Abstract
Genome-wide binding profiles of estrogen receptor (ER) and FOXA1 reflect cancer state in ER+ breast cancer. However, routine profiling of tumor transcription factor (TF) binding is impractical in the clinic. Here, we show that plasma cell-free DNA (cfDNA) contains high-resolution ER and FOXA1 tumor binding profiles for breast cancer. Enrichment of TF footprints in plasma reflects the binding strength of the TF in originating tissue. We defined pure in vivo tumor TF signatures in plasma using ER+ breast cancer xenografts, which can distinguish xenografts with distinct ER states. Furthermore, state-specific ER-binding signatures can partition human breast tumors into groups with significantly different ER expression and mortality. Last, TF footprints in human plasma samples can identify the presence of ER+ breast cancer. Thus, plasma TF footprints enable minimally invasive mapping of the regulatory landscape of breast cancer in humans and open vast possibilities for clinical applications across multiple tumor types.
Transcription factor footprints in cell-free DNA can be used to detect breast cancer phenotypes.
INTRODUCTION
Transcription factors (TFs) are at the apex of gene regulation (1). They usually bind small stretches of DNA in a sequence-specific manner (2). The size of the mammalian genomes is several orders of magnitude greater than the size of TF binding motifs. Hence, there are many more TF binding site (TFBS) sequences that occur by chance compared to functional TFBS (3). Although the question of how TFs discriminate functional binding sites from random motif occurrences is still being actively investigated (4, 5), at least two mechanisms enable us to connect TF binding to cell state. First, the cell type–specific expression of TFs restricts the pool of motifs recognized in a given cell type. Second, most motifs in the genome are occluded by nucleosomes most of the time (6, 7). As a result, the sites in the genome bound by any given TF contribute to the epigenomic signature of a cell type. Furthermore, because functional TF binding drives gene regulation, mapping a TF’s binding sites in a cell also contributes to an understanding of the regulatory landscape of the cell (8). Methods like chromatin immunoprecipitation with DNA sequencing (ChIP-seq), chromatin immunoprecipitation, exonuclease digestion and DNA sequencing (ChIP-exo), and cleavage under target and release using nuclease (CUT&RUN) have been used to identify binding sites of human TFs across cell types (9–11).
Connecting genome-wide TF binding landscape to cell state acquires added significance in TF-driven diseases like estrogen receptor–positive (ER+) breast cancer. In ER+ breast cancer, the subset of all possible binding sites that are occupied by ER reflects disease prognosis (12, 13). Furthermore, endocrine therapies work by either modulating ER binding, removal of ligand, or degrading ER, resulting in global changes in ER binding and, in some cases, even increased ER residence times on chromatin (14). ER binding also depends on the pioneer factor FOXA1 (15). FOXA1 expression and binding might persist after ER removal during endocrine therapy, possibly driving treatment resistance (13). Together, a genome-wide map of the binding profiles of ER and associated factors could be leveraged for defining disease state, predicting treatment outcome, and possibly choosing effective therapy. However, routine and serial mapping of TF binding in tumors is currently not possible in the clinic because of risks involved in obtaining tumor tissues. Here, we leverage an alternate means to obtain the same information one would get from ChIP or CUT&RUN in a minimally invasive manner to define underlying disease biology.
Dying cells in the human body release their content into the bloodstream (16). Genomic DNA that is bound by nucleosomes and TFs escapes endogenous nucleases and so remains protected in plasma (Fig. 1A) (17). Regular turnover of lymphoid and myeloid cells in the human body is the major contributor to the pool of cell-free DNA (cfDNA) in plasma (18). However, in the presence of cancer, a detectable fraction of cfDNA arises from tumors (19, 20). This suggests that cfDNA has the potential to map the tumor epigenome in real time and therefore can help uncover the regulatory landscape of cancer from plasma. Fragmentomics seeks to uncover the tissue of origin of cfDNA using the information in cfDNA fragment length. Fragmentomics had its earliest application in prenatal diagnosis and is now being explored as an alternative to mutations and methylation profiling to identify cfDNA tissue of origin in cancer (21–23). cfDNA properties such as promoter nucleosome dynamics, locus-specific fragment length distribution, nucleosome spacing in gene bodies, and nucleosome depletion at promoters have been used to identify the tissue of origin of cfDNA to aid detection of cancer (17, 24, 25). Because TFs and nucleosomes protect distinctly different lengths of DNA, cfDNA facilitates direct mapping of protein-DNA interactions in their cells of origin (17). TF binding from cfDNA has also been characterized by averaging across thousands of putative sites, either looking at short protections (17) or by inferring TF binding by nucleosome depletion at TFBS (26). However, classifying single binding sites as either TF bound or nucleosome bound to then develop a regulatory signature from cfDNA data has not been attempted before. Such a regulatory signature has the potential to distinguish distinct TF-driven cancer states.
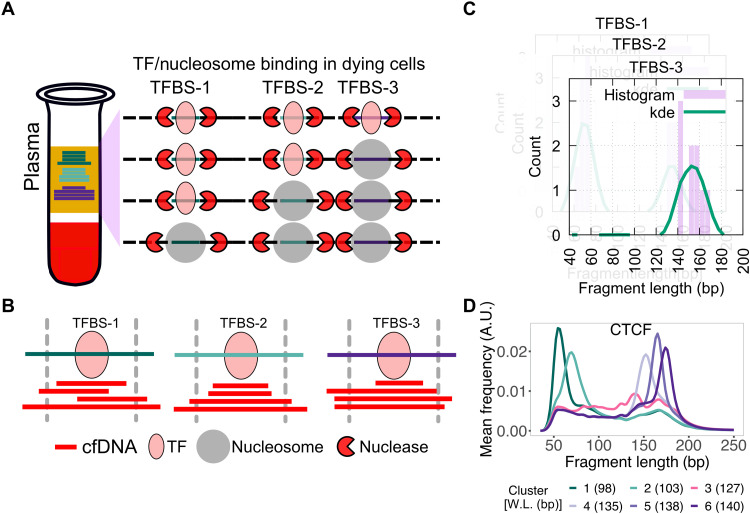
Fig. 1. Schematic for identifying subset of binding sites with TF footprints.
(A) When TFs or nucleosomes are bound at TFBSs, they protect different lengths of DNA from nucleases in dying cells in the human body. (B) When sequenced cfDNA fragments are mapped to TFBSs ± 50 bp, varying numbers of short and long cfDNA fragments are found at the three TFBSs shown in (A). (C) cfDNA fragment length distribution is estimated at each TFBS (purple bars) and smoothed using kernel density estimation (kde) (green line). (D) K-means clustering is performed on smoothed length distribution to group TFBSs with similar cfDNA fragment length distribution. Here, smoothened length distributions of clusters of CTCF TFBS are shown. Weighted length (W.L.) for each CTCF length cluster is shown in parentheses. A.U., arbitrary units.
Here, we map TF binding from plasma at single binding sites to define cfDNA signatures that reflect distinct ER+ disease states. We map TF footprints in plasma cfDNA by combining library protocols that enrich for short fragments with computational methods that identify the subset of TFBS that leave footprints in plasma. We further show that the strength of TF footprints in plasma is proportional to the binding strength of the TF in the tissue of origin of the cfDNA fragments when compared with gold standard methods of CUT&RUN and ATAC-seq (assay for transposase-accessible chromatin using sequencing) datasets from tumor cells. This confirms that we can map regulatory landscapes of tumors from plasma. We further show that different states of ER lead to distinct ER binding profiles inferred from plasma that reflect unique ER-regulated gene expression. We can use the cfDNA signatures of unique ER states to partition ER+ breast tumors in The Cancer Genome Atlas (TCGA) into groups that not only have distinct ER expression but also have significantly different mortality. Last, we identify TFBS where the enrichment of TF footprints in human plasma samples is used to identify the presence of breast cancer. Thus, our results show that plasma cfDNA contains TF binding information that is specific to tumor state.
RESULTS
Unique cfDNA fragment length distributions identify TF binding in the tissue of origin
ChIP-seq and CUT&RUN applied to cell lines and tissue samples represent gold standard methods of determining TF binding across the genome. To study human diseases, it is impractical and nearly impossible to perform repeat analyses using biopsy tissues. We therefore set out to develop an alternative to ChIP-seq and CUT&RUN that can be applied to physiological and pathological states of humans in a minimally invasive manner by inferring specific TF binding from plasma cfDNA. TF footprints [<80 base pairs (bp)] are too short to be captured by standard protocols for preparing genomic libraries, but single-strand library protocol (SSP) for cfDNA can robustly capture short and longer nucleosomal cfDNA fragments (17). In all our analyses, we used cfDNA sequencing datasets generated using SSP in this study as well as from a published study (17).
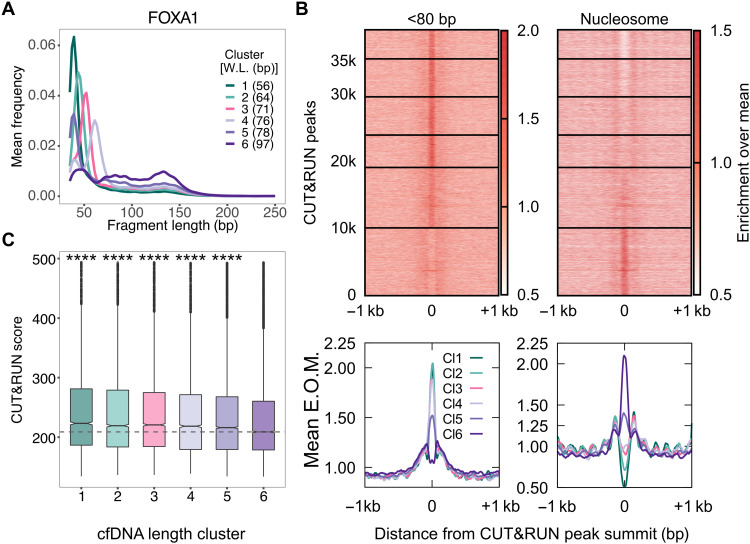
To ask whether we can uncover TF-nucleosome dynamics from plasma cfDNA, we undertook a candidate approach of examining binding sites of specific TFs. We started with CTCF (CCCTC-binding factor) because it is constitutively expressed (27), has a long residence time on DNA (28), and has known binding profiles in a large, diverse set of cell types (9). We aggregated CTCF binding sites from 70 cell lines representing 18 cell types and analyzed fragment length distributions of cfDNA from a healthy donor from the IH02 dataset (17) at these sites. At each TFBS, we mapped cfDNA fragment midpoints (Fig. 1B) and estimated a fragment length distribution (Fig. 1C). K-means clustering of these fragment length distributions identified two types of clusters—one enriched with short cfDNA fragments (<100 bp; clusters 1 and 2) and the other enriched with long cfDNA fragments (>120 bp; clusters 3 to 6) (Fig. 1D). When we mapped enrichment of cfDNA fragments around 1 kb of the TFBS, clusters 1 and 2 showed strong enrichment of short protections at TFBSs relative to 1 kb upstream and downstream of the TFBS (Fig. 2A). Notably, these two clusters also showed strong nucleosome phasing at least 1 kb upstream and downstream of the TFBS (Fig. 2B). It is well known that CTCF binding organizes nucleosomes in its vicinity (29). Thus, the fragment length profile of plasma cfDNA at CTCF binding sites not only identified TF binding but also uncovered the chromatin structure surrounding the bound CTCF. Because most cfDNA in a healthy individual arises from lymphoid and myeloid cells, we asked whether the TFBS clustering based on cfDNA reflected nucleosome positioning in a representative lymphoblastoid cell line, GM12878. MNase-seq (Micrococcal Nuclease digestion followed by sequencing) data (9) from GM12878 showed strong nucleosome phasing for clusters 1 and 2, but not for the rest of clusters (Fig. 2C). This strongly suggests that we are capturing CTCF binding and associated nucleosome landscape from lymphoid/myeloid cells in cfDNA and that the mechanism of DNA release from these cell types gives a signal similar to MNase profiling.
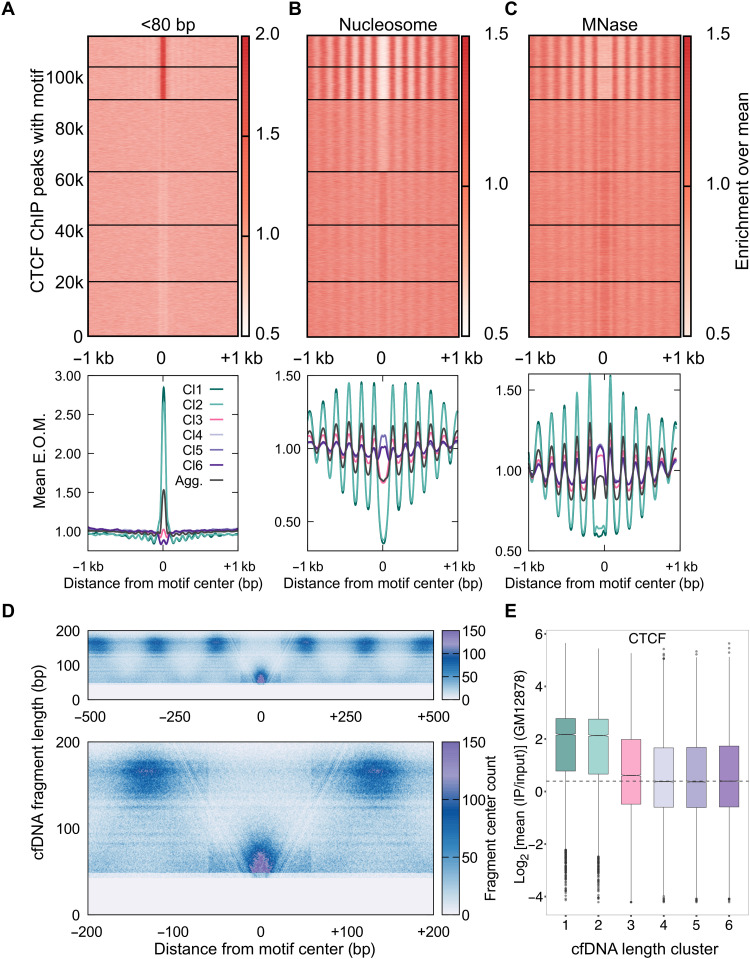
Fig. 2. cfDNA maps CTCF-nucleosome dynamics in plasma from a healthy individual.
(A) Enrichment over the mean signal in TFBS ± 1 kb of cfDNA short (<80 bp) fragments is plotted as a heatmap (top) (117,144 CTCF TFBS) and as metaplots for each cluster (bottom). E.O.M., enrichment over mean. (B) Same as (A) for nucleosome-sized fragments (130 to 180 bp). (C) Same as (B) for MNase-seq dataset from GM12878 cells. (D) Fragment midpoint versus fragment length plot (V plot) of cfDNA fragments centered at CTCF binding sites from clusters 1 and 2. Fragment densities at motif center ± 500 bp (top) and motif center ± 200 bp (bottom) are plotted. (E) Boxplot of CTCF mean ChIP signal from the GM12878 cell line across length clusters. Number of sites (n) in length clusters and P value using Kolmogorov-Smirnov (KS) test with alternative = “greater” option are the following: Cl1: n = 11,978, P(1,6) < 2.2 × 10−16; Cl2: n = 12,811, P(2,6) < 2.2 × 10−16; Cl3: n = 28,132, P(3,6) = 1.1 × 10−31; Cl4: n = 20,839, P(4,6) = 0.95; Cl5: n = 22,087, P(5,6) = 0.96; Cl6: n = 21,297. P(a,b) denotes P values calculated between scores in length cluster a and scores in length cluster b.
To further visualize the chromatin structure around CTCF-bound sites and to identify the minimum protection conferred by CTCF on DNA, we plotted the count of cfDNA fragment midpoints around CTCF-bound sites as V-plots for sites in clusters 1 and 2 (30). With the V-plot spanning TFBS ± 500 bp, we observe strongly positioned nucleosomes with protection length between 140 and180 bp, flanking short protections at the CTCF sites in the center (Fig. 2D, top). In the V-plot spanning TFBS ± 200 bp, a strong “V” is evident at the center, where there is an enrichment of fragments <80 bp. A “V” indicates a well-positioned, strong barrier to nucleases, which further confirms that cfDNA is directly mapping TF binding and its associated nucleosome landscapes from the cells of origin (Fig. 2D, bottom).
The separation of bound and unbound sites by our clustering approach is also apparent when we compare the short and nucleosomal fragment enrichment at individual clusters to the aggregate enrichments across all sites (black lines in Fig. 2A, bottom). TF enrichment, nucleosome occlusion, and nucleosome ordering are substantially weaker in aggregate compared to clusters 1 and 2 as expected. In other words, identifying the subset of sites that are bound could inform us of TF binding strength in cfDNA cells of origin. To test this idea, we calculated the ChIP scores from GM12878 cells at TFBS belonging to each cfDNA length cluster. We found that the ChIP scores of the first two clusters were almost four times higher than those of the other four clusters (Fig. 2E). The fact that hematopoietic ChIP scores correlate with our inferred sites of CTCF binding in cfDNA supports the conclusion that cfDNA length profile at TFBS reports on TF binding strength in cfDNA tissue of origin.
Binding sites of hematopoietic TFs are sensitive to changes in cfDNA tissues of origin
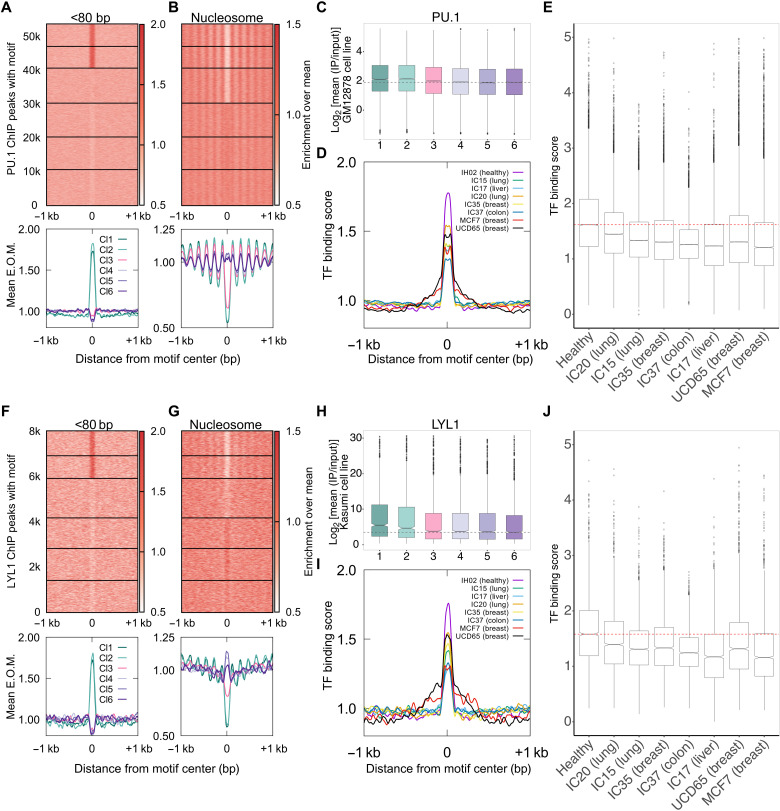
Because most cfDNA in healthy individuals is of lymphoid/myeloid origin, we asked whether we could map protections for lymphoid/myeloid-specific TFs: PU.1, a pioneer factor that plays a crucial role in myeloid and B cell development (31), and LYL1, an important factor for erythropoiesis (32) and development of other hematopoietic cell types (33). Upon clustering the binding sites of PU.1 and LYL1 based on cfDNA length distributions of the healthy donor, IH02 dataset, we found an enrichment of short protections at a subset of binding sites similar to CTCF (clusters 1 and 2; Fig. 3, A and F). Distribution of longer fragments around the binding sites showed strong nucleosomal phasing in clusters 1 and 2 (Fig. 3, B and G). The presence of nucleosome phasing further confirmed specific TF binding, as this is a known outcome of LYL1 and PU.1 binding to DNA (26, 34, 35). Clusters 1 and 2, which had the highest enrichment of short protections, also had significantly higher ChIP scores in lymphoid/myeloid cell lines compared to cluster 6 (nucleosomal) for both PU.1 and LYL1 (Fig. 3, C and H). We observe an even bigger separation in ATAC-seq scores from CD34+ cells (36) between the length clusters of PU.1 sites (short, cluster 1 and long, cluster 6), indicating that plasma cfDNA footprints also reflect ATAC-seq enrichments in cfDNA tissues of origin (fig. S1, A to F). In summary, we can map the binding of hematopoietic TFs in plasma cfDNA in humans.
Fig. 3. cfDNA of lymphoid/myeloid origin contains hematopoietic TF footprints.
(A) Enrichment over the mean signal in PU.1 TFBS ± 1 kb of cfDNA short (<80 bp) fragments is plotted as a heatmap (top) (53,613 PU.1 TFBS) and as metaplots for each cluster (bottom). (B) Same as (A) for nucleosome-sized fragments (130 to 180 bp). (C) Boxplot of PU.1 mean ChIP signal (log2) from GM12878 cell line across length clusters. Number of sites (n) in length clusters and P value using KS test are the following: Cl1: n = 6528, P(1,6) = 9.2 × 10−20; Cl2: n = 6447, P(2,6) = 1.7 × 10−22; Cl3: n = 10,377, P(3,6) = 0.00011; Cl4: n = 10,036, P(4,6) = 0.19; Cl5: n = 9673, P(5,6) = 0.7; Cl6: n = 10,552. Significant string was determined after Bonferroni correction. (D) Enrichment metaplots for short fragments in PU.1 TFBS belonging to clusters 1 and 2 for healthy (IH02), cancer cfDNA (IC15, IC17, IC20, IC35, and IC37), and PDX cfDNA (MCF7 and UCD65). (E) Boxplot of mean of short fragment enrichment (TFBS ± 50 bp) for the samples and TFBS plotted in (D). (F) Same as (A) for LYL1 (7999 TFBS). (G) Same as (B) for LYL1. (H) Same as (C) for LYL1. Number of sites (n) in length clusters and P value using KS test are the following: Cl1: n = 1083, P(1,6) = 4.7 × 10−12; Cl2: n = 1001, P(2,6) = 3 × 10−7; Cl3: n = 1748, P(3,6) = 0.18; Cl4: n = 1351, P(4,6) = 0.15; Cl5: n = 1415, P(5,6) = 0.62; Cl6: n = 1401. Significant string was determined after Bonferroni correction. (I) Same as (D) for LYL1. (J) Same as (E) for LYL1.
In cancer patients, cancer cells also contribute significantly to plasma cfDNA. Hence, we hypothesized that cfDNA derived from cancer cells would dilute lymphoid/myeloid signal. Such dilution would lead to a proportional decrease in the enrichment of short fragments at clusters 1 and 2 of hematopoietic TFBS because of cfDNA contributions from nonhematopoietic cell types where PU.1 and LYL1 are absent. To test this hypothesis, we performed k-means clustering of PU.1 and LYL1 binding sites based on the cfDNA length distributions for cfDNA from donors with cancer using published datasets (17). We found that the short fragment enrichment for the bound clusters 1 and 2 was the highest for healthy human plasma (Fig. 3, D and I). Cancer samples had significantly weaker short fragment enrichment at sites from clusters 1 and 2 for PU.1 and LYL1 (Fig. 3, E and J) and did not have higher ChIP scores compared to cluster 6 (fig. S2, A to D). In addition to using cfDNA from cancer patients, we also used cfDNA from xenograft models derived from human cancer cell lines (Fig. 4A). Because the only source of human cfDNA in mice bearing a xenograft is from cancer cells, fragments that uniquely map to the human genome in this context represent pure circulating tumor DNA (ctDNA). We found no expression of PU.1 or LYL1 in breast tumor model systems, and accordingly, we observed no nucleosome phasing or higher ChIP scores for the top two clusters in the xenograft cfDNA (fig. S2, E to H). In addition, we found an expected decrease in enrichment of short fragments in clusters 1 and 2 from the xenografts (UCD65 and MCF7) compared to healthy donor (Fig. 3, D, E, I, and J). The clear separation between cfDNA from a healthy donor and cfDNA from cancer patients and from xenografts suggests that the length profiles of cfDNA at hematopoietic TFBS when combined with local enrichment of short fragments can identify dilution of lymphoid/myeloid cfDNA across diverse plasma samples.
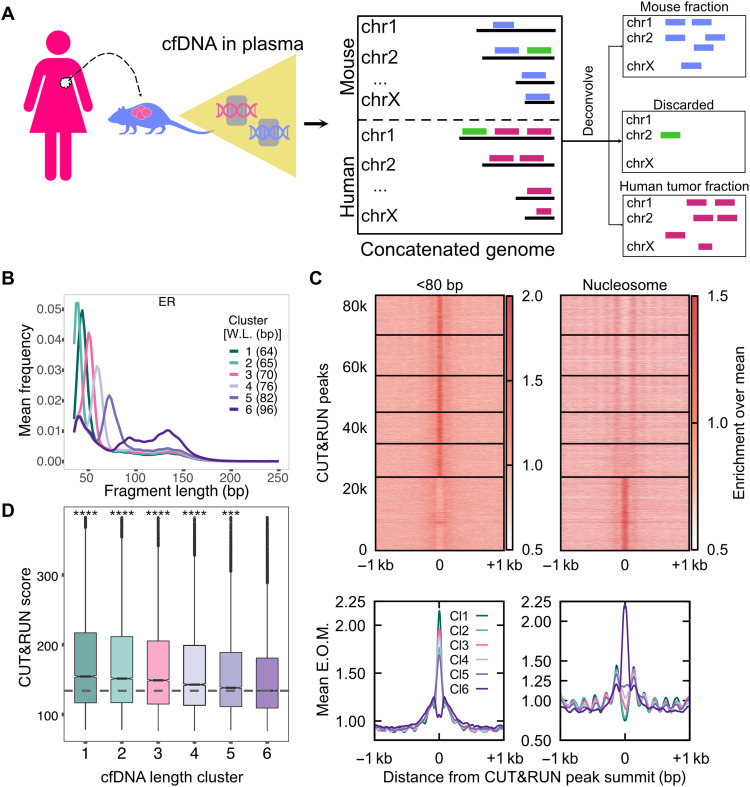
Fig. 4. ER+ breast cancer xenograft models enable identification of pure tumor cfDNA footprints for ER.
(A) Schematic of human cancer cell lines implanted in the mouse and the process of identifying tumor cfDNA by mapping mouse plasma cfDNA to an in silico concatenated genome. Fragments mapped uniquely to the human genome (violet lines) define tumor cfDNA (ctDNA). Fragments mapped uniquely to the mouse genome (blue lines) arise from the tumor microenvironment and from the mouse lymphoid/myeloid cells. Fragments mapped to both genomes were discarded (green lines). (B) Average length distributions at clusters of ER CUT&RUN peaks (summit ± 50 bp) generated by k-means clustering (n = 6) of the ctDNA fragment length distribution. (C) Enrichment over the mean signal in TFBS ± 1 kb of cfDNA short (<80 bp) fragments is plotted as a heatmap (top) (83,311 ER TFBS) and as metaplots for each cluster (bottom). (D) Boxplot of ER CUT&RUN scores for peak summits in k-means clusters. Number of sites (n) in length clusters and P value using KS test are the following: Cl1: n = 12,785, P(1,6) = 1.2 × 10−151; Cl2: n = 13,301, P(2,6) = 7.9 × 10−116; Cl3: n = 11,943, P(3,6) = 1.5 × 10−80; Cl4: n = 10,363, P(4,6) = 1.6 × 10−37; Cl5: n = 10,848, P(5,6) = 1.1 × 10−8; Cl6: n = 24,029. Significant string was determined after Bonferroni correction. ****P < 0.0001 and ***0.0001 < P < 0.001.
ctDNA maps tumor-specific TF binding
We were able to uncover strong signals of CTCF and hematopoietic TF binding in plasma cfDNA because most cells that release cfDNA have these TFs bound in their genome. However, tumor-specific TFs will, by definition, have weaker signals because tumor cfDNA is a minor fraction of total cfDNA even in stage IV disease (37). To develop pure tumor signatures of TF binding in cfDNA, we turned to human cancer xenografts implanted in mice. Because the tumor-derived cfDNA in xenograft models would map to the human genome, whereas the endogenous cfDNA from the mouse would map to the mouse genome, we could identify cfDNA molecules from sequencing that were specifically from the tumor, hence ctDNA, but obtained from an in vivo system (Fig. 4A).
ER+ tumors are driven by the TFs ER and FOXA1 (15). We hypothesized that plasma-derived TF binding profiles could distinguish different ER+ disease states. To model different states of ER+ diseases, we used three types of ER+ breast tumor cells. MCF7 has elevated ER and was isolated from pleural effusion of a metastatic patient. UCD65 cell line (38) was derived from a lymph node metastasis of a 41-year-old woman. UCD65 has ESR1 gene amplification and even higher ER levels than MCF7. Amplified ESR1 has been found in ~20% of ER+ breast cancers, and significantly, amplified ESR1 was associated with longer survival times for patients receiving tamoxifen monotherapy (39). Last, UCD4 was derived from pleural effusion of a 68-year-old woman, with cells harboring the activating mutation D538G in ER that makes it estrogen independent (40). This mutation usually arises as an escape from endocrine therapy and is not observed in primary tumors [for example, it is absent from all samples in TCGA BRCA (Breast Invasive Carcinoma) cohort]. In summary, MCF7, UCD65, and UCD4 represent distinct states of ER-driven disease, enabling us to ask whether plasma-derived binding profiles of ER and associated TFs can distinguish these unique disease states.
We first identified ER and FOXA1 binding sites in tumor cells using CUT&RUN (10). CUT&RUN relies on a protein A–tagged nuclease linked to a primary antibody that binds epitope of choice and is an alternative to ChIP-seq. The nuclease is activated upon addition of calcium to release DNA fragments bound to the protein targeted by the antibody. Because of the absence of cross-linking and release of bound sites rather than enrichment of bound sites, CUT&RUN captures TF binding at higher sensitivity and provides a greater dynamic range of signals compared to ChIP-seq (10). We performed CUT&RUN for ER and FOXA1 in estradiol (E2)–treated MCF7 cells and obtained ~80,000 and ~40,000 CUT&RUN sites for ER and FOXA1, respectively, with sufficient coverage in both MCF7 and UCD65 xenograft cfDNA datasets.
When we performed fragment length distribution analysis of xenograft cfDNA datasets at ER CUT&RUN peaks and defined six clusters, the five clusters with the lowest expected fragment length (Fig. 4B) showed strong short fragment protections and phased nucleosomes (Fig. 4C) as well as significantly higher ER binding measured as CUT&RUN score (Fig. 4D). We observed similar trends for FOXA1 binding sites (Fig. 5, A to C). Positive correlation between ctDNA short fragment enrichment and CUT&RUN scores strongly suggests that we are capturing binding in cancer cells and that the signal from cfDNA release in vivo is similar to CUT&RUN profiling. Thus, defining binding sites in tumor cells using CUT&RUN enables sensitive mapping in plasma of the TF binding that occurs in tumor cells of origin.
Fig. 5. ER+ breast cancer xenograft models enable identification of pure tumor cfDNA footprints for FOXA1.
(A) Average length distributions at clusters of FOXA1 CUT&RUN peaks (summit ± 50 bp) generated by k-means clustering (n = 6) of the ctDNA fragment length distribution. (B) Enrichment over the mean signal in TFBS ± 1 kb of cfDNA short (<80 bp) fragments is plotted as a heatmap (top) (39,500 FOXA1 TFBS) and as metaplots for each cluster (bottom). (C) Boxplot of FOXA1 CUT&RUN scores (see Materials and Methods) for peak summits in k-means clusters. P values from KS test. Number of sites (n) in length clusters and P value using KS test are the following: Cl1: n = 4220, P(1,6) = 3.4 × 10−36; Cl2: n = 5669, P(2,6) = 3.2 × 10−19; Cl3: n = 5699, P(3,6) = 4.5 × 10−15; Cl4: n = 4831, P(4,6) = 3.1 × 10−10; Cl5: n = 9033, P(5,6) = 3.9 × 10−10; Cl6: n = 10,017. Significant string was determined after Bonferroni correction. ****P < 0.0001.
FOXA1, a pioneer factor, binds to nucleosomes and facilitates DNA access and binding for other TFs (41). At low levels of MNase, nucleosomal protections have been identified at FOXA binding sites, pointing to the possibility of a FOXA-bound nucleosome (42). However, this nucleosomal protection is lost at higher levels of MNase. In cfDNA, we found an almost complete lack of nucleosomal-sized particles at FOXA1 sites that had strong short footprints (length clusters 1 to 4). To probe this further, we analyzed the enrichment of nucleosomal fragments (130 to 180 bp) from MNase titration assays in K562 (43) and MCF7 cells (GSE77526) at CTCF and FOXA1 sites. We do not find an enrichment of nucleosomes over mean at low MNase levels at CTCF binding sites (fig. S4E). However, FOXA1 sites feature nucleosomal protection both in healthy cfDNA (fig. S4F) and in UCD65 xenograft cfDNA (fig. S4G). This suggests the presence of fragile nucleosome at FOXA1-bound sites. However, cfDNA nucleosome profile matches that generated from higher levels of MNase, suggesting higher fragmentation during cfDNA generation that does not preserve these fragile nucleosomes.
Unique sets of TFBS display tissue-specific TF protections in plasma
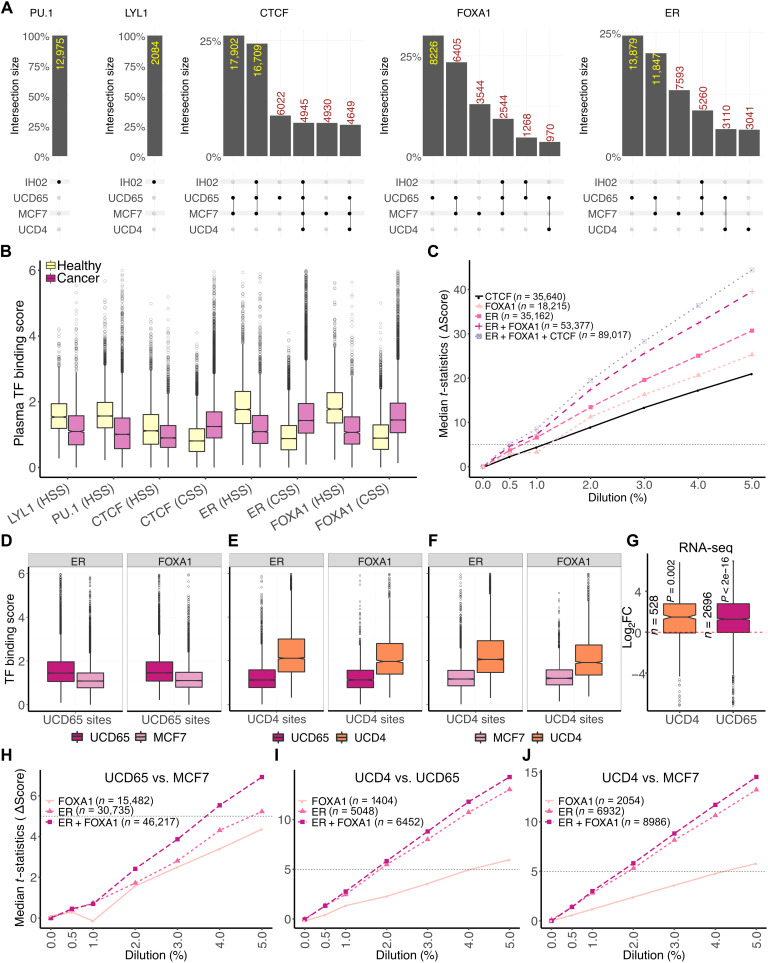
We have defined sets of binding sites that show TF-specific protections in two pure systems: plasma from a healthy human and plasma from xenograft tumor mouse models. We then asked whether we could define subsets of these sites that would be unique to tumor and hematopoietic cells. In addition, to ask whether we could identify sites that uniquely captured ER activity, we compared the ER+ xenografts with three states of ER: MCF7 (elevated ER), UCD65 (ER amplified), and UCD4 (containing activating D538G mutation). To do this, we performed length clustering analysis at all TFBS with both healthy plasma dataset and xenograft plasma datasets to identify binding site clusters with significantly higher ChIP/CUT&RUN binding scores compared to the nucleosomal cluster of binding sites for each cfDNA dataset. We then intersected the significant binding sites between healthy plasma and xenograft plasma. First, we found that PU.1 and LYL1 sites had TF protections that correlated with binding strength only in healthy plasma (Fig. 6A), indicating that all TFBS of PU.1 and LYL1 with significant footprints in plasma could be used to identify hematopoietic contribution to cfDNA. CTCF is a constitutive factor, ER is expressed in T cells and other hematopoietic cells (44), and factors related to FOXA1 that have the same binding motifs are expressed in hematopoietic cells, for example, FOXM1 (45). The partial overlap of binding of these or related factors in hematopoietic and cancer cells led us to find sites with significant TF protections in both healthy plasma and xenograft model plasma for CTCF, FOXA1, and ER (Fig. 6A and figs. S3 and S4). For example, a large fraction of CTCF sites (16,709 in set 2 and 4945 in set 4) are shared between xenograft plasma and healthy plasma, whereas the rest of CTCF sites (17,902 in set 1, 6022 in set 3, 4930 in set 5, and 4649 in set 6; CTCF in Fig. 6A) are cancer specific. In contrast, the top three sets of sites for FOXA1 and ER are xenograft specific, with the largest set of sites specific to UCD65 (8226 for FOXA1 and 13,879 for ER). FOXA1 has sites specific to MCF7 as well (set 3), and ER has sites specific to MCF7 (set 3) and UCD4 (set 6). Thus, despite the overlap in binding between hematopoietic cells and cancer cells, ER and FOXA1 have enough unique sites protected in plasma that distinguish healthy plasma from xenograft plasma. Significantly, we were able to also find sites that differentiated xenografts that differed in ER activity, supporting our premise that cfDNA TF footprints identify regulatory landscape of tumors.
Fig. 6. Tissue-specific TFBSs enable detection of disease states.
(A) Upset plots (67) of cfDNA-inferred bound sites in different plasma samples for LYL1, PU.1, CTCF, FOXA1, and ER (left to right). Plots were generated using the ComplexUpset R package (DOI: 10.5281/zenodo.4661589). (B) Boxplots of TF binding scores measured as mean enrichment of short fragments at CUT&RUN peak summit ± 100 bp for ER and FOXA1 and motif center ± 50 bp for LYL1, PU.1, and CTCF. CSS, cancer-specific sites; HSS, healthy-specific sites. (C) Line plot of median t statistic calculated for change in the binding scores (score in healthy plasma used as baseline) at binding sites of an individual TF or a collection of TFs at different in silico dilutions of healthy cfDNA with PDX ctDNA. At each dilution, 100 bootstrapped samples were generated. Horizontal dashed line is drawn where the t statistic equals 5. (D) Boxplot of TF binding scores in pure ctDNA (UCD65/MCF7) at ER and FOXA1 sites specific to UCD65. (E) Boxplot of TF binding scores in pure ctDNA (UCD65/UCD4) at ER and FOXA1 sites specific to UCD4. (F) Boxplot of TF binding scores in pure ctDNA (MCF7/UCD4) at ER and FOXA1 sites specific to UCD4. (G) Boxplot of log2 fold change (Log2FC) between UCD4/UCD65 and MCF7 in expression of genes adjacent to sites specific to UCD4 and UCD65. (H) Line plot of median t statistic calculated for the change in TF binding scores at UCD65- or MCF7-specific ER, FOXA1, or ER and FOXA1 sites combined at different in silico dilutions of healthy cfDNA with PDX ctDNA. At each dilution, 100 bootstrapped samples were generated. A horizontal dashed line is drawn where the t statistic equals 5. (I) Same as (H) for UCD4-specific ER and FOXA1 sites against UCD65. (J) Same as (H) for UCD4-specific ER and FOXA1 sites against MCF7.
Although FOXA1 is not expressed in lymphoid/myeloid cells, some FOXA1 binding sites identified in MCF7 cells showed significant enrichment of TF footprints in healthy plasma. We asked whether related FOX factors like FOXM1 and FOXK2 that are expressed in lymphoid/myeloid cells may be binding at these sites to give rise to short footprints in cfDNA. We therefore calculated scores for FOXM1 and FOXK2 binding from ChIP experiments conducted in GM12878 cells and found that FOXM1 ChIP scores, but not FOXK2 ChIP scores, strongly correlated with short length clusters in healthy plasma (fig. S4). This suggests that FOXM1 occupies sites in lymphoid/myeloid cells that are a subset of sites bound by FOXA1 in MCF7 cells.
With these collections of sites that were unique to cancer and to the ER status (normal ER–MCF7 versus amplified ER–UCD65 versus mutated ER–UCD4), we calculated a plasma TF binding score, which was the number of short reads (<80 bp) mapped within 50 bp of the TFBS normalized by the number of reads in 1000 bp around the TFBS. This plasma TF score tracks with the identity of the sites in that the sites unique to healthy plasma had a significantly higher TF score for healthy plasma compared to xenograft models and vice versa. Similarly, UCD65, MCF7, and UCD4 had higher plasma TF scores at their respective specific sites when compared to others (Fig. 6, B and D to F). Thus, unique sets of sites identified using cfDNA length clusters also had localized enrichment of short fragments relative to the surrounding 1000 bp in a system-specific manner. Genes adjacent to ER binding sites that were uniquely protected in UCD4 and UCD65 also featured significantly higher expression in the respective tumor cells relative to MCF7 (Fig. 6G). This indicates that the binding sites protected uniquely in UCD4 and UCD65 not only represent cfDNA signatures to differentiate these different disease states but also reflect their altered gene regulation. In summary, our analysis shows the potential of cfDNA length clusters to identify not only the tissue of origin but also the breast cancer disease state.
In the plasma of an individual with cancer, both lymphoid/myeloid cells and tumor cells will contribute to cfDNA, with most of the contribution still being from the lymphoid/myeloid cells. To ask at what dilution of tumor DNA we could detect the presence of cancer using TF footprints, we performed in silico dilutions of xenograft plasma cfDNA, which represents pure tumor DNA, mixed into healthy plasma cfDNA at 0, 0.5, 1, 2, 3, 4, and 5%. We then calculated plasma TF binding score at sites specific to healthy plasma and xenograft plasma. We compared these scores between the in silico diluted plasma samples and nondiluted plasma sample to calculate a paired t statistic. We set a cutoff of 5 for the median paired t statistic to indicate a significant difference between diluted and nondiluted plasma sample (fig. S5). We found ER sites to be strongest in separating tumor diluted cfDNA from pure healthy cfDNA (detection at <1% tumor cfDNA) followed by FOXA1 and CTCF (detection at ~1% of tumor cfDNA; Fig. 6C). PU.1 (detection at 2% tumor cfDNA) and LYL1 had weaker but significant contributions (fig. S6). Combined ER and FOXA1 sites showed a median t statistic greater than 5 between 0.5 and 1% tumor fraction. Because most metastatic diseases have tumor-derived cfDNA fractions higher than 1% (46, 47), our analysis suggests that we would be able to delineate TF binding in metastatic tumors, despite the significant interference from cfDNA of lymphoid/myeloid origin.
UCD65 has ESR1 amplification and expresses much higher levels of ER. UCD4 has a mutated ER (activating D538G mutation) (40). The presence of ESR1 amplification results in better survival with endocrine treatment (39), whereas D538G mutation confers resistance to endocrine therapy. These two xenografts along with MCF7 represent three unique states of ER activity and corresponding disease progression. Thus, this system enables to ask whether cfDNA TF fingerprints can provide clinically actionable information. So, we asked whether we could use cfDNA TF fingerprints to differentiate these three xenograft models. Both ER and FOXA1 sites contribute to differentiating UCD65 from MCF7. Combining sites from both TFs is synergistic and separates UCD65 and MCF7 at 4% of tumor fraction (t statistic > 5; Fig. 6H). Thus, at marginally higher tumor fractions, we can even identify signatures of differences in ER expression levels using TFBS defined by a combination of CUT&RUN and cfDNA length clustering. Notably, ER sites could robustly differentiate UCD4 from UCD65 and MCF7 (Fig. 6, I and J), highlighting the fact that mutated ER leads to differential binding signature that can be identified in plasma cfDNA at 2% tumor fraction. Significantly, FOXA1 sites were much weaker than ER in differentiating UCD4 from UCD65 and MCF7, highlighting that the mutation-specific changes in TF footprints in plasma are strongest for ER. In summary, by identifying the subset of high-resolution TFBS protected in distinct plasma samples, we can define TF signatures unique to ER+ breast cancer and further unique to amplified wild-type ER and D538G ER mutant.
Identified TFBS report on tumor TF binding in individuals with breast cancer
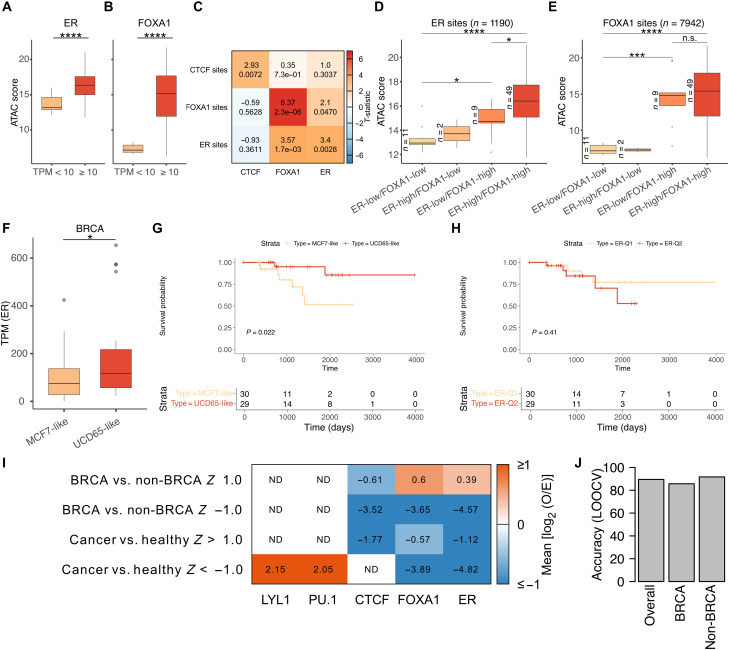
Because our in silico dilution analyses indicate that TF footprints in plasma can identify breast cancer disease at tumor-derived cfDNA fractions of 1 to 4%, we next asked whether the TFBSs we identified to be uniquely protected in xenograft model plasma would reflect disease states in heterogeneous human samples. To test this, we first turned to ATAC-seq datasets generated using primary tumor samples in the TCGA database. ATAC-seq reports DNA accessibility, which highly correlates with TF binding (48). We asked whether tumors exhibited TF-specific accessibility at the TFBSs that we had identified. We separated BRCA based on a specific TF’s expression and then calculated accessibility at sites identified to be UCD65 specific. BRCA with ER expression [transcripts per million (TPM) ≥ 10] were enriched for nonbasal, non-normal–like tumors and vice versa (fig. S7). We found that tumors that express ER had much higher accessibility at UCD65-specific ER sites compared to tumors that do not express ER (TPM < 10; Fig. 7A). Similarly, BRCA with FOXA1 expression were enriched for nonbasal, non-normal–like tumors and vice versa (fig. S7). We found even stronger accessibility differences at UCD65-specific FOXA1 binding sites, with FOXA1-expressing tumors having much higher ATAC scores than FOXA1-nonexpressing tumors at most sites (Fig. 7B).
Fig. 7. Plasma footprints represent TF-specific accessibility in primary tumors and can predict presence of breast cancer.
(A) Boxplot of ATAC scores from TCGA BRCA datasets stratified based on ER expression levels (ER low: TPM < 10; ER high: TPM ≥ 10) at cfDNA-inferred ER CUT&RUN peaks with ER motif. (B) Same as (A) for FOXA1 sites. (C) Heatmap of t statistic calculated at cfDNA-inferred TFBSs (rows) between top 15 and bottom 15 tumors when stratified by a TF’s expression (columns). (D) Boxplot of mean ATAC scores at ER sites for tumors stratified by both ER and FOXA1 expression (FOXA1 low: TPM < 10; FOXA1 high: TPM ≥ 10; ER low: TPM < 10; ER high: TPM ≥ 10). (E) Boxplot of mean ATAC scores at FOXA1 sites for tumors stratified by both ER and FOXA1 expression. Tumor stratification was identical to (D). (F) Boxplot of ER expression in TCGA breast cancer cohorts (excluding basal) segregated into “MCF7-like” and “UCD65-like” based on ATAC scores at cfDNA-inferred MCF7- and UCD65-specific sites. (G) Survival probability curves for TCGA breast cancer cohorts (excluding basal) segregated into MCF7- and UCD65-like. (H) Survival probability curves for TCGA breast cancer cohorts (excluding basal) segregated based on ESR1 expression into two quantiles. (I) Heatmap of enrichment [log2 (observed/expected)] of frequency of TF features selected for a given classification (rows) divided by the overall frequency of TF features. (J) Prediction accuracy of classifying patients to BRCA (breast cancer) and non-BRCA (non–breast cancer) using TF scores from plasma cfDNA using leave-one-out cross-validation (LOOCV). n.s., not significant; ND, not determined. *P < 0.05, ***P < 0.001, and ****P < 0.0001.
FOXA1 is known to act as a pioneer factor, enabling ER binding by establishing accessibility at its binding sites (15). We asked whether we could reproduce this finding at ER and FOXA1 binding sites that we identified by taking advantage of the heterogeneity in ER and FOXA1 expression across TCGA samples. If the ER and FOXA1 sites we identified are representative of ER and FOXA1 function across human breast tumors, then accessibility at ER binding sites should depend on the presence of FOXA1. CTCF is a good control as its expression should not influence accessibility at ER or FOXA1 sites. We first calculated the mean ATAC score for each tumor sample by aggregating the ATAC score across all sites of a given TF. For CTCF, ER, and FOXA1 sites, we performed two-sample t test [sample 1: cohorts with high TF expression (top 15 of 74), sample 2: cohorts with low TF expression (bottom 15 of 74)]. We found that the mean ATAC scores at CTCF, FOXA1, and ER sites were significantly different when tumors were grouped by the expression of the respective TF, with strongest difference seen for FOXA1 (diagonal cells in Fig. 7C). Notably, we observed a strong difference (t statistic = 3.57; P = 1.7 × 10−3) in mean ATAC scores at ER sites when tumors were grouped on the basis of FOXA1 expression. This difference was stronger than at FOXA1 sites when tumors were grouped on the basis of ER expression (t statistic = 2.1; P = 0.047), suggesting that FOXA1 expression has a stronger influence on accessibility at ER sites than vice versa.
To further explore the effect of FOXA1 at ER sites, we stratified BRCA by both ER and FOXA1 expression levels. In tumors with low ER expression, increase in FOXA1 expression led to a significant increase in mean ATAC scores at ER sites, suggesting that FOXA1 keeps the chromatin open at ER sites even in the absence of ER (Fig. 7D). Expression of ER and FOXA1 led to the highest accessibility at ER sites, suggesting further chromatin opening after ER binding (Fig. 7D). In stark contrast, at FOXA1 sites, accessibility increase is seen only due to increase in FOXA1 expression. The presence of ER did not lead to a significant increase in accessibility (Fig. 7E). Our observation of FOXA1 expression driving accessibility at both ER and FOXA1 binding sites recapitulates in human tumors the in vitro findings that FOXA1 is a pioneer factor that opens up ER sites. Together, our analysis shows that sites with tumor-specific plasma protections in xenograft models can define TF-specific accessibility across human breast tumors. These results indicate that TF protections in plasma can define tumor TF binding in humans.
Because UCD65 and MCF7 represent distinct ER states, we asked whether we could use the UCD65- and MCF7-specific sites derived from our cfDNA analysis to classify primary BRCA tumors from TCGA based on their ATAC scores at these sites. If the unique ER sites reflect a disease state, our classification should separate TCGA samples based on ER expression and survival: UCD65 has higher ER expression because of ESR1 amplification, and patients with ESR1-amplified tumors have higher survival. We devised a score based on ATAC-seq enrichment at UCD65- and MCF7-specific sites that classified each TCGA tumor as UCD65- or MCF7-like. Thus, this classification relied only on genome-wide ER binding profile. We then compared the ER expression of UCD65-like tumors to MCF7-like tumors. We observed higher ER expression in UCD65-like tumors, demonstrating that genome-wide ER signature reflects disease state (Fig. 7F). Notably, we also observed significantly higher survival in patients with UCD65-like tumors compared to MCF7-like tumors, recapitulating another aspect of ER-amplified tumors, just with genome-wide ER binding profiles (Fig. 7G). Just separating tumors into two groups based on ER expression alone does not show any significant differences in survival, highlighting the fact that it is the genome-wide ER fingerprint that contains predictive value for ER function (Fig. 7H). In summary, TF protections in plasma reflect ER+ disease state and can be used as prognostic indicators.
Last, we asked whether TF binding scores from plasma cfDNA can distinguish cancer from healthy states and breast cancer from other cancers and healthy states. We compared TF binding scores in 19 human plasma cfDNA sequencing datasets [healthy = 4 (2 male and 2 female), lung cancer = 6 (5 male and 1 female), colorectal cancer = 1 (female), total non-BRCA = 11, and breast cancer (BRCA) = 8 (8 female); table S1]. To take advantage of even samples that were sequenced at low depths, we defined TF features as aggregates of 250 binding sites of the TF after ordering all its binding sites by ChIP/CUT&RUN score. We ended up with a total of 359 features (PU.1 = 43, LYL1 = 7, CTCF = 120, ER = 124, and FOXA1 = 65). We made two classification groups: cancer versus healthy (n = 15 and n = 4, respectively) and BRCA versus non-BRCA (n = 7 and n = 12, respectively). We calculated the Z score for each feature for these two groups of classification. We then filtered for those features with |Z| > 1 in each of the two classifications as features that differentiated the two classes in each classification. We then asked which of the TFs had their features overrepresented or underrepresented in each classification. We found PU.1 features to be over-represented in having higher TF binding scores in healthy samples compared to cancer samples (Fig. 7I). In classifying BRCA and non-BRCA, we found no TFs to be overrepresented in features that had higher binding scores in non-BRCA. However, ER and FOXA1 features were overrepresented with higher binding scores in BRCA compared to non-BRCA (Fig. 7I). The fact that FOXA1 and ER binding sites can separate BRCA from non-BRCA indicates that the sites identified from xenografts are transferrable to human samples. Furthermore, despite dilution by cfDNA from lymphoid and myeloid cells, cancer-specific TF protections in plasma are sensitive markers of disease presence. To ask how accurate these features are in identifying the presence of breast cancer, we resorted to leave-one-out cross-validation. We identified features that significantly separated BRCA from non-BRCA using all but one of the samples (18 of 19) and then used these features to predict the status of the left-out sample. We observed an overall prediction accuracy of 89.5%, prediction accuracy of 85.7% for BRCA (6 of 7 predicted correctly), and accuracy of 91.7% for non-BRCA (11 of 12 predicted correctly; Fig. 7J). Notably, these 19 samples have diverse sequencing depth ranging from ~41 million to ~600 million sequenced cfDNA fragments (table S2). Thus, our analysis of datasets consisting of low to intermediate depth sequencing of 19 human plasma samples shows potential for plasma TF footprints to identify breast cancer tissue of origin.
DISCUSSION
Interaction of TFs with DNA is fundamental to gene regulation, and distinct cell types are defined by unique TF binding profiles. It is known that cfDNA fragments in plasma maintain information regarding chromatin dynamics and TF binding (49). Previous approaches have averaged the coverage of short and long cfDNA fragments from thousands of sites to infer binding of a single TF (17, 26). These analyses on aggregated sites can be used to build diagnostic classifiers but lack the granularity to generate binding profiles of TFs specific to the cells releasing cfDNA. We hypothesized that if TF protections in tissue of origin lead to short footprints in plasma, then mapping these footprints at individual sites would allow us to study the TF’s function in cfDNA tissues of origin. To this end, we first defined the subset of all possible sites of a TF that give rise to short footprints in cfDNA. Unexpectedly, we found that enrichment of short footprints in plasma for a TFBS correlates with strength with which the TF binds that site in the cells from which the cfDNA originated. We observed this for both constitutive factors with long residence times, such as CTCF, and tumor-specific dynamic factors with short residence times, such as ER. This finding elevates cfDNA from a mere classifier to a means to understand TF binding in living mammals in a minimally invasive manner and in real time.
TFs in the same family use overlapping binding sites in different cell types. For example, FOXA1, which is active in hormone-dependent cancers of the breast and prostate, shares binding motif with other FOX factors like FOXM1, whose expression is enhanced in lymphoid tissues (45), and FOXK2, which is expressed in many tissues including lymphoid/myeloid (50). We show that a subset of FOXA1 sites bound in ER+ tumors also give rise to short cfDNA footprints in healthy plasma. We can predict these protections to arise from FOXM1 rather than FOXK2 binding based on correlation of the enrichment of cfDNA TF footprints with binding strengths of FOXM1 in a lymphoid cell line, underscoring our ability to uncover specific TF binding profiles directly from plasma. Similarly, ER, which drives ER+ breast cancers, is also active in T cells (44), with shared and unique binding sites between tumors and T cells. Our inference of TF footprints at each binding site of ER from xenograft models and healthy plasma has enabled identification of sites in an ER+ tumor also bound by ER in lymphoid/myeloid cells. This careful characterization of ER and FOXA1 has enabled us to identify binding sites of FOXA1 and ER that are not only specific to ER+ breast cancer but also able to distinguish basal levels of ER from ER overexpression and the presence of mutated ER. Starting only with a reference set of CUT&RUN sites from E2-treated MCF7 cells, we have been able to identify ER and FOXA1 binding sites that defined lymphoid/myeloid signatures and ER+ tumor subtypes. Thus, further high-resolution characterization of TFs in the future can only improve our ability to generate binding profiles from plasma in health and disease.
Our results uncover two aspects of cfDNA biology that can vastly expand the information we can gain from its study. First, enrichment of short fragments enables accurate identification of TF footprints at single binding sites in plasma. Second, careful consideration of which sites in the genome to look at can yield cancer-sensitive signatures that enable tissue-of-origin mapping of TF binding profiles. Here, we have shown that analysis of CUT&RUN data on tumor cells combined with cfDNA data from xenograft models can identify tumor TF sites that are bound in a TF-specific manner across human breast tumors. The analytical framework presented in this study is not limited to TFs. It can easily be customized to the question of interest by defining open/closed chromatin using datasets generated by ATAC-seq and DNase I–seq (deoxyribonuclease I sequencing). In addition, regions of interest across organisms could be used by imputation. Depending on the disease biology, probing for gain or loss of TF binding can be of potential use. For example, in the case of acute myeloid leukemia, loss of PU.1 expression could lead to overall loss in binding (51). In the future, putative tumor state–specific TFs, their gene targets, and binding sites where we can analyze cfDNA footprints could be identified by mining TCGA ATAC-seq and RNA-seq (RNA sequencing) datasets (52, 53) and by de novo identification of TFBSs from models of pure tumor cfDNA. Given that most tumors release cfDNA, we believe that our characterization of ER+ breast tumors using cfDNA TF footprints represents the tip of the iceberg for characterizing tumor phenotypes from plasma and is applicable across disease states.
MATERIALS AND METHODS
Plasma samples
The plasma sample information is described in table S1.
ChIP-seq peaks
We collected ChIP-peaks from publicly available datasets (9, 54, 55). We obtained clustered peaks for CTCF and PU.1 from ENCODE (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegTfbsClustered/wgEncodeRegTfbsClusteredV3.bed.gz). For LYL1, we used peaks from ReMap (http://remap.univ-amu.fr/storage/remap2020/hg38/MACS2/TF/LYL1/remap2020_LYL1_all_macs2_hg38_v1_0.bed.gz).
TF motifs
We used TF motifs from JASPAR (CTCF: http://jaspar.genereg.net/matrix/MA0139.1/; PU.1: http://jaspar.genereg.net/matrix/MA0080.5; ER: http://jaspar.genereg.net/matrix/MA0112.1, http://jaspar.genereg.net/matrix/MA0112.2, and http://jaspar.genereg.net/matrix/MA0112.3; FOXA1: http://jaspar.genereg.net/matrix/MA0148.1, http://jaspar.genereg.net/matrix/MA0148.2, and http://jaspar.genereg.net/matrix/MA0148.3) (56) and HOCOMOCO (LYL1: http://hocomoco.autosome.ru/motif/LYL1_HUMAN.H11MO.0.A) (57).
Genome-wide signal
We used publicly available genome-wide signal files in bigwig format to map ChIP and MNase signal to TFBSs and their flanks. CTCF: www.encodeproject.org/files/ENCFF578TBN/@@download/ENCFF578TBN.bigWig, PU.1: www.encodeproject.org/files/ENCFF324NQZ/@@download/ENCFF324NQZ.bigWig, LYL1: GEO: GSE63484.
Xenograft models
All animal experiments were conducted in an AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care International)–accredited facility at the University of Colorado Denver under an Institutional Animal Care and Use Committee–approved protocol. Xenograft tumors for MCF7 and UCD4 cells were established by mixing 1 × 106 cells in Cultrex and injecting them into the mammary fat pad of nonobese diabetic severe combined immunodeficient gamma (NSG) female mice. Xenograft tumors for UCD65 were established by mixing 3 × 106 cells in Cultrex and by injecting them into the mammary fat pad of NSG female mice. All tumors received continuous estrogen supplementation throughout the study, as previously described (38). Tumor measurements were taken weekly throughout the duration of the experiment. Tumor burden was measured as the combined volume of the right and left tumor in each animal. Total tumor burden for MCF7 was 811, 1109, and 2534 mm3. Total tumor burden for UCD65 was 1741, 1805, 2072, 2332, and 2897 mm3. Total tumor burden for UCD4 was 2374, 2509, 2999, and 3014 mm3. Cardiac exsanguination was immediately used following euthanasia by carbon dioxide (using a 50% displacement flow rate). Whole blood was drawn into a 25-gauge syringe primed with anticoagulant acid citrate dextrose (ACD) solution A from BD yellow top ACD tubes. To prevent red blood cell lysis, the syringe needle was carefully removed from the syringe and the whole blood was carefully ejected into 1.5-ml microcentrifuge tubes containing 10 to 20% ACD solution A. The tubes were inverted 10 times to mix well. Whole blood from mice was processed to capture the cfDNA-rich plasma fraction using a series of centrifugation steps. The steps included two spins at 1700g for 10 min and once at 14,000g for 10 min. All centrifugation steps were performed at room temperature, and the top clearer fraction was carefully pipetted and put into a clean tube for the next centrifugation step. Samples were either immediately processed for cfDNA extraction or stored at −80°C until cfDNA extraction could be accomplished.
Human cfDNA datasets
We obtained deidentified human plasma samples from repositories listed in table S1. Each repository obtained samples with informed consent from volunteers following approved Institutional Review Board protocols.
cfDNA extraction
Human plasma (1 to 4 ml) or mouse plasma (0.2 to 0.5 ml) was thawed from −80°C storage. Plasma samples were spun at maximum speed [21,000 rcf (relative centrifugal force)] at 4°C for 5 to 10 min to pellet any cell debris. Supernatant was transferred to new tubes, and cfDNA was extracted using the QIAGEN ccfMinElute kit (catalog no. 55204), eluted in 30 μl of nuclease-free water, and directly added to the SSP or stored at −20°C.
Single-stranded DNA library protocol
The capture of cfDNA fragments from plasma was performed similar to Snyder et al. (17). Briefly, 1 to 10 ng of cfDNA were dephosphorylated using FastAP Thermosensitive Alkaline Phosphatase (Thermo Fisher Scientific, catalog no. EF0651), denatured, and incubated overnight with CircLigase II (Lucigen, catalog no. CL9025K) and 0.093 to 0.125 μM biotinylated CL78 primer (17) at 60°C with shaking every 5 min. Captured cfDNA fragments were denatured and then bound to magnetic streptavidin M-280 beads (Invitrogen, catalog no. 11205D) for 30 min at room temperature with nutation. Beads were washed, and second-strand synthesis was performed using Bst 2.0 DNA polymerase (New England Biolabs, catalog no. M0537) with an increasing temperature gradient of 15° to 31°C with shaking at 1750 rpm. Beads were washed, and a 3′ gap fill was performed using T4 DNA polymerase (Thermo Fisher Scientific, catalog no. EL0011) for 30 min at room temperature. Beads were washed, and a double-stranded adapter was ligated using T4 DNA ligase (Thermo Fisher Scientific, catalog no. EP0062) for 2 hours at room temperature with shaking at 1750 rpm. Beads were washed and resuspended in 30 μl of 10 mM TET buffer [10 mM tris-HCl (pH 8.0), 1 mM EDTA (pH 8.0), and 0.05% Tween 20]. DNA was denatured at 95°C for 3 min, and cfDNA libraries were collected after immediate magnetic separation.
Quantitative real-time polymerase chain reaction (PCR) was performed on cfDNA libraries using iTAQ Supermix (Bio-Rad, catalog no. 1725124), and Ct values were used to determine the number of PCR cycles needed to amplify each library. PCR was performed with KAPA HiFi DNA polymerase (Kapa Biosystems, catalog no. KK2502) using barcoded indexing primers for Illumina. Primer dimers were removed from the libraries using AMPure beads (Beckman Coulter, catalog no. A63881). Libraries were eluted in 0.1× TE (Tris EDTA buffer), and concentrations were determined using Qubit. The length distribution of each library was assessed by the Agilent Bioanalyzer using the D1000 or HSD1000 cassette. Libraries were sequenced for 150 cycles in paired-end mode on NovaSeq 6000 system at University of Colorado Cancer Center Genomics Shared Resource.
Cleavage under target and release using nuclease
We used an immunotethered strategy for profiling the binding of the ERα and FOXA1 TF in human MCF7 breast cancer cells. MCF7 cells were estrogen-withdrawn for 72 hours before being plated and then treated with either ethanol (vehicle control) or 10−10 M E2 for 1 hour before cell collection. The CUT&RUN method uses an antibody to a specific chromatin epitope to tether protein A–MNase at chromosomal binding sites within permeabilized cells. The nuclease is activated by the addition of calcium and cleaves DNA around binding sites (10). Cleaved DNA is isolated and subjected to paired-end Illumina sequencing to map the distribution of the chromatin epitope genome-wide. We used a primary antibody to human ERα (ab3575, Abcam, Cambridge, MA) and human FOXA1 (ab170933) and protein A–MNase fusion (pA-MNase, a gift from S. Henikoff, Fred Hutchinson Cancer Research Center, Seattle WA) (10). CUT&RUN profiling with 5 × 105 cells and library amplification with 13 cycles of PCR were performed as described (10). Libraries were sequenced for 10 million paired-end reads on the Illumina NovaSeq 6000 platform at the University of Colorado Denver Cancer Center Genomics Shared Resource. Paired-end reads were mapped to the GRch38 assembly of the human genome using Bowtie2 (58).
CUT&RUN peaks
To call peaks, we used custom Python script (deposited in GitHub). Briefly, we first normalized a coverage of <120-bp protected fragments in CUT&RUN data at 10-bp resolution and then smoothed the coverage with a Savitzky-Golay filter (59) available as a SciPy (60) method “signal.savgol_filter” with parameters window_length = 9, polyorder = 1. We determined the cutoff for each dataset by iteratively eliminating outliers and used the “find_peaks” method in SciPy to call peaks that were separated by at least 250 bp.
Aligning mouse-extracted cfDNA to in silico concatenated genome
The names of chromosomes of human (hg38; GRCh38 assembly) and mouse (mm10; GRCm38 assembly) reference genomes were first prefixed by hg38 and mm10, respectively, and then the fasta files were concatenated together to represent an in silico human + mouse genome. We then aligned xenograft cfDNA to this concatenated genome using Bowtie2 (58) with parameters “--local --very-sensitive-local --no-unal --no-mixed --no-discordant -I 10 -X 700” (alignment report of both xenograft and human plasma samples can found in table S2). We selected for mapped reads and then filtered out reads with secondary alignment from the bam file using the command: “samtools view -F 4 <bam file> | grep -v ´XS:´” (61).
This filtering ensured that we did not consider any reads that aligned to both human and mouse genomes. To get human aligned reads, we filtered for the hg38 prefix in the reads’ chromosome name.
Defining TFBSs under ChIP-seq peaks
We first selected for ChIP-seq peaks that do not overlap with ENCODE blacklist regions, and we considered all peaks except the ones on chromosome Y. We then used FIMO (Find Individual Motif Occurrences) (62) with parameters “--max-stored-scores 10000000 --oc <output-directory> <motif-file> <fasta-file>” to scan for motifs on sequences underlying ChIP-seq peaks. In case of overlapping peaks in 50-bp span, we keep the motif with higher FIMO score. The final number of motifs under ChIP-peaks used for TFs is tabulated in table S3.
cfDNA length distribution clustering
Length distribution of mapped cfDNA fragments to a TFBS is estimated by “density” function in R with a smoothing bandwidth of 3 at 100 equally spaced points (n = 100) between 35 and 250 bp. Clustering of estimated cfDNA length distribution at individual sites was performed using “kmeans” function in R with the following parameters: centers = 6, iter.max = 250, and nstart = 20. A cluster is visually represented by the mean of fragment length distributions of sites in that cluster. Weighted length of each cluster was calculated by multiplying fragment length to its normalized frequency. Clusters 1 to 6 were assigned by ranking the clusters by their weighted length.
Mapping cfDNA length class to TFBS and its flank
Genome-wide cfDNA read density (bigwig) was generated for short (<80 bp) and nucleosomal-sized fragments (130 to 180 bp). First, a bedgraph (coverage of bases genome-wide; no normalization performed) file was generated using bedtools (63) genomecov utility with the command line option “-bga,” and then the bedgraph file was converted to bigwig using kent tools “bedGraphToBigWig” (64). While creating the bigwig file, we considered the cfDNA fragment center ± 30 bp (if fragment is >60 bp). Bigwig is mapped to TFBS ± 1 kb using pyBigWig module from deeptools (65), and then enrichment over mean (E.O.M.) is calculated. E.O.M. is smoothed using the Savitzky-Golay filter (59) available as a SciPy (60) method “signal.savgol_filter” with parameters window_length = 51 and polyorder = 3.
ChIP-seq score calculation sites in cfDNA length clusters
For a TFBS in a given cluster, log2 of mean fold enrichment over control was calculated for TFBS ± 300 bp. pyBigWig module from deeptools (65) was used to map signal from bigwig file to defined genomic regions.
MNase signal mapping to CTCF sites
MNase data from ENCODE (9) were mapped to CTCF motif center ± 1 kb. E.O.M. and smoothing were performed similar to how it was done for cfDNA length class heatmaps (see the “Mapping cfDNA length class to TFBS and its flank” section).
V plots
For CTCF sites in cfDNA length clusters 1 and 2, cfDNA fragment centers were mapped to the CTCF motif center ± 500 bp. The total number of cfDNA centers of a given length is plotted against the distance of the fragment centers from the CTCF motif center.
CUT&RUN score calculation
CUT&RUN score has been calculated as the read density in regions spanning CUT&RUN peak summit ± 50 bp.
Defining significant sites and specific sites
cfDNA length clusters that have significantly higher binding scores (ChIP scores for CTCF, PU.1, and LYL1 and CUT&RUN scores for ER and FOXA1) compared to cluster 6 are considered significant, i.e., overall, sites in these clusters have stronger binding strength inferred from TF binding experiments compared to cluster 6. Specific sites are identified by subtracting significant sites of one sample from significant sites from another sample. In the case of disease state detection analysis, i.e., healthy versus cancer, cancer-specific sites (CSSs) and healthy-specific sites (HSSs) were defined. CSSs for ER, for example, are defined by subtracting sites in healthy plasma (IH02) (17) significant clusters 1 and 2 from UCD65 clusters 1 to 4. Similarly, HSSs for ER are defined by subtracting sites from UCD65 clusters 1 to 4 from IH02 clusters 1 and 2. In the case of cancer state detection analysis, i.e., separating tumor subtypes (UCD65 versus MCF7, UCD4 versus UCD65, and UCD4 versus MCF7) using tumor TFBSs, tumor-specific sites were defined by a similar approach. We did not observe enrichment at FOXA1 binding sites in UCD4 dataset; hence, tumor-specific sites were not defined for FOXA1 in UCD4.
RNA-seq analysis
We used published RNA-seq datasets (40). We used Salmon (66) with hg38 release 95 transcripts. We first generated the transcriptome indices with the command “salmon index -t Homo_sapiens.GRCh38.cdna.all.fa -i salmon_index.” We then generated TPM table for MCF7 24 hours and UCD65 using the following command: salmon quant -i salmon_index/ -p 8 -l IU -r <sample>.fastq.gz -o quants/<sample>. For UCD4 (paired-end RNA-seq data), we used the following command: “salmon quant -i salmon_index/ -p 8 -l IU -1 ucd4.fq1.trimmred.fastq.gz -2 ucd4.trimmed.fastq.gz -o quants/ucd4.”
Dilution analysis
Disease detection
In silico patient data were generated by diluting healthy sample (IH02) (17) with different fractions of UCD65 cfDNA. For each dilution level, 100 in silico patient datasets were generated by randomly sampling reads from IH02 and UCD65 datasets at the ratio defined by the dilution level. For a given cancer/healthy-specific binding site, the TF binding score was calculated as the ratio of the short fragment coverage in (<80 bp) TFBS ± 50 to the coverage in TFBS ± 1 kb. Reference TF binding score is calculated just in healthy state, and for each in silico patient dataset, scores are calculated in the same fashion. ΔScore (used in Fig. 6C) for CSSs was calculated as the difference between patient and healthy states (gain in score), but for HSSs, the sign was reversed (loss in score). T test was performed on ΔScore values from all sites (healthy-specific + cancer-specific) to reflect how many SDs away the scores are from the healthy reference.
Cancer state detection
For each xenograft (UCD4, UCD65, and MCF7) model, 100 in silico patient data were generated by diluting healthy plasma (IH02) with different fractions of ctDNA. For each of three comparisons of xenograft models, the following were calculated (using UCD65 versus MCF7 as an example): (i) TF binding scores at tumor subtype–specific sites using UCD65 and MCF7 in silico patient data, respectively, and (ii) calculated ΔScore for UCD65-specific sites by subtracting scores of MCF7 dilution from UCD65 dilution. Similarly, ΔScore values for MCF7-specific sites were calculated by subtracting scores of UCD65 dilution from MCF7 dilution, and (iii) calculated t statistics on ΔScore using “ttest_1samp” function from scipy.stats module (60) with expected value in null hypothesis = 0.
TCGA ATAC-seq and expression analysis
FPKM (Fragments Per Kilo Base per Million mapped fragments) files for each cohort were downloaded from the TCGA website. FPKM for a gene was converted to TPM using the following formula
where N is the total number of genes found in the FPKM table.
ATAC insert bigwig files from Corces et al. (48) were used to map ATAC signal around TF sites (peak ± 150 bp).
Classifying TCGA BRCA tumors as MCF7- or UCD65-like
From cfDNA-inferred MCF7- and UCD65-specific ER CUT&RUN sites (Fig. 6A, ER UpSet plot), we selected the subset of sites with ERE (Estrogen Response Element). We found a total of 1603 EREs specific to MCF7 and 2320 for UCD65. For each TCGA BRCA sample, we first calculated the average ATAC enrichment score (mean signal in ERE ± 150 divided by the mean signal in ER ± 1 kb) for MCF7- and UCD65-specific sites. We then Z-scaled MCF7- and UCD65-specific enrichment scores separately across cohorts and subtracted MCF7-specific Z scores from the UCD65-specific scores to define a ΔZ score for each cohort. A negative ΔZ score indicates MCF7-like tumor, whereas a positive ΔZ score indicates a UCD65-like tumor based on accessibility at ER binding sites.
Survival analysis
We collected “days_to_last_follow_up” and “days_to_death” as well as “vital_staus” data from TCGA for 59 breast cancer cohorts with ATAC-seq data and who had PAM50 (Prediction Analysis of Microarray 50) classification other than “Basal.” We assigned a label of “1” for patients with “Alive” status, and “2” for patients with “Died” status. We replaced the days_to_last_follow_up value with days_to_death for Died status. For survival analysis based on ESR1 expression, we formed two groups based on median ESR1 expression (88.256).
Cancer versus healthy and breast cancer versus non–breast cancer prediction analysis
HSSs and CSSs were ordered by their binding strength inferred from ChIP (motif center ± 300 bp; for PU.1, LYL1, and CTCF) or CUT&RUN (summit ± 100 bp; for ER and FOXA1) and grouped in a bin of size 250 to define TF features. cfDNA-inferred binding score at TF features is defined by the following formula
To identify what TF features are class specific (for example, class1 – cancer and class2 – healthy), we defined a Z-score metric using the following formula
where SD stands for standard deviation. Features with ∣Zfeature∣ > 1 were selected and, depending on the sign, were annotated as class1 specific (+ve) or class2 specific (−ve). Enrichment of a TF in particular category (for example, healthy specific) was calculated by abundance of the TF features as log2 (observed frequency/expected frequency).
To predict a class (breast cancer or non–breast cancer) for a cfDNA sample, the leave-one-out cross-validation approach was adopted, where the cfDNA sample of our interest was kept away during the feature selection process described above. Each sample was then assigned a single score by subtracting the sum of binding scores of features with negative Z scores (Zfeature < −1) from the mean of features with positive Z scores (Zfeature > 1) and then dividing by the total number of features (∣Zfeature∣ > 1). For the left-out sample, distances from the median of two classes were calculated and assigned the class label with closest distance.
Acknowledgments
The results shown here are, in part, based on data generated by the TCGA Research Network: www.cancer.gov/tcga. We thank O. Rissland and S. Jagannathan for critical comments on the manuscript.
Funding: This work was supported by the RNA Bioscience Initiative, University of Colorado School of Medicine, ACS IRG #16-184-56 from the American Cancer Society (to S. Ramachandran); Earlier.org—Friends for an Earlier Breast Cancer Test (to S. Ramachandran); National Cancer Institute grants R01CA140985 (to C.A.S.) and R01CA205044 (to P.K.); the Breast Cancer Research Foundation 16-072 (to C.A.S.); the University of Colorado Cancer Center’s Oncology Research Information Exchange Network, University of Colorado Cancer Center Pathology Shared Resource [this resource is supported by the Cancer Center Support Grant (P30CA046934)]; and Colorado Lung Cancer Specialized Program of Research Excellence (P50 CA058187). S. Ramachandran is a Pew-Stewart Scholar for Cancer Research, supported by the Pew Charitable Trusts and the Alexander and Margaret Stewart Trust. A.Z. was supported by an American Cancer Society–Virginia Cochary Award for Excellence in Breast Cancer Research Postdoctoral Fellowship, PF-20-095-01-DMC.
Author contributions: S. Rao, S. Ramachandran, and P.K. conceived and designed the project and wrote the manuscript. E.K. and A.Z. generated cfDNA sequencing datasets. A.L.H. performed the CUT&RUN experiments. S. Rao performed all computational analyses. C.A.S. advised on clinical models. All authors approved the final manuscript.
Competing interests: P.K., S. Ramachandran, S. Rao, A.Z., and A.L.H. are listed as co-inventors on a patent application related to this work (International Publication Number WO2022061080A1). The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw and processed sequencing datasets generated in this study have been deposited in Gene Expression Omnibus under the accession GSE171434. All scripts and pipelines used in this study are available at Zenodo (https://doi.org/10.5281/zenodo.6587679). The xenograft models can be provided by P.K.’s pending scientific review and a completed material transfer agreement. Requests for the xenograft models should be submitted to P.K. (peter.kabos@cuanschutz.edu).
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 and S3
Other Supplementary Material for this manuscript includes the following:
Table S2
REFERENCES AND NOTES
- 1.Spitz F., Furlong E. E., Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Todeschini A. L., Georges A., Veitia R. A., Transcription factors: Specific DNA binding and specific gene regulation. Trends Genet. 30, 211–219 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Wunderlich Z., Mirny L. A., Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 25, 434–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jindal G. A., Farley E. K., Enhancer grammar in development, evolution, and disease: Dependencies and interplay. Dev. Cell 56, 575–587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacQuarrie K. L., Fong A. P., Morse R. H., Tapscott S. J., Genome-wide transcription factor binding: Beyond direct target regulation. Trends Genet. 27, 141–148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voss T. C., Hager G. L., Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 15, 69–81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramachandran S., Henikoff S., Transcriptional regulators compete with nucleosomes post-replication. Cell 165, 580–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee T. I., Young R. A., Transcriptional regulation and its misregulation in disease. Cell 152, 1237–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consortium E. P., An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skene P. J., Henikoff S., An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi M. J., Kuntala P. K., Lai W. K. M., Yamada N., Badjatia N., Mittal C., Kuzu G., Bocklund K., Farrell N. P., Blanda T. R., Mairose J. D., Basting A. V., Mistretta K. S., Rocco D. J., Perkinson E. S., Kellogg G. D., Mahony S., Pugh B. F., A high-resolution protein architecture of the budding yeast genome. Nature 592, 309–314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi D., Singhal H., Li L., Xiao T., Liu W., Pun M., Jeselsohn R., He H., Lim E., Vadhi R., Rao P., Long H., Garber J., Brown M., Estrogen receptor signaling is reprogrammed during breast tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 116, 11437–11443 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross-Innes C. S., Stark R., Teschendorff A. E., Holmes K. A., Ali H. R., Dunning M. J., Brown G. D., Gojis O., Ellis I. O., Green A. R., Ali S., Chin S. F., Palmieri C., Caldas C., Carroll J. S., Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan J., Zhou W., Hafner M., Blake R. A., Chalouni C., Chen I. P., De Bruyn T., Giltnane J. M., Hartman S. J., Heidersbach A., Houtman R., Ingalla E., Kategaya L., Kleinheinz T., Li J., Martin S. E., Modrusan Z., Nannini M., Oeh J., Ubhayakar S., Wang X., Wertz I. E., Young A., Yu M., Sampath D., Hager J. H., Friedman L. S., Daemen A., Metcalfe C., Therapeutic ligands antagonize estrogen receptor function by impairing its mobility. Cell 178, 949–963.e18 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Hurtado A., Holmes K. A., Ross-Innes C. S., Schmidt D., Carroll J. S., FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo Y. M. D., Chan K. C. A., Sun H., Chen E. Z., Jiang P., Lun F. M. F., Zheng Y. W., Leung T. Y., Lau T. K., Cantor C. R., Chiu R. W. K., Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2, 61ra91 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Snyder M. W., Kircher M., Hill A. J., Daza R. M., Shendure J., Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lui Y. Y., Chik K. W., Chiu R. W. K., Ho C. Y., Lam C. W. K., Lo Y. M. D., Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 48, 421–427 (2002). [PubMed] [Google Scholar]
- 19.Schwarzenbach H., Hoon D. S., Pantel K., Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Diehl F., Li M., Dressman D., He Y., Shen D., Szabo S., Diaz L. A. Jr., Goodman S. N., David K. A., Juhl H., Kinzler K. W., Vogelstein B., Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. U.S.A. 102, 16368–16373 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zviran A., Schulman R. C., Shah M., Hill S. T. K., Deochand S., Khamnei C. C., Maloney D., Patel K., Liao W., Widman A. J., Wong P., Callahan M. K., Ha G., Reed S., Rotem D., Frederick D., Sharova T., Miao B., Kim T., Gydush G., Rhoades J., Huang K. Y., Omans N. D., Bolan P. O., Lipsky A. H., Ang C., Malbari M., Spinelli C. F., Kazancioglu S., Runnels A. M., Fennessey S., Stolte C., Gaiti F., Inghirami G. G., Adalsteinsson V., Houck-Loomis B., Ishii J., Wolchok J. D., Boland G., Robine N., Altorki N. K., Landau D. A., Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 26, 1114–1124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M. C., Oxnard G. R., Klein E. A., Swanton C., Seiden M. V., Liu M. C., Oxnard G. R., Klein E. A., Smith D., Richards D., Yeatman T. J., Cohn A. L., Lapham R., Clement J., Parker A. S., Tummala M. K., McIntyre K., Sekeres M. A., Bryce A. H., Siegel R., Wang X., Cosgrove D. P., Abu-Rustum N. R., Trent J., Thiel D. D., Becerra C., Agrawal M., Garbo L. E., Giguere J. K., Michels R. M., Harris R. P., Richey S. L., McCarthy T. A., Waterhouse D. M., Couch F. J., Wilks S. T., Krie A. K., Balaraman R., Restrepo A., Meshad M. W., Rieger-Christ K., Sullivan T., Lee C. M., Greenwald D. R., Oh W., Tsao C. K., Fleshner N., Kennecke H. F., Khalil M. F., Spigel D. R., Manhas A. P., Ulrich B. K., Kovoor P. A., Stokoe C., Courtright J. G., Yimer H. A., Larson T. G., Swanton C., Seiden M. V., Cummings S. R., Absalan F., Alexander G., Allen B., Amini H., Aravanis A. M., Bagaria S., Bazargan L., Beausang J. F., Berman J., Betts C., Blocker A., Bredno J., Calef R., Cann G., Carter J., Chang C., Chawla H., Chen X., Chien T. C., Civello D., Davydov K., Demas V., Desai M., Dong Z., Fayzullina S., Fields A. P., Filippova D., Freese P., Fung E. T., Gnerre S., Gross S., Halks-Miller M., Hall M. P., Hartman A. R., Hou C., Hubbell E., Hunkapiller N., Jagadeesh K., Jamshidi A., Jiang R., Jung B., Kim T. H., Klausner R. D., Kurtzman K. N., Lee M., Lin W., Lipson J., Liu H., Liu Q., Lopatin M., Maddala T., Maher M. C., Melton C., Mich A., Nautiyal S., Newman J., Newman J., Nicula V., Nicolaou C., Nikolic O., Pan W., Patel S., Prins S. A., Rava R., Ronaghi N., Sakarya O., Satya R. V., Schellenberger J., Scott E., Sehnert A. J., Shaknovich R., Shanmugam A., Shashidhar K. C., Shen L., Shenoy A., Shojaee S., Singh P., Steffen K. K., Tang S., Toung J. M., Valouev A., Venn O., Williams R. T., Wu T., Xu H. H., Yakym C., Yang X., Yecies J., Yip A. S., Youngren J., Yue J., Zhang J., Zhang L., Zhang L. (Q.), Zhang N., Curtis C., Berry D. A., Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31, 745–759 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bronkhorst A. J., Ungerer V., Holdenrieder S., The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 17, 100087 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulz P., Thallinger G. G., Auer M., Graf R., Kashofer K., Jahn S. W., Abete L., Pristauz G., Petru E., Geigl J. B., Heitzer E., Speicher M. R., Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 48, 1273–1278 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Ramachandran S., Ahmad K., Henikoff S., Transcription and remodeling produce asymmetrically unwrapped nucleosomal intermediates. Mol. Cell 68, 1038–1053.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulz P., Perakis S., Zhou Q., Moser T., Belic J., Lazzeri I., Wölfler A., Zebisch A., Gerger A., Pristauz G., Petru E., White B., Roberts C. E. S., John J. S., Schimek M. G., Geigl J. B., Bauernhofer T., Sill H., Bock C., Heitzer E., Speicher M. R., Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat. Commun. 10, 4666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holwerda S. J., de Laat W., CTCF: The protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen A. S., Pustova I., Cattoglio C., Tjian R., Darzacq X., CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6, e25776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Y., Sinha M., Peterson C. L., Weng Z., The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLOS Genet. 4, e1000138 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henikoff J. G., Belsky J. A., Krassovsky K., MacAlpine D. M., Henikoff S., Epigenome characterization at single base-pair resolution. Proc. Natl. Acad. Sci. U.S.A. 108, 18318–18323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher R. C., Scott E. W., Role of PU.1 in hematopoiesis. Stem Cells 16, 25–37 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Chiu S. K., Saw J., Huang Y., Sonderegger S. E., Wong N. C., Powell D. R., Beck D., Pimanda J. E., Tremblay C. S., Curtis D. J., A novel role for Lyl1 in primitive erythropoiesis. Development 145, dev162990 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Zhu J., Emerson S. G., Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene 21, 3295–3313 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Barozzi I., Simonatto M., Bonifacio S., Yang L., Rohs R., Ghisletti S., Natoli G., Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol. Cell 54, 844–857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J. N., Pinello L., Yissachar E., Wischhusen J. W., Yuan G. C., Roberts C. W. M., Functionally distinct patterns of nucleosome remodeling at enhancers in glucocorticoid-treated acute lymphoblastic leukemia. Epigenetics Chromatin 8, 53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corces M. R., Buenrostro J. D., Wu B., Greenside P. G., Chan S. M., Koenig J. L., Snyder M. P., Pritchard J. K., Kundaje A., Greenleaf W. J., Majeti R., Chang H. Y., Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J., Anagnostou V., Fiksel J., Cristiano S., Papp E., Speir S., Reinert T., Orntoft M. B. W., Woodward B. D., Murphy D., Parpart-Li S., Riley D., Nesselbush M., Sengamalay N., Georgiadis A., Li Q. K., Madsen M. R., Mortensen F. V., Huiskens J., Punt C., van Grieken N., Fijneman R., Meijer G., Husain H., Scharpf R. B., Diaz L. A. Jr., Jones S., Angiuoli S., Ørntoft T., Nielsen H. J., Andersen C. L., Velculescu V. E., Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9, eaan2415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabos P., Finlay-Schultz J., Li C., Kline E., Finlayson C., Wisell J., Manuel C. A., Edgerton S. M., Harrell J. C., Elias A., Sartorius C. A., Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res. Treat. 135, 415–432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holst F., Stahl P. R., Ruiz C., Hellwinkel O., Jehan Z., Wendland M., Lebeau A., Terracciano L., al-Kuraya K., Jänicke F., Sauter G., Simon R., Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat. Genet. 39, 655–660 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Finlay-Schultz J., Jacobsen B. M., Riley D., Paul K. V., Turner S., Ferreira-Gonzalez A., Harrell J. C., Kabos P., Sartorius C. A., New generation breast cancer cell lines developed from patient-derived xenografts. Breast Cancer Res. 22, 68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaret K. S., Lerner J., Iwafuchi-Doi M., Chromatin scanning by dynamic binding of pioneer factors. Mol. Cell 62, 665–667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwafuchi-Doi M., Donahue G., Kakumanu A., Watts J. A., Mahony S., Pugh B. F., Lee D., Kaestner K. H., Zaret K. S., The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62, 79–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mieczkowski J., Cook A., Bowman S. K., Mueller B., Alver B. H., Kundu S., Deaton A. M., Urban J. A., Larschan E., Park P. J., Kingston R. E., Tolstorukov M. Y., MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 7, 11485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohammad I., Starskaia I., Nagy T., Guo J., Yatkin E., Väänänen K., Watford W. T., Chen Z., Estrogen receptor α contributes to T cell-mediated autoimmune inflammation by promoting T cell activation and proliferation. Sci. Signal. 11, eaap9415 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Sheng Y., Yu C., Liu Y., Hu C., Ma R., Lu X., Ji P., Chen J., Mizukawa B., Huang Y., Licht J. D., Qian Z., FOXM1 regulates leukemia stem cell quiescence and survival in MLL-rearranged AML. Nat. Commun. 11, 928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leary R. J., Sausen M., Kinde I., Papadopoulos N., Carpten J. D., Craig D., ’Shaughnessy J. O., Kinzler K. W., Parmigiani G., Vogelstein B., Diaz L. A. Jr., Velculescu V. E., Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 4, 162ra154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adalsteinsson V. A., Ha G., Freeman S. S., Choudhury A. D., Stover D. G., Parsons H. A., Gydush G., Reed S. C., Rotem D., Rhoades J., Loginov D., Livitz D., Rosebrock D., Leshchiner I., Kim J., Stewart C., Rosenberg M., Francis J. M., Zhang C. Z., Cohen O., Oh C., Ding H., Polak P., Lloyd M., Mahmud S., Helvie K., Merrill M. S., Santiago R. A., O’Connor E. P., Jeong S. H., Leeson R., Barry R. M., Kramkowski J. F., Zhang Z., Polacek L., Lohr J. G., Schleicher M., Lipscomb E., Saltzman A., Oliver N. M., Marini L., Waks A. G., Harshman L. C., Tolaney S. M., van Allen E. M., Winer E. P., Lin N. U., Nakabayashi M., Taplin M. E., Johannessen C. M., Garraway L. A., Golub T. R., Boehm J. S., Wagle N., Getz G., Love J. C., Meyerson M., Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corces M. R., Granja J. M., Shams S., Louie B. H., Seoane J. A., Zhou W., Silva T. C., Groeneveld C., Wong C. K., Cho S. W., Satpathy A. T., Mumbach M. R., Hoadley K. A., Robertson A. G., Sheffield N. C., Felau I., Castro M. A. A., Berman B. P., Staudt L. M., Zenklusen J. C., Laird P. W., Curtis C.; Cancer Genome Atlas Analysis Network, Greenleaf W. J., Chang H. Y., Akbani R., Benz C. C., Boyle E. A., Broom B. M., Cherniack A. D., Craft B., Demchok J. A., Doane A. S., Elemento O., Ferguson M. L., Goldman M. J., Hayes D. N., He J., Hinoue T., Imielinski M., Jones S. J. M., Kemal A., Knijnenburg T. A., Korkut A., Lin D. C., Liu Y., Mensah M. K. A., Mills G. B., Reuter V. P., Schultz A., Shen H., Smith J. P., Tarnuzzer R., Trefflich S., Wang Z., Weinstein J. N., Westlake L. C., Xu J., Yang L., Yau C., Zhao Y., Zhu J., The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zukowski A., Rao S., Ramachandran S., Phenotypes from cell-free DNA. Open Biol. 10, 200119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., Sanli K., von Feilitzen K., Oksvold P., Lundberg E., Hober S., Nilsson P., Mattsson J., Schwenk J. M., Brunnström H., Glimelius B., Sjöblom T., Edqvist P. H., Djureinovic D., Micke P., Lindskog C., Mardinoglu A., Ponten F., A pathology atlas of the human cancer transcriptome. Science 357, eaan2507 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Antony-Debré I., Paul A., Leite J., Mitchell K., Kim H. M., Carvajal L. A., Todorova T. I., Huang K., Kumar A., Farahat A. A., Bartholdy B., Narayanagari S.-R., Chen J., Ambesi-Impiombato A., Ferrando A. A., Mantzaris I., Gavathiotis E., Verma A., Will B., Boykin D. W., Wilson W. D., Poon G. M., Steidl U., Pharmacological inhibition of the transcription factor PU.1 in leukemia. J. Clin. Invest. 127, 4297–4313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han H., Cho J. W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C. Y., Lee M., Kim E., Lee S., Kang B., Jeong D., Kim Y., Jeon H. N., Jung H., Nam S., Chung M., Kim J. H., Lee I., TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The Cancer Genome Atlas Research Network, Weinstein J. N., Collisson E. A., Mills G. B., Shaw K. R. M., Ozenberger B. A., Ellrott K., Shmulevich I., Sander C., Stuart J. M., The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross-Innes C. S., Brown G. D., Carroll J. S., A co-ordinated interaction between CTCF and ER in breast cancer cells. BMC Genomics 12, 593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chèneby J., Ménétrier Z., Mestdagh M., Rosnet T., Douida A., Rhalloussi W., Bergon A., Lopez F., Ballester B., ReMap 2020: A database of regulatory regions from an integrative analysis of human and arabidopsis DNA-binding sequencing experiments. Nucleic Acids Res. 48, D180–D188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fornes O., Castro-Mondragon J. A., Khan A., van der Lee R., Zhang X., Richmond P. A., Modi B. P., Correard S., Gheorghe M., Baranašić D., Santana-Garcia W., Tan G., Chèneby J., Ballester B., Parcy F., Sandelin A., Lenhard B., Wasserman W. W., Mathelier A., JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kulakovskiy I. V., Vorontsov I. E., Yevshin I. S., Sharipov R. N., Fedorova A. D., Rumynskiy E. I., Medvedeva Y. A., Magana-Mora A., Bajic V. B., Papatsenko D. A., Kolpakov F. A., Makeev V. J., HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 46, D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savitzky A., Golay M. J. E., Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627–1639 (1964). [Google Scholar]
- 60.Harris C. R., Millman K. J., van der Walt S. J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N. J., Kern R., Picus M., Hoyer S., van Kerkwijk M. H., Brett M., Haldane A., del Río J. F., Wiebe M., Peterson P., Gérard-Marchant P., Sheppard K., Reddy T., Weckesser W., Abbasi H., Gohlke C., Oliphant T. E., Array programming with NumPy. Nature 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kent W. J., Zweig A. S., Barber G., Hinrichs A. S., Karolchik D., BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramirez F., Dundar F., Diehl S., Gruning B. A., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lex A., Gehlenborg N., Strobelt H., Vuillemot R., Pfister H., UpSet: Visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 20, 1983–1992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 and S3
Table S2