In a phase I dose escalation/expansion study, use of the HER3-DXd antibody drug conjugate showed clinical activity in EGFR TKI-resistant, EGFR-mutant non–small cell lung cancer, presenting a future option for drug-resistant cancers.
Abstract
Receptor tyrosine-protein kinase ERBB3 (HER3) is expressed in most EGFR-mutated lung cancers but is not a known mechanism of resistance to EGFR inhibitors. HER3-DXd is an antibody–drug conjugate consisting of a HER3 antibody attached to a topoisomerase I inhibitor payload via a tetrapeptide-based cleavable linker. This phase I, dose escalation/expansion study included patients with locally advanced or metastatic EGFR-mutated non–small cell lung cancer (NSCLC) with prior EGFR tyrosine kinase inhibitor (TKI) therapy. Among 57 patients receiving HER3-DXd 5.6 mg/kg intravenously once every 3 weeks, the confirmed objective response rate by blinded independent central review (Response Evaluation Criteria in Solid Tumors v1.1) was 39% [95% confidence interval (CI), 26.0–52.4], and median progression-free survival was 8.2 (95% CI, 4.4–8.3) months. Responses were observed in patients with known and unknown EGFR TKI resistance mechanisms. Clinical activity was observed across a broad range of HER3 membrane expression. The most common grade ≥3 treatment-emergent adverse events were hematologic toxicities. HER3-DXd has clinical activity in EGFR TKI–resistant cancers independent of resistance mechanisms, providing an approach to treat a broad range of drug-resistant cancers.
Significance:
In metastatic EGFR-mutated NSCLC, after disease progression on EGFR TKI therapy, treatment approaches include genotype-directed therapy targeting a known resistance mechanism or chemotherapy. HER3-DXd demonstrated clinical activity spanning known and unknown EGFR TKI resistance mechanisms. HER3-DXd could present a future treatment option agnostic to the EGFR TKI resistance mechanism.
See related commentary by Lim et al., p. 16 .
This article is highlighted in the In This Issue feature, p. 1
Introduction
For a subset of patients with locally advanced or metastatic non–small cell lung cancer (NSCLC), initial and subsequent therapies are guided by the identification of oncogenic driver mutations. Approximately 10% to 15% of patients with NSCLC in the United States and Europe and 30% to 40% of those in Asia have an EGFR-activating mutation (termed EGFR-mutated; ref. 1). In these patients, EGFR-targeted tyrosine kinase inhibitors (TKI) result in high response rates [objective response rate (ORR), 76%–80%; ref. 2] and can provide extended disease control (3). Third-generation EGFR TKIs, such as osimertinib, overcome some resistance mechanisms and have been shown to confer improved overall survival (OS) compared with first-generation EGFR TKIs (median OS, 38.6 vs. 31.8 months; ref. 4); however, relapse is typical with the development of resistance to EGFR TKI treatment (5–7). Mechanisms associated with EGFR TKI resistance are diverse and include EGFR, MET, PIK3CA, and BRAF genomic alterations, among other resistance mechanisms (7, 8); alterations commonly associated with resistance to osimertinib are the EGFR C797S mutation and amplifications of HER2 or MET (8–10). However, a significant portion of patients who develop clinical EGFR TKI resistance have tumor genomic alterations associated with EGFR TKI resistance that are yet undefined (11).
After disease progression on EGFR TKI therapy (which may include sequential EGFR TKIs), patients are commonly treated with chemotherapy or investigational genotype-directed therapies targeting an identified resistance mechanism (e.g., MET amplification), if known. Several studies have reported successful treatment (response rates, 30%–47%) of patients with MET amplification as an EGFR TKI resistance mechanism with a combination of an EGFR TKI and MET TKI (12, 13). The recent CHRYSALIS study combined the EGFR–MET bispecific antibody amivantamab with the third-generation EGFR TKI lazertinib in patients with EGFR-mutated NSCLC: the ORR was 40% in patients with disease progression on platinum-based chemotherapy (14) and 36% in chemotherapy-naïve patients with disease progression on osimertinib [median progression-free survival (PFS) was 4.9 months; ref. 15]. For platinum-based chemotherapy in EGFR-mutated NSCLC following disease progression on first-generation EGFR TKI, response rates of 25% to 44% have been reported, with median PFS of 2.7 to 6.4 months and a median OS of 8.1 to 19.2 months (16). Salvage therapies after EGFR TKI and platinum-based chemotherapy have limited efficacy (median PFS, 2.8–3.2 months; median OS, 7.5–10.6 months; ref. 17). Given that resistance mechanisms to EGFR TKIs are diverse and that the efficacy of chemotherapy is limited, there is a need to develop novel treatment approaches for previously treated EGFR-mutated NSCLC that provide salvage therapy across a broad spectrum of resistance-associated genomic alterations.
Receptor tyrosine-protein kinase ERBB3 (HER3) is expressed across a variety of malignant solid tumors and has been found in 83% of primary NSCLC tumors (18, 19). HER3 overexpression is associated with metastatic progression and decreased relapse-free survival in patients with NSCLC (19). EGFR-mutated NSCLC is associated with higher expression of HER3 compared with EGFR wild-type NSCLC (20). Although increases in HER3 expression have been observed in cell lines that have developed acquired resistance to an EGFR TKI in vitro (21), genomic alterations in HER3 are not known to constitute a mechanism of resistance to EGFR TKI in EGFR-mutated NSCLC. Given the broad overexpression of HER3 in NSCLC, this membrane protein provides an attractive molecular target for the treatment of advanced EGFR-mutated NSCLC.
Antibody–drug conjugates (ADC) are a class of cancer therapy in which antibodies specific for tumor-associated antigens are used to selectively target the delivery of cytotoxic drugs (22). Patritumab deruxtecan (U3-1402, HER3-DXd) is a novel, investigational, HER3-directed ADC composed of a human immunoglobulin G1 mAb to HER3 (patritumab) covalently linked to a topoisomerase I inhibitor payload (MAAA-1181a, an exatecan derivative) via a tetrapeptide-based cleavable linker with a drug-to-antibody ratio of approximately 8 (23–26). After trafficking of the ADC to the lysosome, the linker is cleaved by lysosomal enzymes that are upregulated in tumor cells, allowing the cytotoxic payload to be released and to enter the nucleus, leading to cell death (21, 23, 26, 27). The payload is cell membrane–permeable, which enables a bystander antitumor effect resulting in elimination of both target and surrounding tumor cells (25, 28). An ADC using the same linker-payload technology but directed against HER2 [trastuzumab deruxtecan (T-DXd)] was recently approved for the treatment of HER2+ metastatic breast and gastric cancers (29, 30).
The antitumor activity of HER3-DXd was shown in multiple solid tumor murine xenograft models, including EGFR-mutated NSCLC patient-derived xenograft models with known resistance to EGFR TKI therapy (31). On the basis of these nonclinical data, a phase I, dose escalation/expansion study was initiated, and patients with locally advanced or metastatic NSCLC were enrolled (study U31402-A-U102; clinicaltrials.gov NCT03260491; EudraCT 2017-000543-41; JapicCTI 194868). Here, we report the safety, clinical activity, biomarker analyses, and pharmacokinetics of HER3-DXd in patients with advanced or metastatic EGFR-mutated NSCLC treated in this study.
Results
U31402-A-U102 is a phase I, global, multicenter, dose escalation and dose expansion study evaluating HER3-DXd in patients with locally advanced or metastatic NSCLC, including patients with NSCLC harboring an EGFR-activating mutation and prior EGFR TKI therapy. The dose escalation part enrolled 36 patients with EGFR-mutated NSCLC (adenocarcinoma) who had acquired resistance to at least one EGFR TKI; the patients were treated in one of four dose groups [intravenously (i.v.), once every 3 weeks]: 3.2 mg/kg (n = 4), 4.8 mg/kg (n = 15), 5.6 mg/kg (n = 12), and 6.4 mg/kg (n = 5; Supplementary Fig. S1). In the dose escalation part, the recommended dose for expansion (RDE) of HER3-DXd was determined to be 5.6 mg/kg, i.v. once every 3 weeks. Cohort 1 of the dose expansion part enrolled 45 patients with EGFR-mutated NSCLC (adenocarcinoma), prior EGFR TKI treatment, and prior platinum-based chemotherapy who all received a dose of 5.6 mg/kg, i.v. once every 3 weeks. Safety assessments include data for all patients in the dose escalation and dose expansion parts (N = 81); efficacy measures are presented for the pooled population of patients who received HER3-DXd 5.6 mg/kg (n = 57; Supplementary Fig. S1). Safety and efficacy data for each dose group in the dose escalation part are presented in Supplementary Tables S1 to S3. At the data cutoff date (September 24, 2020), 22% of all patients (18/81) treated were continuing study treatment. Data for dose expansion cohort 2 (squamous or nonsquamous NSCLC without EGFR-activating mutations) and cohort 3 (squamous or nonsquamous EGFR-mutated NSCLC with any histology other than combined small cell and non–small cell; uptitration study) will be presented in the future.
Patient Demographics and Baseline Characteristics
For dose escalation and dose expansion cohort 1, reported here, study eligibility criteria required that all patients had advanced or metastatic EGFR-mutated NSCLC with adenocarcinoma histology. Fifty-three percent of enrolled patients (43/81) had a history of stable central nervous system (CNS) metastases, and 19% of patients (15/81) had received prior treatment for CNS metastases (Table 1). Patients had a median of 4 (range, 1–9) prior lines of systemic therapy for locally advanced or metastatic NSCLC (Table 1). All patients had been previously treated with an EGFR TKI, with 89% of patients (72/81) having received prior osimertinib and 80% of patients (65/81) having received prior platinum-based chemotherapy (Table 1).
Table 1.
Demographics, baseline characteristics, and prior therapies
| Dose escalation | Dose expansion | Pooled RDEa | All patients | ||||
|---|---|---|---|---|---|---|---|
| Characteristics | 3.2 mg/kg (n = 4) | 4.8 mg/kg (n = 15) | 5.6 mg/kg (n = 12) | 6.4 mg/kg (n = 5) | 5.6 mg/kg (n = 45) | 5.6 mg/kg (n = 57) | All doses (N = 81) |
| Age, median (range), years | 59.5 (51–67) | 56.0 (44–76) | 63.0 (50–77) | 68.0 (60–80) | 66.0 (40–80) | 65.0 (40–80) | 64.0 (40–80) |
| Female, n (%) | 3 (75) | 10 (67) | 8 (67) | 3 (60) | 28 (62) | 36 (63) | 52 (64) |
| Time since first NSCLC diagnosis, median (range), months | 32 (19.8–60.4) | 50 (15.9–121.9) | 31 (11.1–72.2) | 37 (12.7–50.8) | 57 (13.4–141.1)b | 45 (11.1–141.1)c | 45 (11.1–141.1)d |
| ECOG performance status, n (%) | |||||||
| 0 | 2 (50) | 7 (47) | 4 (33) | 2 (40) | 19 (42) | 23 (40) | 34 (42) |
| 1 | 2 (50) | 8 (53) | 8 (67) | 3 (60) | 26 (58) | 34 (60) | 47 (58) |
| Sum of target lesion diameters at baseline, median (range), mme | 37 (30–58) | 44 (10–111) | 50.5 (15–195) | 56 (27–89) | 55 (13–152) | 54 (13–195) | 51.5 (10–195) |
| History of CNS metastases, n (%) | 4 (100) | 8 (53) | 6 (50) | 4 (80) | 21 (47) | 27 (47) | 43 (53) |
| Patients with previously treated CNS metastases | 0 | 0 | 2 (17) | 0 | 13 (29) | 15 (26) | 15 (19) |
| Brain metastasis at baseline | 3 (75) | 5 (33) | 1 (8) | 0 | 8 (18) | 9 (16) | 17 (21) |
| EGFR-activating mutations,fn (%) | |||||||
| Ex19del | 2 (50) | 8 (53) | 5 (42) | 4 (80) | 28 (62) | 33 (58) | 47 (58) |
| L858R | 2 (50) | 5 (33) | 6 (50) | 1 (20) | 14 (31) | 20 (35) | 28 (35) |
| G719X | 0 | 1 (7) | 0 | 0 | 5 (11) | 5 (9) | 6 (7) |
| L861X | 0 | 2 (13) | 0 | 0 | 2 (4) | 2 (4) | 4 (5) |
| Ex19ins | 0 | 0 | 1 (8) | 0 | 0 | 1 (2) | 1 (1) |
| No. of prior lines of systemic therapy in the locally advanced or metastatic setting, median (range) | 4 (2–5) | 4 (2–5) | 2.5 (1–9) | 2 (1–5) | 4 (1–9) | 4 (1–9) | 4 (1–9) |
| Prior cancer regimens, n (%)g | |||||||
| Prior EGFR TKI therapyh | 4 (100) | 15 (100) | 12 (100) | 5 (100) | 45 (100) | 57 (100) | 81 (100) |
| Prior osimertinib | 3 (75) | 15 (100) | 12 (100) | 5 (100) | 37 (82) | 49 (86) | 72 (89) |
| Prior PBC | 2 (50) | 9 (60) | 7 (58) | 2 (40) | 45 (100) | 52 (91) | 65 (80) |
| Prior PBC and osimertinib | 1 (25) | 9 (60) | 7 (58) | 2 (40) | 37 (82) | 44 (77) | 56 (69) |
| Prior immunotherapyi | 2 (50) | 3 (20) | 2 (17) | 0 | 21 (47) | 23 (40) | 28 (35) |
Abbreviation: PBC, platinum-based chemotherapy.
aPooled patients who received the HER3-DXd RDE, including 12 patients in the 5.6-mg/kg group in the dose escalation part and 45 patients in the dose expansion part.
b n = 43.
c n = 55.
d n = 79.
eBy BICR per RECIST 1.1.
fPatients with multiple EGFR-activating mutations are listed in more than one row.
gNo patients received prior treatment with a topoisomerase I inhibitor.
hEGFR TKI therapies included afatinib, dacomitinib, erlotinib, gefitinib, olmutinib, and osimertinib.
iPrior immunotherapies included atezolizumab, ipilimumab, nivolumab, and pembrolizumab.
Genomic alterations known to be associated with EGFR TKI resistance were identified in 78% of patients [62/80; identified in assays of tumor tissue or circulating tumor DNA (ctDNA) in blood collected prior to treatment with HER3-DXd for 80 of 81 patients or identified by local testing as captured by electronic case report forms]. The most commonly occurring genomic alterations were additional mutations in EGFR [61 instances, including T790M (28 instances) and C797X (12 instances)], mutations in PIK3CA (17 instances), and mutations in KRAS (7 instances). There were 24 instances of amplification (including 10 instances of EGFR amplification) and 5 instances of fusion.
Dose Determination and Safety
Among the 36 patients in the dose escalation part, 5 patients experienced dose-limiting toxicities: 1 patient in the 5.6 mg/kg dose group experienced grade 3 febrile neutropenia and grade 4 platelet count decrease; 1 patient in the 4.8 mg/kg dose group experienced grade 3 neutrophil count decreased, grade 4 platelet count decrease, and white blood cell count decrease; and 3 patients in the 6.4 mg/kg dose group experienced grade 4 platelet count decrease. Both the MTD and the RDE were determined to be 5.6 mg/kg.
In the combined dose escalation and dose expansion parts, a total of 81 patients received one or more doses of HER3-DXd and were included in the safety analysis. The median treatment duration was 5.7 (range, 0.7–28.3) months [Supplementary Table S1; in the pooled group of patients who were treated with HER3-DXd at 5.6 mg/kg, i.v. once every 3 weeks (n = 57), the median treatment duration was 5.5 months (range, 0.7–18.6 months)]. All 81 patients had at least one treatment-emergent adverse event (TEAE; Table 2), with the most common (≥50%) being fatigue (64%, 52/81) and nausea (60%, 49/81; Supplementary Table S4). Grade ≥3 TEAEs occurred in 64% of patients (52/81), with the most common (≥10%) being thrombocytopenia (26%, 21/81; grade 4, 13.6%, 11/81; grade 5, 0), neutropenia (15%, 12/81; grade 4, 9.9%, 8/81; grade 5, 0), and fatigue (10%, 8/81; Table 2). Occurrences of grade ≥3 thrombocytopenia and neutropenia were generally early and transient; in the 45 patients in dose expansion cohort 1 who were treated with prior EGFR TKI and prior platinum-based chemotherapy, the median time to first onset was 8 days for thrombocytopenia and 12 days for neutropenia; the median duration was 8 days for both thrombocytopenia and neutropenia. Treatment was discontinued due to TEAEs in 9% of patients (7/81), and no patient discontinued study treatment due to thrombocytopenia. TEAEs were associated with dose reduction in 22% of patients (18/81) and with dose interruption in 37% of patients (30/81). TEAEs were associated with death in 6% of patients (5/81), but none were judged by the investigator to be related to study treatment [disease progression, 2; respiratory failure, 2; and shock (infection induced), 1; Table 2]. Safety outcomes for each dose group are shown in Supplementary Table S2.
Table 2.
Adverse events summary
| Pooled RDE 5.6 mg/kg | All patients 3.2/4.8/5.6/6.4 mg/kg | |
|---|---|---|
| TEAEs | (n = 57), n (%) | (n = 81), n (%) |
| Any TEAE | 57 (100) | 81 (100) |
| Grade ≥3 TEAEs | 42 (74) | 52 (64) |
| Serious TEAEs | 25 (44) | 32 (40) |
| TEAEs associated with treatment discontinuation | 6 (11)a | 7 (9)b |
| TEAEs associated with dose reduction | 12 (21) | 18 (22) |
| TEAEs associated with dose interruption | 21 (37) | 30 (37) |
| TEAEs associated with death | 4 (7)c | 5 (6)d |
| Treatment-related TEAEs | 55 (96) | 78 (96) |
| Grade ≥3 treatment-related TEAEs | 31 (54) | 38 (47) |
| Treatment-related TEAEs associated with death | 0 | 0 |
| Serious treatment-related TEAEs | 12 (21) | 15 (19) |
| Grade ≥3 TEAEs occurring in ≥5% of patients | ||
| Platelet count decrease/thrombocytopenia | 17 (30) | 21 (26) |
| Neutrophil count decrease/neutropenia | 11 (19) | 12 (15) |
| Fatigue | 8 (14) | 8 (10) |
| Anemia/hemoglobin decrease | 5 (9) | 6 (7) |
| Dyspnea | 5 (9) | 5 (6) |
| Febrile neutropenia | 5 (9) | 5 (6) |
| Hypoxia | 4 (7) | 5 (6) |
| White blood cell count decrease/leukopenia | 4 (7) | 5 (6) |
| Hypokalemia | 3 (5) | 4 (5) |
| Lymphocyte count decrease/lymphopenia | 3 (5) | 4 (5) |
| Adjudicated ILD | 5 (9)e | 5 (6)e |
| Adjudicated treatment-related ILD | 4 (7)f | 4 (5)f |
aFatigue (two patients); decreased appetite, interstitial lung disease (ILD), neutrophil count decrease, pneumonitis, and upper respiratory tract infection (one patient each).
bFatigue (two patients); nausea, decreased appetite, ILD, neutrophil count decrease, pneumonitis, and upper respiratory tract infection (one patient each).
cTEAEs associated with death were respiratory failure (two patients) and disease progression and shock (one patient each).
dTEAEs associated with death were respiratory failure and disease progression (two patients each) and shock (one patient).
eTwo grade 1, one grade 2, one grade 3, and one grade 5.
fTwo grade 1, one grade 2, and one grade 3.
Interstitial lung disease (ILD) has been identified as an important risk in previous studies of T-DXd, which is also an ADC with a DXd-based topoisomerase I payload (29). In study U31402-A-U102 reported here, a predefined list of adverse events (AE) suggestive of potential ILD triggered review by an independent adjudication committee to determine ILD diagnosis, grade, time of ILD onset, and relation to study drug. Treatment-related ILDs occurred in 5% of patients (4/81; Table 2). All cases of adjudicated treatment-related ILD resolved after drug discontinuation. One patient in the dose expansion part experienced a grade 5 ILD event that was adjudicated to be unrelated to study treatment. The protocol recommendations for management of ILD are shown in Supplementary Table S5.
Antitumor Activity
Within the pooled population of patients (n = 57) who received the HER3-DXd RDE (5.6 mg/kg, i.v. once every 3 weeks), the median follow-up was 10.2 months (range, 5.2–19.9), and 32% of patients (18/57) were continuing study treatment at the time of the analysis (Supplementary Table S1).
The confirmed ORR by blinded independent central review (BICR) was 39% [95% confidence interval (CI), 26.0–52.4] in patients who received HER3-DXd at a dose of 5.6 mg/kg i.v. once every 3 weeks (Table 3). There was 1 complete response (CR) and 21 partial responses (PR); 19 patients had stable disease (SD) as a best response. The median time to the first documentation of objective response (CR or PR) was 2.6 (range, 1.2–5.4) months. The median duration of response (DOR) was 6.9 [95% CI, 3.1–not evaluable (NE)] months. Among patients who had at least one evaluable tumor assessment after the initiation of study treatment (51/57 patients), 53% of patients (27 of 51) had a best reduction in sum of diameters of ≥30% with HER3-DXd treatment (Fig. 1). Among the 52 of 57 patients with prior treatment that included platinum-based chemotherapy, those with a history of brain metastases (25 of 52) had a confirmed ORR of 32% [95% CI, 15.0–53.5; 1 CR, 7 PR, 12 SD, 4 progressive disease (PD), 1 NE] compared with 41% (95% CI, 22.4–61.2; 11 PR, 7 SD, 4 PD, 5 NE) in patients without a history of brain metastases (27 of 52).
Table 3.
Responses by BICR per RECIST 1.1
| Pooled RDE (5.6 mg/kg) | ||
|---|---|---|
| Characteristics | All pooled (n = 57) | Prior PBC and osimertinib (n = 44) |
| Confirmed ORR, % (n) [95% CI] | 39 (22) | 39 (17) |
| [26.0–52.4] | [24.4–54.5] | |
| BOR, n (%) | ||
| CR | 1 (2) | 1 (2) |
| PR | 21 (37) | 16 (36) |
| SD | 19 (33) | 13 (30) |
| PD | 9 (16) | 8 (18) |
| NE | 7 (12) | 6 (14) |
| DCR,a % (n) [95% CI] | 72 (41) | 68 (30) |
| [58.5–83.0] | [52.4–81.4] | |
| TTR, median (range), months | 2.6 (1.2–5.4) | 2.7 (1.2–5.4) |
| Duration of response, median (95% CI), months | 6.9 (3.1–NE) | 7.0 (3.1–NE) |
| Progression-free survival, median (95% CI), months | 8.2 (4.4–8.3) | 8.2 (4.0–NE) |
| Overall survival, median (95% CI), months | NE (9.4–NE) | NE (8.2–NE) |
Abbreviation: PBC, platinum-based chemotherapy.
aDCR = rate of confirmed BOR of CR, PR, or SD.
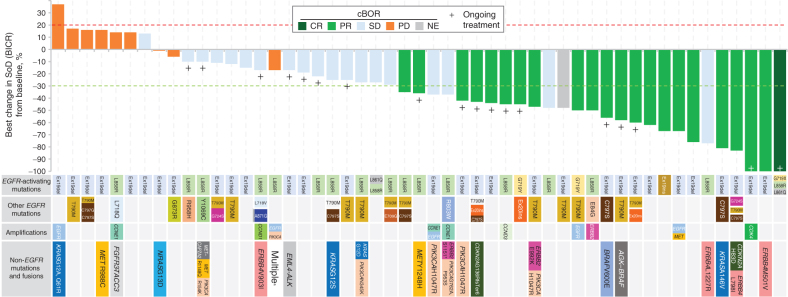
Figure 1.
Best percentage change in the tumor sum of diameters (SoD) from baseline for the pooled HER3-DXd (5.6 mg/kg, i.v. once every 3 weeks) population. Tumor genomic alterations prior to treatment with HER3-DXd are provided for each patient. Six patients could not be evaluated for BOR due to lack of adequate post–baseline tumor assessment and are not shown; one patient had a BOR of NE due to achieving SD too early (<5 weeks) and is shown with hatched markings. Genomic analysis was performed centrally using Oncomine Comprehensive Assay v3 (Thermo Fisher Scientific). Results from local testing are included, if available, together with any additional mutations detected using GuardantOMNI assay of ctDNA in blood collected prior to treatment with HER3-DXd. For ctDNA analysis, a minor allelic frequency of ≥0.1% was used as a threshold for detection of mutations. aPatient had multiple tumor mutations comprising CDKN2A A143V; PIK3CA E542K, E545K, E726K; ERBB2 K200N; and ERBB3 Q847*, Q849*. cBOR, Confirmed BOR.
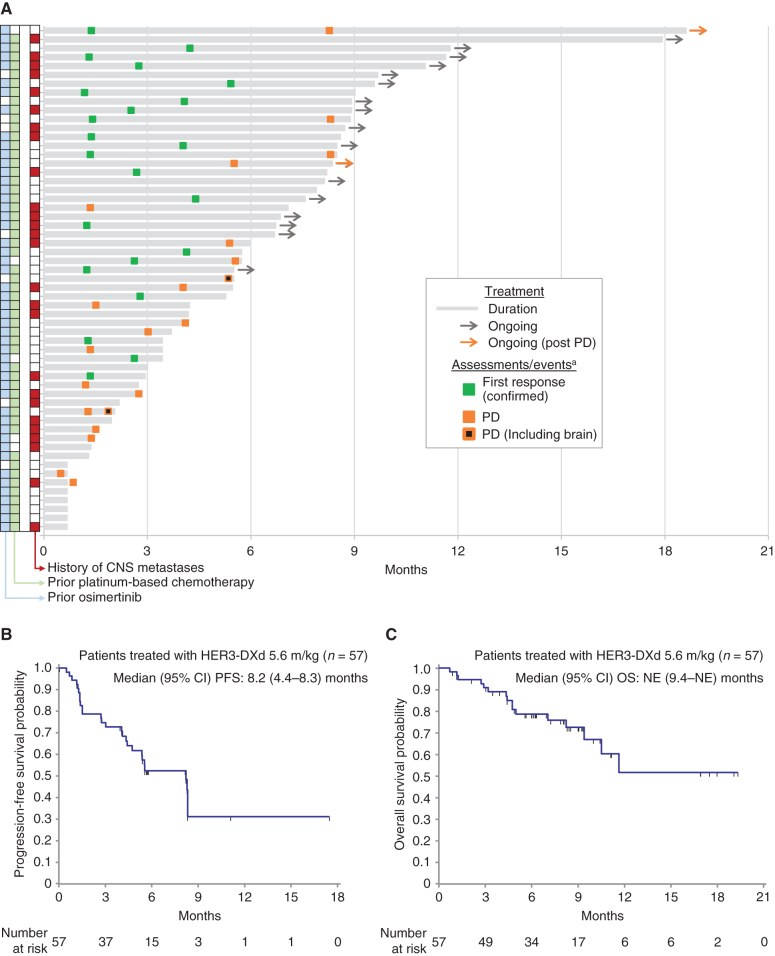
Treatment durations for each patient in the pooled HER3-DXd (5.6 mg/kg, i.v. once every 3 weeks) population are shown with the times of first responses in Fig. 2A. Two patients continued study treatment following radiographic disease progression.
Figure 2.
Tumor response as assessed by BICR in patients treated with HER3-DXd at 5.6 mg/kg (n = 57). A, Swimmer plot showing treatment duration, first occurrence of confirmed tumor response, and progression. Prior treatment and history of CNS metastases are indicated for each patient. B, Kaplan–Meier plot of progression-free survival probability. C, Kaplan–Meier plot of overall survival probability. aMarkers show the time of the initial response for confirmed responses. Two patients continued treatment after progression. In one case (top of swimmer plot), the patient had an equivocal (small) lesion and continued on treatment, but the lesion was later assessed as unequivocal—the swimmer plot is marked PD at the date of the lesion's first appearance. In the other case, treatment continued because PD was determined by BICR but not by the local investigator (the study treatment discontinuation criteria were based on local tumor assessment).
At a median follow-up of 10.2 months, median PFS was 8.2 (95% CI, 4.4–8.3) months (Fig. 2B; 16 of 57 patients were ongoing without events), and the median OS was not reached at the time of data cutoff (95% CI, 9.4–NE months; Fig. 2C; 35 of 57 patients were ongoing without events).
Confirmed responses occurred at a similar frequency among the subgroups of patients who had received EGFR TKI and platinum-based chemotherapy prior to study entry (n = 52; Supplementary Table S3; Supplementary Fig. S2A and S2B) and patients who had received both osimertinib and platinum-based chemotherapy prior to study entry (n = 44; Table 3; Supplementary Fig. S2C and S2D). Sixteen patients in the dose escalation part had prior EGFR TKI (including osimertinib) but had not received prior platinum-based chemotherapy; in these patients, the confirmed ORR was 63% (95% CI, 35.4–84.8), and median PFS was 7.6 (95% CI, 5.4–13.7) months.
Biomarker Analysis
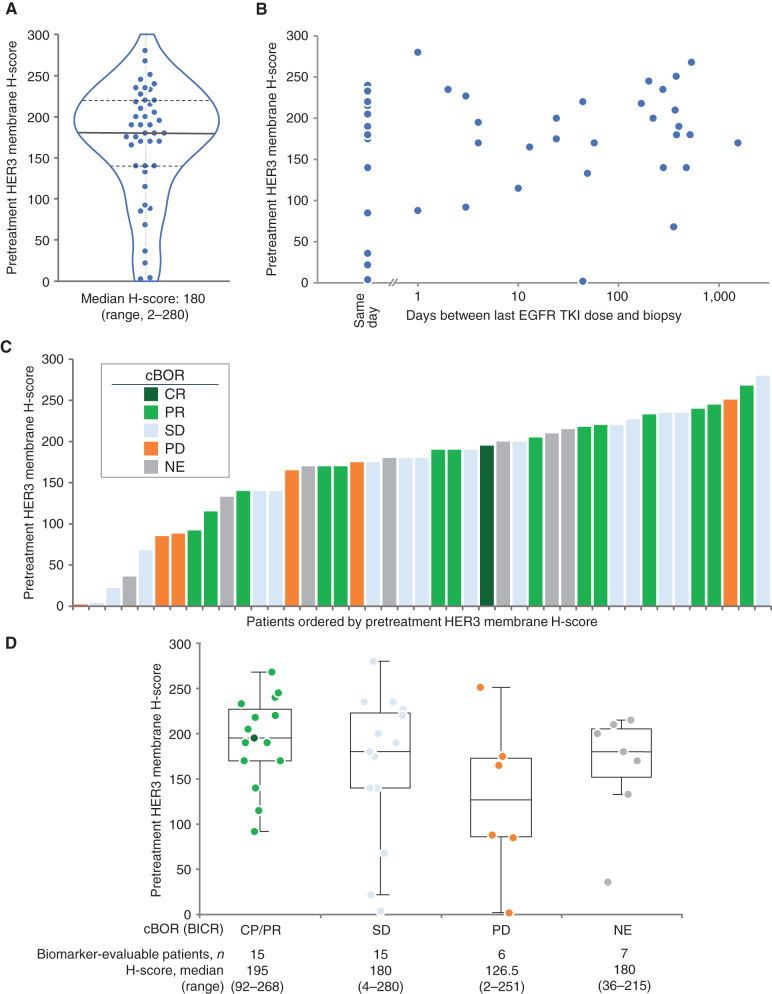
HER3 membrane expression was quantified using an IHC-based H-score (0–300 scale). Representative examples of IHC staining and associated H-scores are shown in Supplementary Fig. S3A–S3D. The analysis of pretreatment biopsy tissue, which was available for 43 of 57 patients in the pooled HER3-DXd (5.6 mg/kg, i.v. once every 3 weeks) population, demonstrated that tumor HER3 expression was observed in all patients; the median H-score was 180 (range, 2–280; Fig. 3A). There appeared to be no correlation between HER3 membrane H-score in the tumor and the time between a patient's last EGFR TKI dose and their analyzed tumor biopsy specimen (Fig. 3B). The confirmed best overall response (BOR) in patients according to tumor HER3 membrane expression level by H-score is shown in Fig. 3C and D. Confirmed responses were seen across a wide range of baseline tumor HER3 membrane H-scores, but there was a trend toward enrichment of confirmed responses in patients with higher baseline H-scores.
Figure 3.
Pretreatment HER3 membrane expression and association with BOR. BOR was assessed by BICR in patients treated with HER3-DXd at 5.6 mg/kg (43 of 57 patients evaluable for HER3 membrane expression). A, Distribution of pretreatment HER3 membrane H-score (0–300). B, Pretreatment HER3 membrane H-score and association with time since last treatment with EGFR TKI. C and D, Pretreatment HER3 membrane H-score and confirmed BOR (cBOR).
EGFR-activating mutations were detected in tumor tissue or ctDNA in blood (collected prior to treatment with HER3-DXd) from all patients in the pooled HER3-DXd (5.6 mg/kg, i.v. once every 3 weeks) population. Most patients had the common EGFR-activating mutations Ex19del or L858R, but the enrollment criteria also allowed the inclusion of patients with atypical EGFR-activating mutations (specifically G719X and L861Q, with others possible following discussion with the sponsor). Among the 7 of 57 patients with atypical EGFR-activating mutations (G719X, L861Q, and Ex19ins), the BORs were one CR and three PRs (confirmed); one patient had SD, and two were not evaluable for response. Genomic alterations known to be associated with EGFR TKI resistance were identified in 77% of patients (44/57). Diverse known mechanisms of EGFR TKI resistance were detected, including EGFR C797S, MET or HER2 amplification, and BRAF fusion; furthermore, several patients had no known EGFR TKI resistance mechanism (Supplementary Fig. S4). HER3-DXd treatment resulted in antitumor activity across this spectrum of known and unknown EGFR TKI resistance mechanisms (Fig. 1). In the 23 of 57 patients with known EGFR-related resistance mechanisms (excluding T790M), the confirmed ORR was 35% (CR/PR, 8; SD, 7; PD, 5; NE, 3). In the 13 of 57 patients with known EGFR-independent resistance mechanisms, the confirmed ORR was 46% (CR/PR, 6; SD, 4; PD, 2; NE, 1). In the 21 of 57 patients with other/unknown resistance mechanisms, the confirmed ORR was 38% (CR/PR, 8; SD, 8; PD, 2; NE, 3).
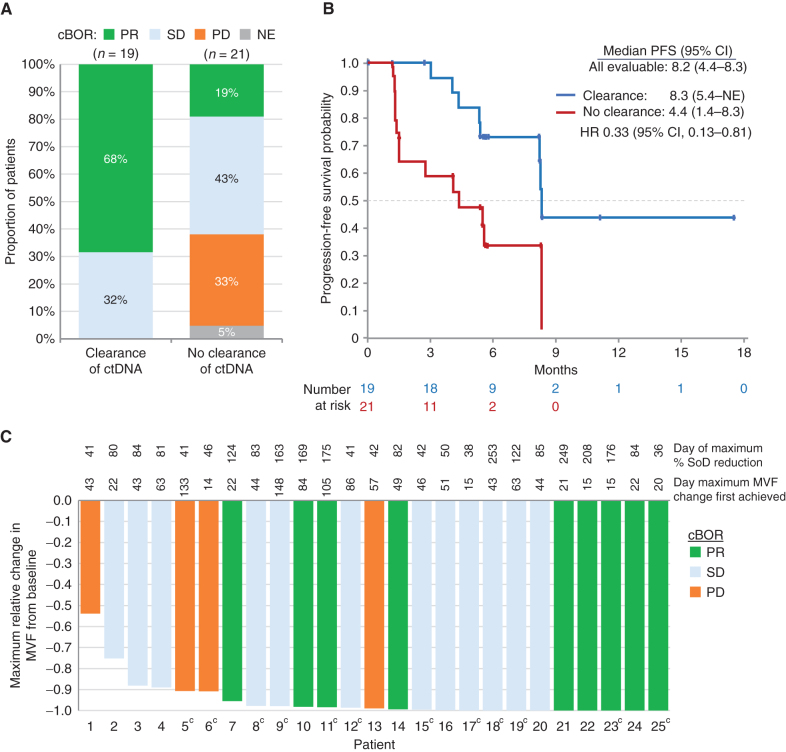
The association between response to HER3-DXd and clearance of EGFR-activating mutations was evaluated using ctDNA obtained at baseline and during therapy. Early clearance of ctDNA was defined by undetectable Ex19del or L858R at week 3 or week 6 following detection of either mutation prior to treatment. Early clearance of ctDNA was evaluable in 40 of 57 patients from the pooled HER3-DXd (5.6 mg/kg, i.v. once every 3 weeks) population. Although the confirmed ORR was 43% (17/40) in these patients, the confirmed ORR was 68% (13/19) in patients with early clearance of ctDNA compared with 19% (4/21) in patients without early clearance. No patient with early clearance of ctDNA had a confirmed BOR of PD (Fig. 4A). Although median PFS in all 40 patients evaluable for early clearance of ctDNA was 8.2 (95% CI, 4.4–8.3) months, median PFS was longer in patients with early clearance of ctDNA versus those without [8.3 (95% CI, 5.4–NE) months vs. 4.4 (95% CI, 1.4–8.3) months; HR, 0.33; 95% CI, 0.13–0.81; Fig. 4B]. In all patients in whom the allelic frequency of the EGFR-activating mutations Ex19del or L858R could be quantified in pretreatment ctDNA (where the minor variant frequency was >1%; 25 of 57 patients), reductions relative to baseline were observed (Fig. 4C).
Figure 4.
Analyses of ctDNA changes. A, Proportions of confirmed BOR (cBOR; BICR per RECIST 1.1) by early clearance of ctDNA.aB, Kaplan–Meier plots showing PFS by early clearance of ctDNA.aC, Waterfall plot showing maximum change relative to baseline in the minor variant frequency (MVF) of EGFR-activating mutations in ctDNA and confirmed BOR by BICR (25 of 57 patients were evaluableb). aEarly clearance of ctDNA was defined as nondetectable plasma of both EGFR Ex19del and EGFR L858R at week 3 or week 6, where one or more allele comprising EGFR Ex19del or EGFR L858R was detectable at baseline. Serial ctDNA samples were collected from 45 of 57 patients, but 5 did not have evaluable ctDNA data for either week 3 or 6. In the 17 patients not evaluable for early clearance of ctDNA, responses were CR/PR, 29%; SD, 24%; PD, 17%; and NE, 35%. bFewer patients (25 of 57) were evaluable for relative change in ctDNA than for early clearance of ctDNA (40 of 57), as the former required ctDNA levels to be above the limit of quantification (MVF > 1%); the latter required levels to be above the limit of detection, which was lower (MVF > 0.02%). cPatients without GeneStrat data at C1D1 were analyzed if they had GuardantOMNI data at C1D1 when the MVF was >1%; this is because GeneStrat and GuardantOMNI data were concordant, except for the low MVF range (<1%). SoD, sum of diameters.
Pharmacokinetics
Patients in the dose escalation part received HER3-DXd infusions prepared from a frozen liquid formulation, whereas patients in the dose expansion part received HER3-DXd infusions prepared from a lyophilized powder formulation. Pharmacokinetic (PK) parameters for released MAAA-1181a (the HER3-DXd payload) and MAAA-1181a–conjugated antibody for the patients in dose escalation and dose expansion cohort 1 are summarized in Supplementary Table S6. The mean (standard deviation) serum concentration–time profiles of the MAAA-1181a–conjugated antibody (for 3.2, 4.8, 5.6, and 6.4 mg/kg doses) and released payload MAAA-1181a (5.6 mg/kg) are shown in Supplementary Fig. S5A and S5B.
In the 3.2 to 6.4 mg/kg dose range, the maximum serum concentration (Cmax) of the MAAA-1181a–conjugated antibody increased approximately dose proportionally, whereas the AUC for the MAAA-1181a–conjugated antibody increased slightly greater than dose proportionally (Supplementary Fig. S6A and S6B).
Twenty-four patients from the dose escalation 5.6 mg/kg group (n = 10; frozen liquid formulation) and dose expansion cohort 1 (n = 14; lyophilized powder formulation) were included in a PK comparability analysis. The geometric mean ratios (GMR) of lyophilized powder/frozen liquid for the PK parameters of Cmax, AUClast, and AUC0–21d at cycle 1 were in the range of 93.4% to 96.4% for the MAAA-1181a–conjugated antibody (Supplementary Table S7), indicating that MAAA-1181a–conjugated antibody exposure is similar for the lyophilized powder and frozen liquid formulations at 5.6 mg/kg, i.v. once every 3 weeks and that no dose adjustment is warranted between frozen liquid and lyophilized powder formulations of HER3-DXd. The GMRs for the PK parameters of Cmax, AUClast, and AUC0–21d of the released payload MAAA-1181a are shown in Supplementary Table S7.
Discussion
Treatment options remain limited for patients with EGFR-mutated NSCLC who develop resistance to EGFR TKI therapy. Although targeting specific resistance mechanisms has demonstrated clinical efficacy, these molecular alterations are diverse; development and deployment of specific therapeutic approaches against each resistance mechanism might be impractical. In addition, most first-line osimertinib-resistant lung cancers do not harbor an obvious, currently targetable genomic mechanism of resistance (11). In this study, we explored a ubiquitous biological feature of EGFR-mutated lung cancers, the expression of HER3, and clinically evaluated the targeting of this tumor marker by HER3-DXd. In contrast to most previous studies in EGFR-mutated NSCLC that focused on targeting specific genomic alterations, HER3-DXd provides a newer investigational treatment strategy that might provide benefit to a broader patient population with HER3 protein expression. The findings of this study show that HER3-DXd provides a feasible therapeutic approach with clinically meaningful antitumor activity, with an ORR of 39% and median PFS of 8.2 months.
Most patients with EGFR-mutated NSCLC have tumors that express membrane HER3 and, because of the breadth of its expression, HER3 is an attractive molecular target for therapeutic intervention (19). A previous strategy to block the ligand-binding site of HER3 with an unconjugated antibody (such as patritumab or seribantumab) in combination with erlotinib did not confer clinically meaningful efficacy in EGFR-mutated NSCLC (19, 32, 33). HER3-DXd has a mechanism of action that is distinct from mAb therapies; as an ADC, it is designed to produce antitumor activity through targeted delivery of its cytotoxic payload. The mechanism of action of an ADC is complex, and multiple factors are relevant to its antitumor activity: target antigen expression on the cell surface, ADC internalization and trafficking to the lysosome, enzymatic cleavage of the ADC (in the case of HER3-DXd, by cathepsin L and B), and the cytotoxic activity of the delivered payload (in the case of HER3-DXd, a topoisomerase I inhibitor; refs. 22, 26).
Because of the antigen specificity of HER3-DXd, we explored the association of HER3 expression in pretreatment tumor tissues with clinical response. Although confirmed responses were observed across tumors with a broad range of HER3 membrane H-scores, there was a slight enrichment of confirmed responses in patients with higher HER3 membrane H-scores at baseline. An important area of future investigation is to understand the utility of alternative measures of HER3 expression, other biomarkers related to the expression of HER3 dimerization partners, and the dynamics of HER3 expression to predict clinical benefit in patients treated with HER3-DXd. These characteristics will be further explored in this study (cohort 3; Supplementary Fig. S1) and in additional clinical evaluations of HER3-DXd. In addition, analysis is ongoing into identifying markers beyond HER3 expression, including internalization, drug processing, and DNA damage repair, using patient samples from this study.
Beyond radiographic and clinical measures of antitumor activity, this study also demonstrated an increased response rate and prolonged PFS in the subset of patients who achieved early clearance of ctDNA; this has been observed previously for a broad range of therapies, including EGFR TKIs and immune checkpoint inhibitors (34–36). In this study, reduction of ctDNA for the EGFR-activating mutations Ex19del or L858R was observed in all patients with quantifiable ctDNA levels at baseline, with maximum reductions occurring between day 15 and day 148. Although the analysis of ctDNA is an investigational approach to monitor tumor dynamics and response to therapy, the clinical utility of these data at the current time has not yet been determined.
The safety profile of HER3-DXd was manageable, with a low rate of discontinuation due to TEAEs (9%, 7/81). Dose reduction and dose delay were successfully used in individual circumstances to mitigate toxicity and to avoid permanent discontinuation of study treatment. The most common toxicities comprised those related to gastrointestinal toxicity or cytopenia, and the most common grade ≥3 TEAEs comprised cytopenia (most commonly thrombocytopenia and neutropenia). No single TEAE was identified as a major cause of treatment discontinuation. No patient discontinued study treatment due to thrombocytopenia; when it occurred, the onset of grade ≥3 thrombocytopenia was typically early during study treatment (median time to first onset, 8 days) and was transient (median duration, 8 days). Drug-related ILD is an identified risk with DXd-based ADCs (ref. 29; T-DXd is approved in the United States with boxed warnings for ILD and embryofetal toxicity), and surveillance and early management are important in the treatment of patients with this class of therapy (Supplementary Table S5). However, in this study of HER3-DXd, the frequency of adjudicated treatment-related ILD was low (5%, 4/81), analogous to the incidences reported in trials of EGFR TKIs for patients with NSCLC (ILD rate, 0%–5.7%; ref. 37). Overall, the safety profile of HER3-DXd in this study was similar to that observed for HER3-DXd in patients with metastatic breast cancer (38).
The data presented in this report support further study of HER3-DXd in this setting, and a pivotal, phase II, global study of HER3-DXd in patients with EGFR-mutated NSCLC after disease progression on EGFR TKI and platinum-based chemotherapy has been initiated (HERTHENA-Lung01; NCT04619004). A phase I, dose escalation and dose expansion study has been initiated to evaluate HER3-DXd in combination with osimertinib (and also as monotherapy) in patients with EGFR-mutated NSCLC with tumor progression after treatment with osimertinib monotherapy and no platinum-based chemotherapy (NCT04676477). In addition, HER3-DXd is being evaluated in HER3-expressing metastatic breast cancer (NCT02980341) and metastatic colorectal cancer (NCT04479436).
Methods
Trial Design and Patients
This is a global, open-label, multiple-dose, two-part, phase I study conducted at 22 sites in the United States, Japan, Asia, and Europe (Supplementary Fig. S1). In the dose escalation part, HER3-DXd was assessed in patients with metastatic or unresectable EGFR-mutated NSCLC who had acquired resistance to EGFR TKI (per Jackman criteria; ref. 39). In the dose expansion part, HER3-DXd is being assessed in three cohorts: cohort 1, EGFR-mutated (G719X, Ex19del, L858R, or L861Q; other EGFR-activating mutations may be eligible following discussion with the sponsor) adenocarcinoma NSCLC treated with one or more prior EGFR TKIs and one or more prior platinum-based chemotherapy regimens; cohort 2, squamous or nonsquamous NSCLC without EGFR-activating mutations and with prior anti–PD-1/PD-L1 therapy (unless unable or unwilling); and cohort 3: EGFR-mutated (G719X, Ex19del, L858R, or L861Q; other EGFR-activating mutations may be eligible following discussion with the sponsor) NSCLC, including any histology other than combined small cell and non–small cell with one or more prior EGFR TKIs and one or more prior platinum-based chemotherapy regimens.
General eligibility criteria included age ≥18 years, Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1, and adequate bone marrow, organ, and cardiac function. Patients with a history of or suspected ILD/pneumonitis were excluded, as were patients with any evidence of small cell histology or combined small cell and non–small cell histology. Stable brain metastases were allowed. Eligibility criteria for the dose escalation part included metastatic/unresectable, EGFR-mutated, EGFR TKI-resistant NSCLC after progression on osimertinib or T790M negative after progression on erlotinib, gefitinib, or afatinib, as well as available pretreatment tumor tissue (after progression on EGFR TKIs) for retrospective analysis of HER3 expression. Eligibility criteria for the dose expansion part cohort 1 included EGFR-mutated NSCLC adenocarcinoma, one or more prior EGFR TKIs, and one or more prior platinum-based chemotherapy regimens (patients with EGFR T790M following treatment with erlotinib, gefitinib, afatinib, or dacomitinib must have received and have progressed following treatment with osimertinib, unless unable or unwilling). Investigators obtained written informed consent from all patients prior to study participation. The study was conducted in compliance with the protocol, the ethical principles that have their origin in the Declaration of Helsinki, the International Council for Harmonisation consolidated Guideline E6 for Good Clinical Practice, and applicable local regulatory requirements. The study was approved by the institutional review board or ethics committee for each site.
Procedures
Patients received HER3-DXd, i.v. once every 3 weeks on day 1 of each cycle of a 21-day cycle. Patients in the dose escalation part received 3.2 to 6.4 mg/kg HER3-DXd, i.v. once every 3 weeks, which was guided by a Bayesian logistic regression model following the escalation with overdose control principle. In the dose expansion part, patients in cohorts 1 and 2 received the RDE of 5.6 mg/kg HER3-DXd, i.v. once every 3 weeks (40); patients in cohort 3 were randomized 1:1 to receive the RDE (cohort 3a) or an uptitration of HER3-DXd (cohort 3b). Two formulations were dosed in this study: frozen liquid drug product and lyophilized drug product. The frozen liquid drug product (used in dose escalation) was 50 mg HER3-DXd in a 2.5-mL solution (20 mg/mL) in a single-use vial. The lyophilized drug product (used in dose expansion cohort 1) was 100 mg HER3-DXd in lyophilized powder dosage form in a single-use vial to be reconstituted with 5 mL of water for injection to 20 mg/mL.
Analysis of Genomic Alterations and Exploratory Biomarkers
Patients provided archival tumor tissue or consented to a fresh tumor biopsy as a pretreatment sample. End-of-treatment biopsies were optional; on-study biopsies were optional for all cohorts except for cohort 3. Blood samples were analyzed for ctDNA to validate findings from tumor tissue samples as part of these exploratory analyses.
Analysis of genomic alterations was performed centrally using Oncomine Comprehensive Assay v3 (Thermo Fisher Scientific) from formalin-fixed, paraffin-embedded tumor tissue. If available, results from local testing were included in the list of detected genomic alterations. Any additional mutations detected using GuardantOMNI assays (Guardant Health) in ctDNA from blood collected prior to treatment with HER3-DXd were also included. For the ctDNA-based data, a minor allelic frequency of 0.1% was used as a threshold for detection of mutations.
Analyses of HER3 membrane H-score and clearance of ctDNA were performed for the patients in dose escalation and dose expansion cohort 1 who received the HER3-DXd RDE (5.6 mg/kg, i.v. once every 3 weeks).
HER3 membrane expression was assessed by IHC in pretreatment tumor samples. HER3 IHC was performed on formalin-fixed, paraffin-embedded tissue using the BenchMark ULTRA IHC/ISH system (Roche Diagnostics). Staining was conducted with an anti-HER3 recombinant rabbit mAb (clone SP438) after antigen retrieval in CC1 buffer followed by detection with the OptiView DAB IHC Detection Kit (Ventana Medical Systems).
HER3 IHC staining of tumor cells was evaluated by determining the percentage of tumor cells with membrane and cytoplasmic staining at specific intensities as follows: 0 (absence of staining), 1+ (weak staining), 2+ (moderate-intensity staining), and 3+ (strong staining). From these data, the H-scores (a weighted summed score) were calculated for each patient sample for both cytoplasmic and membrane components.
Assessment of EGFR-mutated (Ex19del and L858R) ctDNA by GeneStrat (Biodesix) was conducted from C1D1 to C5D1 for every cycle, at C7D1 and C9D1, and then on day 1 of every third cycle through end of treatment. Early clearance of ctDNA was defined as nondetectable plasma of both EGFR Ex19del and EGFR L858R at week 3 or week 6, where at least one of either EGFR Ex19del or EGFR L858R was detectable at baseline.
PK Assay and Analysis
The PK assay methods for measuring MAAA-1181a–conjugated antibody and the released payload MAAA-1181a were developed and validated at PPD Laboratories. The method for measuring MAAA-1181a–conjugated antibody was an ELISA in human serum with a quantitation range of 100 to 4,000 ng/mL. The method for measuring the released payload MAAA-1181a was a liquid chromatography–mass spectrometry assay in human serum with a quantitation range of 10 to 2,000 pg/mL. Samples with analyte concentrations above the respective assay's upper limit of quantitation are diluted into each assay's quantitation range and analyzed.
Noncompartmental analysis was conducted using Phoenix WinNonlin (Version 8.1; Certara). The power model to assess dose proportionality has the following form: Y = a•(dose)b. “Y” is the PK parameter, and “a” and “b” are the coefficient and exponent of the power equation, respectively. Following logarithmic transformation, the power model can be analyzed using linear regression.
Objectives and Endpoints
For the dose escalation part, the primary objective was to assess the safety and tolerability of HER3-DXd and determine the RDE. Primary endpoints included AEs, serious AEs (SAE), and other safety assessments. Secondary objectives in the dose escalation part were to investigate the antitumor activity of HER3-DXd, as well as characterize the PK of MAAA-1181a–conjugated antibody and released MAAA-1181a. Secondary endpoints included confirmed responses by BICR per Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1, ORR, disease control rate (DCR), DOR, time to response (TTR), PFS, OS, and serum concentration and PK parameters of MAAA-1181a–conjugated antibody and released MAAA-1181a. Exploratory objectives included identification of biomarkers that correlate with HER3-DXd activity. Exploratory endpoints included correlation of biomarkers with clinical activity of HER3-DXd.
For the dose expansion part, the primary objective was to assess the antitumor activity of HER3-DXd. Primary endpoints included confirmed ORR by BICR per RECIST 1.1. Secondary endpoints included ORR by investigator per RECIST 1.1, DCR, DOR, TTR, PFS by BICR and by investigator per RECIST 1.1, and OS. Secondary objectives in dose expansion were to assess safety and tolerability of HER3-DXd, as well as characterize the PK of MAAA-1181a–conjugated antibody and released MAAA-1181a. Secondary endpoints included SAEs, TEAEs, physical examination findings, vital signs, ophthalmologic findings, standard clinical laboratory parameters, electrocardiogram parameters, echocardiogram/multigated acquisition scan findings, and serum concentration and PK parameters of MAAA-1181a–conjugated antibody and released MAAA-1181a. Exploratory objectives included the identification of biomarkers that correlate with HER3-DXd activity. Exploratory endpoints included correlation of biomarkers (e.g., ctDNA, HER3 IHC) with clinical activity of HER3-DXd.
Statistical Analysis
Descriptive statistics were provided for selected demographic, efficacy, safety, and PK data from both dose escalation and dose expansion parts. Assessments of change from baseline to posttreatment included only patients with both baseline and posttreatment measurements. Safety analyses were performed on the basis of the safety analysis set, which included all patients who were enrolled in the dose escalation part and dose expansion part in cohort 1 and received at least one dose of HER3-DXd (n = 81). Efficacy analyses were performed based on the efficacy analysis set, which is identical to the safety analysis set. PK analyses were performed on the basis of the PK analysis set, which included all patients in the safety analysis set who had at least one PK sample with measurable concentration of MAAA-1181a–conjugated antibody or MAAA-1181a. ORR and DCR and their two-sided 95% exact CI (using the Clopper–Pearson method) were provided. Distribution of the time-to-event endpoints was estimated using the Kaplan–Meier method. TEAEs were defined as AEs starting or worsening during the on-treatment period (from the start date of study treatment to 47 days after the end date of study treatment). SAEs starting or worsening after 47 days, if reported as related to the study treatment, were also defined as TEAEs.
Authors' Disclosures
P.A. Jänne reports grants and personal fees from Daiichi Sankyo during the conduct of the study; grants and personal fees from AstraZeneca, Boehringer Ingelheim, Eli Lilly, Takeda; personal fees from Pfizer, Roche/Genentech, Chugai, LOXO Oncology, SFJ Pharmaceuticals, Voronoi, Biocartis, Novartis, Sanofi, Mirati Therapeutics, Transcenta, Silicon Therapeutics, Syndax, Nuvalent, Bayer, Esai, Allorion Therapeutics, Accutar Biotech, AbbVie; grants from Astellas, PUMA, and grants from Revolution Medicines outside the submitted work; in addition, P.A. Jänne has a patent for EGFR mutations issued and licensed to LabCorp. C. Baik reports other support from Daiichi during the conduct of the study; AbbVie, AstraZeneca, Lilly, Jansen, Spectrum, Pfizer, Blueprint, TurningPoint, Loxo; personal fees from AstraZeneca, Blueprint, Daiichi, Takeda, TurningPoint, and personal fees from Guardant outside the submitted work. M.L. Johnson reports grants and other support from AbbVie, Amgen, AstraZeneca, Atreca, Calithera Biosciences, Checkpoint Therapeutics, CytomX, Daiichi Sankyo, EMD Serono, Genentech/Roche, GlaxoSmithKline, Gritstone Oncology, Guardant Health, Incyte, Janssen, Loxo Oncology, Merck, Mirati Therapeutics, Novartis, Pfizer, Ribon Therapeutics, Sanofi, WindMIL, and Boehringer Ingelheim; grants from Acerta, Adaptimmune, Apexigen, Arcus Biosciences, Array BioPharma, Artios Pharma, BeiGene, BerGenBio, Corvus Pharmaceuticals, Curis, Dracen Pharmaceuticals, Dynavax, Lilly, Genmab, Genocea Biosciences, Harpoon, Hengrui Therapeutics, Immunocore, Jounce Therapeutics, Kadmon Pharmaceuticals, Lycera, Neovia Oncology, OncoMed Pharmaceuticals, PMV Pharmaceuticals, Regeneron Pharmaceuticals, Rubius Therapeutics, Seven and Eight Biopharmaceuticals/Birdie Biopharmaceuticals, Shattuck Labs, Silicon Therapeutics, Stem CentRx, Syndax Pharmaceuticals, Takeda Pharmaceuticals, Tarveda, TCR2 Therapeutics, TMUNITY Therapeutics, and University of Michigan; other support from Association of Community Cancer Centers, Lilly, Achilles Therapeutics, Bristol-Myers Squibb, Editas Medicine, Eisai, G1 Therapeutics, and other support from Ideaya Biosciences outside the submitted work. H. Hayashi reports personal fees from Daiichi Sankyo Co., Ltd, AstraZeneca K.K., Boehringer Ingelheim Japan Inc., Bristol-Myers Squibb Co. Ltd., Chugai Pharmaceutical Co. Ltd., Eli Lilly Japan K.K., Kyorin Pharmaceutical Co. Ltd, Merck Biopharma Co., Ltd., MSD K.K., Novartis Pharmaceuticals K.K., Ono Pharmaceutical Co. Ltd., Shanghai Haihe Biopharm, Taiho Pharmaceutical Co. Ltd., and Takeda Pharmaceutical Company Limited; personal fees from AstraZeneca K.K., Boehringer Ingelheim Japan Inc., Bristol-Myers Squibb Co. Ltd., Chugai Pharmaceutical Co. Ltd., Eli Lilly Japan K.K, Pfizer Japan Inc., Shanghai Haihe Biopharm, Takeda Pharmaceutical Company Limited and Merck Biopharma Co., Ltd.; and grants from AstraZeneca K.K., Astellas Pharma Inc., MSD K.K., Ono Pharmaceutical Co., Ltd., Nippon Boehringer Ingelheim Co., Ltd., Novartis Pharma K.K., grants, Pfizer Japan Inc., Bristol Myers Squibb Company, Eli Lilly Japan K.K., Chugai Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd., Merck Serono Co., Ltd./Merck Biopharma Co., Ltd., Takeda Pharmaceutical Co., Ltd., Taiho Pharmaceutical Co., Ltd., SymBio Pharmaceuticals Limited, AbbVie Inc, inVentiv Health Japan, ICON Japan K.K., GRITSONE ONCOLOGY, INC, PAREXEL International Corp., Kissei Pharmaceutical Co., Ltd., EPS Corp., Syneos Health, Pfizer R&D Japan G.K., A2 Healthcare Corp., Quintiles Inc./IQVIA Services JAPAN K.K., EP-CRSU CO., LTD., Linical Co., Ltd., Eisai Co., Ltd., CMIC Shift Zero K.K., Kyowa Hakko Kirin Co., Ltd, Bayer Yakuhin, Ltd., EPS International Co., Ltd.; grants from Daiichi Sankyo Co., Ltd during the conduct of the study; and grants from Otsuka Pharmaceutical Co., Ltd., outside the submitted work. M. Nishio reports grants and personal fees from Ono Pharmaceutical, Bristol Myers Squibb, Pfizer, Chugai Pharmaceutical, Eli Lilly, Taiho Pharmaceutical, AstraZeneca, personal fees from Boehringer-Ingelheim, MSD, Novartis, Daiichi Sankyo, and Takeda Pharmaceutical Company Limited; personal fees from Merck Biopharma, TEIJIN PHARMA LIMITED., and personal fees from AbbVie, outside the submitted work; D.-W. Kim reports grants, nonfinancial support, and other support from Daiichi-Sankyo during the conduct of the study; grants and other support from Amgen; grants from Alpha Biopharma, AstraZeneca, Boehringer-Ingelheim, Hanmi, Janssen, Merus, Mirati Therapeutics, MSD, Novartis, ONO Pharmaceutical, Pfizer, Roche/Genentech, Takeda, TP Therapeutics, Xcovery, Yuhan, Chong Keun Dang, Bridge BioTherapeutics, and grants from GSK outside the submitted work. K.A. Gold reports personal fees from AstraZeneca, Takeda, and Rakuten; grants from Pharmacyclics, grants from Daiichi Sankyo during the conduct of the study, and grants from Pfizer outside the submitted work. C.E. Steuer reports advisory board membership from Boehringer Ingelheim, EMD Serono, Dava Oncology, Bergen Bio, Armo, AbbVie, Eli Lilly, Sanofi, and Mirati. H. Murakami reports grants and personal fees from Daiichi Sankyo during the conduct of the study; AstraZeneca, Chugai Pharma, Takeda; grants from AbbVie and IQVIA; personal fees from Ono Pharmaceutical, Bristol-Myers Squibb Japan, MSD, Pfizer, Novartis, Eli Lilly Japan, and personal fees from Taiho Pharmaceutical outside the submitted work. J.C.-H. Yang reports grants, personal fees, and other support from AstraZeneca, personal fees and other support from Amgen, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi Sankyo; other support from Eli Lilly, Merck KGaA, Merck Sharp & Dohme, Novartis; personal fees from Ono Pharmaceuticals, Pfizer, Roche/Genentech, Takeda Oncology, Yuhan Pharmaceuticals; other support from JNJ, GlaxoSmithKline, and other support from Puma Biotechnology outside the submitted work. Z. Qi reports other support from Daiichi Sankyo Inc, other support from ArticulateScience LLC during the conduct of the study, and other support from Bristol Myers Squibb outside the submitted work. D. Sternberg reports other support from Daiichi Sankyo, Inc. during the conduct of the study and outside the submitted work. C. Yu reports personal fees and other support from Daiichi Sankyo, Inc. during the conduct of the study; other support from Daiichi Sankyo, Inc. outside the submitted work. H.A. Yu reports other support from AstraZeneca, Cullinan, Daiichi, Janssen, Blueprint, Lilly, Novartis, and other support from Pfizer outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
P.A. Jänne: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. C. Baik: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. W.-C. Su: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. M.L. Johnson: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. H. Hayashi: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. M. Nishio: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing. D.-W. Kim: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. M. Koczywas: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. K.A. Gold: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. C.E. Steuer: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. H. Murakami: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. J.C.-H. Yang: Conceptualization, resources, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing, approval. S.-W. Kim: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing, approval. M. Vigliotti: Conceptualization, resources, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing, approval. R. Shi: Conceptualization, resources, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing, approval. Z. Qi: Conceptualization, resources, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing, approval. Y. Qiu: Conceptualization, resources, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing, approval. L. Zhao: Conceptualization, resources, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing, approval. D. Sternberg: Conceptualization, resources, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing, approval. C. Yu: Conceptualization, resources, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing, approval. H.A. Yu: Conceptualization, resources, data curation, formal analysis, supervision, methodology, writing–original draft, writing–review and editing, approval.
Supplementary Material
Acknowledgments
This study was funded by Daiichi Sankyo, Inc. P.A. Jänne received salary support from the American Cancer Society (CRP-17-111-01-CDD). We thank the patients, their families, and their caregivers for their participation. We also thank Masayuki Kanai, Maha Karnoub, Kei Enomoto, Ru Chen, Pang-Dian Fan, Minseok Lee, Ling He, and Pomy Shrestha for their valuable contributions. Medical editorial assistance was provided by Amos Race (ArticulateScience LLC) and funded by Daiichi Sankyo, Inc.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
References
- 1. Zhang YL, Yuan JQ, Wang KF, Fu XH, Han XR, Threapleton Det al. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2016;7:78985–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KHet al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 3. Karachaliou N, Fernandez-Bruno M, Paulina Bracht JW, Rosell R. EGFR first- and second-generation TKIs—there is still place for them in EGFR-mutant NSCLC patients. Transl Cancer Res 2019;8:S23–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Yet al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med 2020;382:41–50. [DOI] [PubMed] [Google Scholar]
- 5. Rebuzzi SE, Alfieri R, La Monica S, Minari R, Petronini PG, Tiseo M. Combination of EGFR-TKIs and chemotherapy in advanced EGFR mutated NSCLC: review of the literature and future perspectives. Crit Rev Oncol Hematol 2020;146:102820. [DOI] [PubMed] [Google Scholar]
- 6. Tan CS, Kumarakulasinghe NB, Huang YQ, Ang YLE, Choo JR, Goh BCet al. Third generation EGFR TKIs: current data and future directions. Mol Cancer 2018;17:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer 2018;17:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Minari R, Bordi P, Tiseo M. Third-generation epidermal growth factor receptor-tyrosine kinase inhibitors in T790M-positive non-small cell lung cancer: review on emerged mechanisms of resistance. Transl Lung Cancer Res 2016;5:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Papdimitrakopoulou V, Wu Y, Han J, Ahn M, Ramalingam SS, John Tet al. Analysis of resistance mechanisms to osimertinib in patients with EGFR T790M advanced NSCLC from the AURA3 study. Ann Oncol 2018;29:viii741. [Google Scholar]
- 10. Ramalingam SS, Cheng Y, Zhou C, Ohe Y, Inamura F, Cho BCet al. Mechanisms of acquired resistance to first-line osimertinib: preliminary data from the phase III FLAURA study. Ann Oncol 2018;29:viii740. [Google Scholar]
- 11. Schoenfeld AJ, Chan JM, Kubota D, Sato H, Rizvi H, Daneshbod Yet al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res 2020;26:2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sequist LV, Han JY, Ahn MJ, Cho BC, Yu H, Kim SWet al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol 2020;21:373–86. [DOI] [PubMed] [Google Scholar]
- 13. Wu YL, Zhang L, Kim DW, Liu X, Lee DH, Yang JCet al. Phase Ib/II study of capmatinib (INC280) plus gefitinib after failure of epidermal growth factor receptor (EGFR) inhibitor therapy in patients with EGFR-mutated, MET factor-dysregulated non-small-cell lung cancer. J Clin Oncol 2018;36:3101–9. [DOI] [PubMed] [Google Scholar]
- 14. Rybrevant (amivantamab) [US package insert]. Horsham, PA: Janssen Biotech, Inc; 2021. [Google Scholar]
- 15. Bauml J, Cho BC, Park K, Lee KH, Cho EK, Kim D-Wet al. Amivantamab in combination with lazertinib for the treatment of osimertinib-relapsed, chemotherapy-naïve EGFR mutant (EGFRm) non-small cell lung cancer (NSCLC) and potential biomarkers for response. J Clin Oncol 2021;39:9006. [Google Scholar]
- 16. Han B, Yang L, Wang X, Yao L. Efficacy of pemetrexed-based regimens in advanced non-small cell lung cancer patients with activating epidermal growth factor receptor mutations after tyrosine kinase inhibitor failure: a systematic review. Onco Targets Ther 2018;11:2121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang CJ, Hung JY, Tsai MJ, Wu KL, Liu TC, Chou SHet al. The salvage therapy in lung adenocarcinoma initially harbored susceptible EGFR mutation and acquired resistance occurred to the first-line gefitinib and second-line cytotoxic chemotherapy. BMC Pharmacol Toxicol 2017;18:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Q, Zhang R, Yan H, Zhao P, Wu L, Wang Het al. Prognostic significance of HER3 in patients with malignant solid tumors. Oncotarget 2017;8:67140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scharpenseel H, Hanssen A, Loges S, Mohme M, Bernreuther C, Peine Set al. EGFR and HER3 expression in circulating tumor cells and tumor tissue from non-small cell lung cancer patients. Sci Rep 2019;9:7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kawano O, Sasaki H, Endo K, Suzuki E, Haneda H, Yukiue Het al. ErbB3 mRNA expression correlated with specific clinicopathologic features of Japanese lung cancers. J Surg Res 2008;146:43–8. [DOI] [PubMed] [Google Scholar]
- 21. Yonesaka K, Takegawa N, Watanabe S, Haratani K, Kawakami H, Sakai Ket al. An HER3-targeting antibody-drug conjugate incorporating a DNA topoisomerase I inhibitor U3-1402 conquers EGFR tyrosine kinase inhibitor-resistant NSCLC. Oncogene 2019;38:1398–409. [DOI] [PubMed] [Google Scholar]
- 22. Drago JZ, Modi S, Chandarlapaty S. Unlocking the potential of antibody-drug conjugates for cancer therapy. Nat Rev Clin Oncol 2021;18:327–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hashimoto Y, Koyama K, Kamai Y, Hirotani K, Ogitani Y, Zembutsu Aet al. A novel HER3-targeting antibody-drug conjugate, U3-1402, exhibits potent therapeutic efficacy through the delivery of cytotoxic payload by efficient internalization. Clin Cancer Res 2019;25:7151–61. [DOI] [PubMed] [Google Scholar]
- 24. Koganemaru S, Kuboki Y, Koga Y, Kojima T, Yamauchi M, Maeda Net al. U3-1402, a novel HER3-targeting antibody-drug conjugate, for the treatment of colorectal cancer. Mol Cancer Ther 2019;18:2043–50. [DOI] [PubMed] [Google Scholar]
- 25. Nakada T, Sugihara K, Jikoh T, Abe Y, Agatsuma T. The latest research and development into the antibody-drug conjugate, [fam-] trastuzumab deruxtecan (DS-8201a), for HER2 cancer therapy. Chem Pharm Bull 2019;67:173–85. [DOI] [PubMed] [Google Scholar]
- 26. Ogitani Y, Aida T, Hagihara K, Yamaguchi J, Ishii C, Harada Net al. DS-8201a, a novel HER2-targeting ADC with a novel DNA topoisomerase I inhibitor, demonstrates a promising antitumor efficacy with differentiation from T-DM1. Clin Cancer Res 2016;22:5097–108. [DOI] [PubMed] [Google Scholar]
- 27. Ueno S, Hirotani K, Abraham R, Blum S, Frankenberger B, Redondo-Muller Met al. U3-1402a, a novel HER3-targeting ADC with a novel DNA topoisomerase I inhibitor, demonstrates a potent tumor efficacy. Cancer Res 2017;77:3092.28377455 [Google Scholar]
- 28. Ogitani Y, Hagihara K, Oitate M, Naito H, Agatsuma T. Bystander killing effect of DS-8201a, a novel anti-human epidermal growth factor receptor 2 antibody-drug conjugate, in tumors with human epidermal growth factor receptor 2 heterogeneity. Cancer Sci 2016;107:1039–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Modi S, Saura C, Yamashita T, Park YH, Kim SB, Tamura Ket al. Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N Engl J Med 2020;382:610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shitara K, Bang YJ, Iwasa S, Sugimoto N, Ryu MH, Sakai Det al. Trastuzumab deruxtecan in previously treated HER2-positive gastric cancer. N Engl J Med 2020;382:2419–30. [DOI] [PubMed] [Google Scholar]
- 31. Haikala HM, Köhler J, Lopez T, Eser P, Xu M, Yu Cet al. EGFR inhibition enhances the cellular uptake and antitumor activity of the novel HER3 antibody drug conjugate U3-1402. Cancer Res 2020;80. Abstract 5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haikala HM, Janne PA. Thirty years of HER3: from basic biology to therapeutic interventions. Clin Cancer Res 2021;27:3528–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schoeberl B, Pace EA, Fitzgerald JB, Harms BD, Xu L, Nie Let al. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci Signal 2009;2:ra31. [DOI] [PubMed] [Google Scholar]
- 34. Ebert EBF, McCulloch T, Hansen KH, Linnet H, Sorensen B, Meldgaard P. Clearing of circulating tumour DNA predicts clinical response to first line tyrosine kinase inhibitors in advanced epidermal growth factor receptor mutated non-small cell lung cancer. Lung Cancer 2020;141:37–43. [DOI] [PubMed] [Google Scholar]
- 35. Ricciuti B, Jones G, Severgnini M, Alessi JV, Recondo G, Lawrence Met al. Early plasma circulating tumor DNA (ctDNA) changes predict response to first-line pembrolizumab-based therapy in non-small cell lung cancer (NSCLC). J Immunother Cancer 2021;9:e001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ebert EBF, McCulloch T, Holmskov Hansen K, Linnet H, Sorensen B, Meldgaard P. Clearing of circulating tumour DNA predicts clinical response to osimertinib in EGFR mutated lung cancer patients. Lung Cancer 2020;143:67–72. [DOI] [PubMed] [Google Scholar]
- 37. Long K, Suresh K. Pulmonary toxicity of systemic lung cancer therapy. Respirology 2020;25:72–9. [DOI] [PubMed] [Google Scholar]
- 38. Yonemori K, Masuda N, Takahashi S, Kogawa T, Nakayama T, Yamamoto Yet al. Single agent activity of U3-1402, a HER3-targeting antibody-drug conjugate, in HER3-overexpressing metastatic breast cancer: updated results from a phase I/II trial. Ann Oncol 2019;30:iii48. [Google Scholar]
- 39. Jackman D, Pao W, Riely GJ, Engelman JA, Kris MG, Janne PAet al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol 2010;28:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu HA, Johnson M, Steuer CE, Vigliotti M, Chen S, Kamai Yet al. Preliminary phase 1 results of U3-1402—a Novel HER3-targeted antibody-drug conjugate-in EGFR TKI-resistant EGFR-mutant NSCLC. J Thorac Oncol 2019;14:S336–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.