Abstract
EmrR, the repressor of the emrRAB operon of Escherichia coli, was purified to 95% homogeneity. EmrR was found to bind putative ligands of the EmrAB pump—2,4-dinitrophenol, carbonyl cyanide m-chlorophenylhydrazone, and carbonyl cyanide p-(trifluoro-methoxy)phenylhydrazone—with affinities in the micromolar range. Equilibrium dialysis experiments suggested one bound ligand per monomer of the dimeric EmrR.
Bacteria have evolved mechanisms for the neutralization or extrusion of toxic compounds from the cell (6). Multidrug resistance pumps are integral membrane proteins that transport a broad range of structurally diverse compounds from the cell by using energy from either the proton motive force or ATP (7, 17, 18).
In three cases described so far, expression of a multidrug pump is increased by its chemically unrelated substrates via a transcriptional regulator (reviewed in reference 8). In Bacillus subtilis, BmrR, a MerR family transcriptional activator, binds chemically unrelated hydrophobic cations, such as tetraphenylphosphonium and rhodamine, and activates transcription of the BMR pump (1). The structure of the C-terminal ligand binding domain (13) of BmrR was recently resolved (20). The design of the binding site agrees well with its ability to accommodate a broad range of hydrophobic cations. In Staphylococcus aureus, QacR, belonging to the TetR family of repressors, also binds various hydrophobic cations and controls the expression of the QacA MDR pump (5).
In Escherichia coli, transcription of the EmrAB pump is controlled by EmrR, a 20.6-kDa protein that is encoded by the first gene of the emrRAB operon (11) and belongs to the MarR family of transcriptional repressors (16, 19). EmrA and EmrB form a multidrug pump that traverses the cell envelope and extrudes the antibiotic thiolactomycin, uncouplers of the proton motive force, and possibly other hydrophobic compounds (4, 10). Repression of the emrRAB operon by EmrR is relieved in the presence of inducers such as uncouplers of oxidative phosphorylation, salicylic acid, 2,4-dinitrophenol (DNP), carbonyl cyanide m-chlorophenylhydrazone (CCCP), and carbonyl cyanide p-(trifluoro-methoxy)phenylhydrazone (FCCP) (11). In addition to controlling the emr operon, EmrR (formerly MprA) regulates expression of the mcb operon coding for microcin B17 production (3, 9, 12).
The structural information gained from MDR regulators will be very useful in understanding the general principles of multidrug recognition that might be common to soluble multidrug sensors and the larger multidrug pumps they regulate. Toward this aim, we have undertaken the present study of ligand binding with purified EmrR.
Expression and purification of EmrR.
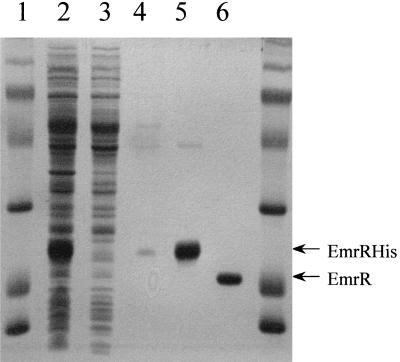
The emrR coding sequence was cloned into pET-14b vector (Novagen, Madison, Wis.) forming an N-terminal fusion with a histidine tag. EmrR was expressed and purified by immobilized metal affinity chromatography (IMAC) to 95% homogeneity. After the His tag was cleaved with thrombin, the purified protein migrated as a single band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with an apparent molecular mass of 20 kDa (Fig. 1), in good agreement with the calculated molecular mass of 20.6 kDa. A band corresponding to a possible EmrR dimer was observed under nonreducing conditions. Western blotting confirmed that the purified protein is EmrR (data not shown). Gel filtration of the IMAC-purified EmrR indicated the presence of two fractions with apparent molecular masses of 54 kDa (calculated molecular mass of EmrR dimer is 41.5 kDa) and 89 kDa, respectively (data not shown). EmrR contains four cysteine residues that may form intermolecular disulfide bonds and contribute to formation of high-molecular-weight aggregates. The high-molecular-weight species was not observed when gel filtration analysis was conducted in the presence of 100 mM dithiothreitol, suggesting that the putative tetramer was formed by disulfide cross-linking (EmrR has four cysteines). Reducing conditions did not affect the dimer, suggesting that it is not held together by disulfide bonds. The cytoplasm is strongly reduced, which suggests that the dimer is the natural form of EmrR.
FIG. 1.
Affinity purification of EmrR. Protein was purified as described above. Samples were loaded onto a 12.5% acrylamide SDS-PAGE gel, run at 200 V until the dye front was at the bottom of the gel, and then stained with Coomassie brilliant blue R-250. Lanes 1 and 7, molecular mass markers; lane 2, crude lysate; lane 3, material passed through the IMAC column; lane 4, wash with 50 mM imidazole in binding buffer; lane 5, elution with 200 mM imidazole in binding buffer; lane 6, EmrR after cleavage with thrombin.
Ligand binding properties of EmrR.
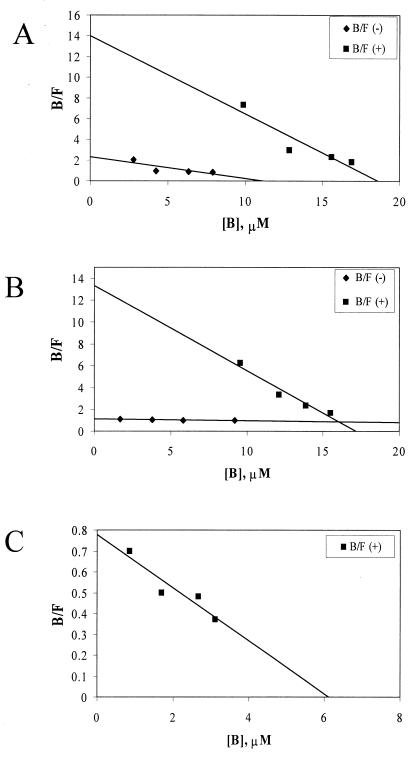
We examined three putative ligands of EmrR—DNP, CCCP, and FCCP—all of which are easily assayed spectrophotometrically. The substances absorb in the broad range of 260 to 400 nm, and absorbance at 380 nm was used to prevent interference from protein absorbance. To determine whether these ligands bind specifically to EmrR, crude extracts were prepared from cultures that had been either induced (+EmrR) or not (−EmrR) with isopropyl-β-d-thiogalactopyranoside (IPTG). EmrR without a histidine tag was expressed for those experiments by using a pET-21a vector (Novagen). Slide-a-Lyzer cassettes (Pierce) were loaded with 400 μl of 1-mg/ml extract and dialyzed ∼16 h against 400 ml of a buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 5% glycerol [pH 7.4]) containing various concentrations of ligand. Absorbance of protein was determined from dialysis against buffer without ligand and subtracted from all samples. Concentrations of ligand in both the bath and the cassette were measured by calculating from a standard curve for each ligand. To ensure that binding to protein had no effect on absorbance by the ligand, samples were measured both before and after treatment with proteinase K (data not shown). Bound ligand was determined by subtracting the measured concentration of a given bath from the concentrations measured from cassettes in that bath. Data were then plotted according to Scatchard (Fig. 2). The KS and the n[E]t were calculated according to the following formula: [bound ligand]/[free ligand] = (n[E]t/KS) − ([bound ligand]/KS). CCCP showed specific binding, but nonspecific binding was considerable, with a KNS-CCCP of 4.8 μM relative to a KS-CCCP of 1.3 μM, a 3.5-fold difference. FCCP showed a higher degree of specificity (KNS-FCCP = 62.5 μM versus KS-FCCP = 1.3 μM, nearly a 50-fold difference). DNP had no detectable nonspecific binding (the points clustered around the origin of the plot) and a KS-DNP of 11.1 μM in the crude extract (Fig. 2).
FIG. 2.
Equilibrium dialysis of EmrR bound to ligands. Ligand concentrations were obtained by measuring absorbance at 380 nm with a microtiter plate reader and referencing a corresponding calibration curve that produced a linear absorbance/concentration relationship in the 1 to 100 μM concentration range for each ligand. The data from equilibrium dialysis measurements were plotted to determine the KS (squares), KNS (diamonds), and n[E]t (x axis intercepts) for CCCP (A), FCCP (B), and DNP (C). The nonspecific data for DNP clustered around the origin and are not shown. Each data point is an average of three independent determinations. B, bound; F, free ligand.
Once ligand binding to EmrR was determined to be specific, additional ligand binding experiments were conducted with the IMAC-purified protein (Table 1). The concentration of binding sites (n[E]t, i.e., n, the number of binding sites per monomer, multiplied by the total amount of specific protein [E]t) can be calculated from the above formula or by examination of the x axis intercept. Binding parameters for EmrR in crude extract and isolated form were very similar. In crude extracts of 1 mg of total protein/ml, with an estimated [E]t of 5.8 μM, the n[E]t values were 18.6 μM for CCCP, 17.3 μM for FCCP, and 6.8 μM for DNP. These data do not take into account nonspecific binding, although for DNP, which shows essentially no nonspecific binding, they indicate one binding site per monomer. In a 1-mg/ml solution of IMAC-purified protein (estimated [E]t = 50 μM), n[E]t = 43 μM for CCCP. This would suggest approximately one binding site per monomer of EmrR. Similar data were obtained for FCCP, n[E]t = 43 μM, and for DNP, n[E]t = 41 μM, i.e., one binding site per monomer. This comparison between the behavior of EmrR in crude extract and in purified form suggests that the protein does not require cofactor(s) for ligand binding.
TABLE 1.
Ligand binding properties of purified EmrRa
| Ligand | KS (μM) | No. of sites/monomer |
|---|---|---|
| CCCP | 2.0 | 0.88 |
| FCCP | 3.0 | 0.82 |
| DNP | 15.0 | 0.81 |
Scatchard plots were generated from measurements performed as described in the legend for Fig. 2, and calculations were performed as described in the text.
Our data indicate that CCCP, FCCP, and DNP all bind to EmrR specifically and with micromolar affinities. It appears that one ligand binds per EmrR monomer. A homologous MarR repressor binds salicylate and controls the expression of a global regulator, MarA, in E. coli (14–16, 19). The helix-turn-helix motif of the MarR family regulators is well defined and is located at the center of the protein (2), suggesting that ligand binding is localized to the C- and/or N-terminal domain. We have expressed and purified the C-terminal domain (amino acids 77 to 173) and found that it has no ligand binding activity. The N-terminal domain (amino acids 1 to 88) formed an insoluble protein, suggesting that work toward crystallization and understanding the mechanism of ligand binding will have to be performed with the full-length dimeric protein. In collaboration with G. Petsko’s group, we recently obtained crystals of EmrR (1a), and future structural work should reveal the basis of drug recognition by this interesting multidrug sensor.
Acknowledgments
We thank Victoria L. Haynes and Natalia Yakovleva for help with some experiments.
This work was supported by NIH grant GM54412 and NSF grant MCB-9317013.
REFERENCES
- 1.Ahmed M, Borsch C M, Taylor S S, Vazquez-Laslop N, Neyfakh A A. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J Biol Chem. 1994;269:28506–28513. [PubMed] [Google Scholar]
- 1a.Clifton, J., A. Brooun, K. Lewis, and G. Petsko. Unpublished data.
- 2.Dalrymple B P, Swadling Y. Expression of a Butyrivibrio fibrisolvens E14 gene (cinB) encoding an enzyme with cinnamoyl ester hydrolase activity is negatively regulated by the product of an adjacent gene (cinR) Microbiology. 1997;143:1203–1210. doi: 10.1099/00221287-143-4-1203. [DOI] [PubMed] [Google Scholar]
- 3.del Castillo I, Gonzalez-Pastor J E, San Millan J L, Moreno F. Nucleotide sequence of the Escherichia coli regulatory gene mprA and construction and characterization of mprA-deficient mutants. J Bacteriol. 1991;173:3924–3929. doi: 10.1128/jb.173.12.3924-3929.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furukawa H, Tsay J T, Jackowski S, Takamura Y, Rock C O. Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J Bacteriol. 1993;175:3723–3729. doi: 10.1128/jb.175.12.3723-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grkovic S, Brown M H, Roberts N J, Paulsen I T, Skurray R A. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J Biol Chem. 1998;273:18665–18673. doi: 10.1074/jbc.273.29.18665. [DOI] [PubMed] [Google Scholar]
- 6.Levy S B. The challenge of antibiotic resistance. Sci Am. 1998;278:46–53. doi: 10.1038/scientificamerican0398-46. [DOI] [PubMed] [Google Scholar]
- 7.Lewis K. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem Sci. 1994;19:119–123. doi: 10.1016/0968-0004(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 8.Lewis K. Multidrug resistance: versatile drug sensors of bacterial cells. Curr Biol. 1999;9:R403–R407. doi: 10.1016/s0960-9822(99)80254-1. [DOI] [PubMed] [Google Scholar]
- 9.Lomovskaya O, Kawai F, Matin A. Differential regulation of the mcb and emr operons of Escherichia coli: role of mcb in multidrug resistance. Antimicrob Agents Chemother. 1996;40:1050–1052. doi: 10.1128/aac.40.4.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomovskaya O, Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci USA. 1992;89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lomovskaya O, Lewis K, Matin A. EmrR is a negative upstream regulator of the E. coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao W, Siegele D A. Genetic analysis of the stationary phase-induced mcb operon promoter in Escherichia coli. Mol Microbiol. 1998;27:415–424. doi: 10.1046/j.1365-2958.1998.00690.x. [DOI] [PubMed] [Google Scholar]
- 13.Markham P N, Ahmed M, Neyfakh A A. The drug-binding activity of the multidrug-responding transcriptional regulator BmrR resides in its C-terminal domain. J Bacteriol. 1996;178:1473–1475. doi: 10.1128/jb.178.5.1473-1475.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin R G, Jair K W, Wolf R E, Jr, Rosner J L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin R G, Rosner J L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller P F, Sulavik M C. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 17.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 18.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seoane A S, Levy S B. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol. 1995;177:3414–3419. doi: 10.1128/jb.177.12.3414-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheleznova E A, Markham P N, Neyfakh A A, Brennan R G. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell. 1999;96:353–362. doi: 10.1016/s0092-8674(00)80548-6. [DOI] [PubMed] [Google Scholar]