Abstract
Purpose:
Macrophages are critical in driving an immunosuppressive tumor microenvironment that counteracts the efficacy of T-cell–targeting therapies. Thus, agents able to reprogram macrophages toward a proinflammatory state hold promise as novel immunotherapies for solid cancers. Inhibition of the macrophage scavenger receptor Clever-1 has shown benefit in inducing CD8+ T-cell–mediated antitumor responses in mouse models of cancer, which supports the clinical development of Clever-1–targeting antibodies for cancer treatment.
Patients and Methods:
In this study, we analyzed the mode of action of a humanized IgG4 anti–Clever-1 antibody, FP-1305 (bexmarilimab), both in vitro and in patients with heavily pretreated metastatic cancer (n = 30) participating in part 1 (dose-finding) of a phase I/II open-label trial (NCT03733990). We studied the Clever-1 interactome in primary human macrophages in antibody pull-down assays and utilized mass cytometry, RNA sequencing, and cytokine profiling to evaluate FP-1305–induced systemic immune activation in patients with cancer.
Results:
Our pull-down assays and functional studies indicated that FP-1305 impaired multiprotein vacuolar ATPase–mediated endosomal acidification and improved the ability of macrophages to activate CD8+ T-cells. In patients with cancer, FP-1305 administration led to suppression of nuclear lipid signaling pathways and a proinflammatory phenotypic switch in blood monocytes. These effects were accompanied by a significant increase and activation of peripheral T-cells with indications of antitumor responses in some patients.
Conclusions:
Our results reveal a nonredundant role played by the receptor Clever-1 in suppressing adaptive immune cells in humans. We provide evidence that targeting macrophage scavenging activity can promote an immune switch, potentially leading to intratumoral proinflammatory responses in patients with metastatic cancer.
Translational Relevance.
Successfully overcoming innate immune checkpoints remains elusive in cancer therapy. We demonstrate that adaptive immune activation in advanced cancer can be achieved by modulating endolysosomal acidification and antigen degradation in macrophages. Thus, targeting scavenger receptors may provide an attractive immunotherapeutic strategy in cancer.
Introduction
Immunotherapies targeting CTLA-4 and PD-1/-L1 checkpoint molecules have revolutionized cancer treatment in the last few years and are offering unforeseen prospects of cure for some patients. Despite these advances, most patients remain refractory or develop therapeutic resistance for unspecified reasons (1). In light of current knowledge from prognostic and preclinical studies, tumor-associated macrophages (TAM) are proving to be major contributors to therapeutic resistance by effectively suppressing antitumor immune responses (2). Strategies under clinical development for stimulating the antitumor properties of these cells include Toll-like receptor (TLR), STING, and CD40 agonists, and inhibitors of PI3Kγ and the CD47/SIRPα “don't eat me” signaling axis, which promotes phagocytosis of cancer cells (3). Preclinical studies have shown that Clever-1 (also known as Stabilin-1 and FEEL-1) is also a potential target for promoting macrophage reprogramming as Clever-1 impairs the secretion of proinflammatory cytokines, leading to ineffective activation of antigen-specific T-cell responses (4).
Clever-1 is a multifunctional scavenger and adhesion receptor expressed by human monocytes, subsets of immunosuppressive macrophages, lymphatic endothelial cells, and sinusoidal endothelial cells in the liver and spleen. In macrophages, Clever-1 is involved in receptor-mediated endocytosis and recycling, intracellular sorting, and transcytosis of altered and normal self-components (5). Immunotherapeutic targeting of Clever-1 in various tumor models delays tumor growth (6) by activating cytotoxic CD8+ T-cells and renders refractory tumors more responsive to anti–PD-1 therapy (7). To elucidate the significance of Clever-1 in suppressing antitumor immune responses in cancer patients, a phase I/II first-in-man clinical trial (MATINS; NCT03733990) was initiated to study the safety, tolerability, and early efficacy of FP-1305 (bexmarilimab), a humanized anti–Clever-1 antibody, in patients with selected advanced or metastatic solid tumors (8). The FP-1305 antibody contains IgG4(S241P) heavy-chain and κ light-chain constant regions and has further been optimized by introducing the L248E mutation to avoid Fcγ receptor binding (9). Thus, FP-1305 has very low antibody-dependent cell cytotoxicity and complement-mediated effector functions, which is important to circumvent any unwanted effects on lymphatic and sinusoidal endothelial cells that express Clever-1 (10).
In this study we describe the systemic immune signatures induced by FP-1305 treatment in patients with advanced cancer and provide a mechanistic understanding of how a macrophage-targeted approach can promote robust activation of T-cells and lead to promising antitumor immune responses.
Patients and Methods
Study design, patients, and procedures
This multi-institutional, first-in-human, open-label, nonrandomized phase I dose-escalation study was approved by local institutional review boards (IRB). All participating patients signed informed consent. Patients in part 1 of the MATINS trial were dosed between December 2018 and April 2020 in the study sites at Helsinki, Oulu, and Tampere University Hospitals, Erasmus MC/Cancer Institute, Royal Marsden London, and University Hospitals Birmingham. The data cut-off date for protocol-specified part 1 analysis was May 7, 2020. Participation was offered for subjects with advanced (inoperable or metastatic), treatment-refractory, histologically confirmed hepatobiliary, pancreatic, colorectal, or ovarian cancer or cutaneous melanoma without standard treatment options. Melanoma patients had to be immunotherapy-refractory (progression on or after anti–PD-1/CTLA-4 therapy). Patients meeting the inclusion and none of the exclusion criteria specified in the study protocol (EudraCT No. 2018--002732--24) were consented, prescanned (CT) for metastatic lesions, and prebiopsied before FP-1305 administration. Eligible patients had no cancer therapy for at least 3 weeks before first FP-1305 administration. Patients were e18 years old with a life expectancy of more than 12 weeks, an Eastern Cooperative Oncology Group performance status of 0 or 1, adequate organ function, and no ongoing systemic infections or active autoimmune diseases. Concurrent antineoplastic therapies or systemic steroids were not permitted. Patients with measurable or nonmeasurable disease were allowed to be enrolled in part 1.
Sample randomization and collection
The patient samples were analyzed in a blinded fashion not knowing patient clinical characteristics or drug responses. Since only 9 mL of blood was obtained from each patient at indicated timepoints, all analyses could not be performed uniformly on all patient samples. Thus, these were also randomly assigned per patient (Supplementary Table S1). Signed informed consent was obtained from healthy donors donating cells. The samples were stored anonymously and handled according to the ethical guidelines set by the University of Turku (Turku, Finland).
Sample collection and peripheral blood mononuclear cell isolation
Heparin blood was collected at indicated timepoints at the clinical study sites. Blood was delivered overnight to MediCity at room temperature. Peripheral blood mononuclear cells (PBMC) were isolated with Ficoll-Paque density gradient centrifugation (GE Healthcare) and used fresh for flow cytometry, monocyte RNA sequencing (RNA-seq), and lipopolysaccharide (LPS) induction assays, while the remaining cells were frozen for later use.
Human macrophage differentiation and polarization
Monocytes were enriched from buffy-coat PBMCs from healthy donors with CD14 Microbeads (Miltenyi Biotec). Monocytes were cultured in Iscove's modified Dulbecco's medium (IMDM; 21980–032; Thermo Fisher) supplemented with 10% FBS, penicillin/streptomycin, and 50 ng/mL human M-CSF (574806; BioLegend) and incubated for 7 days with one medium change. After differentiation, the macrophages were M2-polarized with 100 nmol/L dexamethasone (D-2915; Merck) and 20 ng/mL of human IL-4 (200–04–50UG; PeproTech) for 2 days.
Coimmunoprecipitation
Macrophages were detached with 5 mmol/L EDTA in PBS and gentle scraping. Collected cells were lysed with immunoprecipitation (IP) lysis buffer (20 mmol/L Tris-HCl, pH 8.0; 137 mmol/L NaCl; 1% Triton X-100; 2 mmol/L EDTA) supplemented with 2× complete Protease Inhibitor Cocktail (Merck). The lysates were precleared and incubated with 10 μg of 9-11 (InVivo Biotech), FP-1305 (clone CP12; Abzena), or isotype-control antibodies (rat IgG2a, clone eBR2a; Thermo Fisher and human IgG4, clone QA16A15; BioLegend, respectively). Immune complexes were precipitated with Dynabeads Protein G beads (10003D; Thermo Fisher) and eluted with 2× nonreducing sample buffer (120 mmol/L Tris-HCl, pH 6.8; 20% glycerol; 4% SDS) at 70°C for 10 minutes. For mass spectrometry, DTT was added to 50 mmol/L, after which the samples were incubated at 95°C for 5 minutes and run approximately 1.5 cm into 10% hand-cast–resolving Tris-glycine gels. The gels were stained with Coomassie Brilliant Blue and the protein lanes were cut and submitted for mass spectrometry.
Mass spectrometry
Mass spectrometry was performed at the Turku Proteomics Facility (University of Turku, Turku, Finland), using standard protocols. Briefly, samples were in-gel–digested and dissolved in 0.1% formic acid. LC-ESI-MS/MS analysis was performed on a nanoflow high-performance liquid chromatography (HPLC) system (Easy-nLC1200) coupled with the Q Exactive HF mass spectrometer (both from Thermo Fisher) and a nano-electrospray ionization source. Peptides were loaded on trapping columns and separated inline on a 15 cm C18 column (Dr. Maisch HPLC GmbH). The mobile phase consisted of 0.1% formic acid or 80:20 acetonitrile:water (v/v) in 0.1% formic acid (solvent). Peptides were eluted with 30- or 60-minute gradients from 8% to 43% solvent followed with 100% solvent wash.
LC-MS/MS data acquisition and analysis
Mass spectrometric data was acquired automatically with Thermo Xcalibur v4.1 software (Thermo Fisher). An information-dependent acquisition method consisted of an Orbitrap MS survey of the mass range 300–2000m/z followed by HCD fragmentation. Data files were searched for protein identification using Proteome Discoverer v2.3 software (Thermo Fisher) connected to an in-house server running the Mascot v2.6.1 software (Matrix Science). Data were searched against the SwissProt database with the taxonomy filter Homo sapiens, trypsin as the enzyme, oxidation and acetylation as variable modifications, carbamidomethylation as a fixed modification, ±10ppm as the peptide mass tolerance, ±0.02Da as the fragment mass tolerance, maximum missed cleavages as 2, and the instrument type as ESI-telomeric repeat amplification protocol (TRAP). A minimum of two peptides per protein were used to filter the results. The criteria used for including a protein as a 9-11– or FP-1305–specific hit were that the protein was present in at least two of three biological replicates, the protein was identified by at least three unique peptides, and that the protein was not identified in the IgG control or it had an over 3-fold enrichment of uniquely identified peptides over the IgG control. To detect likely contaminants among the mass spectrometry hits, we uploaded the spectral count data of the 9-11– and FP-1305–specific proteins into CRAPome (11) and analyzed them against 16 selected CRAPome controls (cell type HEK293, total cell lysate, Dynabeads magnetic affinity support) using default settings. FC-A>4 was used as the cut-off value. Reported high-confidence (>0.7) protein–protein interactions between the hit proteins were downloaded from the STRING database (12) and visualized with Cytoscape 3.7.2 (13). The yFiles Organic layout was applied and clusters identified with the Markov Clustering (MCL) algorithm of the clusterMaker app using default settings (14). Gene Ontology Biological Process terms enriched in the five largest clusters were identified with the STRING Enrichment app (redundancy cutoff 0.7; ref. 15). Functional enrichment among differently precipitated proteins was determined by performing Ingenuity Pathway Analysis (IPA; ref. 16).
Western blot analysis
Primary antibodies used for Western blotting were: anti-ATP6V0A1 (NBP1–59949; Novus Biologicals), anti-ATP6V1A (PA5–29191; Invitrogen), anti-TCIRG1 (PA5–90425; Invitrogen), and anti–Clever-1 (clone 3-372; InVivo Biotech).
KG-1 cell culture, differentiation, and RNAi
The Clever-1high acute myelogenous leukemia cell line KG-1 (CCL-246; ATCC) was cultured in IMDM supplemented with 20% FBS and penicillin/streptomycin. To induce KG-1 macrophage differentiation, phorbol 12-myristate 13-acetate (PMA; P8139; Merck) was added to 300 nmol/L for three days. On the day of transfection, KG-1 cells were resuspended in OPTI-MEM reduced serum medium (51985026; Thermo Fisher). siRNA (2 μmol/L) were used per electroporation: pooled scramble control siRNA (ON-TARGETplus Control Pool D-001810–10–20) and single siRNAs targeting the human Clever-1 RNA sequences AUGAUGAGCUCACGUAUAA (siR1) or UCAAGUCGCUGCCUGCAUA (siR2; J-014103–05–0020 and J-014103–08–0020, respectively; all from Dharmacon). After electroporation with program U-001 on the Nucleofector II device (Amaxa Biosystems) and overnight recuperation, the KG-1 cells were differentiated into macrophages.
Endocytosis and acidification experiments
LysoSensor Green DND-189 (L7535) was added to prewarmed culture medium, after which the cells were incubated for the indicated timepoints before FACS. For DQ-OVA antigen degradation (D12053) and acetylated low-density lipoprotein (acLDL) uptake and acidification, the cells were resuspended in CO2-independent medium (18045054) and the indicated treatments added to 10 μg/mL. The cells were incubated at 16°C for 30 minutes to allow initial surface binding, washed, and incubated for the indicated timepoints before FACS. Native human acLDL was purchased from Bio-Rad (5685–3404) or Thermo Fisher (L35354) and labeled with Alexa Fluor (AF) 488 Microscale, AF647 Protein Labeling, or pHrodo iFL Green Microscale Protein Labeling Kit (A30006, A20173, and P36015, respectively) according to manufacturer's instructions. To quench extracellular fluorescence, Trypan blue was added to 0.1% before FACS. Standard curves for cells treated with LysoSensor Green or acLDL labeled with pHrodo Green were prepared with the Intracellular pH Calibration Buffer Kit (P35379) according to manufacturer's instructions (all reagents from Thermo Fisher).
Immunofluorescence
FP-1305, 9-11, and their isotype-control antibodies were conjugated with AF488 or AF647 Protein Labeling kits (A20173 and A10235; Thermo Fisher). M2-polarized primary human macrophages were plated in 8-well ibiTreat chambers (80826; Ibidi) and treated with the conjugated antibodies for 5 minutes or 2 hours. To assess ATP6V0A1 localization, KG-1 cells were PMA differentiated and incubated with 10 μg/mL AF647-conjugated human acLDL for 3 hours. After the indicated treatments cells were fixed with 4% PFA, permeabilized with 0.1% Triton X-100, and stained with the following antibodies: anti–LAMP-1 (clone D2D11, 9091; Cell Signaling Technology), anti-ATP6V0A1 (H00000535-A01; Abnova), anti–Clever-1 (9-11), rabbit IgG (BE0095; BioxCell), mouse IgG (010–0102–0005; Rockland), and rat IgG2a (clone 2A3; InVivoPlus). Z-stacks of macrophages from the 2-hour timepoint were captured with an LSM 880 confocal microscope with Airyscan detector and a 63×/1.4 objective (C Plan-Apochromat, Carl Zeiss). Images were acquired and processed with Zen software (2.3 SP1 black edition, Carl Zeiss). NIH ImageJ software (v.1.52p) was used for all image analyses. After background subtraction with median filtering, the Colocalization Threshold plug-in was used to calculate Manders' colocalization coefficients above Costes threshold within each macrophage. Z-stacks of macrophages (5 minutes) and KG-1 cells were acquired with Marianas spinning disk (Intelligent Imaging Innovations) connected to CSU-W1 scanning unit (Yokogawa) using a 63×/1.4 objective (Plan-Apochromat, Carl Zeiss) and SlideBook 6.1 software (Intelligent Imaging Innovations). Z-stack mid-slices of non-transfected or Scr-transfected Clever-1+ cells and siR1-transfected Clever-1− cells were used in analyses. Clever-1 and ATP6V0A1 intensity profiles were generated from background-subtracted images using the Plot Profile function. A fixed threshold was applied to LAMP-1 channel images to determine LAMP-1+ vesicle regions from which the mean fluorescence intensity of ATP6V0A1 was measured.
Mixed leukocyte reaction
At 24-hour macrophage polarization, the indicated antibody treatments were added to 100 μg/mL and the polarization was continued another 24 hours. For M1 polarization, macrophages were first incubated for 24 hours with 20 ng/mL IFNγ, then with 100 ng/mL LPS for another 24 hours. Lymphocytes were isolated from buffy-coat PBMCs with CD3 Microbeads (Miltenyi Biotec), labelled with 1 μmol/L CellTrace Violet (C34557; Invitrogen), and added on macrophages at a 5:1 ratio. For positive control reactions, wells were coated with 10 μg/mL anti-CD3 (clone OKT3) and medium supplemented with 2 μg/mL anti-CD28 (clone CD28.3) and 20 ng/mL recombinant human IL-2 (589104; all from BioLegend). MLR reactions were incubated for 4 days before staining with anti–CD4-PE (555347) anti–CD8-APC (555369) antibodies (both from BD) and 7-AAD (00–6993–50; Invitrogen) for dead cell exclusion. FACS was run on the LSRFortessa (BD) and analyzed with FlowJo software v. 10.7.1 (TreeStar).
Cytometry time-of-flight and data analysis
The used antibodies and their clones, metal tags, and providers are listed in Supplementary Materials and Methods with detailed sample handling and staining procedures. The data analysis was performed similarly as by Kimball and colleagues (17) with R studio version 1.2.1335 and cytofkit package (Bioconductor). Manually gated events (FlowJo) were imported into cytofkit and subjected to PhenoGraph analysis. For panel 1, clustering was performed by using CD4, CD8, CD45RA, CCR7, CD45RO, CD127, CD25, CCR6, and CXCR3 with the following additional settings: merge method as minimum, transformation as CytofAsinh, cluster method as Rphenograph, visualization method as tSNE, and cellular progression as “NULL”. For panel 2, clustering was performed by using CD3, CD11b, CD64, CD14, CD16, CD56, CD19, CD11c, and HLA-DR with the same additional settings. The clusters were displayed on tSNE blots by using cytofkit shinyAPP to visualize different patients before and after treatment. For each patient, the relative size of each cluster was calculated at both timepoints. A cluster×patient matrix was formed from the arithmetic differences in relative cluster sizes between timepoints. The matrix was clustered hierarchically with Euclidean distance and complete linkage and plotted as a heatmap (R package ComplexHeatmap, function Heatmap; ref. 18) along with black circles representing relative cluster sizes at each timepoint, with circle radiuses proportional to the square root of the relative cluster sizes. Median marker expression in each cell cluster was calculated separately at both timepoints and for each patient. The change in marker expressions for each cluster between the timepoints was assessed by applying a one-sample t test on the differences of medians. P values were corrected for multiple testing using false discovery rate correction (Benjamini–Hochberg, R function p.adjust; ref. 19). A marker×cluster matrix was formed from the FDR-corrected negative log10-transformed P values multiplied by the sign of the effect (positive or negative), clustered, and displayed as a heatmap as described above (18).
Flow cytometry
PBMCs were plated at 0.2×106 cells/well in round-bottom 96-well plates. All wells were stained with anti–human CD14-Pacific Blue (clone M5E2; BD Pharmingen) together with 10 ng/μL in-house conjugated (AF647) anti–Clever-1 antibodies 9-11 or FP-1305. Irrelevant isotype control antibodies rat IgG2a or human in-house conjugated IgG4 (S241/L248E) were used for signal normalization, respectively. Blood neutrophils were identified by anti–CD11b-AF488 (clone ICRF44; BioLegend) and anti–CD15-APC (clone HI98; BD) after red blood cell (RBC) lysis with PharmLyse. Lymphocyte FACS was performed by Covance according to their standard protocols.
RNA sequencing
CD14+ monocytes were enriched from patients as previously described. Monocyte purity (>97%) was confirmed by FACS after anti-CD14 (clone M5E2; BD Pharmingen) and anti-CD11b (clone ICRF44; BioLegend) staining. Monocyte RNA was extracted with TRIsure (BioLine). cDNA library preparation and sequencing were performed by Novogene according to their standard protocols on the Illumina NovaSeq 6000 platform, where 150-bp paired-end reads were generated. High-quality reads were mapped to GRCh38 reference genome with HISAT2 (20). Transcript abundancy was quantified using HTSeq (21) and gene-expression levels calculated as FPKM (fragments per kilobase million). Regularized-logarithm transformed raw read counts were used for unsupervised hierarchical clustering (Euclidean distance, complete linkage) and principal component analysis (PCA), which were performed with R v.3.6.2 on genes with FPKM >1. For differential expression analysis, read counts were trimmed mean of M-values–normalized in edgeR (22) and differentially expressed genes in D1 and D7 samples were identified in each patient with DEGseq (23) as genes with FDR-adjusted P < 0.005 and |log2(fold change)| >1. Functional enrichment among differentially expressed genes (FPKM >1) was determined by performing IPA core analysis for each patient and subsequent IPA comparison analysis. Heatmaps were plotted with R package pheatmap. The RNA-seq data has been submitted to Gene Expression Omnibus (GEO) under the accession code GSE151085.
qPCR
Total RNA was reverse transcribed with the SuperScript VILO cDNA synthesis kit (11754–250; Thermo Fisher). qPCR was performed with predesigned TaqMan gene expression assays (Applied Biosystems) for IL1B (Hs01555410_m1), IL1R2 (Hs00174759_m1), JUN (Hs01103582_s1), TNFRSF1A (Hs01042313_m1), and GAPDH (Hs02758991_g1), and TaqMan universal master mix II (Applied Biosystems) in triplicate reactions (4 ng of cDNA per reaction) following manufacturer's instructions. The reactions were run with QuantStudio 3 and analyzed with the QuantStudio software v1.5.1 (Applied Biosystems). Relative quantification was calculated with the ∆∆CT method using GAPDH as an endogenous control.
T-cell activation assay
PBMCs were plated at 1–3×106 cells per well in ultra-low-attachment 96-well plates (Corning) in IMDM supplemented with brefeldin A (420601; BioLegend), 50 ng/mL PMA, and 0.5 μg/mL ionomycin (Sigma) and incubated at 37°C for 4 hours. The cells were washed, stained with Zombie Red viability dye (423109; BioLegend), and surface stained with anti–CD8-FITC (clone RPA-T8; BD). The cells were fixed with 4% PFA and permeabilized using 0.3% Triton X-100 in PBS and thereafter stained with anti–IFNγ-APC (clone B27; BD) and anti–IL-2-BV421 (clone MQ1-17H12; BioLegend).
IFNγ and CXCL10
Serum samples were analyzed using V-PLEX Proinflammatory panel-1 (Meso Scale Diagnostics) according to the kit instructions.
LPS induction assay
PBMCs (0.1×106) were plated in ultra-low-attachment 96-well plates and incubated with or without TLR4-specific LPS (20 ng/mL; LPS-EB Ultrapure, InvivoGen) in IMDM for 24 hours. The cleared supernatant was analyzed with Bio-Plex Pro Human Inflammation Panel-1, 37-Plex (#171AL001M; Bio-Rad) according to the manufacturer's protocol.
IHC
Formalin-fixed paraffin-embedded (FFPE) tissues were stained with Vectastain Elite ABC-HRP rat kit (PK-6104; Vector Laboratories) using rat anti–Clever-1 (clone S2–7/H7) as previously described (24). CD8 and CD163 staining was performed at Covance according to their validated protocols. For immunofluorescence (IF) analysis, rabbit anti-Granzyme B (PA1–26616; Invitrogen), mouse anti-CD8 (LS-C311966; LSbio), or irrelevant isotype controls and the secondary antibodies anti–rabbit-AF488 (A11034; Life Technologies) and anti–mouse-AF546 (A11030; Invitrogen) were used. The slides were mounted with Prolong Gold containing DAPI (Thermo Fisher).
Single-cell RNA and T-cell receptor sequencing
T-cells were enriched using CD3 Microbeads (Miltenyi Biotec). Single cells were partitioned using a Chromium Controller and scRNA-seq and TCRαβ libraries were prepared using Chromium Next GEM Single Cell 52 Library & Gel Bead Kit (all from 10X Genomics) per manufacturer's instructions (CG000207 Rev D). Full-length cDNA was amplified using 14 cycles of PCR (Veriti, Applied Biosystems). T-cell receptor (TCR) cDNA was further amplified in a hemi-nested PCR reaction using Chromium Single Cell Human T Cell V(D)J Enrichment Kit (10X Genomics). Finally, the total cDNA and the TCR-enriched cDNA was subjected to fragmentation, end repair and A-tailing, adapter ligation, and sample index PCR (14 and 9 cycles, respectively). The libraries were sequenced using an Illumina NovaSeq, S1 flowcell with the following read length configuration: Read1 = 26, i7 = 8, i5 = 0, Read2 = 91.
Single-cell RNA-seq data analysis
Postprocessing including demultiplexing, read alignment, and quality control was performed at the Institute for Molecular Medicine Finland (FIMM) using the 10X Genomics Cell Ranger package (v3.1.0) with GRCh38 as the reference genome. The Cell Ranger outputs were analyzed by Seurat (v3.1) for graph-based clustering, analysis of differentially expressed genes, and extraction of TCR sequences from cell subsets. TCR clonotype data including clonotype ID and CDR3 amino acid sequences were added to the metadata for each cell; C1D1, C4D8, and C4D15 data were merged. For quality control, genes expressed in less than 5 cells were removed. We also filtered out cells that had less than 200 genes and cells with greater than 4,000 genes and 10% mitochondrial genes. Data were normalized by the function of NormalizeData, and variable genes across the single cells were detected by the FindVariableFeatures. A linear transformation was applied on the data by the function ScaleData prior to dimensional reduction. Then, FindNeighbors and FindClusters were used to apply nearest neighbor graph-based clustering to the cells using 15 dimensions and 0.6 resolution. The clustering was visualized with Uniform Manifold Approximation and Projection (UMAP) using RunUMAP with 15 dimensions. To analyze CD8+ T-cell subsets and the clonality during the treatment, CD8A-expressing CD8+ T-cell clusters were selected from initial clusters and subclustered by reapplying FindVariableFeatures, ScaleData, FindNeighbors, FindClusters, and RunUMAP with 15 dimensions and resolution 0.6. Metadata of each CD8+ T-cell were extracted and frequencies of each clonotype at each timepoint were calculated. The changes in CD8+ T-cell clones during the treatment were chased by analyzing the CDR3 amino acid sequences. The scRNA-seq data has been submitted to GEO under the accession code: GSE152169.
Statistical analysis
Data are presented as mean ± SD unless otherwise noted with violin plots additionally showing individual data points. Comparisons between repeated measures were performed with one-way ANOVA (parametric), Friedman test (nonparametric), or two-way ANOVA with Sidak multiple comparisons test. Group analyses between two timepoints (treatments, genotypes) were performed with two-sided (un)paired Student t test (parametric) or Mann–Whitney U test and Wilcoxon matched-pairs signed rank test (non-parametric). For linear regression analyses we visually inspected residual plots to validate linearity and homoscedasticity. P < 0.05 was considered statistically significant. Statistical analyses were performed with Prism 9 (GraphPad).
Results
Clever-1 harbors an interaction motif for the multiprotein vacuolar ATPase complex
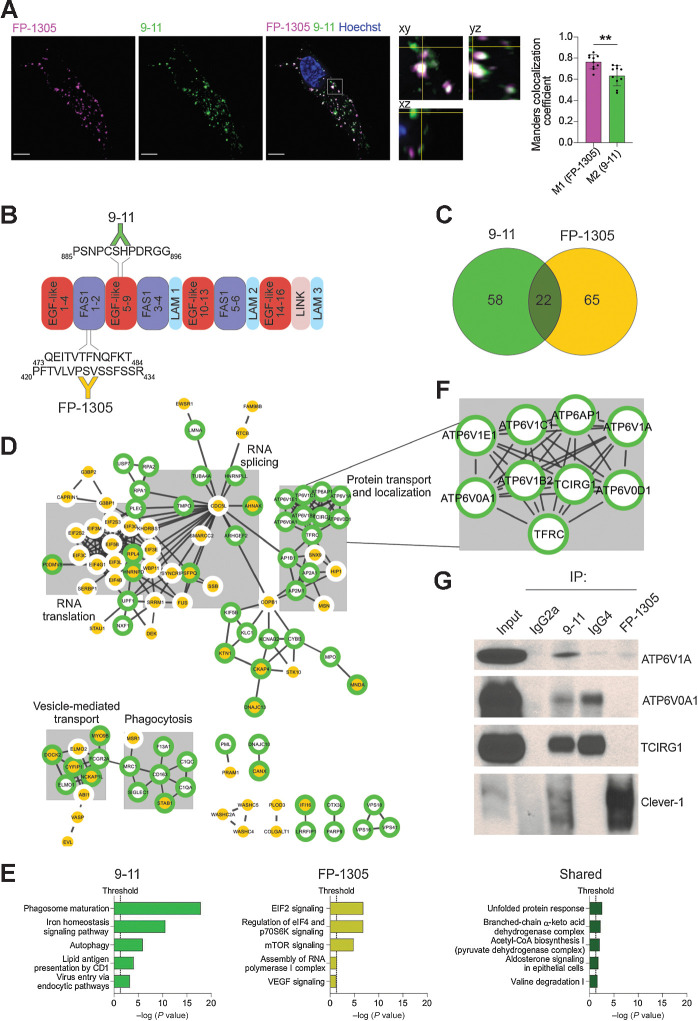
Clever-1 is a very large (approximately 280 kDa) type-I transmembrane protein with a large extracellular portion containing seven fasciclin-like domains, multiple EGF-like domains, and one X-link homology region (25). Thus, antibodies targeting Clever-1 at different sites most likely produce a multitude of biological responses related to its scavenging and adhesion functions. To gain insight into the mode of action of FP-1305 and uptake by Clever-1 on myeloid cells, primary human M2 macrophages polarized with IL-4 and dexamethasone were given fluorochrome-conjugated FP-1305 (AF647) and a noncompeting antibody, 9-11 (AF488) for intracellular colocalization analysis. Both antibodies were rapidly internalized (Supplementary Fig. S1A) and found to colocalize in intracellular vesicles. However, some structures were positive only for either one of the antibodies, suggesting antibody-related differences in the endosomal trafficking of Clever-1 (Fig. 1A). To further explore this phenomenon, we performed Co-IP/LC-MS/MS with FP-1305 or 9-11 (Fig. 1B). Co-IP/LC-MS/MS analysis of three biological replicates resulted in a list of 244 proteins enriched by either or both antibodies (Supplementary Data File 1). To identify and remove common background contaminants, we analyzed the list of 244 proteins against the CRAPome database with FC-A>4 as the cut-off value for high-scoring interactions. This resulted in the removal of most nuclear, ribosomal, and ribonucleoproteins and pruned the hits to 145 proteins of which 58 were specific for 9-11, 65 were specific for FP-1305, and 22 were common between the two antibodies (Fig. 1C and Supplementary Data File 1). We mapped all high-confidence (score>0.7) protein–protein interactions reported in the STRING database between the 145 proteins in CytoScape and identified protein clusters with the MCL tool of the clusterMaker app (Fig. 1D). We used the STRING Enrichment tool to search for significant non-redundant (cut-off>0.7) Gene Ontology Biological Process (GO BP) terms shared by proteins in the five largest clusters. The proteins within these clusters were thematically connected by GO BP terms related to protein transport and localization, RNA splicing, RNA translation, vesicle-mediated transport, and phagocytosis (Fig. 1D and Supplementary Data File 1). We then performed IPA separately on proteins precipitated with 9-11, proteins precipitated with FP-1305, and proteins that were immunoprecipitated with both antibodies. The most significant canonical pathway identified by IPA was phagosome maturation, which was obtained with 9-11 (Fig. 1E). The proteins constituting this pathway were subunits of the lysosomal membrane V0 and the cytosolic V1 sectors of the vacuolar ATPase (v-ATPase; Fig. 1F). Specificities of the v-ATPase subunits ATP6V1A, ATP6V0A1, and TCIRG1 for the 9-11 antibody were validated by co-IP/Western blot (Fig. 1G). Because FP-1305 was unable to precipitate Clever-1 when bound to these subunits, it can be postulated that FP-1305 can interfere in the acidification of phago-lysosomes during phagosome maturation.
Figure 1.
Clever-1 harbors an interaction motif for the multiprotein vacuolar ATPase complex. A, Representative confocal images of FP-1305 and 9-11 internalization into primary human macrophages after 2-hour antibody treatment (N = 2 healthy donors). A single focal plane and orthogonal view of the indicated magnified area shows vesicles containing either one or both of the antibodies. Colocalization was assessed with Manders colocalization coefficients M1 (FP-1305) and M2 (9-11); N = 10 cells from two independent experiments with the same donor. Scale bar, 5 μm. Statistical comparison of co-localization was conducted using Wilcoxon matched-pairs signed rank test. B, Schematic of the main 9-11 and FP-1305 antibody epitopes mapped on the human Clever-1 protein primary structure. FAS1, fasciclin domain; LAM, laminin-type EGF-like domain; LINK, C-type lectin-like hyaluronan-binding LINK module. C, Venn diagram showing the number of 9-11– and FP-1305–specific proteins and proteins shared by both antibodies in the CRAPome-pruned Clever-1 interactome. D, High-confidence protein–protein interactions in the Clever-1 interactome retrieved from the STRING database and mapped using Cytoscape. Proteins present in the 9-11 interactome have a green border and proteins present in the FP-1305 interactome a yellow center. The five largest clusters are highlighted with grey boxes and titled after common themes in their significantly enriched GO Biological Process terms. E, Significantly enriched pathways from IPA performed separately on 9-11–specific, FP-1305–specific, and shared proteins. F, Magnification of the Clever-1 interactome showing subunits of the v-ATPase complex, which immunoprecipitated specifically with 9-11. G, Coimmunoprecipitation/Western blot validation showing that the v-ATPase subunits ATP6V1A, ATP6V0A1, and TCIRG1 are immunoprecipitated with 9-11 but not with FP-1305. Clever-1 signal was detected with the 3-372 antibody, which is the parent antibody of FP-1305. Rat IgG2a and human IgG4 served as isotype controls in coimmunoprecipitation for 9-11 and FP-1305, respectively. **, P < 0.01.
Clever-1 interference impairs lysosomal acidification and promotes cross-presentation to induce CD8+ T-cell proliferation
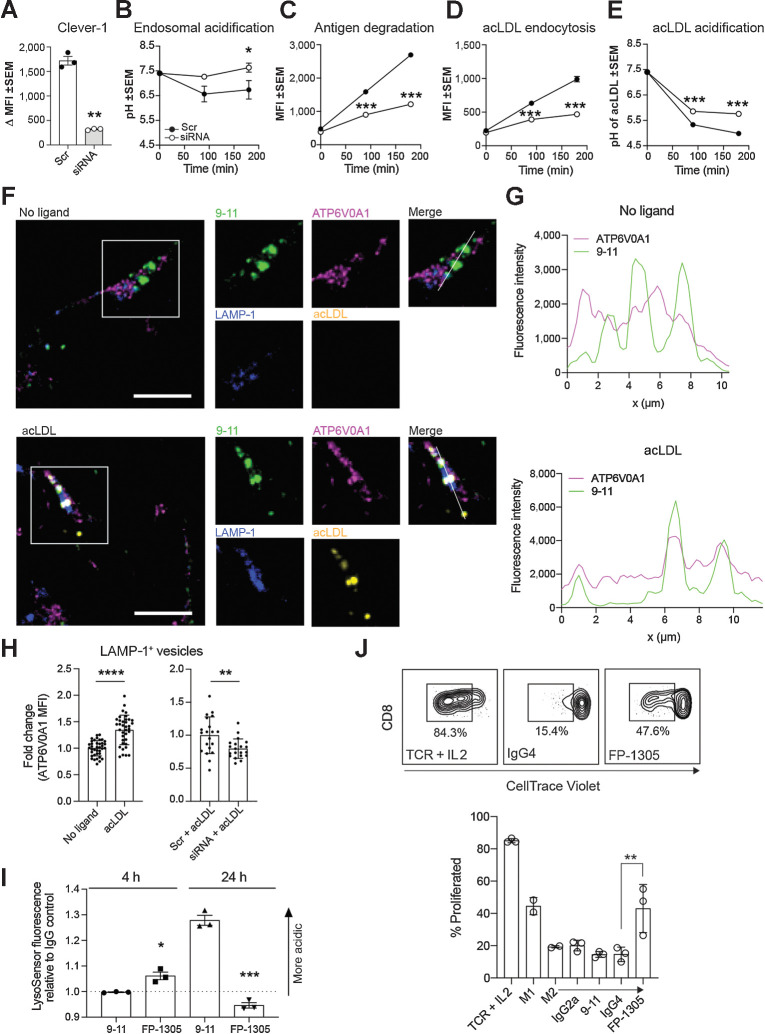
The v-ATPase is an ATP-dependent proton pump that regulates lysosomal pH through the reversible assembly of its V0 and V1 sectors on the lysosomal surface (26). To investigate the functional effects of the observed interaction between v-ATPase and Clever-1, we utilized the Clever-1high acute myelogenous leukemia cell line KG-1, which differentiates into macrophage-like cells after PMA treatment (27–29). We used RNA interference to knock down Clever-1 expression (Fig. 2A) and kinetically monitored steady-state endosomal acidification and DQ-ovalbumin (OVA) antigen degradation in KG-1 macrophages. Clever-1 knockdown significantly impaired both endolysosomal acidification and antigen degradation (Fig. 2B and C). To more specifically interrogate the contribution of Clever-1 to these processes, we performed endocytosis and acidification experiments with acLDL, a ligand of Clever-1 and other scavenger receptors. Predictably, Clever-1 knockdown significantly reduced but did not completely inhibit acLDL endocytosis (Fig. 2D). However, Clever-1 knockdown significantly impaired the acidification of endocytosed acLDL (Fig. 2E). These results were independently verified using another siRNA (Supplementary Fig. S1B). Confocal microscopy further verified the colocalization of Clever-1 and ATP6V0A1 in LAMP-1+ lysosomes (Fig. 2F). Treatment with acLDL increased the lysosomal recruitment of ATP6V0A1, which was inhibited by Clever-1 knockdown (Fig. 2G and H). FP-1305 treatment of primary human macrophages initially lowered endolysosomal pH (4 hours) but converted to increased endolysosomal pH after 24 hours, opposite to the endolysosomal pH changes observed with 9-11 (Fig. 2I). While the FP-1305 epitope on Clever-1 is important for acLDL scavenging (30), FP-1305 treatment did not affect the potential of primary human macrophages to phagocytose apoptotic cancer cells (Supplementary Fig. S1C).
Figure 2.
Clever-1 regulates the acidification and degradation of endocytosed antigens. A, Clever-1 knockdown efficiency in KG-1 macrophages quantified by flow cytometry with 9-11; N = 3, Student's paired two-tailed t test. B–E, Total endo-lysosomal acidification (B), antigen degradation (C), acLDL endocytosis (D), and acLDL acidification (E) kinetics in KG-1 macrophages transfected with scramble (Scr) or Clever-1 siRNA; N = 3, two-way ANOVA with Sidak's multiple comparisons test. F, Confocal microscopy images of Clever-1 (green), ATP6V0A1 (magenta), and LAMP-1 (blue) localization in KG-1 macrophages treated with or without 10 μg/mL acLDL (yellow) for 3 hours. Scale bar, 10 μm. G, Clever-1 (green) and ATP6V0A1 (magenta) intensity profiles generated from the KG-1 macrophage confocal images. Intensities were obtained across the white lines displayed in the corresponding merged images. H, ATP6V0A1 mean fluorescence intensity (MFI) in LAMP-1+ lysosomes of non-transfected KG-1 macrophages treated with or without 10 μg/mL acLDL (n > 35 cells from three independent experiments) and scramble (Scr) or Clever-1 siRNA-transfected KG-1 macrophages treated with 10 μg/mL acLDL (n = 20 cells). MFI normalized to each experiment's no-ligand or Scr-mean. Student's unpaired two-tailed t test. I, M2-polarized primary human macrophages (N = 3) were treated with 50 μg/mL of 9-11 or FP-1305 for the indicated timepoints. Changes in endosomal pH were measured by LysoSensor Green fluorescence intensity relative to cells treated with control IgGs; Student's unpaired two-tailed t test. J, Representative flow cytometry plots and quantification of CD8+ T-cell proliferation in mixed leukocyte reactions with M2-polarized primary human macrophages treated with 9-11 or FP-1305 or their respective isotype controls in comparison to TCR (anti-CD3/CD28) activation or M1 polarization. Each dot represents one healthy donor; one-way ANOVA with Holm–Sidak multiple comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
It has been suggested that reduced lysosomal acidification preserves potential MHC class I–compatible epitopes, thus rescuing antigens from excessive degradation for cross-presentation (31). To analyze how Clever-1 can impact antigen cross-presentation, we first used bone marrow–derived macrophages from wildtype and Clever-1 knockout (Clever-1−/−) mice that were TAM-polarized in vitro with Lewis lung carcinoma–conditioned medium and fed OVA SIINFEKL peptide, full-length OVA protein, or cell-associated OVA on irradiated EG.7 cells. We detected higher antigen cross-presentation on Clever-1−/− macrophages in all instances, suggesting that the absence of Clever-1 not only elevated macrophages' ability to cross-present peptide antigen (OVA SIINFEKL peptide) but also directed antigen processing to generate cross-presentable peptides (OVA protein, irradiated EG.7 cells; Supplementary Fig. S2A). The addition of FP-1305 to mixed leukocyte reactions (MLR) with M2-polarized primary human macrophages and allogeneic T-cells significantly increased the proliferation of CD8+ T-cells compared to the IgG4 isotype control (Fig. 2J). Indeed, FP-1305 treatment increased CD8+ T-cell proliferation to a comparable level with M1-polarized macrophages. CD4+ T-cell proliferation was also increased by FP-1305, but the effect was less robust (Supplementary Fig S2B). While MLR does not measure antigen presentation, data by Palani and colleagues (4) show that treatment with 3-372, the parent antibody of FP-1305, significantly increases IFNγ secretion in antigen recall assays, suggesting that antigen-specific T-cell activation and, by inference, antigen presentation are also improved by Clever-1 blockade. This, along with the previously reported increases in TNFα (32), CCL3 (30), and IL-12 secretion and activation of the NF-κB and mTOR signaling pathways (7) in monocytes and macrophages by Clever-1 interference shows an important role for Clever-1 in suppressing adaptive immune activation. To elucidate this role even more, we incubated whole blood from healthy donors with FP-1305 to mimic monocyte-mediated T-cell activation in the circulation. Mass cytometry time-of-flight (CyTOF) showed an increase in the proportion of CD4+ and CD8+ effector T (TEFF)-cells over naïve cells and reduced levels of CTLA-4 expression on CD4+ T-cells 24 hours after FP-1305 treatment (Supplementary Fig. S3A–C). Altogether, these results suggest that FP-1305–specific binding to Clever-1 can support monocyte/macrophage-mediated T-cell activation.
Figure 3.
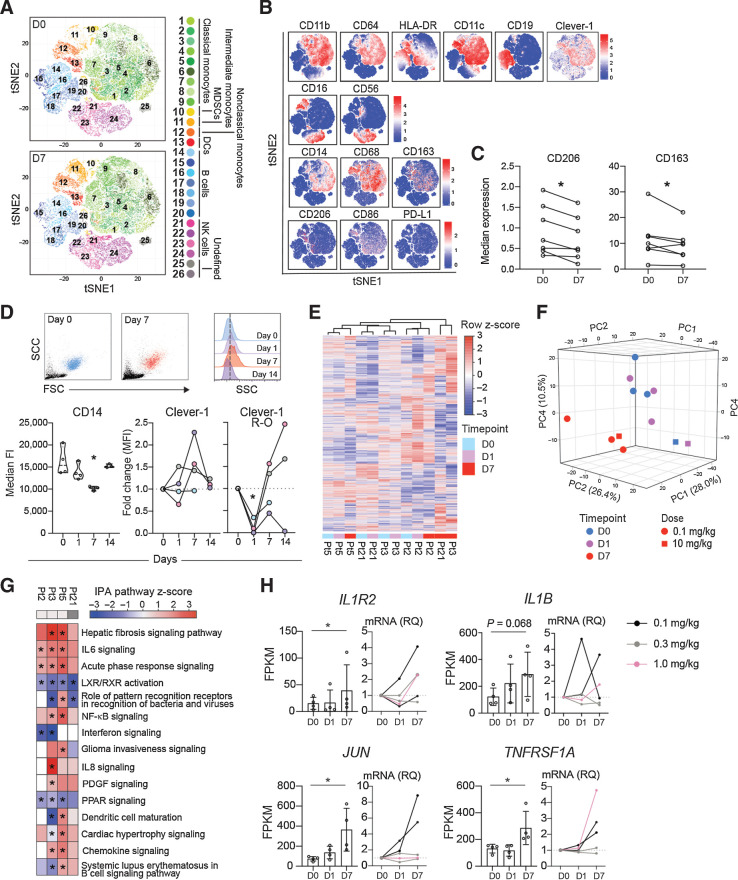
FP-1305 binds circulating CD14+ monocytes and suppresses LXR/RXR and PPAR signaling pathways. A, T-distributed stochastic neighbor embedding (tSNE) of CD3-excluded circulating mononuclear cells from D0 (N = 7) and D7 (N = 7) samples pre-gated for viability, singlets, and CD45+ cells. B, tSNE heatmaps showing expression of indicated markers on cell clusters of a representative patient (D0 and D7). C, Median expression of CD206 and CD163 on CD14+ monocytes on D0 and D7; paired Student's t test. D, Representative flow cytometry plots on D0 (blue) and D7 (red) showing size (FSC) and granularity (SSC) of CD14high monocytes (dashed line in half-offset histograms shows median SSC fluorescence on D0) and median expression of CD14 and Clever-1 (9-11 antibody) at different timepoints during treatment cycle 1. Pt2 (gray), Pt3 (pink), Pt5 (lilac), Pt21 (light blue). Clever-1 R-O by administered FP-1305 was detected by competitive binding of fluorochrome-conjugated FP-1305. One-way ANOVA performed between D0, D1, and D7 samples. E, Unsupervised hierarchical clustering of 13,589 genes expressed in CD14+ monocytes obtained from 4 patients across D0, D1, and D7 samples. F, Principal component analysis of patient samples across different timepoints. G, Ingenuity Pathway analysis of patient gene expression changes on D7 compared with predose (D0). Red color indicates predicted pathway activation and blue color inhibition. Light gray denotes 0.1 mg/kg dose and dark grey 10 mg/kg. H, FPKM values and qPCR validation of gene expression changes involved in relevant pathways. RNA sequenced Pt2 and Pt3 are marked in black. *, P < 0.05.
Clever-1 targeting by FP-1305 promotes phenotypic changes in circulating monocytes
To analyze FP-1305 functions in human, patients with various solid tumors [immunotherapy-refractory melanoma, pancreatic–ductal adenocarcinoma, cholangio, hepatocellular, ovarian, and colorectal (CRC) carcinoma] received intravenous FP-1305 every 3 weeks in part 1 of a phase I/II open-label trial (MATINS). The tumor types were selected on the basis of the association of Clever-1 mRNA expression with survival data obtained from the The Cancer Genome Atlas (TCGA) repository, analysis of biobank materials for macrophage Clever-1 positivity (Auria Biobank, University of Turku), and published research (33). Full clinical data were presented at the American Society of Clinical Oncology (ASCO) 2020 virtual meeting (8). FP-1305 was well tolerated and no dose-limiting toxicity was observed during the dose-escalation phase with a maximum dose of 10 mg/kg.
In the circulation, Clever-1 is expressed by CD14+ classical monocytes (4), the precursors of tissue macrophages, and are the first to encounter intravenously administered FP-1305. In accordance, Clever-1 was most abundant on CD14high monocytes in the MATINS patients. No positivity was observed on lymphocyte populations and blood neutrophils were only faintly stained (Supplementary Fig. S4A). Cell-surface Clever-1 expression on CD14high blood monocytes in the patients (normalized median fluorescence intensity 5,965 range 13,622) was not significantly different from healthy donors (4,422 range 5,741; Supplementary Table S1). CyTOF of different myeloid cell populations at predose (D0) and 7 days after FP-1305 administration (D7; Fig. 3A and B; Supplementary Fig. S4B and Supplementary Table S2) revealed that FP-1305 induced a significant downregulation of CD206 and CD163 on CD14high monocytes (Fig. 3C). CD206 and CD163 are the most common markers used to classify alternatively activated M2 macrophages, which suggests that FP-1305 was able to reduce the immunosuppressive phenotype of circulating monocytes. Interestingly, the monocytes increased in size and granularity by D7. We also observed a transient decrease in cell-surface CD14 on D7 (Fig. 3D). FP-1305 did not induce depletion of Clever-1+ monocytes (detected by the non-competitive 9-11 antibody) since cell-surface Clever-1 remained constant or even increased over 2 weeks post-administration. Median Clever-1 receptor occupancy (R-O) of FP-1305 24 hours (D1) after administration was 71% (Supplementary Table S1) as measured by the decreased binding of fluorochrome-conjugated FP-1305 (AF647) competitor antibody on monocytes. On D7, R-O was cleared to some extent, possibly because of the release of new monocytes from the bone marrow and trafficking of the targeted ones into tissues (Fig. 3D).
FP-1305 blocks tolerogenic gene expression related to LXR/RXR and PPAR pathways
To understand monocyte responses induced by Clever-1 targeting, we performed RNA-seq on the circulating CD14+ population on D0, D1, and D7 samples. Unsupervised hierarchical clustering demonstrated a close relationship of the transcriptome between each patient's D0 and D1 samples (Fig. 3E). However, the D7 samples from different patients were more similar by timepoint, showing separation from other samples in principal component analysis (Fig. 3F) and overlapping gene expression changes (Supplementary Fig. S4C). IPA on the differentially expressed genes between D7 and D0 samples revealed a significant downregulation of the LXR/RXR and PPAR pathways (Fig. 3G), possibly related to the FP-1305–induced impairment in modified LDL scavenging, as shown previously in monocytes cultured ex vivo (30). At the same time, inflammation-related pathways were activated, as observed by the upregulation of IL1R2, IL1B, JUN, and TNFRSF1A (Fig. 3H and Supplementary Data File 2). Changes in the expression of these genes were verified by qPCR from the two sequenced patients (Pt2 and Pt3, black) and expanded to other patient samples to show a rather heterogenous response related to dose and timepoint (Fig. 3H). The increased monocyte activation seemed to coincide with the magnitude of FP-1305 R-O. Patients showing the activation pattern in RNA-seq had high Clever-1 occupancy (Pt2 100%, Pt3 90%, Pt5 100%, and Pt21 66%) on D1 (Fig. 3D), whereas patients with low occupancy (ranging from 0% to 10%) did not show similar gene expression changes in monocytes (Supplementary Fig. S4D and S4E). A similar observation was seen when the gene expression changes were validated by qPCR (Fig. 3H; Supplementary Fig. S4E). The low occupancy of monocyte Clever-1 in some patients most likely relates to the availability and abundance of FP-1305 epitope on monocytes or preference of FP-1305 binding to other cellular sources of Clever-1.
FP-1305 induces robust T-cell activation and downregulation of immune checkpoint expression
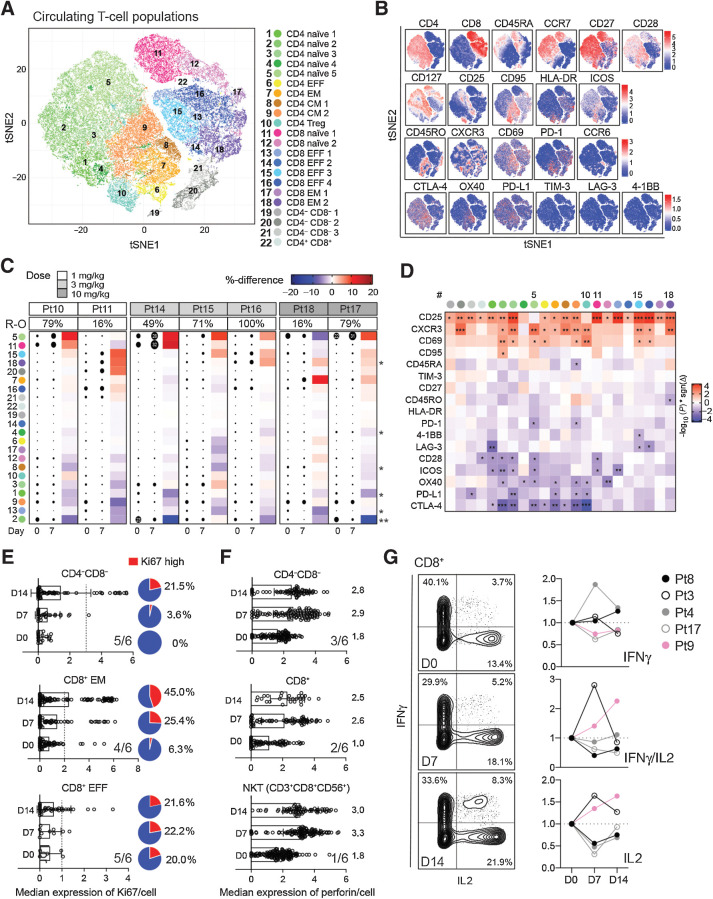
To investigate how FP-1305–induced changes in monocyte and macrophage function contribute to adaptive immune responses in vivo, we used CyTOF to analyze the T-cell populations in MATINS study patients' D0 and D7 samples. Unbiased PhenoGraph clustering of T-cells identified 22 different CD3+ T-cell metaclusters across all patient samples (Fig. 4A and B; Supplementary Table S3). The analysis showed changes in several T-cell populations across patients receiving different doses of FP-1305 (1, 3, or 10 mg/kg), indicating that this transition was not dose dependent (Fig. 4C). The most pronounced effects were seen within the naïve CD4+ (#5) and CD8+ T-cell (#11) and CD8+ TEFF-cell (#15) clusters with increased CXCR3 and CD25 expression, respectively (Fig. 4C and D; Supplementary Fig. S5). In addition, we observed a significant induction of a CD8+ effector memory T (TEM)-cell population (#18) with upregulation of CD25 and CXCR3 (Fig. 4C and D). In contrast, CD25 was not significantly upregulated in the regulatory T (TREG)-cell population (#10; Fig. 4D). In T-cells, CD25 (IL2-receptor ± chain) expression is induced by IL-2 a few days after TCR ligation, enabling clonal expansion of antigen-experienced cells. Consequently, CXCR3 expression is rapidly induced on naïve T-cells and facilitates their migration into the tumor tissue and differentiation into TEFF-cells (34). The upregulation of activation markers in most clusters occurred regardless of the changes in cluster size, proposing a general induction of peripheral T-cell activation (Fig. 4D). In support of this, the expression of several regulatory immune checkpoint molecules CTLA-4, LAG-3, PD-1, and PD-L1 were downregulated on CD4+ T-cell populations (Fig. 4D). The expression of the costimulatory receptor CD28 but not CD27 was also diminished on activated naïve CD4+ T-cells, possibly suggesting a transition to terminally differentiated TEFF-cells (35). The peripheral T-cell activation was accompanied by increased Ki67 expression in CD8+ TEFF-cells (5/6 of the patients), TEM (4/6), and double negative (CD4−CD8−; 5/6) T-cells, indicative of their proliferation (Fig. 4E). Moreover, the CD4−CD8− and CD8+ T-cells, together with natural killer T-cells (NKT-cell), increased levels of perforin 7 or 14 days after FP-1305 administration in a subset of patients (Fig. 4F), providing evidence of their higher cytotoxic potential. In addition, two patients (2/5) demonstrated increased secretion of IL-2 and IFNγ in peripheral CD8+ T-cells 7 or 14 days after FP-1305 administration (Fig. 4G).
Figure 4.
FP-1305 elicits activation and proliferation of circulating cytotoxic T-cells. A, tSNE plots of circulating T-cells pre-gated for viability, singlets, and CD3+ cells; 5,000 cells/sample (N = 7). CM, central memory; EFF, effector; EM, effector memory; TREG, regulatory T-cell. B, tSNE heatmaps showing expression of indicated markers on T-cell clusters of a representative patient. C, Heatmap of cluster sizes on D0 and D7. Dot size indicates the relative percent of the cluster per sample. Red color points to positive and blue to negative change between D0 and D7 samples within a patient. The asterisk indicates statistical significance of the change across all patients. D, Heatmap of marker differences between D0 and D7 samples in each T-cell cluster (N = 7); paired Student's t test (C and D). E and F, Median expression of Ki67 (E) and perforin (F) in selected T-cells from a representative patient (Pt10). Overall response across 6 patients with at least >10% increase in Ki67 and perforin is indicated in the right corner of each graph. Cutoff for Ki67high cells is depicted as a dashed line for each cell type. The median expression of perforin is indicated for each timepoint on the right side of the bars. G, Expression of IFNγ and IL-2 in PMA/ionomycin-stimulated peripheral CD8+ T-cells obtained D0, D7, or D14. Plots are shown for Pt9 (pink). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FP-1305 restores patient responses to proinflammatory stimuli
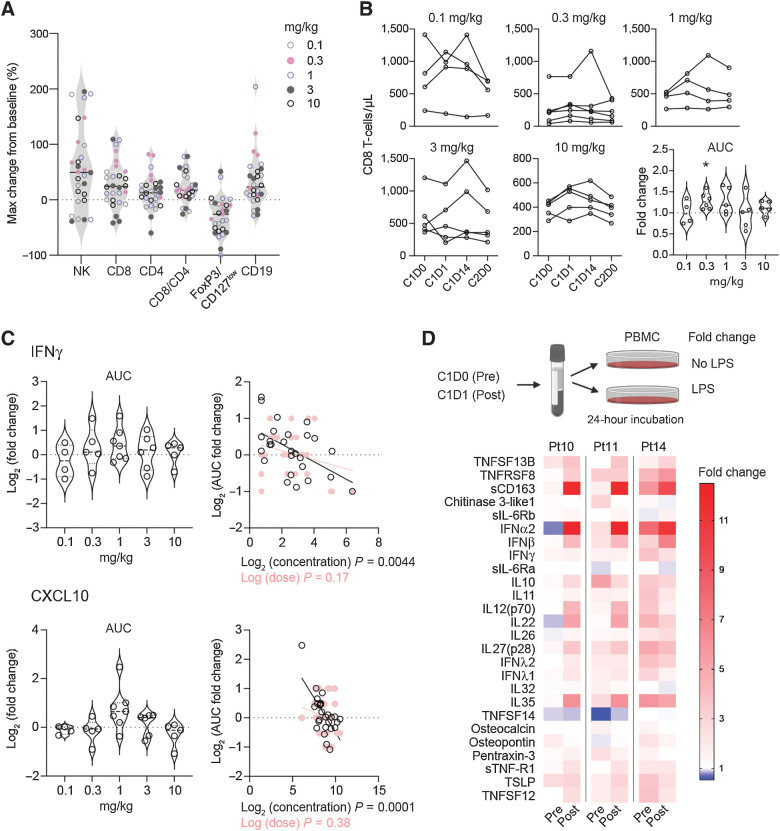
To have an overall view of FP-1305–induced systemic immune activation in the entire part 1 cohort, we looked at changes in absolute lymphocyte numbers during cycle 1 of FP-1305 treatment. Most patients demonstrated an increase in NK-, CD8+ T-, and B-cells, whereas the FOXP3+CD127low TREG population decreased (Fig. 5A). The apparent difficulty in combining all patient data is the difference in their FP-1305 dose, cancer type, and previous lines of therapy. Most likely the lowest dose of 0.1 mg/kg is subtherapeutic and cannot produce as effective a response as an optimal dose would. Thus, to statistically validate the increase in CD8+ T-cells, we calculated the AUC for cycle 1 across the different dose levels. While all dose levels included patients with increased circulating CD8+ T-cells in response to FP-1305 therapy, the increase was significant only with the 0.3 mg/kg dose (Fig. 5B).
Figure 5.
FP-1305 reinvigorates patient responses to inflammatory stimuli by increasing interferon secretion. A, Maximum relative change in the absolute numbers of blood lymphocyte populations during cycle 1 in all patients. B-cell changes were measured during cycles 1–4 due to slower kinetics. B, Absolute CD8+ T-cell numbers and AUC during cycle 1, shown by dose level. The AUCs were analyzed from patients with samples from all timepoints and a fold change was calculated against an AUC using each patient's predose cell number, which was considered to remain constant throughout cycle 1. Statistical analysis was performed with Wilcoxon's matched-pairs signed rank test. C, AUC fold change for systemic IFNγ and CXCL10 in MATINS patients during cycle 1. No significant differences between doses were observed with Wilcoxon's matched-pairs signed rank test. The right-hand plots show linear regression analysis of log2 transformed values of AUC fold change with predose concentrations (pg/mL) and dose (0.1–10 mg/kg). Each dot represents one patient (A–C). D, Schematic of TLR4-specific LPS stimulation of patient PBMCs ex vivo on D0 and D1 and heatmap of LPS-induced secreted factors on D0 and D1 shown as fold change relative to unstimulated cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
IFNγ is critical for T-, NKT-, and NK-cell trafficking into tumors via induction of CXCR3 ligands CXCL9, CXCL10, and CXCL11 (36). Therefore, we measured the AUCs for systemic IFNγ and CXCL10 after FP-1305 administration. The increase in both IFNγ and CXCL10 was not dose dependent but, in fact, significantly associated with low predose levels of these cytokines/chemokines. (Fig. 5C). This suggests that FP-1305 would be more effective in immunosuppressed patients. Indeed, since most patients with cancer have a TH2-skewed immune profile, we wanted to test whether FP-1305 can reinvigorate patient responses to TLR4-mediated innate immune activation. PBMCs isolated from D1 samples showed improved IFNα/β secretion after LPS exposure ex vivo compared with PBMCs from respective D0 samples (Fig. 5D). At the same time, the levels of sCD163 increased in postadministration samples, supporting increased immune activation not only of T- and NK-cells but also of peripheral monocytes (37), further supporting the CyTOF and RNA-seq data.
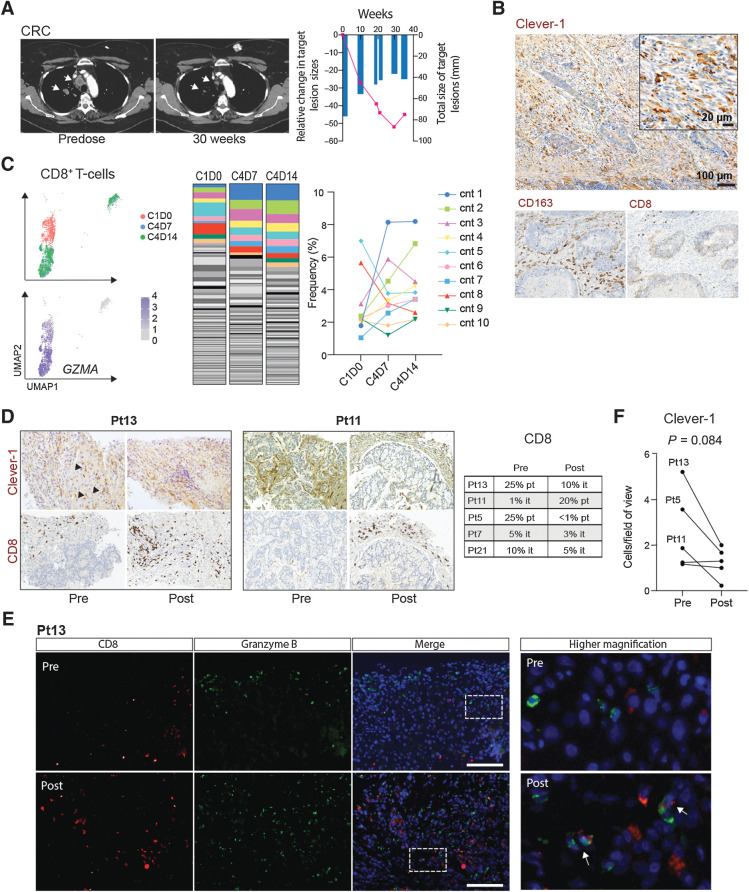
FP-1305 promotes clonal expansion of CD8+ TEFF-cells in a patient with partial response
To understand the relevant peripheral changes that would be indicative of an effective FP-1305–induced antitumor response, we looked more closely at data obtained from one patient with CRC with a partial response according to Response Evaluation Criteria in Solid Tumors Version 1.1 (RECIST 1.1; Fig. 6A; ref. 38). IHC staining of the primary tumor showed very high numbers of Clever-1+ and CD163+ macrophages and few intratumoral CD8+ T-cells (Fig. 6B). scRNA-seq together with TCR-seq of the responding patient's circulating CD8+ T-cells showed clonal expansion of GZMAhigh (granzyme A) T-cell clonotypes (cnt) at cycle 4 (Fig. 6C; Supplementary Fig. S6A and S6B). These T-cells differed from D0 T-cells by higher expression of FCGR3A (CD16), PFN1 (perforin), DUSP1 (MKP-1), and IL32 (Supplementary Fig. S6C). Both CD16 and MKP-1 have been shown to contribute to T-cell activation and cytotoxicity (39, 40). These data indicate that FP-1305 is able to induce proliferation of antigen-specific cytotoxic CD8+ T-cells. Moreover, the appearance of a TCF7high (TCF1) CD8+ T-cell population at cycle 4 (Supplementary Fig. S6A and S6B) suggests induction of a memory response with self-renewing potential (41). Unfortunately, we were not able to study whether the identified T-cell clonotypes were present in the tumor due to unavailable posttreatment biopsy material and therefore cannot conclude whether the clones are linked to antitumor T-cells. Additionally, the patient developed autoimmune dermatitis grade (Gr) 2, myositis (Gr 2), thyroiditis (Gr 2), and pneumonitis (Gr 1) at week 18. The significant shrinkage of the metastatic lesions coupled with the delayed onset of immune-related systemic features suggests that FP-1305 was able to boost clonal expansion of antigen-specific T-cells against the tumor but also against self-antigens.
Figure 6.
FP-1305–induced antitumor responses are reflected by clonal expansion of systemic effector CD8+ T-cells in a responding patient. A, CT images of a patient (Pt6) with microsatellite-stable (MSS) colorectal carcinoma (six previous lines of therapy). Arrows highlight shrinking lung metastases. Size of the target lesions and change over time are presented in the right-hand graph. B, IHC analyses of Clever-1, CD163, and CD8 (all in brown) in archived rectal carcinoma tissue from the colorectal cancer (CRC) patient. C, Uniform manifold approximation and projection (UMAP) plots of CD8+ T-cell transcriptomes colored by timepoint and expression of granzyme A (GZMA). Abundance of diverse CD8+ T-cell clonotypes (cnt) at cycle 1, D0 (predose), and cycle 4 timepoints D7 and D14. The ten most abundant TCR sequences are colored to show dynamics of specific clones across the different timepoints. D, IHC staining of Clever-1 and CD8 in patient tumor biopsies pre- and post-treatment. Posttreatment biopsies were taken after cycle 3. Arrows point to Clever-1+ liver sinusoids. Quantification of peri- and intratumoral CD8+ T-cells in biopsies are reported by predominant immune cell location. it, intratumoral; pt, peritumoral. E, Immunofluorescence staining of CD8 (red) and granzyme B (green) in pre- and posttreatment biopsies of Pt13 with DAPI nuclear stain (blue). Arrows point to double positive cells. Scale bar, 100 μm. F, Quantification of Clever-1+ macrophages in tumor biopsies pre- and posttreatment. Student's paired two-tailed t test.
FP-1305 treatment leads to reduced numbers of Clever-1+ TAMs and changes in intratumoral CD8+ T-cells
To analyze whether the immune activation and increase in circulating lymphocytes was associated with immune infiltration into the tumor, we studied the numbers of CD8+ T-cells in pre- and posttreatment tumor biopsies. Paired tumor samples were possible only in 5 patients who demonstrated progressive disease and therefore may not adequately recapitulate the responses occurring in responding tumor lesions. However, in one patient (Pt13), a peritumoral CD8+ T-cell profile changed to an intratumoral CD8+ T-cell profile 9 weeks postadministration (Fig. 6D). The intratumoral CD8+ T-cells were granzyme B+ to some extent (Fig. 6E). In another patient (Pt11), the presence of peri- and/or intratumoral CD8+ T-cells was only very limited prior to FP-1305 administration and increased substantially posttreatment. While these are promising signs of antitumor CD8+ T-cell activation, validation of these results should preferably be done from the posttreatment biopsies of responding patients. It is also worth noting that many of the pretreatment biopsies showed very low numbers of Clever-1+ macrophages (Supplementary Table S4), as can be seen in the 2 patients not showing CD8+ T-cell increase in their posttreatment biopsies (Fig. 6D). In tissues with a higher number of Clever-1+ macrophages, Clever-1 staining did not appear to change dramatically (moderate weakening of signal intensity) but the number of Clever-1+ macrophages tended to reduce in posttreatment samples (Fig. 6F). This suggests that either the existing macrophages were phenotypically changing so that they would express less Clever-1 or that the recruitment of new monocyte-derived macrophages was impaired. The latter possibility relates to unpublished observations and data published by Karikoski and colleagues (42).
Discussion
The tumor microenvironment remains a major barrier to successful cancer eradication, limiting the efficacy of current checkpoint inhibitors. A range of preclinical studies confirm that macrophages are key orchestrators of this immunosuppressive niche, yet harnessing their therapeutic capability remains a significant challenge. Clever-1 is a macrophage scavenger receptor that is expressed on blood monocytes and is markedly upregulated on macrophage subsets by, for example, glucocorticoids, supporting tissue tolerance (25, 43). Macrophages express a range of scavenger receptors and, historically, it has been assumed that significant redundancy exists within this large receptor family. However, previous human in vitro and murine in vivo studies have highlighted the potential of inhibiting Clever-1 to skew macrophages towards a proinflammatory phenotype (4, 7).
The notable immunologic finding from this early-phase clinical trial is that anti–Clever-1 treatment can induce robust peripheral T-cell activation in patients with advanced cancer. Systemic immune activation is a promising feature of the clinical antitumor activity of FP-1305, because peripheral T-cell expansion has been reported to predict tumor infiltration and clinical response to anti–PD-L1 therapy (44). Moreover, recent findings indicate that newly generated, transitory CD8+ TEFF-cells emerging from lymphoid tissue-resident stem-like TCF1+ CD8+ T-cells play a critical role in PD-1–targeted immunotherapy (45), a subset of which was detectable after four cycles of FP-1305 treatment in a responding patient. The downregulation of negative immune checkpoints on CD4+ T-cell subsets one week after FP-1305 administration further supports the induction of newly activated lymphocytes from peripheral lymphoid organs. Where pre- and posttreatment tumor biopsies were available, there were also features of increased CD8+ T-cell infiltration after treatment in select cases, suggesting that FP-1305 could also have a direct impact on the tumor microenvironment. Our previous results in mouse tumor models have, however, shown that CD8+ T-cells are indispensable for tumor control following Clever-1 inhibition (7). Because many of the standard lines of therapy include drugs with lymphodepleting properties, a possible low baseline level of circulating CD8+ T-cells may be insufficient to eradicate the tumor even if properly activated.
The rationale of including various tumor types for anti–Clever-1 therapy was mostly based on survival data and tissue staining of larger cohorts (5). However, analysis of MATINS pretreatment biopsies showed that several patients had very few Clever-1+ macrophages in their tumors. Thus, by improving patient preselection to include higher tumor Clever-1 expression could possibly result in more consistent clinical responses. While we only studied the mode of action of FP-1305 in patient monocytes, we cannot rule out the possibility that the T-cell responses reported here could also be partly mediated by Clever-1 targeting on (lymphatic) endothelial cells. Indeed, our recent findings show that genetic deletion of Clever-1 in mice reduces lymphatic endothelial cell tolerance (46). However, the MLR together with the analyses of FP-1305 treated blood ex vivo indicates a direct contribution from monocytes and macrophages to the improved T-cell activation sequential to Clever-1 targeting in MATINS patients.
Clever-1 contains multiple binding sites for various ligands (25). Therefore, it is not surprising that antibodies targeting nonoverlapping epitopes on Clever-1 can have different effects on known Clever-1 activities. Indeed, coimmunoprecipitation of Clever-1 with 9-11 identified an interaction of Clever-1 with several proteins of the v-ATPase complex that were not identified with FP-1305. This clearly suggests that FP-1305 might out-compete v-ATPase complex binding to Clever-1 and inhibit endolysosomal maturation. This seemed to be the case when endolysosomal acidification was measured in primary human macrophages treated with FP-1305 but not 9-11. The recruitment of ATP6V0A1 to lysosomes is critical for lysosomal acidification and degradation of proteins. The duration of this process determines how obtainable phagocytosed antigens are for loading onto MHC class I molecules and cross-presented on the cell surface to CD8+ T-cells (31). When we tested the ability of tumor-conditioned macrophages to cross-present phagocytosed OVA, the cells genetically devoid of Clever-1 outperformed those of wildtype origin. Based on our in vitro studies, Clever-1 is required for the rapid delivery of its ligands into lysosomes for degradation. Without Clever-1, this process is delayed, thus possibly freeing the antigen to enter alternative pathways that lead to cross-presentation (47). Furthermore, allogeneic T-cells that were incubated with FP-1305–treated M2 macrophages demonstrated increased proliferation. Taken together, we conclude that Clever-1 inhibition enhances macrophages' ability to cross-present antigens and, consequently, to activate CD8+ T-cells, although the possible contribution of other cellular or soluble factors cannot be dismissed. This also supports our clinical findings of the increased adaptive immune activation in patients receiving FP-1305.
The observation that FP-1305 can inhibit the scavenging of acLDL (30) suggests that this is the primary cause for the downregulation of the LXR/RXR and PPAR nuclear receptor pathways in the monocytes of MATINS patients. The LXR/RXR and PPAR receptors regulate lipid metabolism in monocytes (48), and it is well established that these pathways shape the immune responses of myeloid cells (49). Their downregulation potentially drives the adaptive immune activation in heavily pretreated patients. Recently, Donadon and colleagues identified a morphologically distinct TAM subset in colorectal liver metastases that associated with poor disease-free survival. The authors identified LXR/RXR as the most enriched pathway in these large macrophages, which was related to genes involved in cholesterol metabolism, scavenger receptors, MERTK, and complement (50). From a clinical point of view, indirectly regulating lipid metabolism by targeting Clever-1 may be beneficial because the actions are cell-specific and, therefore, prevent divergent responses or side effects from targeting these pathways systemically.
In conclusion, therapeutic blockade of Clever-1 unveils a novel nonredundant pathway linking the innate and adaptive immune system in humans. Anti-scavenger receptor therapy therefore has the capability to convert immunologically ignorant tumors to an immune-activated state. Our deep immunophenotyping of patient samples provides a strong scientific rationale to further explore the efficacy of Clever-1 as an immunotherapeutic drug target.
Authors' Disclosures
R. Virtakoivu reports grants from Emil Aaltonen Foundation and K. Albin Johansson Foundation during the conduct of the study; in addition, R. Virtakoivu has a patent for Anti–Clever-1 Agents for Controlling Cell Surface Markers Expression on Leucocytes, and Using These Expression Levels to Guide Anti–Clever-1–based Cancer Treatment pending. J. Koivunen reports other support from Faron Pharmaceuticals during the conduct of the study. J. Koivunen also reports grants from AstraZeneca, Boehringer Ingelheim, and Roche; non-financial support from AstraZeneca, BMS, and Pfizer; and personal fees from AstraZeneca, Boehringer Ingelheim, Roche, BMS, Merck, Pfizer, Takeda, Amgen, Pierre-Fabre, Novartis, and Eli Lilly outside the submitted work. P. Jaakkola reports personal fees from Faron Pharmaceuticals during the conduct of the study. A. Pasanen reports personal fees from Faron Pharmaceuticals during the conduct of the study. A. Pasenen also reports personal fees from Novartis (advisory board), Janssen-Cilag (advisory board, travel grant, and lecture fee), Roche (advisory board, travel grant, and lecture fee), and Gilead (advisory board and travel grant) outside the submitted work. S. Shetty reports personal fees from Faron Pharmaceuticals during the conduct of the study. M.J.A. de Jonge reports personal fees from Faron Pharmaceuticals during the conduct of the study. D. Robbrecht reports personal fees from Faron Pharmaceuticals outside the submitted work. Y.T. reports personal fees from Faron Pharmaceuticals during the conduct of the study, as well as personal fees from Roche, Eisai, AstraZeneca, Ipsen, and Bayer outside the submitted work. T. Skyttä reports personal fees from Faron Pharmaceuticals during the conduct of the study, as well as personal fees from BMS, MSD, AstraZeneca, Pierre Fabre, Novartis, Incyte, Boehringer Ingelheim, and Roche outside the submitted work. A. Minchom reports other support and personal fees from Faron Pharmaceuticals during the conduct of the study. A. Minchom also reports personal fees from Chugai Pharmaceutical, Janssen Pharmaceutical, Merck Pharmaceuticals, Novartis Oncology, and Bayer Pharmaceuticals, as well as non-financial support from Amgen Pharmaceuticals and Loxo Oncology outside the submitted work. S. Jalkanen reports grants from Finnish Academy during the conduct of the study; in addition, S. Jalkanen has a patent for US 7354577 issued and owns stocks of Faron Pharmaceuticals. M.K. Karvonen reports being an employee of Faron Pharmaceuticals Ltd and stock ownership of Faron Pharmaceuticals Ltd. J. Mandelin reports personal fees and other support from Faron Pharmaceuticals during the conduct of the study, as well as other support from Faron Pharmaceuticals outside the submitted work. In addition, J. Mandelin has a patent for Method for Determining Potency of Therapeutic Anti–Clever-1 Antibody pending; a patent for Anti–Clever-1 Agents for Controlling the Expression of Cell Surface Markers on Leucocytes, and Using These to Guide Anti–Clever-1–based Cancer Treatment pending; a patent for Stable Anti–Clever-1 Antibody Formation pending; and a patent for Treatment of Diseases with Clever-1 Inhibition in Combination with an Interleukin Inhibitor and/or Type I Interferon pending. P. Bono reports personal fees from Faron Pharmaceuticals during the conduct of the study. P. Bono also reports personal fees from MSD, Oncorena, Ipsen, and Herantis Pharma; personal fees and other support from TILT Biotherapeutics; and other support from Terveystalo outside the submitted work. M. Hollmén reports personal fees and non-financial support from Faron Pharmaceuticals during the conduct of the study, as well as grants from Academy of Finland, Sigrid Jusélius Foundation, the Finnish Cancer Foundations, and Business Finland outside the submitted work. In addition, M. Hollmén has a patent for Anti–Clever-1 Agents for Controlling Cell Surface Markers Expression on Leucocytes, and Using These Expression Levels to Guide Anti–Clever-1–based Cancer Treatment Pending, and owns shares of Faron Pharmaceuticals. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Figures
Tables
Excel spreadsheets of mass spectrometry analysis
Excel spreadsheet of differentially expressed genes in CD14+ monocytes
Acknowledgments
We thank Mari Parsama, Teija Kanasuo, Sari Mäki, and Riikka Sjöroos for excellent technical assistance, and the Cell Imaging and Cytometry Core Facilities at Turku Bioscience Center for their help in mass cytometry and imaging. Mass spectrometry analysis was performed at the Turku Proteomics Facility, University of Turku and Åbo Akademi University. The facility is supported by Biocenter Finland. The study was supported by Finnish Functional Genomics Centre, University of Turku, Åbo Akademi, and Biocenter Finland. We also want to thank all the patients for participating in the clinical trial. A humble recognition should be addressed to Maria Lahtinen and Mari Kimpanpää for managing patient sample logistics at Faron Pharmaceuticals Ltd., and Maria Jokinen, Maria Oliveira, and Jarna Hannukainen at Faron Pharmaceuticals Ltd. and Laura Gardner at Simbec-Orion for clinical management of the MATINS study. This study was funded by the Academy of Finland (A. Takeda, T. Lönnberg, S. Jalkanen, and M. Hollmén), Emil Aaltonen Foundation (R. Virtakoivu), Maud Kuistila Memorial Foundation, Oskar Öflund Foundation, Ida Montin Foundation (all to M. Viitala), Cancer Research UK fellowship C53575/A29959 (S. Shetty), Sigrid Jusélius Foundation (M. Hollmén), and the Finnish Cancer Foundations (M. Viitala and M. Hollmén). This project has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement no. 960914. Faron Pharmaceuticals sponsored the MATINS trial.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article is featured in Highlights of This Issue, p. 4131
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Contributions
R. Virtakoivu: Conceptualization, data curation, formal analysis, funding acquisition, investigation, visualization, methodology, writing–review and editing. J.H. Rannikko: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. M. Viitala: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. F. Vaura: Data curation, software, formal analysis, validation, visualization, methodology, writing–review and editing. A. Takeda: Data curation, software, funding acquisition, visualization, writing–review and editing. T. Lönnberg: Resources, funding acquisition, methodology, writing–review and editing. J. Koivunen: Resources, investigation, writing–review and editing. P. Jaakkola: Resources, investigation, writing–review and editing. A. Pasanen: Resources, investigation, writing–review and editing. S. Shetty: Resources, investigation, writing–review and editing. M.J.A. de Jonge: Resources, investigation, writing–review and editing. D. Robbrecht: Resources, investigation, writing–review and editing. Y.T. Ma: Resources, investigation, writing–review and editing. T. Skyttä: Resources, investigation, writing–review and editing. A. Minchom: Resources, investigation, writing–review and editing. S. Jalkanen: Resources, funding acquisition, writing–review and editing. M.K. Karvonen: Conceptualization, resources, data curation, project administration, writing–review and editing. J. Mandelin: Conceptualization, resources, data curation, validation, investigation, project administration, writing–review and editing. P. Bono: Conceptualization, supervision, project administration, writing–review and editing. M. Hollmén: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy.Cell 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer.Nat Rev Drug Discov 2018;17:887–904. [DOI] [PubMed] [Google Scholar]
- 3. Jahchan NS, Mujal AM, Pollack JL, Binnewies M, Sriram V, Reyno L, et al. Tuning the tumor myeloid microenvironment to fight cancer.Front Immunol 2019;10:1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palani S, Elima K, Ekholm E, Jalkanen S, Salmi M. Monocyte stabilin-1 suppresses the activation of Th1 lymphocytes.J Immunol 2016;196:115–23. [DOI] [PubMed] [Google Scholar]
- 5. Hollmén M, Figueiredo CR, Jalkanen S. New tools to prevent cancer growth and spread: a ‘Clever’ approach.Br J Cancer 2020;123:501–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karikoski M, Marttila-Ichihara F, Elima K, Rantakari P, Hollmén M, Kelkka T, et al. Clever-1/stabilin-1 controls cancer growth and metastasis.Clin Cancer Res 2014;20:6452–64. [DOI] [PubMed] [Google Scholar]
- 7. Viitala MK, Virtakoivu R, Tadayon S, Rannikko J, Jalkanen S, Hollmén M. Immunotherapeutic blockade of macrophage clever-1 reactivates the CD8+ T cell response against immunosuppressive tumors.Clin Cancer Res 2019;25:3289–303. [DOI] [PubMed] [Google Scholar]
- 8. Bono P, Virtakoivu R, Vaura F, Jaakkola P, Shetty S, Thibault A, et al. Immune activation in first-in-human anti-macrophage antibody (anti-Clever-1 mAb; FP-1305) phase I/II MATINS trial: Part 1 dose-escalation, safety and efficacy results.J Clin Oncol 2020;38:(suppl; abstr 3097). [Google Scholar]
- 9. Reddy MP, Kinney CA, Chaikin MA, Payne A, Fishman-Lobell J, Tsui P, et al. Elimination of Fc receptor-dependent effector functions of a modified IgG4 monoclonal antibody to human CD4.J Immunol 2000;164:1925–33. [DOI] [PubMed] [Google Scholar]
- 10. Irjala H, Elima K, Johansson EL, Merinen M, Kontula K, Alanen K, et al. The same endothelial receptor controls lymphocyte traffic both in vascular and lymphatic vessels.Eur J Immunol 2003;33:815–24. [DOI] [PubMed] [Google Scholar]
- 11. Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data.Nat Methods 2013;10:730–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible.Nucleic Acids Res 2017;45:D362–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks.Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, et al. clusterMaker: a multi-algorithm clustering plugin for cytoscape.BMC Bioinformatics 2011;12:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape stringapp: network analysis and visualization of proteomics data.J Proteome Res 2019;18:623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis.Bioinformatics 2014;30:523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kimball AK, Oko LM, Bullock BL, Nemenoff RA, van Dyk LF, Clambey ET. A beginner's guide to analyzing and visualizing mass cytometry data.J Immunol 2018;200:3–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data.Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 19. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing.J R Statistic Society B 1995;57:289–300. [Google Scholar]
- 20. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements.Nat Methods 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data.Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data.Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data.Bioinformatics 2010;26:136–8. [DOI] [PubMed] [Google Scholar]
- 24. Boström MM, Irjala H, Mirtti T, Taimen P, Kauko T, Ålgars A, et al. Tumor-associated macrophages provide significant prognostic information in urothelial bladder cancer.PLoS One 2015;10:e0133552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kzhyshkowska J, Gratchev A, Goerdt S. Stabilin-1, a homeostatic scavenger receptor with multiple functions.J Cell Mol Med 2006;10:635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collins MP, Forgac M. Regulation and function of V-ATPases in physiology and disease.Biochim Biophys Acta Biomembr 2020:183341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ackerman AL, Cresswell P. Regulation of MHC class I transport in human dendritic cells and the dendritic-like cell line KG-1.J Immunol 2003;170:4178–88. [DOI] [PubMed] [Google Scholar]
- 28. St Louis DC, Woodcock JB, Franzoso G, Blair PJ, Carlson LM, Murillo M, et al. Evidence for distinct intracellular signaling pathways in CD34+ progenitor to dendritic cell differentiation from a human cell line model.J Immunol 1999;162:3237–48. [PubMed] [Google Scholar]
- 29. Teobald I, Dunnion DJ, Whitbread M, Curnow SJ, Browning MJ. Phenotypic and functional differentiation of KG-1 into dendritic-like cells.Immunobiology 2008;213:75–86. [DOI] [PubMed] [Google Scholar]
- 30. Rantakari P, Patten DA, Valtonen J, Karikoski M, Gerke H, Dawes H, et al. Stabilin-1 expression defines a subset of macrophages that mediate tissue homeostasis and prevent fibrosis in chronic liver injury.Proc Natl Acad Sci U S A 2016;113:9298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alloatti A, Kotsias F, Pauwels AM, Carpier JM, Jouve M, Timmerman E, et al. Toll-like receptor 4 engagement on dendritic cells restrains phago-lysosome fusion and promotes cross-presentation of antigens.Immunity 2015;43:1087–100. [DOI] [PubMed] [Google Scholar]
- 32. Dunkel J, Viitala M, Karikoski M, Rantakari P, Virtakoivu R, Elima K, et al. Enhanced antibody production in clever-1/stabilin-1-deficient mice.Front Immunol 2018;9:2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Algars A, Irjala H, Vaittinen S, Huhtinen H, Sundström J, Salmi M, et al. Type and location of tumor-infiltrating macrophages and lymphatic vessels predict survival of colorectal cancer patients.Int J Cancer 2012;131:864–73. [DOI] [PubMed] [Google Scholar]
- 34. De Simone G, Mazza EMC, Cassotta A, Davydov AN, Kuka M, Zanon V, et al. CXCR3 identifies human naive CD8.J Immunol 2019;203:3179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mou D, Espinosa J, Lo DJ, Kirk AD. CD28 negative T cells: is their loss our gain? Am J Transplant 2014;14:2460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Melero I, Rouzaut A, Motz GT, Coukos G. T-cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy.Cancer Discov 2014;4:522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Etzerodt A, Rasmussen MR, Svendsen P, Chalaris A, Schwarz J, Galea I, et al. Structural basis for inflammation-driven shedding of CD163 ectodomain and tumor necrosis factor-α in macrophages.J Biol Chem 2014;289:778–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1).Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 39. Björkström NK, Gonzalez VD, Malmberg KJ, Falconer K, Alaeus A, Nowak G, et al. Elevated numbers of Fc gamma RIIIA+ (CD16+) effector CD8 T cells with NK cell-like function in chronic hepatitis C virus infection.J Immunol 2008;181:4219–28. [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y, Reynolds JM, Chang SH, Martin-Orozco N, Chung Y, Nurieva RI, et al. MKP-1 is necessary for T cell activation and function.J Biol Chem 2009;284:30815–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kratchmarov R, Magun AM, Reiner SL. TCF1 expression marks self-renewing human CD8.Blood Adv 2018;2:1685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karikoski M, Irjala H, Maksimow M, Miiluniemi M, Granfors K, Hernesniemi S, et al. Clever-1/Stabilin-1 regulates lymphocyte migration within lymphatics and leukocyte entrance to sites of inflammation.Eur J Immunol 2009;39:3477–87. [DOI] [PubMed] [Google Scholar]
- 43. Palani S, Maksimow M, Miiluniemi M, Auvinen K, Jalkanen S, Salmi M. Stabilin-1/CLEVER-1, a type 2 macrophage marker, is an adhesion and scavenging molecule on human placental macrophages.Eur J Immunol 2011;41:2052–63. [DOI] [PubMed] [Google Scholar]
- 44. Wu TD, Madireddi S, de Almeida PE, Banchereau R, Chen YJ, Chitre AS, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response.Nature 2020;579:274–8. [DOI] [PubMed] [Google Scholar]
- 45. Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1.Immunity 2019;51:1043–58. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tadayon S, Dunkel J, Takeda A, Eichin D, Virtakoivu R, Elima K, et al. Lymphatic endothelial cell activation and dendritic cell transmigration is modified by genetic deletion of clever-1.Front Immunol 2021;12:602122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burgdorf S, Kautz A, Böhnert V, Knolle PA, Kurts C. Distinct pathways of antigen uptake and intracellular routing in CD4 and CD8 T cell activation.Science 2007;316:612–6. [DOI] [PubMed] [Google Scholar]
- 48. Nagy L, Szanto A, Szatmari I, Széles L. Nuclear hormone receptors enable macrophages and dendritic cells to sense their lipid environment and shape their immune response.Physiol Rev 2012;92:739–89. [DOI] [PubMed] [Google Scholar]
- 49. Kidani Y, Bensinger SJ. Liver X receptor and peroxisome proliferator-activated receptor as integrators of lipid homeostasis and immunity.Immunol Rev 2012;249:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Donadon M, Torzilli G, Cortese N, Soldani C, Di Tommaso L, Franceschini B, et al. Macrophage morphology correlates with single-cell diversity and prognosis in colorectal liver metastasis.J Exp Med 2020;217:e20191847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Tables
Excel spreadsheets of mass spectrometry analysis
Excel spreadsheet of differentially expressed genes in CD14+ monocytes