Abstract
Purpose:
Human papillomavirus (HPV) DNA offers a convenient circulating tumor DNA (ctDNA) marker for HPV-associated malignancies, but current methods, such as digital PCR (dPCR), provide insufficient accuracy for clinical applications in patients with low disease burden. We asked whether a next-generation sequencing approach, HPV sequencing (HPV-seq), could provide quantitative and qualitative assessment of HPV ctDNA in low disease burden settings.
Experimental Design:
We conducted preclinical technical validation studies on HPV-seq and applied it retrospectively to a prospective multicenter cohort of patients with locally advanced cervix cancer (NCT02388698) and a cohort of patients with oropharynx cancer. HPV-seq results were compared with dPCR. The primary outcome was progression-free survival (PFS) according to end-of-treatment HPV ctDNA detectability.
Results:
HPV-seq achieved reproducible detection of HPV DNA at levels less than 0.6 copies in cell line data. HPV-seq and dPCR results for patients were highly correlated (R2 = 0.95, P = 1.9 × 10–29) with HPV-seq detecting ctDNA at levels down to 0.03 copies/mL plasma in dPCR-negative posttreatment samples. Detectable HPV ctDNA at end-of-treatment was associated with inferior PFS with 100% sensitivity and 67% specificity for recurrence. Accurate HPV genotyping was successful from 100% of pretreatment samples. HPV ctDNA fragment sizes were consistently shorter than non–cancer-derived cell-free DNA (cfDNA) fragments, and stereotyped cfDNA fragmentomic patterns were observed across HPV genomes.
Conclusions:
HPV-seq is a quantitative method for ctDNA detection that outperforms dPCR and reveals qualitative information about ctDNA. Our findings in this proof-of-principle study could have implications for treatment monitoring of disease burden in HPV-related cancers. Future prospective studies are needed to confirm that patients with undetectable HPV ctDNA following chemoradiotherapy have exceptionally high cure rates.
Translational Relevance.
HPV-associated cancers often present with locoregionally confined disease and are treated with curative intent. An emerging treatment paradigm includes chemoradiotherapy or surgery followed by adjuvant treatment. An accurate test of minimal residual disease (MRD) could enable personalized use of adjuvant therapy by identifying patients who are cured after radical therapy and therefore would not benefit from additional treatment. We asked whether a new sequencing approach for HPV ctDNA detection would enable more accurate MRD detection in HPV-associated cancers. In a cohort of patients with locally advanced cervix cancer treated with chemoradiotherapy, HPV-seq revealed HPV genotype and detected 20-fold lower levels of HPV ctDNA than dPCR while achieving 100% sensitivity and 67% specificity at the end-of-treatment timepoint for PFS. Thus, HPV-seq is a promising tool for monitoring disease burden in HPV-associated cancers and could open the door to new potential clinical applications of HPV ctDNA analysis.
Introduction
Recent methodologic developments based on high throughput sequencing have expanded the potential applications for circulating tumor DNA (ctDNA) analysis (1, 2). Many clinical settings – particularly those involving patients with low tumor burden – rely on accurate detection of minute levels of ctDNA within a much more abundant pool of cell-free DNA (cfDNA) fragments. Further improvements in low-level ctDNA detection/analysis will have a tremendous impact on future cancer diagnostics for screening, prognostication, and treatment monitoring.
To drive ultrasensitive detection of ctDNA, we and others have developed approaches to increase the number of cancer-specific markers that can be simultaneously assessed in a given patient (3–7). By doing so, simulated data has suggested that dramatic gains may be achieved in the probability of detecting less than 1 genome equivalent of ctDNA. This threshold has been shown to be clinically meaningful in low disease burden settings (8, 9) and can therefore be viewed as an important benchmark for ctDNA technological development.
Cancer types that are driven by oncogenic viruses, such as human papillomavirus (HPV)–associated cancers of the cervix and oropharynx, represent approximately 13% of the global cancer burden (10). In such cancers, the viral genome can be leveraged to distinguish ctDNA from other cellular sources of cfDNA (11–22). Most previous efforts have achieved this through quantitative or digital PCR (qPCR or dPCR; refs. 23, 24), but robust detection of less than 1 copy has not been observed using these methods (12, 13, 18, 24, 25). Viral genome hybrid-capture sequencing has also recently shown promise as a means for ctDNA quantification while simultaneously providing qualitative information regarding sequenced cfDNA fragments, such as fragment length, that may improve the specificity of ctDNA detection (26).
Here, we developed an optimized framework for viral genome hybrid-capture sequencing that provides robust quantitative and qualitative information about the sequenced cfDNA fragments. Using HPV sequencing (HPV-seq) as a model, we show that full-length viral genome dual-strand capture allows for reliable detection of less than 1 copy of ctDNA while preserving HPV genotype and cfDNA fragment-length information (Fig. 1A). For HPV-related malignancies, HPV-seq represents a promising ultrasensitive approach for ctDNA detection and analysis.
Figure 1.
Overview of HPV-seq and dual-strand hybrid capture. A, HPV-seq conducted on plasma cfDNA is designed to provide quantitative and qualitative information about ctDNA in patients with HPV-associated cancers. In addition to being highly sensitive and quantitative, HPV-seq can report on ctDNA fragment size and HPV genotype. Each full-length viral genome (episome or linearized genome) is expected to yield approximately 50 distinct cfDNA fragments. B, HPV-seq is conducted using hybrid-capture sequencing with single-stranded [sense (+) and/or antisense (−)] biotinylated baits tiled across the HPV genome: (i) single-strand viral genome hybrid capture, (ii) sequential dual-strand hybrid capture, and (iii) simultaneous dual-strand hybrid capture. C, Compared with single-strand hybrid capture (i), dual-strand hybrid capture using either a sequential (ii) or simultaneous (iii) approach recovers more HPV molecules. Results were normalized to the degree of HPV sequence enrichment with a single round of capture using single-stranded baits (left-most bar). Subjecting the unbound library to another round of hybrid capture using baits targeting the same strand (second bar from left) did not improve HPV sequence enrichment. The degree of HPV DNA enrichment in postcapture libraries was determined using HPV-16 E6 and E7 dPCR assays. N = 4 per condition. Error bars represent SD. Asterisk indicates statistical significance (P < 0.05) in comparison with single-strand capture conditions.
Materials and Methods
Cell lines
The SiHa cervix cancer cell line, which harbors 1 to 2 integrated copies of HPV-16 (27), was obtained from ATCC (catalog no. HTB-35, RRID:CVCL_0032). The HeLa cervix cancer cell line (RRID:CVCL_0030), which harbors 10 to 50 integrated copies of HPV-18 (28–31), was a kind gift from Dr. Richard Hill (Princess Margaret Cancer Centre). The FaDu head and neck squamous cell carcinoma cell line (RRID:CVCL_1218), which is HPV-negative, was a kind gift from Dr. Bradly Wouters (Princess Margaret Cancer Centre). The SW48 colorectal adenocarcinoma cell line was obtained from ATCC (catalog no. CCL-231, RRID:CVCL_1724). The identity of all cell lines was confirmed via short tandem repeat (STR) profiling, and mycoplasma testing was performed prior to their utilization. Genomic DNA was purified using DNeasy kits (Qiagen) and quantified by Qubit (Life Technologies).
Participants and samples
Patients with locally advanced cervix cancer treated with standard-of-care chemoradiotherapy were accrued onto a prospective clinical trial (NCT02388698) for correlative biomarker analysis. This study was approved by the Ontario Cancer Research Ethics Board and performed in accordance with the principles of the Declaration of Helsinki. All participants provided written informed consent. Results of the primary analysis were previously reported (15). Patients who consented to optional biobanking and with sufficient stored biospecimens were included in this study. Progression-free survival (PFS) was measured from the date of diagnosis. The final cohort included 17 patients with HPV-positive cervix cancer. Patients diagnosed with oropharynx cancer between 2014 and 2016 within the Princess Margaret Cancer Centre's Head and Neck Translational Research program were identified from a prospective Anthology of Clinical Outcomes (32). The final cohort included 13 patients with HPV-positive oropharynx cancer. Control individuals' plasma samples (21 female, 29 male) were obtained following the University Health Network's Research Ethics Board approval. Plasma processing, DNA extraction, and quality assessment were performed as previously described (15). Four cervix cancer samples showed relatively high plasma cfDNA yields (>40 ng/mL; Supplementary Table S6). Analysis of fragment lengths revealed no genomic DNA contamination and confirmed accurate cfDNA yields within these samples (Supplementary Fig. S8).
Targeted panel design
For hybrid capture, panels of 5′-biotinylated 120 nt single-stranded DNA baits (xGen Lockdown probes) were designed and synthesized by Integrated DNA Technologies. Each panel consisted of baits targeting HPV sequences as well as regions of 12 human genes frequently mutated in squamous cell carcinomas (CASP8, CDKN2A, EP300, FBXW7, HRAS, MAPK1, NFE2L2, NOTCH1, PIK3CA, PTEN, TERT, TP53; Supplementary File). Nine different HPV panels were used in this study: (i) HPV genotyping panel consisting of (+) strand baits targeting the E6 and E7 genes from each of 38 HPV types; (ii) HPV-16 (+) & (−) strand baits targeting the full-length viral genome; (iii) HPV-18 (+) & (−) strand baits targeting the full-length viral genome; (iv) HPV-33 (+) strand baits targeting the full-length viral genome; (v) HPV-45 (+) strand baits targeting the full-length viral genome; (vi) HPV-52 (+) strand baits targeting the full-length viral genome; (vii) HPV-31 (+) & (−) strand baits targeting the full-length viral genome; (viii) HPV-33 (+) & (−) strand baits targeting the full-length viral genome; (ix) HPV-35 (+) & (−) strand baits targeting the full-length viral genome. Baits were 1X tiled (end-to-end) for all full-length viral-genome panels. For all 5 HPV dual-strand panels, the HPV (−) strand baits were 50% (60nt) offset from the HPV (+) strand baits.
Library preparation
Illumina-compatible sequencing libraries were prepared from 20 ng of sheared cell line genomic DNA (i.e., SiHa and HeLa) or cfDNA from the cervix cancer cohort. Cell line genomic DNA was sheared using a Covaris M220 sonicator (Covaris) followed by cleanup and size selection using Agencourt AMPure XP beads (Beckman Coulter). DNA concentration was assessed by Qubit (Life Technologies). Sheared cell line genomic DNA was spiked into sheared FaDu genomic DNA at 10% for initial methodologic development and then as an 8-point dilution series (3%, 1%, 0.3%, 0.1%, 0.03%, 0.01%, 0.003%, and 0%) to test analytic sensitivity. Libraries were constructed using KAPA Hyper Prep kits (catalog no. KK8504, Kapa Biosystems) with custom unique molecular identifier (UMI)–containing adapters (33). Following end repair and A-tailing, adapter ligation was performed overnight using 100-fold molar excess of adapters. Agencourt AMPure XP beads were used for library clean-up and ligated fragments were amplified between 4 to 8 cycles using 0.5 μmol/L Illumina universal and sample-specific index primers. Final library quality control was performed using a Bioanalyzer 2100 (Agilent), and concentration was measured using Qubit. The same preparation was applied to the oropharynx cancer cohort (10 ng cfDNA input) and control cohort (10–30 ng cfDNA input).
Hybrid capture
For initial methodologic development, hybrid capture was conducted on approximately 500 ng of amplified library without multiplexing and carried out with technical duplicates for each condition. Using the 10% SiHa DNA mixture, we evaluated the effect of individual probe concentration and hybridization temperature/duration on target-sequence enrichment. Probes were included at either 4,000 or 40,000 amol per probe. Hybridization was carried out at either 47°C for 72 hours or at 65°C for 18 hours. Thereafter, all other hybrid capture reactions were carried out with 4,000 amol per probe at 65°C for 18 hours. Hybrid capture reactions contained 5 μL of 1 mg/mL Cot-1 DNA and 1 nmol each of the IDT xGen Universal Blocking Oligos (TS-p5 and TS-p7, 8nt). The mixture was dried using a SpeedVac and then re-suspended in 1.1 μL water, 8.5 μL NimbleGen 2x hybridization buffer and 3.4 μL NimbleGen hybridization component A. The mixture was heat denatured at 95°C for 10 minutes before adding 4 μL of IDT xGen Lockdown Probes (1,000 amol/probe/μL). After an 18-hour hybridization at 65°C, targets of interest were captured by incubation with 5′-biotinylated oligos, and then were pulled down by Streptavidin M-270 Dynabeads (Thermo Fisher Scientific), followed by buffer wash steps to remove unbound DNA. Captured DNA was subjected to on-bead PCR amplification with 25 μL of 2X KAPA HiFi HotStart ReadyMix, 5 μL of 10 μmol/L Illumina Primermix, and 20 μL of beads captured DNA. Amplified captured DNA was cleaned up with 1X Agencourt AMPure XP beads. Enriched libraries were eluted with Tris-EDTA (TE) buffer (pH 8.0) for downstream analysis. For sequential hybrid capture reactions, following initial hybridization and Streptavidin M-270 Dynabeads incubation, DNA bound to the Dynabeads was separated from the supernatant on a magnetic rack. The supernatant containing the unbound DNA was quickly transferred into another PCR tube for a second round of probe hybridization. Washing of the Dynabeads, on-beads PCR amplification, and DNA clean up was conducted as described above. Enriched libraries from sequential hybrid-capture reactions were eluted with TE pH 8.0 into individual tubes for downstream analysis. With the optimized protocol built, hybrid capture was conducted with multiplexing on samples for non–HPV-associated colon cancer cell line, patients, and controls.
Target sequence enrichment analysis
In order to determine the degree of target sequence enrichment in postcapture versus precapture libraries, we designed E6 and E7 primers/probe sets for dPCR. For HPV-16 E6: HPV16_E6_Forward, 5′-ACTGTGTCCTGAAGAAAAGCA; HPV16_E6_Reverse, 5′-GTCCACCGACCCCTTATATT; and a double quenched probe 5′-/56-FAM/ACATCTGGA/ZEN/CAAAAAGCAAAGATTCCA/3IABkFQ/. For HPV-16 E7: HPV16_E7_Forward, 5′-GAGGAGGATGAAATAGATGGTC; HPV16_E7_Reverse, 5′-CCGAAGCGTAGAGTCACA; and a double quenched probe 5′-/5HEX/TGGACAAGC/ZEN/AGAACCGGACA/3IABkFQ/. First, pre- and postcapture libraries were quantified using the dPCR Library Quantification Kit for Illumina Truseq (Bio-Rad, catalog no. 1863040). All dPCR reactions were carried out using a QX200 Droplet Digital PCR system (Bio-Rad). Thermocycling for 40 cycles was performed on a C1000 Touch Thermal Cycler with 96-Deep Well Reaction Module (Bio-Rad). Droplet analysis and copy-number quantification was performed using QuantaSoft software (Bio-Rad). We prepared dilutions of 2 ng/μL for precapture libraries and 0.02 pg/μL for postcapture libraries; 5 μL of each diluted library was used for E6 and E7 absolute quantification by dPCR. The degree of enrichment of the E6 and E7 sequences was determined by dividing the concentration in postcapture libraries by the concentration in the precaptured libraries.
Sequencing analysis
High throughput DNA sequencing was performed on Illumina NextSeq500, NovaSeq6000, or MiSeq platforms with paired-end reads of ≥75 bp. A 2-nt UMI and a 1 nt invariant spacer sequence were removed from each read (33). A thymine base was encoded in the third position for adapter ligation and a spacer filter was enforced to remove reads not compliant with this design. For the remaining reads (>92%), the UMIs were appended into the header for unique molecular identification. Next, reads were aligned to the human reference genome hg19 using Burrows–Wheeler Aligner (BWA)-mem (v 0.7.15; ref. 34) and SAMtools (v 1.3.1; ref. 35), and recalibrated for base quality score using the Genome Analysis ToolKit (GATK) BaseRecalibrator (v 3.4–46) according to best practice (36). The aligned reads were input to ConsensusCruncher (https://github.com/pughlab/ConsensusCruncher) to identify unique molecules. ConsensusCruncher is a Python-based tool that amalgamates reads derived from the same DNA template labeled with the same UMI into a consensus read (33).
HPV genotyping
Human-unmapped reads were extracted and aligned to a database of 38 HPV reference genomes (Supplementary Table S5). The aligned reads were input to ConsensusCruncher to identify unique molecules. From these unique molecules, properly-paired reads with high mapping quality (≥30) were considered for genotyping. High mapping quality score was included in our pipeline to improve resolution of genotyping by filtering reads mapping to multiple genomes. Following these steps, the HPV genotype was assigned according to the virus with the most reads. Mutations in HPV sequences were evaluated within Integrated Genomics Viewer (37). For Fig. 3A, comparisons were made with sequences in the HPV-33 dPCR primers and probe: HPV33_E6_Forward, 5′-ATATTTCGGGTCGTTGGGCA; double quenched probe, 5′-/56-FAM/GCGCTGTGC/ZEN/GGCGTG/3IABkFQ/; HPV33_E6_Reverse, 5′-CTACGTCGGGACCTCCAA.
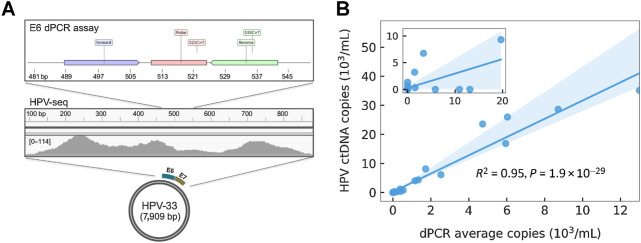
Figure 3.
HPV genotyping and ctDNA quantification from plasma cfDNA using HPV-seq. A, HPV-seq correctly genotyped the baseline (pretreatment) plasma cfDNA sample from P19 as HPV-33. HPV-mapping reads were found across the E6 and E7 genes of HPV-33 (100% match to correct genotype). Comparison with dPCR results from the same sample, in which the E6 assay was false negative, reveals mutations in the reverse primer and probe sequences that likely affected dPCR assay performance. B, Comparison of HPV ctDNA copies/mL plasma evaluated by dPCR (x-axis; average of E6 and E7 copies) and HPV-seq (y-axis) for 33 plasma cfDNA samples of patients with cervix cancer obtained at end-of-treatment or posttreatment and 13 plasma cfDNA samples of patients with OPC obtained at pretreatment. Linear regression and its 95% CI (shaded) are shown.
HPV quantification
The HPV copy number was calculated using the following formula, Copies (HPV) = DepthHPV/DepthHuman × Copies (Human), where Depth is the depth of coverage sequenced on the given panel, and Copies (Human) is evaluated using N (ng) × 1,000/3.3 and then converted to Copies/mL based on the cfDNA concentration of a plasma sample. The estimation of depth of coverage is based on the sequenced unique molecules of HPV and human DNAs. First, the human-unmapped reads were aligned to the HPV genotype-specific reference genome (e.g., HPV-16). Then, aligned reads were input to ConsensusCruncher to identify unique molecules. From the all unique molecules output of ConsensusCruncher, the number of HPV-mapping and (on-target) human-mapping properly paired reads were determined. Here, no mapping quality control was applied to the HPV-mapping reads in order to maximize the recovery of all HPV-mapping reads (i.e., even reads that might map to multiple HPV genotypes).
Fragment length analysis
Lengths of sequenced fragments were evaluated using CollectInsertSizeMetrics of the Picard tool (http://broadinstitute.github.io/picard/). In this study, the input bam files were ‘all unique molecules’ generated by ConsensusCruncher. Human-mapping and HPV-mapping reads were analyzed separately for comparison.
Small nucleotide variant calling
Mutation data from the SiHa and FaDu cell lines were obtained from the Catalogue of Somatic Mutations in Cancer (COSMIC) database (38). Within the 9-kb human gene capture panel that was part of HPV-seq, SiHa harbors one unique mutation within the CASP8 gene (chr2:202131411; c.202C>T). In the cell line dilution series, variants were called by intersecting output from Ides (7) and Vardict 2 (39) with requirement of ≥2 supporting reads.
Software and statistical analysis
Pearson correlation coefficients and p values were calculated for dPCR and HPV-seq results using SciPy (40) in Python v3.7.2 (41). The Student t test was used to test statistical significance of the degree of HPV sequence enrichment as well as differences in fragment lengths between human and HPV DNA fragments using R v3.5.1 (http://www.R-project.org/). Error bars and ranges indicate SD. The probability of HPV detection in Fig. 2 was estimated based on 100-times downsamplings on each coverage level. For Fig. 3B, the linear regression with associated 95% confidence interval (CI) was generated using seaborn (https://doi.org/10.5281/zenodo.12710). For Fig. 2, data was plotted using brokenaxes (https://github.com/bendichter/brokenaxes) and matplotlib v2.2.3 (https://doi.org/10.5281/zenodo.1343133). For Fig. 3A, primers/probes were plotted using DNA Features Viewer (https://github.com/Edinburgh-Genome-Foundry/DnaFeaturesViewer). The Mann–Whitney test was used to test differences in end-of-treatment HPV ctDNA levels between recurrent and nonrecurrent patients. Kaplan–Meier analysis and log-rank test were performed in SAS v9.3 (SAS Institute). All tests were two-tailed. A P value of 0.05 was considered statistically significant.
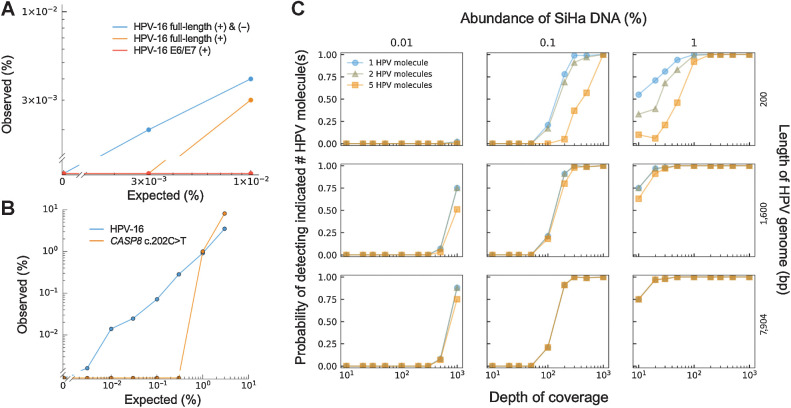
Figure 2.
Analytic sensitivity of HPV-seq. A, HPV-seq was conducted on fragmented SiHa genomic DNA at the indicated dilution. Hybrid capture baits targeted the indicated HPV-16 sequences. The lower limit of detection (LLOD) of HPV-seq was dependent on the use of dual-strand hybrid capture and the length of HPV-16 genome targeted by the baits. B, HPV-seq with full-length dual-strand hybrid capture (blue) provided an improvement in analytic sensitivity and LLOD (0.003%) as compared with hybrid capture for a single mutation (1%). C, Influence of multiple markers and sequencing depth on LLOD. Downsampling of HPV-seq data from full-length dual-strand hybrid capture demonstrates the dependence of the LLOD on the targeted length of HPV-16 genome (i.e., number of markers; right y-axis) and the sequencing depth (x-axis). The probability of detecting the indicated number of HPV molecules (1, blue circles; 2, gray triangles; 5, yellow squares) is shown (left y-axis).
Results
Enhanced enrichment of viral DNA from sequencing libraries
Efficient recovery of target sequences from cfDNA is necessary for robust ctDNA detection by hybrid-capture sequencing. We first evaluated the degree of enrichment of HPV sequences using a range of hybrid capture conditions. Fragmented cell line genomic DNA containing 10% SiHa (cervix cancer cell line with single integrated full-length HPV-16 genome) was used to simulate cfDNA. Following construction of Illumina-compatible sequencing libraries, single-stranded sense (+) biotinylated DNA baits were used at a range of concentrations for hybrid capture. Increasing concentrations up to 4,000 amol per bait produced greater enrichment (data not shown), but concentrations above 4,000 amol per bait did not improve target recovery as measured by dPCR (Supplementary Fig. S1). Moreover, at ≥4,000 amol per bait, there was no significant impact on target recovery from increasing hybrid-capture incubation time from 18 hours at 65°C to 72 hours at 47°C. Thus, 4,000 amol per bait with 18 hours of incubation at 65°C provided optimal experimental conditions that were utilized for all subsequent analyses.
Next, we asked whether the hybrid capture reaction had harvested all of the target sequences in the library. To test this, we examined the unbound library following hybrid capture to see whether there were retained HPV sequences. Subjecting the unbound library to another round of hybrid capture using the same single-stranded (+) baits had a minor impact on total target recovery (Fig. 1B and C), suggesting that the hybrid capture reaction had approached saturation as expected. However, substituting the (+) baits for staggered antisense (−) baits (see Methods) in the second round of hybrid capture resulted in a dramatic improvement in the degree of enrichment of target sequences (98 ± 16% increase; P = 3.1 × 10–4). This effect was maintained when the (+) and (−) baits were all combined in a single-capture reaction (67 ± 14% increase; P = 1.8 × 10–3). Based on these results, dual-strand hybrid capture may improve upon standard target enrichment procedures for robust ctDNA detection.
HPV-seq enables ultrasensitive detection of cancer DNA
Having established our hybrid capture methodology for HPV-seq, we next sought to evaluate its analytic sensitivity and lower limit of detection (LLOD). Reported qPCR/dPCR approaches for HPV ctDNA detection have LLOD of ≥1 copy (13, 18), which may not be sufficient for low disease burden settings, so we tested the ability of HPV-seq to detect HPV DNA at levels less than 1 copy. HPV-seq with dual-strand full-length viral capture was able to detect SiHa DNA at 0.01% and 0.003%, corresponding to 0.6 and 0.2 copies, respectively, with no signal in the negative control (0%) condition (Fig. 2A), indicating robust LLOD at less than 1 copy. Of note, 16 unique HPV DNA fragments were recovered at the 0.003% dilution level, indicating that even lower levels might still be detectable. In contrast, single-strand full-length viral capture failed to detect SiHA DNA at 0.003%, and single-strand partial-length (360 bps within the E6 and E7 genes) viral capture did not detect SiHa DNA at either level. Similar results were observed for detection of HeLa DNA, with both single-strand and dual-strand full-length capture able to detect HeLa DNA at 0.003% (0.7 copies; Supplementary Fig. S3).
We next compared the analytic sensitivity of HPV-seq with that of mutation detection following hybrid capture. The baits used for HPV-seq encompassed a single clonal heterozygous mutation in SiHa within the CASP8 gene, allowing us to evaluate LLOD in a dilution series. The LLOD using just the CASP8 mutation was only 1% (60 copies), whereas HPV detection again provided a LLOD of 0.003% (0.2 copies) and displayed a high correlation with the expected levels (R2 = 0.994, 95% CI 0.988–1.00, P = 4.2 × 10–08; Fig. 2B; Supplementary Fig. S2). These results highlight the potential of utilizing HPV DNA fragments to drive analytic sensitivity.
When fragmented to the typical size of cfDNA (approximately 140–180 bp), the 7.9 kb HPV-16 genome should yield approximately 50 independent cancer-specific markers. We reasoned that the LLOD in the SiHa dilution series would be dependent on this large number of independent markers of HPV-16. Indeed, the smaller E6/E7 capture baits failed to produce robust detection of SiHa DNA at less than 1 copy (Fig. 2A). To further test this hypothesis, we subsampled the sequencing reads from HPV-seq with dual-strand full-length viral capture conducted on 0.01%, 0.1%, and 1% fragmented SiHa DNA (Fig. 2C). Reads were subsampled at various depths of coverage ranging from 10- to 1,000-fold. We evaluated reads mapping to 200 bp, 1,600 bp, or the full-length 7,904 bp HPV-16 genome to simulate the effect of variable numbers of independent markers on LLOD. As expected, the bait lengths for the HPV-16 genome increased the likelihood of HPV detection, with improved analytic sensitivity observed across the three dilution levels. Taken together, these results show the impact of dual-strand full-length viral capture on ultrasensitive detection of HPV.
HPV-seq for ctDNA detection and genotyping in patients with HPV-associated cervix and oropharynx cancer
Uterine cervix cancer is one of several cancer types that is commonly HPV-associated. We previously evaluated the clinical utility of HPV genotype-matched dPCR from plasma cfDNA for treatment response monitoring in a cohort of patients with locally advanced cervix cancer (15). In this prospective multicenter series, baseline (pretreatment) sensitivity was high, but the small dynamic range in longitudinal samples presented challenges for interpreting results at end-of-treatment or posttreatment. We therefore asked whether HPV-seq could improve upon the quantitative and qualitative information obtained from these plasma cfDNA samples. Of the 57 samples from the previous study, 38 samples with remaining plasma were subjected to HPV-seq (Supplementary Table S1) using either (i) an HPV genotyping panel (single-strand baits targeting the E6 and E7 genes from each of 38 HPV types; N = 5), (ii) dual-strand full-length viral capture for HPV-16 (N = 25), or (iii) single-strand full-length viral capture for other HPV types (N = 8). The HPV genotyping panel allowed us to test whether HPV-seq could provide HPV genotype information directly from plasma, while the full-length viral capture panels were used to test HPV-seq as a monitoring tool for detecting residual disease. To more broadly evaluate the utility of HPV-seq for ctDNA detection and genotyping, a cohort of 13 HPV-associated patients with oropharynx cancer was subjected to HPV-seq (Supplementary Table S1) using either (i) a HPV genotyping panel (as described above; N = 13), or (ii) dual-strand full-length viral capture for HPV-16, HPV-18, HPV-31, HPV-33, and HPV-35 (N = 13).
We evaluated the performance of HPV-seq for HPV genotyping. This analysis was conducted by applying the HPV genotyping panel to available pretreatment cervix cancer plasma samples (Supplementary Table S1) and compared with cervical swab genotyping results (by dPCR) previously reported for this cohort (15), pretreatment oropharynx cancer plasma samples, 50 control plasma samples from donors (female, n = 21; male, n = 29), and 5 non–HPV-associated colon cancer cell line samples (SW48). Among 5 patients with cervix cancer patients harboring tumors positive for HPV-16 (N = 2), HPV-33, HPV-45, and HPV-52 and 13 patients with oropharynx cancer patients harboring tumors positive for HPV-16, HPV ctDNA was detected in all samples. Among the 50 control plasma samples, HPV cfDNA was detected in 6 samples (2 female and 4 male), but was not detected in any colon cancer cell line samples (Table 1; Supplementary Table S1; Supplementary Fig. S4). Levels in patients were highly correlated with dPCR results (R2 = 0.93; 95% CI, 0.88–0.99; P = 1.73 × 10–10). Altogether, more than 99.7% of HPV-mapping reads (80%–100% for each patient) were assigned to the same HPV genotype that was observed in the corresponding tumor tissue (Table 1; Supplementary Fig. S5). A few reads mapped to alternative HPV types (<0.004%), which could be consistent with the occurrence of multiple infections in these patients (42–45). Considering the HPV genotype with the largest number of mapped reads, HPV-seq provided an overall accuracy of 100% for genotyping in this patient cohort. This was consistent with our findings from SiHa (HPV-16 positive) and HeLa (HPV-18 positive) cell line DNA, in which 100% of HPV reads mapped to the correct genotype. Based on 89 samples evaluated with the genotyping panel, the accuracies for HPV-16 and HPV-18 were 94.38% and 98.88%, respectively (Supplementary Table S2).
Table 1.
HPV genotyping using HPV-seq for plasma cfDNA samples from 5 cervix cancer and 13 OPC patients prior to treatment, 50 control individuals, and 5 non–HPV-associated SW48 cell line samples.
| dPCR copies/mL | HPV molecules from plasma HPV-seqa | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Tissue HPV type | E6 | E7 | HPV-16 | HPV-33 | HPV-45 | HPV-52 | HPV-67 | Other typesd | Plasma HPV typeb | HPV-seq genotype accuracyc | HPV-seq copies/mL | ||||
| Controls | ||||||||||||||||
| 5 SW48 | NA | — | — | 0 | 0 | 0 | 0 | 0 | NA | — | 0 | |||||
| 44 Controls | NA | — | — | 0 | 0 | 0 | 0 | 0 | NA | — | 0 | |||||
| Control 6 | NA | — | — | 34 | 0 | 0 | 0 | 0 | HPV-16 | — | 0.56 | |||||
| Control 13 | NA | — | — | 0 | 0 | 0 | 0 | 28 | HPV-35 (2), HPV-83 (2) | HPV-67 | — | 0.34 | ||||
| Control 14 | NA | — | — | 292 | 0 | 0 | 0 | 0 | HPV-16 | — | 3.15 | |||||
| Control 33 | NA | — | — | 2 | 0 | 0 | 0 | 0 | HPV-16 | — | 0.05 | |||||
| Control 35 | NA | — | — | 162 | 126 | 0 | 0 | 0 | HPV-35 (170), HPV-31 (122), HPV-18 (108) |
HPV-35 |
— | 8.39 | ||||
| Control 37 | NA | — | — | 22 | 0 | 0 | 0 | 0 |
HPV-16 |
— | 0.34 | |||||
| Patients with cervix cancer | ||||||||||||||||
| P2 | HPV-16 | 1.01 | 1.01 | 8 | 0 | 0 | 0 | 0 | HPV-18 (2) | HPV-16 | 80% | 0.55 | ||||
| P6 | HPV-16 | 9.11 | 32.52 | 84 | 0 | 0 | 0 | 0 | HPV-16 | 100% | 14.84 | |||||
| P10 | HPV-45 | 2.83 | 5.67 | 0 | 0 | 112 | 2 | 0 | HPV-45 | 98.25% | 8.20 | |||||
| P14 | HPV-52 | 901.91 | 851.07 | 0 | 0 | 0 | 1,572 | 0 | HPV-12 (2) | HPV-52 | 99.87% | 117.83 | ||||
| P19 | HPV-33 | 0 | 113.49 | 0 | 545 | 0 | 0 | 0 | HPV-33 | 100% | 79.95 | |||||
| Patients with oropharynx cancer | ||||||||||||||||
| OPC1 | HPV-16 | 9,491.00 | 7,968.38 | 211,621 | 0 | 0 | 0 | 0 | HPV-16 | 100% | 28,130.32 | |||||
| OPC2 | HPV-16 | 83.93 | 46.08 | 3,879 | 0 | 0 | 0 | 0 | HPV-16 | 100% | 989.33 | |||||
| OPC3 | HPV-16 | 4.19 | 2.49 | 174 | 0 | 0 | 0 | 0 | HPV-16 | 100% | 29.39 | |||||
| OPC4 | HPV-16 | 385.86 | 372.46 | 37,461 | 0 | 0 | 0 | 0 | HPV-83 (2) | HPV-16 | 99.99% | 4,130.60 | ||||
| OPC5 | HPV-16 | 5,021.90 | 4,432.08 | 569,541 | 0 | 0 | 0 | 0 | HPV-35 (8) | HPV-16 | >99.99% | 50,836.63 | ||||
| OPC6 | HPV-16 | 1,227.26 | 1,104.35 | 69,339 | 0 | 0 | 0 | 0 | HPV-16 | 100% | 12,121.82 | |||||
| OPC7 | HPV-16 | 1,317.51 | 1,361.13 | 60,319 | 0 | 2 | 0 | 0 | HPV-35 (8), HPV-83 (2), HPV-6 (2), HPV-42 (2) | HPV-16 | 99.97% | 6,750.78 | ||||
| OPC8 | HPV-16 | 12,925.02 | 13,048.90 | 75,696 | 0 | 0 | 0 | 0 | HPV-16 | 100% | 37,955.25 | |||||
| OPC9 | HPV-16 | 1,860.36 | 3,199.94 | 14,379 | 0 | 0 | 0 | 0 | HPV-16 | 100% | 10,571.01 | |||||
| OPC10 | HPV-16 | 6,052.22 | 6,026.24 | 432,456 | 0 | 0 | 0 | 0 | HPV-35 (12), HPV-31 (2), HPV-71 (2) | HPV-16 | >99.99% | 44,812.17 | ||||
| OPC11 | HPV-16 | 23.92 | 16.64 | 241 | 0 | 0 | 0 | 0 | HPV-16 | 100% | 62.93 | |||||
| OPC12 | HPV-16 | 5,605.84 | 6,269.95 | 319,930 | 0 | 0 | 0 | 0 | HPV-35 (4), HPV-31 (2) | HPV-16 | >99.99% | 54,848.97 | ||||
| OPC13 | HPV-16 | 1,592.93 | 1,847.60 | 121,607 | 0 | 0 | 0 | 0 | HPV-83 (2), HPV-39 (2) | HPV-16 | >99.99% | 14,257.68 | ||||
Abbreviation: OPC, oropharynx cancer.
aThe number of properly paired reads with high mapping quality score (≥30) on the HPV genome.
bPlasma HPV genotype based on the viral genome with the highest number of properly paired mapped reads.
cDefined as the proportion of detected HPV molecules represented by the HPV genotype found in tumor tissue.
dClade A9, HPV-16, HPV-31, HPV-33, HPV-35, HP-V52; Clade A7, HPV-18, HPV-39, HPV-45; Clade A3, HPV-83; Clade A1, HPV-42; Clade A10, HPV-6; Clade A14, HPV-71; Clade B1, HPV-12.
Of note, genotyping was possible even for samples with relatively low ctDNA levels (<10 copies/mL by dPCR). Moreover, in one sample with discordant results between E6 and E7 dPCR assays, HPV-mapping reads were found across both the E6 and E7 loci (Fig. 3A). Closer examination of the mapped reads in the region of the false negative HPV-33 E6 dPCR assay revealed mutations likely affecting primer/probe annealing. While these mutations likely prevented detection of E6 by dPCR, HPV-seq maintained robust performance. These results suggest that HPV-seq using a broad genotyping panel could replace tumor tissue analysis for accurate HPV genotyping.
HPV ctDNA quantification in patients with cervix and oropharynx cancer
With the success of HPV-seq using a genotyping panel on baseline samples, we next asked whether HPV-seq using full-length virus capture would provide sensitive and accurate quantification and enable treatment response monitoring. We subjected a total of 33 samples of patients with cervix cancer and 13 of oropharynx cancer to HPV-seq using full-length virus capture (Supplementary Table S1). All samples of patients with cervix cancer were obtained at either end-of-treatment, 3 months posttreatment, or at the time of recurrence, and all oropharynx cancer samples were obtained at pretreatment. HPV ctDNA levels according to HPV-seq and dPCR were highly correlated (R2 = 0.95, 95% CI 0.91–0.99, P = 1.9 × 10–29; Fig. 3B). Five samples contained less than 1 copy/mL plasma of HPV with the lowest detected level by HPV-seq at 0.03 copies/mL (Supplementary Table S3).
Of the 11 cervix cancer and 13 oropharynx cancer samples with dPCR-positive results (cervix cancer: 6 at end-of-treatment, 5 at posttreatment/recurrence; oropharynx cancer: 13 at pretreatment), HPV-seq produced positive results in 8 (73%) and 13 (100%) samples, respectively (Supplementary Table S3). The 3 samples with dPCR-positive HPV-seq–negative results had low dPCR values (approximately 1 copy). Interestingly, these samples were from patients with cervix cancer who were disease-free with at least 30 months of follow up.
Of the 22 samples of patients with cervix cancer with dPCR-negative results, 17 (77%) were also negative by HPV-seq (Supplementary Table S3). The 5 dPCR-negative HPV-seq–positive results were further analyzed to assess whether they represented false positive HPV-seq results. Of note, each of these 5 samples were at the end-of-treatment timepoint using the dual-strand full-length viral capture baits. Interestingly, one of these samples (from P5) displayed a low level of ctDNA (0.12 copies/mL) 12 months prior to recurrence with a biopsy-proven solitary 1.2-cm lung metastasis, although the 3 months posttreatment sample from this patient was falsely negative by both HPV-seq and dPCR (Supplementary Table S3). None of the other 4 patients with dPCR-negative HPV-seq–positive results have experienced disease relapse after more than 36 months of follow up. Supporting the veracity of the HPV-seq results, the HPV-mapping reads were distributed across the HPV-16 genome (Supplementary Fig. S6) and displayed high mapping quality scores (Supplementary Table S3).
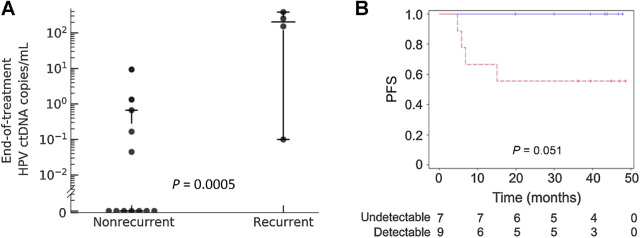
Four (P4, P5, P7, and P14) of the 17 patients with cervix cancer included in this study experienced disease recurrence. The end-of-treatment timepoint was analyzed by HPV-seq for 16 of 17 patients including all 4 of the patients who relapsed. HPV ctDNA levels were significantly higher at the end-of-treatment timepoint among those 4 patients compared with the 12 patients who did not relapse (P = 0.0005; Fig. 4A). With a median follow up of 27.5 months (range 4.8–48.6), detectable HPV ctDNA at the end-of-treatment timepoint was associated with poor PFS (log-rank P = 0.051; Fig. 4B). Sensitivity and specificity of HPV-seq for predicting recurrence at the end-of-treatment timepoint was 100% and 67%, respectively.
Figure 4.
End-of-treatment detectable HPV ctDNA is associated with disease recurrence. A, HPV ctDNA levels at the end-of-treatment timepoint obtained using HPV-seq are significantly higher among patients who subsequently relapsed (N = 4) versus patients who remained disease-free (N = 12). Horizontal bars indicate the median value and 1.5 times the interquartile range. B, PFS according to HPV ctDNA status at the end-of-treatment timepoint. Data represent HPV copies/mL plasma. Detectable HPV ctDNA, dashed red line. Undetectable HPV ctDNA, solid blue line. Vertical hash marks indicate censoring.
ctDNA fragment lengths in patients with cancer
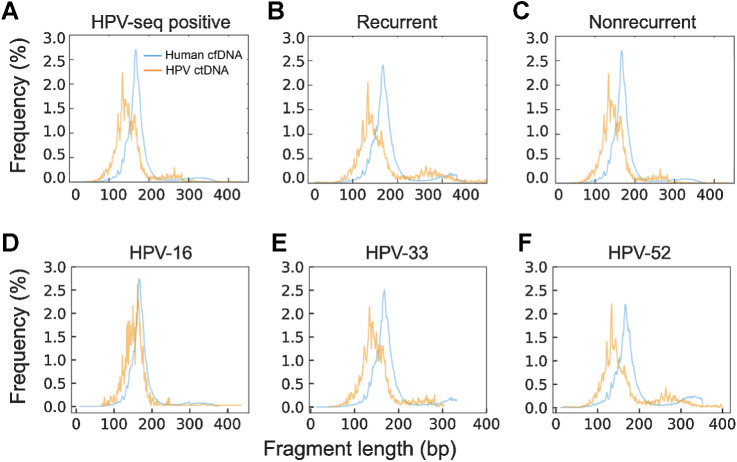
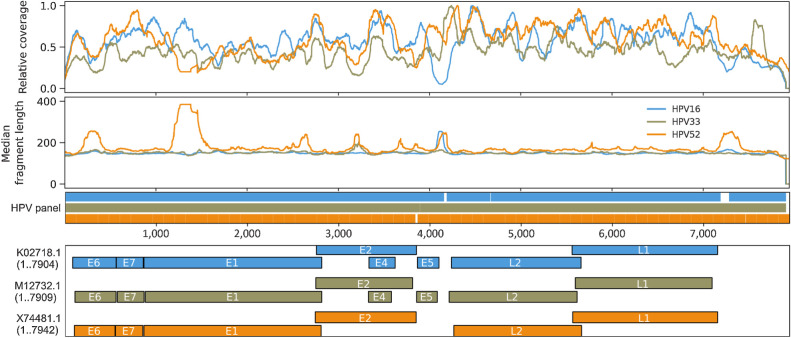
Fragment lengths of sequenced cfDNA fragments may be useful for discriminating between healthy and malignant tissues-of-origin (6, 26, 46–49). Shorter fragments have been observed in ctDNA from a variety of cancer types, including nasopharyngeal carcinoma and hepatocellular carcinoma, but this has not been previously evaluated in HPV-associated cancers. We compared HPV-mapping ctDNA fragment lengths with human-mapping cfDNA fragments from cervix cancer samples analyzed with full-length viral capture HPV-seq. HPV ctDNA displayed a median fragment size of 146 bp, which was 22 bp shorter than the median fragment size of human-mapping cfDNA (Fig. 5A; Supplementary Table S4). Of note, similar results were seen among samples taken at the time of recurrence (Fig. 5B) as well as samples taken from patients who have not recurred (Fig. 5C). Regardless of HPV genotype, median HPV ctDNA fragment length was shorter than human-mapping cfDNA (Fig. 5D–F). Similar results were observed in the oropharynx cancer cohort among HPV-16 ctDNA fragments (Supplementary Table S4; Supplementary Fig. S7). Analysis of genome-wide fragmentomic patterns revealed consistent features across multiple HPV genotypes (Fig. 6). Oscillations in relative-read depth coverage were largely conserved, and regions of higher median fragment lengths were often associated with a local nadir in coverage. Taken together, along with HPV genotyping and quantification, these results illustrate the capability to perform qualitative analyses of ctDNA fragments using HPV-seq.
Figure 5.
Length distributions of human-mapping cfDNA and HPV-mapping ctDNA fragments for 13 patients with cervix cancer. A, Sequenced fragment insert sizes merged from HPV-seq positive patients analyzed at end-of-treatment or posttreatment (N = 13). Samples (N = 3) from patients with recurrence (B) and samples (N = 10) from patients without recurrence (C) display similar ctDNA fragment-size distributions. Samples from tumors harboring HPV-16 (N = 9; D), HPV-33 (N = 2; E), and HPV-52 (N = 2; F) are shown.
Figure 6.
Genome-wide fragmentomic patterns of HPV-mapping ctDNA fragments across 3 HPV genotypes. First track: Relative read depth coverage for HPV-mapping paired-end reads. Coverage is normalized relative to the maximum for each genome. Relative coverage of 0 indicates no properly paired mapping reads. Samples from tumors harboring HPV-16 (blue; N = 22 samples from 21 patients), HPV-33 (gold; N = 2 samples from 1 patient), and HPV-52 (orange; N = 2 samples from 1 patient) are shown. Second track: Median HPV ctDNA fragment lengths (bp) inferred from sequenced fragment insert sizes. Third track: Capture probes for the 3 HPV genotypes. Fourth track: HPV gene organization from GenBank.
Discussion
High analytic sensitivity is a prerequisite for a ctDNA test applied to oncology settings with low disease burden (1). Here, we validated the concept that simultaneous assessment of multiple cancer-specific markers in a given patient has a major impact on analytic performance of ctDNA testing. Specifically, we used HPV-associated cervix and oropharynx cancer as two highly relevant models, showing for the first time that HPV-seq using dual-strand full-length viral capture enables robust detection of less than 1 copy of ctDNA. We also showed that HPV-seq reveals qualitative information regarding ctDNA fragments such as HPV genotype, mapping location along the HPV genome, and ctDNA fragment length distribution that cannot be easily inferred from qPCR/dPCR.
Virus-associated cancers have long served as a model for ctDNA methodology development and clinical utility testing (50). Epstein–Barr virus (EBV) provides a convenient marker for nasopharyngeal and other cancers, but sensitivity and specificity shortfalls of the established qPCR test present barriers to its use for early detection and response monitoring (51–53). Recently, full-length viral-capture sequencing was reported to improve the specificity of EBV detection in a prospective screening study (26). Ours is the first study to show the potential for this approach to also drive greater sensitivity in virus-associated cancers.
Despite its limitations, the plasma EBV DNA qPCR test continues to be employed clinically and in trials (24). The sensitivity of this test is aided by targeting of a conserved amplicon in the EBV genome (54). The HPV genome, however, does not harbor any repeated sequences that lend themselves to more sensitive qPCR/dPCR assays. Moreover, distinct high-risk oncogenic HPV genotypes have widely divergent sequences (55). These features make HPV detection ideal for hybrid-capture sequencing methods due to the flexibility for incorporating hundreds of baits spanning the entire genome of multiple viral genotypes.
We found that dual-strand capture was a key element for enabling robust detection of less than 1 copy of ctDNA. While single-strand and partial-genome viral capture were also able to produce quantitative and qualitative HPV-seq results in our study, the most robust ultrasensitive ctDNA detection was achieved using dual-strand full-length viral capture. Sequential capture of the complementary strands was feasible but laborious, so we instead implemented an approach for simultaneous dual-strand capture using staggered reverse-complementary baits. This approach is applicable to any hybrid-capture sequencing workflow and, importantly, could therefore have an immediate impact for ctDNA applications in other cancer types as well.
Locally advanced cervix cancer is managed with concurrent chemoradiotherapy. The role of subsequent adjuvant therapy is currently being evaluated in multiple international clinical trials (e.g., NCT00980954, NCT02036164, NCT0270396, NCT02853604, and NCT03468010). Notably, the results of the OUTBACK phase III trial (NCT01414608) were recently reported at the American Society of Clinical Oncology (ASCO) conference and found no improvement in survival with adjuvant chemotherapy (56). These results suggest either adjuvant chemotherapy is ineffective against metastasizing tumor cells and/or identification of patients at highest risk of recurrence was not achieved by current clinical factors, highlighting the need for novel biomarkers such as HPV DNA.
We previously reported on the capability of HPV genotype-specific dPCR from end-of-treatment plasma cfDNA to pre-date clinical recurrence in patients with locally advanced cervix cancer. From that study, we found low levels of HPV ctDNA (approximately 1 copy/10 ng) using dPCR in the end-of-treatment timepoint from 3 patients who have remained disease-free. These low dPCR values became less pronounced when evaluating dPCR levels as HPV copies/mL due to high plasma cfDNA yields. Using HPV-seq, 2 of these 3 samples had no detectable HPV fragments, suggesting that they may have been false positive dPCR results. Single positive dPCR droplets within these samples highlighted their low HPV DNA levels or the possibility of false positives (57). Conversely, 5 (23%) of the dPCR-negative surveillance samples harbored residual HPV ctDNA fragments detectable by HPV-seq. This again occurred specifically in the end-of-treatment samples, including from 4 patients who have remained disease-free. Importantly, there were no qualitative attributes (mapping quality, fragment length) of HPV DNA fragments in these samples that would easily distinguish them from the dPCR-positive samples, suggesting that these represented bona fide ctDNA molecules.
There is no current standard timepoint for MRD ctDNA testing in chemoradiotherapy-treated patients. Based on our results, at the end of treatment timepoint sensitivity exceeded specificity (100% vs. 67%) for MRD detection using HPV-seq on the cervix cancer cohort. If this high sensitivity is confirmed in larger studies, patients with undetectable end-of-treatment ctDNA levels using HPV-seq could be candidates for observation and thus spared adjuvant therapy or intensive surveillance. However, a positive end-of-treatment result might not guarantee subsequent recurrence and could necessitate repeat testing or else alternative surveillance procedures (e.g., medical imaging). Although we defined any detectable HPV sequences as a positive result in this proof-of-principle study, larger studies may establish an optimal end-of-treatment cut-off value that would maximize the trade-off between sensitivity and specificity. Future studies should also consider later timepoints to allow for additional clearance of residual ctDNA from effectively treated tumor cells. Recently, in a prospective cohort of patients with locally advanced cervix cancer, 2 patients displayed residual detectable HPV DNA after chemoradiotherapy based on dPCR, but only 1 patient experienced relapse (57). The patient with positive HPV DNA without relapse had a low HPV DNA level (10 copies/mL) at the end of chemoradiotherapy and negative HPV DNA at the following blood collection (6 months; ref. 57). Furthermore, posttreatment longitudinal surveillance (3 to 6 months) using dPCR in a cohort of HPV-associated patients with oropharynx cancer also demonstrated accurate detection of recurrence (58). These findings highlight that evaluating HPV DNA at repeated/later timepoints may provide more accurate detection of molecular relapse.
The findings of this study will require validation in larger cohorts to confirm the clinical utility for quantitative and qualitative ctDNA analysis using HPV-seq. The inclusion of two HPV-associated cancers with distinct disease sites highlights the generalizability of our assay; however, future studies will need to expand the utility of HPV-seq to other HPV-associated cancers including anogenital cancer types. The addition of other orthogonal markers of HPV in the circulation such as antibodies or cfDNA methylation patterns may lead to more refined and robust biomarker panels for measuring disease burden and response to therapy in HPV-associated cancers. In this study we assessed HPV-seq in patients with a cancer diagnosis and a group of controls. Future investigations of our methods to cancer screening settings would need to include large numbers of appropriately matched at-risk controls and standard clinical evaluations (e.g., oropharyngeal and pelvic exams, imaging, and/or biopsy) to evaluate specificity and positive predictive value in an intended use population. Such studies could evaluate whether shorter plasma HPV DNA fragment lengths or genome-wide fragmentomic patterns revealed by HPV-seq could boost specificity of cancer detection, as was recently demonstrated for plasma EBV DNA detection in nasopharyngeal cancer (26).
In conclusion, HPV-seq enables ultrasensitive detection and qualitative analysis of cervix and oropharynx cancer ctDNA. This study will have implications for treatment monitoring of HPV-related cancers and could open the door to new potential clinical applications of HPV-ctDNA analysis, including for early detection and MRD testing.
Authors' Disclosures
K. Han reports grants from Cancer Research Society during the conduct of the study; in addition, K. Han has a patent for Detection of circulating tumor DNA pending. J. Zou reports a patent for Detection of circulating tumor DNA pending. Z. Zhao reports a patent for Detection of circulating tumor DNA pending. L.L. Siu reports personal fees from Merck, Pfizer, AstraZeneca, Roche, Morphosys, Oncorus, Symphogen, Seattle Genetics, GlaxoSmithKline, Voronoi, Arvinas, Tessa, Navire, Relay Therapeutics, Rubius, Janpix, and Daiichi Sanyko; other support from Agios and Treadwell Therapeutics; and grants from Merck, Novartis, Bristol-Myers Squibb, Pfizer, Boerhinger-Ingelheim, GlaxoSmithKline, Roche/Genentech, Karyopharm, AstraZeneca, Astella, Bayer, AbbVie, Amgen, Symphogen, Intensity Therapeutics, Mirati Therapeutics, Shattucks, and Avid Therapeutics outside the submitted work. T.J. Pugh reports personal fees and other support from Illumina Inc.; personal fees from Merck, Canadian Pension Plan Investment Board, and Chrysalis Biomedical Advisors; and grants from Roche/Genentech outside the submitted work. S.V. Bratman reports grants from Conquer Cancer Foundation of ASCO, Cancer Research Society, and Princess Margaret Cancer Foundation during the conduct of the study, as well as grants from Nektar Therapeutics, personal fees from Bristol-Myers Squibb, and personal fees and nonfinancial support from Adela outside the submitted work; in addition, S.V. Bratman has a patent for Identification and use of circulating nucleic acid tumor markers issued, licensed, and with royalties paid from Roche Molecular Diagnostics; a patent for Cancer detection and classification using methylome analysis pending and licensed to Adela; a patent for Cancer detection, classification, prognostication, therapy prediction and therapy monitoring using methylome analysis pending and licensed to Adela; a patent for Use of cell-free DNA fragment length to improve detection of circulating tumor DNA pending and licensed to Adela; and a patent for Detection of circulating tumor DNA pending. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
We thank Marco Di Grappa for technical assistance and Jim Dimitroulakos for critical review of the manuscript. This work was supported by the University of Toronto Department of Radiation Oncology Collaborative Seed Grant (K. Han, E. Leung, and S.V. Bratman); Princess Margaret Cancer Foundation, with philanthropic support from Sean and Peta Boyd and from the Joe and Cara Finley Centre for Head & Neck Cancer Research; Cancer Research Society Operating Grant (K. Han); and Conquer Cancer Foundation of ASCO Career Development Award (S.V. Bratman). Any opinions, findings, and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of the American Society of Clinical Oncology or the Conquer Cancer Foundation. S.V. Bratman and T.J. Pugh are supported by the Gattuso-Slaight Personalized Cancer Medicine Fund.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
E. Leung: Conceptualization, resources, data curation, funding acquisition, writing–review and editing. K. Han: Conceptualization, resources, data curation, funding acquisition, writing–review and editing. J. Zou: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft. Z. Zhao: Conceptualization, data curation, formal analysis, investigation, methodology. Y. Zheng: Data curation. T.T. Wang: Software, methodology. A. Rostami: Project administration, writing–review and editing. L.L. Siu: Writing–review and editing. T.J. Pugh: Supervision, methodology, writing–review and editing. S.V. Bratman: Conceptualization, resources, supervision, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med 2018;379:1754–65. [DOI] [PubMed] [Google Scholar]
- 2. Burgener JM, Rostami A, De Carvalho DD, Bratman SV. Cell-Free DNA as a post-treatment surveillance strategy: Current status. Semin Oncol 2017;44:330–46. [DOI] [PubMed] [Google Scholar]
- 3. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563:579–83. [DOI] [PubMed] [Google Scholar]
- 5. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA Profiling. Cancer Discov 2017;7:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McDonald BR, Contente-Cuomo T, Sammut S-J, Odenheimer-Bergman A, Ernst B, Perdigones N, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med 2019;11:eaax7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health 2020;8:e180–90. [DOI] [PubMed] [Google Scholar]
- 11. Pornthanakasem W, Shotelersuk K, Termrungruanglert W, Voravud N, Niruthisard S, Mutirangura A. Human papillomavirus DNA in plasma of patients with cervical cancer. BMC Cancer 2001;1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lo YM, Chan LY, Lo KW, Leung SF, Zhang J, Chan AT, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res 1999;59:1188–91. [PubMed] [Google Scholar]
- 13. Cao H, Banh A, Kwok S, Shi X, Wu S, Krakow T, et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. Int J Radiat Oncol Biol Phys 2012;82:e351–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim KY, Le Q-T, Yom SS, Ng RHW, Chan KCA, Bratman SV, et al. Clinical utility of epstein-barr virus DNA testing in the treatment of nasopharyngeal carcinoma patients. Int J Radiat Oncol Biol Phys 2017;98:996–1001. [DOI] [PubMed] [Google Scholar]
- 15. Han K, Leung E, Barbera L, Barnes E, Croke J, Di Grappa MA, et al. Circulating human papillomavirus DNA as a biomarker of response in patients with locally advanced cervical cancer treated with definitive chemoradiation. JCO Precis Oncol 2018;2:1–8. [DOI] [PubMed] [Google Scholar]
- 16. Higginson DS, Damerla R, Scher ED, Gutinov SI, Thompson C, Cleary C, et al. HPV cell free DNA as a marker of response to chemoradiation for HPV-positive oropharngeal cancers. J Clin Oncol 2016;34:6083. [Google Scholar]
- 17. Damerla RR, Lee NY, You D, Soni R, Shah R, Reyngold M, et al. Detection of early human papillomavirus–associated cancers by liquid biopsy. JCO Precision Oncology 2019;3:PO.18.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chera BS, Kumar S, Beaty BT, Marron D, Jefferys S, Green R, et al. Rapid clearance profile of plasma circulating tumor HPV Type 16 DNA during chemoradiotherapy correlates with disease control in HPV-Associated Oropharyngeal Cancer. Clin Cancer Res 2019;25:4682–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bernard-Tessier A, Jeannot E, Guenat D, Debernardi A, Michel M, Proudhon C, et al. Clinical Validity of HPV Circulating Tumor DNA in Advanced Anal Carcinoma: An Ancillary Study to the Epitopes-HPV02 Trial. Clin Cancer Res 2019;25:2109–15. [DOI] [PubMed] [Google Scholar]
- 20. Cabel L, Jeannot E, Bieche I, Vacher S, Callens C, Bazire L, et al. Prognostic Impact of Residual HPV ctDNA Detection after Chemoradiotherapy for Anal Squamous Cell Carcinoma. Clin Cancer Res 2018;24:5767–71. [DOI] [PubMed] [Google Scholar]
- 21. Hanna GJ, Supplee JG, Kuang Y, Mahmood U, Lau CJ, Haddad RI, et al. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann Oncol 2018;29:1980–6. [DOI] [PubMed] [Google Scholar]
- 22. Kang Z, Stevanović S, Hinrichs CS, Cao L. Circulating cell-free DNA for metastatic cervical cancer detection, genotyping, and monitoring. Clin Cancer Res 2017;23:6856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olmedillas-López S, García-Arranz M, García-Olmo D. Current and Emerging applications of droplet digital PCR in oncology. Mol Diagn Ther 2017;21:493–510. [DOI] [PubMed] [Google Scholar]
- 24. Kim KY, Le Q-T, Yom SS, Pinsky BA, Bratman SV, Ng RHW, et al. Current state of PCR-based epstein-barr virus DNA testing for nasopharyngeal cancer. J Natl Cancer Inst 2017;109:djx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Le Q-T, Zhang Q, Cao H, Cheng A-J, Pinsky BA, Hong R-L, et al. An international collaboration to harmonize the quantitative plasma Epstein-Barr virus DNA assay for future biomarker-guided trials in nasopharyngeal carcinoma. Clin Cancer Res 2013;19:2208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lam WKJ, Jiang P, Chan KCA, Cheng SH, Zhang H, Peng W, et al. Sequencing-based counting and size profiling of plasma Epstein-Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc Natl Acad Sci U S A. 2018;115:E5115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res 2014;24:185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Siadat-Pajouh M, Periasamy A, Ayscue AH, Moscicki AB, Palefsky JM, Walton L, et al. Detection of human papillomavirus type 16/18 DNA in cervicovaginal cells by fluorescence based in situ hybridization and automated image cytometry. Cytometry 1994;15:245–57. [DOI] [PubMed] [Google Scholar]
- 29. Mincheva A, Gissmann L, zur Hausen H. Chromosomal integration sites of human papillomavirus DNA in three cervical cancer cell lines mapped by in situ hybridization. Med Microbiol Immunol 1987;176:245–56. [DOI] [PubMed] [Google Scholar]
- 30. Meissner JD. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol 1999;80:1725–33. [DOI] [PubMed] [Google Scholar]
- 31. Hart KW, Williams OM, Thelwell N, Fiander AN, Brown T, Borysiewicz LK, et al. Novel method for detection, typing, and quantification of human papillomaviruses in clinical samples. J Clin Microbiol 2001;39:3204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wong K, Huang SH, O'Sullivan B, Lockwood G, Dale D, Michaelson T, et al. Point-of-care outcome assessment in the cancer clinic: audit of data quality. Radiother Oncol 2010;95:339–43. [DOI] [PubMed] [Google Scholar]
- 33. Wang TT, Abelson S, Zou J, Li T, Zhao Z, Dick JE, et al. High efficiency error suppression for accurate detection of low-frequency variants. Nucleic Acids Res 2019;47:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 2016;44:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones E, Oliphant T, Peterson P. SciPy: Open source scientific tools for Python, 2001.
- 41. van Rossum G. Python tutorial. Amsterdam, Netherlands: Centrum voor Wiskunde en Informatica; 1995. [Google Scholar]
- 42. Salazar KL, Zhou HS, Xu J, Peterson LE, Schwartz MR, Mody DR, et al. Multiple human papilloma virus infections and their impact on the development of high-risk cervical lesions. Acta Cytol 2015. Dec 15;59:391–8. [DOI] [PubMed] [Google Scholar]
- 43. Fakhry C, Blackford AL, Neuner G, Xiao W, Jiang B, Agrawal A, et al. Association of Oral Human Papillomavirus DNA Persistence With Cancer Progression After Primary Treatment for Oral Cavity and Oropharyngeal Squamous Cell Carcinoma. JAMA Oncol 2019;5:985–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bruno MT, Scalia G, Cassaro N, Boemi S. Multiple HPV 16 infection with two strains: a possible marker of neoplastic progression. BMC Cancer 2020;20:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mollers M, Vriend HJ, van der Sande MAB, van Bergen JEAM, King AJ, Lenselink CH, et al. Population- and type-specific clustering of multiple HPV types across diverse risk populations in the Netherlands. Am J Epidemiol 2014 May 15;179:1236–46. [DOI] [PubMed] [Google Scholar]
- 46. Jiang P, Chan CW, Chan KC, Cheng SH, Wong J, Wong VW, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018;10.eaat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016;164:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PLoS Genet 2016;12:e1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lam WKJ, Chan KCA, Lo YMD. Plasma Epstein-Barr virus DNA as an archetypal circulating tumour DNA marker. J Pathol 2019;247:641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, et al. Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med 2017;377:513–22. [DOI] [PubMed] [Google Scholar]
- 52. Lin J-C, Wang W-Y, Chen KY, Wei Y-H, Liang W-M, Jan J-S, et al. Quantification of plasma Epstein–Barr Virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med 2004;350:2461–70. [DOI] [PubMed] [Google Scholar]
- 53. Chan ATC, Lo YMD, Zee B, Chan LYS, Ma BBY, Leung S-F, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst 2002;94:1614–9. [DOI] [PubMed] [Google Scholar]
- 54. Le Q-T, Jones CD, Yau T-K, Shirazi HA, Wong PH, Thomas EN, et al. A comparison study of different PCR assays in measuring circulating plasma epstein-barr virus DNA levels in patients with nasopharyngeal carcinoma. Clin Cancer Res 2005;11:5700–7. [DOI] [PubMed] [Google Scholar]
- 55. Bernard H-U, Calleja-Macias IE, Dunn ST. Genome variation of human papillomavirus types: phylogenetic and medical implications. Int J Cancer 2006;118:1071–6. [DOI] [PubMed] [Google Scholar]
- 56. Mileshkin LR, Moore KN, Barnes E, Gebski V, Narayan K, Bradshaw N, et al. Adjuvant chemotherapy following chemoradiation as primary treatment for locally advanced cervical cancer compared to chemoradiation alone: the randomized phase III OUTBACK trial (ANZGOG 0902, RTOG 1174, NRG 0274). J Clin Oncol 39:18s, 2021(suppl abstr LBA3). [Google Scholar]
- 57. Cabel L, Bonneau C, Bernard-Tessier A, Héquet D, Tran-Perennou C, Bataillon G, et al. HPV ctDNA detection of high-risk HPV types during chemoradiotherapy for locally advanced cervical cancer. ESMO Open 2021;6:100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chera BS, Kumar S, Shen C, Amdur R, Dagan R, Green R, et al. Plasma circulating Tumor HPV DNA for the surveillance of cancer recurrence in HPV-Associated oropharyngeal cancer. J Clin Oncol 2020;38:1050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Figure 1. Overview of HPV-seq and dual-strand hybrid capture. A, HPV-seq conducted on plasma cfDNA is designed to provide quantitative and qualitative information about ctDNA in patients with HPV-associated cancers. In addition to being highly sensitive and quantitative, HPV-seq can report on ctDNA fragment size and HPV genotype. Each full-length viral genome (episome or linearized genome) is expected to yield approximately 50 distinct cfDNA fragments. B, HPV-seq is conducted using hybrid-capture sequencing with single-stranded [sense (+) and/or antisense (−)] biotinylated baits tiled across the HPV genome: (i) single-strand viral genome hybrid capture, (ii) sequential dual-strand hybrid capture, and (iii) simultaneous dual-strand hybrid capture. C, Compared with single-strand hybrid capture (i), dual-strand hybrid capture using either a sequential (ii) or simultaneous (iii) approach recovers more HPV molecules. Results were normalized to the degree of HPV sequence enrichment with a single round of capture using single-stranded baits (left-most bar). Subjecting the unbound library to another round of hybrid capture using baits targeting the same strand (second bar from left) did not improve HPV sequence enrichment. The degree of HPV DNA enrichment in postcapture libraries was determined using HPV-16 E6 and E7 dPCR assays. N = 4 per condition. Error bars represent SD. Asterisk indicates statistical significance (P < 0.05) in comparison with single-strand capture conditions.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/6535/9401563/353ae4e6e4d3/5857fig1.jpg)