Abstract
Purpose:
Patient-derived organoids (PDO) of lung cancer has been recently introduced, reflecting the genomic landscape of lung cancer. However, clinical relevance of advanced lung adenocarcinoma organoids remains unknown. Here, we examined the ability of PDOs to predict clinical responses to targeted therapies in individual patients and to identify effective anticancer therapies for novel molecular targets.
Experimental Design:
Eighty-four organoids were established from patients with advanced lung adenocarcinoma. Formalin-fixed, paraffin-embedded tumor specimens from corresponding patients were analyzed by whole-exome sequencing (n = 12). Organoids were analyzed by whole-exome sequencing (n = 61) and RNA sequencing (n = 55). Responses to mono or combination targeted therapies were examined in organoids and organoid-derived xenografts.
Results:
PDOs largely retained somatic alterations including driver mutations of matching patient tumors. PDOs were able to recapitulate progression-free survival and objective responses of patients with non–small cell lung cancer receiving clinically approved tyrosine kinase inhibitors. PDOs recapitulated activity of therapeutic strategies under clinical investigation. YUO-071 harboring an EGFR exon 19 deletion and a BRAF G464A mutation and the matching patient responded to dabrafenib/trametinib combination therapy. YUO-004 and YUO-050 harboring an EGFR L747P mutation was sensitive to afatinib, consistent with the response in the matching patient of YUO-050. Furthermore, we utilized organoids to identify effective therapies for novel molecular targets by demonstrating the efficacy of poziotinib against ERBB2 exon 20 insertions and pralsetinib against RET fusions.
Conclusions:
We demonstrated translational relevance of PDOs in advanced lung adenocarcinoma. PDOs are an important diagnostic tool, which can assist clinical decision making and accelerate development of therapeutic strategies.
Translational Relevance.
We demonstrated that in vitro drug responses in patient-derived organoids (PDO) are correlated to clinical responses to targeted therapies in individual patients with advanced lung adenocarcinoma and PDOs can be used to identify effective anticancer therapies for novel molecular targets. PDOs recapitulated progression-free survival and objective responses of non–small cell lung cancer patients receiving clinically approved targeted agents. PDOs also predicted the activity of therapeutic strategies under clinical investigation. YUO-071 harboring EGFR exon 19 deletion/BRAF G464A mutation and the matching patient responded to dabrafenib/trametinib combination therapy. YUO-004 and YUO-050 harboring an EGFR L747P mutation was sensitive to afatinib, consistent with the response in the matching patient of YUO-050. Furthermore, we utilized organoids to demonstrate preclinical efficacy of poziotinib against ERBB2 exon 20 insertions and pralsetinib against RET fusions. Our findings suggest the utility of PDOs in clinical decision making and development of therapeutic strategies.
Introduction
Non–small cell lung cancer (NSCLC) is a leading cause of cancer-related mortality worldwide. Over the last decade, precision medicine tailored to individual patients has greatly improved survival and disease control in patients with advanced NSCLC. Implementation of precision medicine requires identification of actionable molecular targets and treatment with therapies targeting the specific genetic aberrations (1). However, molecular profiling-based drug selection has several limitations. Only a portion of NSCLC benefits from targeted therapies, including molecular subsets such as EGFR activating mutations, T790 mutation, BRAF V600E mutation, MET exon 14 skipping mutations, and ALK, ROS1, RET, and NTRK1/2/3 fusions (2). In addition, responses to targeted therapies are heterogeneous in patients harboring the identical driver mutation (3–5). Rarer molecular subtypes among the driver oncogenes display diverse clinical and biological characteristics, further complicating the clinical decision making for patients with advanced NSCLC (6–9).
Recent studies have focused on patient-derived organoids (PDO) as preclinical models to investigate tumor biology. PDOs are tissue-specific stem cells derived from various adult human organs and cancers and have been cultured in three-dimensional (3D) conditions utilizing extracellular matrix components (10). Although 2D conventional cell lines have been widely used in cancer research, they may not represent the complex biological characteristics of patient tumors. Furthermore, establishment and utilization of 2D patient-derived cells which represent their parental tumors has been hampered by a low success rate of model establishment (11). In addition, classical in vivo models are time-consuming to generate and labor-intensive, limiting high-throughput studies (12). Notably, PDOs have a short establishment time and retain biological features of the patient tumors, presenting a unique value for precision medicine.
Previous studies on NSCLC PDOs have utilized primary patient tumors and patient-derived xenografts of early-stage NSCLC including lung squamous cell carcinoma, lung adenocarcinoma, and large cell carcinoma (13–16). In this study, we establish PDOs using malignant effusions and metastatic surgical specimens of advanced lung adenocarcinoma and demonstrate that PDOs are a clinically-relevant platform which can be used for patient-specific drug testing and proof of concept preclinical studies.
Materials and Methods
Patient consent and samples
This study was approved by Yonsei University Hospital Institutional Review Board (Seoul, Korea; IRB no.: 4–2016–0788) and conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent. Pleural effusion and surgically resected metastatic tumors were collected from patients with advanced lung adenocarcinoma at Yonsei Cancer Center. FISH, direct sequencing, TruSight Oncology 500 (Illumina), or TruSight Tumor 170 (Illumina) was performed for molecular profiling of NSCLC at initial diagnosis or at recurrence. Cell-free DNA (cfDNA) of matching patient for YUO-071 (Guardant Health) was analyzed using Guardant360 assay.
Processing of malignant effusions
To standardize the experimental protocol, only 200 mL of malignant effusion was used to collect tumor cells. Red blood cells in the cell pellet were removed by hypotonic lysis in sterile MilliQ H2O (Merk Millipore) followed by adding Advanced DMEM/F12 medium (Gibco), being strained over a 100 μm filter with retained debris, and centrifuged at 500 × g for 5 minutes.
Surgical tumor tissue processing
Tumor tissue was washed three times with advanced DMEM/F12 supplemented with antibiotics (Invivogen) and chopped with sterile blades. Tumor pieces were dissociated in 2 mg/mL collagenase (Sigma-Aldrich) on shaker at 37°C for 1 to 2 hours. After incubation, the suspensions were added with advanced DMEM/F12 medium, passed through 100 μm cell strainers, and centrifuged at 500 × g for 5 minutes.
Establishment of organoids
Organoids were established as described previously (15). In brief, cells were counted under a microscope and centrifuged at 500 × g for 5 minutes. Then, cells were resuspended in ice-cold 500 μL Matrigel (Corning) and 20 μL drops of Matrigel cell suspension were seeded on prewarmed 48-well culture plates (Corning) at a density of ∼2 × 103 cells per 20 μL Matrigel/well. The Matrigel was solidified for 15 minutes at 37°C and overlaid with 250 μL airway organoid medium [AO; AdDF+++, 20% conditioned R-spondin1 medium supplemented with B27 (Invitrogen), 1.25 mmol/L N-acetylcystein (Sigma-Aldrich), 5 mmol/L nicotinamide (Sigma), 25 ng/mL human fibroblast growth factor 7 (Peprotech), 100 ng/mL human noggin (Peprotech), 100 ng/mL human FGF 10 (Peprotech), 500 nmol/L A83–01 (Tocris), and 500 nmol/L SB202190 (Sigma)]. AdDF+++ medium is advanced DMEM/F12 medium (Invitrogen) supplemented with 10 nmol/L HEPES (Invitrogen), 1× GlutaMax (Invitrogen), and 1× antibiotic–antimycotic (Invitrogen). Y-27632 (10 μmol/L; Enzo Life Science) was added for the first 2 days. Cultures were kept at 37°C, 5% CO2 in a humidified incubator. Medium was replenished every 2 to 3 days.
Histology and IHC
Organoids and their parental tumors were fixed in 4% paraformaldehyde for 24 hours at 4°C, washed with PBS, and then transferred to 70% ethanol, processed for paraffin embedding, sectioning, deparaffinization, dehydration, and hematoxylin–eosin staining. IHC was performed using the antibody against thyroid transcription factor 1 (TTF-1, Clone EP1584Y; Abcam), Calretinin (Clone DAK-Calret-1; Agilent Technologies), and p53 (Leica Biosystems). Organoid imaging was performed on OLYMPUS BX51 microscope (Olympus) using a 20× magnification. Images were processed using Olympus cellSens software and Photoshop CS4 (8 bit).
Next-generation sequencing
RNA was isolated from organoids using TRizol (Invitrogen) following the manufacturer's instructions. RNA libraries were generated from 55 PDOs using TruSeq-Stranded mRNA Sample Prep Kit (Illumina). The libraries were subjected to paired-end sequencing with a 150 bp read length using NovaSeq 6000 (Illumina). Quality scores for over 75% of raw reads were >Q30. We used Arriba (https://github.com/suhrig/arriba/), which is based on the STAR aligner, to detect fusion genes in organoids. A minimum coverage fraction of Arriba is 0.15, ignoring the fusion events that are not fully expressed. GENCODE19, hs37d5, blacklist_hg19_hs37d5_GRCh37 assembly, and annotation files were used. The circos plots from RNA-seq data of fusion transcript candidates with highest coverage was drawn.
Genomic DNA (gDNA) was isolated from organoids, matching normal blood samples and formalin-fixed, paraffin-embedded (FFPE) tumor specimens using the DNeasy Blood & Tissue Kits (Qiagen). Concentration and purity of gDNA were assessed by agarose gel electrophoresis and PicoGreen dsDNA assay (Invitrogen). Exome libraries were generated from 61 PDOs and 58 matching normal blood samples using SureSelect v6 Kit (Agilent Technologies) and sequenced on NovaSeq (Illumina). Sequencing reads were mapped to the human chromosome (hg19) using the Burrows-Wheeler alignment tool (17, 18). Somatic mutations in each tumor specimen or organoid were detected using MuTect2 and annotated with Oncotator (19). High-quality somatic mutations were acquired by (i) filtering out germline mutations with allele frequencies >0.01 in the Exome Aggregation Consortium (ExAC) database, (ii) filtering out somatic mutations with allele frequencies <0.01, and (iii) including mutations in the cosmic database. Copy-number variations were detected using CNVkit (20).
Clinically relevant somatic alterations were selected on the basis of the TARGET database (21). “Actionable targets” and “Clinically relevant driver genes” were selected on the basis of the NCCN guideline (version 8, 2020) and the lung adenocarcinoma The Cancer Genome Atlas (TCGA) database (2). Specifically, “Actionable targets” were defined as driver oncogenes in lung adenocarcinoma, which clinically respond to FDA-approved targeted agents, including EGFR L858R mutation, exon 19 deletions, T790M mutation, G719X mutation, S768I mutation, L861Q mutation, ROS1 fusions, RET fusions, and BRAF V600E mutation. We were not able to detect ALK- and NTRK fusions in our organoid library. “Clinically relevant driver genes” were defined as driver oncogenes in lung adenocarcinoma that are under clinical investigation for druggability, including MET amplification, ERBB2 amplification, and mutations in kinase domains of ERBB2 (exon 19–21), KRAS (exon 2–3), BRAF (exon 11–18), and EGFR (exon 18–21) other than the aforementioned EGFR and BRAF actionable targets.
The next-generation sequencing (NGS) data are available at the Sequence Read Archive (accession no.: PRJNA725056).
Cell viability assay
Organoids were cultured in AO medium for 5 to 10 days after organoids were dissociated into single cells using TrypLE (Thermo Fisher Scientific). Dispase solution (1 mg/mL) was added to plates and incubated at 37°C for 10 minutes before cell suspension was pipetted with a 1 mL tip a few times, washed twice with cold PBS, and centrifuged at 1300 rpm for 3 minutes. Organoid pellets were resuspended in AdDF+++ medium containing 5% Matrigel, seeded on 96-well ultra-low attachment plates (Corning; 2,500 organoids/well), and treated with drugs after 1 hour (22). At indicated time points, CellTiterGlo 3D (Promega) was used to measure luminescence according to the manufacturer's protocol.
For 2D cultures, cells were seeded on 96-well plates in AdDF+++ medium (5,000 cells/well; ref. 7). After overnight incubation, AdDF+++ medium containing drugs were added to the wells. After 3 days, CellTiterGlo 3D was used to measure luminescence according to the manufacturer's protocol.
IC50 values were calculated from three biological replicates (three technical replicates per biological replicate) using GraphPad Prism version 7. In vitro response to a targeted therapy was defined as sensitive (IC50 value < 100 nmol/L) or resistant (IC50 value > 100 nmol/L; ref. 7).
Measuring change of cell viability after drug exposure
A protocol for drug exposure was based on Sharma and colleagues (23). Briefly, organoids were suspended in Matrigel and seeded on 48-well plates (∼250 organoids/well, three wells per biological replicate). Organoids were replenished with fresh AdDF+++ medium containing a drug at the indicated concentration every 3 days up to 15 days. At indicated time points, medium was decanted and 200 μL CellTiterGlo 3D was added to each well. After 30 minutes incubation at room temperature, 100 μL was used to measure luminescence. Relative cell viability was calculated as follows: (luminescence at the indicated time point)/(luminescence at day 0). Percentage change of cell viability was calculated as follows: [(luminescence at the indicated time point) − (luminescence at day 0)]/(luminescence at day 0) × 100.
Direct sequencing
Direct sequencing of EGFR exon 18, 19, 20, and 21 was performed as described previously (7).
Model switching between 3D cultures and 2D cultures
To generate 2D cultures from 3D organoids, which we termed 2D PDO, approximately 1 × 106 organoid fragments were suspended in AO medium and seeded on a 100Φ collagen-coated plate. Successful 2D PDO was defined as a culture that could be passaged in monolayer condition for 1 month. An additional attempt was made for each 3D PDO that failed to generate 2D cultures. Cells were replenished with fresh AO medium twice a week. To minimize clonal selection in 2D cultures, 2D PDOs generated within 1 month were used for cell viability assays and immunoblot analysis. To generate 3D cultures from 2D PDOs, which we termed 2D-3D PDO, 2D cells were dissociated, resuspended in Matrigel, seeded at 1 × 105 cells per well in 24-well plates, and cultured in a similar manner to 3D PDOs.
Immunoblots
Organoids were suspended in Matrigel, plated in 24-well plates, and overlaid with AdDF+++ medium. After overnight incubation, medium was replaced with AdDF+++ medium containing a drug and incubated for the indicated times. Organoids were harvested using Cell Recovery Solution (Corning) and washed three times with ice-cold PBS according to the manufacturer's instructions. pEGFR (#2234), EGFR (#2232), pAKT (#4060), AKT (#4691), pERK (#4370), ERK (#4696), pMEK1/2 (#9154), MEK1/2 (#4694), pS6 (#4858), S6 (#2217), pERBB2 (#2243), ERBB2 (#2165), Src (#2108), pSTAT3 (#9145), STAT3 (#9139), pRET (#3221), RET (#3223), pShc (#2434), Shc (#2432), and secondary antibodies (#7074 and #7076) were purchased from Cell Signaling Technology. pSrc (MAB2685) was purchased from R&D systems and Actin (MAB1501R) was from Merck Millipore.
Immunofluorescence
Cell strainers were used to obtain organoids with a size ranging from 20 to 70 μm. Organoids were suspended in Matrigel and plated on glass-bottom 24-well plates and overlaid with AdDF+++ medium containing DMSO or 100 nmol/L trametinib plus 100 nmol/L dabrafenib. After 5 days, organoids were stained with Ki-67 (Cell Signaling Technology, #9449), cleaved caspase 3 (Cell Signaling Technology, #9661), and Hoechst 33258 (Invitrogen). Organoids were imaged on a Leica TCS SP8 confocal microscope (25× objective). Secondary antibodies (711–296–152 and 715–096–151) were purchased from Jackson ImmunoResearch.
Xenograft
Animal experiments were performed in accordance with the guidelines of Institutional Animal Care and Use Committee (IACUC) and Animal Research Committee at Yonsei University College of Medicine. To generate PDO-derived xenografts, organoids were harvested from 12 wells of 24-well plates, mechanically dissociated, resuspended in 100 μL Matrigel, and subcutaneously injected into 6-week-old female BALB/c nude mice purchased from Saeronbio (14). Our preliminary success rates for generating xenografts from different PDO models were 62.5% (5/8) using nude mice and 91.7% (11/12) using NOG immunodeficient mice. Tumor-bearing mice (n = 6, randomly allocated to each group) were treated once daily with vehicle, gefitinib (25 mg/kg), afatinib (25 mg/kg), or osimertinib (25 mg/kg). Tumor samples were collected 2 hours after 30 days of treatment and subjected to immunoblot analysis. Percentage change in tumor volume was calculated as follows: (Vt −V0)/V0 × 100. Vt is the tumor volume of mouse treated with a drug for time t and V0 is the tumor volume of mouse at the beginning of the study. Tumor growth inhibition (TGI) was calculated as follows: [1 – (TVt – TV0)/(CVt − CV0)] × 100. TV is the tumor volume of mouse treated with a drug and CV is the tumor volume of mouse treated with vehicle (24).
Drugs
Drugs were purchased from SelleckChem. Most drugs were dissolved in DMSO whereas cetuximab was dissolved in phosphate buffered saline for in vitro cell viability assays.
Statistical analysis
Data are presented as the mean ± SEM unless indicated otherwise. Data were analyzed using the Student t test, the Mann–Whitney U test, or one-way ANOVA followed by the Dunnett test.
Results
Establishment and genomic characterization of PDOs from advanced lung adenocarcinoma
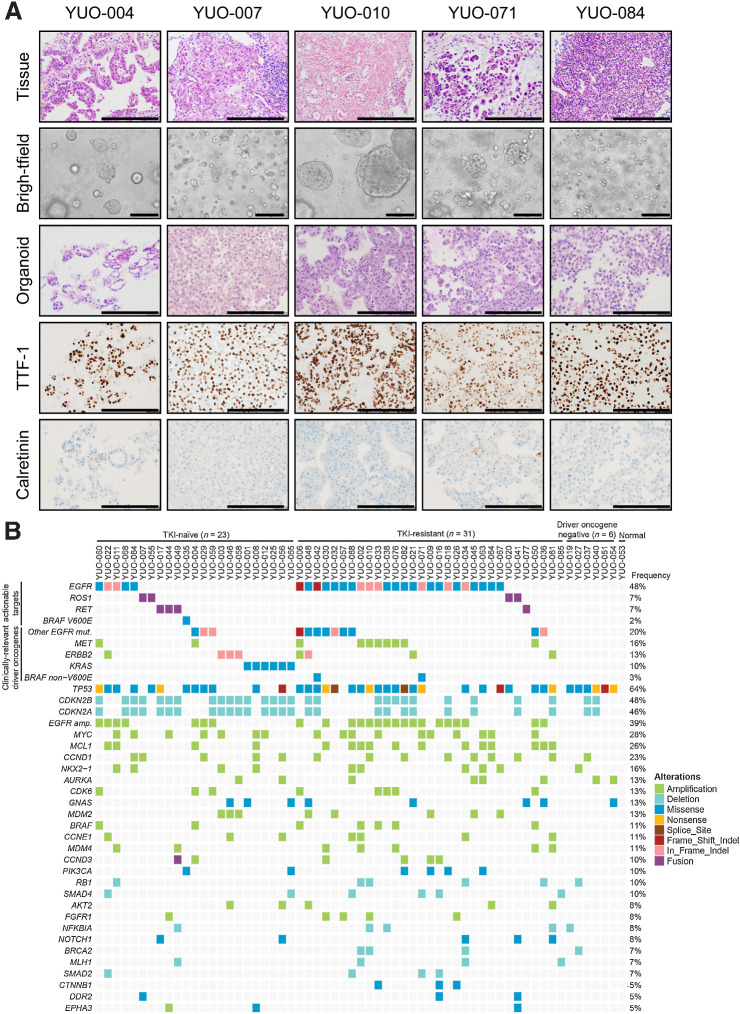
From June 2018 to March 2020, we established 83 tumor organoids (77 from malignant effusions, 3 from brain metastasis, 1 from bone metastasis, and 2 from lung primary tumor) at a success rate of 83.0% (83/100) and 1 normal-like organoid from patients with NSCLC. The organoids could be maintained for 2 to 3 months without changes in morphology (Fig. 1A; Supplementary Fig. S1). Seventeen samples failed in expansion due to lack of tumor cells in effusions, fungi contaminations, and excessive immune cells.
Figure 1.
Establishment and characterization of patient-derived organoids from advanced lung adenocarcinoma. A, Representative H&E and IHC stained images of NSCLC organoids and their parental tumor tissues. The tumor organoids were positive for TTF-1, an adenocarcinoma marker, and negative for Calrectinin, a mesothelial cell marker. NSCLC organoids recapitulated morphologic and histologic features of original tumor tissues. H&E, brightfield, and IHC images are shown. Scale bar, 100 μm. B, Genomic landscape in 61 patient-derived organoids of advanced lung adenocarcinoma. Organoids were derived from TKI-naïve NSCLC, TKI-resistant NSCLC, NSCLC without driver oncogenes, and normal tissue. Clinically relevant somatic alterations selected from the TARGET database are shown. Actionable targets and clinically relevant driver genes based on the NCCN guideline (version 8.2020) and the lung adenocarcinoma TCGA database are indicated (left). Type of alteration is indicated by color codes. The percentage of organoids harboring the indicated alterations are shown (right).
To determine the genomic landscape, whole-exome sequencing (WES) and RNA-sequencing were performed on organoids with robust cell growth (n = 61 and 55, respectively; Fig. 1B). Clinical annotations of these organoids are summarized in Supplementary Table S1. Of 60 tumor organoids, 54 harbored lung cancer driver oncogenes including EGFR mutations (n = 34), MET amplification (n = 10), ERBB2 mutations/amplification (n = 8), and KRAS mutations (n = 6; ref. 22). EGFR and MET amplification co-occurred with EGFR mutations and were enriched in tyrosine kinase inhibitor (TKI)-resistant models compared with TKI-naïve models. WES also revealed FGFR1 amplification and PIK3CA mutations, known mechanisms of TKI resistance, as well as potential candidates including MYC and MCL-1 amplification (21, 25). YUO-053, a normal-like organoid, did not harbor bona fide tumorigenic mutations.
We examined whether PDOs were able to recapitulate the genetic alterations of corresponding tumors. Of 41 cases where genetic tests were performed, driver mutation status of organoids (38/41; 92.7%) was largely concordant to that of corresponding tumors detected by routine testing (n = 35) and/or targeted NGS (n = 9). In two out three discordant cases, organoids (YUO-048 and YUO-055) harbored additional driver mutations (HER2 exon 20 insertion and ROS1 fusion, respectively) that were not detected in matching patient tumors, possibly due to cross-cell contamination. Notably, YUO-004 harbored an EGFR L747P mutation, suggesting that the matching patient had been misdiagnosed with EGFR exon 19 deletion due to limitations of PCR genetic tests (26). YUO-020, YUO-041, and YUO-077 retained ROS1- or RET-fusion genes detected at the initial diagnosis (Supplementary Table S1; Supplementary Fig. S2A). In addition, we performed WES on 12 archival FFPE tumor specimens and compared with matching PDOs (Supplementary Figs. S2B and S2C). Somatic alterations found in the archival materials were largely preserved in PDOs (Supplementary Figs. S2B and S2C). Despite similar genetic alterations, some (3/11; 27.3%) tumors contained low numbers of single-nucleotide variants (SNV), insertion/deletion (Indel), and obscure copy-number variations (Supplementary Figs. S2C and S2D). In two archival samples, EGFR driver mutations were missed possibly due to low tumor contents, although they were detected by routine testing and organoids (YUO-006 and YUO-016; Supplementary Figs. S2D and S2E). These results show that PDOs reflect genetic characteristics of advanced lung adenocarcinoma.
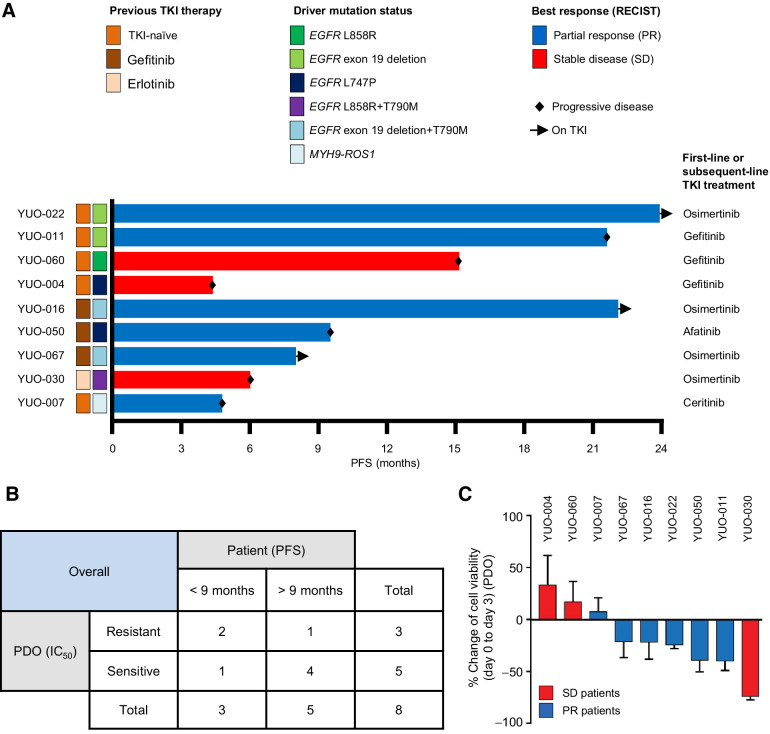
Predictive values of advanced lung adenocarcinoma PDOs
Patients with NSCLC harboring an actionable mutation generally progress to TKI treatment within 9 to 10 months (3, 4, 27). We tested if PDOs can recapitulate clinical outcomes in patients receiving clinically approved TKIs. First, we compared in vitro responses (IC50) to TKI monotherapy with a progression-free survival (PFS) in patients with EGFR-mutant-positive (n = 8) or ROS1-positive NSCLC (n = 1), where follow-up was available (Fig. 2A; Supplementary Fig. S3; Supplementary Table S2; ref. 28). Five patients achieved a PFS of >9 months and four of five matching PDOs were sensitive to TKI treatment (IC50 < 100 nmol/L). Three patients had a PFS of <9 months and two of three matching PDOs were resistant to TKI treatment (IC50 > 100 nmol/L). One patient (YUO-067) was still on osimertinib at the data cutoff. Overall, PDOs were able to predict PFS at an accuracy of 75.0% (Fig. 2B; Supplementary Table S2).
Figure 2.
Advanced lung adenocarcinoma organoids can predict patient treatment responses to a TKI monotherapy. A, Swimmers' plot showing clinical annotations of 9 patients with NSCLC who received subsequent TKI therapy after their tumor specimens were obtained to generate organoids. Each bar represents an individual patient. Subsequent TKI therapy each patient received is indicated on the right. B, Supplementary Table summarizing correlations between clinical responses (PFS) in patients and in vitro responses (mean IC50 value from three independent experiments at 3 days) in matching PDOs. C, Bar graphs showing percentage change of cell viability in PDOs after exposure to each TKI at 100 nmol/L for 3 days. Bar colors represent each patient whose best response was stable disease (red) or partial response (blue) to the TKI. Data are presented as the mean ± SEM (n = 3). PR, partial response; SD, stable disease. See also Supplementary Table S2.
Next, we assessed the ability of PDOs to reflect the RECISTs, an important indicator of tumor burden change and drug efficacy in the clinic (29). We exposed each organoid to a single dose of the relevant drugs for 3 days and measured changes in cell proliferation (Fig. 2C). Interestingly, regression of cell growth was observed in most organoids (5/6; 83.3%) established from patients who achieved a partial response, with the exception of YUO-007 that was established from a patient with a short PFS of 4.8 months on ceritinib. On the other hand, cell growth was observed in most organoids (2/3; 66.0%) established from patients who achieved stable disease. We noted that in vitro drug responses in YUO-030 were not correlated to both PFS and RECIST of the matching patient (Supplementary Table S2). The median change of cell viability was distinguishable between the two groups (−23.3% vs. 16.8%). Together, these data demonstrate that in vitro drug responses in PDOs are correlated to clinical outcomes in patients with oncogene-driven NSCLC treated with systemic therapy.
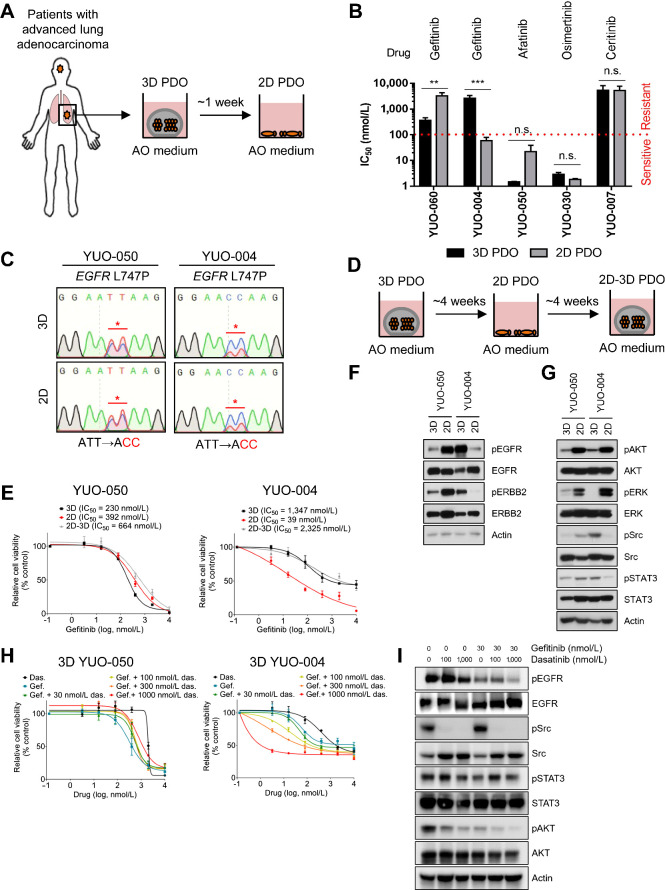
Comparison of clinical relevance in 3D and 2D culture conditions
2D cultures have been widely used for translational research (7, 11). To directly compare clinical relevance of 2D cultures and organoids, we attempted to generate 2D cultures from nine PDOs with known clinical responses to TKIs (2D PDO, designated with a “2D” prefix to the model identifier; Fig. 3A). We established 2D PDOs with a success rate of 55.5% and assessed IC50 values of the relevant drugs in these models (Supplementary Fig. S4A). Intriguingly, some 2D PDOs (2/5; 40.0%) exhibited differential TKI sensitivity to their 3D counterparts, of which one (1/2; 50.0%) failed to capture the clinical response (Fig. 3B; Supplementary Table S2). We examined whether a selection of driver mutations in the 2D culture influenced TKI sensitivity. Sanger sequencing analysis revealed that 2D YUO-004 retained the driver mutation at a similar mutation allele frequency (Fig. 3C). To confirm that culture condition is the determinant of drug sensitivity in YUO-004, we switched the 2D cultures into 3D cultures (2D–3D PDO, designated with a “2D–3D” prefix to the model identifier; Fig. 3D; ref. 30). 2D–3D YUO-004 displayed increased IC50 value of gefitinib, recapitulating the clinical response. Conversely, YUO-050 maintained an EGFR L747P mutation in 2D culture condition and was resistant to gefitinib across different culture conditions (Fig. 3C and E). In addition, 3D and 2D cultures of YUO-050 and YUO-004 were sensitive to afatinib, although the IC50 value of afatinib in 2D culture was slightly increased (YUO-050) or decreased (YUO-004) compared with the corresponding 3D culture (Supplementary Fig. S4B).
Figure 3.
Drug sensitivity to gefitinib is associated with culture condition in YUO-004. A, Procedure for generating 2D PDOs. 3D PDOs were plated on collagen-coated plates and cultured in AO medium for more than a week up to 4 weeks. B, Comparison of IC50 values to each TKI (top) between 3D and 2D PDOs (two-tailed Student t test: n.s., not significant; **, P < 0.01; ***, P < 0.005). Red line denotes sensitive (IC50 value < 100 nmol/L) or resistant (IC50 value > 100 nmol/L) response to a drug. C, DNA chromatograms showing EGFR L747P mutation in 3D culture and 2D culture of YUO-050 and YUO-004. D, Scheme for model switching. 2D PDOs that were maintained as monolayer less than 4 weeks were switched to 3D culture condition and cultured for up to 4 weeks. All models were maintained in AO medium. E, 3D, 2D, and 2D–3D cultures of YUO-050 and YUO-004 were treated with the indicated concentrations of gefitinib for 3 days. IC50 value of gefitinib is indicated for each culture condition (top). F, Representative immunoblots of indicated molecules in YUO-050 and YUO-004 at baseline. G, Representative immunoblots of indicated molecules in YUO-050 and YUO-004 at baseline. H, 3D YUO-050 and YUO-004 were treated with dasatinib alone, gefitinib alone, or gefitinib in combination with the indicated concentrations of dasatinib for 3 days. I, Representative immunoblots of indicated molecules in YUO-004 treated with the indicated concentration of gefitinib with or without dasatinib. In B, E, and H, data are presented as the mean ± SEM (n = 3).
Culture condition modulates the expression or activation of HER family kinases and impacts drug sensitivity in cancer cell lines driven by EGFR or ERBB2 (31, 32). Therefore, we examined whether HER family signaling components are associated with culture conditions in YUO-004. Compared with 3D cultures, EGFR and ERBB2 phosphorylation was decreased in 2D YUO-004 and the converse was observed for YUO-050 (Fig. 3F). Surprisingly, Src and STAT3 phosphorylation was significantly decreased in 2D YUO-004 compared with the 3D counterpart, whereas the difference was negligible between 2D and 3D culture of YUO-050 (Fig. 3G). Src cross-activates with EGFR and combined EGFR and Src inhibition has synergistic anticancer effects on EGFR-dependent cancers (33, 34). Thus, we examined the effect of Src activation in YUO-004. Although dasatinib, a highly selective Src inhibitor, had no effects on YUO-050, treatment of dasatinib sensitized YUO-004 to gefitinib (Fig. 3H and I; ref. 35). Similar results were observed for 2D–3D YUO-004, which regained high phosphorylation levels of Src and STAT3 (Supplementary Figs. S4C and S4D). These data illustrate that YUO-004 requires 3D culture-dependent Src activation to predict the clinical response.
Predictive values of PDOs in diverse clinical settings
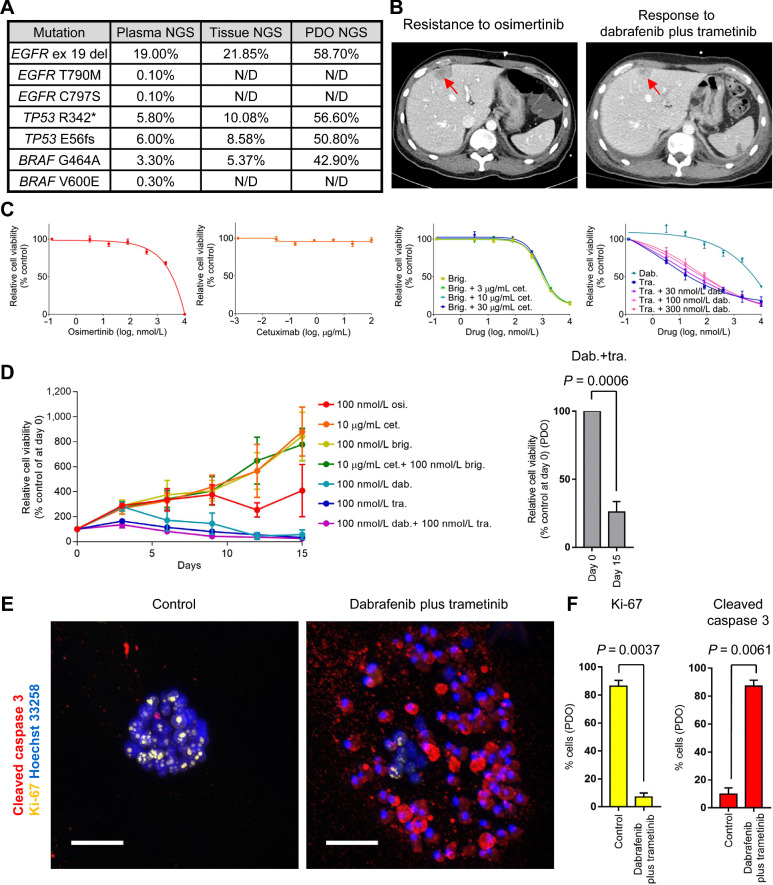
Next, we tested the ability of organoids to predict activity of novel therapeutic strategies that are under clinical investigation. Patients with EGFR-mutant NSCLC progressing to osimertinib acquire heterogeneous mechanisms of resistance including a BRAF V600E mutation (25). Effective treatments for NSCLC harboring both EGFR mutation and BRAF mutation remain to be elucidated. In our study, a patient with NSCLC harboring an EGFR exon 19 deletion and T790M mutation progressed to osimertinib (Supplementary Table S1). To select subsequent treatments, targeted NGS analysis was performed on tissue and liquid biopsies and identified BRAF G464A, BRAF V600E, and EGFR C797S mutations as potential druggable targets (Fig. 4A). On the basis of these results, the patient was initiated on dabrafenib/trametinib combination therapy, an approved treatment for BRAF V600E-mutant NSCLC (5). Compared with a CT scan prior to the combination therapy, a follow-up CT scan obtained after 1 week of the treatment demonstrated marked reduction in tumor size (Fig. 4B). Unfortunately, the patient suddenly expired because of a cerebrovascular accident unrelated to disease progression after 2.5 weeks on the treatment. Simultaneously, we performed WES and cell viability assays on YUO-071 generated from malignant effusion resistant to osimertinib. YUO-071 retained the EGFR activating mutation and BRAF G464A mutation similar to the tissue NGS (Fig. 4A). Interestingly, YUO-071 responded to trametinib with or without dabrafenib but was resistant to osimertinib, cetuximab, brigatinib, and cetuximab/brigatinib combination, which had demonstrated preclinical efficacy against EGFR exon 19 deletion/T790M/C797S mutations (Fig. 4C; ref. 36). Long-term exposure to trametinib and dabrafenib achieved 74% organoid growth inhibition, whereas osimertinib, cetuximab, brigatinib, and cetuximab plus brigatinib resulted in organoid growth (Fig. 4D). In addition, the trametinib/dabrafenib combination drastically decreased Ki-67 and increased cleaved caspase 3 (Fig. 4E and F). Considering the presence of multiple BRAF clones in the patient tumors, it remains to be determined which BRAF mutation (V600E and/or G464A) is responding to the combination therapy in this patient (Fig. 4A). Together, these findings demonstrate that PDOs capture the clinical response to the dabrafenib/trametinib combination therapy in a patient with NSCLC harboring EGFR plus BRAF mutations.
Figure 4.
PDOs recapitulate a clinical response to dabrafenib/trametinib combination therapy against EGFR exon 19 deletion plus BRAF G464A mutation. A, Summary of NGS analyses in liquid and tissue biopsies and YUO-071. B, CT scans showing tumor (red arrows) at disease progression to osimertinib (left) and after dabrafenib plus trametinib combination therapy (right) in a patient from which YUO-071 was generated. C, YUO-071 was treated with the indicated concentrations of osimertinib (far left), cetuximab (left), brigatinib with or without cetuximab at the indicated concentrations (right), and dabrafenib alone, trametinib alone, or trametinib plus dabrafenib at the indicated concentrations (far right) for 5 days. D, YUO-071 was exposed to osimertinib, dabrafenib, trametinib, dabrafenib plus trametinib, cetuximab, brigatinib, cetuximab plus brigatinib at the indicated concentrations for 15 days (left). Relative cell viability of YUO-071 before (day 0) and after the long-term exposure (day 15) to dabrafenib plus trametinib is shown on the right panel. E, Representative immunofluorescence images of indicated molecules in YUO-071 treated with control or 100 nmol/L dabrafenib in combination with 100 nmol/L trametinib for 5 days. Scale bar, 100 μmol/L. F, Bar graphs showing quantification of Ki-67–positive cells (left) and cleaved caspase 3–positive cells (right) in each group from E. In C, D, and F, data are presented as the mean ± SEM (n = 3; two-tailed Student t test). N/D, none detected.
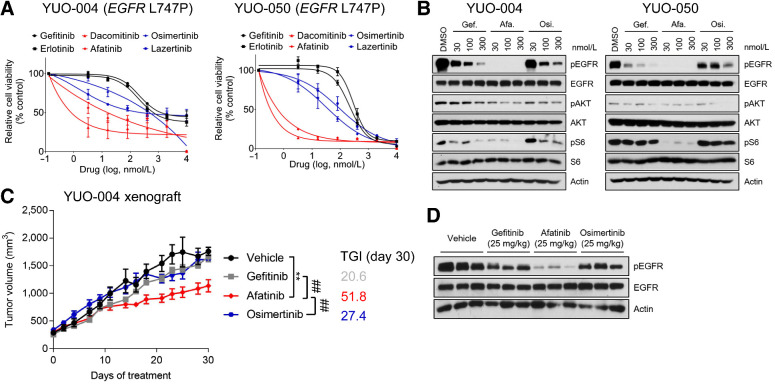
EGFR activating mutations respond to first-generation EGFR-TKIs, whereas rare EGFR mutations exhibit differential sensitivity to therapies (4, 6, 7). Clinical and preclinical data regarding an EGFR L747P mutation are sparse (37). To identify effective therapies against the EGFR L747P mutation, we screened clinically available EGFR-TKIs in YUO-004 and YUO-050 established from EGFR L747P-mutant NSCLCs resistant to gefitinib (Supplementary Tables S1 and S2). YUO-004 and YUO-050 were sensitive to second-generation EGFR-TKIs (dacomitinib and afatinib) but resistant to first-generation (gefitinib and erlotinib) and third-generation EGFR-TKIs (osimertinib and lazertinib; Fig. 5A). Compared with gefitinib and osimertinib, afatinib potently inhibited EGFR downstream signaling components (Fig. 5B). Afatinib induced modest growth delay with 51.8% TGI in YUO-004 xenografts accompanied by marked inhibition of EGFR phosphorylation, whereas gefitinib and osimertinib had no anticancer effects (TGI = 20.6% and 27.4%, respectively; Fig. 5C and D). In alignment with these findings, the matching patient for YUO-050 responded to afatinib and achieved PFS of 9.5 months (Supplementary Table S2). Together, these results demonstrate that afatinib is more potent than first-generation and third-generation EGFR-TKIs against the EGFR L747P mutation.
Figure 5.
PDOs predict clinical activity of afatinib against EGFR L747P mutation. A, YUO-004 and YUO-050 were treated with the indicated concentrations of gefitinib, erlotinib, dacomitinib, afatinib, osimertinib, and lazertinib for 3 days. First-generation EGFR-TKIs are colored in dark, second-generation EGFR-TKIs are in red, and third-generation EGFR-TKIs are in blue. Data are presented as the mean ± SEM (n = 3). B, Representative immunoblots of indicated molecules in YUO-004 and YUO-050 treated with the indicated concentrations of gefitinib, afatinib, and osimertinib for 6 hours. C, Tumor growth curve of YUO-004 xenografts treated with indicated drugs at 25 mg/kg once daily (n = 6 per group; one-way ANOVA with Dunnett's posttest: n.s., not significant; **, P < 0.005 vs. vehicle; ##, P < 0.01 vs. afatinib). D, Immunoblots of indicated molecules in tumor samples obtained from YUO-004 xenografts treated with vehicle and 25 mg/kg gefitinib, afatinib, and osimertinib for 30 days.
PDOs can identify effective anti-cancer therapies for novel molecular targets
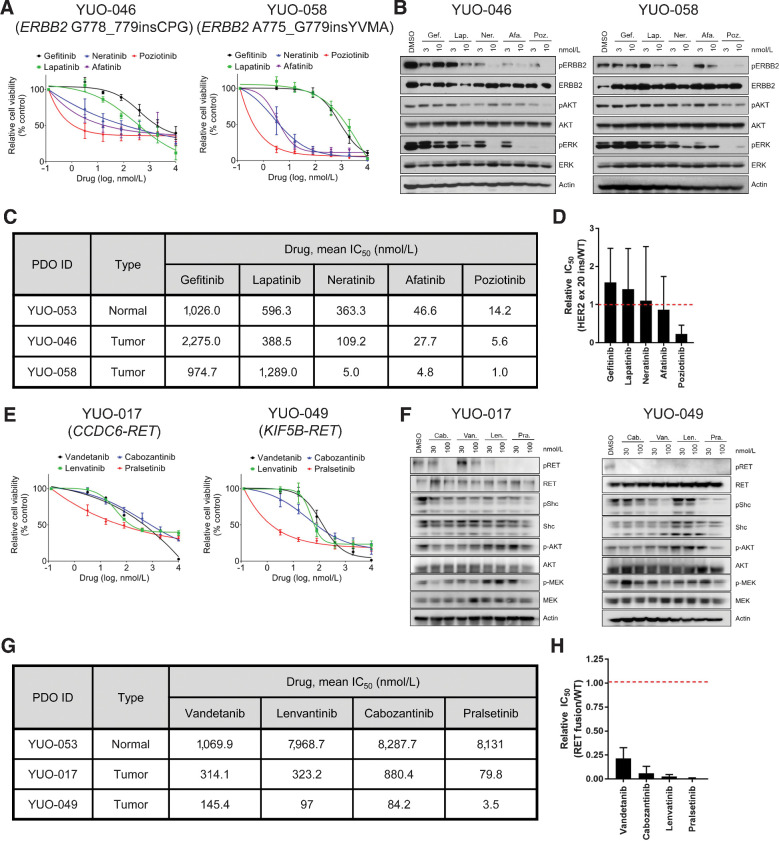
ERBB2 mutations and RET fusions are emerging targets for targeted therapies, which are found in approximately 2% of patients with NSCLC (1). We utilized organoids to investigate effective targeted therapies for ERBB2-mutant and RET-rearranged NSCLC. Poziotinib, an experimental drug for treatment of ERBB2-mutant NSCLC, was the most potent TKI against ERBB2 G778_779insCPG and A775_G779insYVMA insertions (Fig. 6A; ref. 38). Compared with other ERBB2 inhibitors, poziotinib potently suppressed the ERBB2-ERK-AKT signaling pathway (Fig. 6B). Cells expressing wild-type target proteins can be used to determine selectivity of a targeted therapy for mutant target proteins (14, 38). Using the normal-like organoid (YUO-053), which is devoid of tumorigenic mutations (Fig. 1B), we found that poziotinib was more mutant-selective than other drugs (Fig. 6C and D). In addition, pralsetinib, a highly potent inhibitor for RET-mutant and RET-rearranged tumors, was more effective than vandetanib, lenvatinib, and cabozantinib against CCDC6-RET fusion and KIF5B–RET fusion in cell viability assays and immunoblot analysis (Fig. 6E and F; ref. 39). Pralsetinib was more mutant-selective than other RET-targeted therapies (Fig. 6G and H). These findings underline preclinical efficacy of poziotinib and pralsetinib against NSCLCs harboring ERBB2 exon 20 insertions and RET fusions, respectively.
Figure 6.
PDOs can identify effective therapies for advanced lung adenocarcinoma harboring ERBB2 exon 20 insertions or RET rearrangements. A, YUO-046 and YUO-058 harboring ERBB2 exon 20 insertions were treated with the indicated concentrations of gefitinib, lapatinib, neratinib, afatinib, and poziotinib for 5 days. B, Representative immunoblots of indicated molecules in YUO-046 and YUO-058 treated with the indicated concentrations of gefitinib, lapatinib, neratinib, afatinib, and poziotinib for 6 hours. C, IC50 values of gefitinib, lapatinib, neratinib, afatinib, and poziotinib in YUO-053, a normal-like organoid, and tumor organoids harboring ERBB2 exon 20 insertions. D, Bar graphs showing mean relative IC50 values of the ERBB2 inhibitors in ERBB2-mutant organoids to the normal organoid. E, YUO-017 and YUO-049 harboring RET fusions were treated with the indicated concentrations of vandetanib, lenvatinib, cabozantinib, and pralsetinib for 5 days. F, Representative immunoblots of indicated molecules in YUO-017 and YUO-049 treated with the indicated concentrations of cabozantinib, pralsetinib, vandetanib, and lenvatinib for 2 hours. G, IC50 values of vandetanib, lenvatinib, cabozantinib, and pralsetinib in a normal-like organoid and tumor organoids harboring RET rearrangements. H, Bar graphs showing mean relative IC50 values of the RET inhibitors in RET fusion positive organoids to the normal organoid. In A and E, data are presented as the mean ± SEM (n = 3). In C and G, mean IC50 values were calculated from three biological replicates (three technical replicates per independent experiment) using GraphPad Prism. In D and H, data are presented as the mean ± SD (n = 2).
Discussion
Tumor organoids reflect the genetic alterations of tumors they were derived from and can be used to investigate drug–gene interactions (14–16, 28, 40). Importantly, PDOs of colorectal cancer, head and neck cancer, gastrointestinal cancer, and rectal cancer have been shown to predict clinical responses to not only chemoradiotherapies but also targeted therapies, bringing new insight into the clinical utility of organoids (28, 29, 40, 41). Compared with previous studies on lung cancer PDOs (14–16), we correlated in vitro drug responses in PDOs to clinical responses in matching patients and assessed preclinical efficacy of targeted therapies under clinical development, demonstrating clinical relevance of NSCLC PDOs.
It is generally perceived that 3D cultures better reflect in vivo physiology than 2D cultures. In the context of EGFR- or ERBB2-driven cancer, previous studies used conventional cell lines to investigate the physiological differences between 2D and 3D cultures (31, 32). For example, Breslin and colleagues showed that 3D cultures of ERBB2-overexpressing cancer cells, when compared with their 2D counterparts, display increased expression and activation of HER family kinases and resistance to HER targeted drugs (31). However, in vivo or clinical relevance of these observations were not demonstrated. In our study, we compared predictive values between 2D and 3D cultures of clinically annotated patient-derived models and showed potential advantages of 3D organoids in translational research. Particularly, we show that 3D organoids may capture unique information such as Src activation, which is not represented by either genetic tests or 2D cultures. We noted that some 3D PDOs (4/9; 44.4%) cannot be cultured in the monolayer condition, indicating that some NSCLCs may grow only as suspension cells or require extracellular matrix for optimal growth (42, 43).
In this study, we utilized organoids to assess the clinical activity of novel therapeutic strategies. We demonstrate that dabrafenib/trametinib combination therapy elicits in vitro and clinical responses in a NSCLC harboring an EGFR exon 19 deletion and a BRAF G464A mutation. Accordingly, Ho and colleagues have shown that cancer cell lines harboring both EGFR activating mutation and BRAF V600E mutation is dependent on BRAF–MEK pathway and responds to a BRAF inhibitor monotherapy (44). BRAF G464A mutation belongs to a non-V600E BRAF mutation which may exhibit different clinical and molecular characteristics to the BRAF V600E mutation (8, 9). Our findings are in keeping with several preclinical studies and case reports, which have demonstrated the efficacy of trametinib with or without dabrafenib against non-V600E BRAF mutation (7, 9, 45). Moreover, we report preclinical and clinical efficacy of afatinib against the rare EGFR L747P mutation (37). Our findings and few case reports suggest that the EGFR L747P mutation is resistant to gefitinib and osimertinib but sensitive to afatinib (37, 46). These results demonstrate that organoids in addition to the molecular profiling can be a powerful diagnostic tool for precision medicine in diverse clinical settings.
Finally, we identify poziotinib as the most potent agent, among ERBB2 targeted therapies tested, against ERBB2 exon 20 insertions which lack clinically approved inhibitors. Poziotinib has demonstrated significant preclinical and clinical efficacy compared to erlotinib, lapatinib, neratinib, and afatinib against the ERBB2 exon 20 insertions (38, 47, 48). In addition, we show that pralsetinib is more effective than multi-kinase inhibitors vandetanib, cabozantinib, and lenvatinib. The multi-kinase inhibitors had limited clinical efficacy in RET-rearranged tumors, whereas pralsetinib has demonstrated promising results in an ongoing phase I clinical trial (NCT03037385; refs. 39, 49). These data and our recent work on amivantamab, an EGFR-MET bispecific antibody for treatment of EGFR exon 20 insertions, underline the feasibility of PDO-based preclinical studies (50).
This study had several limitations. It was a retrospective study based on extensive organoid biobanking and NGS. To expand the clinical utility of organoids, future studies need to be prospective and determine the time frame for organoids to be informative for clinical decision making. We also acknowledge that the predictive value of organoids needs to be validated in a large cohort.
In summary, we demonstrate that advanced lung adenocarcinoma organoids can recapitulate clinical responses to targeted therapies and facilitate development of novel therapeutic strategies. The clinical relevance of organoids will contribute to implementation of precision medicine.
Authors' Disclosures
S.-Y. Kim reports grants and non-financial support from Interpark Bio Convergence Corp. and Ministry of Science and ICT during the conduct of the study. B.C. Cho reports grants from Novartis, Bayer, AstraZeneca, MOGAM Institute, Dong-A ST, Champions Oncology, Janssen, Yuhan, Ono, Dizal Pharma, MSD, AbbVie, Medpacto, GIInnovation, Eli Lilly, Blueprint Medicines, and Interpark Bio Convergence Corp.; personal fees from Novartis, AstraZeneca, Boehringer Ingelheim, Roche, BMS, Ono, Yuhan, Pfizer, Eli Lilly, Janssen, Takeda, MSD, Medpacto, Blueprint Medicines, TheraCanVac Inc., Gencurix Inc, Bridgebio therapeutics, Kanaph Therapeutics, Cyrus Therapeutics, Interpark Bio Convergence Corp., Guardant Health, and Oscotec Inc.; and other support from Interpark Bio Convergence Corp., Champions Oncology, and DAAN Biotherapeutics outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
We thank the patients for their contributions to this study. Patient blood and FFPE samples were provided by the Biobank, Severance Hospital, Seoul, Korea. This work was supported by Interpark Bio Convergence Corp., Seoul, Korea, and Science Research Program through the NRF funded by the Ministry of Science and ICT (2016R1A2B3016282). We also thank Dr. Koo (IMBA, Austria) for his advice on establishing organoids. The selection of clinically relevant driver genes are in whole or in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
S.-Y. Kim: Conceptualization, resources, data curation, software, formal analysis, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration. S.-M. Kim: Resources, formal analysis, validation, investigation, visualization, methodology, writing–original draft. S. Lim: Resources, validation, investigation. J.Y. Lee: Resources, validation. S.-J. Choi: Resources, data curation, software, formal analysis, visualization. S.-D. Yang: Data curation, software, formal analysis, visualization. M.R. Yun: Formal analysis. C.G. Kim: Formal analysis, investigation, visualization. S.R. Gu: Visualization. C. Park: Resources, data curation. A.-Y. Park: Resources. S.M. Lim: Data curation. S.G. Heo: Resources, data curation, software, formal analysis. H. Kim: Conceptualization, data curation, formal analysis, supervision, investigation, methodology, writing–review and editing. B.C. Cho: Conceptualization, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing.
References
- 1. Yang C-Y, Yang JC-H, Yang P-C. Precision management of advanced non–small cell lung cancer. Annu Rev Med 2020;71:117–36. [DOI] [PubMed] [Google Scholar]
- 2. National Comprehensive Cancer Network. Non-small cell lung cancer (version 8. 2020). Available from: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf.
- 3. Lim SM, Kim HR, Lee J-S, Lee KH, Lee Y-G, Min YJ, et al. Open-label, multicenter, phase II study of ceritinib in patients with non–small-cell lung cancer harboring ROS1 rearrangement. J Clin Oncol 2017;35:2613–8. [DOI] [PubMed] [Google Scholar]
- 4. Mok TS, Wu Y-L, Thongprasert S, Yang C-H, Chu D-T, Saijo N, et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–57. [DOI] [PubMed] [Google Scholar]
- 5. Planchard D, Besse B, Groen HJ, Souquet P-J, Quoix E, Baik CS, et al. Dabrafenib plus trametinib in patients with previously treated BRAFV600E-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17:984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Galli G, Corrao G, Imbimbo M, Proto C, Signorelli D, Ganzinelli M, et al. Uncommon mutations in epidermal growth factor receptor and response to first and second generation tyrosine kinase inhibitors: a case series and literature review. Lung Cancer 2018;115:135–42. [DOI] [PubMed] [Google Scholar]
- 7. Kim S-Y, Lee JY, Kim DH, Joo H-S, Yun MR, Jung D, et al. Patient-derived cells to guide targeted therapy for advanced lung adenocarcinoma. Sci Rep 2019;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marchetti A, Felicioni L, Malatesta S, Grazia Sciarrotta M, Guetti L, Chella A, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574–9. [DOI] [PubMed] [Google Scholar]
- 9. Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clevers H. Modeling development and disease with organoids. Cell 2016;165:1586–97. [DOI] [PubMed] [Google Scholar]
- 11. Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014;346:1480–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res 2013;73:5315–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dijkstra KK, Monkhorst K, Schipper LJ, Hartemink KJ, Smit EF, Kaing S, et al. Challenges in establishing pure lung cancer organoids limit their utility for personalized medicine. Cell Rep 2020;31:107588. [DOI] [PubMed] [Google Scholar]
- 14. Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ, Chun SM, et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun 2019;10:3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, Heo I, Böttinger L, Klay D, et al. Long-term expanding human airway organoids for disease modeling. EMBO J 2019;38:e100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi R, Radulovich N, Ng C, Liu N, Notsuda H, Cabanero M, et al. Organoid cultures as preclinical models of non-small cell lung cancer. Clin Cancer Res 2020;26:1162–74. [DOI] [PubMed] [Google Scholar]
- 17. Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tso K-Y, Lee SD, Lo K-W, Yip KY. Are special read alignment strategies necessary and cost-effective when handling sequencing reads from patient-derived tumor xenografts? BMC Genomics 2014;15:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: cancer variant annotation tool. Hum Mutat 2015;36:E2423-E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med 2014;20:682–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 2018;556:457–62. [DOI] [PubMed] [Google Scholar]
- 23. Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yun MR, Kim DH, Kim S-Y, Joo H-S, Lee YW, Choi HM, et al. Repotrectinib exhibits potent antitumor activity in treatment-naïve and solvent-front–mutant ROS1-rearranged non–small cell lung cancer. Clin Cancer Res 2020;26:3287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walsh K, Wallace W, Butler R, Mackean M, Harrison D, Stirling D, et al. A cautionary lesson on the use of targeted methods for EGFR mutation analysis: a case report. J Clin Pathol 2014;67:734–5. [DOI] [PubMed] [Google Scholar]
- 27. Jänne PA, Yang JC-H, Kim D-W, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor–resistant non–small-cell lung cancer. N Engl J Med 2015;372:1689–99. [DOI] [PubMed] [Google Scholar]
- 28. Tiriac H, Belleau P, Engle DD, Plenker D, Deschenes A, Somerville TDD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov 2018;8:1112–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van Werkhoven E, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med 2019;11:eaay2574. [DOI] [PubMed] [Google Scholar]
- 30. Fujita-Sato S, Galeas J, Truitt M, Pitt C, Urisman A, Bandyopadhyay S, et al. Enhanced MET translation and signaling sustains K-Ras–driven proliferation under anchorage-independent growth conditions. Cancer Res 2015;75:2851–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Breslin S, O'Driscoll L. The relevance of using 3D cell cultures, in addition to 2D monolayer cultures, when evaluating breast cancer drug sensitivity and resistance. Oncotarget 2016;7:45745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pickl M, Ries C. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene 2009;28:461–8. [DOI] [PubMed] [Google Scholar]
- 33. Moro L, Dolce L, Cabodi S, Bergatto E, Erba EB, Smeriglio M, et al. Integrin-induced epidermal growth factor (EGF) receptor activation requires c-Src and p130Cas and leads to phosphorylation of specific EGF receptor tyrosines. J Biol Chem 2002;277:9405–14. [DOI] [PubMed] [Google Scholar]
- 34. Ochi N, Takigawa N, Harada D, Yasugi M, Ichihara E, Hotta K, et al. Src mediates ERK reactivation in gefitinib resistance in non-small cell lung cancer. Exp Cell Res 2014;322:168–77. [DOI] [PubMed] [Google Scholar]
- 35. Song L, Morris M, Bagui T, Lee FY, Jove R, Haura EB. Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res 2006;66:5542–8. [DOI] [PubMed] [Google Scholar]
- 36. Uchibori K, Inase N, Araki M, Kamada M, Sato S, Okuno Y, et al. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nat Commun 2017;8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liang S-K, Ko J-C, Yang JC-H, Shih J-Y. Afatinib is effective in the treatment of lung adenocarcinoma with uncommon EGFR p. L747P and p. L747S mutations. Lung Cancer 2019;133:103–9. [DOI] [PubMed] [Google Scholar]
- 38. Robichaux JP, Elamin YY, Vijayan R, Nilsson MB, Hu L, He J, et al. Pan-cancer landscape and analysis of ERBB2 mutations identifies poziotinib as a clinically active inhibitor and enhancer of T-DM1 activity. Cancer Cell 2019;36:444–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Subbiah V, Gainor JF, Rahal R, Brubaker JD, Kim JL, Maynard M, et al. Precision targeted therapy with BLU-667 for RET-driven cancers. Cancer Discov 2018;8:836–49. [DOI] [PubMed] [Google Scholar]
- 40. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018;359:920–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell 2020;26:17–26. [DOI] [PubMed] [Google Scholar]
- 42. Fridman R, Benton G, Aranoutova I, Kleinman HK, Bonfil RD. Increased initiation and growth of tumor cell lines, cancer stem cells and biopsy material in mice using basement membrane matrix protein (Cultrex or Matrigel) co-injection. Nat Protoc 2012;7:1138. [DOI] [PubMed] [Google Scholar]
- 43. Zheng C, Sun YH, Ye XL, Chen HQ, Ji HB. Establishment and characterization of primary lung cancer cell lines from Chinese population. Acta Pharmacol Sin 2011;32:385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ho C-C, Liao W-Y, Lin C-A, Shih J-Y, Yu C-J, Yang JC-H. Acquired BRAF V600E mutation as resistant mechanism after treatment with osimertinib. J Thorac Oncol 2017;12:567–72. [DOI] [PubMed] [Google Scholar]
- 45. Marconcini R, Galli L, Antonuzzo A, Bursi S, Roncella C, Fontanini G, et al. Metastatic BRAF K601E-mutated melanoma reaches complete response to MEK inhibitor trametinib administered for over 36 months. Exp Hematol Oncol 2017;6:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang J, Wang Y, Zhai Y, Wang J. Non-small cell lung cancer harboring a rare EGFR L747P mutation showing intrinsic resistance to both gefitinib and osimertinib (AZD9291): a case report. Thorac Cancer 2018;9:745–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, et al. HER kinase inhibition in patients with HER2-and HER3-mutant cancers. Nature 2018;554:189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mazieres J, Peters S, Lepage B, Cortot AB, Barlesi F, Beau-Faller M, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013;31:1997–2003. [DOI] [PubMed] [Google Scholar]
- 49. Drilon A, Rekhtman N, Arcila M, Wang L, Ni A, Albano M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yun J, Lee S-H, Kim S-Y, Jeong S-Y, Kim J-H, Pyo K-H, et al. Antitumor activity of amivantamab (JNJ-61186372), an EGFR–MET bispecific antibody, in diverse models of EGFR exon 20 insertion–driven NSCLC. Cancer Discov 2020;10:1194–209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.