Abstract
Cofactors are required for almost half of all enzyme reactions, but their functions and binding partners are not fully understood even after decades of research. Functionalised cofactor mimics that bind in place of the unmodified cofactor can provide answers, as well as expand the scope of cofactor activity. Through chemical proteomics approaches such as activity‐based protein profiling, the interactome and localisation of the native cofactor in its physiological environment can be deciphered and previously uncharacterised proteins annotated. Furthermore, cofactors that supply functional groups to substrate biomolecules can be hijacked by mimics to site‐specifically label targets and unravel the complex biology of post‐translational protein modification. The diverse activity of cofactors has inspired the design of mimics for use as inhibitors, antibiotic therapeutics, and chemo‐ and biosensors, and cofactor conjugates have enabled the generation of novel enzymes and artificial DNAzymes.
Keywords: Chemical Probes, Cofactors, Photoaffinity Labelling, Protein Modifications, Proteomics
Cofactors are essential, but their functions and binding partners are not fully understood even after decades of research. Functionalised cofactor mimics that bind in place of the unmodified cofactor can provide answers, as well as expand the scope of cofactor activity. This Review outlines the growing use of cofactor mimics in interactome discovery, drug development, the study of post‐translational modifications, and other novel biotechnologies.

1. Introduction
Cofactors are essential small molecules or metal ions bound by enzymes to assist catalysis and function. Nearly half of all enzyme reactions are cofactor‐dependent. [1] Cofactors may be tightly bound (prosthetic groups) or loosely associated (coenzymes). Typically, cofactors are functionally diverse and capable of a range of roles depending on the enzyme they are bound to and the substrates available. Cofactors are critically important for the biological function of proteins, with roles including group transfer, catalysis, and redox chemistry. Their catalytically active forms are then regenerated in situ or via subsequent recycling. Although the subject of study for decades, the scope of cofactor functions, binding modes, and binding proteins is not fully understood. [2] In silico prediction can reveal cofactor binding proteins based on sequence and structure similarity to known binding proteins. [3] However, the high structural and functional diversity of cofactor binding proteins remains a challenge for bioinformatic prediction. Moreover, in silico methods require experimental validation, for which a high‐throughput solution is desired.
Functionalised cofactor mimics that bind in place of the unmodified cofactor are increasingly used to address these unanswered questions. The cofactor biotin could be considered the archetypal case here since it has been used countless times to functionalise other cofactors and small molecules as a purification tag, owing to its exceptional affinity to avidin.
The introduction of a small clickable handle (e.g. alkyne or azide) to the cofactor structure can often be tolerated by cofactor binding proteins, and enable the expansion of its activity. Incorporation of photoaffinity groups can be used to capture noncovalently bound interacting partners upon irradiation. Subsequent bioorthogonal attachment of affinity tags such as biotin to click‐reactive handles can allow for enrichment of cofactor binding or target proteins. Enriched proteins can then be identified by mass spectrometry, and proteins of unknown function annotated with cofactor binding after confirmatory validation. In this way, functionalised cofactor mimics can be used to profile a cofactor's interactome (Section 2).
Additionally, cofactors that behave as co‐substrates to supply functional groups to substrate biomolecules can be hijacked by mimics which transfer enrichment tags, click handles, or photoaffinity groups which in turn can be used to reveal those substrates. By this strategy, functionalised cofactor mimics can be used to discover the substrates of cofactor‐binding enzymes and thereby study post‐translational modifications like acetylation, methylation, AMPylation, and PARylation (Section 3).
Synthetic cofactors have also been exploited beyond the field of chemical proteomics profiling, including in myriad successful drug discovery and biotechnology applications. For example, cofactor mimics have been functionalised with fluorophores for inhibitor development, real‐time cellular imaging, or biochemical sensors of the cellular environment (Section 4). Artificial cofactor mimics have also been developed to expand their intrinsic functionality, resulting in the development of artificial enzymes including electroenzymes and DNAzymes (Section 5).
This Review covers synthetic cofactors modified with moieties that extend their functionality, such as click chemistry handles, photoaffinity labels, fluorophores, and photosensitisers. Although cofactors conjugated to solid supports, electrodes, or nanoparticles have been described e.g. for PQQ, [4] FAD, [5] heme,[ 6 , 7 ] and coenzyme A, [8] they are not the focus of this Review. In addition, synthetic cofactor analogues that do not incorporate additional functionality, such as those developed as inhibitors for medicinal chemistry or biochemical studies (see Table 1, for examples) are not discussed. Meanwhile, the reader is directed elsewhere[ 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 ] for discussion of (nonfunctionalised) cofactor analogues employed to generate artificial metalloenzyme or biohybrid catalysts with improved stability, efficiency, chemoselectivity, or reactivity scope.
Table 1.
Summary table. Examples of functionalised cofactor mimics and cofactor‐based inhibitors developed for chemical biology or medicinal chemistry applications.
|
Cofactor |
Examples of functionalised cofactor mimics |
Biomimetic inhibitor, (indication, target) |
|---|---|---|
|
Adenosine triphosphate (ATP) |
Various biotinylated,[ 27 , 28 ] alkynylated, [63] and photoaffinity‐functionalised [53] probes. Various fluorophore probes. [93] |
|
|
Biotin |
Alkyne‐ and photoaffinity‐functionalised probes [47] |
|
|
Cobalamin and methylcobalamin (vitamin B12) |
Alkyne‐, azide‐, and biotin‐functionalised probes, [134] alkyne‐ and photoaffinity‐functionalised probes, [48] fluorophore probe, [92] protoporphyrin IX conjugate, [125] ampicillin conjugate, [126] antisense RNA oligonucleotide conjugate [128] |
|
|
Coenzyme A (panthothenic acid) |
Various biotin‐, [44] alkyne‐, [88] azide‐, [89] and photoaffinity‐functionalised [43] probes. Various fluorophore probes. [97] |
|
|
Flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) |
Azidoflavins[ 41 , 42 ] and alkyne‐ and photoaffinity‐functionalised probes, [47] redox sensitiser conjugate, [117] cofactor conjugate [121] |
|
|
Glutathione |
Alkyne‐ and photoaffinity‐functionalised probes [45] |
TLK199/ezatiostat hydrochloride/Telintra[ 131 , 132 , 133 ] (myelodysplastic syndromes, glutathione S‐transferase 1) |
|
Heme |
Fluorescent analogues,[ 109 , 110 , 135 ] alkyne derivatives,[ 136 , 137 ] redox sensitiser conjugate, [118] cofactor conjugates,[ 121 , 125 ] G‐quadruplex conjugates[ 122 , 123 , 124 ] |
Non‐iron porphyrins (bacterial/protozoal parasite infection [138] and hyperbilirubinemia) |
|
Lipoamide |
Alkyne‐ and photoaffinity‐functionalised probes [51] |
|
|
Menaquinone (vitamin K) |
Fluorogenic redox sensor analogue [116] |
Ref. [139] (SXR‐mediated transcriptional activity) |
|
Molybdopterin |
|
Ref. [140] (cofactor deficiency) |
|
NAD+ and NADP+ |
Various biotin‐, [82] alkyne‐, [78] azide‐ [84] and photoaffinity‐functionalised probes.[ 86 , 87 ] Various fluorophore probes[ 83 , 100 , 104 , 105 , 106 , 107 , 108 ] |
Ref. [141] NCB10 (cardiovascular disease, S‐adenosyl‐l‐homocysteine hydrolase) |
|
Pyridoxal phosphate (PLP) |
Alkyne and electrophilic trap probes,[ 21 , 25 , 35 ] fluorescent chemosensor analogues[ 111 , 112 , 113 ] |
|
|
S‐Adenosyl methionine (SAM) |
Various alkyne‐, [75] azide‐ [72] and photoaffinity‐functionalised probes. [76] Various fluorophore probes.[ 75 , 98 , 99 ] |
|
|
Tetrahydrofolic acid (TFA) |
Fluorophore probe [91] |
|
|
Tetrahydrobiopterin |
Redox sensitiser conjugate [119] |
|
|
Thiamine pyrophosphate (TPP) |
Alkyne and photoaffinity‐labelled probe [47] |
|
|
Ubiquinone |
2. Cofactor Interactome Discovery Using Functionalised Cofactor Mimics
Much is unknown about the diversity of cofactors, but the continuous discovery of novel cofactor‐dependent enzymes and their substrate proteins indicates a still large number yet to be characterised. Moreover, the role of some cofactors as allosteric modulators and signalling molecules means difficult‐to‐predict noncanonical cofactor binding modes that are still not fully understood. Functionalised cofactor mimics can be profitably employed to profile such cofactor interactomes by leveraging their affinity for their binding proteins and their activity towards native reactive nucleophiles.
2.1. Cofactor Interactome Discovery Using Activity‐Based Cofactor Probes
Chemical proteomics approaches using cofactor mimics functionalised with enrichment tags or click handles have been broadly and successfully applied for cofactor interactome discovery. Where the native cofactor is covalently bound by its interacting partners, this activity can be leveraged by functional mimics that bind under physiological conditions enabling subsequent isolation of these partners for identification. Exemplifying this approach, a suite of clickable probes (1.2–1.4, Figure 1) of the cofactor pyridoxal phosphate (1.1, PLP) was used to interrogate the Staphylococcus aureus PLP‐binding proteome. [21] Inspired by activity‐based protein profiling (ABPP),[ 22 , 23 , 24 ] pyridoxal (PL) was equipped with a small alkyne tag that did not impair cellular uptake, phosphorylation by PL kinase, or activity of PLP‐dependent enzymes (PLP‐DEs). Probe‐treated cells were lysed, the transient aldimine bond between the probe and active site lysine of PLP‐DEs reduced, and the alkyne tag clicked to biotin‐azide to enable enrichment of proteins bound by the cofactor probe on avidin beads (Figure 1 A). After elution, tryptic digest, and analysis via label‐free mass spectrometry, 73 % of the known as well as several previously uncharacterised PLP‐dependent enzymes were identified, demonstrating the utility of this strategy. Applying the principle in human cancer cell lines led to the identification of one‐third of the human PLP‐dependent enzymes. [25] More recently, additional probes (e.g. 1.5 and 1.6) have been developed, resulting in a total of 13 diverse PL mimics. [26] PLPome profiling of different pathogenic bacteria using this probe library revealed enhanced labelling coverage and five novel PLP‐DEs were further characterised by a series of in vitro activity assays. Additionally, a library of small molecules was screened for their antibiotic activity and two PLP‐dependent enzymes were verified as targets of the marketed drug phenelzine. [26]
Figure 1.
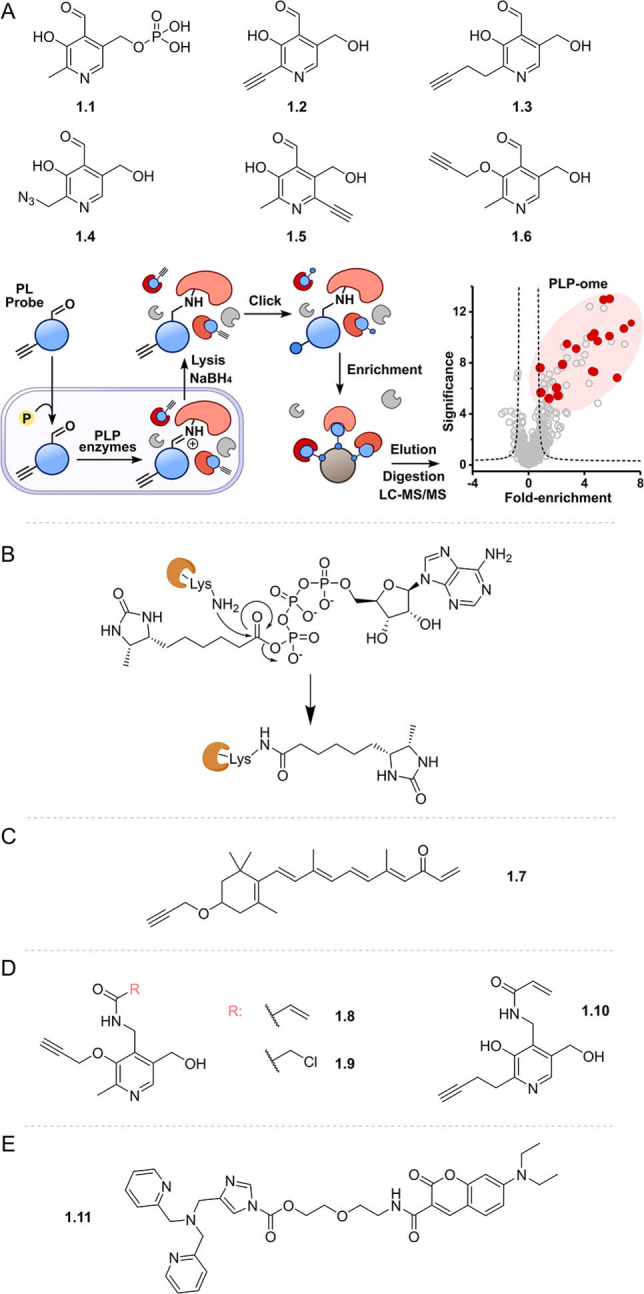
Examples of activity‐based cofactor probes designed to bind covalently to binding proteins to enable identification. A) Suite of clickable pyridoxal probes and schematic for profiling the PLP interactome. PL probes are taken up by bacterial cells, phosphorylated, and incorporated into PLP‐dependent enzymes. Following cell lysis, sodium borohydride‐mediated reduction of the imine bond and click ligation to enrichment tags enables identification of labelled enzymes by mass spectrometry (adapted from Hoegl et al. [21] ); B) mechanism of transfer of the biotin tag from an ATP mimic to a kinase active site lysine residue; C) vitamin A probe and D) electrophile trap pyridoxal mimics; e) AIZin‐1, a zinc‐responsive protein labelling reagent; PL: pyridoxal; PLP: pyridoxal phosphate; LC‐MS/MS: liquid chromatography tandem mass spectrometry.
Profiling a cofactor's interactome can also be accomplished using cofactor mimics functionalised with reactive electrophiles to transfer tags to nucleophilic residues in cofactor binding pockets. Some of the most established probes using this approach are ATP and ADP analogues developed to study protein kinases by leveraging conserved active site lysine residues. Examples include reactive (desthio)biotinylated acyl phosphates of ATP or ADP probes,[ 27 , 28 ] which, upon target binding and attack from the lysine ϵ‐amino group, release ATP or ADP and covalently attach the biotin moiety to the kinase through an amide bond (e.g. Figure 1 B), enabling isolation and identification by mass spectrometry. Impressively, at least 75 % of the known human kinases have been engaged with these probes, [27] demonstrating improved sensitivity over alternative approaches like affinity chromatography using sepharose beads equipped with ATP or kinase inhibitor‐immobilized beads (kinobeads).[ 29 , 30 ] However, these reactive ATP probes have poorer kinase selectivity than kinobeads, due to reaction with abundant non‐kinase ATP‐binding proteins such as ATPase heat shock proteins. [31] Nonspecific binding proteins, however, can be reduced using ATP competition, [32] which has been applied to both identify off‐targets and profile selectivity of ATP‐competitive kinase inhibitors. [33] Similarly, functionalising cofactors with electrophilic warheads to enable covalent linkage of the cofactor itself to nucleophilic binding site residues also allows for activity‐based identification of binding enzymes. Incorporating a Michael acceptor into a vitamin A mimic in place of an aldehyde or carboxylic acid moiety (1.7) made it possible to study retinal aldehyde dehydrogenases (ALDHs). [34] The authors showed that their substrate‐based probe is not only able to detect endogenous retinal ALDHs and retinoid‐interacting proteins but also suitable for competitive ABPP experiments in order to find (off‐)targets of inhibitors. [34] However, detection of retinal binding proteins correlated strongly with abundance, indicating difficulties in identifying low‐abundance targets.
A similar trap approach was adopted in the design of PLP cofactor probes bearing either a vinyl amide (1.8, 1.10) or a chloromethyl amide (1.9) to target pyridoxal kinases (PLKs) containing a catalytically active Cys residue. Interestingly, different probe structures led to different binding efficiencies in Gram‐positive and Gram‐negative bacteria, enabling a tailored approach for the study of this enzyme class. [35]
Metal ion cofactors play essential roles in protein structure, catalysis, and signal transduction. Like small‐molecule cofactors, chemical proteomics methods have been developed to map metal cofactor binding proteomes. [36] For example, metalloproteins that bind Zn2+ and Fe/S cluster cofactors via cysteine residues have been profiled through competition with cysteine reactive probes.[ 37 , 38 ] Other work instead focused on identifying proteins localised in Zn2+‐rich environments using an electrophilic protein labelling probe (1.11), which is activated in the presence of zinc, enabling transfer of a fluorophore to nucleophilic residues. [39] This strategy enabled characterisation of the proteomes of Zn2+‐rich vesicles generated by oxidative stress and was also subsequently extended to visualise copper flux in brain cells. [40] Although this approach does not profile proteins directly binding metal cofactors, it can offer insights into metal homeostasis and their dynamic roles as signal transducers.
2.2. Cofactor Interactome Discovery Using Photoaffinity‐Functionalised Cofactor Mimics
When cofactor mimics do not covalently bind to their protein partners through their own activity or the use of electrophilic traps, a photoaffinity label can be introduced to cement the cofactor binding protein interactions upon irradiation. Early examples of photoaffinity‐functionalised cofactor mimics include a series of azidoflavins which were used as active site probes of flavin enzymes in vitro.[ 41 , 42 ]
The use of bifunctionalised cofactor mimics containing both a photoaffinity group and a click handle is a powerful strategy for the chemical proteomic profiling of binding partners based solely on the affinity of the binding, rather than intrinsic cofactor activity. Cell‐permeable mimics enable profiling in situ, i.e. in live cells or tissues, affording greater biological relevance. This greatly increases the scope of cofactor profiling, although the success of the approach hinges on the development of bifunctionalised mimics that maintain activity and cellular permeability. Optimisation of the biological system under investigation can also provide challenges. For example, a benzophenone photoaffinity label and an alkyne were introduced in the development of bisubstrate probes based on the cofactor CoA (e.g. 2.1, Figure 2) that were designed to target both the substrate and cofactor binding sites of lysine acetyltransferases. [43] Chemical proteomic profiling with these probes identified two lysine acetyltransferase enzymes out of a total of 34, with noted limitations including low crosslinking yields and non‐cell permeability of the probe. Improved enrichment (≈50 %) of lysine acetyltransferases was later achieved with a modified approach, using biotinylated and amine‐functionalised Lys‐CoA probes, which were immobilised on resins prior to incubation with cell lysates. [44]
Figure 2.
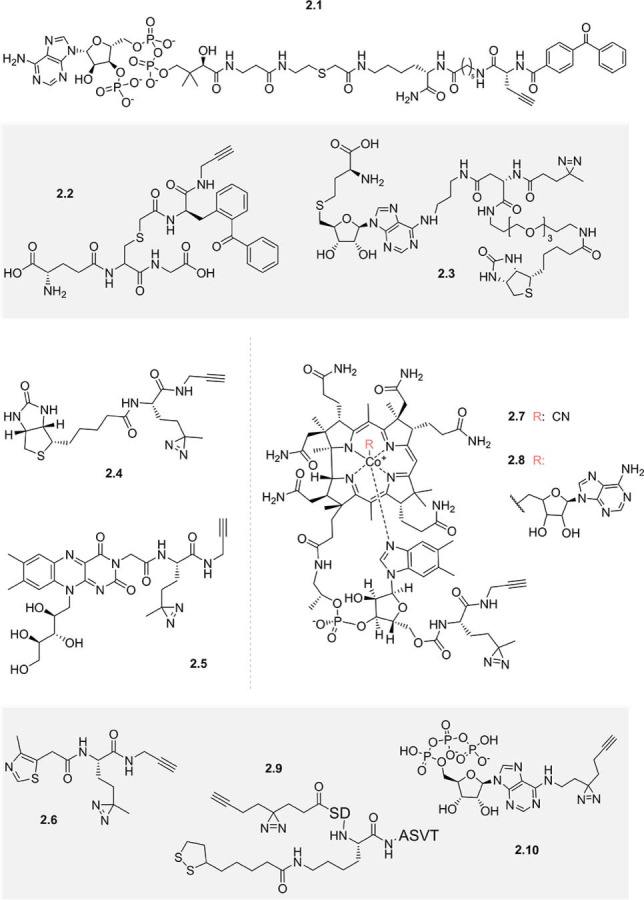
Examples of photoaffinity‐functionalised cofactor mimics designed to identify substrate or binding proteins.
Similarly, a benzophenone and terminal alkyne were appended to the thiol of glutathione to generate a photoaffinity‐functionalised cofactor mimic for proteomic profiling of glutathione S‐transferases (GSTs) 2.2. [45] When the tissues with the highest GST activity (liver and lung) were selected for use in proteomic profiling, the probe then identified 9 out of 19 cytosolic mammalian glutathione S‐transferases. Besides the choice of tissue type, cellular fractionation can also aid proteomic detection of low‐abundance target proteins. [46] These experiments underline the importance of optimising target protein abundance for successful chemical proteomic profiling.
In another study, a series of biotinylated S‐adenosyl‐l‐homocysteine (SAH) photoaffinity probes (e.g. 2.3) were used to profile methyltransferases in three human cancer cell lysates. [46] Comparing probes bearing an aliphatic diazirine, a benzophenone, or an aryl azide, the authors concluded that the diazirine led to the broadest and most specific profiling of human methyltransferases, at ≈25 % of the 200+ known and predicted.
Diazirine photoaffinity labels have been frequently exploited in conjunction with alkynes for the generation of functionalised cofactor mimics. This approach was adopted in the synthesis of chemical probes of vitamin B derived cofactors thiamine, riboflavin, and biotin [47] (2.4–2.6). These probes enriched B vitamin transporters and other proteins in the filamentous anoxygenic photoheterotroph Chloroflexus aurantiacus J‐10‐fl, though inclusion of a control using the unmodified cofactor as a competitor could have verified the specificity of binding. The same research group developed a photoaffinity cobalamin (vitamin B12) cofactor mimic 2.7. [48] This probe was found to support the growth of B12‐auxotrophic bacteria and archaea and be taken up and adenosylated (2.8) by E. coli. [49] Proteomic profiling with the probe led to five significantly enriched proteins in E. coli, of which four were known cobalamin binders, although no components of the B12 transporter system were found.
The development of a minimalist diazirine alkyne linker [50] has greatly facilitated the synthesis of photoaffinity‐labelled and clickable probes. This linker was incorporated into a lipoylated peptide to serve as a functionalised mimic of the cofactor lipoamide (2.9) in order to identify potential regulators of lysine lipoylation. [51] This probe was used in chemical proteomic profiling experiments, uncovering a new delipoylation function of Sirt2. Recently, proteomic profiling of lipoylation has also been performed by functionalising lipoamide in situ, utilising the reactivity of the lipoamide 1,2‐dithiolane ring to chemoselectively conjugate cyclooctyne probes to lipoamide‐bound proteins in bacterial and mammalian lysates. [52]
The minimalist photocrosslinker was also exploited in the recently developed ATP mimetic 2.10, which was found to enrich 59 known ATP‐binding proteins in A549 cell membrane fractions, as well as highly abundant cytosolic ATP binders from cell lysates. [53] A contrasting chemical proteomics method, thermal proteome profiling (TPP), [54] which examines the stabilisation of proteins induced by ligand binding, has also been recently used to profile ATP‐binding proteins, finding 315 of 7859 of proteins annotated with GO terms relating to this cofactor. [55] One significant advantage of TPP is that there is no requirement for ligand functionalisation which may alter bioactivity, and thus binding proteins can be identified using only the native cofactor. However, TPP depends on full proteome profiling, without enrichment of binding proteins, unlike a workflow using functionalised cofactor mimics (Figure 1 A). Thus, low‐abundance proteins are more liable to escape detection by the mass spectrometer, meaning many cofactor‐dependent enzymes may remain unknown. Given the respective advantages and disadvantages of the two methods, it may be that, similar to target deconvolution, [56] the two orthogonal methods complement each other to improve the scope of cofactor proteome profiling.
3. Cofactor Mimics Functionalised to Transfer Tags to Substrate Biomolecules
Certain cofactors are employed by enzymes as co‐substrates to supply a moiety to be transferred to a substrate protein or other biomolecule. Profiling these modifications is of great interest due to their crucial roles in cell signaling and regulation of enzyme activity, stability, and localization. An ideal readout would exploit the native modification, for example in the case of phosphorylation, a PTM using ATP as a co‐substrate, complexing agents can be used to enrich the negatively charged phosphate group. [57]
Where this is not feasible, functionalised cofactor mimics can be leveraged to transfer artificial tags onto these substrates, to enable their identification or visualisation. For example, ATP is also used in the post‐translational modification of proteins through the transfer of AMP to serine, tyrosine, or threonine residues (AMPylation) of substrate proteins. Alkyne‐functionalised ATP derivatives have been used to profile AMPylated substrates in mammalian lysates using a chemical proteomics workflow.[ 58 , 59 , 60 ] This methodology was recently extended with the development of cell‐permeable alkyne‐functionalised pro‐AMP probes to study proteins that are AMPylated during human neurogenesis [61] and bacterial infection. [62] Functionalised ATP mimics also have utility beyond the proteome: an alkyne‐bearing analogue was used to supplement media supporting HeLa cells, with the resultant incorporation into nascent cellular RNAs enabling localisation and structural studies. [63]
The methyl donor S‐adenosyl‐l‐methionine (SAM) is another cofactor that has been modified in various ways to site‐specifically label and functionalise substrate biomolecules including DNA,[ 64 , 65 , 66 , 67 ] RNA,[ 68 , 69 , 70 , 71 ] proteins,[ 72 , 73 ] and small molecules. [74] Examples of such SAM mimics, which include alkenyl and alkynyl selenium analogues [73] (3.1–3.3, Figure 3) with increased stability and reactivity towards nucleophiles, are reviewed elsewhere. [75] Three different photoaffinity‐functionalised SAM probes (3.4–3.6) have also been developed, and were accepted by a promiscuous methyl transferase, even though they are much bulkier than unmodified SAM. This enabled the efficient transfer of the photoaffinity labels from the cofactor mimics to the 5′ caps of mRNA in vitro. [76] The generated aryl azide‐ and diazirine‐functionalised mRNAs were then capable of crosslinking cap interacting protein eIF4E, whereas the bulkier benzophenone sterically hindered protein binding.
Figure 3.
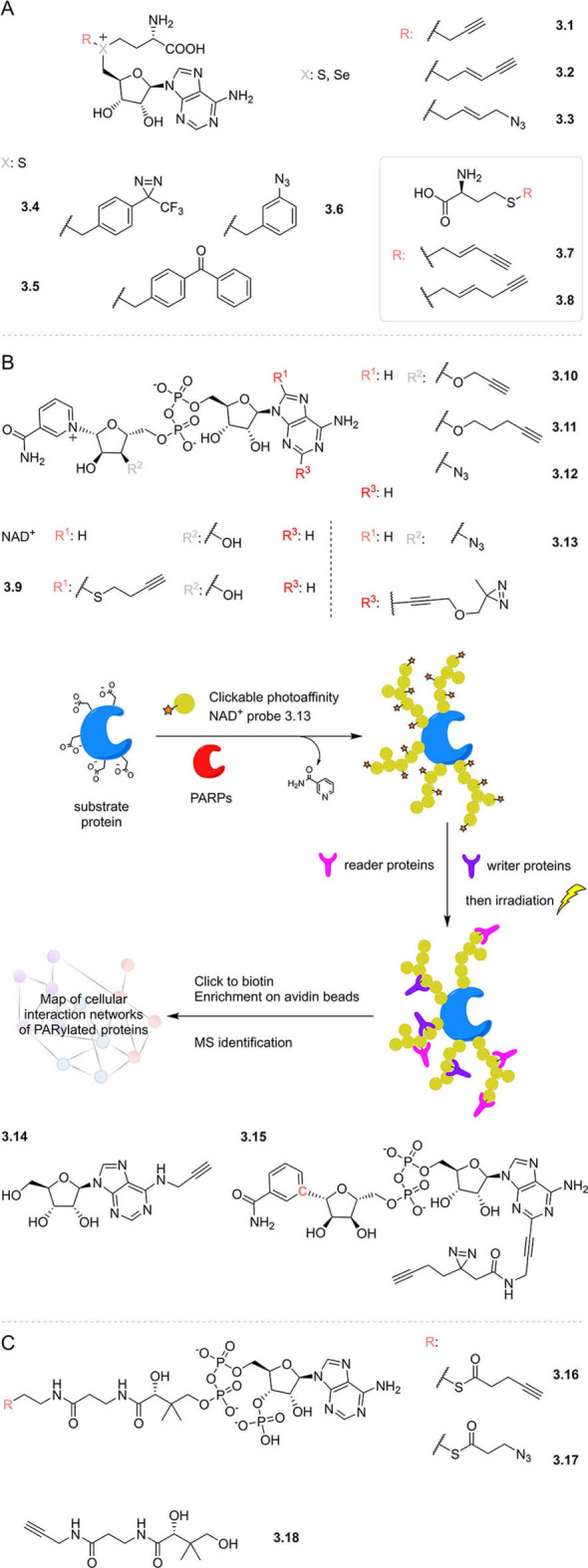
Examples of functionalised cofactor mimics designed to transfer labels onto substrate proteins. A) Alkyne‐ and photoaffinity‐labelled SAM mimics; B) alkyne‐ and photoaffinity‐labelled NAD+ mimics. Schematic for profiling the interaction network of PARylated proteins; C) probes based on the structure of coenzyme A; NAD+: nicotinamide adenine dinucleotide; PARPs: poly‐ADP‐ribose polymerases; MS: mass spectrometry; PARylation: polyADP‐ribosylation.
Elsewhere, to facilitate studies in live cells, cell‐permeable alkynylated methionine analogues (3.7–3.8) have been supplied exogenously for uptake and in situ conversion to SAM analogues by an engineered SAM‐synthetase. [77] Engineering specific protein methyltransferases to accept the alkynylated SAM allowed the analysis of specific targets (e.g. histones) in live cells in the presence of native SAM.
A similar chemical genetic approach has been used in the development of functionalised NAD mimics to study cellular protein ADP‐ribosylation, the transfer of up to 200 ADP‐ribosyl groups from NAD to substrate proteins (PARylation), by poly‐ADP‐ribose polymerases (PARPs). Implementing a “bump‐hole” strategy, NAD analogues containing a range of bulky substituents (e.g. 3.9) have been used as substrates only by engineered PARPs containing “holes” in their binding pocket to accommodate the “bumps”.[ 78 , 79 , 80 ] Many other biotinylated, alkyne and azide derivatives of NAD have also been synthesized to investigate PARylation using native PARPs,[ 81 , 82 ] including modifications to the purine base [83] and the ribose alcohol moieties which led to improved activity and selectivity (3.10–3.12). [84]
An alternative way to study PARylation has also been established, using simple alkynylated adenosine analogues (e.g. 3.14) for in situ metabolism. ADP‐ribosylation was then quantitatively measured via TMT isobaric mass spectrometry. This robust strategy led to the identification of thousands of protein targets, the largest number to date. Moreover, it enabled quantification of responses of PARylated proteins to the clinical PARP inhibitors olaparib and rucaparib. [85]
Further functionalised NAD cofactor mimics include recently reported clickable photoaffinity probes with diazirine moieties on different positions of the adenine.[ 86 , 87 ] These probes were used in different applications: one report focused on the profiling of reader and eraser proteins that bind to PARylated proteins in a PARylation‐dependent manner, by using the synthesised NAD mimic (3.13) as a PARP substrate (Figure 3 B). Proteins PARylated with the NAD mimic were then photoaffinity labelled, and trapped interacting partners after UV irradiation, enabling enrichment and identification. [87] The other study used a photoaffinity NAD mimic with an enzymatically stable nicotinamide glycosidic bond (3.15) to preclude consumption of the probe as a PARylation substrate, and instead enable identification of proteins that directly interact with the NAD cofactor itself. [86]
Cofactor co‐substrate acetyl coenzyme A (Ac‐CoA) supplies the acetyl group for lysine acetyltransferase (KAT) mediated acetylation of ϵ‐amino groups of certain Lys residues of target proteins. This post‐translational modification facilitates epigenetic programming, the cell cycle, apoptosis, metabolism, and signal transduction. [88] Ac‐CoA analogues designed to transfer functionalized alkyne‐ or azide‐modified acyl groups to substrate proteins (3.16–3.17), mediated by either engineered KATs [88] and/or wild‐type KATs, [89] led to the identification of hundreds of KAT target proteins, including histone proteins as the primary substrates for both enzymes investigated. [89]
Coenzyme A is also used as a co‐substrate for the transfer of 4′‐phosphopantetheine to substrate proteins. Recently, alkyne‐bearing panthetheine probes (e.g. 3.18) were developed and supplied to live HepG2 cells. [90] Probe uptake and metabolic incorporation yielded functional alkyne‐substituted CoA mimics, enabling the profiling of 4′‐phosphopantetheinylated proteins. Thus, the exact modification sites of all five known 4′‐phosphopantetheinylated proteins and an additional putative 4′‐phosphopantetheinylation site in the protein DHRS2 were identified by mass spectrometry.
4. Fluorescent Cofactor Mimics and Cofactor‐Based Sensors
Profiling of cofactor binding proteins and their targets aside, functionalised cofactor mimics with fluorophores have been exploited in many important applications. These include studying enzyme mechanisms, covalent labelling of target proteins and DNA, real‐time monitoring of the cellular environment, and discovering novel inhibitors in screening assays. [81]
Fluorophores are often bulky hydrophobic moieties; nevertheless, conjugates to cofactors can make effective probes, often with binding constants highly similar to that of the unmodified cofactor. This was the case for fluorescein derivatives of folic acid (vitamin B9) and dihydrofolate, which bound human dehydrofolate reductase with dissociation constants of 115 vs. 111 nM and 47 vs. 44 nM (comparison to the unmodified compounds), respectively. [91] Conjugation to fluorophores can also be tolerated by cofactor uptake mechanisms across different organisms, as shown by fluorophore derivatives of cobalamin (vitamin B12), modified either on the main tetrapyrrole corrin ring (biosynthesised using an allyl‐functionalised SAM mimic) or on the ribose moiety of the lower nucleotide loop, and successfully uptaken by bacteria, C. elegans, and garden cress. [92]
Perhaps the most established fluorophore‐functionalised cofactor mimics derive from ATP, of which modifications to the ribose, base, or phosphate chain are well established[ 93 , 94 , 95 ] and in many cases commercially available. These ATP analogues not only serve to investigate ATP binding but also can be used for high‐throughput‐screening approaches. In a recent example, a fluorescence polarization‐based assay employing ATP‐γ‐S BODIPY FL was used to screen a small set of predicted inhibitors of the Protein Kinase A ATP‐pocket. [96] Fluorescence polarisation was also recently applied to identify competitive inhibitors of KATs, using a fluorescein‐functionalised Ac‐CoA probe (4.1, Figure 4), illustrating its versatility for inhibitor discovery. [97]
Figure 4.
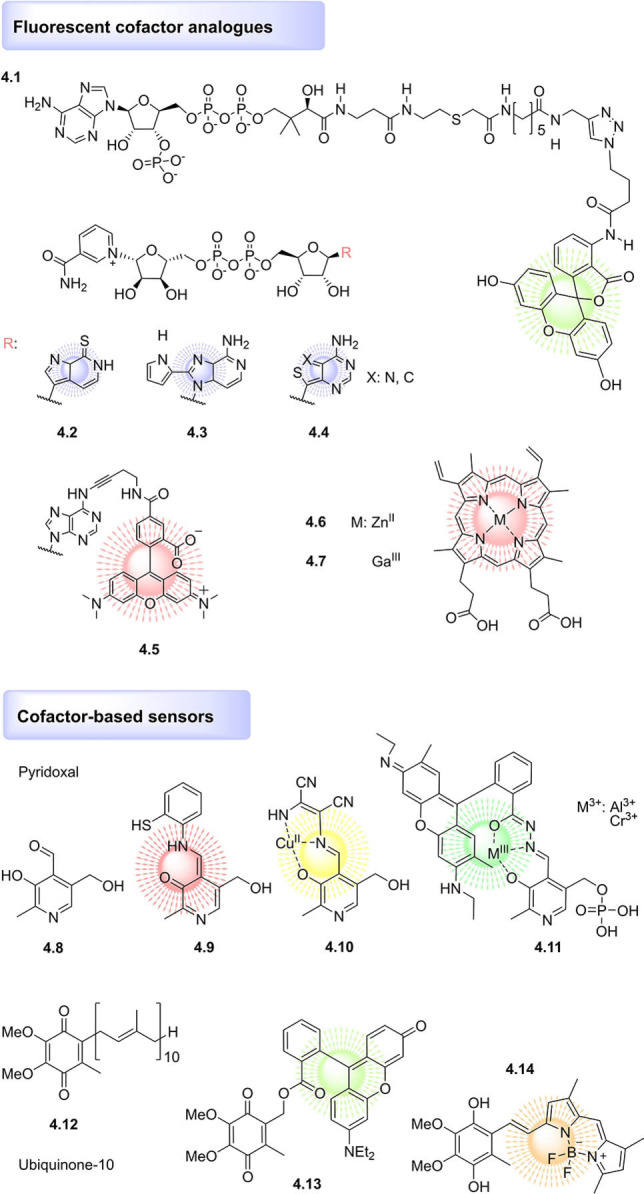
Examples of fluorophore‐functionalised cofactor mimics and cofactor‐based sensors.
Fluorescent cofactor analogues often make use of linkers to minimise disruption to protein binding; indeed, inclusion of a linker was recently found to be necessary to generate rhodamine‐SAM mimics recognised by the methyltransferase M.TaqI and thus capable of labelling the enzyme's DNA targets.[ 98 , 99 ] The linker approach was also utilised in the design of a TAMRA‐functionalised NAD+ mimic (4.5) which was accepted as a competitive substrate for protein PARylation in live cells, allowing real‐time visualisation of the process by FRET‐FLIM microscopy. [100]
However, fluorophores can also be nested in the structure of the cofactor rather than appended via a linker. Classic examples of this include the numerous fluorescent adenosine analogues developed and applied to generate fluorescent mimics of ATP,[ 93 , 101 ] SAM,[ 102 , 103 ] and NAD[ 104 , 105 , 106 , 107 , 108 ] (4.2–4.4; reviewed elsewhere [81] ). The most successful analogues maintain similar size and H‐bonding pattern to adenosine to minimise functional perturbation and thereby enable the real‐time monitoring of the respective cofactor‐mediated transformations.
Another facile method of generating fluorescent cofactor mimics with minimal perturbation to the cofactor structure and binding mode is achieved by exchanging the metal ion in heme. [109] For example, ZnII protoporphyrin IX (4.6) is a fluorescent heme analogue, and was recently incorporated into different vertebrate globins, allowing the investigation of the heme‐binding pocket, electron transfer processes, and even protein–protein interactions. [109] Another fluorescent hemin analogue, GaIII‐protoporphyrin IX (4.7), has been studied as a photosensitizer for antimicrobial photodynamic therapy, a proposed topical treatment for multiresistant bacterial infections. [110] GaIII‐protoporphyrin IX was shown to be taken up by MRSA, in a manner attributed to the expression of high‐affinity cell‐surface hemin receptors. GaIII‐protoporphyrin IX exerted antimicrobial activity upon 10 s irradiation with visible light, while exhibiting low cytotoxicity towards HEK293 cells. [110] Thus, fluorescent cofactor mimics are not only useful tools for chemical biology, but may even have utility in clinical applications.
Besides fluorophore‐functionalised cofactor mimics, cofactor derivatives have been designed to serve as turn‐on fluorescent chemosensors to report on cellular redox or pH status, or the presence of particular metal ions. Synthetically tractable Schiff bases derived from pyridoxal (4.8) are particularly frequent chemosensor scaffolds. Recent examples of pyridoxal‐based chemosensors include a fluorescent pH sensor [111] (4.9) and a colorimetric CuII sensor (4.10), which additionally served as a fluorescent sensor for hypochlorite ions with a potential application in monitoring contaminants in tap and pond water samples. [112] In addition, the cofactor pyridoxal phosphate has been conjugated to a rhodamine spirolactam to create a chemosensor that is colourless and nonfluorescent until binding of a AlIII or CrIII ion triggers a ring‐opening reaction, generating strongly fluorescent 4.11. [113] This cofactor‐based chemosensor then successfully detected AlIII and CrIII ions in living HeLa cells.
The redox‐sensitive cofactor ubiquinone (coenzyme Q10, 4.12) has been functionalised with rhodamine [114] (4.13) and BODIPY [115] (4.14) to generate fluorogenic on/off redox sensors, with the fluorescent signal switched off by photoinduced electron transfer between the fluorophore and ubiquinone. However, these ubiquinone‐based sensors were characterised by low sensitivity, limiting their utility in cellular studies. Adapting the strategy by replacing ubiquinone with a truncated version of cofactor menaquinone (vitamin K) resulted in a more sensitive redox sensor, which was further capable of mimicking a native quinone substrate of NAD(P)H:quinone oxidoreductase, resulting in similar rates of reaction. [116]
5. Artificial Cofactors Developed with Novel Functionality
Beyond studying the interacting partners of cofactors or cofactor binding proteins, cofactor mimics have been developed to expand their intrinsic functionality. For example, the cofactor FAD was modified with redox‐active ferrocene (5.1, Figure 5) to facilitate electron exchange between protein‐bound FAD and an electrode. [117] Reconstitution of glucose oxidase with the functionalised FAD mimic resulted in a semisynthetic electroenzyme with 60 % of the native enzyme's activity, for application as a glucose biosensor. Similarly, a heme ferrocene was developed and used to generate an electrochemically active horseradish peroxidase. [118] Redox functionality has also been incorporated in mimics of the cofactor tetrahydrobiopterin via conjugation to a ruthenium(II)–diimine redox‐active sensitizer (5.2). [119] The resultant fluorescent probe competed with the native pterins for binding to the heme domain of murine iNOS. Pterins conjugated with lipophilic decyl chains have also been developed (e.g. 5.3), resulting in cofactor mimics with increased singlet oxygen quantum yields which intercalate in unilamellar vesicles, for potential use as photosensitizers in biomembranes. [120]
Figure 5.
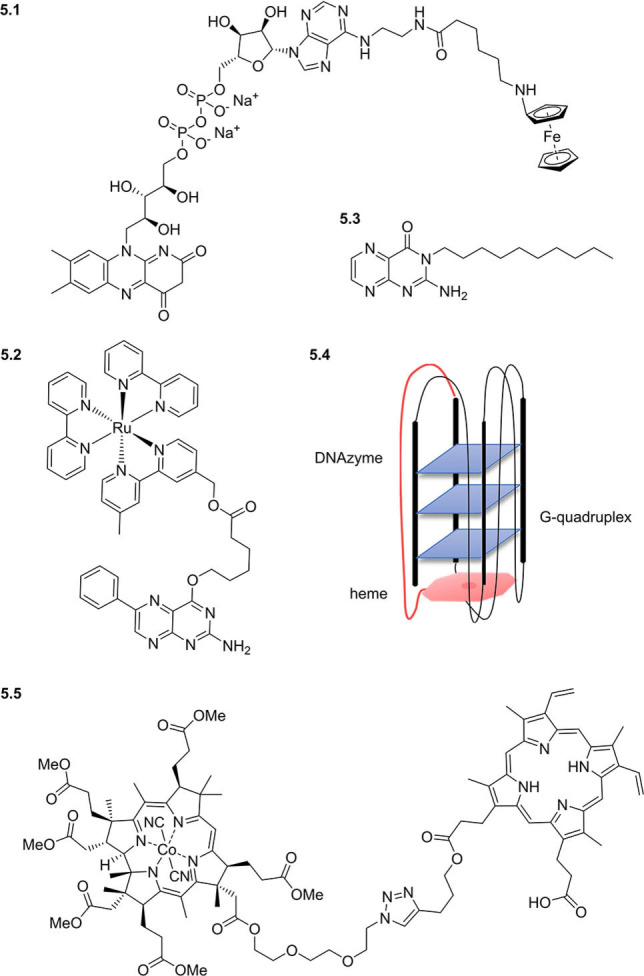
Examples of cofactor mimics with artificial activity.
Cofactors have also been functionalised with other biomolecules to elicit novel activity. For example, two cofactors—heme and flavin—have been conjugated together and used to replace native heme in myoglobin, to transform it from an oxygen‐storage hemoprotein to an oxygen‐activating one, capable of generating FeIII‐peroxoanions. [121] Heme has been further functionalised via covalent attachment to DNA G‐quadruplex structures to form peroxidase‐mimicking DNAzymes (5.4), enhancing activity compared to noncovalently attached unmodified hemin.[ 122 , 123 , 124 ] In addition to conferring novel activity, hybrid cofactor mimics have been reported to synergistically improve binding: conjugation of protoporphyrin IX with vitamin B12 derivative dicyanocobinamide (each an activator of soluble guanylyl cyclase by targeting the regulatory and catalytic domains, respectively) resulted in a hybrid cofactor conjugate (5.5) more potent than the individual cofactors. [125] Cofactor conjugation can improve uptake as well as potency: so‐called “Trojan Horse” cofactor–drug conjugates exploit dedicated cofactor transporters to mediate the active co‐transport of the drugs. [126] Indeed, a recently reported vitamin B12–ampicillin conjugate exhibited more than 500 times improved activity against E. coli compared with ampicillin itself. [127] Furthermore, conjugation to vitamin B12 enabled the unprecedented delivery of antisense RNA oligonucleotides into E. coli and S. Typhimurium cells, [128] highlighting the potential power of the approach. Thus through conjugation to biomolecules of interest, the diverse biochemical functions of cofactors can be harnessed to create novel tools with capabilities greater than the sum of their parts.
6. Summary and Outlook
Myriad synthetic cofactors have now been developed to replace unmodified cofactors in their cognate binding sites and provide additional functionality through click chemistry handles, photoaffinity labels, fluorophores, or photosensitisers. The diverse activity of cofactors has further inspired the design of mimics for use as inhibitors, antibiotic therapeutics, and chemo‐ and biosensors. By lending their activity to other biomolecules through the synthesis of conjugates, cofactors have also enabled the generation of novel enzymes and artificial DNAzymes. Examples of these synthetic cofactors are summarised in Table 1. Thus far, many functionalised analogues are reported in the literature for some cofactors, while others, such as PQQ, coenzyme B/M/Q, and ascorbic acid are yet to be explored in this way.
Cofactor mimics have been particularly used to great effect to reveal the interacting partners, functions, mechanisms, and localisation of the native cofactor in its physiological environment. They have also been used to study and site‐specifically label the substrate proteins, RNAs, and DNA of cofactor‐binding enzymes. They therefore represent tools to study many important post‐translational modifications including methylation, PARylation, acetylation, phosphopantetheinylation, and AMPylation. These tools could be applied in the future to study changing patterns of modification in response to environmental stimuli or disease.
The utility of cofactor mimics for interactome discovery is impressive, with many reports of annotation of previously uncharacterised proteins. In this regard, chemical proteomics goes hand in hand with bioinformatic prediction towards the goal of interactome annotation. When combined with quantitative mass spectrometry, interactome discovery has been particularly fruitful, indeed up to 73 % of known PLP‐binding enzymes in S. aureus were identified using cofactor probes. [21] However, challenges remain for increasing the proportion of known (and previously unknown) binding partners identified by functionalised mimics, particularly in higher organisms. Issues that have arisen include limited cell permeability and solubility of particular functionalised cofactor mimics, as well as limited uptake in the presence of the native cofactor. Reported circumventions of these problems include design of truncated or otherwise simplified cofactor analogues or prodrugs to mask charged moieties, and alternative administration of more permeable precursors that are converted to the active cofactor mimic in situ. Uptake of cofactor mimics can also be enhanced by the choice of organisms which are auxotrophic for the cofactor and thus rely on uptake, by suppression of endogenous cofactor synthesis via knockout of biosynthesis genes, or by chemical inhibition of biosynthetic enzymes. Other important considerations include the optimisation of the biological sample, since the low abundance of target proteins is a common issue preventing identification even with sensitive mass spectrometry. Remediation can include selection of tissues, organisms, or organelles which highly express the target protein class.
Nevertheless, modification of the native cofactor may inevitably impede binding to some of its cognate partners, depending on the particular binding mode, meaning that the full interactome is not captured. In order to maximise the number of identified interactors, the following is recommended: 1) the use of minimally modified mimics (e.g. small diazirine photocrosslinker in the case of bifunctionalised probes); 2) evaluation in parallel of a suite of probes modified at different positions; and 3) parallel use of a complementary method for interactome discovery such as thermal proteome profiling or limited proteolysis–mass spectrometry,[ 129 , 130 ] which obviates the need for synthetic functionalisation.
In addition to interactome profiling, electrophilic trap and photoaffinity‐functionalised cofactor mimics can be used to reveal the cofactor binding pocket by identifying via MS the residue(s) that become modified. This can further provide structural and mechanistic insights into the role of cofactors in previously uncharacterised binding proteins.
Cofactor mimics are powerful tools not only for studying biological systems but also for drug discovery and development. Indeed, biomimetic cofactor‐based inhibitors have been developed for medicinal chemistry applications (see Table 1), for example, antibiotic heme analogue Ga‐PPIX or clinical glutathione S‐transferase 1 inhibitor ezatiostat (formerly Telintra and TLK199).[ 131 , 132 , 133 ] Furthermore, identification of novel cofactor‐binding proteins in bacteria can lead to the discovery of biomarkers or therapeutic targets for future antibiotic development. Cofactors can also be employed to enhance antibiotic uptake through the use of “Trojan horse” conjugates. Cofactor mimics also have utility in the drug development pipeline since fluorescent analogues aid high‐throughput lead optimisation of cofactor competitive inhibitors.
All in all, cofactors are extraordinarily versatile small molecules, which are multifunctional in nature and now also in scientific research. Through diversifying their scope with artificial activities, they promise even greater utility.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Isabel V. L. Wilkinson is an EMBO Postdoctoral Fellow hosted in the lab of Prof. Stephan A. Sieber at the Technical University of Munich (Germany). Prior to this, she completed a Doctoral Prize Fellowship and DPhil (2019) at the University of Oxford (UK), under the supervision of Prof. Angela Russell. She obtained her MSci in Natural Sciences (Chemistry) at the University of Cambridge in 2015. Her research focuses on chemical proteomics and affinity‐based profiling for target deconvolution and cofactor interactome discovery.

Biographical Information
Martin Pfanzelt studied chemistry at the Technical University Munich (Germany), with special focus on organic and biological chemistry. After his MSc, which he received in 2017, he started his PhD under the supervision of Prof. Stephan A. Sieber, where he focuses on the investigation of pyridoxal phosphate‐dependent enzymes in bacteria.

Biographical Information
Stephan A. Sieber completed his graduate studies at the University of Marburg (Germany) and Harvard Medical School, Boston (USA), before joining the Cravatt group at the Scripps Research Institute, La Jolla (USA) as a postdoc. In 2006 he began his independent research career at the University of Munich (LMU) (Germany) funded by a DFG Emmy‐Noether grant. In 2009 he was appointed full professor at the Technical University of Munich. His research focus is in the field of chemical biology and chemical proteomics with emphasis on bioactive small molecules to decipher novel antibiotic targets in pathogenic bacteria.

Acknowledgements
I.V.L. Wilkinson would like to thank the European Molecular Biology Organization for financial support (ALTF 484‐2020). M.P. acknowledges support from the Studienstiftung des deutschen Volkes. S.A.S. acknowledges the European Research Council (ERC) and the European Union's Horizon 2020 research and innovation program (grant agreement no. 725085, CHEMMINE, ERC consolidator grant). Open Access funding enabled and organized by Projekt DEAL.
I. V. L. Wilkinson, M. Pfanzelt, S. A. Sieber, Angew. Chem. Int. Ed. 2022, 61, e202201136; Angew. Chem. 2022, 134, e202201136.
References
- 1. Fischer J. D., Holliday G. L., Rahman S. A., Thornton J. M., J. Mol. Biol. 2010, 403, 803–824. [DOI] [PubMed] [Google Scholar]
- 2. Richter M., Nat. Prod. Rep. 2013, 30, 1324–1345. [DOI] [PubMed] [Google Scholar]
- 3. Zhao J., Cao Y., Zhang L., Comput. Struct. Biotechnol. J. 2020, 18, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katz E., Schlereth D. D., Schmidt H. L., Olsthoorn A. J. J., J. Electroanal. Chem. 1994, 368, 165–171. [Google Scholar]
- 5. Xiao Y., Patolsky F., Katz E., Hainfeld J. F., Willner I., Science 2003, 299, 1877–1881. [DOI] [PubMed] [Google Scholar]
- 6. Zimmermann H., Lindgren A., Schuhmann W., Gorton L., Chem. Eur. J. 2000, 6, 592–599. [DOI] [PubMed] [Google Scholar]
- 7. Mukherjee S., Sengupta K., Das M. R., Jana S. S., Dey A., J. Biol. Inorg. Chem. 2012, 17, 1009–1023. [DOI] [PubMed] [Google Scholar]
- 8. Jing Y., Montano J. L., Levy M., Lopez J. E., Kung P. P., Richardson P., Krajewski K., Florens L., Washburn M. P., Meier J. L., ACS Chem. Biol. 2021, 16, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prier C. K., Arnold F. H., J. Am. Chem. Soc. 2015, 137, 13992–14006. [DOI] [PubMed] [Google Scholar]
- 10. Mordhorst S., Andexer J. N., Nat. Prod. Rep. 2020, 37, 1316–1333. [DOI] [PubMed] [Google Scholar]
- 11. Oohora K., Hayashi T., Dalton Trans. 2021, 50, 1940–1949. [DOI] [PubMed] [Google Scholar]
- 12. Natoli S. N., Hartwig J. F., Acc. Chem. Res. 2019, 52, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanifuji K., Sickerman N., Lee C. C., Nagasawa T., Miyazaki K., Ohki Y., Tatsumi K., Hu Y., Ribbe M. W., Angew. Chem. Int. Ed. 2016, 55, 15633–15636; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 15862–15865. [Google Scholar]
- 14. Richtar J., Heinrichova P., Apaydin D. H., Schmiedova V., Yumusak C., Kovalenko A., Weiter M., Sariciftci N. S., Krajcovic J., Molecules 2018, 23, 2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drenth J., Trajkovic M., Fraaije M. W., ACS Catal. 2019, 9, 6435–6443. [Google Scholar]
- 16. Pan H. J., Huang G., Wodrich M. D., Tirani F. F., Ataka K., Shima S., Hu X., Nat. Chem. 2019, 11, 669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oohora K., Onoda A., Hayashi T., Acc. Chem. Res. 2019, 52, 945–954. [DOI] [PubMed] [Google Scholar]
- 18. Zachos I., Nowak C., Sieber V., Curr. Opin. Chem. Biol. 2019, 49, 59–66. [DOI] [PubMed] [Google Scholar]
- 19. Zachos I., Döring M., Tafertshofer G., Simon R. C., Sieber V., Angew. Chem. Int. Ed. 2021, 60, 14701–14706; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 14822–14828. [Google Scholar]
- 20. Liu Y., Guo X., Liu W., Wang J., Kent Zhao Z., ChemBioChem 2021, 22, 1765–1768. [DOI] [PubMed] [Google Scholar]
- 21. Hoegl A., Nodwell M. B., Kirsch V. C., Bach N. C., Pfanzelt M., Stahl M., Schneider S., Sieber S. A., Nat. Chem. 2018, 10, 1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barglow K. T., Cravatt B. F., Nat. Methods 2007, 4, 822–827. [DOI] [PubMed] [Google Scholar]
- 23. Berger A. B., Vitorino P. M., Bogyo M., Am. J. PharmacoGenomics 2004, 4, 371–381. [DOI] [PubMed] [Google Scholar]
- 24. Keller L. J., Babin B. M., Lakemeyer M., Bogyo M., Curr. Opin. Chem. Biol. 2020, 54, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fux A., Pfanzelt M., Kirsch V. C., Hoegl A., Sieber S. A., Cell Chem. Biol. 2019, 26, 1461–1468.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfanzelt M., Maher T. E., Absmeier R. M., Schwarz M., Sieber S. A., Angew. Chem. Int. Ed. 2022, 61, e202117724; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2022, 134, e202117724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Patricelli M. P., Szardenings A. K., Liyanage M., Nomanbhoy T. K., Wu M., Weissig H., Aban A., Chun D., Tanner S., Kozarich J. W., Biochemistry 2007, 46, 350–358. [DOI] [PubMed] [Google Scholar]
- 28. Qiu H., Wang Y., Anal. Chem. 2007, 79, 5547–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., Drewes G., Nat. Biotechnol. 2007, 25, 1035–1044. [DOI] [PubMed] [Google Scholar]
- 30. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M., Mol. Cell 2008, 31, 438–448. [DOI] [PubMed] [Google Scholar]
- 31. Xiao Y., Wang Y., Mass Spectrom. Rev. 2016, 35, 601–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adachi J., Kishida M., Watanabe S., Hashimoto Y., Fukamizu K., Tomonaga T., J. Proteome Res. 2014, 13, 5461–5470. [DOI] [PubMed] [Google Scholar]
- 33. Patricelli M. P., Nomanbhoy T. K., Wu J., Brown H., Zhou D., Zhang J., Jagannathan S., Aban A., Okerberg E., Herring C., Nordin B., Weissig H., Yang Q., Lee J. D., Gray N. S., Kozarich J. W., Chem. Biol. 2011, 18, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koenders S. T. A., Wijaya L. S., Erkelens M. N., Bakker A. T., Van Der Noord V. E., Van Rooden E. J., Burggraaff L., Putter P. C., Botter E., Wals K., Van Den Elst H., Den Dulk H., Florea B. I., Van De Water B., Van Westen G. J. P., Mebius R. E., Overkleeft H. S., Le Dévédec S. E., Van Der Stelt M., ACS Cent. Sci. 2019, 5, 1965–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hübner I., Dienemann J. N., Friederich J., Schneider S., Sieber S. A., ACS Chem. Biol. 2020, 15, 3227–3234. [DOI] [PubMed] [Google Scholar]
- 36. Zeng X., Cheng Y., Wang C., Biochemistry 2021, 60, 3507–3514. [DOI] [PubMed] [Google Scholar]
- 37. Pace N. J., Weerapana E., ACS Chem. Biol. 2014, 9, 258–265. [DOI] [PubMed] [Google Scholar]
- 38.D. W. Bak, E. Weerapana, bioRxiv 2021, 10.1101/2021.04.01.438105. [DOI]
- 39. Miki T., Awa M., Nishikawa Y., Kiyonaka S., Wakabayashi M., Ishihama Y., Hamachi I., Nat. Methods 2016, 13, 931–937. [DOI] [PubMed] [Google Scholar]
- 40. Lee S., Chung C. Y. S., Liu P., Craciun L., Nishikawa Y., Bruemmer K. J., Hamachi I., Saijo K., Miller E. W., Chang C. J., J. Am. Chem. Soc. 2020, 142, 14993–15003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fitzpatrick P. F., Ghisla S., Massey V., J. Biol. Chem. 1985, 260, 8483–91. [PubMed] [Google Scholar]
- 42. Massey V., Ghisla S., Yagi K., Biochemistry 1986, 25, 8095–8102. [DOI] [PubMed] [Google Scholar]
- 43. Montgomery D. C., Sorum A. W., Meier J. L., J. Am. Chem. Soc. 2014, 136, 8669–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Montgomery D. C., Garlick J. M., Kulkarni R. A., Kennedy S., Allali-Hassani A., Kuo Y. M., Andrews A. J., Wu H., Vedadi M., Meier J. L., J. Am. Chem. Soc. 2016, 138, 6388–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stoddard E. G., Killinger B. J., Nair R. N., Sadler N. C., Volk R. F., Purvine S. O., Shukla A. K., Smith J. N., Wright A. T., J. Am. Chem. Soc. 2017, 139, 16032–16035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Horning B. D., Suciu R. M., Ghadiri D. A., Ulanovskaya O. A., Matthews M. L., Lum K. M., Backus K. M., Brown S. J., Rosen H., Cravatt B. F., J. Am. Chem. Soc. 2016, 138, 13335–13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Anderson L. N., Koech P. K., Plymale A. E., Landorf E. V., Konopka A., Collart F. R., Lipton M. S., Romine M. F., Wright A. T., ACS Chem. Biol. 2016, 11, 345–354. [DOI] [PubMed] [Google Scholar]
- 48. Romine M. F., Rodionov D. A., Maezato Y., Anderson L. N., Nandhikonda P., Rodionova I. A., Carre A., Li X., Xu C., Clauss T. R. W., Kim Y. M., Metz T. O., Wright A. T., Proc. Natl. Acad. Sci. USA 2017, 114, E1205–E1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosnow J. J., Hwang S., Killinger B. J., Kim Y. M., Moore R. J., Lindemann S. R., Maupin-Furlow J. A., Wright A. T., Appl. Environ. Microbiol. 2018, 84, 955–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Z., Hao P., Li L., Tan C. Y. J., Cheng X., Chen G. Y. J., Sze S. K., Shen H.-M., Yao S. Q., Angew. Chem. Int. Ed. 2013, 52, 8551–8556; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 8713–8718. [Google Scholar]
- 51. Xie Y., Chen L., Wang R., Wang J., Li J., Xu W., Li Y., Yao S. Q., Zhang L., Hao Q., Sun H., J. Am. Chem. Soc. 2019, 141, 18428–18436. [DOI] [PubMed] [Google Scholar]
- 52. Tang Q., Guo Y., Meng L., Chen X., Angew. Chem. Int. Ed. 2021, 60, 4028–4033; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 4074–4079. [Google Scholar]
- 53. Jelcic M., Wang K., Hui K. L., Cai X. C., Enyedi B., Luo M., Niethammer P., Cell Chem. Biol. 2020, 27, 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Franken H., Mathieson T., Childs D., Sweetman G. M. A., Werner T., Tögel I., Doce C., Gade S., Bantscheff M., Drewes G., Reinhard F. B. M., Huber W., Savitski M. M., Nat. Protoc. 2015, 10, 1567–1593. [DOI] [PubMed] [Google Scholar]
- 55. Sridharan S., Kurzawa N., Werner T., Günthner I., Helm D., Huber W., Bantscheff M., Savitski M. M., Nat. Commun. 2019, 10, 1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilkinson I. V. L., Terstappen G. C., Russell A. J., Drug Discovery Today 2020, 25, 1998–2005. [DOI] [PubMed] [Google Scholar]
- 57. Qiu W., Evans C. A., Landels A., Pham T. K., Wright P. C., Anal. Chim. Acta 2020, 1129, 158–180. [DOI] [PubMed] [Google Scholar]
- 58. Grammel M., Luong P., Orth K., Hang H. C., J. Am. Chem. Soc. 2011, 133, 17103–17105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu X., Woolery A. R., Luong P., Hao Y. H., Grammel M., Westcott N., Park J., Wang J., Bian X., Demirkan G., Hang H. C., Orth K., LaBaer J., Mol. Cell. Proteomics Mol. Cell. Proteom 2014, 13, 3164–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Broncel M., Serwa R. A., Bunney T. D., Katan M., Tate E. W., Mol. Cell. Proteomics Mol. Cell. Proteom 2016, 15, 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kielkowski P., Buchsbaum I. Y., Kirsch V. C., Bach N. C., Drukker M., Cappello S., Sieber S. A., Nat. Commun. 2020, 11, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rauh T., Brameyer S., Kielkowski P., Jung K., Sieber S. A., ACS Infect. Dis. 2020, 6, 3277–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zheng Y., Beal P. A., Bioorg. Med. Chem. Lett. 2016, 26, 1799–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dalhoff C., Lukinavičius G., Klimašauskas S., Weinhold E., Nat. Chem. Biol. 2006, 2, 31–32. [DOI] [PubMed] [Google Scholar]
- 65. Pljevaljčić G., Schmidt F., Weinhold E., ChemBioChem 2004, 5, 265–269. [DOI] [PubMed] [Google Scholar]
- 66. Lukinavičius G., Lapiene V., Staševskij Z., Dalhoff C., Weinhold E., Klimašauskas S., J. Am. Chem. Soc. 2007, 129, 2758–2759. [DOI] [PubMed] [Google Scholar]
- 67. Kim S., Gottfried A., Lin R. R., Dertinger T., Kim A. S., Chung S., Colyer R. A., Weinhold E., Weiss S., Ebenstein Y., Angew. Chem. Int. Ed. 2012, 51, 3578–3581; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 3638–3641. [Google Scholar]
- 68. Motorin Y., Burhenne J., Teimer R., Koynov K., Willnow S., Weinhold E., Helm M., Nucleic Acids Res. 2011, 39, 1943–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tomkuvienė M., Clouet-D'Orval B., Černiauskas I., Weinhold E., Klimašauskas S., Nucleic Acids Res. 2012, 40, 6765–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Holstein J. M., Stummer D., Rentmeister A., Chem. Sci. 2015, 6, 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schulz D., Holstein J. M., Rentmeister A., Angew. Chem. Int. Ed. 2013, 52, 7874–7878; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 8028–8032. [Google Scholar]
- 72. Islam K., Bothwell I., Chen Y., Sengelaub C., Wang R., Deng H., Luo M., J. Am. Chem. Soc. 2012, 134, 5909–5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Blum G., Bothwell I. R., Islam K., Luo M., Curr. Protoc. Chem. Biol. 2013, 5, 67–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Singh S., Zhang J., Huber T. D., Sunkara M., Hurley K., Goff R. D., Wang G., Zhang W., Liu C., Rohr J., Van Lanen S. G., Morris A. J., Thorson J. S., Angew. Chem. Int. Ed. 2014, 53, 3965–3969; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 4046–4050. [Google Scholar]
- 75. Deen J., Vranken C., Leen V., Neely R. K., Janssen K. P. F., Hofkens J., Angew. Chem. Int. Ed. 2017, 56, 5182–5200; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 5266–5285. [Google Scholar]
- 76. Muttach F., Mäsing F., Studer A., Rentmeister A., Chem. Eur. J. 2017, 23, 5988–5993. [DOI] [PubMed] [Google Scholar]
- 77. Wang R., Islam K., Liu Y., Zheng W., Tang H., Lailler N., Blum G., Deng H., Luo M., J. Am. Chem. Soc. 2013, 135, 1048–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Carter-O'Connell I., Jin H., Morgan R. K., Zaja R., David L. L., Ahel I., Cohen M. S., Cell Rep. 2016, 14, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Carter-O'Connell I., Jin H., Morgan R. K., David L. L., Cohen M. S., J. Am. Chem. Soc. 2014, 136, 5201–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gibson B. A., Zhang Y., Jiang H., Hussey K. M., Shrimp J. H., Lin H., Schwede F., Yu Y., Kraus W. L., Science 2016, 353, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Depaix A., Kowalska J., Molecules 2019, 24, 4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Vivelo C. A., Leung A. K. L., Proteomics 2015, 15, 203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wallrodt S., Buntz A., Wang Y., Zumbusch A., Marx A., Angew. Chem. Int. Ed. 2016, 55, 7660–7664; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 7790–7794. [Google Scholar]
- 84. Zhang X. N., Cheng Q., Chen J., Lam A. T., Lu Y., Dai Z., Pei H., Evdokimov N. M., Louie S. G., Zhang Y., Nat. Commun. 2019, 10, 4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kalesh K., Lukauskas S., Borg A. J., Snijders A. P., Ayyappan V., Leung A. K. L., Haskard D. O., DiMaggio P. A., Sci. Rep. 2019, 9, 6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Šileikytė J., Sundalam S., David L. L., Cohen M. S., J. Am. Chem. Soc. 2021, 143, 6787–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lam A. T., Zhang X. N., Courouble V. V., Strutzenberg T. S., Pei H., Stiles B. L., Louie S. G., Griffin P. R., Zhang Y., ACS Chem. Biol. 2021, 16, 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yang C., Mi J., Feng Y., Ngo L., Gao T., Yan L., Zheng Y. G., J. Am. Chem. Soc. 2013, 135, 7791–7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Han Z., Chou C. W., Yang X., Bartlett M. G., Zheng Y. G., ACS Chem. Biol. 2017, 12, 1547–1555. [DOI] [PubMed] [Google Scholar]
- 90. Chen N., Liu Y., Li Y., Wang C., Angew. Chem. Int. Ed. 2020, 59, 16069–16075; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 16203–16209. [Google Scholar]
- 91. McAlinden T. P., Hynes J. B., Patil S. A., Bobbin Westerhof G., Jansen G., Schornagel J. H., Kerwar S. S., Freisheim J. H., Biochemistry 1991, 30, 5674–5681. [DOI] [PubMed] [Google Scholar]
- 92. Lawrence A. D., Nemoto-Smith E., Deery E., Baker J. A., Schroeder S., Brown D. G., Tullet J. M. A., Howard M. J., Brown I. R., Smith A. G., Boshoff H. I., Barry C. E., Warren M. J., Cell Chem. Biol. 2018, 25, 941–951.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bagshaw C. R., J. Cell Sci. 2001, 114, 459–460. [DOI] [PubMed] [Google Scholar]
- 94. Hiratsuka T., Eur. J. Biochem. 2003, 270, 3479–3485. [DOI] [PubMed] [Google Scholar]
- 95. LaConte L. E. W., Srivastava S., Mukherjee K., Methods Mol. Biol. 2017, 1647, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Moreira B. P., Armstrong T., Batista I. C. A., Clemente Tavares N., Pires C. V., De Moraes Mouraõ M., Falcone F. H., Dekker L. V., ACS Omega 2020, 5, 9064–9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Simon R. P., Rumpf T., Linkuviene V., Matulis D., Akhtar A., Jung M., J. Med. Chem. 2019, 62, 2582–2597. [DOI] [PubMed] [Google Scholar]
- 98. Goyvaerts V., Van Snick S., D'Huys L., Vitale R., Helmer Lauer M., Wang S., Leen V., Dehaen W., Hofkens J., Chem. Commun. 2020, 56, 3317–3320. [DOI] [PubMed] [Google Scholar]
- 99. Long Y., Ubych K., Jagu E., Neely R. K., Bioconjugate Chem. 2021, 32, 192–198. [DOI] [PubMed] [Google Scholar]
- 100. Buntz A., Wallrodt S., Gwosch E., Schmalz M., Beneke S., Ferrando-May E., Marx A., Zumbusch A., Angew. Chem. Int. Ed. 2016, 55, 11256–11260; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 11423–11428. [Google Scholar]
- 101. Secrist J. A., Barrio J. R., Leonard N. J., Science 1972, 175, 646–647. [DOI] [PubMed] [Google Scholar]
- 102. Ottink O. M., Nelissen F. H. T., Derks Y., Wijmenga S. S., Heus H. A., Anal. Biochem. 2010, 396, 280–283. [DOI] [PubMed] [Google Scholar]
- 103. Zhang J., Zheng Y. G., ACS Chem. Biol. 2016, 11, 583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Moreau C., Kirchberger T., Zhang B., Thomas M. P., Weber K., Guse A. H., Potter B. V. L., J. Med. Chem. 2012, 55, 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hallé F., Fin A., Rovira A. R., Tor Y., Angew. Chem. Int. Ed. 2018, 57, 1087–1090; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 1099–1102. [Google Scholar]
- 106. Feldmann J., Li Y., Tor Y., Chem. Eur. J. 2019, 25, 4379–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Pergolizzi G., Butt J. N., Bowater R. P., Wagner G. K., Chem. Commun. 2011, 47, 12655–12657. [DOI] [PubMed] [Google Scholar]
- 108. Rovira A. R., Fin A., Tor Y., J. Am. Chem. Soc. 2017, 139, 15556–15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tangar A., Derrien V., Lei R., Estevez M. J. S., Sebban P., Bernad S., Miksovska J., Metallomics 2019, 11, 906–913. [DOI] [PubMed] [Google Scholar]
- 110. Morales-De-Echegaray A. V., Maltais T. R., Lin L., Younis W., Kadasala N. R., Seleem M. N., Wei A., ACS Infect. Dis. 2018, 4, 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bamnavat K., Bhardwaj V., Anand T., Kumar S. A., Sahoo S. K., Dyes Pigm. 2021, 184, 108844. [Google Scholar]
- 112. Li X., Wen Q., Gu J., Liu W., Wang Q., Zhou G., Gao J., Zheng Y., J. Mol. Liq. 2020, 319, 114124. [Google Scholar]
- 113. Upadhyay Y., Paira P., Ashok Kumar S. K., Choi H. J., Kumar R., Sahoo S. K., Inorg. Chim. Acta 2019, 489, 198–203. [Google Scholar]
- 114. Komatsu H., Shindo Y., Oka K., Hill J. P., Ariga K., Angew. Chem. Int. Ed. 2014, 53, 3993–3995; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 4074–4076. [Google Scholar]
- 115. Greene L. E., Godin R., Cosa G., J. Am. Chem. Soc. 2016, 138, 11327–11334. [DOI] [PubMed] [Google Scholar]
- 116. Belzile M. N., Godin R., Durantini A. M., Cosa G., J. Am. Chem. Soc. 2016, 138, 16388–16397. [DOI] [PubMed] [Google Scholar]
- 117. Riklin A., Katz E., Wiliner I., Stocker A., Bückmann A. F., Nature 1995, 376, 672–675. [DOI] [PubMed] [Google Scholar]
- 118. Ryabov A. D., Goral V. N., Gorton L., Csöregi E., Chem. Eur. J. 1999, 5, 961–967. [Google Scholar]
- 119. Glazer E. C., Le Nguyen Y. H., Gray H. B., Goodin D. B., Angew. Chem. Int. Ed. 2008, 47, 898–901; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2008, 120, 912–915. [Google Scholar]
- 120. Vignoni M., Walalawela N., Bonesi S. M., Greer A., Thomas A. H., Mol. Pharm. 2018, 15, 798–807. [DOI] [PubMed] [Google Scholar]
- 121. Matsuo T., Hayashi T., Hisaeda Y., J. Am. Chem. Soc. 2002, 124, 11234–11235. [DOI] [PubMed] [Google Scholar]
- 122. Kosman J., Stanislawska A., Gluszynska A., Juskowiak B., Int. J. Biol. Macromol. 2017, 101, 799–804. [DOI] [PubMed] [Google Scholar]
- 123. Liu Y., Lai P., Wang J., Xing X., Xu L., Chem. Commun. 2020, 56, 2427–2430. [DOI] [PubMed] [Google Scholar]
- 124. Zhang J., Song X., Xia M., Xue Y., Zhou M., Ruan L., Lu H., Chen J., Wang D., Chai Z., Hu Y., Chem. Commun. 2021, 57, 3038–3041. [DOI] [PubMed] [Google Scholar]
- 125. Chromiński M., Proinsias K. Õ., Martin E., Gryko D., Eur. J. Org. Chem. 2013, 1530–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Klahn P., Brönstrup M., Nat. Prod. Rep. 2017, 34, 832–885. [DOI] [PubMed] [Google Scholar]
- 127. Zhao S., Wang Z. P., Wen X., Li S., Wei G., Guo J., He Y., Org. Lett. 2020, 22, 6632–6636. [DOI] [PubMed] [Google Scholar]
- 128. Giedyk M., Jackowska A., Równicki M., Kolanowska M., Trylska J., Gryko D., Chem. Commun. 2019, 55, 763–766. [DOI] [PubMed] [Google Scholar]
- 129. Piazza I., Kochanowski K., Cappelletti V., Fuhrer T., Noor E., Sauer U., Picotti P., Cell 2018, 172, 358–372.e23. [DOI] [PubMed] [Google Scholar]
- 130. Schopper S., Kahraman A., Leuenberger P., Feng Y., Piazza I., Müller O., Boersema P. J., Picotti P., Nat. Protoc. 2017, 12, 2391–2410. [DOI] [PubMed] [Google Scholar]
- 131. Wu J. H., Batist G., Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3350–3353. [DOI] [PubMed] [Google Scholar]
- 132. Raza A., Galili N., Smith S., Godwin J., Lancet J., Melchert M., Jones M., Keck J. G., Meng L., Brown G. L., List A., Blood 2009, 113, 6533–6540. [DOI] [PubMed] [Google Scholar]
- 133. Zhang J., Ye Z. W., Janssen-Heininger Y., Townsend D. M., Tew K. D., in Handb. Exp. Pharmacol., Springer Science And Business Media Deutschland GmbH, 2021, pp. 71–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Jackowska A., Gryko D., Org. Lett. 2021, 23, 4940–4944. [DOI] [PubMed] [Google Scholar]
- 135. Lemon C. M., Marletta M. A., Inorg. Chem. 2021, 60, 2716–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Woodward J. J., Martin N. I., Marletta M. A., Nat. Methods 2007, 4, 43–45. [DOI] [PubMed] [Google Scholar]
- 137. Nierth A., Marletta M. A., Angew. Chem. Int. Ed. 2014, 53, 2611–2614; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 2649–2652. [Google Scholar]
- 138. McLean K. J., Munro A. W., Drug Discovery Today 2017, 22, 566–575. [DOI] [PubMed] [Google Scholar]
- 139. Suhara Y., Hanada N., Okitsu T., Sakai M., Watanabe M., Nakagawa K., Wada A., Takeda K., Takahashi K., Tokiwa H., Okano T., J. Med. Chem. 2012, 55, 1553–1558. [DOI] [PubMed] [Google Scholar]
- 140. Chrysochos N., Ahmadi M., Wahlefeld S., Rippers Y., Zebger I., Mroginski M. A., Schulzke C., Dalton Trans. 2019, 48, 2701–2714. [DOI] [PubMed] [Google Scholar]
- 141. Kailing L. L., Bertinetti D., Paul C. E., Manszewski T., Jaskolski M., Herberg F. W., Pavlidis I. V., Front. Genet. Front. Microbiol. 2018, 9, 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Pande V., Sun W., Beke L., Berthelot D., Brehmer D., Brown D., Corbera J., Irving S., Meerpoel L., Nys T., Parade M., Robinson C., Sommen C., Viellevoye M., Wu T., Thuring J. W., ACS Med. Chem. Lett. 2020, 11, 2227–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Bonday Z. Q., Cortez G. S., Grogan M. J., Antonysamy S., Weichert K., Bocchinfuso W. P., Li F., Kennedy S., Li B., Mader M. M., Arrowsmith C. H., Brown P. J., Eram M. S., Szewczyk M. M., Barsyte-Lovejoy D., Vedadi M., Guccione E., Campbell R. M., ACS Med. Chem. Lett. 2018, 9, 612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]


