Abstract
Methyl‐coenzyme M reductase, which is responsible for the production of the greenhouse gas methane during biological methane formation, carries several unique posttranslational amino acid modifications, including a 2‐(S)‐methylglutamine. The enzyme responsible for the Cα‐methylation of this glutamine is not known. Herein, we identify and characterize a cobalamin‐dependent radical SAM enzyme as the glutamine C‐methyltransferase. The recombinant protein from Methanoculleus thermophilus binds cobalamin in a base‐off, His‐off conformation and contains a single [4Fe‐4S] cluster. The cobalamin cofactor cycles between the methyl‐cob(III)alamin, cob(II)alamin and cob(I)alamin states during catalysis and produces methylated substrate, 5′‐deoxyadenosine and S‐adenosyl‐l‐homocysteine in a 1 : 1 : 1 ratio. The newly identified glutamine C‐methyltransferase belongs to the class B radical SAM methyltransferases known to catalyze challenging methylation reactions of sp3‐hybridized carbon atoms.
Keywords: MCR Modification, Metalloproteins, Methanogenesis, Radical Reactions, Radical S-Adenosylmethionine
The enzyme responsible for the unique Cα‐methylation of a glutamine residue within methyl‐coenzyme M reductase was identified. Biochemical, spectroscopic and mass spectrometric characterization clearly classify the newly identified glutamine C‐methyltransferase as a cobalamin‐dependent radical SAM enzyme and, thus, as a new member of the class B radical SAM methyltransferase family.
Introduction
Methane plays an important role in the global carbon cycle, but also acts as a potent greenhouse gas, when excessively released into the atmosphere. The formation of methane in methanogenic archaea as well as the reverse reaction in anaerobic methanotrophic archaea relies on the action of the enzyme methyl‐coenzyme M reductase (MCR). [1] In most cases, MCR is a hexameric protein with the subunit composition (αβγ)2 containing two molecules of the nickel‐tetrapyrrole coenzyme F430 as essential prosthetic groups (Figure 1a).[ 2 , 3 ] The first crystal structure of an MCR (MCR I from Methanothermobacter marburgensis) surprisingly revealed the presence of five very unusual posttranslational amino acid modifications within the α subunit: thioglycine, S‐methylcysteine, 2‐(S)‐methylglutamine, 1‐N‐methyl‐histidine, and 5‐(S)‐methylarginine. [3] Subsequently, these modifications were confirmed by mass spectrometry and also found in MCRs from other methanogens, as well as two additional modifications, namely a didehydroaspartate and a 6‐hydroxytryptophan.[ 4 , 5 , 6 , 7 , 8 ] Although most of the MCR modifications have been known for some time, knowledge about the enzymes responsible for their introduction is still limited, and restricted to the formation of the thioglycine, [9] methylcysteine, [10] and methylarginine[ 11 , 12 , 13 , 14 ] modifications.
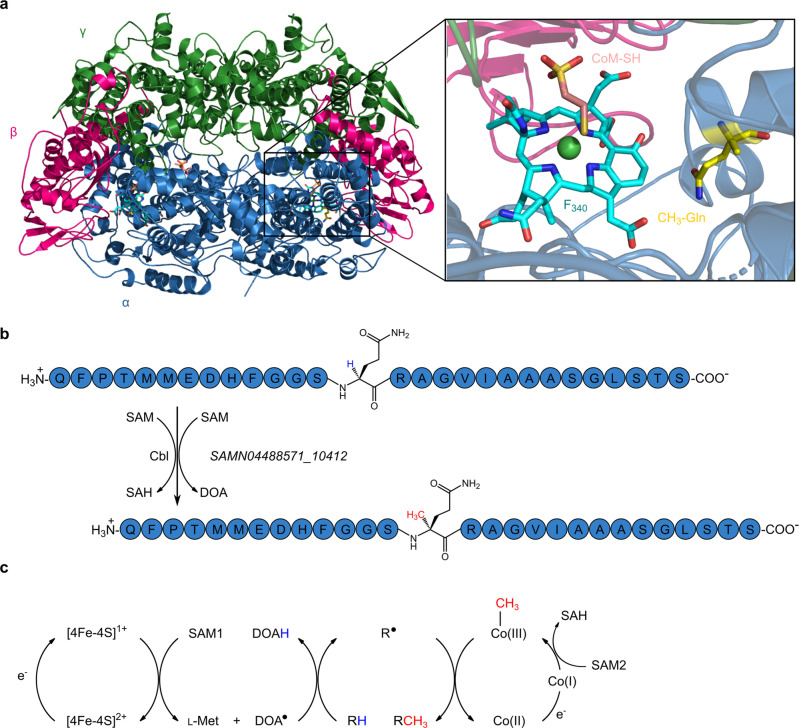
Figure 1.
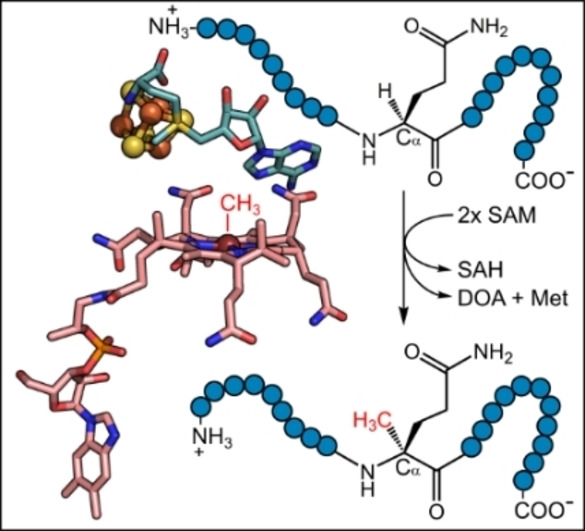
The 2‐(S)‐methylglutamine modification within methyl‐coenzyme M reductase (MCR) is introduced by a cobalamin‐dependent radical SAM enzyme. a) Crystal structure of MCR from Methanothermobacter marburgensis (PDB ID 1MRO). The close‐up view of the active site of MCR on the right shows the modified amino acid 2‐(S)‐methylglutamine in yellow. The figure was prepared using PyMOL. b) A 28 amino acid peptide of the MCR α subunit from Methanoculleus thermophilus. The glutamine residue that is subjected to methylation is highlighted. The methylation reaction is proposed to be catalyzed by a cobalamin (Cbl)‐dependent radical SAM methyltransferase encoded by the SAMN04488571_10412 gene in M. thermophilus. c) General reaction mechanism proposed for class B radical SAM methyltransferases (modified from Zhang et al. 2012). SAM=S‐adenosyl‐l‐methionine, SAH=S‐adenosyl‐l‐homocysteine, DOA⋅=5′‐deoxyadenosyl radical, DOAH=5′‐deoxyadenosine, R⋅=substrate radical, RH=substrate.
The methylations of glutamine at C2 and arginine at C5 are rather unusual and have only been reported for MCR so far.[ 4 , 5 , 6 , 7 , 8 , 15 ] From a chemical perspective, the introduction of methyl groups at these carbon positions is challenging, because they represent sp3‐hybridized electrophilic carbons that are not amenable to the conventional S‐adenosyl‐l‐methionine (SAM)‐dependent methylation via an SN2 mechanism. Rather, these C‐methylation reactions were hypothesized to proceed via radical chemistry. [16] Indeed, the arginine C‐methyltransferase (RCMT), encoded by the mm10 gene, is a radical SAM enzyme.[ 12 , 14 ] Radical SAM enzymes use a [4Fe‐4S] cluster, which is coordinated by three cysteine residues, and a SAM molecule to generate a 5′‐deoxyadenosyl radical (DOA⋅). [17] This radical initiates further radical reactions to achieve some of the most challenging substrate conversions observed in nature. [18] Thus, RCMT utilizes a radical mechanism in order to achieve the difficult activation and methylation of the C5 position of arginine. Surprisingly, RCMT uses cobalamin as an additional cofactor.[ 12 , 14 ]
So far, the glutamine C‐methyltransferase (QCMT) remained elusive. In a previous study, we identified a gene encoding a putative radical SAM methyltransferase, which is often located near the mcr gene cluster and the mm10 gene, [11] and we considered whether the encoded protein could be the so far unknown QCMT (Figure 1b). Based on amino acid sequence analysis, the putative QCMT belongs to the subclass B within the radical SAM methyltransferase family. [11] As such, it exhibits an N‐terminal cobalamin‐binding domain followed by the radical SAM domain containing the SAM binding site and the characteristic cysteine motif CX3CX2C providing the three cysteine ligands for the [4Fe‐4S] cluster.[ 19 , 20 ] For class B radical SAM methyltransferases a general reaction mechanism was proposed (Figure 1c). The reaction starts with an electron transfer from the reduced [4Fe‐4S]1+ cluster to SAM, which is reductively cleaved into l‐methionine and a 5′‐deoxyadenosyl radical (DOA⋅). The latter abstracts a hydrogen atom from the substrate yielding a substrate radical, which in turn reacts with methylcob(III)alamin (MeCbl) to the methylated product and cob(II)alamin. The cob(II)alamin must be reduced to cob(I)alamin before a second SAM molecule can carry out the regeneration of MeCbl. During this reaction, S‐adenosyl‐l‐homocysteine (SAH) is released. [19]
In this work, we present the first biochemical and biophysical characterization of a putative QCMT. Purification of the recombinant enzyme from Methanoculleus thermophilus and in vitro enzyme activity assays showed that QCMT catalyzes the methylation of a specific glutamine residue within an MCR‐derived peptide substrate. QCMT binds cobalamin in a base‐off, His‐off mode and contains a single [4Fe‐4S] cluster, both required for the enzymatic reaction. The methylation of the glutamine residue proceeds with the formation of DOA and SAH in an approximately 1 : 1 ratio, indicating the consumption of two SAM molecules during product formation. Supported by isotope labeling, it was shown that methyl group transfer proceeds from SAM via the enzyme‐bound cobalamin cofactor. QCMT carries out a remarkably high number of in vitro turnovers for a radical SAM enzyme. Altogether, our results show that the newly identified QCMT is a class B radical SAM methyltransferase catalyzing the challenging Cα‐methylation of a glutamine residue of MCR.
Results and Discussion
Production and Purification of QCMT
In order to investigate the function of the putative QCMT, we aimed to produce the recombinant protein in E. coli. For this purpose, we cloned the gene SAMN04488571_10412 encoding the putative QCMT from Methanoculleus thermophilus (DSM 2373) into the expression vector pET21a(+) yielding a construct for the production of a C‐terminally His6‐tagged fusion protein. The recombinant QCMT was produced in E. coli BL21(DE3) Star pLysS with coproduction of the E. coli cobalamin‐uptake system encoded on pBAD42‐BtuCEDFB. [21] Soluble QCMT was anaerobically purified by immobilized metal affinity chromatography (IMAC) followed by preparative size exclusion chromatography (SEC) (Figure S1a). On average, we obtained around 11 mg of QCMT per 1 liter of bacterial culture after preparative SEC.
QCMT Contains One [4Fe‐4S] Cluster and Cobalamin
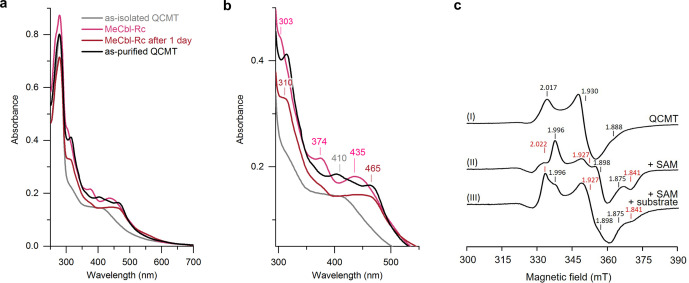
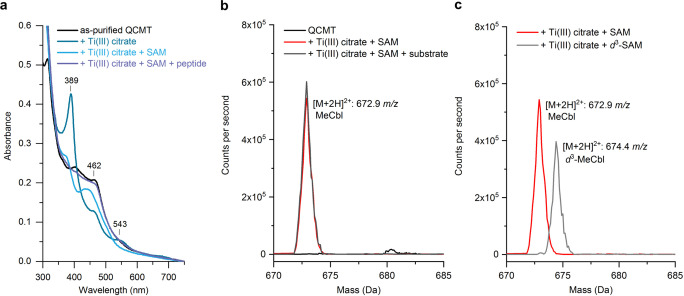
Analysis of the as‐isolated QCMT (after IMAC) by UV/Visible absorption spectroscopy (UV/Vis) revealed a broad absorption feature around 410 nm indicating the presence of iron‐sulfur clusters (Figure 2a, b). However, the UV/Vis absorption spectrum did not suggest the presence of a cobalamin cofactor in as‐isolated QCMT. Also, no released cobalamin cofactor was detectable by UV/Vis upon acid/heat treatment of the protein. Although it did not contain detectable amounts of cobalamin after IMAC, QCMT was produced as a soluble protein, which is in contrast to other members of the class B radical SAM methyltransferase family. [21] Next, we reconstituted the enzyme with methylcobalamin after IMAC ‐ henceforth referred to as “MeCbl‐Rc” QCMT. Immediately after MeCbl reconstitution, the UV/Vis absorption spectrum of MeCbl‐Rc QCMT exhibited absorption maxima at about 435 nm and 374 nm as well as a shoulder around 303 nm (Figure 2a, b) indicating the presence of enzyme‐bound MeCbl possessing neither the dimethylbenzimidazole moiety (base‐off) nor a histidine residue (His‐off) as the lower axial ligand.[ 22 , 23 , 24 , 25 ] After one day of storage under anaerobic conditions, the UV/Vis absorption spectrum of MeCbl‐Rc QCMT had changed, now exhibiting a rather broad absorption band around 465 nm and a shoulder at 310 nm. These spectral changes indicate that photolytic degradation of the MeCbl had taken place resulting in formation of cob(II)alamin. [26] The determination of the iron and sulfide contents of as‐isolated QCMT revealed the presence of 2.1±1.1 mol iron and 2.5±0.8 mol sulfide per mol protein. Since these values indicated that the proposed [4Fe‐4S] cluster was not fully assembled, chemical reconstitution of the iron‐sulfur cluster was performed. The comparison of the UV/Vis absorption spectra of as‐isolated QCMT before and after iron‐sulfur cluster reconstitution suggested the improvement of iron‐sulfur cluster occupancy in the reconstituted protein (Figure S1b). Therefore, for all further experiments, the as‐isolated QCMT was reconstituted with MeCbl and iron‐sulfur clusters, unless stated otherwise. After cofactor reconstitution, QCMT was further purified by preparative SEC followed by a second iron‐sulfur cluster reconstitution in order to ensure intact [4Fe‐4S] clusters. The final, purified and fully reconstituted protein is hereinafter referred to as “as‐purified” QCMT. Iron/sulfide analyses revealed the presence of 3.1±0.6 mol iron and 4.3±0.7 mol sulfide per mol protein, which indicated the presence of a single [4Fe‐4S] cluster. The UV/Vis absorption spectrum of as‐purified QCMT (Figure 2a, b) showed absorption features around 314, 403 and 462 nm. The absorption feature at 462 nm of as‐purified QCMT suggested the presence of cob(II)alamin.[ 23 , 24 ] Therefore, after cobalamin reconstitution, the cofactor remained stably bound in the protein during the diverse steps of purification. When as‐isolated QCMT was reconstituted with hydroxocobalamin (OHCbl‐Rc QCMT) instead of MeCbl followed by SEC and reconstitution of the iron‐sulfur cluster, the as‐purified QCMT exhibited a UV/Vis absorption spectrum apparently identical to that of the MeCbl‐reconstituted version (Figure S1c). As‐purified QCMT was found to be a monomer in solution when analyzed by analytical SEC displaying a relative molecular weight of 53 500 (theoretical mass=49 070 Da for the His6‐tagged protein) (Figure S1d).
Figure 2.
Characterization of QCMT‐bound cofactors by UV/Vis and EPR spectroscopy. a) UV/Vis absorption spectra of as‐isolated QCMT, QCMT reconstituted with MeCbl, MeCbl‐reconstituted QCMT after one day of storage and as‐purified QCMT. b) Same as in panel (a) with focus on the 300–550 nm region. Wavelengths of characteristic absorption features are indicated in corresponding color for as‐isolated QCMT (410 nm), MeCbl‐Rc QCMT (303, 374 and 435 nm) and MeCbl‐Rc QCMT after 1 day (310 and 465 nm). Absorption bands of as‐purified QCMT are at 314, 403 and 462 nm. c) EPR spectra at 13 K of dithionite‐reduced (5 mM) (I) as‐purified QCMT (200 μM), (II) as‐purified QCMT (200 μM) after addition of SAM (1 mM), and (III) as‐purified QCMT (200 μM) after addition of peptide substrate (300 μM) and SAM (1 mM). The g‐values of the individual signals are indicated. For spectra II and III, g(1)‐values are marked in red and g(2)‐values in black. Other EPR conditions: microwave power, 10 mW (non‐saturating), single scan.
Characterization of the QCMT‐Bound Cofactors by EPR
Electron paramagnetic resonance (EPR) spectroscopy was used to investigate the ligation state and electronic properties of the QCMT‐bound cofactors as well as the influence of SAM and the peptide substrate (PS‐24, see below and Methods section). In order to determine the cob(II)alamin coordination and possible changes thereof in the presence of SAM and/or peptide substrate, EPR spectra were recorded at 65 K for QCMT samples containing different supplements (Figure S2). The as‐purified QCMT showed EPR signals typical of a low spin Co2+ with g ∥ at 2.006 and around 2.25. Due to the interaction of the unpaired electron with the Co2+ nucleus, the g ∥ signal is split into eight lines, which is typical for corrinoid systems. [27] The hyperfine splitting (A//,Co) amounts to 14.7 mT and lacks superhyperfine splittings from nitrogen nuclei indicating a base‐off (or His‐off) conformation. The spectra of the as‐purified QCMT and the one supplemented with peptide substrate are very similar. The addition of both SAM and SAM together with peptide substrate substantially altered the spectra in the same way. The difference mainly derives from the strongly diminished signals of the hyperfine splitting from the interaction with the cobalt nucleus, which are barely detectable. Nevertheless, the hyperfine splitting constants remain unchanged (14.7 mT). All spectra are consistent with a five‐coordinated cob(II)alamin in base‐off, His‐off conformation, potentially with a water as the fifth ligand to the cobalt.[ 27 , 28 , 29 ]
In order to characterize the QCMT‐bound iron‐sulfur cluster, EPR spectra of samples reduced with sodium dithionite were recorded at 13 K, which revealed the presence of a [4Fe‐4S] cluster (Figure 2c). The spectral features of the iron‐sulfur cluster in the different samples were identified by simulation of the experimental spectra using EasySpin (Figure S3). [30] The spectrum of reduced as‐purified QCMT exhibited rhombic symmetry with g x,y,z =1.888, 1.930 and 2.017. Addition of SAM to reduced as‐purified QCMT led to a substantial spectral change with appearance of two clearly distinguishable species. Two spectra of rhombic symmetry were detected with g(1) x,y,z =1.841, 1.927 and 2.022 and with g(2) x,y,z =1.875, 1.898 and 1.996. The overall spectrum is dominated by the signals from the first species (ratio 1.0 : 0.56). Because the intensity of the total signal of the preparation without additions was equal to the intensity of the two signals obtained with the preparation with added SAM, it is concluded that binding of SAM resulted in the presence of two populations of the same tetranuclear cluster. When SAM and the peptide substrate were added to the preparation, the spectrum changed again. However, this spectral change was due to a change of the relative proportions of the two species already observed in the sample with SAM. Now, the overall spectrum shows an increased share of the second species (ratio 1.0 : 0.81). These observations indicate that the cluster‐bound SAM might adopt two different conformations depending on the presence or absence of the peptide substrate.
One of the fascinating aspects of class B radical SAM methyltransferases is the dual role of SAM during catalysis. On the one hand SAM is used for initiation of radical catalysis, and on the other hand it serves as a methyl group donor to regenerate MeCbl. Intriguingly, the crystal structure of the cobalamin‐dependent radical SAM enzyme OxsB revealed two different SAM binding modes. [31] One of the orientations corresponds to the classical binding mode of SAM observed in almost all radical SAM enzymes, which allows for reductive cleavage of SAM. In the second orientation, the sulfur atom of SAM is shifted in such a way that the attached methyl group is positioned to allow for methylation of cobalamin. Two distinct SAM binding modes were also assumed in the mechanistic proposal for the cobalamin‐dependent radical SAM methyltransferase CysS. [32] In the case of QCMT, the EPR spectroscopic data of the dithionite‐reduced enzyme also suggest the presence of two different SAM binding modes. Thus, the two distinct species of the [4Fe‐4S]1+ cluster observed by EPR after addition of SAM might reflect these two different binding modes. While in the absence of the substrate the “methylation conformation” might dominate, the addition of the substrate might induce the “radical SAM conformation” of the SAM molecule (Figure S4). A substrate‐induced stabilization of the “radical SAM conformation” of SAM was also observed in the crystal structure of pyruvate formate‐lyase activating enzyme (PFL‐AE), [33] although in this case there is no need for a “methylation conformation”. An alternative interpretation of the EPR spectra takes into account that SAM is cleaved to a certain extent in the EPR sample containing dithionite, SAM and peptide substrate (not shown, but see also below). As a consequence, DOA (≈200 μM) and methionine are formed and the cluster is oxidized, but immediately reduced again due to the excess of dithionite in the sample. In this scenario, DOA and methionine might remain bound within the active site with methionine still acting as a cluster ligand giving rise to one of the cluster species observed by EPR. The crystal structure of the cobalamin‐dependent radical SAM methyltransferase TokK shows the binding geometry of methionine and DOA to the [4Fe‐4S] cluster (Figure S4). [34] However, we consider this explanation for the occurrence of two cluster species as less likely, since both species are also observed for the EPR sample without peptide substrate (spectrum II), in which no SAM cleavage takes place (not shown, but see also below).
QCMT Methylates Glutamine in MCR‐Derived Peptides
In order to test our hypothesis that QCMT is one of the methyltransferases modifying the M. thermophilus MCR protein, we established an in vitro enzyme activity assay using two peptide substrates with different lengths derived from the M. thermophilus MCR α subunit. The peptide QS‐28 comprises 28 amino acids (QFPTMMEDHFGGSQRAGVIAAASGLSTS), a subsection of which is contained in the shorter 24mer PS‐24 (PTMMEDHFGGSQRAGVIAAASGLS). Both potential substrates include the glutamine residue corresponding to the methylated residue previously identified in MCR I of M. thermophilus by mass spectrometry. [6] The initial in vitro reaction mixtures contained as‐purified QCMT, peptide QS‐28, SAM, and sodium dithionite as a reducing agent. After incubation of the mixture under anaerobic conditions, the formation of DOA was observed by HPLC analysis. However, the detected amounts of SAH were not substantially above those of the negative control lacking the enzyme (Figure S5a). Mass spectrometric (MS) analyses also did not show evidence of QS‐28 peptide methylation (Figure S5b and S5c). One possible reason for the lack of peptide methylation by the dithionite‐reduced enzyme might be the formation of cobalamin species with SO2 − coordinated to the cobalt ion preventing methyl group transfer from SAM to cobalamin. [35]
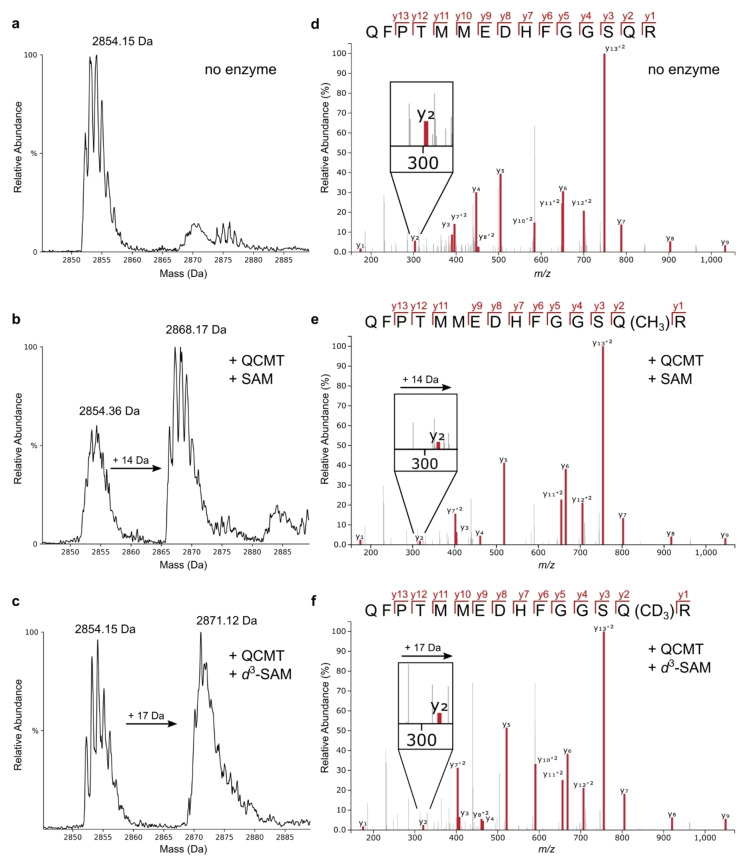
Therefore, in subsequent assays, we used titanium(III) citrate instead of sodium dithionite as reducing agent in analogy to other class B radical SAM methyl‐transferases.[ 12 , 36 ] Using the new conditions, formation of both co‐products, DOA and SAH, was detected by HPLC analysis (see below), and the LC‐MS analysis revealed a +14 Da shift of the QS‐28 peptide, consistent with enzymatic methylation of the substrate (Figure 3b). In contrast, for reaction mixtures lacking QCMT only the unmodified peptide was observed at 2854.15 Da (Figure 3a). To confirm these assignments, we developed a method to expose QCMT to deuterium‐labeled d 3‐SAM. This was achieved by replacing SAM in the assay mixture with an enzymatic SAM generation system consisting of methionine adenosyltransferase (MAT), ATP and d 3‐methionine. The results of this isotopic labeling assay confirm that deuterium‐labeled methyl groups are provided to the QCMT enzyme, resulting in an increased mass shift of +17 Da for the peptide substrate (Figure 3c). When the QCMT activity assay was performed with peptide PS‐24, methylation and d 3‐methylation were similarly observed when incubated with SAM and d 3‐SAM, respectively (Figure S6).
Figure 3.
Deconvoluted EMS LC‐MS spectra of peptide QFPTMMEDHFGGSQRAGVIAAASGLSTS after QCMT in vitro assay (a)–(c) and EPI MS/MS spectra of peptide QFPTMMEDHFGGSQRAGVIAAASGLSTS after purification from QCMT assay and trypsinolysis, resulting in peptide QFPTMMEDHFGGSQR as precursor ion (d)–(f). a) EMS‐spectrum obtained from an assay mixture without QCMT. b) EMS‐spectrum obtained from an assay mixture with QCMT and SAM showing a mass shift of +14 Da. c) MS‐spectrum obtained from an assay mixture with QCMT and d 3‐SAM showing a mass shift of +17 Da. d) MS/MS spectrum of unmodified peptide from an assay without QCMT. e) MS/MS spectrum of enzymatically methylated peptide. f) MS/MS spectrum of enzymatically d 3‐methylated peptide. Data in (e) and (f) are consistent with methylation occuring on Q14 in peptide QS‐28.
To determine which amino acid within the peptide substrate was methylated by QCMT, MS/MS fragmentation analysis was performed after tryptic digestion of the HPLC‐purified QS‐28 peptide. The resulting shorter length fragment (QR‐15), which had better fragmentation characteristics than the full‐length peptide, was then analyzed via ion‐trap supported MS/MS fragmentation in enhanced product ion scan mode. Trypsin digestion resulted in a dominant [M+3 H]3+ precursor peak of the QR‐15 peptide, with m/z values consistent with the methylation and d 3‐methylation being present within QR‐15 (Figure S7). The EPI‐MS/MS fragmentation spectra showed a complete y‐ion series from y1 to y13 of the tryptic QR‐15 peptide (Figure 3d, f), which corresponds to the LC‐MS peptide substrate spectra (Figure 3a–c). Upon methylation, all y‐ions except for y1 are observed with a +14 Da mass shift, consistent with methylation at the glutamine on position 14 of the QR‐15 peptide. Because the intensity of peaks y1 to y3 was relatively low, the MS/MS fragmentation analysis was also performed with the peptide resulting from assays with d 3‐labeled SAM (Figure 3f). Consistently, the y1 peak remained unmodified whereas y2 and all subsequent y‐ion series peaks showed a mass shift of +17 Da. Therefore, MS/MS analysis showed that QCMT catalyzes the site‐specific methylation at Q14 of the QS‐28 peptide substrate, corresponding to Q418 in the MCR α subunit from M. thermophilus.
To determine whether QCMT‐catalyzed glutamine methylation using a peptide substrate in vitro indeed occurs at the Cα‐position, we performed LC‐MS analysis of the amino acids resulting from complete peptide hydrolysis; conditions in which glutamine is converted to glutamate. In the presence of QCMT, the glutamate peak decreased concomitant with a new peak with an altered retention time that co‐elutes with a synthetic standard of 2‐methylglutamate (Figures 4 and S8). These results are consistent with 2‐methylglutamine as the product of QCMT‐catalyzed methylation.
Figure 4.
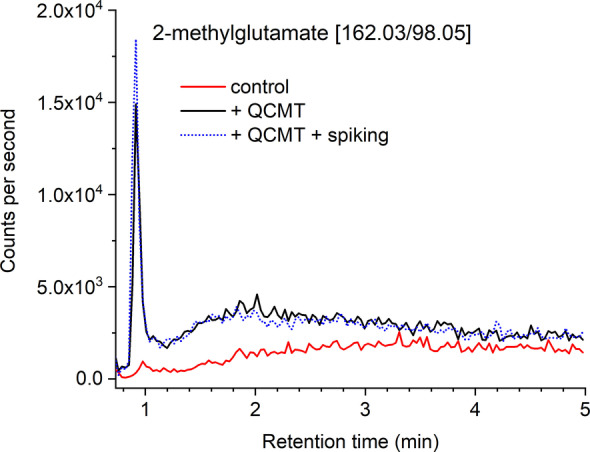
LC‐MS chromatogram showing the 2‐methylglutamate MRM trace in acid‐hydrolyzed QS‐28 peptides. In the presence of QCMT, a new peak was identified as 2‐methylglutamate as confirmed by spiking with a synthetic standard and corresponding retention time changes (see Figure S8). Note that under acid hydrolysis glutamine is converted to glutamate.
With these results we have identified the glutamine C‐methyltransferase responsible for the posttranslational Cα‐methylation of glutamine in MCR. Several crystal structures of MCRs from different methanogens carrying the methylglutamine modification show that the methyl group is introduced at the Cα position with retention of configuration of Cα (2‐(S)‐methylglutamine).[ 3 , 7 , 8 , 37 ] In contrast, for most of the cobalamin‐dependent radical SAM methyltransferases characterized so far, the methylation reactions for non‐proteinogenic substrates proceed with inversion of configuration, except for GenK, which was reported to catalyze its reaction under retention of configuration. [38] The arrangement of cofactors and substrates within the active sites of QCMT and GenK that leads to retention of configuration remains to be elucidated through structure determination.
QCMT Produces DOA and SAH in a 1 : 1 Ratio during Turnover
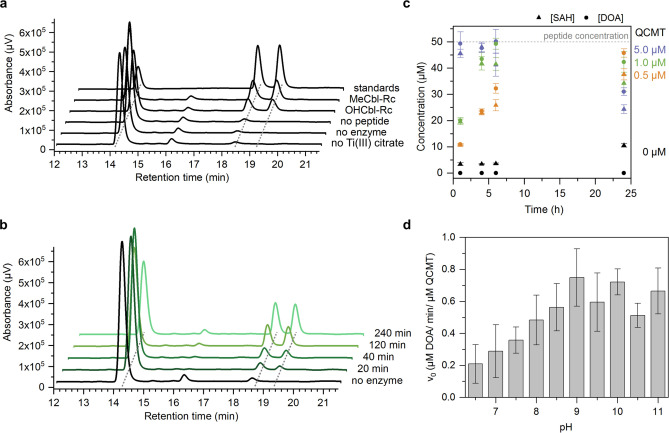
After the demonstration that the putative QCMT is indeed able to catalyze the methylation of glutamine, the formation of DOA and SAH as co‐products of the reaction was investigated in more detail by HPLC analysis of the reaction mixtures. As already mentioned above, full activity with production of both, DOA (retention time 19.2 min) and SAH (retention time 18.4 min), was observed only in the presence of the strong reductant titanium(III) citrate (Figure 5a). Furthermore, only completely reconstituted as‐purified QCMT exhibited full activity, while production of DOA and SAH by as‐isolated or MeCbl‐Rc QCMT before iron‐sulfur cluster reconstitution was very low (Figure S9). As‐purified QCMT reconstituted with either MeCbl or OHCbl were equally active (Figure 5a). Supplementation of the assay mixture with additional MeCbl was not required in order to obtain full activity. Control reactions lacking either titanium(III) citrate, peptide substrate or as‐purified QCMT did not show substantial DOA or SAH production above background levels (Figure 5a). The observation that the reductive cleavage of SAM by the reduced enzyme only takes place in the presence of the peptide substrate supports the idea of substrate‐induced conformational changes of the QCMT‐bound SAM as discussed above. For PFL‐AE, it was suggested that SAM adopts a stable catalytic conformation only in the presence of the substrate in order to avoid any unproductive SAM cleavage, [33] which apparently also applies for QCMT.
Figure 5.
Analysis of DOA and SAH formation during the QCMT‐catalyzed methylation reaction. a) HPLC chromatograms of standard solutions and in vitro activity assay mixtures. From top to bottom: standard solution with SAM (R t=14.1 min), SAH (R t=18.4 min) and DOA (R t=19.2 min), standard activity assay mixture with as‐purified QCMT (MeCbl‐Rc), with as‐purified QCMT (OHCbl‐Rc), standard activity assay mixture without peptide, without enzyme and without titanium(III) citrate. b) HPLC chromatograms of a standard activity assay mixture (1 μM QCMT, 200 μM SAM, 50 μM peptide, 1 mM titanium(III) citrate) showing the formation of DOA and SAH at different time points. c) Formation of DOA and SAH at different time points depending on the concentration of QCMT. Shown are mean values with standard deviation of n=3 independent activity assays with the same enzyme preparation. The SAH concentration detected in the negative control (0 μM QCMT) was subtracted from the SAH concentrations measured in the presence of 0.5, 1.0 and 5.0 μM QCMT, respectively. The decrease of DOA and SAH during prolonged incubation for 24 h in the assays containing either 5 μM or 1 μM enzyme might be due to minor enzyme impurities (such as MTA/SAH nucleosidase) or chemical degradation, which was not further investigated. d) pH optimum of QCMT activity. Shown are mean values with standard deviation of n=3–4 enzyme preparations. With each enzyme preparation the activity assay was repeated three times.
In order to quantify the final amounts of DOA and SAH and to determine the ratio in which these co‐products are formed, assay mixtures containing different amounts of enzyme (0, 0.5, 1 and 5 μM), but identical amounts of SAM (200 μM) and peptide (50 μM) were analyzed by HPLC at different time points (Figure 5b). The formation of DOA and SAH was then quantified for the different assay mixtures after 1, 4, 6 and 24 h of incubation (Figure 5c). In each case, about equal amounts of DOA and SAH were produced throughout the time‐course. The reactions seemed to be completed after 1 h with 5 μM enzyme, after 6 h with 1 μM enzyme and after 24 h with 0.5 μM enzyme (Figure 5c). The average final amounts of the co‐products calculated from these three time points are 48.2±2.1 μM DOA and 41.6±4.0 μM SAH. Therefore, during the methylation reaction catalyzed by QCMT, DOA and SAH are formed in a 1 : 1 ratio with respect to each other. Since peptide methylation was nearly complete after the mentioned reaction time points (Figure S10), SAH and DOA are also formed in a 1 : 1 ratio with respect to the peptide substrate. Moreover, the activity assay with 0.5 μM enzyme shows that QCMT is capable of almost 100 turnovers under the employed in vitro assay conditions.
In order to obtain insights about ionizable amino acid residues potentially involved in QCMT catalysis and/or substrate binding, the pH‐optimum for QCMT activity was determined. For this purpose, the initial velocity v 0 of DOA formation was measured in different buffer systems from pH 6.5 to pH 11.0 (Figure 5d). The highest velocity was observed at pH 9.0, at which a specific activity of 0.75±0.18 μM DOA/min/μM QCMT was determined. Even at pH values above 9.0 (pH 9.5–11.0) the QCMT activity remained high varying around 0.62 μM DOA/min/μM QCMT. The alkaline pH‐optimum of QCMT indicates that either a lysine or arginine residue might be involved in catalysis and/or substrate binding. Future studies using appropriate enzyme variants will shed further light on the identity of catalytically important amino acid residues.
QCMT‐Catalyzed Methylation Proceeds via MeCbl
Finally, in order to follow the path of QCMT‐catalyzed methyl group transfer, additional UV/Vis absorption spectra were recorded during a single turnover experiment. When as‐purified, cob(II)alamin‐containing QCMT was treated with the strong reductant titanium(III) citrate, the UV/Vis absorption spectrum of the enzyme changed and a sharp absorption maximum at 389 nm appeared together with a minor maximum around 543 nm and a shoulder around 462 nm (Figure 6a). These features are characteristic for cob(I)alamin.[ 22 , 24 , 25 ] Next, a twofold excess of SAM was added to the reduced QCMT, again resulting in an altered UV/Vis absorption spectrum. Now, the spectrum exhibited maxima around 374 and 435 nm (Figure 6a) similar to the spectrum of MeCbl‐Rc QCMT measured immediately after reconstitution (see above and Figure 2a, b) indicating that methyl group transfer from SAM to cob(I)alamin yielding MeCbl had taken place. The methyl group transfer from SAM to cob(I)alamin was confirmed by HPLC analysis, which revealed the formation of SAH (Figure S11a). Finally, after addition of an equimolar amount of peptide QS‐28, the spectrum changed again, now exhibiting similar features to that of as‐purified QCMT (Figure 6a). This observation suggests that the methyl group was transferred from MeCbl to the substrate with concomitant formation of cob(II)alamin. Indeed, HPLC analysis revealed the formation of DOA upon addition of substrate, and MS analysis demonstrated that the peptide was methylated (Figure S11a). In contrast, no change of the UV/Vis spectrum of MeCbl, no DOA formation and no peptide methylation were observed in a control reaction containing a different peptide not related to the substrate (Figure S11b). Finally, in a single turnover experiment that was conducted as described above, but without addition of SAM, cob(I)alamin was formed, but no MeCbl, SAH, DOA or methylated peptide were detected (Figure S11c). When as‐purified QCMT was reduced with sodium dithionite, an absorption maximum at about 386 nm appeared, again suggesting the reduction of cob(II)alamin to cob(I)alamin. However, in this case, the addition of SAM did not result in a further change of the UV/Vis absorption spectrum (Figure S5d).
Figure 6.
Characterization of QCMT‐bound cobalamin. a) UV/Vis absorption spectra of 20 μM as‐purified QCMT, 20 μM as‐purified QCMT treated with 80 μM titanium(III) citrate, 20 μM as‐purified QCMT treated with 80 μM titanium(III) citrate and 40 μM SAM, 20 μM as‐purified QCMT treated with 80 μM titanium(III) citrate, 40 μM SAM and 20 μM peptide substrate. Characteristic absorption bands of the titanium(III) citrate reduced QCMT are indicated. b) QCMT contains MeCbl in the presence of SAM under reducing conditions. LC‐MS/MS spectrum showing all precursor ions that fragment to the mass of unmethylated cobalamin (665.3 m/z, [M+2 H]2+). QCMT‐bound MeCbl was detected after addition of titanium(III) citrate and excess SAM from an enzymatic regeneration system. c) Isotopic labeling shows that QCMT‐bound cobalamin is methylated from externally supplied SAM. LC‐MS/MS spectrum showing all precursor ions that fragment to the mass of unmethylated cobalamin (665.3 m/z, [M+2 H]2+). In the presence of externally supplied d 3‐SAM produced from d 3‐methionine, QCMT‐bound d 3‐MeCbl was detected, as evidenced by a mass shift of +1.5 Da for the [M+2 H]2+ peak.
In addition to the described UV/Vis, HPLC and MS analyses under single turnover conditions, the cobalamin species bound to the enzyme under different assay conditions were characterized by mass spectrometry. In these experiments, an excess of SAM, which was enzymatically synthesized in situ, was used. The enzyme‐bound cobalamin was isolated using a C4 reversed phase HPLC column under denaturing conditions, followed by precursor ion MS/MS. For as‐purified QCMT the cobalamin cofactor exhibited an m/z of 665.3 Da corresponding to the [M+2 H]2+ ion of cobalamin. In contrast, after reduction of as‐purified QCMT with titanium(III) citrate and supply of SAM, enzyme‐bound MeCbl with an m/z of 672.9 Da for the [M+2 H]2+ ion was detected irrespective of the presence of the peptide substrate (Figure 6b). When d 3‐Met, ATP and MAT were used as SAM supply system, the formation of d 3‐MeCbl with an m/z of 674.4 Da for the [M+2 H]2+ ion was observed (Figure 6c). Therefore, the reduced cob(I)alamin is methylated by externally provided SAM yielding enzyme‐bound MeCbl as also observed by UV/Vis and HPLC. Altogether, the experiments described above show that the enzyme‐bound cob(II)alamin is reduced to cob(I)alamin in the presence of titanium(III) citrate. Cob(I)alamin is then methylated to methyl‐cob(III)alamin by the addition of SAM, which goes along with the formation of SAH as a co‐product. Finally, the subsequent addition of the peptide substrate leads to methylation of the substrate with concomitant formation of cob(II)alamin. Substrate methylation is initiated by SAM cleavage and radical catalysis yielding DOA as the second co‐product. The QCMT‐catalyzed transfer of the methyl group from SAM to cobalamin to the peptide substrate with concomitant formation of SAH and DOA clearly classifies the enzyme as following a typical class B radical SAM methyltransferase mechanism (Figure 1c).
Based on the observed UV/Vis absorption spectra of QCMT‐bound MeCbl (Figures 2 and 6) and the EPR spectra of cob(II)alamin (Figure S2) as well as by comparison of the spectral data with previously reported data of the corrinoid/iron‐sulfur protein (CoFeSP)[ 22 , 24 , 25 ] and several variants of methionine synthase (MetH variant H759G [23] and variant I690C/G743C [29] ), we conclude that the cobalamin cofactor of QCMT is bound in a base‐off, His‐off conformation. This cobalamin conformation was also reported for the enzyme TsrM, [39] which is annotated as a class B radical SAM methyltransferase, but does not employ radical chemistry.[ 39 , 40 ] In all these enzymes (CoFeSP, MetH, TsrM), the cobalamin cofactor cycles between the methyl‐cob(III)alamin and cob(I)alamin forms during turnover. However, sporadically the cob(I)alamin is oxidized to catalytically inactive cob(II)alamin and, thus, requires reductive reactivation. In this context, it was reported, that the base‐off, His‐off conformation facilitates the reduction of cob(II)alamin to cob(I)alamin by about 100 mV. [41] Class B radical SAM methyltransferases employing the radical mechanism shown in Figure 1c must reduce the cob(II)alamin intermediate after each substrate methylation during turnover. Thus, for these enzymes the base‐off, His‐off conformation appears to be an absolute requirement in order to facilitate cob(II)alamin reduction under physiological conditions. In agreement with this, all class B radical SAM methyltransferases including QCMT lack the “classical” DXHXXG motif, which indicates the replacement of the dimethylbenzimidazole by a histidine ligand (base‐off, His‐on) in cobalamin‐binding proteins. [42] Moreover, the crystal structures of TokK, [34] Mmp10 [14] and OxsB [31] revealed the presence of the cobalamin cofactor bound in base‐off, His‐off conformation supporting our proposal of a similar cobalamin binding mode within QCMT. However, details in the cobalamin coordination, such as coordinated water or more distant amino acid residues at the lower axial position vary for each enzyme and are potentially tailored to meet the requirements of the catalyzed reaction. [43]
Conclusion
In this study, we identified and characterized the previously unknown methyltransferase responsible for the unique Cα‐methylation of a glutamine residue in MCR. QCMT contains a cobalamin cofactor and one [4Fe‐4S] cluster, which is the canonical radical SAM cluster, as essential prosthetic groups. The cobalamin cofactor cycles between three different states: methyl‐cob(III)alamin, cob(II)alamin and cob(I)alamin. The QCMT‐bound cob(II)alamin is reduced to cob(I)alamin, which is then methylated to methyl‐cob(III)alamin by the addition of SAM. Finally, the peptide substrate is methylated site‐specifically at glutamine and regio‐specifically at Cα with concomitant formation of cob(II)alamin. During turnover, DOA and SAH are formed as co‐products in a 1 : 1 ratio. Altogether, these results clearly classify the enzyme as following a typical class B radical SAM methyltransferase mechanism. The characterization of the so far unknown QCMT responsible for Cα glutamine methylation adds a new member to the fascinating family of cobalamin‐dependent radical SAM methyltransferases and provides a further step toward recombinant production of MCR carrying its natural amino acid modifications in non‐methanogenic hosts such as E. coli.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work was funded by the Deutsche Forschungsgemeinschaft (grant 235777276/GRK 1976 to G.L., J.N.A. and T.F.), the German Scholars Organization and Carl Zeiss Foundation (GSO/CZS 20 to C.L.) and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (Grant agreement No. 716966, J.N.A.). We thankfully acknowledge Dr. Dipali Mhaindarkar for initial experiments and Katharina Strack for skillful technical assistance. We thank Prof. Dr. Squire J. Booker for the kind gift of plasmid pBAD42‐BtuCEDFB. [21] Open Access funding enabled and organized by Projekt DEAL.
J. Gagsteiger, S. Jahn, L. Heidinger, L. Gericke, J. N. Andexer, T. Friedrich, C. Loenarz, G. Layer, Angew. Chem. Int. Ed. 2022, 61, e202204198; Angew. Chem. 2022, 134, e202204198.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Thauer R. K., Kaster A.-K., Seedorf H., Buckel W., Hedderich R., Nat. Rev. Microbiol. 2008, 6, 579. [DOI] [PubMed] [Google Scholar]
- 2.
- 2a. Ellefson W. L., Wolfe R. S., J. Biol. Chem. 1981, 256, 4259; [PubMed] [Google Scholar]
- 2b. Thauer R. K., Biochemistry 2019, 58, 5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ermler U., Grabarse W., Shima S., Goubeaud M., Thauer R. K., Science 1997, 278, 1457. [DOI] [PubMed] [Google Scholar]
- 4. Selmer T., Kahnt J., Goubeaud M., Shima S., Grabarse W., Ermler U., Thauer R. K., J. Biol. Chem. 2000, 275, 3755. [DOI] [PubMed] [Google Scholar]
- 5. Grabarse W., Mahlert F., Shima S., Thauer R. K., Ermler U., J. Mol. Biol. 2000, 303, 329. [DOI] [PubMed] [Google Scholar]
- 6. Kahnt J., Buchenau B., Mahlert F., Krüger M., Shima S., Thauer R. K., FEBS J. 2007, 274, 4913. [DOI] [PubMed] [Google Scholar]
- 7. Wagner T., Kahnt J., Ermler U., Shima S., Angew. Chem. Int. Ed. 2016, 55, 10630; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 10788. [Google Scholar]
- 8. Wagner T., Wegner C.-E., Kahnt J., Ermler U., Shima S., J. Bacteriol. 2017, 199, e00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a. Nayak D. D., Mahanta N., Mitchell D. A., Metcalf W. W., eLife 2017, 6, e29218; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b. Mahanta N., Liu A., Dong S., Nair S. K., Mitchell D. A., Proc. Natl. Acad. Sci. USA 2018, 115, 3030; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9c. Liu A., Si Y., Dong S.-H., Mahanta N., Penkala H. N., Nair S. K., Mitchell D. A., Nat. Chem. Biol. 2021, 17, 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nayak D. D., Liu A., Agrawal N., Rodriguez-Carerro R., Dong S.-H., Mitchell D. A., Nair S. K., Metcalf W. W., PLoS Biol. 2020, 18, e3000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deobald D., Adrian L., Schöne C., Rother M., Layer G., Sci. Rep. 2018, 8, 7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Radle M. I., Miller D. V., Laremore T. N., Booker S. J., J. Biol. Chem. 2019, 294, 11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lyu Z., Shao N., Chou C.-W., Shi H., Patel R., Duin E. C., Whitman W. B., J. Bacteriol. 2020, 202, e00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fyfe C. D., Bernardo-García N., Fradale L., Grimaldi S., Guillot A., Brewee C., Chavas L. M. G., Legrand P., Benjdia A., Berteau O., Nature 2022, 602, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kurth J. M., Müller M.-C., Welte C. U., Wagner T., Microorganisms 2021, 9, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buckel W., Thauer R. K., Angew. Chem. Int. Ed. 2011, 50, 10492; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011, 123, 10676. [Google Scholar]
- 17. Sofia H. J., Chen G., Hetzler B. G., Reyes-Spindola J. F., Miller N. E., Nucleic Acids Res. 2001, 29, 1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Broderick J. B., Duffus B. R., Duschene K. S., Shepard E. M., Chem. Rev. 2014, 114, 4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Q., van der Donk W. A., Liu W., Acc. Chem. Res. 2012, 45, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bauerle M. R., Schwalm E. L., Booker S. J., J. Biol. Chem. 2015, 290, 3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lanz N. D., Blaszczyk A. J., McCarthy E. L., Wang B., Wang R. X., Jones B. S., Booker S. J., Biochemistry 2018, 57, 1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ragsdale S. W., Lindahl P. A., Münck E., J. Biol. Chem. 1987, 262, 14289. [PubMed] [Google Scholar]
- 23. Jarrett J. T., Amaratunga M., Drennan C. L., Scholten J. D., Sands R. H., Ludwig M. L., Matthews R. G., Biochemistry 1996, 35, 2464. [DOI] [PubMed] [Google Scholar]
- 24. Goetzl S., Jeoung J.-H., Hennig S. E., Dobbek H., J. Mol. Biol. 2011, 411, 96. [DOI] [PubMed] [Google Scholar]
- 25. Kung Y., Ando N., Doukov T. I., Blasiak L. C., Bender G., Seravalli J., Ragsdale S. W., Drennan C. L., Nature 2012, 484, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schrauzer G. N., Lee L. P., Sibert J. W., J. Am. Chem. Soc. 1970, 92, 2997. [DOI] [PubMed] [Google Scholar]
- 27. Bayston J. H., Looney F. D., Pilbrow J. R., Winfield M. E., Biochemistry 1970, 9, 2164. [DOI] [PubMed] [Google Scholar]
- 28. Liptak M. D., Fleischhacker A. S., Matthews R. G., Telser J., Brunold T. C., J. Phys. Chem. B 2009, 113, 5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liptak M. D., Datta S., Matthews R. G., Brunold T. C., J. Am. Chem. Soc. 2008, 130, 16374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stoll S., Schweiger A., J. Magn. Reson. 2006, 178, 42. [DOI] [PubMed] [Google Scholar]
- 31. Bridwell-Rabb J., Zhong A., Sun H. G., Drennan C. L., Liu H.-w., Nature 2017, 544, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.
- 32a. Wang Y., Schnell B., Baumann S., Müller R., Begley T. P., J. Am. Chem. Soc. 2017, 139, 1742; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32b. Wang Y., Begley T. P., J. Am. Chem. Soc. 2020, 142, 9944. [DOI] [PubMed] [Google Scholar]
- 33. Vey J. L., Yang J., Li M., Broderick W. E., Broderick J. B., Drennan C. L., Proc. Natl. Acad. Sci. USA 2008, 105, 16137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Knox H. L., Sinner E. K., Townsend C. A., Boal A. K., Booker S. J., Nature 2022, 602, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salnikov D. S., Silaghi-Dumitrescu R., Makarov S. V., van E. R., Boss G. R., Dalton Trans. 2011, 40, 9831. [DOI] [PubMed] [Google Scholar]
- 36.
- 36a. Parent A., Guillot A., Benjdia A., Chartier G., Leprince J., Berteau O., J. Am. Chem. Soc. 2016, 138, 15515; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36b. Wang B., Blaszczyk A. J., Knox H. L., Zhou S., Blaesi E. J., Krebs C., Wang R. X., Booker S. J., Biochemistry 2018, 57, 4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grabarse W., Mahlert F., Duin E. C., Goubeaud M., Shima S., Thauer R. K., Lamzin V., Ermler U., J. Mol. Biol. 2001, 309, 315. [DOI] [PubMed] [Google Scholar]
- 38. Kim H. J., Liu Y.-n., McCarty R. M., Liu H.-w., J. Am. Chem. Soc. 2017, 139, 16084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blaszczyk A. J., Silakov A., Zhang B., Maiocco S. J., Lanz N. D., Kelly W. L., Elliott S. J., Krebs C., Booker S. J., J. Am. Chem. Soc. 2016, 138, 3416. [DOI] [PubMed] [Google Scholar]
- 40. Knox H. L., Chen P. Y., Blaszczyk A. J., Mukherjee A., Grove T. L., Schwalm E. L., Wang B., Drennan C. L., Booker S. J., Nat. Chem. Biol. 2021, 17, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harder S. R., Lu W. P., Feinberg B. A., Ragsdale S. W., Biochemistry 1989, 28, 9080. [DOI] [PubMed] [Google Scholar]
- 42. Marsh E. N. G., Holloway D. E., FEBS Lett. 1992, 310, 167. [DOI] [PubMed] [Google Scholar]
- 43. Bridwell-Rabb J., Li B., Drennan C. L., ACS Bio. Med. Chem. Au 2022, 10.1021/acsbiomedchemau.1c00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.