudoroff pathway.[ 39 , 42 ] Furthermore, R5P is regenerated from F6P by reactions occurring in the pentose‐phosphate cycle. [40] It has been demonstrated that the RuMP pathway can play an important role in the degradation of methoxylated lignin monomers by non‐methylotrophic bacteria. [44] However, most knowledge about the RuMP pathway originates from methylotrophic bacteria that grow on reduced C1 components such as methane or methanol, which they oxidize to formaldehyde. [40] In addition to these components, numerous methylated sugars are present in the marine ecosystem [43] and are thus a potential source of formaldehyde. However, the RuMP pathway has not yet been investigated in the context of marine carbohydrate degradation, we therefore aimed to investigate whether this pathway plays a role in the degradation of porphyran by Bacteriodetes.
Figure 2.

HPS and PHI catalyze the incorporation of formaldehyde to produce fructose‐6‐phosphate. A protein concentration of 10 μg mL−1 for HPS and PHI were used in the biocatalysis. For substrates, 0.75 mm d‐ribulose‐5‐phosphate disodium salt and 0.5 mm formaldehyde were used. The reactions were performed in a 50 mm sodium phosphate buffer pH 7.5 supplemented with 5 mm MgCl2 for 5 min, at an incubation temperature of 30 °C and an agitation of 1,000 rpm. The formaldehyde concentration was then determined using the Nash reagent. Mean values are shown, error bars present±s.d. (n=3).
We reasoned that the presence of the RuMP pathway in Z. galactanivorans should lead to an increased resistance to formaldehyde compared to F. agariphila. To test this hypothesis, we cultivated each organism in the presence of increasing formaldehyde concentrations. For F. agariphila a significant decrease in growth rate was observed at formaldehyde concentrations greater than 100 μm, while no growth was seen at 500 μm (Figure 1a). In contrast, the presence of formaldehyde concentrations up to 500 μm revealed just minor effects on the growth of Z. galactanivorans. However, no growth was detected in the presence of a formaldehyde concentration of 1,000 μm (Figure S2, Supporting Information). In order to prove that the increased resistance towards formaldehyde is caused by the enzymes HPS and PHI, hxlA and hxlB gene knockout strains of Z. galactanivorans were created and the influence of formaldehyde on their growth was investigated again. In the presence of 500 μm formaldehyde, the ΔhxlA‐hxlB strain was unable to grow, whereas the wild‐type (WT) and a control knock‐out strain lacking the P450 monooxygenase encoding gene (Δmgd) were able to grow normally (Figure 1b). Knockout of the hxlA and hxlB genes thus resulted in a formaldehyde‐sensitive strain, which displayed normal growth behavior in the absence of formaldehyde. Both findings supported our assumption that these genes were responsible for the detoxification of formaldehyde.
Figure 1.
The genes encoding for the key enzymes of the RuMP pathway are crucial for formaldehyde resistance of Z. galactanivorans and are upregulated in the presence of porphyran. a) Effect of increasing concentrations of formaldehyde on the growth of F. agariphila and Z. galactanivorans. For each bacterial strain, the growth rate obtained in the absence of formaldehyde was taken as 100 %. b) Growth curve of WT, Δmgd (cytochrome P450 monooxygenase) and ΔhxlA‐hxlB (HPS and PHI) mutant strains of Z. galactanivorans in ZoBell 2216 medium containing no or 500 μm formaldehyde. c) Expression of genes encoding cytochrome P450 monooxygenase (mgd), 3‐hexulose‐6‐phosphate synthase (hxlA) and 6‐phospho‐3‐hexulose isomerase (hxlB) in Z. galactanivorans grown with laminarin, agar or porphyran as sole carbon source. The effect of substrate on gene expression was tested by one‐way ANOVA on log‐transformed data, followed by a post‐hoc Tukey test (*, P<0.05). Expression data from the publicly available GEO dataset GSE99940. For a)–c) Values are mean±s.e.m (n=3).
After demonstrating their role for formaldehyde resistance, we were interested to know whether the genes encoding HPS and PHI were also upregulated in the presence of porphyran, considering that this is the origin of formed formaldehyde due to the oxidative demethylation of G6Me. In order to evaluate gene regulation, Z. galactanivorans was grown with the marine carbohydrates laminarin, agar or porphyran as a sole carbon source. The β‐glucan laminarin was selected as a control considering that it is the most abundant polysaccharide in the marine ecosystem [45] and agar was chosen as control because it may contain G6Me. [46] The genes encoding the P450 monooxygenase (mgd), HPS (hxlA), and PHI (hxlB) were upregulated in the presence of porphyran compared to laminarin and agar (Figure 1c). No upregulation in the presence of agar was observed, a possible explanation for this is the absence of G6Me in agar. Upregulation of mgd in the presence of porphyran indicates that there is a possible source of formaldehyde, while at the same time, the upregulation of hxlA and hxlB suggests that formaldehyde detoxification via the RuMP pathway can occur.
Following this demonstration that the genes of HPS and PHI were upregulated in the presence of porphyran, we wanted to verify whether they encode enzymes that catalyze the key reactions of the RuMP pathway. Therefore, we expressed the enzymes in Escherichia coli and purified them (Figure S3, Supporting Information). In order to determine the activity of the enzymes, the R5P‐dependent disappearance of formaldehyde was determined using the Nash reagent [47] and the formation of F6P was monitored by an enzyme‐coupled assay. [48] In the presence of d‐ribulose‐5‐phosphate, a decrease in the formaldehyde concentration (Figure 2) and the formation of F6P (Figure S4, Supporting Information) was observed for the reaction mixture that contained both enzymes. After 5 min incubation at 30 °C, approximately 0.34 mm of the initial formaldehyde concentration of 0.5 mm was removed from the solution, which corresponds to a conversion of 68.5 %. Meanwhile, in the control reactions without d‐ribulose‐5‐phosphate and in the absence of either HPS or both enzymes no incorporation of formaldehyde and thus no formation of F6P was observed. For the reaction mixture containing HPS but not PHI, a decrease in formaldehyde could also be detected, which is reasonable considering that HPS catalyzes the reaction of R5P to AH6P independently of PHI. Moreover, the enzymes were able to catalyze the reverse reaction, since very low formation of formaldehyde was observed when F6P was used at a substrate concentration of 20 mm (Figure S5, Supporting Information). In conclusion, Z. galactanivorans harbors the active key enzymes of the RuMP pathway.
Since we could prove that Z. galactanivorans utilizes the RuMP pathway for formaldehyde detoxification, we were interested in the distribution of this pathway in marine ecosystems. We therefore queried approximately 5,500 marine bacterial genomes from the MarDB and MarRef databases for the key enzymes of the RuMP pathway and identified 197 genomes (equivalent to ∼3.58 %) encoding HPS‐ and PHI‐ gene pairs (Figure 3). Among the 197 genomes, only 16 contain similar cytochrome P450 monooxygenase, ferredoxin reductase, and ferredoxin encoding clusters like Z. galactanivorans (Figure 3). The key enzymes of the RuMP pathway as well as the enzymes of the cytochrome P450 cluster were highly similar to those of Z. galactanivorans, which is exemplarily shown for five selected reference genomes, including Cellulophaga, Maribacter, and Zobellia in Figure 4.
Figure 3.
Taxonomic distribution of the RuMP pathway in marine prokaryotes. The colored outer rings indicate the occurrence of the HPS/PHI pairs (dark blue) and the P450 cluster (dark orange). Genomes that encode homologous sequences are shown independently (lighter colors). The intersection of genomes encoding both clusters is shown in green.
Figure 4.
Key enzymes of the RuMP pathway and the enzymes of the P450 cluster from Zobellia galactanivorans are highly similar to those in five selected reference genomes of other marine taxa. The similarity is indicated by the opacity of each link as well as the given percentage within each coding sequence (CDS). The outer scale shows the genomic region of the CDS in kbp.
In addition, these 16 bacterial genomes also featured CAZymes belonging to the GH86 and GH117 families which can catalyze the degradation of agar and porphyran. This supports the hypothesis that the RuMP pathway may be responsible for the detoxification of formaldehyde, which is produced during the degradation of marine carbohydrates and thus may provide growth advantages for these marine bacteria over others. Besides the marine strains with genomically clustered RuMP‐based detoxification genes, we found 104 additional marine isolates where putative HPS and PHI homologs are distributed over the genomes. Interestingly the best hits are found for some Zobellia, a few Maribacter, and Cellulophaga as well as Arenibacter strains, which are bacterial genera commonly isolated at the surface of macroalgae.[ 49 , 50 ] This suggests that marine RuMP‐based detoxification is mainly specific to bacteria living on multicellular algae, reminiscent of methylotrophic bacteria of the phyllosphere. [51]
In summary, we demonstrated in this work that Z. galactanivorans exhibited higher resistance to formaldehyde than F. agariphila and that this was based on the presence of the RuMP pathway. Consequently, the knockout of the genes, encoding the key enzymes of this pathway, led to a formaldehyde‐sensitive strain. We could also demonstrate that in the presence of porphyran the genes encoding the cytochrome P450 monooxygenase and the RuMP pathway were upregulated. This revealed that there is a potential source of formaldehyde through the oxidative demethylation of G6Me and simultaneously a possibility for its detoxification via the RuMP pathway. By verifying the enzyme activity of expressed and purified HPS and PHI, we could demonstrate that the genes encoding the enzymes are indeed responsible for the fixation of formaldehyde. As a result, we were able to provide evidence for a connection between porphyran degradation and formaldehyde detoxification. Genomic analyses in marine genome databases revealed that this pathway is the exception rather than the rule in marine microbes. It may thus provide growth advantages for some marine bacteria over others in the competition for marine polysaccharides.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
The authors thank Nolwen Le Duff for technical assistance with microbiology experiments. F.T. acknowledges support from the French government via the National Research Agency program ALGAVOR (ANR‐18‐CE02‐0001). D.B. was supported by a scholarship from the Institute of Marine Biotechnology e.V. We thank the German Research Foundation (DFG) for funding through the Research Unit FOR2406 “Proteogenomics of Marine Polysaccharide Utilization” (POMPU) (grant# BO 1862/17‐2 to U.T.B. and SCHW 595/10‐2 to T.S.). Open Access funding enabled and organized by Projekt DEAL.
S. Brott, F. Thomas, M. Behrens, K. Methling, D. Bartosik, T. Dutschei, M. Lalk, G. Michel, T. Schweder, U. T. Bornscheuer, ChemBioChem 2022, 23, e202200269.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
References
- 1. Krause-Jensen D., Duarte C. M., Nat. Geosci. 2016, 9, 737–742. [Google Scholar]
- 2. Arnosti C., Wietz M., Brinkhoff T., Hehemann J. H., Probandt D., Zeugner L., Amann R., Annu. Rev. Mar. Sci. 2021, 13, 81–108. [DOI] [PubMed] [Google Scholar]
- 3. Usman A., Khalid S., Usman A., Hussain Z., Wang Y., in Algae Based Polymers, Blends and Compositions: Chemistry, Biotechnology and Material Sciences (Eds.: Zia K. M., Zuber M., Ali M.), Elsevier, Amsterdam, 2017, pp. 115–153. [Google Scholar]
- 4. Bäumgen M., Dutschei T., Bornscheuer U. T., ChemBioChem 2021, 22, 2247–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thomas F., Hehemann J. H., Rebuffet E., Czjzek M., Michel G., Front. Microbiol. 2011, 2, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grondin J. M., Tamura K., Déjean G., Abbott D. W., Brumer H., J. Bacteriol. 2017, 199, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Munoz R., Rosselló-Móra R., Amann R., Syst. Appl. Microbiol. 2016, 39, 281–296. [DOI] [PubMed] [Google Scholar]
- 8. Lapébie P., Lombard V., Drula E., Terrapon N., Henrissat B., Nat. Commun. 2019, 10, 2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., Kassabgy M., Huang S., Mann A. J., Waldmann J., Weber M., Klindworth A., Otto A., Lange J., Bernhardt J., Reinsch C., Hecker M., Peplies J., Bockelmann F. D., Callies U., Gerdts G., Wichels A., Wiltshire K. H., Glöckner F. O., Schweder T., Amann R., Science 2012, 336, 608–611. [DOI] [PubMed] [Google Scholar]
- 10. Brunet M., de Bettignies F., Le Duff N., Tanguy G., Davoult D., Leblanc C., Gobet A., Thomas F., Environ. Microbiol. 2021, 23, 1638–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barbeyron T., L'Haridon S., Corre E., Kloareg B., Potin P., Int. J. Syst. Evol. Microbiol. 2001, 51, 985–997. [DOI] [PubMed] [Google Scholar]
- 12. Barbeyron T., Thomas F., Barbe V., Teeling H., Schenowitz C., Dossat C., Goesmann A., Leblanc C., Glöckner F. O., Czjzek M., Amann R., Michel G., Environ. Microbiol. 2016, 18, 4610–4627. [DOI] [PubMed] [Google Scholar]
- 13. Thomas F., Bordron P., Eveillard D., Michel G., Front. Microbiol. 2017, 8, 1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas F., Barbeyron T., Tonon T., Génicot S., Czjzek M., Michel G., Environ. Microbiol. 2012, 14, 2379–2394. [DOI] [PubMed] [Google Scholar]
- 15. Thomas F., Lundqvist L. C. E., Jam M., Jeudy A., Barbeyron T., Sandström C., Michel G., Czjzek M., J. Biol. Chem. 2013, 288, 23021–23037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dudek M., Dieudonné A., Jouanneau D., Rochat T., Michel G., Sarels B., Thomas F., Nucleic Acids Res. 2020, 48, 7786–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jouanneau D., Klau L. J., Larocque R., Jaffrennou A., Duval G., Le Duff N., Roret T., Jeudy A., Aachmann F. L., Czjzek M., Thomas F., Glycobiology 2021, 31, 1364–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Labourel A., Jam M., Jeudy A., Hehemann J., Czjzek M., Michel G., J. Biol. Chem. 2014, 289, 2027–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Labourel A., Jam M., Legentil L., Sylla B., Hehemann J. H., Ferrières V., Czjzek M., Michel G., Acta Crystallogr. Sect. D 2015, 71, 173–184. [DOI] [PubMed] [Google Scholar]
- 20. Ficko-Blean E., Duffieux D., Rebuffet É., Larocque R., Groisillier A., Michel G., Czjzek M., Acta Crystallogr. 2015, 71, 209–223. [DOI] [PubMed] [Google Scholar]
- 21. Ficko-Blean E., Préchoux A., Thomas F., Rochat T., Larocque R., Zhu Y., Stam M., Génicot S., Jam M., Calteau A., Viart B., Ropartz D., Pérez-Pascual D., Correc G., Matard-Mann M., Stubbs K. A., Rogniaux H., Jeudy A., Barbeyron T., Médigue C., Czjzek M., Vallenet D., McBride M. J., Duchaud E., Michel G., Nat. Commun. 2017, 8, 1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hehemann J. H., Correc G., Thomas F., Bernard T., Barbeyron T., Jam M., Helbert W., Michel G., Czjzek M., J. Biol. Chem. 2012, 287, 30571–30584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rebuffet E., Groisillier A., Thompson A., Jeudy A., Barbeyron T., Czjzek M., Michel G., Environ. Microbiol. 2011, 13, 1253–1270. [DOI] [PubMed] [Google Scholar]
- 24. Rees D. A., Conway E., Biochem. J. 1962, 84, 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anderson N. S., Rees D. A., J. Chem. Soc. 1965, 5880–5887. [Google Scholar]
- 26. Correc G., Hehemann J. H., Czjzek M., Helbert W., Carbohydr. Polym. 2011, 83, 277–283. [Google Scholar]
- 27. Ropartz D., Giuliani A., Fanuel M., Hervé C., Czjzek M., Rogniaux H., Anal. Chim. Acta 2016, 933, 1–9. [DOI] [PubMed] [Google Scholar]
- 28. Reisky L., Büchsenschütz H. C., Engel J., Song T., Schweder T., Hehemann J. H., Bornscheuer U. T., Nat. Chem. Biol. 2018, 14, 342–344. [DOI] [PubMed] [Google Scholar]
- 29. Robb C. S., Reisky L., Bornscheuer U. T., Hehemann J. H., Biochem. J. 2018, 475, 3875–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen N. H., Djoko K. Y., Veyrier F. J., McEwan A. G., Front. Microbiol. 2016, 7, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kamps J. J. A. G., Hopkinson R. J., Schofield C. J., Claridge T. D. W., Commun. Chem. 2019, 2, 126. [Google Scholar]
- 32. Tayri-Wilk T., Slavin M., Zamel J., Blass A., Cohen S., Motzik A., Sun X., Shalev D. E., Ram O., Kalisman N., Nat. Commun. 2020, 11, 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shishodia S., Zhang D., El-Sagheer A. H., Brown T., Claridge T. D. W., Schofield C. J., Hopkinson R. J., Org. Biomol. Chem. 2018, 16, 4021–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu K., Ye W., Zhou L., Collins L. B., Chen X., Gold A., Ball L. M., Swenberg J. A., J. Am. Chem. Soc. 2010, 132, 3388–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mann A. J., Hahnke R. L., Huang S., Werner J., Xing P., Barbeyron T., Huettel B., Stüber K., Reinhardt R., Harder J., Glöckner F. O., Amann R. I., Teeling H., Appl. Environ. Microbiol. 2013, 79, 6813–6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Strom T., Ferenci T., Quayle J. R., Biochem. J. 1974, 144, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goldberg I., Rock J. S., Ben-Bassat A., Mateles R. I., Biotechnol. Bioeng. 1976, 18, 1657–1668. [DOI] [PubMed] [Google Scholar]
- 38. Whitaker W. B., Sandoval N. R., Bennett R. K., Fast A. G., Papoutsakis E. T., Curr. Opin. Biotechnol. 2015, 33, 165–175. [DOI] [PubMed] [Google Scholar]
- 39. Müller J. E. N., Meyer F., Litsanov B., Kiefer P., Potthoff E., Heux S., Quax W. J., Wendisch V. F., Brautaset T., Portais J. C., Vorholt J. A., Metab. Eng. 2015, 28, 190–201. [DOI] [PubMed] [Google Scholar]
- 40. Kato N., Yurimoto H., Thauer R. K., Biosci. Biotechnol. Biochem. 2006, 70, 10–21. [DOI] [PubMed] [Google Scholar]
- 41. Desmons S., Fauré R., Bontemps S., ACS Catal. 2019, 9, 9575–9588. [Google Scholar]
- 42. He H., Edlich-Muth C., Lindner S. N., Bar-Even A., ACS Synth. Biol. 2018, 7, 1601–1611. [DOI] [PubMed] [Google Scholar]
- 43. Panagiotopoulos C., Repeta D. J., Mathieu L., Rontani J. F., Sempéré R., Mar. Chem. 2013, 154, 34–45. [Google Scholar]
- 44. Mitsui R., Kusano Y., Yurimoto H., Sakai Y., Kato N., Tanaka M., Appl. Environ. Microbiol. 2003, 69, 6128–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Becker S., Tebben J., Coffinet S., Wiltshire K., Iversen M. H., Harder T., Hinrichs K. U., Hehemann J. H., Proc. Natl. Acad. Sci. USA 2020, 117, 6599–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chiovitti A., Bacic A., Craik D. J., Kraft G. T., Liao M. L., Carbohydr. Res. 2004, 339, 1459–1466. [DOI] [PubMed] [Google Scholar]
- 47. Nash T., Biochem. J. 1953, 55, 416–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hohorst H.-J., Methods Enzym. Anal. 1965, 134–138. [Google Scholar]
- 49. Martin M., Portetelle D., Michel G., Vandenbol M., Appl. Microbiol. Biotechnol. 2014, 98, 2917–2935. [DOI] [PubMed] [Google Scholar]
- 50. Martin M., Barbeyron T., Martin R., Portetelle D., Michel G., Vandenbol M., Front. Microbiol. 2015, 6, 1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vorholt J. A., Nat. Rev. Microbiol. 2012, 10, 828–840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.





