Abstract
Purpose:
In REFLECT, lenvatinib demonstrated an effect on overall survival (OS) by confirmation of noninferiority to sorafenib in unresectable hepatocellular carcinoma. This analysis assessed correlations between serum or tissue biomarkers and efficacy outcomes from REFLECT.
Experimental Design:
Serum biomarkers (VEGF, ANG2, FGF19, FGF21, and FGF23) were measured by ELISA. Gene expression in tumor tissues was measured by the nCounter PanCancer Pathways Panel. Pharmacodynamic changes in serum biomarker levels from baseline, and associations of clinical outcomes with baseline biomarker levels, were evaluated.
Results:
Four hundred and seven patients were included in the serum analysis set (lenvatinib n = 279, sorafenib n = 128); 58 patients were included in the gene-expression analysis set (lenvatinib n = 34, sorafenib n = 24). Both treatments were associated with increases in VEGF; only lenvatinib was associated with increases in FGF19 and FGF23 at all time points. Lenvatinib-treated responders had greater increases in FGF19 and FGF23 versus nonresponders at cycle 4, day 1 (FGF19: 55.2% vs. 18.3%, P = 0.014; FGF23: 48.4% vs. 16.4%, P = 0.0022, respectively). Higher baseline VEGF, ANG2, and FGF21 correlated with shorter OS in both treatment groups. OS was longer for lenvatinib than sorafenib [median, 10.9 vs. 6.8 months, respectively; HR, 0.53; 95% confidence interval (CI), 0.33–0.85; P-interaction = 0.0397] with higher baseline FGF21. In tumor tissue biomarker analysis, VEGF/FGF-enriched groups showed improved OS with lenvatinib versus the intermediate VEGF/FGF group (HR, 0.39; 95% CI, 0.16–0.91; P = 0.0253).
Conclusions:
Higher baseline levels of VEGF, FGF21, and ANG2 may be prognostic for shorter OS. Higher baseline FGF21 may be predictive for longer OS with lenvatinib compared with sorafenib, but this needs confirmation.
Translational Relevance.
Biomarker identification in hepatocellular carcinoma (HCC) is challenging because of the molecular heterogenicity of HCC. Advanced disease is primarily diagnosed by radiologic criteria rather than by tumor biopsy. This paucity of available tumor tissue further hampers biomarker discovery, leading to an increased focus on evaluating serum biomarkers in advanced HCC. Identifying biomarkers predictive of treatment effect could be clinically useful in guiding therapeutic decisions. The baseline serum level of FGF21 is a candidate predictive biomarker for longer overall survival with lenvatinib versus sorafenib. This analysis aimed to further elucidate pharmacodynamic differences in the mechanism of action of lenvatinib from that of sorafenib. In contrast to sorafenib, lenvatinib demonstrated evidence of inhibition of the FGFR family and a greater magnitude of VEGFR inhibition. These results suggest the inhibitory activity of lenvatinib against FGFR may contribute to its increased tumor response.
Introduction
Lenvatinib is a multikinase inhibitor targeting vascular endothelial growth factor (VEGF) receptors 1–3, fibroblast growth factor (FGF) receptors 1–4, platelet-derived growth factor receptor-α (PDGFRα), KIT, and RET, with a distinct in vitro kinase inhibitory profile and kinase binding mode compared with sorafenib (1–5). Until the approval of lenvatinib in 2018, sorafenib remained the only approved first-line systemic treatment for patients with unresectable hepatocellular carcinoma (uHCC; ref. 6). Results from the IMbrave150 study showed that atezolizumab plus bevacizumab improved outcomes versus sorafenib (7), indicating that new options are widening the treatment landscape for uHCC. Presently, recommended and preferred first-line standard-of-care systemic therapies for hepatocellular carcinoma (HCC) include sorafenib and lenvatinib, and atezolizumab plus bevacizumab (8).
In the global, randomized, open-label, phase III REFLECT study, lenvatinib demonstrated noninferiority versus sorafenib for the first-line treatment of patients with uHCC in overall survival (OS) outcomes [hazard ratio (HR), 0.92; 95% confidence interval (CI), 0.79–1.06]. The median OS was 13.6 months (95% CI, 12.1–14.9) in the lenvatinib arm versus 12.3 months (95% CI, 10.4–13.9) in the sorafenib arm. In addition, treatment with lenvatinib significantly (P < 0.0001) improved progression-free survival [median: 7.4 months (95% CI, 6.9–8.8) vs. median: 3.7 months (95% CI, 3.6–4.6)], time to progression [median: 8.9 months (95% CI, 7.4–9.2) vs. median: 3.7 months (95% CI, 3.6–5.4)], and objective response rate [24.1% (95% CI, 20.2–27.9) vs. 9.2% (95% CI, 6.6–11.8)] based on modified Response Evaluation Criteria in Solid Tumors (mRECIST; ref. 9).
Lenvatinib is a distinct type V kinase inhibitor; in preclinical models, it inhibits both VEGF- and FGF-driven angiogenesis and has demonstrated antiproliferative activity against HCC cell lines dependent on the FGF signaling pathway (4, 10). Additionally, lenvatinib has been shown to induce HCC cell death via FGF receptor (FGFR) inhibition under nutrient- and oxygen-starved conditions, which mimic the tumor microenvironment under angiogenesis inhibition (11). Lenvatinib has also demonstrated increased antitumor activity compared with sorafenib against HCC xenograft tumors overexpressing VEGF (12).
The molecular heterogeneity of HCC provides a challenge in biomarker identification (13). One of the main targets for treatment in HCC is the angiogenic activity of the disease (14). Therefore, prognostic and predictive biomarkers can be found not only in the cancer cell itself, but also in the tumor microenvironment or serum. Among many cancer-associated signaling pathways, the VEGF signaling cascade pathway has been implicated in the pathogenesis of HCC (15). Additionally, the FGF signaling pathway may be involved as FGFR4 is mainly expressed in liver tissue. Studies suggest overexpression of the FGFR4 receptor and amplification of FGF19 contribute to HCC progression (16, 17). In this analysis, we evaluated serum and tumor tissue biomarkers using samples collected from the phase III REFLECT study and assessed correlations between those biomarkers and clinical outcomes, in order to identify potential biomarkers associated with clinical benefit and/or resistance, and to elucidate pharmacodynamic differences between sorafenib and lenvatinib.
Materials and Methods
Study design
The study design for the phase III open-label, multicenter, noninferiority REFLECT study (ClinicalTrials.gov identifier NCT01761266) has been reported previously (9). Briefly, from March 1, 2013 to July 30, 2015, 954 eligible patients with uHCC who had not received prior therapy for advanced disease were randomized (1:1) to receive oral lenvatinib (12 mg/day if bodyweight ≥ 60 kg or 8 mg/day if bodyweight < 60 kg) or sorafenib 400 mg twice daily in 28-day cycles. Patients provided written informed consent before undergoing any study-specific procedures. Relevant institutional review boards approved the study, which was done in accordance with the Declaration of Helsinki and local laws. The primary endpoint was OS. Secondary endpoints were progression-free survival, time to progression, objective response rate, patient-reported outcomes, and plasma pharmacokinetics. The data cutoff for the primary analysis was November 13, 2016.
Serum biomarker analysis
Serum samples were taken from patients in the REFLECT study who consented to serum biomarker assessment. Samples were collected at baseline, cycle 1 day 15, and cycles 2–4 day 1, and were stored at −20°C or below until assayed. Biomarker assays for VEGF, angiopoietin-2 (ANG2), FGF19, FGF21, and FGF23 were performed on serum samples using enzyme-linked immunosorbent assays (ELISA). VEGF, ANG2, FGF19, FGF21, and FGF23 were assayed at baseline and posttreatment (cycle 1 day 15, and cycles 2–4 day 1). Details on the ELISAs used can be found in the Supplementary Appendix.
Tumor tissue biomarker analysis
Archival tumor tissue samples were collected from patients in the REFLECT study who consented to tumor tissue biomarker assessment. Archival tumor samples from the most recent surgery or biopsy were collected at any time during the study, unless material was not available. If the entire tumor block could not be provided, 10 slides of 5-μm-thick tissue sections and 3–5 slides of 10-μm-thick tissue sections were provided. Procedures and medications that patients received between therapy initiation and tissue sample collection are listed in Supplementary Table S1.
Total RNA was isolated from tumor tissue samples for gene-expression analysis. Macrodissection was performed before extraction of total RNA if tumor content in the sample was less than 50%. Tumor samples for gene expression were analyzed using the nCounter PanCancer Pathways Panel (NanoString Technologies Inc.; Supplementary Excel) with 29 custom genes [ANGPT2, TEK, KDR, FLT4, CDH2, KRT18, SNAI1, SNAI2, TWIST1, TWIST2, VIM, ZEB1, ZEB2, GAS6, AXL, SDF1 (CXCL12), CXCR4, CSF1, GLUT1 (SLC2A1), MCT1 (SLC16A1), MCT4 (SLC16A4), PDH (PDP1), PDK1, LDHA, NANOG, SOX2, NOTCH4, PD-L1 (CD274), and PD-1 (PDCD1)]. Pathway enrichment analysis was conducted on the sets of genes identified as having expression levels that were nominally significantly associated with OS in the lenvatinib and sorafenib treatment arms. For this analysis, 12 canonical pathways and 1 cancer driver panel predefined by the PanCancer Pathways Panel, along with 1 additional angiogenic and growth factor pathway defined by our group, were used. Thirty-six genes were selected given their roles in the angiogenic and growth factor pathway (ANGPT1, ANGPT2, FGF1, FGF2, FGF3, FGF4, FGF5, FGF6, FGF7, FGF8, FGF9, FGF10, FGF11, FGF12, FGF13, FGF14, FGF16, FGF17, FGF18, FGF19, FGF20, FGF21, FGF22, FGF23, FGFR1, FGFR2, FGFR3, FGFR4, FIGF, FLT1, FLT4, KDR, PGF, TEK, VEGFA, and VEGFC).
Clustering analysis was performed using baseline expression levels of these 36 genes, the predefined cancer driver panel and other genes involved in transcriptional regulation, and Wnt and DNA-repair pathways. In the clustering analysis, the distance matrix was calculated using the Manhattan method, and the dendrogram was generated using Ward's method. From the dendrogram generated in the clustering analysis, three subgroups (VEGF-enriched, FGF-enriched, and intermediate) were identified in each treatment arm. The OS for each group identified in the clustering analysis were compared by plotting Kaplan–Meier curves. Baseline serum biomarker levels were also compared among subgroups identified by the clustering analysis in either lenvatinib, sorafenib, or combined arms.
Statistical analysis
The intent-to-treat (ITT) population included all patients randomized to treatment. The serum analysis set included all patients with at least 1 serum biomarker measurement at any time point. The gene-expression analysis set included all patients with a tumor gene-expression biomarker measurement.
All analyses were noted in the statistical analysis plan, and all statistical analyses were performed using SAS (version 9.3). Clustering analysis of gene-expression profiles was performed using R version 3.3.2 by R Foundation for Statistical Computing. Percentage changes in serum biomarker levels from baseline to cycle 1 day 15 and cycles 2–4 day 1 were summarized using medians and analyzed using the one-sample Wilcoxon signed-rank test for each treatment arm. The two-sample Wilcoxon rank-sum test was used to compare the distribution of changes in levels between the treatment arms.
Correlation analyses of pharmacodynamic changes of serum biomarker levels from baseline with best overall response [(complete response/partial response) vs. (stable disease/progressive disease/other)] based on mRECIST using independent imaging review assessments were performed using two-sample Wilcoxon rank-sum tests for each treatment arm. Correlation analyses of baseline serum biomarker levels with OS were performed using univariate Cox regression for each treatment arm. HRs were expressed as an increase in 1 standard deviation in baseline values.
OS was assessed using Kaplan–Meier estimates. OS differences between treatment arms were examined using the univariate Cox regression model for subgroups divided on the basis of baseline and changes in biomarker levels: low (0%−<25%), middle (≥25%−<75%), or high (≥75%−100%). Cutoffs were determined by visual inspection of Kaplan–Meier curves or minimum P values from a log-rank test. Dichotomized analyses (cutoff at third quartiles) of baseline levels of each serum biomarker and OS were performed using univariate Cox regression and log-rank test to investigate possible prognostic biomarkers for OS. Correlation analysis of baseline levels of ANG2 and FGF21 with OS by HCC etiology was performed using the univariate Cox regression analysis, and subsequent dichotomized analysis using the third quartile cutoff. Also, multivariate Cox regression analysis with treatment arm, biomarker category, and their interaction was conducted to explore predictive biomarkers for OS.
Gene lists were generated using the results of the univariate Cox regression for OS for the pathway enrichment analysis. The significance of pathway/panel enrichment was evaluated by Fisher exact test based on both the 13 pathways/panels in the PanCancer Pathways Panel and the additional angiogenic and growth factor pathway defined by our group. Correlation analysis of gene-expression levels of FGF ligands with progression-free survival was performed using univariate Cox regression for each treatment arm. False discovery rate adjustments (Benjamini–Hochberg) were utilized for all gene-expression analyses. Given the exploratory nature of our study, all statistical significance demonstrated and reported in this analysis was nominal.
Results
Patients
Eligible patients (N = 954) were randomized to receive lenvatinib (n = 478) or sorafenib (n = 476) in the ITT population of the REFLECT study. In the serum analysis set (n = 407), 279 patients received lenvatinib and 128 patients received sorafenib (Fig. 1). In the gene-expression analysis set (n = 58), 34 patients received lenvatinib and 24 patients received sorafenib; this sample size was decreased from 114 patients due to insufficient amounts of DNA/RNA extracted from available tumor tissue. Demographic and baseline characteristics of all patients are shown in Supplementary Table S2. There were differences in demographic and baseline characteristics between the serum analysis set and the ITT population. Approximately 33% of the ITT population was from the western region in both treatment arms, whereas 48% and 56% of the serum analysis set were from the western region in the lenvatinib and sorafenib arms, respectively. In the ITT population, compared with the serum analysis set, the percentage of White patients was lower (28% in lenvatinib and 30% in sorafenib vs. 42% in lenvatinib and 48% in sorafenib), whereas the percentage of Asian patients was higher (70% in lenvatinib and 69% in sorafenib vs. 55% in lenvatinib and 48% in sorafenib). The percentage of patients with hepatitis B virus was higher in the ITT population than in the serum analysis set (53% in lenvatinib and 48% in sorafenib vs. 36% in lenvatinib and 26% in sorafenib).
Figure 1.
CONSORT diagram. aA total of 41.7% (n = 398) of patients in the overall study (n = 954) had tissue confirmation of HCC histology. In the overall study, 95.0% (n = 378) of patients who had tissue confirmation of HCC histology consented for tissue analyses. bFewer blood serum samples were obtained from the sorafenib arm until the clinical study protocol was amended to clarify that samples were to be collected uniformly across both arms. cBaseline samples were unavailable for 12 patients in the lenvatinib arm, who were therefore excluded from the correlation analyses. dOnly tissues available (n = 114) at the cutoff date for sample collection (July 15, 2016) were used for biomarker analyses. eSample sizes were decreased due to the insufficient amounts of DNA/RNA extracted from tumor tissues. HCC, hepatocellular carcinoma; ITT, intent-to-treat.
The proportion of Asian patients was larger in the ITT population versus the gene-expression analysis set (approximately two thirds vs. one third), which, in turn, likely led to a difference in the percentage of patients with hepatitis B virus (approximately 50% in the ITT population vs. approximately 30% in the gene-expression analysis set) as hepatitis B is highly prevalent in Asia (18). In addition, the percentage of patients with baseline alpha-fetoprotein (AFP) levels ≥200 ng/mL was approximately 39% to 46% in the ITT population in the sorafenib and lenvatinib arms, respectively, versus 17% to 24% in the gene-expression analysis set. Differences in median AFP levels were observed between the lenvatinib and sorafenib arms in the gene-expression analysis set (22.1 vs. 3.3 ng/mL), the serum analysis set (74.8 vs. 27.6 ng/mL), and the ITT population (131.1 vs. 71.2 ng/mL).
Pharmacodynamic changes in serum biomarkers
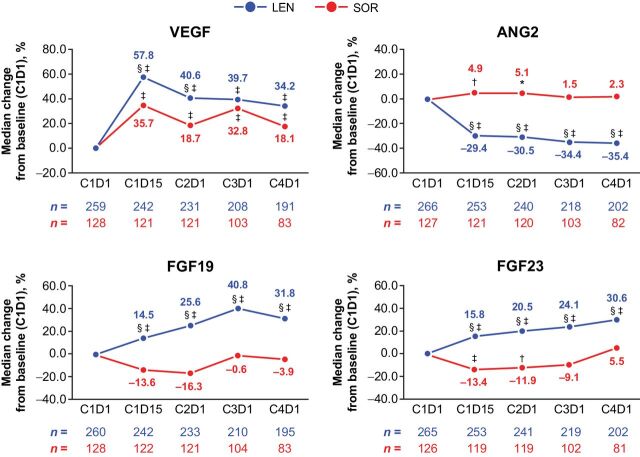
Overall, pharmacodynamic analysis confirmed different target engagement between lenvatinib and sorafenib. Both lenvatinib and sorafenib treatments resulted in increases in VEGF levels versus baseline, which reflect inhibition of the VEGF receptors (VEGFR) by both agents, although the magnitude of change was greater for lenvatinib compared with sorafenib. There was a median change from baseline of 57.8% at cycle 1, day 15 to 34.2% at cycle 4, day 1 with lenvatinib versus 35.7% to 18.1% at the respective time points with sorafenib (Fig. 2; Supplementary Table S3). Conversely, only lenvatinib treatment resulted in increases in FGF19 (median change from baseline of 14.5% at cycle 1, day 15 to 31.8% at cycle 4, day 1), and FGF23 levels (median change from baseline of 15.8% at cycle 1, day 15 to 30.6% to cycle 4, day 1) from baseline during treatment at all time points, which highlights the differential activity of lenvatinib versus sorafenib against the FGFR family. Additionally, ANG2 levels were also seen to decrease from baseline only in the lenvatinib group, which suggested that TIE-2 signaling was influenced by lenvatinib and not sorafenib. No significant changes in FGF21 levels from baseline were observed in either treatment arm (data not shown).
Figure 2.
Percentage changes in levels of serum biomarkers from baseline. *, P < 0.05 vs. baseline; †, P < 0.01 vs. baseline; ‡, P < 0.0001 vs. baseline; §, P < 0.05 between the LEN and SOR arms. ANG2, angiopoietin-2; C, cycle; D, day; FGF, fibroblast growth factor; LEN, lenvatinib; SOR, sorafenib; VEGF, vascular endothelial growth factor.
Associations of changes in levels of pharmacodynamic biomarkers and tumor response
In the lenvatinib arm, pharmacodynamic changes in FGF19 and FGF23 were associated with tumor responses (complete response/partial response vs. stable disease/progressive disease/other; Fig. 3; Supplementary Fig. S1). Patients who achieved an objective response in the lenvatinib arm had a greater median percentage increase from baseline in FGF19 (55% vs. 18%; P = 0.014) and FGF23 levels (48% vs. 16%; P = 0.002) compared with patients who did not achieve an objective response at cycle 4, day 1. A weaker trend was observed at cycle 3, day 1 (Supplementary Table S4). There were no significant observable trends in FGF19 and FGF23 between patients with objective response compared with patients with no objective response in the sorafenib arm, with the exception of FGF23 at cycle 2 day 1 (−21% vs. −7.5%; P = 0.0128) and at cycle 4, day 1 (−20% vs. 9.3%; P = 0.0269). At these time points, there was a decrease in FGF23 levels in the responders compared with the nonresponders. Larger decreases in ANG2 at cycle 3, day 1 and cycle 4, day 1 were observed in responders compared with nonresponders in the lenvatinib arm. There were no observable trends in ANG2 levels between responders and nonresponders in the sorafenib arm.
Figure 3.
![Figure 3. Percentage change in pharmacodynamics biomarker levels and association with objective response. *Median percentage changes of biomarker levels at cycle 4, day 1 from baseline are shown at the top of each box plot. Objective response was assessed by mRECIST using independent imaging review. Figures exclude outliers outside the y-axis range: VEGF [lenvatinib complete response/partial response (n = 4; 358.4, 366.1, 393.2, 642.5); lenvatinib noncomplete response/partial response (n = 7; 391.9, 407.7, 467.7, 469.9, 482.7, 526.3, 692.5); sorafenib complete response/partial response (n = 3; 436.0, 475.7, 803.1); sorafenib noncomplete response/partial response (n = 4; 399.9, 461.0, 505.1, 524.3)]; ANG2 [lenvatinib noncomplete response/partial response (n = 2; 132.9, 193.5); sorafenib noncomplete response/partial response (n = 1; 184.4)]; FGF19 [lenvatinib complete response/partial response (n = 6; 614.3, 640.1, 831.4, 1,097.6, 1,233.3, 1,462.5); lenvatinib noncomplete response/partial response (n = 5; 532.8, 533.2, 555.7, 606.7, 843.4); sorafenib noncomplete response/partial response (n = 2; 773.4, 776.6)]; FGF23 [lenvatinib complete response/partial response (n = 1; 258.1); lenvatinib noncomplete response/partial response (n = 1; 207.7)]. All other groups had 0 outliers. ANG2, angiopoietin-2; C, cycle; D, day; FGF, fibroblast growth factor; LEN, lenvatinib; SOR, sorafenib; VEGF, vascular endothelial growth factor.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8bd5/9401497/0afc7ffc52d4/4848fig3.jpg)
Percentage change in pharmacodynamics biomarker levels and association with objective response. *Median percentage changes of biomarker levels at cycle 4, day 1 from baseline are shown at the top of each box plot. Objective response was assessed by mRECIST using independent imaging review. Figures exclude outliers outside the y-axis range: VEGF [lenvatinib complete response/partial response (n = 4; 358.4, 366.1, 393.2, 642.5); lenvatinib noncomplete response/partial response (n = 7; 391.9, 407.7, 467.7, 469.9, 482.7, 526.3, 692.5); sorafenib complete response/partial response (n = 3; 436.0, 475.7, 803.1); sorafenib noncomplete response/partial response (n = 4; 399.9, 461.0, 505.1, 524.3)]; ANG2 [lenvatinib noncomplete response/partial response (n = 2; 132.9, 193.5); sorafenib noncomplete response/partial response (n = 1; 184.4)]; FGF19 [lenvatinib complete response/partial response (n = 6; 614.3, 640.1, 831.4, 1,097.6, 1,233.3, 1,462.5); lenvatinib noncomplete response/partial response (n = 5; 532.8, 533.2, 555.7, 606.7, 843.4); sorafenib noncomplete response/partial response (n = 2; 773.4, 776.6)]; FGF23 [lenvatinib complete response/partial response (n = 1; 258.1); lenvatinib noncomplete response/partial response (n = 1; 207.7)]. All other groups had 0 outliers. ANG2, angiopoietin-2; C, cycle; D, day; FGF, fibroblast growth factor; LEN, lenvatinib; SOR, sorafenib; VEGF, vascular endothelial growth factor.
Association of baseline biomarker levels and OS
The distribution of baseline levels of serum biomarkers appeared similar, with only minor variations observed between the two treatment arms (Supplementary Fig. S2). Higher baseline levels of VEGF, ANG2, and FGF21 were associated with shorter OS in both treatment arms by Cox regression analysis (Table 1). Upon evaluation of three groups (low, 0%−<25%; middle, ≥25%−<75%; or high, ≥75%−100%) with differing serum biomarker levels, a significant difference in OS was observed between the lenvatinib arm and the sorafenib arm in patients with high FGF21 (HR, 0.53; 95% CI, 0.33–0.85; Fig. 4A). Additionally, a difference in OS was also observed between treatment arms in patients with high ANG2 (HR, 0.64; 95% CI, 0.41–1.00; Fig. 4A). A cutoff level of 75% was used for the dichotomized analysis for baseline ANG2 and FGF21 levels because differences in OS were observed between the lenvatinib arm and the sorafenib arm in patients with high FGF21 (≥75%–100%; Fig. 4A). Median OS for patients with high baseline ANG2 levels was numerically longer in the lenvatinib arm versus the sorafenib arm (median 9.4 months; 95% CI, 7.0–13.6; vs. 7.7 months; 95% CI, 6.1–9.6, respectively; Supplementary Fig. S3; Fig. 4A). Dichotomized analysis also showed different associations between OS and treatment for baseline ANG2 levels (P-interaction = 0.075; Supplementary Table S5); ANG2 levels trend as predictive of response, although not reaching statistical significance.
Table 1.
Association of baseline biomarker levels with OS.a
| Lenvatinib | Sorafenib | |||||
|---|---|---|---|---|---|---|
| Marker | n | HR (95% CI) | P | n | HR (95% CI) | P |
| VEGF | 259 | 1.181 (1.055–1.321) | 0.0037 | 128 | 1.306 (1.036–1.647) | 0.0240 |
| ANG2 | 266 | 1.436 (1.283–1.607) | <0.0001 | 127 | 1.466 (1.269–1.693) | <0.0001 |
| FGF19 | 260 | 1.103 (0.946–1.286) | 0.2094 | 128 | 1.179 (0.981–1.417) | 0.0795 |
| FGF21 | 261 | 1.275 (1.130–1.438) | <0.0001 | 126 | 1.467 (1.213–1.774) | <0.0001 |
| FGF23 | 265 | 0.861 (0.738–1.005) | 0.0572 | 126 | 0.849 (0.665–1.086) | 0.1921 |
Abbreviations: ANG2, angiopoietin-2; CI, confidence interval; FGF, fibroblast growth factor; VEGF, vascular endothelial growth factor.
aCox proportional hazard model including standardized baseline value was used for each treatment group. Hazard ratios are based on comparison between high and low baseline biomarker levels and are expressed as an increase in 1 standard deviation in baseline values.
Figure 4.
![Figure 4. Associations between baseline serum biomarkers and OS. A, Associations between treatment arms and OS by baseline serum biomarker-level group. B, Kaplan–Meier plot showing associations by high or low baseline serum biomarker levels of FGF21 [cutoff level of 0.75 (= 688.0 ng/L)] and OS. ANG2, angiopoietin-2; C, cycle; CI, confidence interval; D, day; FGF, fibroblast growth factor; HR, hazard ratio; LEN, lenvatinib; OS, overall survival; SOR, sorafenib; VEGF, vascular endothelial growth factor.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8bd5/9401497/6bb0d0784777/4848fig4.jpg)
Associations between baseline serum biomarkers and OS. A, Associations between treatment arms and OS by baseline serum biomarker-level group. B, Kaplan–Meier plot showing associations by high or low baseline serum biomarker levels of FGF21 [cutoff level of 0.75 (= 688.0 ng/L)] and OS. ANG2, angiopoietin-2; C, cycle; CI, confidence interval; D, day; FGF, fibroblast growth factor; HR, hazard ratio; LEN, lenvatinib; OS, overall survival; SOR, sorafenib; VEGF, vascular endothelial growth factor.
For patients with high baseline FGF21 levels, median OS was longer in the lenvatinib arm versus the sorafenib arm (median 10.9 months; 95% CI, 8.2–13.1 vs. 6.8 months; 95% CI 4.6–10.3, respectively; Fig. 4B). Multivariate analysis of baseline serum biomarker levels with OS demonstrated that high FGF21 levels are correlated with shorter median OS regardless of the treatment arm (HR, 2.475; 95% CI, 1.565–3.922 with lenvatinib, P = 0.0001 vs. HR, 2.475; 95% CI, 1.279–4.808 with sorafenib, P = 0.0072; Supplementary Table S6).
When serum biomarker analyses were conducted in subgroups of HCC etiology [hepatitis B virus (HBV), hepatitis C virus (HCV), and alcohol], both ANG2 and FGF21 were significantly correlated with OS in patients with an etiology of HBV (Supplementary Table S7). By dichotomized analysis at the third quartile cutoff point, high baseline FGF21 levels were associated with shorter median OS regardless of HBV or HCV etiology in the lenvatinib arm (Supplementary Fig. S4). Additionally, high baseline ANG2 levels were associated with shorter median OS in the HBV etiology subgroup, but not in the HCV etiology subgroup. In the sorafenib arm, high baseline ANG2 levels were associated with shorter median OS regardless of HBV or HCV etiology; high baseline FGF21 levels were associated with shorter median OS in the HBV etiology subgroup, but not in the HCV etiology subgroup.
Tumor tissue gene-expression analysis of the angiogenesis and growth factor pathway
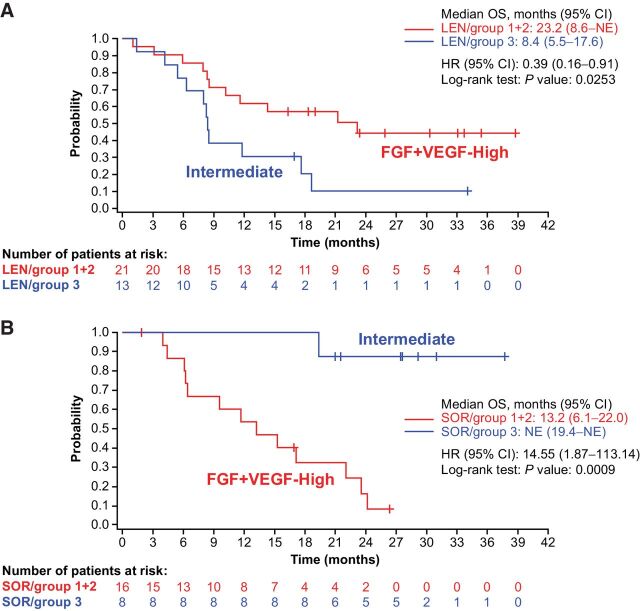
Clustering analysis was performed on baseline expression profiles of all available tumor samples from lenvatinib and sorafenib combined arms (Supplementary Fig. S5), and three subgroups with unique patterns of gene expression were identified; one had high expression levels of a family of FGF ligands, another had high expression levels of VEGF-A, and the third had relatively lower expressions for both FGF ligands and VEGF-A. Among the three groups, one subgroup (high FGF ligand) showed longer OS compared with the other subgroups, suggesting association of the gene-expression signature with OS. From this analysis, patients in the lenvatinib and sorafenib arms were divided into three groups by clustering analysis using the Manhattan method (19, 20) based on expression levels of the same set of 36 genes consisting of the angiogenic and growth factor pathway: Group 1 [VEGF-enriched, lenvatinib arm (41.2%) and sorafenib arm (45.8%)], group 2 [FGF-enriched, lenvatinib arm (20.6%) and sorafenib arm (20.8%)], and group 3 [intermediate expression of VEGF and FGF, lenvatinib arm (38.2%) and sorafenib arm (33.3%); Supplementary Fig. S6]. Although the groups were small, the distribution of patients among all groups was similar in both treatment arms. The Kaplan–Meier plot identified which of these subgroups had the longest OS in either the lenvatinib or sorafenib arm. In the lenvatinib arm, patients in group 2 (high expression of FGF ligands at baseline) appeared to experience longer OS, followed by patients in group 1 (the VEGF-enriched group). In patients treated with sorafenib, patients in group 3 with intermediate expression levels of VEGF and FGF ligands appeared to experience longer OS. No statistical analyses were performed for each of the three groups due to the small group sizes. In an effort to generate larger groups, these three groups were condensed into two groups [e.g., groups 1 and 2 (VEGF- and FGF-enriched) vs. group 3 (intermediate expression levels of FGF and VEGF)] and log-rank tests of OS between the two groups were conducted for each treatment arm (Fig. 5). The group enriched for higher expression of VEGF and FGF genes was associated with improved OS in the lenvatinib arm compared with the intermediate group (23.2 months vs. 8.4 months; HR, 0.39; 95% CI, 0.16–0.91; P = 0.0253). On the contrary, the VEGF- and FGF-enriched groups showed shorter OS compared with the intermediate group in the sorafenib arm (13.2 months vs. not estimable; HR, 14.55; 95% CI, 1.87–113.14; P = 0.0009).
Figure 5.
Association between molecular subgroups (FGF- and VEGF-enriched groups vs. FGF- and VEGF-intermediate groups) identified using the angiogenic and growth factor gene-expression profile and OS in the lenvatinib (A) and sorafenib (B) arms. ANG2, angiopoietin-2; C, cycle; CI, confidence interval; D, day; FGF, fibroblast growth factor; HR, hazard ratio; LEN, lenvatinib; OS, overall survival; SOR, sorafenib; VEGF, vascular endothelial growth factor.
Baseline serum biomarker levels were compared among groups (VEGF-enriched, FGF-enriched, and intermediate) identified by the clustering analysis of tumor gene-expression profiles in the lenvatinib and sorafenib combined arms (Supplementary Table S8). VEGF levels were high in the VEGF-enriched group, and FGF19 levels were high in the FGF-enriched group. In addition, both ANG2 and FGF21 levels were high in the VEGF-enriched group.
In addition, associations between FGF ligand gene-expression levels with progression-free survival as assessed by mRECIST using independent imaging review are shown as forest plots in Supplementary Fig. S7. In the lenvatinib arm, HRs for all FGF ligands except FGF2 and FGF7 were <1, suggesting high expression levels of FGF ligand genes were associated with prolonged progression-free survival.
Tumor tissue gene-expression analysis using 13 canonical cancer pathways
Pathway enrichment analysis was conducted for the sets of genes where expression levels were significantly associated with OS by Cox regression analysis in the separate lenvatinib and the sorafenib treatment arms. Associations with OS were identified in transcriptional regulation, Wnt, and DNA-repair pathways for the lenvatinib arm, and in the cancer driver gene panel for the sorafenib arm (Supplementary Table S9). For the suggested associations identified in the pathway enrichment analysis, clustering analysis was conducted. Patient subgroups identified by clustering analysis in the lenvatinib arm appeared to have different resulting OS benefits (Supplementary Fig. S8; red, blue, and green bars on the heat map) in the Wnt and DNA-repair pathways.
Discussion
The REFLECT study, which was the first positive phase III study in first-line uHCC since sorafenib was approved, demonstrated lenvatinib was noninferior to sorafenib in terms of OS (HR, 0.92; 95% CI, 0.79–1.06). This subsequent exploratory biomarker analysis suggests that the clinical activity of lenvatinib involves a distinct mechanism of action compared with sorafenib in patients with advanced uHCC, with unique inhibition of FGFR, and more potent inhibition of VEGFR with lenvatinib.
The identification of biomarkers with predictive value is crucial in guiding appropriate therapy selection in patients with uHCC. Biomarker studies are generally limited due to the lack of available tumor tissue samples because a tissue biopsy is typically not required to diagnose HCC (21). In our analysis, serum biomarker assays for VEGF, ANG2, FGF19, FGF21, and FGF23 were conducted both at baseline and post-treatment. No consistent changes in FGF21 levels from baseline were found in either treatment arm. The observed differences in serum biomarker changes between the lenvatinib and sorafenib arms support the distinct target kinase-inhibitory profiles of each agent. Although both lenvatinib and sorafenib treatments resulted in increases in VEGF levels, the magnitude of change was greater with lenvatinib, which has been more potent against VEGFR in preclinical studies (1). Additionally, the lenvatinib arm showed decreases in ANG2 levels, as well as increases in levels of FGF19 and FGF23, which is supportive of lenvatinib inhibition of FGFR4 and FGFR1, respectively (22–24). FGF19 and FGF23 levels increased more in patients who achieved an objective response compared with those who did not achieve an objective response in the lenvatinib arm. A subsequent analysis, evaluating the relationship between OS and objective response in the REFLECT study, demonstrated that objective response was an independent predictor of OS in patients with HCC regardless of the treatment arm (25).
Results of this analysis were consistent with the antitumor activity of lenvatinib observed in HCC xenograft models (12) consisting of aggressive VEGF overexpressing tumors. In addition, lenvatinib inhibited tumor FGF signaling pathways in HCC xenograft models and suppressed proliferation of HCC cell lines with an activated FGF signaling pathway (10). Also, lenvatinib induced cell death of HCC cell lines overexpressing FGF19 under a nutrient-depleted culture condition used to mimic the tumor microenvironment after angiogenesis inhibition (11).
In this analysis, results showed that higher levels of baseline VEGF, ANG2, and FGF21 may be prognostic for shorter OS, in patients with uHCC, regardless of treatment. Results from dichotomized analyses suggest that high baseline ANG2 and FGF21 levels were associated with shorter OS in both HBV and HCV etiologies for HCC in the sorafenib arm except for baseline FGF21 levels in the HCV subgroup. In the lenvatinib arm, high baseline ANG2 and FGF21 levels were associated with shorter OS in both HCC etiologies except for baseline ANG2 levels in the HCV subgroup. Additionally, high baseline ANG2 and FGF21 levels were associated with shorter OS in the HBV etiology subgroups across both arms. Clinical studies have demonstrated that ANG2 is an independent biomarker of poor prognosis in patients with HCC, with high ANG2 levels being associated with shorter OS (26, 27). High levels of VEGF at baseline were also associated with shorter OS (26).
Median OS was numerically longer in patients who had high baseline levels of ANG2 with lenvatinib compared with sorafenib, which may be related to the decrease in ANG2 levels seen only with lenvatinib. In addition, higher baseline levels of FGF21 may be predictive of longer OS with lenvatinib compared with sorafenib. This observation was not seen with FGF19 or FGF23. Although this may be due to a small sample size, it is important to note that FGF19, FGF21, and FGF23 each have distinct roles in biology and more specifically, the pathogenesis of HCC (28). FGF19 has been identified as a cancer driver in a subgroup of patients with uHCC (16), and FGF23 has been described as a pharmacodynamic biomarker for FGFR inhibitors in patients with cancer (29). A significant association was reported between baseline serum FGF19 levels and treatment response to lenvatinib in a biomarker analysis of uHCC patients treated with lenvatinib or sorafenib (30). However, the sample size (n = 27) included in the study is small, emphasizing the importance for further biomarker analysis. In another HCC study involving lenvatinib treatment (31), median serum FGF19 levels at baseline were similar between patients with objective response (n = 35) and patients with no objective response (n = 39). However, increases in serum FGF19 levels during treatment were associated with a response in patients receiving lenvatinib. These results are consistent with our findings that pharmacodynamic increases in FGF19 levels were associated with tumor responses in the lenvatinib arm. The details of FGF21′s interactions in HCC are largely unknown (28). FGFR inhibition by lenvatinib may contribute to improved OS in patients with high baseline levels of FGF21 compared with sorafenib. Multivariate analysis of baseline FGF21 levels with OS suggests that FGF21 may be an independent prognostic factor for OS. These results are hypothesis generating and warrant further investigation to evaluate the role of FGF21 in HCC.
Analysis of gene-expression patterns with an emphasis on angiogenesis and growth factor signaling pathway genes identified three subgroups in HCC. Interestingly, the longest median OS among the three subgroups in the lenvatinib arm was observed in patients with high expression levels of FGF ligands, followed by those with high expression levels of VEGF. Conversely, the longest median OS in the sorafenib arm was observed in patients with intermediate expression levels of VEGF and FGF ligands. Although these results should be interpreted with caution given the small number of evaluable samples, they support the distinct mechanism of action of lenvatinib from sorafenib and are consistent with the changes in levels of pharmacodynamic biomarkers (VEGF, FGF ligands, and ANG2) in the lenvatinib arm. These results further support that targeting the FGF and VEGF signaling pathways is important in the treatment of HCC.
This analysis was limited by the small sample of patients available for the biomarker analyses. Moreover, more patients in the lenvatinib arm than in the sorafenib arm were included in the biomarker analysis set. The number of patients was particularly small in the gene-expression analysis set, which was further complicated by any procedures and medications that patients may have received between therapy initiation and sample collection. These procedures and medications were not controlled for and could have affected results, as intra-arterial therapies can alter the microenvironment and biomarker expression patterns. The efficacy of lenvatinib may involve additional signaling pathways, and it is important to continue to investigate its mechanism of action. Of note, lenvatinib has demonstrated immunomodulatory activity and potentiation of the antitumor activity of programmed death receptor-1 (PD-1) inhibitors in syngeneic HCC mouse models, which supports combination treatment with pembrolizumab, a PD-1 inhibitor (32). This observation is supported by clinical data demonstrating a significant response rate (36% by RECIST v1.1) with the combination of lenvatinib and pembrolizumab in advanced HCC (33). Further hypotheses could be raised from the differences in tumor gene-expression patterns of genes involved in transcriptional regulation, Wnt, and DNA-repair pathways in the lenvatinib arm, and the cancer driver gene panel in the sorafenib arm; these factors may affect OS benefits for each drug treatment and may incite future study of novel combinations.
In conclusion, serum biomarker and gene-expression levels appeared to correlate with survival outcomes among patients with evaluable samples from REFLECT. This analysis was limited by the small number of patients with evaluable samples, and the variation in baseline characteristics between the gene-expression analysis set and the ITT population. However, the differences in baseline characteristics between these groups were understandable and most likely due to the small size of the gene-expression analysis set. Despite these limitations, multivariate analysis of important clinical prognostic factors in HCC supports our results. Of note, data were analyzed by mRECIST per a blinded independent review. In patients with unresectable HCC who had not received prior systemic therapy for advanced disease, lenvatinib demonstrated clinical activity based on a mechanism of action that is distinct from sorafenib. Specifically, lenvatinib demonstrated clinical evidence of FGFR inhibition and stronger inhibition of angiogenesis pathways (VEGFR and TIE-2/ANG2). ANG2 and TIE-2 are selectively expressed on endothelial cells and are increased with enhanced tumor angiogenesis. Lenvatinib could lead to decreases in both ANG2 and TIE-2, without direct TIE-2 inhibition, based on its potent angiogenesis inhibition and resultant decrease in endothelial cells.
These results suggest that the inhibitory activity of lenvatinib against FGFR may contribute to the increased tumor response, and FGF21 may be a candidate biomarker predictive of longer OS with lenvatinib. Interestingly, it appears that lenvatinib may perform better in the poor prognosis subgroups independent of the specific pathway, due to its increased activity overall and similarity to the overall study population results. These results are hypothesis generating and warrant further study. The ongoing phase III LEAP-002 study (NCT03713593), evaluating lenvatinib versus lenvatinib plus pembrolizumab in advanced HCC, will provide further material for investigation.
Authors' Disclosures
R.S. Finn reports grants and personal fees from Eisai, Merck, Bayer, Bristol Myers Squibb, Eli Lilly, Pfizer, and Roche/Genentech, as well as personal fees from AstraZeneca and CStone during the conduct of the study. M. Kudo reports personal fees, grants, and other support from Eisai during the conduct of the study. M. Kudo also reports personal fees from Bayer, MSD, Bristol Myers Squibb, Chugai, and Eli Lilly; grants from Gilead Sciences, Taiho, Sumitomo Dainippon Pharma, Takeda, Otsuka, EA Pharma, and AbbVie; and other support from Ono, MSD, Bristol Myers Squibb, Roche, and AstraZeneca outside the submitted work. A.-L. Cheng reports personal fees from Bristol Myers Squibb, Bayer, Eisai, Ono Pharmaceutical, AstraZeneca, Genentech/Roche, MSD, BeiGene Ltd., Exelixis Ltd., Ipsen Innovation, F. Hoffmann-La Roche Ltd., Bayer Yakuhin Ltd., Chugai Pharmaceutical, IQVIA, Novartis, and Amgen Taiwan outside the submitted work. L. Wyrwicz reports other support from Eisai during the conduct of the study. J.-F. Blanc reports personal fees from Eisai, Bayer, Ipsen, Roche, and AstraZeneca during the conduct of the study. A.D. Baron reports other support from Eisai during the conduct of the study, as well as personal fees from Eisai, Bristol Myers Squibb, Janssen, and Lilly outside the submitted work. A. Vogel reports personal fees from Eisai, Roche, MSD, BMS, Bayer, AstraZeneca, Lilly, Ipsen, Incyte, and Pierre Fabre outside the submitted work. M. Ikeda reports grants and personal fees from Bayer, AstraZeneca, BMS, Chugai, Eli Lilly, Eisai, MSD, and Takeda, as well as grants from Merck Serono during the conduct of the study. M. Ikeda also reports personal fees from Astellas, Gilead, Otsuka, Sumitomo Dainippon, Taiho, and Teijin Pharma; grants and personal fees from Yakult, ASLAN, EA Pharma, Nihon Servier, and Novartis; and grants from Chiome Bioscience, GlaxoSmithKline, J-Pharma, Ono, and Pfizer outside the submitted work. F. Piscaglia reports personal fees from Eisai during the conduct of the study, as well as personal fees from Bayer, Bracco, Tiziana Life Sciences, Alkermes, Roche, MSD, Ipsen, AstraZeneca, LaForce Guerbet, and BMS outside the submitted work. K.-H. Han reports grants from Eisai during the conduct of the study. Y. Minoshima reports a patent for WO2020071451A1 issued to Eisai Co., Ltd. M. Kanekiyo reports being an employee of Eisai Inc. M. Ren reports being an employee of Eisai Inc. T. Tamai reports other support from Eisai during the conduct of the study. C.E. Dutcus reports personal fees from Eisai Inc. during the conduct of the study. H. Ikezawa reports personal fees from Eisai Co., Ltd., during the conduct of the study. Y. Funahashi reports personal fees from Eisai Co., Ltd., during the conduct of the study; in addition, Y. Funahashi has a patent for WO02/032872 issued. T.R.J. Evans reports grants and personal fees from Eisai, as well as grants, personal fees, and non-financial support from Merck (MSD) during the conduct of the study. T.R.J. Evans also reports grants, personal fees, and non-financial support from Bayer, Roche and Genentech, and BMS; grants and personal fees from AstraZeneca and Medivir; and grants from MiNA Therapeutics, Adaptimmune, and Cancer Research UK outside the submitted work. T.R.J. Evans also reports reimbursement of study costs for commercially sponsored clinical trials (payable to employing institution) to support non-HCC clinical trials; honorarium from Cancer Research UK for co-editorship, clinical subjects, British Journal of Cancer; and grant support from Cancer Research UK and Chief Scientist's Office, Scotland, to support Glasgow Experimental Cancer Medicine Centre. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Excel_nCounter PanCancer Human Pathways Panel Gene List
Supplementary appendix, supplementary tables s1 - s9, supplementary figures s1 - s8
Acknowledgments
This study was sponsored by Eisai Inc., Woodcliff Lake, NJ, and Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ. Medical writing support was provided by Oxford PharmaGenesis Inc., Newtown, PA, and was funded by Eisai Inc., Woodcliff Lake, NJ, USA, and Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
R.S. Finn: Conceptualization, supervision, investigation, methodology, writing–original draft, writing–review and editing. M. Kudo: Conceptualization, supervision, investigation, writing–original draft, writing–review and editing. A.-L. Cheng: Conceptualization, investigation, writing–original draft, writing–review and editing. L. Wyrwicz: Conceptualization, investigation, writing–original draft, writing–review and editing. R.K.C. Ngan: Conceptualization, investigation, writing–original draft, writing–review and editing. J.-F. Blanc: Conceptualization, investigation, writing–original draft, writing–review and editing. A.D. Baron: Conceptualization, investigation, writing–original draft, writing–review and editing. A. Vogel: Conceptualization, investigation, writing–original draft, writing–review and editing. M. Ikeda: Conceptualization, investigation, writing–original draft, writing–review and editing. F. Piscaglia: Conceptualization, investigation, writing–original draft, writing–review and editing. K.-H. Han: Conceptualization, investigation, writing–original draft, writing–review and editing. S. Qin: Conceptualization, investigation, writing–original draft, writing–review and editing. Y. Minoshima: Conceptualization, formal analysis, validation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M. Kanekiyo: Conceptualization, formal analysis, validation, visualization, methodology, writing–original draft, writing–review and editing. M. Ren: Conceptualization, software, formal analysis, validation, visualization, methodology, writing–original draft, writing–review and editing. R. Dairiki: Conceptualization, software, formal analysis, methodology, writing–original draft, writing–review and editing. T. Tamai: Conceptualization, formal analysis, writing–original draft, writing–review and editing. C.E. Dutcus: Conceptualization, resources, supervision, writing–original draft, project administration, writing–review and editing. H. Ikezawa: Conceptualization, software, formal analysis, validation, visualization, methodology, writing–original draft, writing–review and editing. Y. Funahashi: Conceptualization, resources, formal analysis, supervision, methodology, writing–original draft, project administration, writing–review and editing. T.R.J. Evans: Conceptualization, supervision, investigation, writing–original draft, writing–review and editing.
References
- 1. Tohyama O, Matsui J, Kodama K, Hata-Sugi N, Kimura T, Okamoto K, et al. Antitumor activity of lenvatinib (e7080): an angiogenesis inhibitor that targets multiple receptor tyrosine kinases in preclinical human thyroid cancer models. J Thyroid Res 2014;2014:638747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matsui J, Funahashi Y, Uenaka T, Watanabe T, Tsuruoka A, Asada M. Multi-kinase inhibitor E7080 suppresses lymph node and lung metastases of human mammary breast tumor MDA-MB-231 via inhibition of vascular endothelial growth factor-receptor (VEGF-R) 2 and VEGF-R3 kinase. Clin Cancer Res 2008;14:5459–65. [DOI] [PubMed] [Google Scholar]
- 3. Matsui J, Yamamoto Y, Funahashi Y, Tsuruoka A, Watanabe T, Wakabayashi T, et al. E7080, a novel inhibitor that targets multiple kinases, has potent antitumor activities against stem cell factor producing human small cell lung cancer H146, based on angiogenesis inhibition. Int J Cancer 2008;122:664–71. [DOI] [PubMed] [Google Scholar]
- 4. Yamamoto Y, Matsui J, Matsushima T, Obaishi H, Miyazaki K, Nakamura K, et al. Lenvatinib, an angiogenesis inhibitor targeting VEGFR/FGFR, shows broad antitumor activity in human tumor xenograft models associated with microvessel density and pericyte coverage. Vasc Cell 2014;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okamoto K, Ikemori-Kawada M, Jestel A, von König K, Funahashi Y, Matsushima T, et al. Distinct binding mode of multikinase inhibitor lenvatinib revealed by biochemical characterization. ACS Med Chem Lett 2015;6:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kudo M. Targeted and immune therapies for hepatocellular carcinoma: predictions for 2019 and beyond. World J Gastroenterol 2019;25:789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 2020;382:1894–905. [DOI] [PubMed] [Google Scholar]
- 8. National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology (NCCN Guidelines®). Hepatobiliary cancers. Version 5. 2020. Available from: https://www.nccn.org/professionals/physician_gls/pdf/hepatobiliary.pdf. Accessed October 23, 2020. [DOI] [PubMed]
- 9. Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018;391:1163–73. [DOI] [PubMed] [Google Scholar]
- 10. Matsuki M, Hoshi T, Yamamoto Y, Ikemori-Kawada M, Minoshima Y, Funahashi Y, et al. Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models. Cancer Med 2018;7:2641–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoshi T, Miyano S, Watanabe H, Sonobe RMK, Seki Y, Ohta E, et al. Lenvatinib induces death of human hepatocellular carcinoma cells harboring an activated FGF signaling pathway through inhibition of FGFR-MAPK cascades. Biochem Biophys Res Commun 2019;513:1–7. [DOI] [PubMed] [Google Scholar]
- 12. Adachi Y, Matsuki M, Watanabe H, Takase K, Kodama K, Matsui J, et al. Antitumor and antiangiogenic activities of lenvatinib in mouse xenograft models of vascular endothelial growth factor-induced hypervascular human hepatocellular carcinoma. Cancer Invest 2019;37:185–98. [DOI] [PubMed] [Google Scholar]
- 13. Lu LC, Hsu CH, Hsu C, Cheng AL. Tumor heterogeneity in hepatocellular carcinoma: facing the challenges. Liver Cancer 2016;5:128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morse MA, Sun W, Kim R, He AR, Abada PB, Mynderse M, et al. The role of angiogenesis in hepatocellular carcinoma. Clin Cancer Res 2019;25:912–20. [DOI] [PubMed] [Google Scholar]
- 15. Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology 2008;48:1312–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell 2011;19:347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raja A, Park I, Haq F, Ahn SM. FGF19-FGFR4 signaling in hepatocellular carcinoma. Cells 2019;8:536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liaw YF, Leung N, Kao JH, Piratvisuth T, Gane E, Han KH, et al. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2008 update. Hepatol Int 2008;2:263–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dudoit S, Fridlyand J. A prediction-based resampling method for estimating the number of clusters in a dataset. Genome Biol 2002;3:research0036.1–0036.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mardia KV, Kent JT, Bibby JM. Multivariate analysis. San Diego, CA: Academic Press; 1979. [Google Scholar]
- 21. Hennedige T, Venkatesh SK. Imaging of hepatocellular carcinoma: diagnosis, staging and treatment monitoring. Cancer Imaging 2013;12:530–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Itoh N. Hormone-like (endocrine) Fgfs: their evolutionary history and roles in development, metabolism, and disease. Cell Tissue Res 2010;342:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim KB, Chesney J, Robinson D, Gardner H, Shi MM, Kirkwood JM. Phase I/II and pharmacodynamic study of dovitinib (TKI258), an inhibitor of fibroblast growth factor receptors and VEGF receptors, in patients with advanced melanoma. Clin Cancer Res 2011;17:7451–61. [DOI] [PubMed] [Google Scholar]
- 24. Yu XX, Watts LM, Manchem VP, Chakravarty K, Monia BP, McCaleb ML, et al. Peripheral reduction of FGFR4 with antisense oligonucleotides increases metabolic rate and lowers adiposity in diet-induced obese mice. PLoS One 2013;8:e66923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Cheng AL, et al. Analysis of survival and objective response (OR) in patients with hepatocellular carcinoma in a phase 3 study of lenvatinib (REFLECT). Presented at: Gastrointestinal Cancers Symposium of the American Society of Clinical Oncology, January 17–19, 2019, San Francisco, CA. [Google Scholar]
- 26. Llovet JM, Peña CE, Lathia CD, Shan M, Meinhardt G, Bruix J, et al. Plasma biomarkers as predictors of outcome in patients with advanced hepatocellular carcinoma. Clin Cancer Res 2012;18:2290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pestana RC, Hassan MM, Abdel-Wahab R, Abugabal YI, Girard LM, Li D, et al. Clinical and prognostic significance of circulating levels of angiopoietin-1 and angiopoietin-2 in hepatocellular carcinoma. Oncotarget 2018;9:37721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yie J, Hecht R, Patel J, Stevens J, Wang W, Hawkins N, et al. FGF21 N- and C-termini play different roles in receptor interaction and activation. FEBS Lett 2009;583:19–24. [DOI] [PubMed] [Google Scholar]
- 29. Katoh M. FGFR inhibitors: effects on cancer cells, tumor microenvironment and whole-body homeostasis (review). Int J Mol Med 2016;38:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shigesawa T, Suda G, Kimura M, Shimazaki T, Maehara O, Yamada R, et al. Baseline angiopoietin-2 and FGF19 levels predict treatment response in patients receiving multikinase inhibitors for hepatocellular carcinoma. JGH Open 2020;4:880–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chuma M, Uojima H, Numata K, Hidaka H, Toyoda H, Hiraoka A, et al. Early changes in circulating FGF19 and Ang-2 levels as possible predictive biomarkers of clinical response to lenvatinib therapy in hepatocellular carcinoma. Cancers (Basel) 2020;12:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kimura T, Kato Y, Ozawa Y, Kodama K, Ito J, Ichikawa K, et al. Immunomodulatory activity of lenvatinib contributes to antitumor activity in the Hepa1–6 hepatocellular carcinoma model. Cancer Sci 2018;109:3993–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Finn RS, Ikeda M, Zhu AX, Sung MW, Baron AD, Kudo M, et al. Phase Ib study of lenvatinib plus pembrolizumab in patients with unresectable hepatocellular carcinoma. J Clin Oncol 2020;38:2960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Excel_nCounter PanCancer Human Pathways Panel Gene List
Supplementary appendix, supplementary tables s1 - s9, supplementary figures s1 - s8