Abstract
Purpose:
Circulating tumor DNA (ctDNA) enables personalized treatment strategies in oncology by providing a noninvasive source of clinical biomarkers. In patients with low ctDNA abundance, tumor-naïve methods are needed to facilitate clinical implementation. Here, using locoregionally confined head and neck squamous cell carcinoma (HNSCC) as an example, we demonstrate tumor-naïve detection of ctDNA by simultaneous profiling of mutations and methylation.
Experimental Design:
We conducted CAncer Personalized Profiling by deep Sequencing (CAPP-seq) and cell-free Methylated DNA ImmunoPrecipitation and high-throughput sequencing (cfMeDIP-seq) for detection of ctDNA-derived somatic mutations and aberrant methylation, respectively. We analyzed 77 plasma samples from 30 patients with stage I–IVA human papillomavirus–negative HNSCC as well as plasma samples from 20 risk-matched healthy controls. In addition, we analyzed leukocytes from patients and controls.
Results:
CAPP-seq identified mutations in 20 of 30 patients at frequencies similar to that of The Tumor Genome Atlas (TCGA). Differential methylation analysis of cfMeDIP-seq profiles identified 941 ctDNA-derived hypermethylated regions enriched for CpG islands and HNSCC-specific methylation patterns. Both methods demonstrated an association between ctDNA abundance and shorter fragment lengths. In addition, mutation- and methylation-based ctDNA abundance was highly correlated (r > 0.85). Patients with detectable pretreatment ctDNA by both methods demonstrated significantly worse overall survival (HR = 7.5; P = 0.025) independent of clinical stage, with lack of ctDNA clearance post-treatment strongly correlating with recurrence. We further leveraged cfMeDIP-seq profiles to validate a prognostic signature identified from TCGA samples.
Conclusions:
Tumor-naïve detection of ctDNA by multimodal profiling may facilitate biomarker discovery and clinical use in low ctDNA abundance applications.
Translational Relevance.
Circulating tumor DNA (ctDNA) detection at low fractional abundance within plasma cell-free DNA can be challenging without confirmation of shared molecular features within the tumor. We evaluated the feasibility of tumor-naïve multimodal molecular profiling of cell-free DNA by interrogating orthogonal genetic and epigenetic features reflective of ctDNA biology. Within a cohort of patients with head and neck cancer, we show that ctDNA-derived mutations and methylation were highly correlated and that their detection was associated with poor prognosis. Furthermore, we show that lack of ctDNA clearance post-treatment by methylation profiling is strongly associated with cancer recurrence. This work will provide a blueprint for future studies in which blood but not tumor is available, enable robust ctDNA detection at low abundance, and lead to the discovery of new biomarkers that can be leveraged for clinical applications.
Introduction
Circulating tumor DNA (ctDNA) within plasma cell-free DNA (cfDNA) provides tumor-derived biomarkers for use in liquid biopsy applications. These tumor-specific features, which can include both genetic (e.g., DNA mutations) and epigenetic (e.g., DNA methylation) aberrations, reflect intrinsic properties derived from the tumor. In clinical scenarios with high ctDNA abundance (e.g., advanced/metastatic solid tumors), a growing body of evidence supports its use to aid with diagnosis, prognostication, or monitoring response to therapy (1). However, these same applications often cannot be extended to clinical scenarios where ctDNA abundance is low, which is the case for most non-metastatic solid tumors. A tumor tissue-informed approach, in which tumor-specific features to be analyzed in cfDNA are first selected by tumor DNA profiling, can improve applicability of ctDNA for non-metastatic solid tumors (2, 3). But this process has two major drawbacks. First, access to tumor tissue may not always be feasible due to increasingly competing demands for tumor tissue in research and clinical practice. Second, practical considerations related to the time and cost required for tumor DNA profiling can be significant obstacles to clinical implementation.
To circumvent the need for paired tumor tissue profiling, alternative strategies have been proposed to improve the confidence of tumor-naïve ctDNA detection. Mutations also found in co-isolated peripheral blood leukocytes (PBL) or in healthy control plasma can be removed, reducing the likelihood of false positive results (2, 4–6). Separately, ctDNA can be enriched by analyzing relatively shorter cfDNA fragments due to intrinsic differences in fragment lengths compared with nonmalignant sources of cfDNA (7, 8). A strategy that has not yet been described involves measurement of multiple molecular features (i.e., multimodal profiling) from both plasma cfDNA and PBLs. Through independent corroboration of ctDNA detection using complementary platforms, this approach could extend liquid biopsy applications and biomarker discover efforts beyond what is currently possible.
Here, we simultaneously assessed multiple orthogonal properties of ctDNA from patients with locoregionally confined human papillomavirus (HPV)-negative head and neck squamous cell carcinoma (HNSCC) and risk-matched healthy controls. We profiled plasma cfDNA mutations, methylation, and fragment lengths, and we integrated PBL profiling to remove contaminating signals. Our results demonstrate the complementarity of measuring each ctDNA property for diagnostic, prognostic, and longitudinal monitoring applications. Our results provide a roadmap for using this integrative multimodal approach for further biomarker discovery and clinical applications in many cancer types.
Materials and Methods
HNSCC and healthy control PBL and plasma acquisition
Patients diagnosed with locoregionally confined HPV-negative HNSCC between 2014 and 2016 were identified from a prospective Anthology of Clinical Outcomes (9). All studies were approved by the Research Ethics Board at University Health Network and performed in accordance with the principles of the Declaration of Helsinki. All participants provided written informed consent. HNSCC patient samples were obtained from the Princess Margaret Cancer Centre's HNC Translational Research program based on the following criteria: (i) presentation of locoregionally confined disease (clinically staged M0) at diagnosis, (ii) collection of blood at diagnosis and at least one timepoint post-treatment, (iii) minimum follow-up time of 2 years after diagnosis. All patients received curative-intent treatment consisting of surgery with or without adjuvant radiotherapy. Healthy controls matched by age, gender, and current smoking status were identified from a prospective lung cancer screening program. A total of 5 to 10 mL of blood was collected in Ethylene-Diamine-Tetraacetic Acid (EDTA) tubes. For patients with HNSCC, blood was collected at diagnosis as well as 3 months after primary surgery. Where applicable, additional blood was collected prior to adjuvant radiotherapy, mid adjuvant radiotherapy, and/or 12 months after primary surgery (Supplementary Fig. S1A). Plasma was isolated from blood within 1 hour of collection and stored at −80°C until further processing. From the same blood collection for patients with HNSCC at diagnosis or healthy controls, PBLs were also isolated.
Cell culture
The HPV-negative HNSCC cell line, FaDu, was kindly provided by Bradly Wouters (Princess Margaret Cancer Center, Toronto, Ontario, Canada) and cultured in DMEM (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin. FaDu cell cultures were incubated in a humidified atmosphere containing 5% CO2 at 37°C. The identity of FaDu cells was confirmed by short tandem repeat profiling. Cells were subjected to Mycoplasma testing (e-MycoVALiD Mycoplasma PCR Detection Kit, Intron Bio) prior to use.
Isolation of cfDNA and PBL genomic DNA
cfDNA was isolated from total plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen) following manufacturer's instructions. Genomic DNA (gDNA) was isolated from PBLs, sheared to 150 to 200 bp using the Covaris M220 Focused-ultrasonicator, and size selected by AMPure XP magnetic beads (Beckman Coulter) to remove fragments above 300 bp. Isolated cfDNA and sheared PBL genomic DNA were quantified by Qubit prior to library generation (Supplementary Fig. S1B).
Sequencing library preparation
A total of 5 to 10 or 10 to 20 ng of DNA was used as input for cell-free Methylated DNA ImmunoPrecipitation and high-throughput sequencing (cfMeDIP-seq) or CAncer Personalized Profiling by deep Sequencing (CAPP-seq), respectively. Input DNA was prepared for library generation using the KAPA HyperPrep Kit (KAPA Biosystems) with some modifications. Library adapters were utilized which incorporate a random 2-bp sequence followed by a constant 1-bp T sequence 5′ adjacent to both strands of input DNA upon ligation (10). To minimize adapter dimerization during ligation, library adapters were added at a 100:1 adapter:DNA molar ratio (∼0.07 μmol/L per 10 ng of cfDNA) and incubated at 4°C for 17 hours overnight. After post-ligation cleanup, input DNA was eluted in 40 μL of elution buffer (EB, 10 mmol/L Tris-HCl, pH 8.0–8.5) prior to library generation.
Generation of CAPP-seq libraries
Generation of CAPP-seq libraries were performed as described from Newman and colleagues (2) with some modification. Libraries were PCR amplified at 10 cycles, and up to 12 indexed amplified libraries were pooled together at 500 to 1,000 ng. After the addition of COT DNA and blocking oligos, pooled libraries were placed into a vacuum concentrator (SpeedVac, Thermo Fisher Scientific) to evaporate all liquids and then resuspended in 13 μL resuspension mix (8.5 μL 2× Hybridization buffer, 3.4 μL Hybridization Component A, 1.1 μL nuclease-free water). 4 μL of hybridization probes (i.e., HNSCC selector) was added to the resuspension mix for a total of 17 μL prior to hybridization. After hybridization and PCR amplification/cleanup, libraries were eluted in 30 μL of IDTE pH 8.0 (1× TE solution). Multiplexed libraries were sequenced as paired-end 75/100/125 paired runs on the Illumina NextSeq/NovaSeq/HiSeq4000, respectively. Design of the HNSCC selector incorporated frequently recurrent genomic alterations in HNSCC from The Cancer Genome Atlas (TCGA) COSMIC database (Supplementary Table S3).
Alignment and quality control of CAPP-seq libraries
The first two base pairs on each 5′ end of unaligned paired reads, corresponding to the incorporated random molecular barcodes, were extracted and collated to generate a 4-bp molecular identifier (UMI). The third T base-pair spacer was also removed prior to alignment. Paired reads were aligned to the human genome (genome assembly GRCh37/hg19) by BWA-MEM (11), sorted and indexed by SAMtools (ref. 12; v 1.3.1) and recalibrated for base quality score using the Genome Analysis ToolKit (GATK) BaseRecalibrator (v 3.8) according to best practices (13). Duplicated sequences from BAM files were collapsed on the basis of their UMIs and labeled as Singletons, Single-Strand Consensus Sequences (SSCS) or Duplex Consensus Sequences (DCS) by ConsensusCruncher (10). Quality control of each library was assessed by various metrics obtained from FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), as well as various scripts to obtain capture efficiency (CollectHsMetrics, Picard 2.10.9), depth of coverage (DepthOfCoverage, GATK 3.8), and base-pair position error rate [ides-bgreport.pl (14)].
Detection of mutations and quantification of ctDNA
Removal of potential sequencing errors was performed by integrated Digital Error Suppression (iDES) as described by Newman and colleagues 2016 (14). Background polishing was performed by utilization of the 20 healthy control cfDNA samples as the training cohort to identify candidate mutations (Supplementary Table S4). To prevent the influence of outliers on downstream analysis, candidate mutations within the lower 15th or upper 85th percentile of sequencing depth (≤ 1,500×, ≥ 5,000×) across HNSCC cfDNA or PBL gDNA samples as well as genes with an average sequencing depth ≤ 500× were excluded from analysis. To account for clonal hematopoiesis, non-germline mutations were defined as having a mutant allele fractions (MAF) below 10% in plasma. Candidate mutations in HNSCC cfDNA samples were identified on the basis of the criteria of ≥ 3 supporting reads with duplex support and complete absence in matched PBL gDNA samples. The MAF of each identified mutation was calculated by the number of reads corresponding to the alternative allele, divided by the sum of reads corresponding to the alternative and reference allele. For each HNSCC cfDNA sample with identifiable mutations, the mean MAF across mutations was calculated and used as a measure of ctDNA abundance. In cfDNA samples with only one identifiable mutation, we used the MAF for that single mutation. We note that many of the detectable cancer-derived mutations may not be homozygous and may not be clonal within the tumor, and for these reasons the mean MAF may be an underestimate of the true ctDNA abundance within cfDNA.
Generation of cfMeDIP-seq libraries
The cfMeDIP-seq protocol was performed as described by Shen and colleagues 2019 with modifications to the library preparation step as described in Sequencing Library Preparation. Multiplexed libraries were sequenced as paired-end 75/100/125 on the Illumina NextSeq/NovaSeq/HiSeq4000, respectively. For generalizability, cfMeDIP-seq libraries are described as any MeDIP-seq preparation method utilizing 5–10 ng of input DNA regardless of source (i.e., cfDNA, gDNA).
Alignment and quality control of cfMeDIP-seq libraries
Unaligned paired reads were processed, aligned, sorted, and indexed as previously described in Alignment and Quality Control of CAPP-seq Libraries. Duplicated sequences from BAM files were collapsed by SAMtools. Quality control of each library was assessed by various metrics obtained from FastQC (Babraham Bioinformatics), as well as various metrics obtained from the R package MEDIPS (15) including CpG coverage (MEDIPS.seqCoverage) and enrichment (MEDIPS.CpGenrich).
Selection of informative regions in cfMeDIP-seq profiles
Fragments generated from paired reads of cfMeDIP-seq libraries were counted within nonoverlapping 300 bp windows by MEDIPS (MEDIPS.createSet), scaled by reads per kilobase per million (RPKM), and exported as WIG format (MEDIPS.exportWIG). WIG files from each sample were imported by R and collated as a matrix. Analysis was limited to cfDNA and PBL samples from the 20 healthy control samples to enable applications within a nondisease context. Informative regions were based on the criteria of CpG density and correlation of RPKM values between cfDNA and matched PBLs. Employing a sliding window based on CpG density (≥n CpGs), a minimum threshold of ≥8 CpGs was selected.
Calculation of absolute methylation from cfMeDIP-seq libraries
Fragments from paired reads of cfMeDIP-seq libraries were counted as previously described in Selection of Informative Regions in cfMeDIP-seq Profiles and scaled to absolute methylation levels by the MeDEStrand R package (16). To calculate absolute methylation from counts, a logistic regression model was used to estimate bias of DNA pulldown based on CpG density (i.e., CpG density bias) (MeDEStrand.calibrationCurve). On the basis of the estimated CpG density bias, methylation within each window was corrected for fragments from the positive and negative DNA strand. Windows with corrected fragments were log transformed and scaled to values between 0 and 1 to describe absolute methylation (MeDEStrand.binMethyl). Absolute methylation levels from each cfMeDIP-seq sample was exported as a WIG-like file (i.e., WIG file format without a track line).
Design of in-silico PBL depletion and evaluation of performance
To enrich for windows within the disease setting, methylation from PBLs was removed by a process termed “in-silico PBL depletion.” Analysis was limited to PBL samples from our cohort of 20 healthy control samples to enable applications within a non-cancer specific context. Our strategy for the in-silico PBL depletion was performed as followed:
(i) For each informative window as described in Selection of Informative Regions in cfMeDIP-seq Profiles, calculate the median absolute methylation value across healthy control PBL samples.
(ii) Define PBL-depleted windows based on the criteria of a median absolute methylation value < 0.1.
(iii) Restrict analysis of cfDNA samples within PBL-depleted windows.
Performance of the in-silico PBL depletion strategy was evaluated by comparing absolute methylation distributions in PBL samples before and after depletion from the healthy control cohort used as the training set, to the HNSCC cohort used as the validation set.
Differential methylation analysis
To enable robust detection of HNSCC-associated differentially methylated regions (DMR), analysis was limited to patients with HNSCC with detectable mutations in plasma by CAPP-seq (n = 20/30). Differential methylation analysis was limited to informative regions after in-silico PBL depletion. A collated matrix of binned fragment counts from HNSCC and healthy control cfDNA samples, generated as previously described in Selection of Informative Regions in cfMeDIP-seq Profiles, were utilized for identification of DMRs by the DESeq2 R package (17). Pre-filtering was performed by removal of regions with < 10 counts across all cfDNA samples. A single factor defined as condition (HNSCC vs. healthy control) was used for contrast during differential methylation analysis. Briefly, differential methylation analysis was performed by scaling samples based on size factors and dispersion estimates, followed by fitting of a negative binomial general linear model. For each window, a P value was calculated between the HNSCC and healthy control conditions by Wald test. P values within regions above the default Cook distance cutoff were omitted from Padjusted calculation [Benjamini-Hochberg (18)]. Significant hypermethylated or hypomethylated regions (hyper-/hypo-DMRs) in HNSCC cfDNA samples are defined as windows with an Padjusted < 0.1.
Enrichment of CpG features within HNSCC cfDNA hypermethylated regions
CpG features such as islands, shores, shelves, and open sea (interCGI) are defined as per the AnnotationHub R package (hg19_cpgs annotation). ID coordinates of each hypermethylated window (i.e., “chr.start.end”) within PBL-depleted regions were labeled with an overlapping CpG feature using an inhouse R package that utilizes the annotatr (19) and GenomicRanges R packages (20) (Supplementary Table S5).
To determine the probability of enrichment for an observed overlap of features versus a null distribution, 1,000 random samplings were performed. For each sampling, an equal number of bins were chosen based on the number hypermethylated windows, while maintaining an identical distribution of CpGs. The observed number of overlaps for each CpG feature across samplings were used to generate their respective null distributions, which were subsequently transformed onto a z-score scale. The observed overlap of hypermethylated regions for each CpG feature were also z-score transformed, deriving summary statistics from the null distribution. The estimated P value of the observed overlap from hypermethylated windows was calculated as the number of random samplings with overlap equal or greater/lesser than the observed overlap of the null distribution.
Enrichment of HNSCC cfDNA hypermethylated regions with cancer-specific hypermethylated cytosines from TCGA
File information from publicly available hm450k profiles of all primary tumors from breast (BRCA), colorectal (COAD), head and neck (HNSC), prostate (PRAD), pancreatic (PAAD), lung adeno (LUAD), and lung squamous (LUSC) were downloaded from TCGA. Because of the majority of our HNSCC cohort presenting with tumors of the oral cavity, files from the HNSC group were limited to patients with primary site at the “floor of mouth” (n = 55). An equal number of hm450k files were randomly selected from each of the remaining cancer types, as well as from a separate database of healthy PBLs (Supplementary Table S6, Gene Expression Omnibus series GSE67393).
To generate “tumor-specific” hypermethylated cytosines, differential methylation analysis by Limma (21) was performed for each cancer type, with individual comparisons with each other cancer type as well as PBLs (i.e., contrast). For a given contrast, a linear model was fitted for each probed cytosine incorporating the residual variance and sample beta value, the P value of observed difference between contrasts was then calculated by empirical Bayes smoothing. Hypermethylated cytosines with elevated methylation in a given cancer type versus an individual comparison was defined by a log fold change ≥ 0.25 and a Padjusted value (Benjamini–Hochberg) < 0.01. Hypermethylated cytosines unique to an individual cancer type were designated as “tumor-specific.” For the cases of LUSC, LUAD, and PAAD, either no or very little tumor-specific hypermethylated cytosines were identified (0, 15, and 18, respectively) and therefore were omitted from subsequent analysis. For comparison with cfMeDIP-seq libraries, base-pair positions from tumor-specific hypermethylated cytosines were overlapped with informative windows after in-silico PBL depletion as described in Design of In-silico PBL Depletion and Evaluation of Performance.
The enrichment of overlap for HNSCC cfDNA hypermethylated regions with tumor-specific regions from TCGA was evaluated by 10,000 random samplings using the same methods described in Enrichment of CpG Features with HNSCC cfDNA Hypermethylated Regions.
ctDNA detection by cfMeDIP-seq
For cfMeDIP-seq libraries from our cohort of 30 HNSCC and 20 healthy control cfDNA samples, ctDNA detection was defined on the basis of the observation of a mean RPKM value across HNSCC cfDNA hypermethylated regions within an individual HNSCC cfDNA sample greater than the max mean RPKM value across healthy control cfDNA samples.
Fragment length analysis of ctDNA detected by CAPP-seq and cfMeDIP-seq
For each HNSCC cfDNA CAPP-seq library, the median fragment length from all supporting paired reads of a specified mutation (i.e., singletons, SCSs, DCSs) as well as for paired reads containing the reference allele was measured (Supplementary Table S7). In cases where the median fragment length was reported for patients with > 1 mutation, the median value across the median fragment length from each mutation was calculated. For each HNSCC cfDNA cfMeDIP-seq library, the median fragment length from all fragments mapping to the previously determined HNSCC cfDNA hypermethylated regions was calculated. Because of the relative absence of methylation within our cohort of 20 healthy controls, the fragment length of each healthy control cfMeDIP-seq library was collated prior to any calculations. In both types of libraries, fragment length analysis was limited to cfDNA within the first peak (i.e., <220 bp).
Enrichment of fragments (100–150 bp or 100–220 bp) within hyper-DMRs was calculated as follows. A null distribution of expected counts was generated from random 300-bp bins within our previously designed PBL-depleted windows at identical number and CpG density distribution, from a total of 30 samplings. Observed counts for each sample were determined on the basis of read counts across hyper-DMRs. For each sample, enrichment was calculated on the basis of the mean observed count divided by the mean expected count.
Metrics of ctDNA detection and quantification on HNSCC patient clinical outcomes
The prognostic association of ctDNA detection for overall survival (defined as from the time of diagnosis to time of death by any cause) was evaluated by three metrics: (i) detection of mutations by CAPP-seq, (ii) detection of increased mean RPKM in hypermethylated regions by cfMeDIP-seq. (iii) detection by both previous metrics. Cox proportional hazards regression analysis was calculated between patients with detectable or nondetectable ctDNA for each metric. Patient characteristics are described in Supplementary Table S1.
Cross-validation of ctDNA-derived methylation by cfMeDIP-seq analysis
To evaluate the robustness of cfMeDIP-seq for identifying ctDNA-derived methylation, ROC curve analysis was performed. To minimize confounding results due to low/absent ctDNA, analysis was limited to HNSCC patients with detectable ctDNA by CAPP-seq. Patient and healthy control cfMeDIP-seq profiles were split into a training set (HNSCC: n = 12/20; healthy control: n = 12/20) and testing set (HNSCC: n = 8/20; healthy control: n = 8/20). Training and testing sets were balanced for ctDNA abundance as determined by CAPP-seq analysis. A total of 50 splits were performed with ROC curve analysis performed on each iteration.
Identification of prognostic regions in HNSCC by TCGA analysis
All available HNSCC cases from TCGA with matched legacy hm450k and RNA expression data were selected (Supplementary Table S8). Survival data were obtained from Liu and colleagues (22). For processing hm450k data, methylation was summarized to 300-bp regions as described previously by calculating the mean beta-value between probe IDs within a particular region. To identify regions hypermethylated in HNSCC primary tumors compared with adjacent normal tissue, independent Wilcoxon tests were performed for each region. We selected regions with a Padjusted < 0.05 [Holm method (23)] as well as a log fold change ≥ 1 in primary tumors compared with adjacent normal tissue for subsequent analysis. To identify hypermethylated regions associated with prognosis, multivariate Cox regression was performed, incorporating age, gender, and clinical stage, selecting regions with P < 0.05. Survival analysis was limited to a maximum follow-up time of 5 years after diagnosis. To further identify prognostic regions associated with changes in gene expression, Spearman correlation was calculated for hm450k primary tumor profiles for each region, to matched RNA expression profiles for transcripts within a 2-Kb window. Regions with absolute R values > 0.3 and a FDR < 0.05 were selected, resulting in the final identification of five prognostic regions associated with ZNF323/ZSCAN31, LINC01395, GATA2-AS1, OSR1, and STK3/MST2 expression.
For TCGA patient profiles, the composite methylation score (CMS) was obtained by calculating the sum of beta values across all five prognostic regions. For cfMeDIP-seq profiles, RPKM values across all 943 hyper-DMRs were scaled to a total sum of 1 and the CMS was obtained by calculating the sum of these scaled RPKM values across all five prognostic regions.
Longitudinal monitoring of post-treatment plasma samples by cfMeDIP-seq
cfMeDIP-seq libraries were successfully generated from post-treatment plasma samples for 28 of 30 patients (Supplementary Table S9). For the remaining 2 patients, insufficient material was isolated from plasma and/or did not pass quality metrics. Eligible patients for longitudinal monitoring were further selected on the basis of the criteria of having detectable ctDNA in baseline samples by CAPP-seq (n = 18/30). ctDNA quantification of post-treatment cfMeDIP-seq libraries was performed as described previously, calculating the mean RPKM values across identified hypermethylated regions by differential methylation analysis. For ease of interpretation, both baseline and post-treatment cfMeDIP-seq libraries were converted to percent DNA values based on linear regression against mean MAF within matched baseline CAPP-seq profiles. To achieve high-confidence detection of residual disease, a minimum ctDNA fraction of 0.2% was required in post-treatment samples, corresponding to the maximum of mean RPKM values observed across all healthy controls.
Data and code availability
MeDIP-seq sequencing data generated from the FaDu cell line is accessible at BioProject ID PRJNA729472. Sequencing data for clinical samples are restricted to processed BED files due to privacy concerns and to comply with institutional ethics regulation. Data of cfMeDIP-seq profiles generated from patients and controls have been processed and deposited on Zenodo (10.5281/zenodo.4698004 and 10.5281/zenodo.4698867; Supplementary Table S10). Source data tables used to generate Fig. 2B–F, Fig. 4A and B, and Fig. 6A and B, are provided in Supplementary Tables S4, S7, S10, and S9, respectively. Scripts used to generate the findings described in this study are available within the “HNSCC multimodal” repository at https://github.com/bratmanlab/multimodal-hnscc. The Code Ocean capsule containing the reproducible analysis used can be found at https://codeocean.com/capsule/7176142/tree/v1 (DOI: 10.24433/CO.6511868.v1).
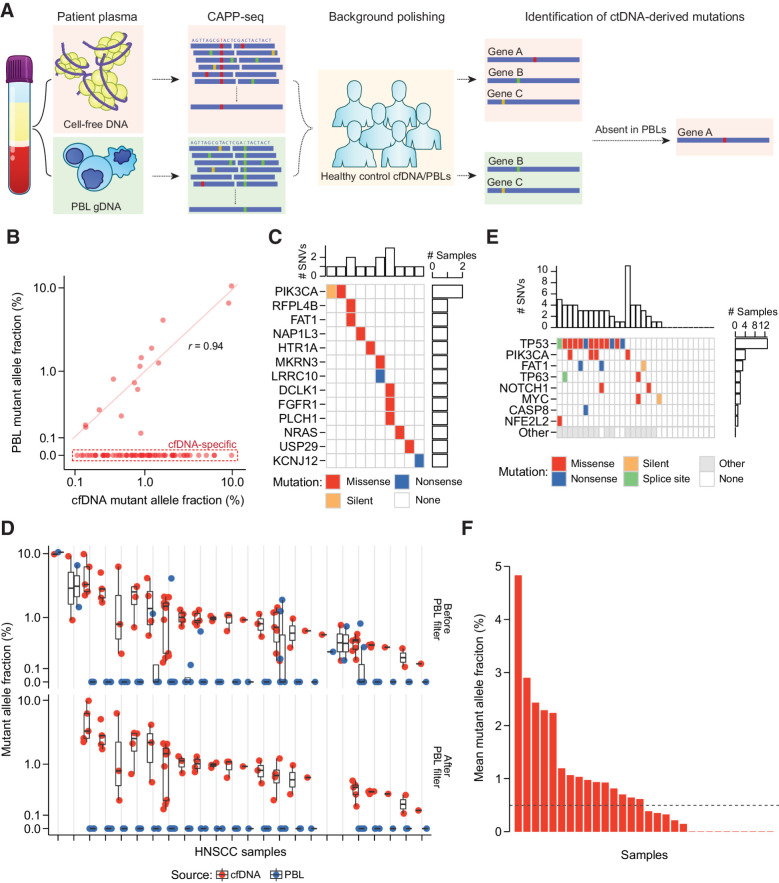
Figure 2.
PBL filtering for detection of ctDNA by CAPP-seq. A, Schematic describing detection and filtering of ctDNA-derived mutations by CAPP-seq. B, MAF of candidate mutations identified in HNSCC patient baseline plasma and PBLs. Dashed red box: 69 of 84 candidate mutations found only in plasma. Mutations found in both matched plasma and PBLs have strongly correlated MAFs (Pearson r = 0.94). C, Oncoprint of 15 candidate mutations identified in both matched patient plasma and PBLs. Top histogram: number of mutations per patient; right histogram: number of patients with one of 13 specified genes mutated. Color indicates type of mutation: missense (red), nonsense (blue), or silent (orange). D, MAF of 69 candidate mutations across HNSCC patient cfDNA (red circle) and PBLs (blue circle) before removal of PBL-associated mutations, and 43 mutations after this removal. The removed mutations represent false positive candidate mutations in ctDNA. Black bar: median of expected overlaps. Box: interquartile range (IQR) of expected overlaps. Whisker: most extreme value within quartile ±1.5 IQR of expected overlaps. E, Oncoprint highlighting frequently mutated genes in HNSCC by TCGA analysis (rows) and their frequency of detection across 30 HNSCC plasma samples (columns). Colors as in B, with additional splice sites (green), and nonhighlighted genes (gray). F, Mutation-based ctDNA levels in HNSCC patient baseline plasma samples as determined by mean MAF of PBL-filtered mutations. Dashed line: median mean allele fraction across all patients with HNSCC.
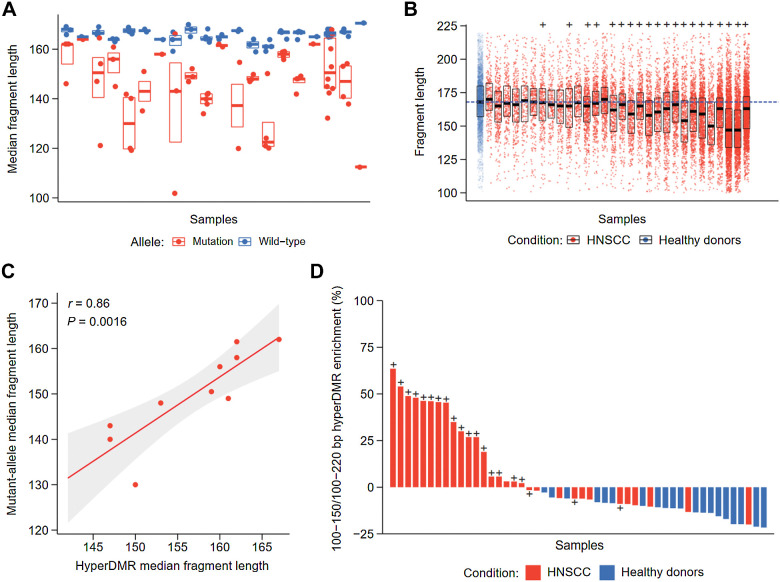
Figure 4.
Fragment length analysis between CAPP-seq and cfMeDIP-seq profiles. A, Median fragment length of detected mutations across patients with HNSCC by CAPP-seq. For each patient, the median fragment length of each mutation and matched reference allele was measured. The distribution of median fragment length for each mutation or matched reference allele is shown per patient. Black bar: median of fragment lengths. Box: IQR of fragment lengths. In cases with a single mutation, the colored line denotes the median length of fragments containing the mutation or matched reference allele, respectively. B, Fragment length distributions within HNSCC hypermethylated regions by cfMeDIP-seq. Fragment lengths from healthy controls were pooled prior to analysis, where each subsequent box denotes an individual HNSCC cfMeDIP-seq profile. + symbols denote HNSCC patients with detectable ctDNA by CAPP-seq (CAPP-seq positive). Black bar: median of fragment lengths. Box: IQR of fragment lengths. Individual HNSCC samples are ordered on the basis of increasing mean methylation (RPKM) within the hypermethylated regions. Dashed blue line defines the median fragment length across all healthy controls. C, Comparison of median fragment lengths from CAPP-seq and cfMeDIP-seq profiles. Points define individual HNSCC samples with methylation values above the median (n = 10). Solid red line: fitted linear regression model. Gray boundaries: 95% CI. D, Ratio of enrichment for hyper-DMR regions by fragments between 100 and 150 bp compared with enrichment for hyper-DMR regions by fragments between 100 and 220 bp. + symbols denote patients with HNSCC with detectable ctDNA by CAPP-seq (CAPP-seq positive).
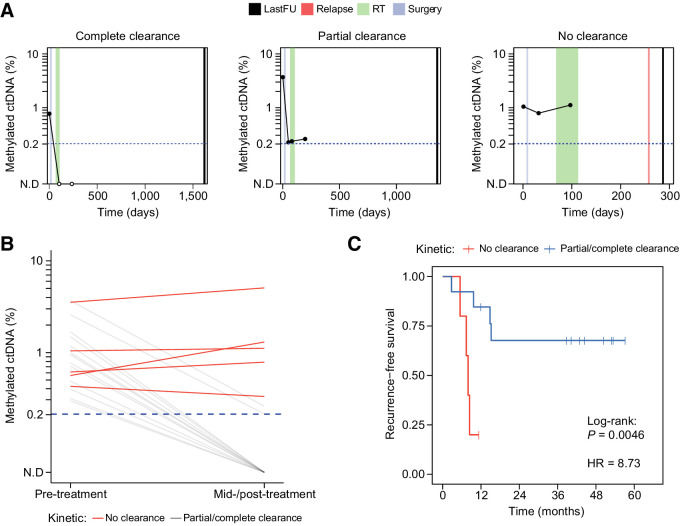
Figure 6.
Clinical utility of ctDNA detection by cfMeDIP-seq for longitudinal monitoring. A, Representative ctDNA kinetics throughout treatment that were observed in this study. Complete clearance was defined as a change from detected ctDNA at diagnosis to a decrease in ctDNA abundance below the detection threshold (i.e., 0.2%) at first available mid-/post-treatment timepoint. Partial clearance was defined as an observed ≥90% decrease in ctDNA abundance from diagnosis compared with last follow-up. No clearance was defined as an observed <90% decrease or ≥0% increase in ctDNA from diagnosis to last follow-up. lastFU = sample collection at last follow-up, RT = radiotherapy. B, Changes in ctDNA abundance pre-treatment (baseline) to first available mid-/post-treatment timepoint across patients with HNSCC (n = 30). Red lines denote patients who demonstrated kinetics of no clearance, whereas gray lines denote patients with kinetics of clearance/partial clearance. C, Kaplan–Meier analysis depicting recurrence-free survival. Patients were stratified on the basis of kinetics of clearance (i.e., no clearance versus clearance/partial clearance).
Results
Multimodal profiling of cfDNA and leukocyte DNA from patients with locoregionally confined HPV-negative HNSCC and healthy controls
To examine the ability of multimodal profiling to characterize ctDNA in the setting of locoregionally confined (non-metastatic) cancer, we recruited 30 newly diagnosed patients with HPV-negative HNSCC into a prospective observational study in which peripheral blood samples were collected at serial timepoints (Supplementary Fig. S1A; Supplementary Table S1). All patients were treated with surgery, with a subset also receiving adjuvant radiotherapy (n = 14) or chemoradiotherapy (n = 11). With a median follow-up of 41.5 months (range: 7–57 months), 9 of 30 patients (30%) developed recurrence (actuarial 2-year recurrence-free survival: 73%).
As the majority of patients exhibited a heavy smoking history, which is well described to alter the genomic/epigenomic landscape of somatic tissues and contribute to premalignant lesions (24–27), we also analyzed blood samples from 20 risk-matched healthy controls (age, sex, smoking status/pack-years; Supplementary Table S1). cfDNA from plasma as well as gDNA from PBLs were co-isolated from blood and subjected to parallel analyses (Supplementary Fig. S1A; Supplementary Table S2). Total plasma cfDNA levels were similar between patients with HNSCC and healthy controls (one-way ANOVA; P = 0.53; F = 0.82; Supplementary Fig. S1B).
We conducted multimodal profiling of cfDNA and PBL gDNA from patients and healthy controls (Fig. 1). Mutations and methylation were independently profiled using CAPP-seq and cfMeDIP-seq, respectively. In addition, we utilized paired-end sequencing for both methodologies to obtain the lengths of sequenced cfDNA fragments.
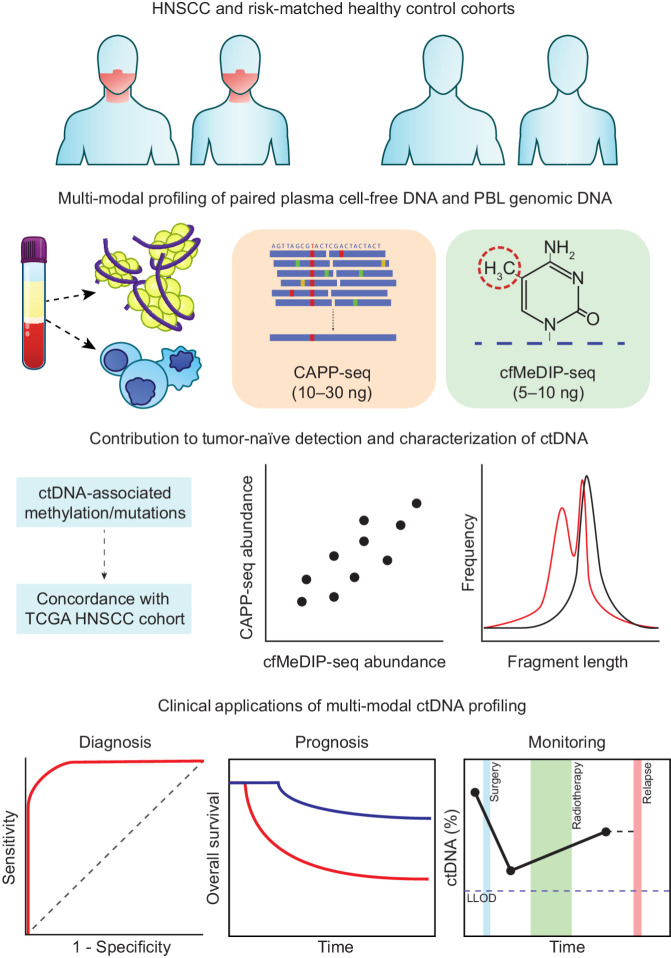
Figure 1.
Multimodal profiling strategy of ctDNA in HNSCC. We identified ctDNA-derived mutations (CAPP-seq) and methylation (cfMeDIP-seq) in a cohort of locoregionally confined HPV-negative HNSCC (n = 30) and risk-matched (age, sex, smoking status, smoking pack-years) healthy controls (n = 20). For both methods, we isolated plasma for ctDNA detection and used matched PBL profiling to remove noninformative regions (i.e., mutations derived from clonal hematopoiesis and regions that are methylated within PBLs). We characterized the mutation- and methylation-based ctDNA results according to their concordance with TCGA datasets, correlation between the two methods, and inferred physical properties (i.e., fragment length). We assessed feasibility of these detection strategies for clinical applications in diagnosis, prognosis, and disease monitoring.
Tumor-naïve detection of mutation-based ctDNA
To evaluate the sensitivity of ctDNA detection in HPV-negative HNSCC without prior knowledge from the tumor, we first measured the abundance of mutations in baseline plasma samples (Fig. 2A). CAPP-seq was conducted with a sequencing panel designed to maximize the number of HNSCC-associated mutations (Supplementary Table S3; Supplementary Fig. S2). We also employed established error suppression methodologies to remove background base substitution errors (10, 14).
Next, we compared candidate mutations detected in cfDNA with matched PBL gDNA profiles to assess the contribution of mutations from clonal hematopoiesis. Of the 24 patients with identifiable cfDNA mutations, 10 demonstrated identical mutations within their matched PBL profile with highly correlated MAFs (Pearson r = 0.94, P = 1.4 × 10−7; Fig. 2B). With the exception of PIK3CA (n = 2), genes within these PBL-derived mutations were unique to each patient (Fig. 2C). Of note, canonical clonal hematopoiesis genes such as DNMT3A, TET2, and ASXL1 (28), were not observed as they were not included within the CAPP-seq capture panel. Attesting to the contribution of clonal hematopoiesis to false positive detection, cfDNA samples from 4 patients only contained PBL-derived mutations (Fig. 2D). Thus, matched PBL profiling greatly minimizes false positive detection of ctDNA at low abundance in HPV-negative HNSCC patient plasma.
After accounting for clonal hematopoiesis, ctDNA was detected within plasma of 20 patients (range = 1–10 mutations per patient; median = 3). To evaluate the plausibility of these mutations being tumor derived, we compared our results with whole-exome sequencing data from 279 HNSCC tumors from TCGA (29). Canonical cancer driver genes in HNSCC primary tumors had similar somatic mutation frequencies in the population when compared with our CAPP-seq data, including TP53 (72% vs. 65%), PIK3CA (21% vs. 20%), FAT1 (23% vs. 15%), and NOTCH1 (19% vs. 10%; Fig. 2E). We also detected mutations in ctDNA that were not found within the canonical HNSCC driver genes (GRIN3A and MYC; Supplementary Fig. S3), illustrating how including genes without known driver effects in a particular cancer type can increase sensitivity of ctDNA detection (30).
The abundance of detectable ctDNA fragments, which provides a relative measure of ctDNA abundance, was calculated from the mean MAF of mutations identified in each patient and ranged from 0.14% to 4.83% (Fig. 2F). The lowest detected ctDNA fraction of 0.14% resembles that previously found utilizing tumor-naïve CAPP-seq analysis (14, 31). Including patients with undetectable ctDNA, our HNSCC cohort had a median ctDNA abundance of 0.49%—similar to previous CAPP-seq observations in locoregional non–small cell lung cancer (14, 30).
Tumor-naïve detection of methylation-based ctDNA from baseline plasma
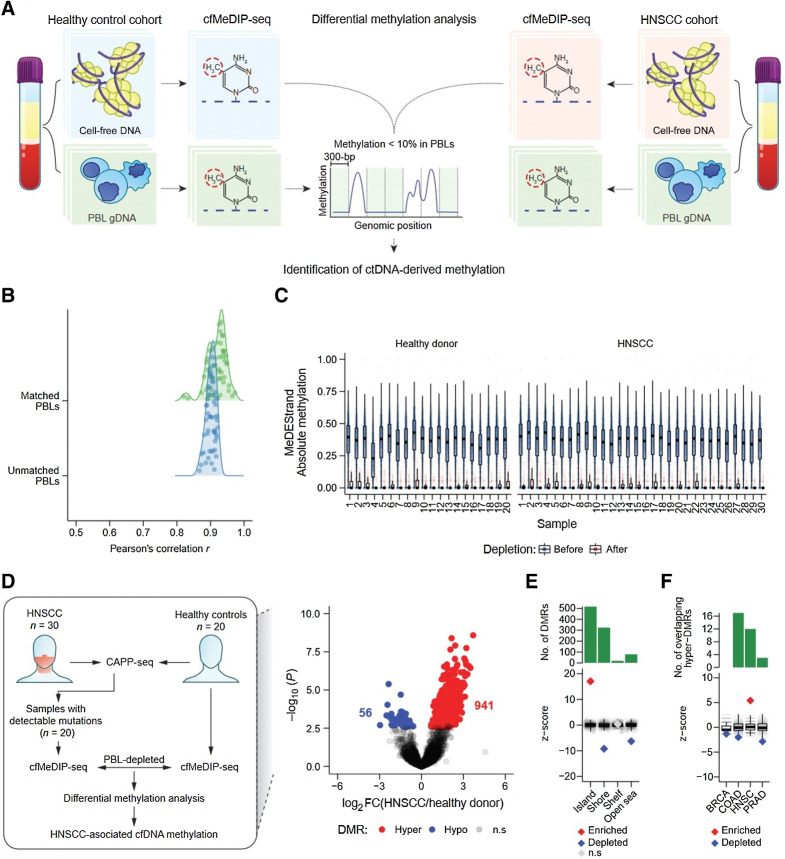
Next, we sought to define ctDNA-associated methylation patterns in the HNSCC and healthy control samples. As the CAPP-seq results illustrated the impact of false positive mutations arising from PBLs, we reasoned that a reduction of false positive ctDNA-associated methylation could be achieved by removal of PBL-derived DNA methylation signals. Therefore, we used matched PBL MeDIP-seq profiles from the HNSCC and healthy control samples to suppress their contribution to the cfDNA methylation signal (Fig. 3A).
Figure 3.
Identification of informative regions for detection of ctDNA by cfMeDIP-seq. A, Schematic of ctDNA-derived methylation detection by DMR analysis following depletion of PBL-derived DNA methylation. B, Kernel density of Pearson correlation coefficients from the comparison between all 50 (patients and healthy controls) baseline cfDNA cfMeDIP-seq profiles (RPKM values within 702,488 300-bp nonoverlapping windows from chromosomes 1–22 with ≥8 CpGs) and either unmatched PBL gDNA (1 vs. 49 comparisons) or matched PBL gDNA (1 vs. 1 comparison) MeDIP-seq profiles. C, MeDEStrand absolute methylation scores in the 702,488 300-bp windows from 20 healthy controls (left) and 30 HNSCC samples (right). Blue: 702,488 windows before PBL depletion. Red: 99,994 windows after both PBL depletion and an additional filter where the median absolute methylation across healthy control PBLs is < 0.1. Black bar: median. Box: IQR. Whisker: most extreme value within quartile ±1.5 IQR. Blue/red circles: individual MeDEStrand absolute methylation scores across windows ≥8 CpGs and PBL-depleted windows, respectively. Absoluate methylation scores were subsampled to a total of 100,000 observations for clarity. D, Left, Workflow of ctDNA detection by differential methylation analysis of HNSCC and healthy control cfMeDIP-seq profiles. We compared cfMeDIP-seq profiles from 20 patients with HNSCC with detectable mutations by CAPP-seq to 20 risk-matched healthy controls within the PBL-depleted windows for each sample. This procedure identified HNSCC-associated cfDNA methylation. Right, Volcano plot of 79,043 genomic regions, with > 10 reads across all samples, displaying −log10P value of differential methylation against log2 fold change of relative methylation (RPKMs) from healthy controls to patients with HNSCC. Gray: regions without significant differential methylation (FDR ≥10%). Blue: 56 hypomethylated 300-bp regions, with significantly lower methylation in the HNSCC cohort compared to healthy controls. Red: 941 300-bp hypermethylated regions, with significantly higher methylation in the HNSCC cohort. E, Permutation analysis measuring the observed overlap of the 941 300-bp hypermethylated regions to 300-bp windows within CpG islands, shores, shelves, or open seas, relative to a distribution of the expected overlap generated by random sampling of the genome. F, Permutation analysis measuring the observed overlap of the 941 300-bp hypermethylated regions to hypermethylated regions within tumor-specific methylated cytosines from TCGA (n = 10,000 permutations total). BRCA: breast invasive carcinoma; COAD: colon adenocarcinoma; HNSC: head and neck squamous cell carcinoma; PRAD: prostate adenocarcinoma. E and F, The expected overlap from 1,000 random samples was transformed to a Z-score distribution (gray circles) and used to calculate the Z-score of the observed overlap (diamond). P values were calculated based on the probability of the observed overlap relative to the distribution generated from the expected overlaps. Red: significant enrichment, P < 0.05. Blue: significant depletion, P < 0.05. Gray: nonsignificant. Black bar: median of expected overlaps. Box: IQR of expected overlaps. Whisker: most extreme value within quartile ±1.5 IQR of expected overlaps. Above: histogram of number of DMRs for each CpG geography.
To enrich for ctDNA-derived methylation in the analysis, we identified 702,488 nonoverlapping 300-bp windows with ≥8 CpGs. These regions showed similar enrichment in MeDIP-seq profiles from both healthy control PBLs and the HNSCC cell line FaDu (Supplementary Fig. S4A). Next, we compared cfMeDIP-seq profiles from the 30 HNSCC samples and 20 healthy controls with MeDIP-seq profiles generated from their PBLs. Specifically, for each cfMeDIP-seq sample, methylation levels across the 300-bp windows with ≥8 CpGs were compared with their paired PBL sample, and to the remaining 49 unpaired PBL samples, resulting in 2,500 total comparisons. The cfMeDIP-seq samples strongly correlated with their matched PBLs (median Pearson r = 0.92) and unmatched PBLs (median of medians r = 0.90; Fig. 3B). The strengths of these correlations likely reflects the known outsize contribution of PBLs to plasma cfDNA.
We used MeDEStrand (16) to convert cfMeDIP-seq enrichment profiles to absolute methylation levels between 0 and 1. We selected the 99,994 300-bp windows with a median absolute methylation < 0.1 (i.e., 10% signal compared with the top methylated 300-bp window) across PBLs from our 20 healthy controls. Importantly, these regions demonstrated a similar range of absolute methylation when applied to the 30 held-out HNSCC PBLs (Fig. 3C; Supplementary Fig. S4B). Taken together, these results confirm that the main source of cfDNA methylation in both control and locoregionally confined HPV-negative HNSCC plasma are derived from PBLs and that bioinformatic removal of PBL-derived methylation may limit signals that confound ctDNA quantification.
Utilizing the 99,994 300-bp windows depleted for methylation in PBLs, we identified ctDNA-derived DMRs by comparing the 20 HNSCC patients with CAPP-seq–detectable ctDNA to the 20 healthy controls. We found 997 DMRs (Fig. 3D): 941 hypermethylated in HNSCC (hyper-DMRs) and 56 hypomethylated in HNSCC (hypo-DMRs).
Of the 300-bp hyper-DMRs, 47.5% resided in contiguous blocks of hypermethylation signals extending up to 1,800 bp in length (Supplementary Fig. S5A), indicative of CpG islands that typically span 300–3,000 bp in length. Indeed, CpG islands were significantly enriched for hyper-DMRs (Fig. 3E). In contrast, CpG islands were significantly depleted for hypo-DMRs (Supplementary Fig. S5B).
Unlike mutation data alone, cfDNA methylation data can distinguish different tissues of origin for a tumor (32–35). Thus, we investigated whether the hyper-DMRs we identified contain regions reflective of HNSCC. Using TCGA primary tumor tissue methylation array data, we identified hypermethylated CpGs specific for HNSCC as well as for breast cancer, colon cancer, and prostate cancer when compared with publicly available PBL methylation array data from an independent study (Supplementary Fig. S6). As expected, the plasma-derived hyper-DMRs from the HNSCC cohort were significantly enriched for HNSCC-specific hypermethylated CpGs as well as significantly depleted for the remaining cancer-specific hypermethylated CpGs (Fig. 3F). The overlap between hyper-DMRs in HNSCC patient plasma and known hyper-DMRs from primary tumors supports the notion that many plasma hyper-DMR regions indeed derive from hypermethylated regions within tumor tissue.
Fragment length–informed analysis of ctDNA by CAPP-seq and cfMeDIP-seq
A growing number of studies have described ctDNA to be associated with decreased fragment length compared with healthy sources of plasma cfDNA, potentially providing an additional metric for robust tumor-naïve detection (4, 7, 8, 36–38). As targeted sequencing has been previously shown to detect ctDNA at reduced fragment length (4, 7), we first utilized CAPP-seq profiles to determine whether we could observe similar trends within HNSCC patients. For each identified mutation per patient, we measured the median length of cfDNA fragments containing the mutant allele as well as the corresponding reference allele (Fig. 2E). For cases where multiple mutations were identified within a patient sample, we determined the median fragment lengths across all mutations and their corresponding reference alleles. In accordance with previous findings, we observed a consistent decrease in ctDNA fragment length compared with healthy cfDNA across patients [median (range) Δ = −17.5 (−58 to −1) bp] (Fig. 4A). There was no significant association between the mean MAF of these mutations and fragment length (Supplementary Fig. S7A).
Unlike bisulfite-based DNA methylation approaches, cfMeDIP-seq does not cause DNA degradation and, therefore, preserves the original fragment size distribution. This provides a novel opportunity to map DNA methylation and fragment lengths concomitantly. Therefore, we assessed the distribution of cfDNA fragment lengths within the previously identified 941 hyper-DMRs for each patient. Because of the nature of these regions having low methylation within healthy controls, we combined the cfDNA fragments across all control samples. Similar to the mutation-based analysis, we observed a relative reduction in fragment length from 19 of 20 CAPP-seq positive patients compared with grouped healthy controls [median (range) Δ = −7 (−21 to −1) bp] (Fig. 4B). This represented a smaller reduction in fragment lengths compared with the mutation-based analysis, possibly due to partial contribution by healthy tissues of cfDNA fragments within the hyper-DMRs. Supporting this notion, the samples with the shortest hyper-DMR fragments displayed higher methylated ctDNA abundance (Pearson r = −0.64, P = 0.002; Supplementary Fig. S7B).
We next investigated whether fragment lengths were concordant between ctDNA molecules identified by both CAPP-seq and cfMeDIP-seq, potentially providing an additional layer of validation toward our multimodal approach. To minimize the possibility of background DNA fragments confounding the calculated fragment length of ctDNA within cfMeDIP-seq profiles, we limited analysis to patients above the median methylation levels across hyper-DMRs (n = 10 patients with HNSCC). Strikingly, ctDNA fragment length was highly concordant between paired CAPP-seq and cfMeDIP-seq profiles for each patient (Pearson r = 0.86, P = 0.0016; Fig. 4C) despite entirely different genomic regions being represented with these two profiling approaches (CAPP-seq: 43 distinct mutations, cfMeDIP-seq: 941 hyper-DMRs).
On the basis of these observations, we evaluated whether we could enrich ctDNA within cfMeDIP-seq profiles by limiting analysis to cfDNA fragments of reduced length. We assessed the proportion of cfDNA fragments within hyper-DMRs consisting of small (100–150 bp) fragments, as similar methods have been described to enrich for ctDNA using non-methylation–based approaches (7, 37). Indeed, this resulted in ctDNA enrichment across the majority of CAPP-seq positive HNSCC samples [median (range) = 28 (−8 to 63) %] but not for any of the healthy controls (Fig. 4D). Thus, in silico size selection of cfDNA fragments enriches for ctDNA within cfMeDIP-seq libraries and could contribute to tumor-naïve multimodal ctDNA analysis.
Application of multimodal ctDNA detection for prognostication
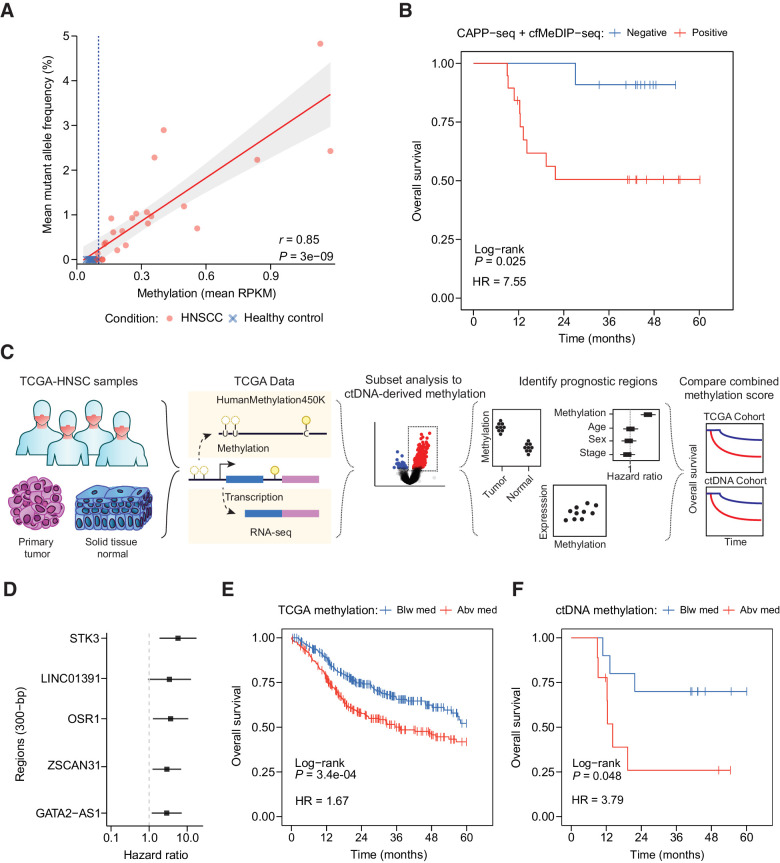
To evaluate the potential clinical applications of tumor-naïve multimodal ctDNA analysis, we compared ctDNA with clinical outcomes in the HNSCC cohort. Fragment length–informed (i.e., limited to 100–150 bp) cfMeDIP-seq profiles were strongly associated with MAFs in matched CAPP-seq profiles (Pearson r = 0.85, P = 3 × 10–9), suggesting that methylation intensity within the 941 hyper-DMRs is indeed reflective of ctDNA abundance (Fig. 5A). Importantly, cross-validation analysis confirmed the robustness of these hyper-DMRs for detecting ctDNA (Supplementary Fig. S8A). Patients with ctDNA detected in baseline plasma by both mutation- and methylation-based methods (n = 19) were significantly more likely to have advanced disease (i.e., stage III–IVA; n = 18/19) when compared with patients with no detectable ctDNA (n = 8/13; Fisher exact test P = 0.028) and displayed dramatically worse overall survival [HR = 7.55, 95% confidence interval (CI) = (0.95–59.94), log-rank P = 0.025] (Fig. 5B). In comparison, stage alone was unable to predict patients with worse overall survival [HR = 2.59, 95% CI = (0.32–20.46), log-rank P = 0.35] (Supplementary Fig. S8B), further demonstrating the potential clinical utility of multimodal ctDNA profiling.
Figure 5.
Prognostic utility of multimodal profiling by CAPP-seq and cfMeDIP-seq. A, Relationship of mean MAF and mean RPKM from identified mutations and hypermethylated regions by CAPP-seq and cfMeDIP-seq (limited to 100–150 bp), respectively. Points denote individual samples from HNSCC or healthy control plasma. Solid red line: fitted linear regression model. Gray boundaries: 95% CI. B, Kaplan–Meier analysis depicting overall survival of patients with detectable ctDNA both by CAPP-Seq and cfMeDIP-seq (mean methylation above healthy controls within hyper-DMRs). C, Schematic describing the analytic framework for evaluating prognostic regions in ctDNA by utilization of TCGA data. D, Forest plot of selected prognostic regions based on multivariate Cox proportional hazards regression and gene co-expression analysis. E, Kaplan–Meier analysis depicting overall survival of patients with HNSCC-TCGA based on a CMS determined from the total methylation across five regions affecting expression of ZSCAN31, LINC01391, GATA-AS1, OSR1, and STK3. Patients were stratified on the basis of either being below (Blw med, blue) or above (Abv med, red) the median total methylation of the five regions previously identified in D across all primary tumors. F, Kaplan–Meier analysis depicting overall survival as described in E for HNSCC plasma cohort with detectable ctDNA by CAPP-seq. To calculate the CMS in this cohort, RPKM values for the five regions were normalized to RPKM values across all previously identified 941 hyper-DMRs.
DNA methylation can impact gene expression and resultant functional activity of cancer drivers (39, 40), so we reasoned that cfMeDIP-seq profiles may provide additional prognostic value independent of ctDNA abundance. To evaluate whether the previously identified hyper-DMRs may be of functional and prognostic significance in HNSCC, we interrogated DNA methylation, RNA expression, and clinical outcome data provided by TCGA for all available patients with HNSCC (n = 520; Fig. 5C). Of eligible plasma-derived hyper-DMRs (n = 764), the majority (n = 483; 63%) overlapped with regions hypermethylated in HNSCC primary tumors compared with adjacent normal tissue. Of note, several of these regions included methylated CpGs targeted by commercially available ctDNA diagnostic tests, including SEPT9, SHOX2, TWIST1, and ONECUT2 (Supplementary Fig. S9A).
Next, we used TCGA HNSCC cohort to identify a subset of the 483 DMRs that were associated with (i) prognosis (22) in multivariable Cox regression and (ii) expression of neighboring gene transcripts. Five DMRs satisfied both criteria, representing regions nearby ZSCAN31, LINC01391, GATA2-AS1, STK3, and OSR1 (Supplementary Fig. S9B–S9F). We constructed a CMS from the sum of beta-values across these five DMRs. In TCGA HNSCC cohort, patients with a higher CMS displayed inferior survival [HR = 1.67, 95% CI = (1.25–2.21), log-rank P = 3.4 × 10–4] (Fig. 5E).
Finally, we evaluated whether the CMS was prognostic when applied to plasma cfDNA methylation profiles from patients with HNSCC with detectable ctDNA. To account for the relative contribution of ctDNA methylation levels provided by the five putative prognostic markers, we normalized fragment-length informed methylation values from these regions to the entire 941 hyper-DMRs. As with TCGA analysis, patients with a higher CMS again displayed inferior survival [HR = 3.79, 95% CI = (0.925–15.5), log-rank P = 0.048] (Fig. 5F). CMS was not associated with ctDNA abundance as determined by methylation or mutations (Welch two sample t test: P = 0.61 and P = 0.82, respectively; Supplementary Fig. S9G and S9H), suggesting that increased methylation of these putative prognostic regions identified from TCGA may also be informative within cfMeDIP-seq profiles. Moreover, these results highlight how plasma cfDNA methylome profiling can be leveraged in combination with existing multi-omic cancer databases for biomarker discovery.
Disease surveillance after definitive treatment by cfMeDIP-seq
As cfMeDIP-seq achieved sensitive and quantitative ctDNA detection in patients with HNSCC, we reasoned that as with CAPP-seq (2, 14, 30, 41), cfMeDIP-seq may also be capable of monitoring therapy-related changes in ctDNA abundance. To quantify percent ctDNA within post-treatment cfMeDIP-seq profiles, we applied a linear transformation of mean RPKM across the previously identified plasma-derived hyper-DMRs (n = 941), limiting fragment size between 100 and 150 bp to further enrich ctDNA. We conservatively calculated the detection threshold of 0.2% ctDNA based on the maximum of mean RPKM values observed across all healthy controls.
Measuring changes in ctDNA abundance by cfMeDIP-seq during and after treatment, we observed a variety of kinetics indicative of complete clearance, partial clearance (>90% reduction), or no clearance (increase or ≤90% reduction) at the last sample collection (i.e., mid-radiotherapy or post-treatment; Fig. 6A; Supplementary Fig. S10). Among 18 eligible patients, 5 (28%) demonstrated no clearance (Fig. 6B). No clearance patients were more likely to experience disease recurrence compared with those with complete or partial clearance [HR = 8.73, 95% CI = (1.5–50.92), log-rank P = 0.0046] (Fig. 6C). Interestingly, all patients with ctDNA abundance greater at last sample collection than at diagnosis demonstrated disease recurrence. In addition, the only patient who did not have documented disease recurrence within this group was lost to follow-up but died within a year after treatment from unknown cause.
For the 13 patients with undetectable post-treatment ctDNA by cfMeDIP-seq, 9 remained disease free with a median of 44.4 months of follow-up (min = 12.2, max = 58.7). Among the other 4 patients, 1 had persistent disease within regional lymph nodes, and the others experienced relapse 3.5 to 7.7 months (median, 7.4 months) after last collection. Of note, these relapses among the patients with undetectable post-treatment ctDNA were considerably more delayed compared to the 4 relapses among the patients with detectable post-treatment ctDNA [median (range): 3.0 (1.7–5.2) months] after last collection. Taken together, these results demonstrate that plasma cfDNA methylome profiling by cfMeDIP-seq can be used to assess response to definitive treatment and identify patients at high risk of rapid recurrence.
Discussion
Broad implementation of ctDNA in clinical settings would be accelerated by methods that can be applied across patients and in the absence of tumor material. Tumor-naïve ctDNA detection currently encounters several limitations due to low ctDNA abundance. Recent studies have profiled paired PBLs and/or healthy control plasma to identify mutations derived from clonal hematopoiesis, a main contributor to false positive detection of ctDNA; however, the incorporation of orthogonal metrics may further improve accuracy and clinical applicability. Here, we evaluated the capabilities of multimodal genome-wide cfDNA profiling techniques for tumor-naïve ctDNA detection within a cohort of HNSCC patients with low ctDNA abundance. We demonstrated a high degree of concordance between ctDNA metrics (abundance and fragment lengths) detected by mutation-based and methylation-based profiling methods. Moreover, we showed that tumor-naïve multimodal ctDNA profiling can provide value by identifying putative prognostic biomarkers independent of ctDNA abundance, as well as by monitoring ctDNA abundance in serial samples.
Tumor-naïve detection of ctDNA has numerous practical advantages in both research and clinical settings. Although tumor mutational profiling can identify patient-specific markers for ctDNA detection at low abundance (42, 43), such personalized approaches rely on high-purity tumor samples from cancer types with sufficient mutational load. Mutational profiling for personalized assay design can be costly and time consuming, and it rarely accounts for genomic heterogeneity within primary tumors or across metastatic clones (3). In addition, ctDNA detection methods that depend on access to tumor tissue diminish a key advantage of noninvasive liquid biopsies. By integrating independent cfDNA properties, we achieved sensitive ctDNA detection in early-stage cancers without the disadvantages of tumor-informed methods.
In our study, the combination of CAPP-seq and cfMeDIP-seq enabled an in-depth molecular characterization of low-abundance ctDNA. Mutation-based ctDNA quantification contributed to the discovery of HNSCC-specific hyper-DMRs in plasma, some of which were confirmed to be prognostic even after adjusting for ctDNA abundance. Thus, simultaneous profiling of mutations and methylation may complement one another by revealing quantitative, tissue-specific, and prognostic ctDNA biomarkers. Moreover, methylome profiling may prove particularly useful in cancer types with few recurrent or clonal mutations.
This study makes multiple notable contributions. It is the first to combine analyses of cfDNA mutations, methylation, and fragment lengths. Moreover, we methodically profiled plasma samples and paired PBLs from both patients with HNSCC and risk-matched healthy controls. These analyses have revealed key insights regarding the optimal handling of multimodal profiling for ctDNA detection and characterization. For instance, our unique approaches to removing the contributing methylation signals from leukocytes and using fragment length characteristics to enrich for tumor-derived methylation will prove useful for future studies. We note that the cohorts in this study were not large enough to rely exclusively on machine learning approaches for signature generation as has been demonstrated in prior cfMeDIP-seq studies (32–34). Instead, we showed in modestly sized cohorts that incorporating mutational and PBL analyses could produce a robust and biologically plausible methylation signature. Finally, to our knowledge no study has previously identified prognostic methylated regions in cfDNA independent of ctDNA abundance. Taken together, our discovery framework could be widely applicable to other clinical settings where tumor tissue availability may be limited.
Future larger studies will be needed to validate specific signatures, thresholds, and prognostic models. Only interventional studies in which the ctDNA result is used in real time to influence patient management can definitively prove its clinical utility as a therapeutic biomarker. Nonetheless, as we have now demonstrated the clinical feasibility and potential use cases for locoregionally confined HPV-negative HNSCC, we envision that multimodal profiling of ctDNA will contribute to accelerated biomarker discovery and ultimately clinal utility for patients with a variety of cancer types. We expect this to be particularly beneficial in cases of low ctDNA abundance where personalized treatment regiments can lead to meaningful improvements in patient outcomes.
Authors' Disclosures
J.M. Burgener reports grants from Terry Fox Research Institute during the conduct of the study; in addition, J.M. Burgener has a patent for cancer detection, classification, prognostication, therapy prediction, and therapy monitoring using methylome analysis pending and licensed to DNAMx and a patent for use of cell-free DNA fragment length to improve detection of circulating tumor DNA pending and licensed to DNAMx. S.Y. Shen reports a patent for synthetic spike-in controls to account for biological and technical bias in MeDIP-seq experiments pending and licensed, a patent for use of methylated cell-free DNA analysis to guide therapy pending and licensed, a patent for cfMeDIP-seq technology: differentiating different types of cancers from within the cfDNA pending and licensed, and a patent for cfMeDIP-seq as a method to determine tissue and/or cell types giving rise to cfDNA & to identify a disease disorder pending and licensed. G. Liu reports grants and personal fees from AstraZeneca and Takeda; grants from Boehringer Ingelheim; and personal fees from Hoffman La Roche, Merck, Bristol Myers Squibb, and Pfizer outside the submitted work. A. Spreafico reports grants and personal fees from Novartis, Merck, BMS, JNJ, and Oncorus and grants from Symphogen, Roche, Northern Biologics, Regeneron, AstraZeneca MedImmune, ArrayBiopharma/Pfizer, GSK, Bayer, Surface Oncology, and Treadwell outside the submitted work. L.L. Siu reports personal fees from Merck, Pfizer, Celgene, AstraZeneca, Morphosys, Roche, Loxo, Oncorus, Symphogen, Seattle Genetics, GlaxoSmithKline, Voronoi, Treadwell Therapeutics, Arvinas, Tessa, Navire, Relay Therapeutics, Rubius, Janpix, Agios, and Treadwell Therapeutics and grants from Merck, Novartis, Bristol-Myers Squibb, Pfizer, Boerhinger-Ingelheim, GlaxoSmithKline, Roche/Genentech, Karyopharm, AstraZeneca, Astellas, Bayer, AbbVie, Amgen, Symphogen, Intensity Therapeutics, Mirati, Shattucks, and Avid outside the submitted work. M.M. Hoffman reports grants from Princess Margaret Cancer Foundation during the conduct of the study; in addition, M.M. Hoffman has a patent for “synthetic spike-in controls for cell-free MeDIP sequencing and methods of using same” pending and licensed to DNAMx. D.D. De Carvalho reports grants from CIHR and Princess Margaret Cancer Foundation during the conduct of the study; other support from DNAMx; grants from Nektar Therapeutics and Pfizer; and personal fees from AstraZeneca outside the submitted work; in addition, D.D. De Carvalho has a patent for cancer detection and classification using methylome analysis pending and licensed to DNAMx, a patent for cancer detection, classification, prognostication, therapy prediction and therapy monitoring using methylome analysis pending and licensed to DNAMx, a patent for use of cell-free DNA fragment length to improve detection of circulating tumor DNA pending and licensed to DNAMx, a patent for methods of capturing cell-free methylated DNA and uses of same pending and licensed to DNAMx, and a patent for synthetic spike-in controls for cell-free MeDIP sequencing and methods of using same pending. S.V. Bratman reports grants and personal fees from Princess Margaret Cancer Foundation and grants from Conquer Cancer Foundation of ASCO during the conduct of the study, as well as grants from Nektar Therapeutics, personal fees from Bristol Myers Squibb, and other support from DNAMx outside the submitted work; in addition, S.V. Bratman has a patent for identification and use of circulating nucleic acid tumor markers pending, licensed, and with royalties paid from Roche Molecular Diagnostics, a patent for cancer detection and classification using methylome analysis pending and licensed to DNAMx, a patent for cancer detection, classification, prognostication, therapy prediction and therapy monitoring using methylome analysis pending and licensed to DNAMx, and a patent for use of cell-free DNA fragment length to improve detection of circulating tumor DNA pending and licensed to DNAMx. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
D.D. De Carvalho and S.V. Bratman are supported by the Gattuso-Slaight Personalized Cancer Medicine Fund at the Princess Margaret Cancer Centre. J.M. Burgener is supported by a fellowship from the Strategic Training in Transdisciplinary Radiation Science for the 21st Century (STARS21) training program. D.D. De Carvalho is supported by the Canadian Institutes of Health Research (CIHR FDN 148430 and CIHR New Investigator Salary award 201512MSH-360794-228629); Ontario Institute for Cancer Research (OICR) with funds from the province of Ontario, Canada Research Chair (950-231346); and the Helen M. Cooke Professorship from Princess Margaret Cancer Foundation. S.V. Bratman is supported by a Career Development Award from Conquer Cancer Foundation of ASCO. Any opinions, findings, and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of the American Society of Clinical Oncology or the Conquer Cancer Foundation. We would like to acknowledge the Princess Margaret Cancer Centre Head & Neck Translational Program, supported by philanthropic funds from the Wharton Family, Joe's Team, Gordon Tozer, and the Reed Fund, as well as the labs of Fei-Fei Liu and Geoffrey Liu, for the provision of plasma samples. We also thank the Princess Margaret Genomics Centre for carrying out the NGS and the Bioinformatics and HPC Core, Princess Margaret Cancer Centre for their expertise in generating the NGS data. We also gratefully acknowledge the support from the Princess Margaret Cancer Foundation. We thank Aaron Newman, Maximilian Diehn, and Ash Alizadeh for input on CAPP-seq panel design and for critical reading of the article.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
J.M. Burgener: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft. J. Zou: Data curation, formal analysis, writing–original draft. Z. Zhao: Investigation, methodology. Y. Zheng: Investigation, methodology. S.Y. Shen: Methodology. S.H. Huang: Data curation. S. Keshavarzi: Formal analysis. W. Xu: Supervision, writing–review and editing. F.-F. Liu: Resources, funding acquisition, writing–review and editing. G. Liu: Resources, writing–review and editing. J.N. Waldron: Resources, writing–review and editing. I. Weinreb: Data curation. A. Spreafico: Writing–review and editing. L.L. Siu: Writing–review and editing. J.R. de Almeida: Writing–review and editing. D.P. Goldstein: Writing–review and editing. M.M. Hoffman: Writing–review and editing. D.D. De Carvalho: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. S.V. Bratman: Conceptualization, resources, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer 2020;1:276–90. [DOI] [PubMed] [Google Scholar]
- 2. Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early stage lung cancer evolution. Nature 2017;545:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chabon JJ, Hamilton EG, Kurtz DM, Esfahani MS, Chen B, Chaudhuri AA, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020;580:245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25:1928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018;10:eaat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang P, Lo YMD. The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet 2016;32:360–71. [DOI] [PubMed] [Google Scholar]
- 9. Wong K, Huang SH, O'Sullivan B, Lockwood G, Dale D, Michaelson T, et al. Point-of-care outcome assessment in the cancer clinic: audit of data quality. Radiother Oncol 2010;95:339–43. [DOI] [PubMed] [Google Scholar]
- 10. Wang TT, Abelson S, Zou J, Li T, Zhao Z, Dick JE, et al. High efficiency error suppression for accurate detection of low-frequency variants. Nucleic Acids Res 2019;47:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014;30:284–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu J, Liu S, Yin P, Bulun S, Dai Y. MeDEStrand: an improved method to infer genome-wide absolute methylation levels from DNA enrichment data. BMC Bioinformatics 2018;19:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 1995;57:289–300. [Google Scholar]
- 19. Cavalcante RG, Sartor MA. annotatr: genomic regions in context. Bioinformatics (Oxford, England) 2017;33:2381–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013;9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018;173:400–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat 1979;6:65–70. [Google Scholar]
- 24. Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 2019;565:312–7. [DOI] [PubMed] [Google Scholar]
- 25. Liu F, Killian JK, Yang M, Walker RL, Hong JA, Zhang M, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. 2010;29:3650–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ryser MD, Lee WT, Ready NE, Leder KZ, Foo J. Quantifying the dynamics of field cancerization in tobacco-related head and neck cancer: a multiscale modeling approach. Cancer Res 2016;76:7078–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tuna M, Amos CI, Mills GB. Genome-wide analysis of head and neck squamous cell carcinomas reveals HPV, TP53, smoking and acquired uniparental disomy. Neoplasia 2019;21:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science 2019;366:eaan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017;7:1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hellmann MD, Nabet BY, Rizvi H, Chaudhuri AA, Danny K, Dunphy M, et al. Circulating tumor DNA analysis to assess risk of progression after long-term response to PD- (L) 1 blockade in NSCLC. Clin Cancer Res 2020;26:2849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563:579–83. [DOI] [PubMed] [Google Scholar]
- 33. Nuzzo PV, Berchuck JE, Korthauer K, Spisak S, Nassar AH, Abou Alaiwi S, et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat Med 2020;26:1041–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nassiri F, Chakravarthy A, Feng S, Yi S. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat Med 2020;26:1044–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, Liu MC, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 2020;31:745–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lam WKJ, Jiang P, Chan KCA, Cheng SH, Zhang H, Peng W. Sequencing-based counting and size profiling of plasma Epstein–Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc Natl Acad Sci U S A 2018;115:E5115–E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Daniel C. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lam WKJ, Jiang P, Chan KCA, Peng W, Shang H, Heung MMS, et al. Methylation analysis of plasma DNA informs etiologies of Epstein-Barr virus-associated diseases. Nat Commun 2019;10:3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484–92. [DOI] [PubMed] [Google Scholar]
- 40. Koch A, Joosten SC, Feng Z, De Ruijter TC, Draht MX, Melotte V, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol 2018;15:459–66. [DOI] [PubMed] [Google Scholar]
- 41. Moding EJ, Liu Y, Nabet BY, Chabon JJ, Chaudhuri AA, Hui AB, et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat Cancer 2020;1:176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zviran A, Schulman RC, Shah M, Hill STK, Deochand S, Khamnei CC, et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat Med 2020;7:1114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McDonald BR, Contente-Cuomo T, Sammut SJ, Odenheimer-Bergman A, Ernst B, Perdigones N, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med 2019;11:eaax7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MeDIP-seq sequencing data generated from the FaDu cell line is accessible at BioProject ID PRJNA729472. Sequencing data for clinical samples are restricted to processed BED files due to privacy concerns and to comply with institutional ethics regulation. Data of cfMeDIP-seq profiles generated from patients and controls have been processed and deposited on Zenodo (10.5281/zenodo.4698004 and 10.5281/zenodo.4698867; Supplementary Table S10). Source data tables used to generate Fig. 2B–F, Fig. 4A and B, and Fig. 6A and B, are provided in Supplementary Tables S4, S7, S10, and S9, respectively. Scripts used to generate the findings described in this study are available within the “HNSCC multimodal” repository at https://github.com/bratmanlab/multimodal-hnscc. The Code Ocean capsule containing the reproducible analysis used can be found at https://codeocean.com/capsule/7176142/tree/v1 (DOI: 10.24433/CO.6511868.v1).
Figure 2.
PBL filtering for detection of ctDNA by CAPP-seq. A, Schematic describing detection and filtering of ctDNA-derived mutations by CAPP-seq. B, MAF of candidate mutations identified in HNSCC patient baseline plasma and PBLs. Dashed red box: 69 of 84 candidate mutations found only in plasma. Mutations found in both matched plasma and PBLs have strongly correlated MAFs (Pearson r = 0.94). C, Oncoprint of 15 candidate mutations identified in both matched patient plasma and PBLs. Top histogram: number of mutations per patient; right histogram: number of patients with one of 13 specified genes mutated. Color indicates type of mutation: missense (red), nonsense (blue), or silent (orange). D, MAF of 69 candidate mutations across HNSCC patient cfDNA (red circle) and PBLs (blue circle) before removal of PBL-associated mutations, and 43 mutations after this removal. The removed mutations represent false positive candidate mutations in ctDNA. Black bar: median of expected overlaps. Box: interquartile range (IQR) of expected overlaps. Whisker: most extreme value within quartile ±1.5 IQR of expected overlaps. E, Oncoprint highlighting frequently mutated genes in HNSCC by TCGA analysis (rows) and their frequency of detection across 30 HNSCC plasma samples (columns). Colors as in B, with additional splice sites (green), and nonhighlighted genes (gray). F, Mutation-based ctDNA levels in HNSCC patient baseline plasma samples as determined by mean MAF of PBL-filtered mutations. Dashed line: median mean allele fraction across all patients with HNSCC.
Figure 4.
Fragment length analysis between CAPP-seq and cfMeDIP-seq profiles. A, Median fragment length of detected mutations across patients with HNSCC by CAPP-seq. For each patient, the median fragment length of each mutation and matched reference allele was measured. The distribution of median fragment length for each mutation or matched reference allele is shown per patient. Black bar: median of fragment lengths. Box: IQR of fragment lengths. In cases with a single mutation, the colored line denotes the median length of fragments containing the mutation or matched reference allele, respectively. B, Fragment length distributions within HNSCC hypermethylated regions by cfMeDIP-seq. Fragment lengths from healthy controls were pooled prior to analysis, where each subsequent box denotes an individual HNSCC cfMeDIP-seq profile. + symbols denote HNSCC patients with detectable ctDNA by CAPP-seq (CAPP-seq positive). Black bar: median of fragment lengths. Box: IQR of fragment lengths. Individual HNSCC samples are ordered on the basis of increasing mean methylation (RPKM) within the hypermethylated regions. Dashed blue line defines the median fragment length across all healthy controls. C, Comparison of median fragment lengths from CAPP-seq and cfMeDIP-seq profiles. Points define individual HNSCC samples with methylation values above the median (n = 10). Solid red line: fitted linear regression model. Gray boundaries: 95% CI. D, Ratio of enrichment for hyper-DMR regions by fragments between 100 and 150 bp compared with enrichment for hyper-DMR regions by fragments between 100 and 220 bp. + symbols denote patients with HNSCC with detectable ctDNA by CAPP-seq (CAPP-seq positive).
Figure 6.
Clinical utility of ctDNA detection by cfMeDIP-seq for longitudinal monitoring. A, Representative ctDNA kinetics throughout treatment that were observed in this study. Complete clearance was defined as a change from detected ctDNA at diagnosis to a decrease in ctDNA abundance below the detection threshold (i.e., 0.2%) at first available mid-/post-treatment timepoint. Partial clearance was defined as an observed ≥90% decrease in ctDNA abundance from diagnosis compared with last follow-up. No clearance was defined as an observed <90% decrease or ≥0% increase in ctDNA from diagnosis to last follow-up. lastFU = sample collection at last follow-up, RT = radiotherapy. B, Changes in ctDNA abundance pre-treatment (baseline) to first available mid-/post-treatment timepoint across patients with HNSCC (n = 30). Red lines denote patients who demonstrated kinetics of no clearance, whereas gray lines denote patients with kinetics of clearance/partial clearance. C, Kaplan–Meier analysis depicting recurrence-free survival. Patients were stratified on the basis of kinetics of clearance (i.e., no clearance versus clearance/partial clearance).