Abstract
We have identified five acyl coenzyme A (CoA) oxidase isozymes (Aox1 through Aox5) in the n-alkane-assimilating yeast Yarrowia lipolytica, encoded by the POX1 through POX5 genes. The physiological function of these oxidases has been investigated by gene disruption. Single, double, triple, and quadruple disruptants were constructed. Global Aox activity was determined as a function of time after induction and of substrate chain length. Single null mutations did not affect growth but affected the chain length preference of acyl-CoA oxidase activity, as evidenced by a chain length specificity for Aox2 and Aox3. Aox2 was shown to be a long-chain acyl-CoA oxidase and Aox3 was found to be active against short-chain fatty acids, whereas Aox5 was active against molecules of all chain lengths. Mutations in Aox4 and Aox5 resulted in an increase in total Aox activity. The growth of mutant strains was analyzed. In the presence of POX1 only, strains did not grow on fatty acids, whereas POX4 alone elicited partial growth, and the growth of the double POX2-POX3-deleted mutant was normal excepted on plates containing oleic acid as the carbon source. The amounts of Aox protein detected by Western blotting paralleled the Aox activity levels, demonstrating the regulation of Aox in cells according to the POX genotype.
The nonconventional yeast Yarrowia lipolytica, for which molecular and genetic tools have been developed, is currently used as a model organism for fundamental studies on protein secretion, dimorphism, and peroxisome biogenesis (for a review, see reference 1). It can utilize n-alkanes, fatty acids, and fats as the sole carbon source. This ability to degrade hydrophobic substrates may explain its presence in certain natural habitats, such as dairy products. Roostita and Fleet have shown that, in cheese, Y. lipolytica is involved in fatty acid (FA) degradation and maturation (34). This yeast type is also found in waste (our wild-type strain was isolated from sewage). The use of Y. lipolytica for the stereospecific conversion of hydroxylated long-chain FA into γ-lactones (5, 11, 30) and for the treatment of olive oil mill wastewater (7) has been reported. Degradation of FA first involves activation of the FA by the synthetase, resulting in an activated fatty acyl coenzyme A (acyl-CoA) molecule. In Y. lipolytica, two long-chain FA synthetases have been identified. One, acyl-CoA synthetase I, is phosphatidylcholine independent and the other, acyl-CoA synthetase II, is phosphatidylcholine dependent. Synthetase I is involved in cellular lipid synthesis, whereas the synthetase II generates acyl-CoA, which is degraded exclusively via β-oxidation (23). In mammalian cells, the activated FA is then degraded in both mitochondria and peroxisomes (29), whereas in yeast cells, such degradation occurs exclusively in peroxisomes (4), as shown for C. tropicalis (16, 38), Y. lipolytica (36), and Saccharomyces strains (18) (see also reference 10 for a review). Peroxisomal β-oxidation is the cyclic degradation of a fatty acid (N), with each cycle yielding an FA that is two carbon atoms shorter (N−2) and an acetyl-CoA molecule. It involves four successive reactions (oxidation, hydration, oxidase, and cleavage) catalyzed by an acyl-CoA oxidase, a bifunctional hydrase-oxidase, and a thiolase. The Y. lipolytica POT1 gene encoding the catabolic 3-oxoacyl-CoA thiolase has been cloned and characterized (3).
Acyl-CoA oxidases catalyze the first step of β-oxidation, the oxidation of the long-chain acyl-CoA thioester to give the corresponding trans-2-enoyl-CoA (40). A number of acyl-CoA oxidase genes have been cloned from plants, animals, and microorganisms (9). There are generally several acyl-CoA oxidase isozymes in a single organism, often with different substrate specificities. Three isozymes are present in plant cells: one is specific for long-chain FA (C16 to C18); the second is active against mid-length FA (C10 to C14); the third is active against a short-chain FA (C6) (15, 17). In yeast cells, one gene has been identified in S. cerevisiae (8), two genes have been identified in Candida maltosa (14, 22), three genes have been identified in Candida tropicalis (24, 27, 28), and five genes have been identified in Yarrowia lipolytica (19; this study). Short-chain specificity (C4 to C10) was demonstrated for the POX4 gene product in C. tropicalis (32) and for the POX3 gene product in Y. lipolytica (39), and long-chain specificity (C10 to C16) was demonstrated for the POX5 gene product in C. tropicalis (32).
We have identified five genes encoding acyl-CoA oxidase in the yeast Y. lipolytica (POX1 through POX5). We evaluated the function of the various isozymes (Aox) by developing mutants lacking one or several isozymes and determining their differential regulation and activity. Mutants completely lacking acyl-CoA oxidase activity, which were unable to grow on oleic acid media, were also developed. Such strains could be used for in vivo studies of Aox peroxisomal import in Y. lipolytica and of the Aox complex structure.
MATERIALS AND METHODS
Strains and growth conditions.
The Y. lipolytica strains used in this study are listed in Table 1 (see also Fig. 1). The genetically tractable strain, PO1d (CLIB139) (1), was derived from the wild-type strain, W29 (ATCC 20460; CLIB89) and was used as the host for transformation. Escherichia coli DH5α (21) was used for gene manipulation, TG1 [supE hsdΔ5 thiΔ(lac-proAB) F′ (traD36 proAB+ lacIq lacZΔM15)] was used for the construction of Aox expression plasmids, and M15(pREP4) (Nals Strs Rifs lac ara gal mtl F− recA+ uvr+) was used for expression experiments. The media and techniques for growing and handling Y. lipolytica were as described by Barth and Gaillardin (2), and those for E. coli were as described by Maniatis et al. (21). The principal yeast media used were rich medium (YPD), minimal glucose medium (YNB), enriched minimal medium (YNBcas), and minimal oleate medium (YNBO). The composition of the media used were as follows (per liter): YPD (10 g of yeast extract [Difco], 10 g of Bacto peptone [Difco], 20 g of glucose), YNB (1.7 g of yeast nitrogen base without amino acid and ammonium sulfate [Difco], 4 g of ammonium chloride, and 20 g of glucose), YNBcas (YNB plus 5 g of Casamino Acids [Difco]), and YNBO (as for YNB but with the glucose replaced by 10 g of oleic acid or methyl oleate). FA media were prepared as previously described (39). Uracil (0.1 g/liter) and leucine (0.3g/liter) were added to the media when required. Cell growth was monitored by measuring light scattering at 600 nm. Cells were collected by centrifugation and washed twice in cold sodium chloride solution (9 g/liter) to eliminate FA droplets before the determination.
TABLE 1.
Y. lipolytica; wild-type, monodisrupted, and multidisrupted strains
| Type | Strain | POX genotypes | Auxotrophies | Phenotype and/or referencea |
|---|---|---|---|---|
| Wild-type | W29 | MatA | 2 | |
| PO1d | MatA ura3-302 leu2-270 xpr2-302 | Leu− Ura− | 2 | |
| Single deletion | MTLY25 | pox1::URA3 | Leu− Ura+ | NG |
| MTLY12 | pox2::URA3 | Leu− Ura+ | NG | |
| MTLY13 | pox3::URA3 | Leu− Ura+ | NG; 39 | |
| MTLY14 | pox4::URA3 | Leu− Ura+ | NG | |
| MTLY15 | pox5::URA3 | Leu− Ura+ | NG | |
| MTLY24 | pox5Δ | Ura− | NG | |
| MTLY26 | pox1::URA3, pINA1192(LEU+) | Ura+ | NG | |
| MTLY16 | pox2::URA3, pINA1192(LEU+) | Ura+ | NG | |
| MTLY17 | pox3::URA3, pINA1192(LEU+) | Ura+ | NG; 39 | |
| MTLY18 | pox4::URA3, pINA1192(LEU+) | Ura+ | NG | |
| MTLY19 | pox5::URA3, pINA1192(LEU+) | Ura+ | NG | |
| Double deletions | MTLY27 | pox1::URA3 pox2::LEU2 | NG | |
| MTLY20 | pox3::URA3 pox2::LEU2 | DG | ||
| MTLY23 | pox4::URA3 pox2::LEU2 | NG | ||
| MTLY22 | pox5::URA3 pox2::LEU2 | NG | ||
| MTLY28 | pox5Δ pox1::URA3 | NG | ||
| MTLY29 | pox5Δ pox2::URA3 | NG | ||
| MTLY30 | pox5Δ pox3::URA3 | NG | ||
| MTLY31 | pox5Δ pox4::URA3 | NG | ||
| MTLY32 | pox5Δ pox2Δ | Ura− | NG | |
| Triple deletions | MTLY35 | pox5Δ pox2Δ pox3::URA3 | DG | |
| MTLY36 | pox5Δ pox2Δ pox3Δ | Ura− | ||
| Quadruple deletions | MTLY37 | pox5Δ pox2Δ pox3Δ pox4::URA3 | No growth |
NG, normal growth; DG, decreased growth.
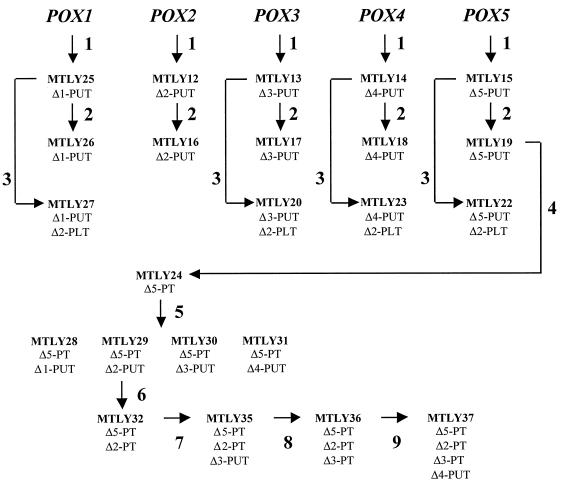
FIG. 1.
Y. lipolytica strains with modified POX genes used in this study. The various strains were obtained by successive transformation of the initial Y. lipolytica strain PO1d (Leu− Ura−) with specific POX-PUT disruption cassettes (arrow 1), pINA1192 for conversion to prototrophy (arrow 2), the POX2-PLT disruption cassettes (arrow 3), strain MTLY19 with POX5-PT (arrow 4), strain MTLY24 with POX2-PUT (arrow 5), strain MTLY29 successively with POX2-PT (arrow 6), POX3-PUT (arrow 7), POX3-PT (arrow 8), or POX4-PUT (arrow 9).
Cloning and sequencing of acyl-CoA oxidase genes.
The plasmids were isolated from a genomic library (41). They contained an insert of 10 kb for pINA-POX1, pINA-POX3, and pINA-POX5 and an insert of 9.6 kb of pINA-POX2. Regions encoding the POX genes were analyzed by inserting into pBluescript II KS(+) (Stratagene) either restriction fragments for POX1, POX2, and POX5 or nebulization fragments of part of the insert for POX3. The sequence coding for the carboxy terminus of Aox5 was absent from pINA-POX5. Only the sequence of an 800-bp amplified fragment for the POX4 gene had been determined (not isolated from the library). Therefore, complete clones were isolated by inverse PCR from a different library (Neuveglise et al. [25]). This library was produced by inserting 2-kb fragments of genomic DNA into the 2-kb pINACN5 E. coli vector carrying a kanamycin resistance gene as a selective marker gene. Divergent oligonucleotide pairs binding within the known sequences were used for amplification. The amplified 4-kb fragments were purified by electrophoresis in agarose gels; they were then blunt ended with T4 DNA polymerase and ligated. The resulting plasmids were used to transform E. coli, and Kanr colonies were isolated and inserts were sequenced by primer walking. A second PCR was performed with oligonucleotide pairs located at the 5′ end of the determined sequence, and cloning and sequencing procedures were repeated. Template preparation, sequencing, and nucleotide sequence analysis were performed as previously described (39).
Construction of disruption cassettes for acyl-CoA oxidase isozyme genes.
We disrupted multiple acyl-CoA oxidase genes by using URA3 as a selectable marker by the SEP procedure (20), which was modified as previously described (39). The promoter and terminator regions of POX genes were amplified by using specific oligonucleotide pairs, thus eliminating the complete open reading frame sequence. A second PCR was performed with the external primers and the promoter and terminator PCR products, which annealed via a common 20-bp extension carrying a site for the rare cutting restriction enzyme I-SceI (37). The resulting PCR product was cloned to give a series of plasmids, designated pPOX-PT, containing the promoter-terminator (PT) module (disruption cassette 2). A URA3 gene was introduced into the I-SceI site of the POX-PT cassette. Thus, a series of plasmids, pPOX-PUT, containing the promoter-URA3-terminator module (PUT) was constructed (disruption cassette 1). These constructs were named pPOX1-PUT, pPOX2-PUT, pPOX3-PUT, pPOX4-PUT, and pPOX5-PUT for plasmids containing disruption cassette 1 and named pPOX1-PT, pPOX2-PT, pPOX3-PT, pPOX4-PT, and pPOX5-PT for plasmids containing disruption cassette 2. Disruption cassettes were amplified by PCR with gene-specific external primers and a gene-specific plasmid. PCR conditions were as follows: template, 50 pmol of primers, 0.2 mM deoxynucleoside triphosphates, 1× reaction buffer, and 5 U of Pfu DNA polymerase (Stratagene, La Jolla, Calif.). In addition, pPOX2-PLT, containing a promoter-LEU2-terminator disruption cassette was constructed by a four-way ligation as follows: the promoter region was isolated as a 1.18-kb BamHI-HindIII fragment from SMT25, the LEU2 gene was carried on a 2.7-kb HindIII-SalI fragment from pINA1192 (6), and the terminator region was isolated as a 1.5-kb SalI-NotI fragment from SMT69. SMT25 and SMT69 are subclones of the pINA-POX2 used for sequencing. These three fragments were inserted into BamHI-NotI digested pBluescript KS(−) vector (Stratagene). pPOX2-PLT was linearized by digestion with BamHI and NotI before being used for transformation.
Transformation of Y. lipolytica by the lithium acetate method.
Y. lipolytica cells were transformed by the lithium acetate method (12), and correct disruption of POX genes was verified by PCR (13) and confirmed by Southern blot hybridization. Disruptions were done in two steps. First, PO1d was transformed with 100 ng of PCR PUT disruption cassette 1, and Ura+ transformants were selected on YNBcas. Colonies appeared within 2 days of incubation at 30°C at a frequency of approximately 104 colonies/μg of DNA. Ura+ clones were then transformed with 200 ng of PCR PT disruption cassette 2 to remove the URA3 gene. Transformants were selected on 5-fluoroorotic acid (5FOA) medium (2). 5FOA-resistant clones appeared within 5 days of incubation at a frequency of 103 colonies/μg of DNA. A clone with the correct PT gene disruption allele was used as the host for the next round of transformation.
Acyl-CoA oxidase induction and preparation of cell extracts.
Strains were grown on YNB and transferred to methyl oleate medium (YNBO) for induction at an initial optical density at 600 nm of 1. After 5 h of induction (or as indicated in the text), cells were collected by centrifugation, washed twice in 3 ml of cold saline (9 g NaCl per liter), pelleted, and suspended in 12 ml of 50 mM potassium phosphate buffer (pH 7.2). They were then disintegrated in a French press (ENERPAC P462) at 108 Pa. Cell extracts were obtained by centrifugation at 6,000 × g for 5 min at 4°C. Acyl-CoA oxidase activity was assayed by monitoring imine quinone formation by the method of Shimizu et al. (35) as previously described (39).
Aox expression in E. coli and antiserum preparation.
For the production of antibodies, the coding domain of the POX genes were amplified by PCR with primer pairs poxF1-poxF2, as described previously for Aox3p (39). Primer poxF1 corresponds to the ATG region and contains a SphI site (n4 GC ATG C n19–23), and primer poxF2 corresponds to the stop region and contains a SmaI site (n3 CCC GGG CTA n19–23), where n corresponds to the POX sequence and the number(s) to the number(s) of bases. The amplified fragments were cloned into pBluescript II KS(+) (Stratagene) and checked by sequencing. The SphI-SmaI fragments were introduced into the expression vector pQE32 (QIAexpressionist kit; Qiagen) in frame after the 6× His-tag domain. Expression, production, and purification were performed as recommended by the manufacturer and as described for Aox3p (39). Antibodies were raised by primary injection of 100 μg of fusion proteins in 0.1 ml of elution buffer emulsified with an equal volume of Freund complete adjuvant (first injection) or incomplete adjuvant (later injections) into rabbits. Booster injections were administered every 2 weeks. One week after the fourth injection, the rabbits were killed and sera were prepared. The specificities of the antisera were determined after 24 h of incubation by double-diffusion Ouchterlony assay on physiological saline buffer set with 1.4% agarose and containing 0.02% NaN3. For Western blotting, cells were centrifuged, suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer containing 50 mM dithiothreitol, and then heated for 10 min at 95°C before being loaded onto SDS-polyacrylamide gels. The proteins were subjected to electrophoresis and electroblotted onto nitrocellulose membranes (Schleicher & Schuell). Preincubation with 2% skim milk powder, antibody incubations (1:1,000 dilutions), and washes were carried out in 10 mM Tris (pH 8)–150 mM NaCl–0.05% Tween. Antigen-antibody complexes were detected with goat anti-rabbit immunoglobulin G conjugated with peroxidase.
RESULTS
Cloning and sequencing of Y. lipolytica POX genes.
We compared yeast acyl-CoA oxidase genes and identified conserved nucleotide blocks that were used to amplify fragments of the Y. lipolytica genes. The amplified fragments were used for the isolation of plasmids containing the corresponding genes by colony hybridization of a genomic library (41), as previously reported for POX3 (39). The full-length POX1, POX2, and POX3 genes and a truncated POX5 gene were isolated. The POX4 gene and the end of the POX5 gene were rescued by divergent PCR as described in Materials and Methods. Sequences have been deposited in the EMBL database under accession numbers AJOO1299 through AJOO1303. Protein sequence analysis was performed and the Y. lipolytica Aoxs were found to be only about 45% identical (50% similar) to Aoxs from the other yeasts, whereas they were 55 to 70% identical to each other (65 to 76% similar). The highest degree of protein identity was observed between Aox3 and Aox5. The acyl CoA oxidases from C. tropicalis and C. maltosa are less conserved; they are 51 to 63% identical to each other (65 to 76% similar) in C. tropicalis and 50% identical (cmPOX1 and cmPOX4; 65 to 76% similar) in C. maltosa. The highest degree of identity was observed between ctPOX2 and cmPOX1 (84% similar) and ctPOX4 and cmPOX4 (83% similar), indicating that they may be a short-chain acyl-CoA oxidase. Such high levels of identity were not observed with any of the Y. lipolytica proteins, providing no information about their potential specific activity.
Development of disruptants of acyl-CoA oxidase isozymes.
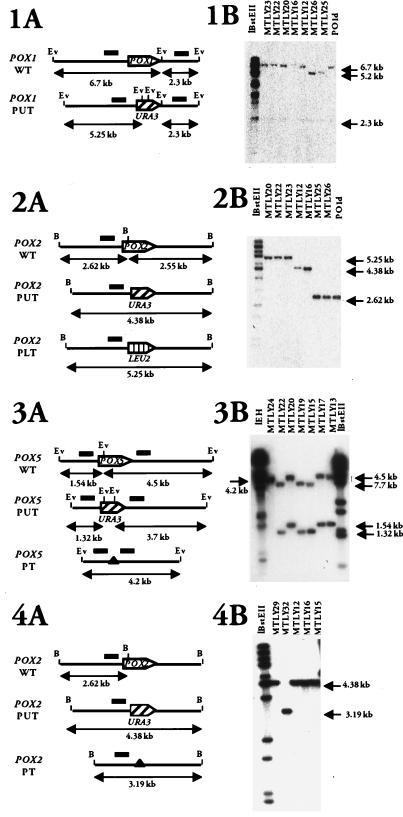
We genetically evaluated the physiological function of acyl-CoA isozymes in Y. lipolytica by creating single- and multidisrupted strains (Fig. 1 and Table 1). Genes were disrupted by the SEP method (20) as previously described for POX3 (39). For the disruption of POX genes, we constructed two cassettes for each gene by PCR: a PUT cassette and a PT cassette (see also Materials and Methods). For the disruption of the POX2 gene, a PLT cassette was constructed as described in Materials and Methods. The nine-step construction of the disrupted strains (Fig. 1) is described below. At each step, gene disruptions were confirmed by Southern blot analysis, as shown in Fig. 2. First, we disrupted each single acyl-CoA oxidase gene separately (Fig. 2, Step 1). Strains MTLY25, MTLY12, MTLY13, MTLY14, and MTLY15 were obtained as Ura+ transformants from the wild-type strain Y. lipolytica PO1d by using disruption cassette 1 in POX1-PUT, POX2-PUT, POX3-PUT, POX4-PUT, and POX5-PUT, respectively. Correct gene disruptions were observed in 50% of the Ura+ clones. Southern blot analysis confirmed that the desired chromosomal regions were correctly replaced, as shown in Fig. 2 for POX1, POX2, and POX5. The POX1 gene was smaller in MTLY25 and MTLY26 than in the wild-type strain, PO1d, or in the strains disrupted in a single other POX gene (MTLY23, MTLY22, MTLY20, MTLY16, and MTLY12), (5.2 versus 6.7 kb). This shows that the POX1 gene was correctly replaced (Fig. 2, panel 1B). Similarly, the POX2 band was 2.62 kb in the wild-type and 4.38 kb in the disrupted pox2ΔPUT strains, MTLY12 and MTLY16 (Fig. 2, panel 2B). The wild-type POX5 bands were 1.54 and 4.5 kb, whereas those in the gene-disrupted mutants were 1.32 and 3.7 kb (Fig. 2, panel 3B). The five monodisrupted strains were transformed with pINA1192, which carries the LEU2 gene, thus rendering the clones prototroph.
FIG. 2.
Checking of POX mutant strains. (A panels) Physical maps of acyl-CoA oxidase wild-type and disrupted genes, giving the sizes of the expected fragments. Open reading frames are represented by an arrowbox indicating the orientation of transcription. Restriction sites are BamHI (B) and EcoRV (Ev). Bars indicate the location of the biding sites for the probes. Black triangles indicate the complete deletion of the specific POX open reading frame obtained with the PT cassettes. (B panels) Southern blot analyses of mutant strains. Genomic DNA was digested with EcoRV (1 and 3) or BamHI (2 and 4). Blots were probed with the POX1-PT cassette (1B), with pSMT44 carrying a fragment of the POX2 promoter region (2B and 4B), and with the POX5-PT cassette (3B). Molecular markers used as standards were lambda DNA digested with EcoRI and HindIII (lEH) or BstEII (lBstEII). The strains from which genomic DNA was obtained are indicated at the top of the gels.
Double acyl-CoA oxidase disruptants were obtained by transforming the monodisruptants MTLY25, MTLY13, MTLY14, and MTLY15 with the POX2-PLT cassette and selecting Leu+ transformants. The pox2 null mutants (Δ2PLT) gave a 5.25-kb fragment (Fig. 2, panel 2). The double mutants MTLY27, MTLY20, MTLY23, and MTLY22 were correctly disrupted with pox1ΔPUT-pox2ΔPLT, pox3ΔPUT-pox2ΔPLT, pox4ΔPUT-pox2ΔPLT, and pox5ΔPUT-pox2ΔPLT, respectively.
The URA3 marker was regenerated in the monodisrupted strain, MTLY19 (pox5ΔPUT), by transformation with POX5 disruption cassette 2 (POX5-PT) (step 4 in Fig. 2) and selection on a 5FOA plate. Strain MTLY24 (pox5ΔPT) was obtained from 20 5FOA-resistant clones and gave a single 4.2-kb band (Fig. 2, panel 3).
Other double disruptants were obtained by transformation of the MTLY24 strain with the four type 1 disruption cassettes. The resulting strains, MTLY28, MTLY29, MTLY30, and MTLY31, were disrupted with pox5ΔPT-pox1ΔPUT, pox5ΔPT-pox2ΔPUT, pox5ΔPT-pox3ΔPUT, and pox5ΔPT-pox4ΔPUT, respectively.
As in step 3, the URA3 marker was regenerated in strain MTLY29 by transformation with POX2 disruption cassette 2 (POX2-PT) and selection on a 5FOA plate. Strain MTLY32 (pox5ΔPT-pox2ΔPT) was obtained and gave a 3.19-kb band (Fig. 2, panel 4).
Finally, the triple disruptants, MTLY35 and MTLY36, and the quadruple disruptant, MTLY37, were as follows: MTLY35, pox5ΔPT-pox2ΔPT-pox3ΔPUT (disruption with the POX3-PUT cassette); MTLY36, pox5ΔPT-pox2ΔPT-pox3ΔPT (disruption with the POX3-PT cassette); and MTLY37, pox5ΔPT-pox2ΔPT-pox3ΔPT-pox4ΔPUT (disruption with the POX4-PUT cassette).
Acyl-CoA oxidase activity in mutant strains.
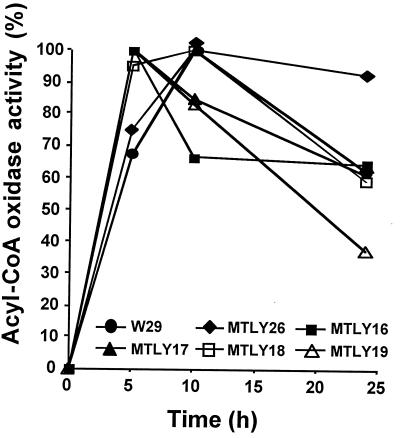
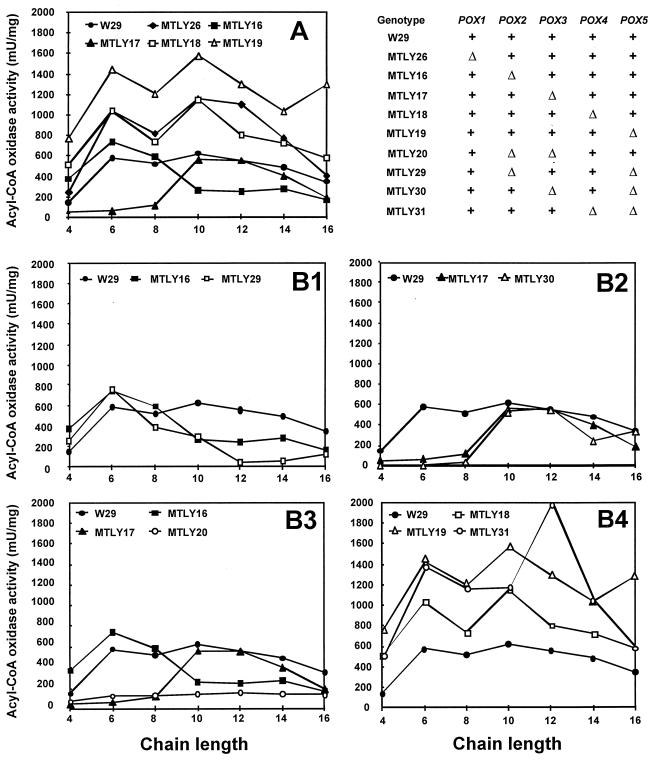
The development of a series of mutants with single gene disruptions made it possible to examine total acyl-CoA oxidase (i.e., Aox) activity and to assess the contribution of each isozyme to Aox activity in cells. Aox activity was followed over a time course after the transfer of growing cells from YNB to YNBO (C18) for induction. The five monodisrupted strains, MTLY26, MTLY16, MTLY17, MTLY18, and MTLY19, and the wild-type strain, W29, were induced as described in Materials and Methods. Aox activity against C10-CoA substrate was measured at t = 0, 5, 10, and 25 h after transfer into YNBO. The six strains had similar growth kinetics (data not shown) and induction patterns (Fig. 3). Little activity was detected at t = 0, and maximum activity was reached by 5 to 10 h after transfer and decreased thereafter. Acyl-CoA activities in mutant strains were further analyzed 5 h after transfer by using substrates of different chain lengths (C4 through C16). The acyl-CoA oxidase activity profiles of strains with single-gene disruption differed according to genotype (Fig. 4A). In strain MTLY26 (Δpox1), no differences in the overall profile were observed, although a slight increase in activity was observed on all substrates, regardless of the chain length. More-pronounced differences were seen in the other four deletion mutants. Both MTLY16 (Δpox2) and MTLY17 (Δpox3) had lower Aox activities against substrates of particular chain lengths. MTLY16 (Δpox2) had a lower activity against C10 to C16 substrates, indicating that POX2 encodes an isozyme with long-chain acyl-CoA specificity. Aox2p is therefore a long-chain acyl-CoA oxidase similar to the POX5ct gene product of C. tropicalis. However, some residual long-chain activity remained in the disrupted strain. In contrast, we have shown that strain MTLY17 has a low level of activity against short-chain molecules (see also Fig. 4A and B2). Confirmation that POX3 encodes an isozyme specific for short-chain molecules was obtained by measuring Aox3p activity when the isozyme was produced in E. coli (39). In the two other mutant strains, MTLY18 (Δpox4) and MTLY19 (Δpox5), Aox activity was twice as high as that in the wild type irrespective of the carbon chain length of the substrate. This finding suggests that Aox4p and Aox5p are involved in the regulation of Aox activity.
FIG. 3.
Time course of acyl-CoA oxidase activity in wild type (W29) and in strains with single deletions. Cells from a glucose-grown culture of the wild type (W29) and single-deletion strains MTLY26 (Δpox1), MTLY16 (Δpox2), MTLY17 (Δpox3), MTLY18 (Δpox4), and MTLY19 (Δpox5) were transferred to methyl oleate induction medium (C18), and the acyl-CoA oxidase activity against C10-CoA substrate was determined as a function of time after transfer. Activity is expressed as a percentage of maximum activity.
FIG. 4.
Acyl-CoA oxidase activity profiles of mutant strains as a function of substrate chain length. Activity was measured independently with C4-CoA through C18-CoA substrates and was standardized for protein concentration. (A) Activity profile of the wild-type strain (W29) compared with the activity in the single-deletion strains MTLY26, MTLY16, MTLY17, MTLY18, and MTLY19 (Δpox1, Δpox2, Δpox3, Δpox4, and Δpox5, respectively). (B) The effect of a second gene deletion on Aox activity in mutant strains was compared with the activity of the corresponding single-deletion strain. The profile of the wild-type strain is included for comparison. Activity in the wild-type strain (W29) was compared with that of MTLY16 (Δpox2) and MTLY29 (Δpox5 Δpox2) (B1); MTLY17 (Δpox3) and MTLY30 (Δpox5 Δpox3) (B2); MTLY16, MTLY17, and the double mutant MTLY20 (Δpox2 Δpox3) (B3); and MTLY18, MTLY19, and double mutant MTLY31 (Δpox4 Δpox5) (B4). Strain names and corresponding POX genotypes are summarized. Wild-type (+) and deleted genes (▵) are as indicated in the upper right corner.
In MTLY20 (Δpox2 Δpox3), total activity was one-fifth that in the wild-type, indicating that Aox2P and Aox3p accounts for most Aox activity (Fig. 4B3). This suggests that one of the other Aox proteins (Aox1p, Aox4p, or Aox5p) is a less-active acyl-CoA oxidase isozyme, with broad substrate chain length specificity. The Aox activity profiles of the double-deleted strains, MTLY29 (Δpox5 Δpox2), MTLY30 (Δpox5 Δpox3), and MTLY31 (Δpox5 Δpox4), were compared with that of the single deleted strains (Fig. 4B2, 4B3, and 4B4, respectively). The lower level of Aox activity against all chain lengths in MTLY29 than in MTLY16 and MTLY19 suggests that Aox5 is responsible for the residual Aox activity observed in MTLY20. Aox activity was only slightly lower in MTLY30 than in MTLY17, and this difference concerned only the short-chain substrates. However, as in strain MTLY19, Aox activity was not high, indicating that the doubling in Aox activity in Δpox4 and Δpox5 strains was abolished by deletion of one of the two major acyl-CoA oxidase isozymes, Aox2p or Aox3p.
Cell growth in mutant strains.
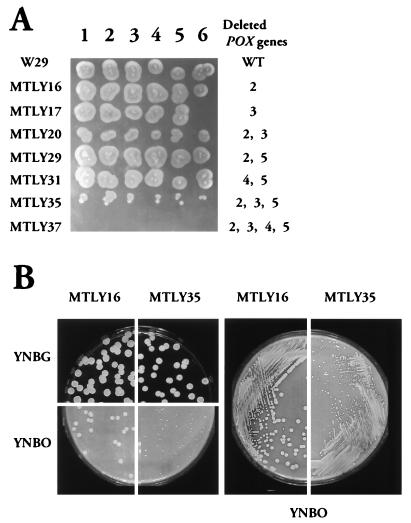
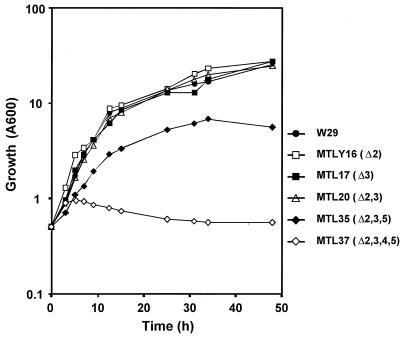
If Y. lipolytica is grown on oleic acid, peroxisome proliferation and enzymes of the peroxisomal β-oxidation system are induced (26). We have shown previously that acyl-CoA oxidase is the rate-limiting step of β-oxidation (31). Therefore, the involvement of acyl-CoA oxidase isozymes in oleic acid β-oxidation was investigated by observation of the growth of Aox disruptants on plates (Fig. 5) and in liquid medium (Fig. 6). All strains grew to a similar extent on glucose (data not shown). There was no significant difference in the growth on oleic acid plates of the wild-type strain (W29) and any single gene null mutant or double null mutant, except for strain MTLY20. A growth defect was observed in strain MTLY20 (Δpox2 Δpox3) and was even more pronounced for the triple null mutant, MTLY35 (Δpox2 Δpox3 and Δpox5), whereas the quadruple null mutant, MTLY37 (Δpox2 Δpox3 Δpox4 Δpox5) did not grow at all on oleic acid plates (Fig. 5). The growth rates on glucose and oleic acid plates of MTLY16, which has a single disruption (Δpox2), and of the triple-deleted mutant MTLY35 (Δpox2 Δpox3 Δpox5) were compared (Fig. 5). There was no colony size difference on glucose plates, whereas growth on oleic acid was clearly affected (Fig. 5).
FIG. 5.
Growth of wild-type (W29) and mutant strains on solid oleic acid medium. Yeast strains were grown in liquid YPD medium at 28°C, centrifuged, washed twice, and suspended at a cell density of 5 × 103 cells/μl. Aliquots (5 μl) in two serial dilutions (A) or 40-μl aliquots of the same dilutions (B) were plated on YNO-agar and YNBG plates.
FIG. 6.
Growth kinetics of wild-type (W29) and POX mutant strains in liquid oleic acid medium. Yeast strains were grown in liquid YPD medium at 28°C, centrifuged, washed twice with saline, and used to inoculate YNO medium at an initial absorbance of 0.5 (A600).
Similar results were obtained if growth was followed in liquid YNO medium (Fig. 6). Except for strain MTLY20, in which the POX2 and POX3 genes encoding the two major short- and long-chain specific Aox were deleted, the phenotypes of the strains differed on plates (reduced growth; Fig. 5A) and in liquid medium (no growth difference; Fig. 6).
Thus, no one individual POX gene is absolutely required for β-oxidation of long-chain FA and POX1 alone cannot support growth on oleic acid plates. POX4 only partially restored growth, and the growth defect was only observed if the POX2 and POX3 genes coding for the major Aox were deleted.
Expression of Aox isozymes in E. coli and mutant strains.
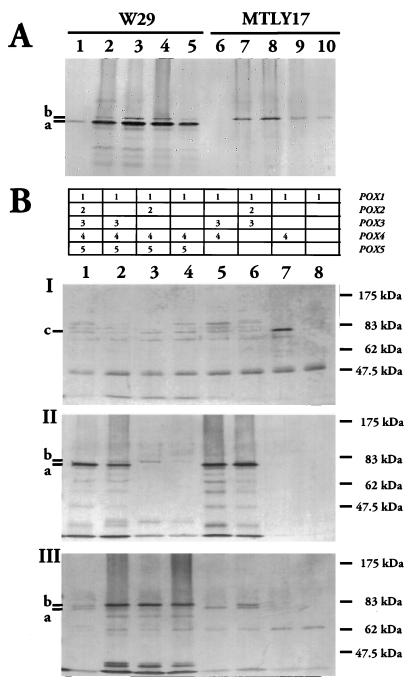
We further analyzed acyl-CoA oxidase isozyme expression by overexpressing the genes in E. coli. We created N-terminal His-tag fusions with Aoxp by using the QIAexpressionist kit (Qiagen). The pQE-Aox plasmids were constructed as described in Materials and Methods. Proteins were only obtained with the POX1, POX3, and POX5 genes (data not shown). Aox1 and Aox5 fusion proteins were purified as previously described for Aox3p. However, we only obtained an active enzyme in E. coli for the Aox3 protein (39). The purified enzymes were used to raise specific antibodies in rabbits for analyzing Aox proteins in wild-type and deleted strains. The expected size of the various acyl-CoA oxidases were 77,232 Da for Aox1, 78,641 Da for Aox2, 77,960 Da for Aox3, 79,241 Da for Aox4, and 78,300 Da for Aox5. On Western blots with anti-Aox3p antibodies, we detected two signals at about 80 kDa (Fig. 7, bands a and b) for the oleic acid-induced wild-type strain (W29) and one band (band b) for MTLY17 (Δpox3). The lower band (band a) corresponds to Aox3p because is not present in MTLY17 (Fig. 7A and B, part II). Similarly, anti-Aox5p antibodies primarily recognizes the upper band (band b) corresponding to Aox5p (Fig. 7B, part II, lanes 2 to 4) and weakly recognizes Aox2p and Aox3p. Anti-Aox1p antibodies also react weakly with Aox4p (Figure 7B, part I). Anti-Aox3p antibodies cross-reacted with Aox5p but not with Aox1p, as shown by Ouchterlony and Western blots with proteins produced in E. coli (data not shown).
FIG. 7.
Western blot analysis of Aox1p, Aox3p, and Aox5p in W29 (wild-type) and mutant strains. (A) Time course of Aox3p accumulation in wild-type W29 and MTLY17 (Δpox3). Strains were grown in minimal glucose and transferred to oleate medium. Samples were taken from YNBO at t = 0 (lanes 1 and 6), 3 h (lanes 2 and 7), 6 h (lanes 3 and 8), 15 h (lanes 4 and 9), and 24 h (lanes 5 and 10) after transfer. Aox proteins were detected with anti-Aox3p antisera. (B) Aox proteins in strains W29 (lane 1), MTLY16 (lane 2), MTLY17 (lane 3), MTLY20 (lane 4), MTLY29 (lane 5), MTLY31 (lane 6), MTLY35 (lane 7), and MTLY37 (lane 8). Samples of wild-type and mutant strains were withdrawn at t = 5 h after induction in YNBO and were probed with anti-Aox1p (I), anti-Aox3p (II), and anti-Aox5p (III) antibodies. Aliquots of cell extracts equivalent to 0.2 A600 U were subjected to SDS-PAGE in 10% acrylamide gels. Molecular mass standards in the prestained protein marker (New England Biolabs) are indicated on the right. The marks (–) on the left indicate the migration positions of Aox1 (c), Aox3 (a), and Aox5 (b).
The band intensities for Aox3p and for Aox5p in Western blot analysis paralleled the levels of Aox activity in the wild-type strain. The low level of activity in cells grown in glucose medium was correlated with the faint signal for the wild type (W29) in this medium with the anti-Aox3p probe (Fig. 7A, lane 1). For cells grown on oleic acid medium, the band intensity was maximal at 6 h after induction, after which it decreased (Fig. 7A, lanes 2 to 5). Similar results were obtained with Aox5p in MTLY17, for glucose-grown (lane 6) and oleate-induced (lanes 7 to 10) cells and if the same samples were probed with anti-Aox5p antibodies (data not shown).
We investigated whether the changes in Aox activity in the various strains resulted from changes in Aox protein levels. Cell extracts of oleic acid-induced cells (t = 5 h) of wild type and mutants MTLY16, MTLY17, MTLY20, MTLY29, MTLY30, MTLY31, MTLY35, and MTLY37 were probed with anti-Aox1p (Fig. 7B, part I), anti-Aox3p (Fig. 7B, part II), and anti-Aox5p antibodies (Fig. 7B, part III). Signals in the 80-kDa region with anti-Aox1p antibodies were faint, except in strain MTLY35 (lane 7). This may reflect weak expression of the POX1 gene on oleic acid medium, which was subsequently confirmed by using a POX1::lacZ fusion. The band obtained for strain MTLY35 indicates induction of the POX1 gene or a strong induction of POX4. The absence of Aox3 and Aox5 signals was correlated with the POX genotypes. The lower-molecular-weight signal (band a [Aox3p]) was not present if POX3 was deleted (Fig. 7B, part II, lanes 3, 4, 7, and 8), whereas it was even stronger in strains deleted of POX5 (lanes 5 and 6). Similarly, the higher-molecular-weight signal (band b [Aox5p]) was not present if POX5 was deleted (Fig. 7B, part III, lanes 6 to 8), whereas it was stronger if one of the major Aox genes (POX2 or POX3) was deleted (lanes 2 to 4). We also detected more Aox2 in strain MTLY35 (lane 6). Thus, Aox protein levels are tightly regulated by POX genotype.
DISCUSSION
Y. lipolytica has a more complex set of Aox isozymes than other yeasts, and this may explain its ability to grow efficiently on hydrophobic substrates such as fat, FA, and alkanes. Indeed, we have identified five POX genes in this yeast, whereas there is only one gene in S. cerevisiae, two in C. maltosa, and three in C. tropicalis. The five genes were cloned and sequenced. They encode proteins of about 80 kDa that are about 45% identical (50% similar) to genes from other yeasts, whereas they are 55 to 70% identical to each other (65 to 76% similar). The level of identity between the Y. lipolytica proteins and other Aox proteins was low, giving no clue as to their potential specificity. These proteins have been shown to be located in peroxisomes, but they have no typical PTS1 or PTS2 motifs.
We investigated the physiological functions of the Aox isozymes by creating strains with various combinations of disruptions. We used sequential gene replacement with disruption cassettes constructed by the SEP method (20). These cassettes were promoter-marker-terminator (PUT) and promoter-terminator (PT) cassettes. Amplified promoter and terminator regions were about 800 bp in length. Correct gene disruption with PUT cassettes was observed in 40 to 50% of the transformants. In contrast, correct gene replacement with PT cassettes was less frequent, in the range of 5% or less, probably reflecting 5FOA selection and the length of the homologous regions. In some cases, we observed integration at the POX locus involving a single crossover in the promoter or terminator region resulting in a large deletion in the nonrecombinant region (from hundreds of base pairs to several kilobases).
We have previously studied a POX3-deleted strain and the production of Aox3 in E. coli and have demonstrated that the POX3 gene encodes a short-chain acyl-CoA oxidase (39). We extended this approach by constructing a set of 23 mutant strains corresponding to 14 combinations of deletions. As in the wild type, Aox activity was induced in the strains with a single deletion if the cells were transferred from glucose to oleate medium, with maximum induction occurring 5 h after transfer (Fig. 3). For two genes, we also observed changes in Aox activity against substrates of particular chain lengths (Fig. 4A), whereas, for the other genes, the overall profile changed. Mutants lacking Aox2 or Aox3 had acyl-CoA oxidase activity with changes in chain-length preference (Fig. 4). Thus, Aox2 is a long-chain Aox and Aox3 a short-chain-specific enzyme. These results are similar to those of Picataggio and coworkers (32, 33) in C. tropicalis, showing that Pxp4 is a short-chain isozyme and Pxp5 is a long-chain isozyme. However, whereas the pxp4-pxp5 double-deleted strain in C. tropicalis cannot grow on FA, a Y. lipolytica Δpox2-Δpox3-deleted strain (MTLY20) had a minor growth defect on plates and no defect in liquid medium (Fig. 5). This suggests that toxic FA intermediates accumulate around colonies on plates, but not in liquid medium. In MTLY20, acyl-CoA oxidase activity is one-fifth that in the wild type, indicating that POX2 and POX3 are the major Aox. Aox activity in the Δpox2-Δpox5 and Δpox3-Δpox5 deletion mutants indicates that POX5 encodes an Aox with broad chain length specificity.
Aox protein levels were analyzed after transfer to YNBO. Aoxp levels paralleled the Aox activity. Two main signals were detected with anti-Aox3p and anti-Aox5p antibodies (Fig. 7, bands a and b). The signals were very weak or absent if the corresponding POX gene was disrupted. Regulation of Aox levels depended on the POX genotype, and the deletion of one of the POX genes resulted in an increase in the remaining Aox levels.
The precise identification of the Aox induced was not possible due to the lack of highly specific antibodies. Instead, partial cross-reactions were obtained. Nevertheless, these results (Aox activity, growth in oleic acid media, and Aox protein levels) strongly suggest that POX1 does not contribute to Aox activity and that the growth reflects the expression levels of the other genes. POX2 and POX3 code for chain-length-specific Aox (long and short chains, respectively). POX5 codes for a nonspecific chain length Aox, and POX4 codes for a minor Aox that facilitates partial growth only and is involved in the regulation of total Aox activity.
These results now open the possibility to investigate more precisely the regulation, the function and the targeting of Aox in Y. lipolytica, as detailed below. The strains with gene deletions obtained should enable us to analyze more specifically the regulation of POX promoters according to POX context. Each promoter could be fused to the β-galactosidase reporter gene and be introduced separately into the various strains, making it possible to determine the regulation of individual promoters as a function of the POX gene deleted.
Aox are multimeric proteins (octamers) and may be homo- or heteromultimers. The structure of the AOX complex may now be analyzed by coimmunoprecipitation studies in the various strains by using anti-Aox antibodies.
Finally, mutants that lack Aox activity and cannot grow on oleic acid could be used to study in vivo import of Aox into Y. lipolytica peroxisomes and to define import signals. The possibility of producing a single Aox protein (Aox2, Aox3, or Aox5) could be tested, and an analysis for Aox activity and growth complementation could be performed.
ACKNOWLEDGMENTS
We thank J. Knight for editing the English version of the text.
This work was supported by the Institut National de la Recherche Agronomique and by the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Barth G, Gaillardin C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol Rev. 1997;19:219–237. doi: 10.1111/j.1574-6976.1997.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 2.Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf K, editor. Nonconventional yeasts in biotechnology. Vol. 1. Berlin, Germany: Springer-Verlag; 1996. pp. 313–388. [Google Scholar]
- 3.Berninger G, Schmidtchen R, Casel G, Knorr A, Rautenstrauss K, Kunau W H, Schweizer E. Structure and metabolic control of the Yarrowia lipolytica peroxisomal 3-oxoacyl-CoA-thiolase gene. Eur J Biochem. 1993;216:607–613. doi: 10.1111/j.1432-1033.1993.tb18180.x. [DOI] [PubMed] [Google Scholar]
- 4.Bühler M, Schindler J. Aliphatic hydrocarbons. In: Rehm H J R, editor. Biotechnology. Weinheim, Germany: Verlag Chemie; 1984. pp. 329–385. [Google Scholar]
- 5.Cardillo R, Fuganti C, Barbeni M, Cabella P, Guarda P A, Allegrone G. European patent 0,412,880. 1991. [Google Scholar]
- 6.Casaregola S, Feynerol C, Diez M, Fournier P, Gaillardin C. Genomic organization of the yeast Yarrowia lipolytica. Chromosoma. 1997;106:380–390. doi: 10.1007/s004120050259. [DOI] [PubMed] [Google Scholar]
- 7.De Felice B, Pontecorvo G, Carfagna M. Degradation of waste waters from olive oil mills by Yarrowia lipolytica ATCC 20255 and Pseudomonas putida. Acta Biotechnol. 1997;3:231–239. [Google Scholar]
- 8.Dmochowska A, Dignard D, Maleszka R, Thomas D Y. Structure and transcriptional control of the Saccharomyces cerevisiae POX1 gene encoding acyl-coenzyme A oxidase. Gene. 1990;88:247–252. doi: 10.1016/0378-1119(90)90038-s. [DOI] [PubMed] [Google Scholar]
- 9.Do Y Y, Huang P L. Characterization of a pollination-related cDNA from Phalaenopsis encoding a protein which is homologous to human peroxisomal acyl-CoA oxidase. Arch Biochem Biophys. 1997;344:295–300. doi: 10.1006/abbi.1997.0212. [DOI] [PubMed] [Google Scholar]
- 10.Endrizzi A, Pagot Y, Le Clainche A, Nicaud J M, Belin J M. Production of lactones and peroxisomal beta-oxidation in yeasts. Crit Rev Biotechnol. 1996;16:301–329. doi: 10.3109/07388559609147424. [DOI] [PubMed] [Google Scholar]
- 11.Ercoli B, Fuganti C, Grasselli P, Servi S, Allegone G, Barbeni M, Pisciotta A. Stereochemistry of the biogeneration of C-10 and C-12 gamma lactones in Yarrowia lipolytica and Pichia ohmeri. Biotechnol Lett. 1992;14:665–668. [Google Scholar]
- 12.Gaillardin C, Ribet A M. LEU2 directed expression of beta-galactosidase activity and phleomycin resistance in Yarrowia lipolytica. Curr Genet. 1987;11:369–375. doi: 10.1007/BF00378179. [DOI] [PubMed] [Google Scholar]
- 13.Gussow D, Clackson T. Direct clone characterization from plaques and colonies by the polymerase chain reaction. Nucleic Acids Res. 1989;17:4000. doi: 10.1093/nar/17.10.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill D E, Boulay R, Rogers D. Complete nucleotide sequence of the peroxisomal acyl CoA oxidase from the alkane-utilizing yeast Candida maltosa. Nucleic Acids Res. 1988;16:365–376. doi: 10.1093/nar/16.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooks M A, Bode K, Couee I. Higher-plant medium- and short-chain acyl-CoA oxidases: identification, purification and characterization of two novel enzymes of eukaryotic peroxisomal beta-oxidation. Biochem J. 1996;320:607–614. doi: 10.1042/bj3200607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawamoto S, Nozaki C, Tanaka A, Fukui S. Fatty acid beta-oxidation system in microbodies of n-alkane-grown Candida tropicalis. Eur J Biochem. 1978;83:609–613. doi: 10.1111/j.1432-1033.1978.tb12130.x. [DOI] [PubMed] [Google Scholar]
- 17.Kirsch T, Loffler H G, Kindl H. Plant acyl-CoA oxidase: purification, characterization, and monomeric apoprotein. J Biol Chem. 1986;261:8570–8575. [PubMed] [Google Scholar]
- 18.Kunau W H, Kionka C, Ledebur A, Mateblowski M, Moreno de la Garza M, Schultz-Borchard U, Thieringer R, Veenhuis M. Beta oxidation systems in eukaryotic microorganisms. In: Fahimi H D S, editor. Peroxisomes in biology and medicine. Berlin, Germany: Springer-Verlag; 1987. pp. 128–140. [Google Scholar]
- 19.Le Clainche A. Ph.D. thesis. Thiverval-Grignon, France: Institut National Agronomique Paris-Grignon; 1997. [Google Scholar]
- 20.Maftahi M, Gaillardin C, Nicaud J M. Sticky-end polymerase chain reaction method for systematic gene disruption in Saccharomyces cerevisiae. Yeast. 1996;12:859–868. doi: 10.1002/(SICI)1097-0061(199607)12:9%3C859::AID-YEA978%3E3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 21.Maniatis E F, Fritsch J, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Masuda Y, Park S M, Ohta A, Takagi M. Cloning and characterization of the POX2 gene in Candida maltosa. Gene. 1995;167:157–161. doi: 10.1016/0378-1119(95)00655-9. [DOI] [PubMed] [Google Scholar]
- 23.Mishina M, Kamiryo T, Tashiro S, Hagihara T, Tanaka A, Fukui S, Osumi M, Numa S. Subcellular localization of two long-chain acyl-coenzyme-A synthetases in Candida lipolytica. Eur J Biochem. 1978;89:321–328. doi: 10.1111/j.1432-1033.1978.tb12533.x. [DOI] [PubMed] [Google Scholar]
- 24.Murray W W, Rachubinski R A. The primary structure of a peroxisomal fatty acyl-CoA oxidase from the yeast Candida tropicalis pK233. Gene. 1987;51:119–128. doi: 10.1016/0378-1119(87)90300-3. [DOI] [PubMed] [Google Scholar]
- 25.Neuveglise C, Nicauda J M, Ross-Macdonald P, Gaillardin C. A shuttle mutagenesis system for tagging genes in the yeast Yarrowia lipolytica. Gene. 1998;213:37–46. doi: 10.1016/s0378-1119(98)00205-4. [DOI] [PubMed] [Google Scholar]
- 26.Nuttley W M, Brade A M, Gaillardin C, Eitzen G A, Glover J R, Aitchison J D, Rachubinski R A. Rapid identification and characterization of peroxisomal assembly mutants in Yarrowia lipolytica. Yeast. 1993;9:507–517. [Google Scholar]
- 27.Okazaki K, Takechi T, Kambara N, Fukui S, Kubota I, Kamiryo T. Two acyl-coenzyme A oxidases in peroxisomes of the yeast Candida tropicalis: primary structures deduced from genomic DNA sequence. Proc Natl Acad Sci USA. 1986;83:1232–1236. doi: 10.1073/pnas.83.5.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okazaki K, Tan H, Fukui S, Kubota I, Kamiryo T. Peroxisomal acyl-coenzyme A oxidase multigene family of the yeast Candida tropicalis; nucleotide sequence of a third gene and its protein product. Gene. 1987;58:37–44. doi: 10.1016/0378-1119(87)90027-8. [DOI] [PubMed] [Google Scholar]
- 29.Osmundsen H, Bremer J, Pedersen J I. Metabolic aspects of peroxisomal beta-oxidation. Biochim Biophys Acta. 1991;1085:141–158. doi: 10.1016/0005-2760(91)90089-z. [DOI] [PubMed] [Google Scholar]
- 30.Pagot Y, Endrizzi A, Nicaud J M, Berlin J M. Utilization of an auxotrophic strain of the yeast Yarrowia lipolytica to improve gamma-decalactone production yields. Lett Appl Microbiol. 1997;25:113–116. doi: 10.1046/j.1472-765x.1997.00182.x. [DOI] [PubMed] [Google Scholar]
- 31.Pagot Y, Le Clainche A, Nicaud J M, Wache Y, Belin J M. Peroxisomal beta-oxidation activities and gamma-decalactone production by the yeast Yarrowia lipolytica. Appl Microbiol Biotechnol. 1998;49:295–300. doi: 10.1007/s002530051172. [DOI] [PubMed] [Google Scholar]
- 32.Picataggio S, Deanda K, Mielenz J. Determination of Candida tropicalis acyl coenzyme A oxidase isozyme function by sequential gene disruption. Mol Cell Biol. 1991;11:4333–4339. doi: 10.1128/mcb.11.9.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picataggio S, Rohrer T, Deanda K, Lanning D, Reynolds R, Mielenz J, Eirich L D. Metabolic engineering of Candida tropicalis for the production of long-chain dicarboxylic acids. Bio/Technology. 1992;10:894–898. doi: 10.1038/nbt0892-894. [DOI] [PubMed] [Google Scholar]
- 34.Roostita R, Fleet G H. Growth of yeasts in milk and associated changes to milk composition. Int J Food Microbiol. 1996;31:205–219. doi: 10.1016/0168-1605(96)00999-3. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu S, Yasui K, Tani Y, Yamada H. Acyl-CoA oxidase from Candida tropicalis. Biochem Biophys Res Commun. 1979;91:108–113. doi: 10.1016/0006-291x(79)90589-8. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka A, Osumi M, Fukui S. Peroxisomes of alkane-grown yeast: fundamental and practical aspects. Ann N Y Acad Sci. 1982;386:183–199. doi: 10.1111/j.1749-6632.1982.tb21416.x. [DOI] [PubMed] [Google Scholar]
- 37.Thierry A, Perrin A, Boyer J, Fairhead C, Dujon B, Frey B, Schmitz G. Cleavage of yeast and bacteriophage T7 genomes at a single site using the rare cutter endonuclease I-Sce I. Nucleic Acids Res. 1991;19:189–190. doi: 10.1093/nar/19.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueda M, Mozzafar S, Tanaka A. Peroxisomal localization of enzymes related to fatty acid beta-oxidation in an n-alkane-grown yeast, Candida tropicalis. Agric Biol Chem. 1985;49:1821–1828. [Google Scholar]
- 39.Wang H, Le Clainche A, Le Dall M T, Wache Y, Pagot Y, Belin J M, Gaillardin C, Nicaud J M. Cloning and characterization of the peroxisomal acyl CoA oxidase ACO3 gene from the alkane-utilizing yeast Yarrowia lipolytica. Yeast. 1998;14:1373–1386. doi: 10.1002/(SICI)1097-0061(199811)14:15<1373::AID-YEA332>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 40.Wang R, Thorpe C. The reductive half-reaction in acyl-CoA oxidase from Candida tropicalis: interaction with acyl-CoA analogues and an unusual thioesterase activity. Arch Biochem Biophys. 1991;286:504–510. doi: 10.1016/0003-9861(91)90072-q. [DOI] [PubMed] [Google Scholar]
- 41.Xuan J W, Fournier P, Declerck N, Chasles M, Gaillardin C. Overlapping reading frames at the LYS5 locus in the yeast Yarrowia lipolytica. Mol Cell Biol. 1990;10:4795–4806. doi: 10.1128/mcb.10.9.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]