Abstract
Purpose:
Almost all cervical cancers are caused by human papillomavirus (HPV) and patients with advanced stage are at high risk for relapse. Circulating HPV DNA (HPV ctDNA) may serve as a residual tumor marker at the end of chemoradiation or to predict relapse during the follow-up period.
Experimental Design:
We analyzed serum samples from 94 HPV16- or HPV18-related CCs from the BioRAIDs prospective cohort. Samples were collected before and after treatment and during an 18-month follow-up period. Using digital droplet PCR (ddPCR), we assessed the relevance of circulating HPV E7 gene as a marker for residual disease compared to HPV integration site and PIK3CA mutations. Finally, the prognostic impact of circulating HPV E7 gene was assessed with its prediction value of relapse.
Results:
HPV E7 gene was the most sensitive tumor marker, superior to both HPV integration sites and PIK3CA mutations in serum. Circulating HPV DNA (HPV ctDNA) was detected in 63% (59/94) of patients, before treatment. HPV ctDNA detection in serum sample was associated with high FIGO stage (P = 0.02) and para-aortic lymph node involvement (P = 0.01). The level of HPV ctDNA was positively correlated with HPV copy number in the tumor (R = 0.39, P < 0.001). Complete clearance of HPV ctDNA by the end of treatment was significantly associated with a longer PFS (P < 0.0001). Patients with persistent HPV ctDNA in serum relapsed with a median time of 10 months (range, 2–15) from HPV ctDNA detection.
Conclusions:
HPV ctDNA detection is a useful marker to predict relapse in cervical cancer.
See related commentary by Wentzensen and Clarke, p. 5733
Translational Relevance.
HPV infection is responsible for almost all cervical cancers and circulating HPV DNA (HPV ctDNA) can be detected into blood of patients. Patients with advanced diseases have high risk to relapse and they are difficult to identify.
In the current study, we observed that HPV ctDNA can be detected before treatment by droplet-digital PCR (ddPCR) in a high proportion of patients with advanced stage (71% of stages III and IV) and persistence of HPV ctDNA after first-line therapy predicted relapse of cervical cancer.
The short turnaround time, high sensitivity, and limited cost of the detection of HPV ctDNA by ddPCR are compatible with clinical practice. This opens the possibility of monitoring patients with cervical cancer and identifying patients with high risk to relapse, allowing studies to evaluate the benefit of salvage surgery or innovative drug combinations for patients with residual HPV ctDNA after treatment.
Introduction
Human papillomavirus (HPV) infection is the etiologic agent of almost all cervical cancers, the second leading cause of gynecological cancer–related death in women worldwide (1, 2). Standard treatment consists of surgery for early-stage disease (stage FIGO ≤ IB2), while chemoradiation is recommended for patients with locally advanced disease (stage FIGO IB3–IIIC/IVA). For locally advanced stages, salvage hysterectomy for suspected residual disease after chemoradiation has been extensively debated. While it improves local control of the tumor, it remains associated with increased morbidity and no impact on improved overall survival was ever documented (3, 4). Salvage hysterectomy after full dose radiotherapy is therefore usually limited to patients with a very high suspicion for residual tumor. However, these patients with high risk of developing local relapse or distant metastases are not easily identifiable as their monitoring remains complex. Radiologic imaging results can be difficult to appreciate and pathologic reports of biopsies may document fibrosis and inflammation, leaving an uncertainty as to whether there are still viable tumor cells (5).
The detection of circulating tumor DNA (ctDNA), a fraction of cell-free circulating DNA released from tumor cells into the bloodstream, is now widely used for monitoring disease in various tumor types (6–8). Somatic mutations may thus serve as ctDNA markers and are commonly detected by digital PCR or NGS techniques (9–11). In patients with HPV-related tumors, circulating HPV DNA (HPV ctDNA) may be similarly detected in blood, either in episomal form or integrated into tumor DNA (12–16). Furthermore, HPV appears to be a stable tumor marker over time, because the same HPV type identified in a primary tumor is also observed in metastatic tissue (15).
The clinical value of HPV ctDNA has already been investigated in patients with HPV-related tumors (17–20). While we previously reported on the use of HPV ctDNA for monitoring the efficacy of immunotherapy or chemotherapy in metastatic anal cancer (21, 22), its use as a predictive and prognostic biomarker has recently been more and more reported by others, both in anal and head and neck cancers (23–25), as well as in a small prospective cohort of cervical cancer (26) and in a small retrospective cohort of metastatic cervical cancer (12).
In the current study from the European prospectively collected BioRAIDs (NCT02428842) cervical cancer dataset (27–31), the main objective was to investigate whether HPV ctDNA may be used for the early prediction of relapse after primary treatment in cervical cancer, similarly to other HPV-related cancers. Beyond HPV E7 detection in serum, we also assessed viral–cellular DNA junction sequences resulting from HPV integration into the tumor genome, together with PIK3CA mutations, a somatic marker of high prevalence in cervical cancer.
Materials and Methods
Patients and samples
The “European funded RAIDs Network (Rational Molecular Assessment and Innovative Drug Selection, www.raids-fp7.eu) prospectively monitored the multicenter trial BioRAIDs (NCT02428842), with the objectives to generate high quality samples and molecular assessments to stratify patient populations with cervical cancer and to identify molecular patterns associated with poor outcome.
Detailed eligibility criteria and treatments have been previously reported elsewhere (28, 29). Main eligibility criteria were: patients with stage IB2–IV disease (2018 FIGO staging system); all histologic subtypes (excluding neuro-endocrine type); no prior treatment for cervical cancer; signed informed consent. The patients (n = 419) included in the study received standard treatment, according to the standards of each center.
The study was approved by the respective local ethics committees: Comité pour la protection des personnes (CPP) Ile de France in France, The Serbian Medical Society Belgrade in Serbia, The Protocol Toetsingscommissie of the Antoni van Leeuwenhoek (PTC) in the Netherlands, The Medizinische Hochschule Hannover Ethik-kommission in Germany, The National Ethics Committee of Health Ministry of Republic of Moldova, Academy of Medical Sciences National Ethics Committee for medicines and medical devices in Romania. The study was conducted according to national requirements and the guidelines of the Declaration of Helsinki.
Tumor samples were collected at baseline before treatment and cryopreserved for molecular analysis. All samples were histologically reviewed at the time they were included into HPV testing. Only tumor samples with at least 30% of tumor cells were selected for the study.
Serum samples were collected at inclusion, at the end of treatment and during follow-up visits at 6, 12, and 18 months. Imaging was performed before treatment for all patients. Depending on the center, additional imaging may be available at the end of treatment and the follow-up visits.
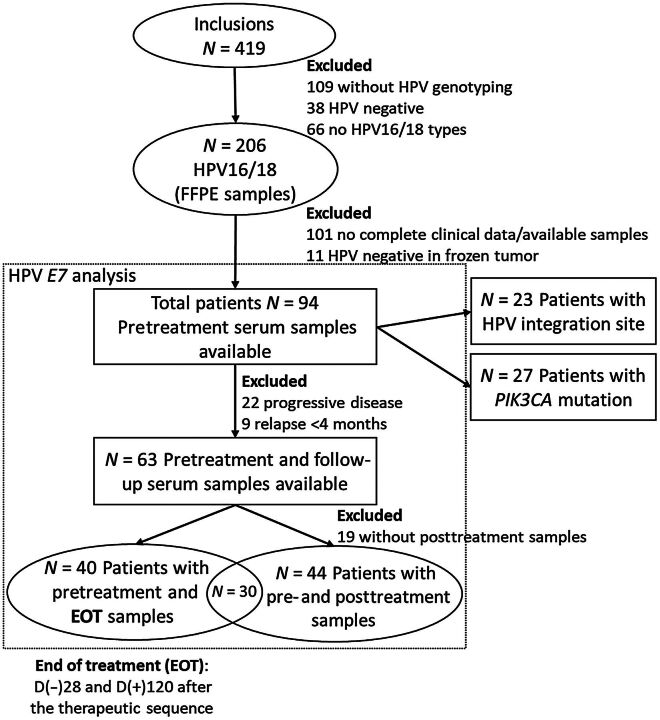
The current study focused on serum samples from 94 patients, selected for HPV16- or HPV18-positive CC, according to the workflow presented in the Fig. 1. Serum samples taken, at range 28 days before and 120 days after the date of the end of treatment (EOT), and 120 days before relapse, were considered as EOT samples.
Figure 1.
Workflow diagram for patient selection and ctDNA analysis.
HPV typing and integration site
HPV typing was previously performed as described previously (30), using the SPF10 primer set Inno-LiPA HPV genotyping extra line probe assay (Fujirebio Europe) according to the manufacturer's protocol. Among the 94 HPV16/18–related cases, 2 squamous cell carcinomas (SCC) and 2 adenocarcinomas were initially identified carrying multiple infections using INNO-LIPA in FFPE samples. One SCC case and 2 adenocarcinoma cases related to HPV16 and HPV18, while the other SCC case was associated with HPV16 and HPV73. However, HPV typing by ddPCR in DNA from frozen tumor confirmed only one HPV type among the three cases initially characterized as HPV16- and HPV18-related. Hence, these 3 cases were considered single-type for further study.
For the 23 patients with an HPV integration site into tumor DNA previously identified by the Capture-HPV method followed by NGS (27), we designed digital droplet PCR (ddPCR) assays to specifically amplify the viral-cellular DNA junction sequences. Primer and probe sequences are listed in the Supplementary Table S1.
HPV ctDNA quantification
Serum samples were stored at −80°C until DNA isolation. Circulating cell-free DNA (cfDNA) was isolated in duplicate from 200 μL of serum using the QIAamp Min Elute Virus Spin Kit (Qiagen), according to the manufacturer's instructions. Elution was performed in 25 μL of supplied elution buffer; eluates from the same sample were pooled and stored at −20°C until HPV ctDNA analysis.
ddPCR reactions were performed using the QX100 system (Bio-Rad Laboratories) according to the manufacturer's protocol. Briefly, 20 μL of MasterMix solution was prepared, containing 10 μL of 2× ddPCR SuperMix for probes without dUTP (Bio-Rad), relevant primers at 450 nmol/L each and relevant TaqMan probes at 250 nmol/L each (Applied Biosystems), DNA template (6 μL), and nuclease-free water. PCR reactions were run on a C1000 thermal cycler (Bio-Rad) using the following program: 95°C 10 minutes, 40 cycles of (94°C 30 seconds, 60°C 1 minute) 98°C 10 minutes. The concentration of HPV DNA copies was estimated with the QuantaSoft v1.7.4 software.
Three replicates were performed to detect HPV16, HPV18, or HPV integration site using HPV16 E7, HPV18 E7, or viral-cellular junction specific primers and TaqMan probes (Supplementary Table S1), multiplexed with a commercial human ddPCR assay targeting RPP30 gene (dHSaCP2500350, Bio-Rad Laboratories), used as human DNA reference. For each patient, genomic DNA from frozen tumor was tested as positive control and a triplicate of no DNA was tested for each plate.
According to our previous findings, serum samples were considered HPV-positive in presence of at least three droplets containing HPV amplicons. A sample was considered HPV-negative when fewer than 3 or no droplets containing HPV amplicons were detected, while more than 500 copies/mL were detected for RPP30 human reference gene. Finally, HPV ctDNA concentration was expressed in copies/mL of serum.
PIK3CA mutation detection
PIK3CA mutation screening in tumor tissue was performed by paired-end whole exome sequencing and paired-end targeted gene panel sequencing as previously described (30).
Serum samples used for PIK3CA analysis were from the same aliquots used for HPV analysis. Circulating cell-free DNA (cfDNA) was isolated from 1 to 2 mL serum with the QIAamp Circulating Nucleic Acid kit (Qiagen) according to the manufacturer's manual. This cfDNA was isolated into 20 μL elution buffer. PIK3CA ctDNA analysis was performed with TaqMan p.E542K, p.E545K, and p.H1047R-specific assays and the QuantStudio 3D Digital PCR system (Thermo Fisher Scientific) according to the manufacturer's specifications. Reaction mixtures, including serum DNA and QuantStudio 3D Digital PCR Master Mix, were loaded on digital PCR chips with 20,000 wells, and cycled under standard conditions: 96°C for 10 minutes, 40 cycles of (60°C 2 minutes, 98°C 30 seconds). QuantStudio 3D analysisSuite determined the proportion of mutant and wild-type templates, and details are given in article by Vitale and colleagues (32).
Tumoral HPV copy number
HPV copy number in tumor cells was determined by ddPCR, using the same specific HPV and RPP30 assays as in HPV ctDNA quantification experiment. Experiments were performed with 5 ng of tumor DNA isolated from frozen tissues, in duplicates for each sample. The RRP30 gene was used as a reference for two copies of DNA sequences per cell. The value retained as corresponding to HPV copy number in tumor cells was twice that of the ratio HPVE7/RPP30 provided by the ddPCR experiment.
Statistical analysis
This report was written in accordance with REMARK criteria (33). For this study, no prespecified power was calculated. Fisher exact test was used for categorical variables and Spearman correlation test was used for continuous variables. Wilcoxon matched pairs signed rank test was used to compare HPV E7 ctDNA levels, HPV integration sites, and PIK3CA assays. Survival curves were compared with the use of an unstratified log-rank test, and HR with 95% confidence interval (CI) was calculated using a univariable and a multivariable Cox model including clinical characteristics such as age, FIGO stage, lymph node involvement, and HPV type. Progression-free survival (PFS) was defined as the time from the start of treatment to tumor recurrence or death. Statistical analyses and figures were performed using GraphPad Prism software version v-6–07 (GraphPad software) and R software version 4.0.2. All statistical tests were two-tailed, and P values < 0.05 were considered statistically significant.
Access to data
All data are available upon request.
Results
Patient characteristics
Ninety-four patients, from the BioRAIDs European study, with a documented HPV16- or HPV18-related tumor were included in the present study. The clinical and biological characteristics as well as their association with detectable HPV ctDNA before treatment are presented in Table 1 and Supplementary Table S2. Briefly, the median age of the patients was 50 years (range, 24–80); the main histologic tumor type was SCC (87%). HPV16 was detected in 81% of the tumors.
Table 1.
Patient clinical and biological characteristics and comparison between patients with detectable versus undetectable HPV ctDNA.
| Characteristics | All patients | Detectable HPV ctDNA at baseline | Undetectable HPV ctDNA at baseline | |
|---|---|---|---|---|
| Total | n = 94 | n = 59 | n = 35 | P |
| Age (in years) | ||||
| Median | 50 (24–80) | 50 (24–80) | 51 (24–78) | |
| FIGO stage (2018) | ||||
| I | 9 (9%) | 4 (7%) | 5 (14%) | 0.02(I/II vs III/IV) |
| II | 17 (18%) | 7 (12%) | 10 (29%) | |
| III | 58 (62%) | 39 (66%) | 19 (54%) | |
| IV | 10 (11%) | 9 (15%) | 1 (3%) | |
| Lymph node | ||||
| (N0) | 32 (34%) | 16 (27%) | 16 (46%) | 0.08 |
| (N+) | 62 (66%) | 43 (73%) | 19 (54%) | |
| Para-aortic lymph node | ||||
| (N0) | 72 (77%) | 40 (68%) | 32 (91%) | 0.01 |
| (N+) | 22 (23%) | 19 (32%) | 3 (9%) | |
| Pelvic lymph node | ||||
| (N0) | 34 (36%) | 18 (31%) | 16 (46%) | 0.18 |
| (N+) | 60 (64%) | 41 (69%) | 19 (54%) | |
| Histology | ||||
| SCC | 82 (87%) | 52 (88%) | 30 (86%) | 0.76(SCC vs ADK+ASC) |
| Adenocarcinoma | 10 (11%) | 6 (10%) | 4 (11%) | |
| ASC | 2 (2%) | 1 (2%) | 1 (3%) | |
| HPV type | ||||
| HPV16 | 76 (81%) | 47 (80%) | 29 (83%) | 0.79 |
| HPV18 | 18 (19%) | 12 (20%) | 6 (17%) | |
| HPV copy number | ||||
| <5 copies/cell | 32 (34%) | 13 (22%) | 19 (64%) | 0.003 |
| ≥5 copies/cell | 62 (66%) | 46 (78%) | 16 (46%) | |
| Initial treatment | Not tested | |||
| Surgery | 19 (20%) | 9 (15%) | 10 (28%) | |
| Chemoradiotherapy | 61 (65%) | 39 (66%) | 22 (63%) | |
| Neo-chemotherapy | 14 (15%) | 11 (19%) | 3 (9%) | |
Abbreviations: ADK, adenocarcinoma; ASC, adenosquamous carcinoma; P, Fisher test; SCC, squamous cell carcinoma.
Almost three quarters of the patients (73%) presented with a FIGO stage III or IV tumor type. Sixty-two patients (66%) had positive lymph nodes (by pathology and/or NMR imaging), predominantly in the pelvis (64%). Most patients (65%) were treated by first line chemo-radiation, 20% underwent primary surgery and 15% received neoadjuvant chemotherapy (Table 1). Median follow-up was 16.7 months and 41% (39/94) patients relapsed during the planned 18-month follow-up period.
Serum had been routinely collected at baseline for all patients (Fig. 1). Additional serum samples collected after treatment were available in 72 patients (77%). Among them, 40 patients had a serum sample taken at the end of treatment and 44 patients had at least one serum sample collected during surveillance (Fig. 1). In 6 patients, serum samples were available at the time of relapse. In this cohort of 94 CC, HPV integration sites and PIK3CA mutations had been previously identified in the tumor tissue of 23 and 27 patients respectively (Fig. 1). Patient characteristics of both subgroups are detailed in Supplementary Table S3.
HPV ctDNA detection before treatment
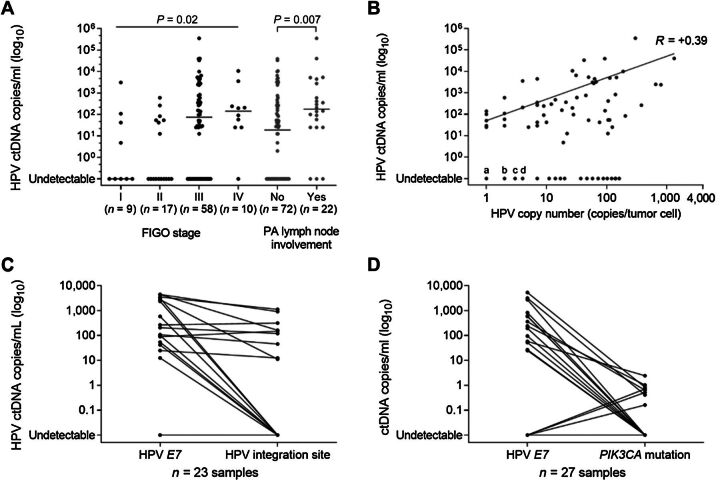
Using E7 gene sequence probes, HPV ctDNA was successfully detected before treatment in serum samples of 63% (59/94) patients (Table 1). The median level of HPV ctDNA was 42 copies/mL serum (range, 0–353,750) in the whole cohort, while the median level of the housekeeping gene, RPP30, was 23,355 copies/mL serum (range, 667–514,167; Supplementary Fig. S1). Among the 59 patients with detectable HPV ctDNA, the median level was 263 copies/mL serum with an interquartile range (IQR) of 53–2,916. There was a positive significant association between the detection of HPV ctDNA and tumor stage (HPV ctDNA detection in 42% (11/26) of stage I–II versus 70% (48/68) of stage III–IV; P = 0.02, Fisher test) and histologically proven para-aortic lymph node involvement [HPV ctDNA detection in 56% (40/72) of node negative versus 86% (19/22) of node positive, P = 0.01, Fisher test; Table 1; Fig. 2A]. In addition, we observed a positive correlation between serum HPV ctDNA level and HPV copy number in tumor cells (R = +0.39, P < 0.001, Spearman; Fig. 2B). There was no significant association between the detection of HPV ctDNA and other clinical characteristics such as patient age, histology, or HPV type.
Figure 2.
ctDNA detection by droplet digital PCR before treatment. A, HPV E7 ctDNA levels according to FIGO stages (P = Kruskal–Wallis test) and para-aortic (PA) lymph node status (P = Mann–Whitney test). B, Positive correlation between HPV E7 ctDNA level and tumor HPV E7 copy number. Spearman correlation r = 0.39 (P < 0.001). For undetectable HPV ctDNA cases, the points “a,” “b,” “c,” and “d” refer to 5, 6, 3, and 5 tumors, respectively. C, HPV ctDNA levels according to HPV E7 gene and HPV integration site (n = 23). D, ctDNA detection according to HPV E7 gene and PIK3CA mutation (n = 27).
ctDNA detection by HPV integration site and PIK3CA mutation
Furthermore, in the serum samples of a subgroup of 23 patients, the detection rates of HPV E7 gene and HPV integration site were compared. The results showed that the HPV integration site was a significantly less sensitive ctDNA marker than serum HPV E7 (P = 0.001, Wilcoxon matched pair signed rank test), with a detection rate of 39% (9/23), while HPV E7 detection rate reached 70% (16/23; Fig. 2C).
Finally, the detection rates of HPV E7 and PIK3CA mutations were compared in serum samples from the 27 patients harboring a PIK3CA mutation in their tumor. HPV E7 detection was significantly more sensitive than PIK3CA mutation (P < 0.001 Wilcoxon matched pair signed rank test) with detection rates of 52% (14/27) and 30% (8/27), respectively (Fig. 2D).
Prognostic value of persistent HPV E7 detection in serum at end of treatment
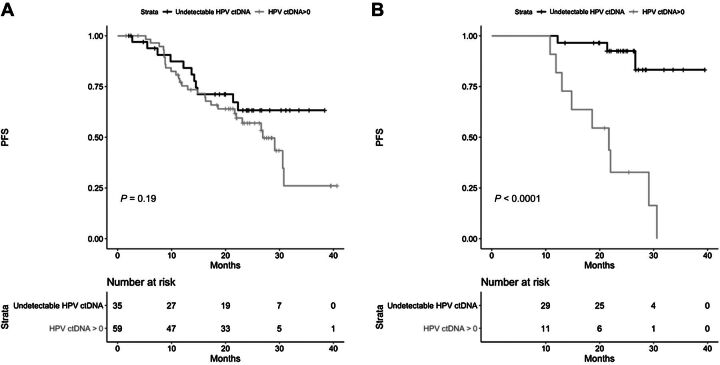
In the sequel, we defined the binary covariate HPV ctDNA detection with value 1 corresponding to HPV E7 positive and value 0 for undetectable HPV E7. Among the 94 patients, 40 experienced PFS. In a univariable Cox landmark regression model, HPV ctDNA detection at baseline was not associated with PFS (HR = 1.59; 95% CI, 0.79–3.20; P = 0.19; Fig. 3A). No significant association was observed with multivariable landmark analysis (HR = 1.08; 95% CI, 0.51–2.30; P = 0.841).
Figure 3.
Progression-free survival of patients according to the positivity of HPV ctDNA detection. A, Before treatment (n = 94). B, At the end of treatment (n = 40).
To estimate the effect of HPV ctDNA detection at the end of the treatment, we used the landmark approach considering only patients at risk of PFS at the end of their treatment. In a univariable Cox landmark regression model, HPV ctDNA detection was associated with a short PFS (HR = 10.95; 95% CI, 2.94–40.7; P < 0.0001; Fig. 3B). Of note 81% (9/11) of the patients with a positive detection experienced a PFS.
Multivariable landmark analysis also showed significant association between HPV ctDNA detection at the end of treatment and PFS (HR = 14.25; 95% CI, 3.10–61.57; P = 0.001).
Diagnosis of relapse by persistence of HPV E7 in serum during follow-up
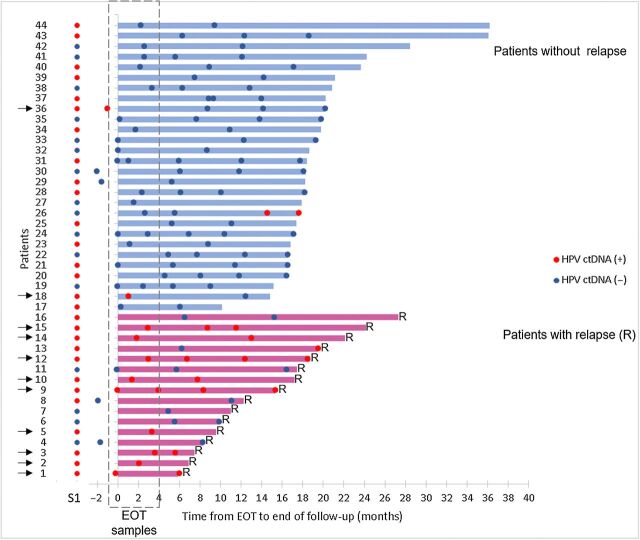
Forty-four of 94 patients had a longitudinal follow-up, among whom 16 patients (36%) did relapse during the follow-up period.
Serum samples from patients with undetectable HPV ctDNA at end of treatment remained HPV negative during follow-up with one exception, patient #26 who presented a serum conversion at 14 months, remaining positive at 17 months (Fig. 4). There was no clinical sign of recurrence at this last follow-up when the patient went off study and could not be traced thereafter. The negative predictive value of undetectable ctDNA for cervical cancer relapse was 95%.
Figure 4.
HPV ctDNA dynamics from the EOT to the end of follow-up. Each line corresponds to a patient (n = 44); patients without relapse and with relapse are identified in blue (top of the figure) and red (bottom), respectively. The length of the lines (top) corresponds to the duration of follow-up. Red ( ) and blue (
) and blue ( ) correspond to HPV-positive and HPV-negative serum samples, respectively. Serum samples in dashed area correspond to EOT samples. Patients with HPV ctDNA in their EOT samples are indicated by a black arrow. Diagnosis of relapse corresponds to the end of the line. S1, serum sample before treatment; R, relapse.
) correspond to HPV-positive and HPV-negative serum samples, respectively. Serum samples in dashed area correspond to EOT samples. Patients with HPV ctDNA in their EOT samples are indicated by a black arrow. Diagnosis of relapse corresponds to the end of the line. S1, serum sample before treatment; R, relapse.
Among the 11 patients with residual HPV ctDNA at end of treatment, 8 could be followed up longitudinally. HPV ctDNA remained detectable in 6 of 8 patients (75%) while there was no clinical sign of recurrence. All 6 patients (patients #3, #9, #10, #12, #14, and #15) relapsed eventually (Fig. 4). HPV ctDNA was detected on average 10 months before a clinical diagnosis of relapse (range, 2–15 months).
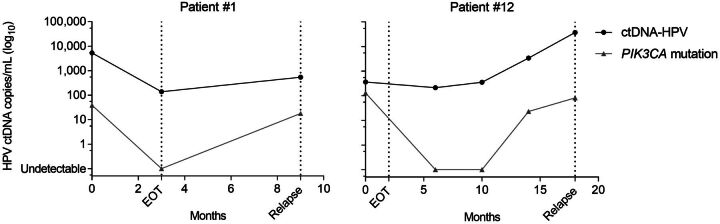
Serum samples at the time of relapse were available for 6 patients (patients #1, #4, #6, #9, #12, and #13), and HPV ctDNA was detected in 4 of 6 patients (patients #1, #9, #12, and #13). It is interesting to note that in these 4 patients, HPV ctDNA detection remained consistently positive throughout the follow-up, before and after treatment. Among these 4 patients, two (patients #1, #12) also harbored a somatic PIK3CA mutation, which became undetectable by the end of the treatment, contrary to HPV ctDNA (Fig. 5). The PIK3CA mutation in serum became detectable again at the time of relapse in both patients. In patient #12, the PIK3CA mutation was detected 6 months before clinical diagnosis of relapse.
Figure 5.
ctDNA dynamics according to HPV E7 (black line) and PIK3CA mutation (gray line).
Discussion
In this European prospective cohort of cervical cancers, we confirmed (i) that HPV E7 sequences are a sensitive marker to detect ctDNA, and (ii) that HPV ctDNA detection after treatment is strongly pronostic of an early recurrence.
Our 63% serum detection rate by ddPCR before treatment is consistent with previous studies that reported HPV detection rates of 56% to 100%, using HPV E7 gene primers in primary cervical cancers (14, 26, 34), or in other HPV-related cancers (14, 17, 18, 20).
HPV-related tumor cells usually contain many copies (up to hundreds) of the HPV genome, in episomal form as well as integrated into tumor DNA. Hence, the release of multiple HPV copies from dying tumor cells into the bloodstream, contributes to an increased sensitivity of the detection method compared with that of a single point mutation (one copy/tumor cell). In addition, HPV E7 ctDNA detection rate is also superior to that of HPV integration site (Fig. 2C) because it includes both the episomal and the integrated HPV (16, 27). This is consistent with another retrospective study of 21 serum samples showing a poor detection rate (33%) of HPV integration site when analyzed by NGS before tumor resection (15). Hence, our results prioritize ctDNA detection by using HPV E7 sequences as a predictive biomarker of relapse when compared with HPV integration site or PIK3CA point mutations (16, 27, 30). However, it is well-known that HPV integration site is unique for each tumor and can be considered as a highly specific tumor marker (35, 36). Depending on center resources, circulating HPV integration site screening may be performed and help for detection of relapse.
A main finding of the present prospective study is that patients with residual HPV ctDNA at the end of treatment have a high risk of relapse. Only 2 of 11 patients with a positive HPV ctDNA sample at the end of the treatment were able to clear the HPV ctDNA sequences in the following sample and did not relapse by the end of follow-up. Similar findings have recently been described in another study in HPV-associated oropharyngeal cancer (25). Transient residual HPV ctDNA after treatment was observed for a subgroup of patients who cleared HPV ctDNA in the following sample and did not relapse (follow-up up to 5 years), suggesting that 2 consecutive serum samples may be required to predict relapse after treatment (25).
The ability to anticipate the presence of residual/persistent diseases up to 15 months before a diagnosis of relapse has a high clinical value (3, 4). In particular, it may enable to propose a salvage surgery to patients by confirming the likelihood of residual pelvic disease after chemoradiation (37, 38). The early detection of persistent/recurrent disease is particularly important in irradiated tissues to minimize the radicality of the surgery which can go as far as total exenteration if the recurrence invades neighboring organs. Similar conclusions were reported in head and neck cancers, the immediate posttreatment HPV ctDNA detection being helpful for the post-radiotherapy management of patients with HPV-related tumors (39, 40).
A new prospective clinical trial comparing the value of imaging versus repeated HPV serum detection by integrating sequential ddPCR would be useful to better define the role and the cost effectiveness of these tests. Because the prognosis of patients with detectable HPV ctDNA at the end of the chemoradiation is poor, this population may benefit from innovative drugs or maintenance therapy, in particular immunotherapy. Monitoring of HPV ctDNA could help future clinical trial to target a high-risk population.
The analysis was focused on patients with HPV16- and HPV18-related tumors, which represent approximately 70% of patients with cervical cancer. Further analyses, including patients with other HPV-type tumors would be useful. Moreover, because the cohort was led at a European level, it was not possible to pursue the follow-up beyond 18 months, as initially planned in the RAIDs consortium. For this reason, it was not possible to obtain additional information on patient (#26) with two consecutive HPV positive serum samples at the end of her follow-up period.
In conclusion, our study confirms that the HPV E7 gene is a sensitive and a suitable tumor marker for treatment monitoring and long-term follow-up of patients with cervical cancer. To our knowledge, this is the first large prospective study to show that detection of HPV ctDNA by ddPCR may predict relapse in cervical cancer at the end of first-line therapy. Further studies are needed to confirm the clinical value of HPV ctDNA for an early detection of residual disease/relapse and to evaluate the role/purpose of salvage surgery as compared with that of innovative drug combinations for patients with residual HPV ctDNA after treatment. Two studies, Circa HPV (NCT03739775) and the PEVO basket trial (NCT04357873), are actually ongoing and will allow a comparison between imaging and HPV ctDNA and evaluate its predictive value for relapse detection and for efficacy of innovative drug combination (41).
Authors' Disclosures
M. Popovic reports grants from FP7 project Rational molecular Assessment and Innovative Drug Selection - RAIDs during the conduct of the study. H. von der Leyen reports grants from EU FP7 program: "PRIORITY" during the conduct of the study. C. Le Tourneau reports personal fees from MSD, BMS, Merck Serono, Nanobiotix, Rakuten, Seattle Genetics, AstraZeneca, Roche, GSK, and Celgene outside the submitted work. M. Kamal reports grants from European commission and Agence de la Recherche Contre le Cancer during the conduct of the study; personal fees from F. Hoffmann-La Roche Ltd outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Figure S1. Serum cell free DNA (cfDNA) detection by droplet digital PCR before treatment. cfDNA levels before treatment according to HPV E7 gene (tumor marker) and RPP30 gene (total cell free DNA marker).
Supplemental Table 1: ddPCR assays targeting the HPV E7 genes and the HPV integration sites: primer and probe sequences, amplicon sizes and annealing temperatures.
Table S2. HPV ctDNA detection according to HPV copy number and histological type.
Table S3. Clinical and biological characteristics of patients with HPV integration site and PIK3CA mutations.
Acknowledgments
We thank the RAIDs consortium (http://www.raids-fp7.eu/consortium): Gemma Kenter, Pierre Fumoleau, Aljosa Mandic, Nina Samet, Choumouss Kamoun, Windy Luscap-Rondoff, Sebastien Armanet, Alexandra Rohel, Souhir Neffati, Sinette Ngoumou Mabiala, Coralie Errera, Marius Craina, Madalin Margan, Sanne Samuels, Henry Zijlmans, Peter Hillemanns, Sorin Dema, Alis Dema, Goran Malenkovic, Branislav Djuran, Anne Floquet, Frédéric Guyon, Pierre Emmanuel Colombo, Michel Fabbro, Christine Kerr, Eleonor Rivin del Campo, Charles Coutant, Frédéric Marchal, Nathalie Mesgouez-Nebout, Jean Guillaume Feron, Philippe Morice, Eric Deutsch, Pauline Wimberger, Jean-Marc Classe, Mathieu Minsat, Istvan Nagy, Attila Kerezt, Balazs Balint, Nicolas de Saint-Jorre, Alexia Savignoni, Patricia Tresca, Noreen Gleeson, Philippe Hupe, Sergio Roman-Roman, Emmanuel Barillot, Fanny Coffin, Bastiaan Nuijen, Alexandre Boissonnas, Marc Billaud, Laurence Lafanechere, Kirsten Ruigrok, Andrea Slocker, Michele Mondini, Maud Bossard, Sjoerd Rodenhuis, Rene Medema, Anika Havemeier, Thomas Fink, Amelie Michon, Christine Kubiak, Judit Cseklye, Dora Latinovics, Peter Bihari, Isabel Brito, Bérengère Ouine, Leanne De Koning, Vincent Puard, Elaine Del Nery, Jos Beijnen, Dominique Koensgen, Daniela Bruennert, Slavica Knezevic, Milos Lucic, and Natalja ter Haar for their precious help in the conduct of the RAIDs project.
European Union's Seventh Program for research, technological development and demonstration (agreement N°304810), the Fondation ARC pour la recherche contre le cancer.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
E. Jeannot: Conceptualization, formal analysis, writing–original draft, writing–review and editing. A. Latouche: Formal analysis, methodology, writing–original draft, writing–review and editing. C. Bonneau: Writing–original draft, writing–review and editing, acquisition of data. M.-A. Calméjane: Writing–original draft, writing–review and editing, acquisition of data. C. Beaufort: Writing–original draft, writing–review and editing. K. Ruigrok-Ritstier: Writing–original draft, writing–review and editing, acquisition of data. G. Bataillon: Writing–original draft, writing–review and editing, acquisition of data. L. Larbi Chérif: Writing–original draft, writing–review and editing, acquisition of data. C. Dupain: Resources, writing–original draft, writing–review and editing. C. Lecerf: Resources, writing–review and editing, acquisition of data. M. Popovic: Writing–original draft, writing–review and editing, acquisition of data. A. de la Rochefordière: Writing–original draft, writing–review and editing, acquisition of data. F. Lecuru: Writing–original draft, writing–review and editing, acquisition of data. V. Fourchotte: Writing–original draft, writing–review and editing. E.S. Jordanova: Writing–original draft, writing–review and editing, acquisition of data. H. von der Leyen: Writing–original draft, writing–review and editing, acquisition of data. C. Tran-Perennou: Writing–original draft, writing–review and editing, acquisition of data. M.-E. Legrier: Resources, writing–original draft, writing–review and editing, acquisition of data. S. Dureau: Resources, writing–original draft, writing–review and editing, acquisition of data. L. Raizonville: Resources, writing–original draft, writing–review and editing, acquisition of data. D. Bello Roufai: Writing–original draft, writing–review and editing, acquisition of data. C. Le Tourneau: Supervision, writing–original draft, writing–review and editing. I. Bièche: Supervision, writing–original draft, writing–review and editing. R. Rouzier: Supervision, writing–original draft, writing–review and editing. E.M.J.J. Berns: Supervision, writing–original draft, writing–review and editing. M. Kamal: Formal analysis, supervision, writing–original draft, writing–review and editing. S. Scholl: Formal analysis, supervision, writing–original draft, writing–review and editing.
References
- 1. de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer 2017;141:664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, et al. Global burden of human papillomavirus and related diseases. Vaccine 2012;30:F12–23. [DOI] [PubMed] [Google Scholar]
- 3. Keys HM, Bundy BN, Stehman FB, Okagaki T, Gallup DG, Burnett AF, et al. Radiation therapy with and without extrafascial hysterectomy for bulky stage IB cervical carcinoma: a randomized trial of the Gynecologic Oncology Group. Gynecol Oncol 2003;89:343–53. [DOI] [PubMed] [Google Scholar]
- 4. Marth C, Landoni F, Mahner S, McCormack M, Gonzalez-Martin A, Colombo N, et al. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;28:iv72–83. [DOI] [PubMed] [Google Scholar]
- 5. Kim JY, Byun SJ, Kim YS, Nam JH. Disease courses in patients with residual tumor following concurrent chemoradiotherapy for locally advanced cervical cancer. Gynecol Oncol 2017;144:34–9. [DOI] [PubMed] [Google Scholar]
- 6. Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Babayan A, Pantel K. Advances in liquid biopsy approaches for early detection and monitoring of cancer. Genome Med 2018;10:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cabel L, Proudhon C, Mariani P, Tzanis D, Beinse G, Bieche I, et al. Circulating tumor cells and circulating tumor DNA: What surgical oncologists need to know? Eur J Surg Oncol 2017;43:949–62. [DOI] [PubMed] [Google Scholar]
- 9. Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [DOI] [PubMed] [Google Scholar]
- 10. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kang Z, Stevanovic S, Hinrichs CS, Cao L. Circulating cell-free DNA for metastatic cervical cancer detection, genotyping, and monitoring. Clin Cancer Res 2017;23:6856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campitelli M, Jeannot E, Peter M, Lappartient E, Saada S, de la Rochefordiere A, et al. Human papillomavirus mutational insertion: specific marker of circulating tumor DNA in cervical cancer patients. PLoS One 2012;7:e43393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeannot E, Becette V, Campitelli M, Calmejane MA, Lappartient E, Ruff E, et al. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. J Pathol Clin Res 2016;2:201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carow K, Golitz M, Wolf M, Hafner N, Jansen L, Hoyer H, et al. Viral-cellular DNA junctions as molecular markers for assessing intra-tumor heterogeneity in cervical cancer and for the detection of circulating tumor DNA. Int J Mol Sci 2017;18:2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holmes A, Lameiras S, Jeannot E, Marie Y, Castera L, Sastre-Garau X, et al. Mechanistic signatures of HPV insertions in cervical carcinomas. NPJ Genom Med 2016;1:16004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cabel L, Jeannot E, Bieche I, Vacher S, Callens C, Bazire L, et al. Prognostic impact of residual HPV ctDNA detection after chemoradiotherapy for anal squamous cell carcinoma. Clin Cancer Res 2018;24:5767–71. [DOI] [PubMed] [Google Scholar]
- 18. Lee JY, Garcia-Murillas I, Cutts RJ, De Castro DG, Grove L, Hurley T, et al. Predicting response to radical (chemo)radiotherapy with circulating HPV DNA in locally advanced head and neck squamous carcinoma. Br J Cancer 2017;117:876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanna GJ, Supplee JG, Kuang Y, Mahmood U, Lau CJ, Haddad RI, et al. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann Oncol 2018;29:1980–6. [DOI] [PubMed] [Google Scholar]
- 20. Chera BS, Kumar S, Beaty BT, Marron D, Jefferys S, Green R, et al. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin Cancer Res 2019;25:4682–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabel L, Bidard FC, Servois V, Cacheux W, Mariani P, Romano E, et al. HPV circulating tumor DNA to monitor the efficacy of anti-PD-1 therapy in metastatic squamous cell carcinoma of the anal canal: A case report. Int J Cancer 2017;141:1667–70. [DOI] [PubMed] [Google Scholar]
- 22. Bernard-Tessier A, Jeannot E, Guenat D, Debernardi A, Michel M, Proudhon C, et al. Clinical validity of HPV circulating tumor DNA in advanced anal carcinoma: an ancillary study to the epitopes-HPV02 Trial. Clin Cancer Res 2019;25:2109–15. [DOI] [PubMed] [Google Scholar]
- 23. Veyer D, Wack M, Mandavit M, Garrigou S, Hans S, Bonfils P, et al. HPV circulating tumoral DNA quantification by droplet-based digital PCR: A promising predictive and prognostic biomarker for HPV-associated oropharyngeal cancers. Int J Cancer 2020;147:1222–7. [DOI] [PubMed] [Google Scholar]
- 24. Lee JY, Cutts RJ, White I, Augustin Y, Garcia-Murillas I, Fenwick K, et al. Next generation sequencing assay for detection of circulating HPV DNA (cHPV-DNA) in Patients undergoing radical (Chemo)Radiotherapy in anal squamous cell carcinoma (ASCC). Front Oncol 2020;10:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chera BS, Kumar S, Shen C, Amdur R, Dagan R, Green R, et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-Associated oropharyngeal cancer. J Clin Oncol 2020;38:1050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han K, Leung E, Barbera L, Barnes E, Croke J, Di Grappa MA, et al. Circulating human papillomavirus DNA as a biomarker of response in patients with locally advanced cervical cancer treated with definitive chemoradiation. JCO Precision Oncology 2018;2:1–8. [DOI] [PubMed] [Google Scholar]
- 27. Kamal M, Lameiras S, Deloger M, Morel A, Vacher S, Lecerf C, et al. Human papilloma virus (HPV) integration signature in Cervical Cancer: identification of MACROD2 gene as HPV hot spot integration site. Br J Cancer 2021;124:777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ngo C, Samuels S, Bagrintseva K, Slocker A, Hupe P, Kenter G, et al. From prospective biobanking to precision medicine: BIO-RAIDs - an EU study protocol in cervical cancer. BMC Cancer 2015;15:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Samuels S, Balint B, von der Leyen H, Hupe P, de Koning L, Kamoun C, et al. Precision medicine in cancer: challenges and recommendations from an EU-funded cervical cancer biobanking study. Br J Cancer 2016;115:1575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scholl S, Popovic M, de la Rochefordiere A, Girard E, Dureau S, Mandic A, et al. Clinical and genetic landscape of treatment naive cervical cancer: Alterations in PIK3CA and in epigenetic modulators associated with sub-optimal outcome. EBioMedicine 2019;43:253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scholl SM, Beal J, de Koning L, Girard E, Popovic M, de la Rochefordiere A, et al. Genetic markers and phosphoprotein forms of beta-catenin pbeta-Cat552 and pbeta-Cat675 are prognostic biomarkers of cervical cancer. EBioMedicine 2020;61:103049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vitale SR, Groenendijk FH, van Marion R, Beaufort CM, Helmijr JC, Dubbink HJ, et al. TP53 mutations in serum circulating cell-free tumor DNA As Longitudinal biomarker for high-grade serous ovarian cancer. Biomolecules 2020;10:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 2005;97:1180–4. [DOI] [PubMed] [Google Scholar]
- 34. Cheung TH, Yim SF, Yu MY, Worley MJ Jr, Fiascone SJ, Chiu RWK, et al. Liquid biopsy of HPV DNA in cervical cancer. J Clin Virol 2019;114:32–36. [DOI] [PubMed] [Google Scholar]
- 35. Bodelon C, Untereiner ME, Machiela MJ, Vinokurova S, Wentzensen N. Genomic characterization of viral integration sites in HPV-related cancers. Int J Cancer;139:2001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vinokurova S, Wentzensen N, Einenkel J, Klaes R, Ziegert C, Melsheimer P, et al. Clonal history of papillomavirus-induced dysplasia in the female lower genital tract. J Natl Cancer Inst 2005;97:1816–21. [DOI] [PubMed] [Google Scholar]
- 37. Berek JS, Howe C, Lagasse LD, Hacker NF. Pelvic exenteration for recurrent gynecologic malignancy: survival and morbidity analysis of the 45-year experience at UCLA. Gynecol Oncol 2005;99:153–9. [DOI] [PubMed] [Google Scholar]
- 38. Li H, Wu X, Cheng X. Advances in diagnosis and treatment of metastatic cervical cancer. J Gynecol Oncol 2016;27:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanaka H, Takemoto N, Horie M, Takai E, Fukusumi T, Suzuki M, et al. Circulating tumor HPV DNA complements PET-CT in guiding management after radiotherapy in HPV-related squamous cell carcinoma of the head and neck. Int J Cancer 2021;14:995–1005. [DOI] [PubMed] [Google Scholar]
- 40. Rutkowski TW, Mazurek AM, Snietura M, Hejduk B, Jedrzejewska M, Bobek-Billewicz B, et al. Circulating HPV16 DNA may complement imaging assessment of early treatment efficacy in patients with HPV-positive oropharyngeal cancer. J Transl Med 2020;18:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Guillebon E, Jimenez M, Mazzarella L, Betsou F, Stadler P, Petak I, et al. Combining immunotherapy with an epidrug in squamous cell carcinomas of different locations: rationale and design of the PEVO basket trial. ESMO Open 2021;6:100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Serum cell free DNA (cfDNA) detection by droplet digital PCR before treatment. cfDNA levels before treatment according to HPV E7 gene (tumor marker) and RPP30 gene (total cell free DNA marker).
Supplemental Table 1: ddPCR assays targeting the HPV E7 genes and the HPV integration sites: primer and probe sequences, amplicon sizes and annealing temperatures.
Table S2. HPV ctDNA detection according to HPV copy number and histological type.
Table S3. Clinical and biological characteristics of patients with HPV integration site and PIK3CA mutations.