Abstract
Immunoglobulin class switch recombination (CSR) plays critical roles in controlling infections and inflammatory tissue injuries. Here, we show that AFF3, a candidate gene for both rheumatoid arthritis and type 1 diabetes, is a molecular facilitator of CSR with an isotype preference. Aff3-deficient mice exhibit low serum levels of immunoglobulins, predominantly immunoglobulin G2c (IgG2c) followed by IgG1 and IgG3 but not IgM. Furthermore, Aff3-deficient mice show weak resistance to Plasmodium yoelii infection, confirming that Aff3 modulates immunity to this pathogen. Mechanistically, the AFF3 protein binds to the IgM and IgG1 switch regions via a C-terminal domain, and Aff3 deficiency reduces the binding of AID to the switch regions less efficiently. One AFF3 risk allele for rheumatoid arthritis is associated with high mRNA expression of AFF3, IGHG2, and IGHA2 in human B cells. These findings demonstrate that AFF3 directly regulates CSR by facilitating the recruitment of AID to the switch regions.
AFF3 directly regulates immunoglobulin class switch recombination by facilitating the recruitment of AID to the switch regions.
INTRODUCTION
Genome-wide association studies (GWASs) are approaches used in genetic research to link single-nucleotide polymorphisms (SNPs) with susceptibility to diseases or quantitative traits (1, 2). GWASs have identified disease-associated loci and their neighboring genes across the human genome. GWASs of autoimmune and inflammatory diseases have identified various candidates, but the roles of these candidates in the onset and progression of immune-mediated diseases are largely unknown, except for those of genes that have been associated with immune responses.
More than 10 GWAS have identified the AFF3 gene as being associated with autoimmune diseases (3–11). Although the mRNA expression of AFF3 is known to be restricted to the lymphocytes and brains of mice and humans (12, 13), the functions of AFF3 in the immune system have not been studied. The vertebrate AF4/FMR2 (AFF) gene family (also known as the ALF family) consists of four genes (Aff1 to Aff4) (14). AFF1 and AFF4 are components of the super elongation complex (SEC), which consists of positive transcription elongation factor b [P-TEFb; a complex of cyclin-dependent kinase 9 (CDK9) and CYCLIN T], ELL (eleven-nineteen lysine-rich leukemia), AF9 (ALL-1 fused gene from chromosome 9), and ENL (eleven-nineteen leukemia) in addition to AFF1 and AFF4 (14, 15). The SEC regulates the elongation of transcription by RNA polymerase II (RNAPII) and is critical for general or environmental cue-induced mRNA expression (16, 17). AFF3 is reported to form a complex with P-TEFb, AF9, and ENL but not with ELL (14) and regulates the expression of a set of genes different from that regulated by AFF4 (14). AFF3 also reportedly binds to the DNA-methylated regions of genetically imprinted genes and regulates their transcription (18, 19).
Immunoglobulin-mediated humoral immunity is critical for host defense against pathogens and toxins, and the dysregulation of this process leads to immunodeficiency and autoimmune diseases (20, 21). Immunoglobulins can be divided into several isotypes based on differences in their C-terminal heavy chain regions. Immunoglobulin class switch recombination (CSR) is conducted via the exchange of exons positioned 3′ adjacent to the exon encoding the antigen recognition domain (22–24). Activation-induced cytidine deaminase (AID) is the enzyme that initiates CSR (24), which functions by mutagenizing the G-rich genomic regions (called switch regions) that precede the exons encoding the Fc domain and inducing DNA repair, leading to chromosomal recombination between two transcriptionally active switch regions to complete CSR. In addition, AID is required for somatic hypermutations (SHMs) in the exons encoding the antigen recognition domains of immunoglobulin genes to increase antibody affinity. The regulation of AID expression, localization of AID to targets, and enzymatic activity of AID are central to the proper execution of CSR and SHM, but the precise underlying mechanisms are under investigation (6, 23–26).
In this study, we searched for autoimmune-associated genes identified by GWAS that met the following three criteria: (i) identified as candidate genes for at least two autoimmune diseases, (ii) exhibited relatively restricted expression in immune cells, and (iii) exerted unknown functions in the immune system. Through this screening strategy, we selected the AFF3 gene among nine genes meeting the criteria because its expression is enriched in lymphocytes, and we analyzed the function of AFF3 in the immune system. We found reduced serum levels of immunoglobulin G2c (IgG2c), IgG1, and IgG3 and impaired antibody-mediated clearance of Plasmodium yoelii parasites in Aff3-deficient mice. Mechanistically, AFF3 was shown to regulate CSR by controlling the mutagenesis of the switch regions through the recruitment of AID. Our data demonstrate a previously unrecognized role of AFF3 in regulating CSR and suggest a mechanistic explanation for the GWAS-based association of AFF3 with autoimmune diseases.
RESULTS
Selection of genes associated with immune-mediated diseases using a GWAS database
We aimed to identify genes associated with autoimmune diseases using a previously reported GWAS database (GWAS Catalog; www.ebi.ac.uk/gwas/home). We focused on genes whose expression is largely restricted to lymphocytes and whose functions in the immune system are unknown. We chose rheumatoid arthritis, type 1 diabetes (T1D), and multiple sclerosis among immune-mediated diseases because the candidate genes for these diseases show a high correlation (27, 28). We first selected candidate genes from the GWAS data of each disease and focused on the genes that were associated with at least two diseases (fig. S1). Then, we chose the genes with higher mRNA expression in T and B cells than in other hematopoietic cell types. To evaluate mRNA expression, we used three databases: the Human Protein Atlas, the Immunological Genome Project, and the RCAI RefDIC. We excluded noncoding RNAs and pseudo-genes and genes that had previously been associated with immune responses. Last, we selected AFF3 as the candidate gene for further analysis (fig. S1). The GWAS data suggest that AFF3 is associated with both rheumatoid arthritis and T1D (3–11) and that its expression is restricted to the lymphocytes and brain in mice and humans (12, 13). While the function of AFF3 is thought to be related to transcriptional elongation based on the structural and biochemical similarity of AFF3 to AFF1/4 (14), the role of AFF3 in the immune system has not been investigated.
Aff3-deficient mice show reduced serum immunoglobulin levels
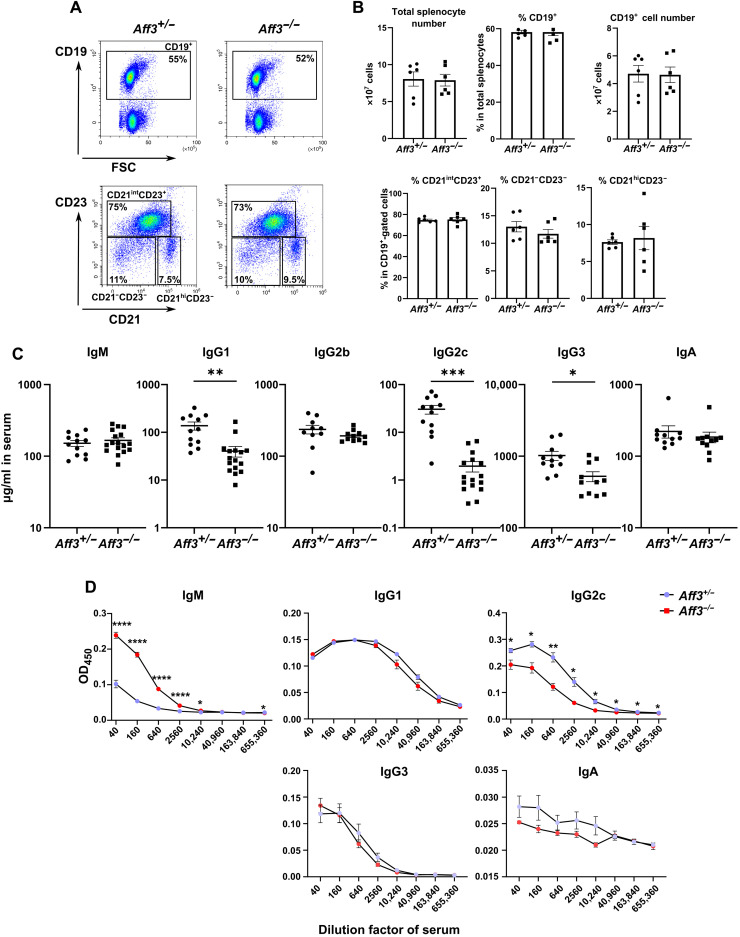
To elucidate the function of Aff3, we generated and analyzed mice in which the gene was disrupted. Using a CRISPR-Cas9 system, we generated mouse strains that were Aff3 deficient and Aff3 floxed (fig. S2). The Aff3-deficient mice were healthy and fertile, with no visible abnormalities up to at least 5 months of age. We examined the cell numbers and frequencies of the T subpopulations in the thymus (fig. S3A) and the cell numbers (fig. S3B) and frequencies of TCRβ+ (β T cell receptor–positive; fig. S3C) and CD25+ cells (fig. S3F) and the CD4/CD8 (fig. S3D) and CD62L/CD44 ratios (fig. S3E) in the spleen, and no differences were observed between the Aff3-deficient and control mice. As CDK9, CYCLIN T1, and AFF4 are reported to be involved in T helper cell 1 (TH1) and cytotoxic T lymphocyte differentiation (29, 30), we assessed CD44 expression and interferon-γ (IFN-γ) secretion in CD4+ and CD8+ T cells from Aff3-deficient mice upon stimulation with anti-CD3 and anti-CD28 antibodies. The expression levels of IFN-γ in CD4+ (fig. S4A) and CD8+ T cells (fig. S4B) from Aff3-deficient mice were equivalent to those in cells from control mice. No abnormalities in the total number of splenocytes or the frequency of the B cell subpopulation were observed in the spleen (Fig. 1, A and B). However, we found that the serum levels of IgG1, IgG2c, and IgG3 were significantly decreased in the Aff3-deficient mice (Fig. 1C). We also examined antigen-specific antibody production. Two weeks after immunization with ovalbumin (OVA)/complete Freund’s adjuvant (CFA), the anti-OVA antibody titers in the serum were measured, and a significant decrease in the IgG2c level and an increase in the IgM level were observed in Aff3-deficient mice (Fig. 1D). The levels of IgG1, IgG3, and IgA were comparable between control and Aff3-deficient mice (Fig. 1D). These results demonstrate that the production of IgG subtypes is decreased in Aff3-deficient mice and that the extent of the reduction varies depending on the isotypes and immune conditions.
Fig. 1. Aff3 deficiency reduces serum antibody levels.
(A) Flow cytometry analysis of splenocytes from Aff3+/− and Aff3−/− mice. Top: Percentage of CD19+ cells. Bottom: CD23/CD21 profiles among CD19+-gated cells. CD21intCD23+ cells are follicular or T2 B cells; CD21hiCD23− cells are marginal zone B cells, and CD21−CD23− cells are B1a or T1 B cells. (B) Quantification of total splenocytes and the percentages of B cell subpopulations (n = 6 in each group). (C) The serum antibody titers in unimmunized mice were measured by ELISA (n = 12 in each group). (D) The relative amounts of antibodies in the OVA-immunized mice were measured by ELISA [n = 5 in the Aff3+/− group (purple) and n = 7 in the Aff3−/− group (red)]. The mice were immunized with OVA/CFA, and their sera were collected at 2 weeks after immunization. The serum was serially diluted and subjected to measurements. In (B to D), the data are shown as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001. The P values were calculated using unpaired t tests with Welch’s correction (B and C) or multiple Welch’s t tests with Holm-Sidak correction (D). The data shown in this figure are representative of at least two experiments.
We further examined the roles of AFF3 in the affinity maturation of immunoglobulins. We performed enzyme-linked immunosorbent assay (ELISA) using 4-hydroxy-3-nitrophenyl acetyl (NP)=2 and NP=25 antigens to examine the effect of AFF3 on antigen-specific high- and low-affinity antibodies, respectively. We found that Aff3 deficiency did not significantly affect either the anti-NP=2 or anti-NP=25 serum titers of IgG2b and IgA (fig. S5A). The ratio of NP=25 to NP=2 was also not significantly affected (fig. S5B). These results suggest that Aff3 deficiency does not affect the affinity maturation of immunoglobulins.
Aff3-deficient mice are highly susceptible to P. yoelii infection
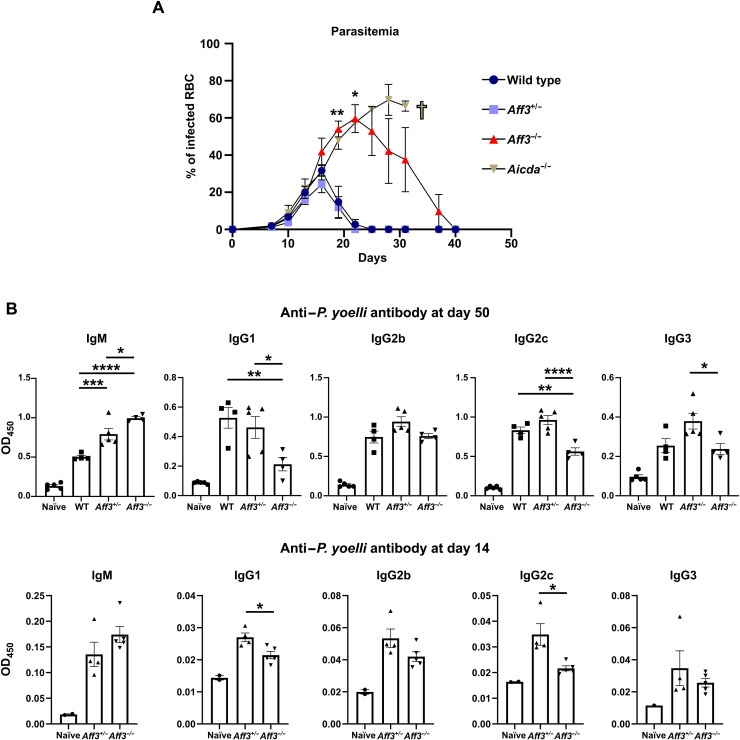
As Aff3 deficiency impaired but did not completely abolish IgG production, we examined whether the impairment was significant enough to affect host defense against pathogens. We chose an infection model involving the mouse malaria parasite P. yoelii because we found that IgG2c was most robustly affected by Aff3 deficiency, and this isotype or its equivalent, IgG2a, has been reported to be the most critical anti-malaria parasite antibody in mice (31–34). We infected mice with P. yoelii and measured the infected erythrocytes in peripheral blood (parasitemia) and anti–P. yoelii antibodies in serum. Aicda (AID-encoding gene)–deficient mice were used to compare the effect of the reduced antibody levels induced by Aff3 deficiency with the effect of the complete absence of CSR induced by Aicda deficiency. Compared with control mice, Aicda-deficient mice showed higher parasitemia levels, and all four Aicda-deficient mice died approximately 1 month after being infected, while none of the control mice died (Fig. 2A). The Aff3-deficient mice showed higher parasitemia levels than the control mice but lower parasitemia levels than the Aicda-deficient mice (Fig. 2A). One of four Aff3-deficient mice died within 1 month after being infected, indicating that Aff3 deficiency moderately increases sensitivity to the parasite. Serum antibody titers against P. yoelii were measured on days 14 and 50, and we observed decreased IgG2c and IgG1 titers and increased IgM titers in Aff3-deficient mice on days 14 and 50 (Fig. 2B). These results indicate that the regulation of IgG production by AFF3 plays a substantial role in host defense against the pathogen. We also generated an influenza virus–infected mouse model. We found that the antiviral IgG level was reduced in Aff3-deficient mice (fig. S6, A and B), indicating that IgG production is impaired in Aff3-deficient mice under various conditions. The body weights of the mice were not affected, probably because of the use of a virulent influenza strain.
Fig. 2. Aff3 deficiency increases the sensitivity of mice to P. yoelii infection.
(A) Wild-type (dark blue), Aff3+/− (light blue), Aff3−/− (red), and Aicda−/− (brown) mice were infected with P. yoelii, and the level of parasitemia was monitored every 4 days (n = 4 to 5 in each group). Aicda−/− mice died after (B) anti–P. yoelii antibodies in serum were measured by ELISA at 50 and 14 days after inoculation (n = 4 to 5 in each group, except n = 2 in naïve group of day 14). In (A and B), the data are shown as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001. The P values were calculated using multiple Welch’s t test with Holm-Sidak correction (A), one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test [(B), day 50] or unpaired t tests with Welch’s correction [(B), day 14]. The asterisks indicating significant differences compared with naïve mice are not shown in (B).
CSR is disturbed in Aff3-deficient mice in a B cell–intrinsic manner
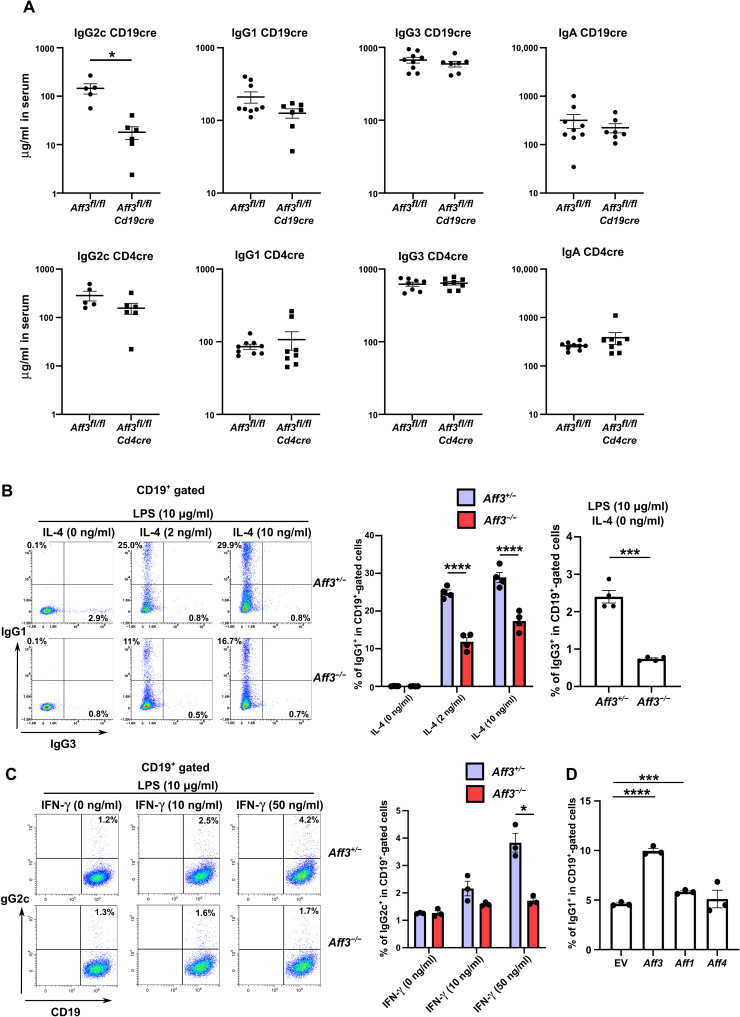
We sought to evaluate whether AFF3 regulates IgG production in B cells or T cells by analyzing the offspring of Aff3-floxed mice crossed with Cd4-cre or Cd19-cre mice. When we analyzed their antibody titers in serum, we found that the IgG2c titers were decreased in the mice generated from the cross with Cd19-cre mice but not in those from the cross with Cd4-cre mice, indicating that Aff3 expression in B cells is necessary to regulate IgG production (Fig. 3A). On the other hand, even the Cd19-cre–mediated deletion of the Aff3 gene showed no effect on IgG1 and IgG3, presumably because of the insufficient deletion of the Aff3 gene mediated by Cd19-cre. We found that approximately 10% of the original Aff3 mRNA content remained in B cells in which Aff3 was deleted by Cd19-cre, whereas only approximately 0.3 and 2% of the mRNA remained in the B cells with the null and deletion mutants generated by Cd4-cre, respectively (fig. S2, B and C).
Fig. 3. Aff3 regulates CSR in a B cell–intrinsic manner.
(A) Serum antibodies from Aff3flox/flox Cd4cre and Cd19cre mice were measured by ELISA (n = 5 in each group). (B) Flow cytometry analysis of class switching to IgG1. CD43− B cells were harvested from Aff3+/− (purple) or Aff3−/− (red) mice and cultured with LPS and IL-4 (10 μg/ml). After 4 days, the cells were analyzed by flow cytometry. The panels show the IgG1 and IgG3 expression profiles and the statistical analysis (n = 4 in each group). (C) Flow cytometry analysis of class switching to IgG2c. CD43− B cells were cultured with LPS and IFN-γ (10 μg/ml). The panels show the IgG2c expression profiles and the statistical analysis (n = 3 in each group). (D) Overexpression of the AFF3 compensated for the Aff3−/− phenotype. CD43− B cells were harvested from Aff3−/− mice and cultured with LPS and IL-4. The cDNAs of Aff3, Aff1, and Aff4 were transduced with a retroviral vector carrying the green fluorescent protein (GFP) marker. The panel shows the percentage of IgG1+ cells in GFP+CD19+ cells (n = 3 in each group). EV, empty vector. The data are shown as means ± SEM. *P < 0.05, ***P < 0.005, and ****P < 0.001. The P values were calculated using unpaired t test with Welch’s correction in (A to C) and via one-way ANOVA with Dunnett’s multiple comparisons test in (D). The data shown in this figure are representative of at least two experiments.
To determine whether AFF3 regulates CSR, we purified naïve CD43− B cells from the spleens of Aff3+/− and Aff3−/− mice and cultured them in the presence of lipopolysaccharide (LPS) together with interleukin-4 (IL-4) or IFN-γ. The cell surface expression levels of IgG1, IgG3 (Fig. 3B), and IgG2c (Fig. 3C) were reduced in Aff3-deficient B cells, strongly suggesting that AFF3 in B cells regulates CSR. To further determine whether AFF3 regulates CSR in a B cell–intrinsic manner, we transduced Aff3 cDNA into B cells with a retroviral vector and assessed IgG1 expression. The expression of IgG1 in Aff3-deficient B cells transduced with Aff3 was recovered (Fig. 3D). In contrast, little recovery was observed in the cells transduced with Aff1, and no recovery was observed following Aff4 transduction (Fig. 3D). These results indicate that AFF3 regulates CSR in a B cell–intrinsic manner and suggest that AFF3 and AFF1/4 are not functionally redundant.
AFF3 directly binds to switch regions
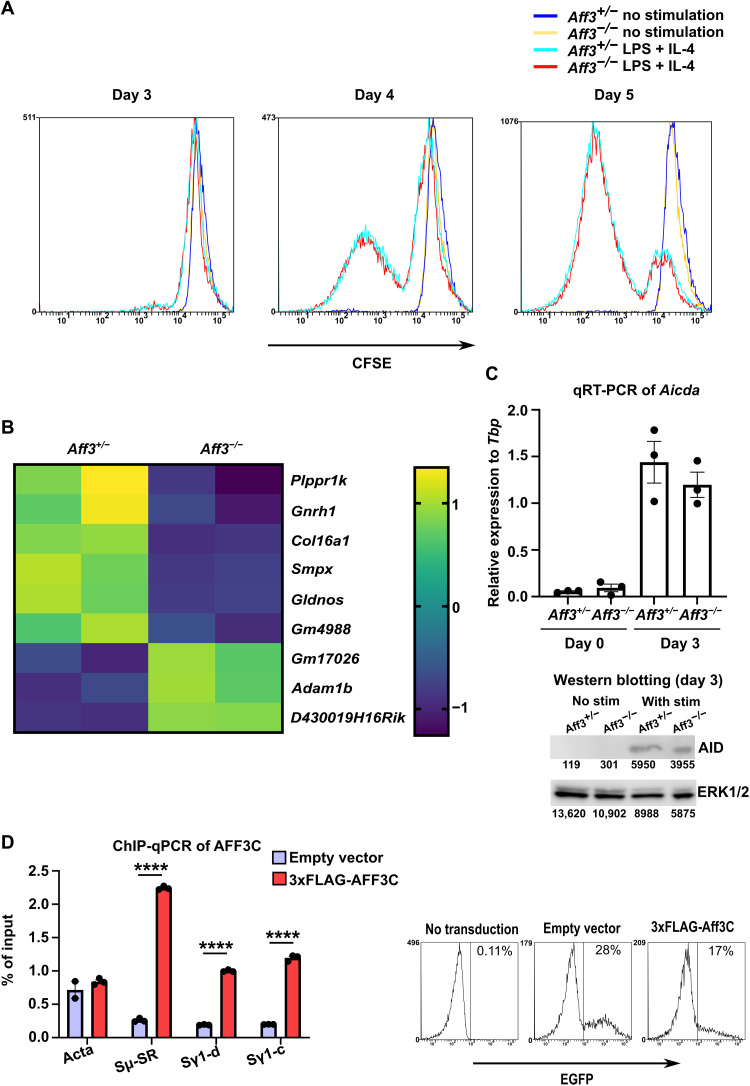
As B cell proliferation and activation are necessary for CSR (23, 35), we analyzed B cell activation and proliferation following stimulation. The applied carboxyfluorescein succinimidyl ester (CFSE) dilutions did not differ between the control and Aff3-deficient B cells (Fig. 4A). We evaluated activation by analyzing the transcriptomes of Aff3-deficient B cells on day 3 of culture with LPS and IL-4. Only slight changes in the expression levels of several genes were observed, none of which were considered to be related to B cell activation or proliferation (Fig. 4B). Moreover, the mRNA and protein expression of AID was not altered in the absence of Aff3 (Fig. 4C). These results suggest that the major function of AFF3 in B cells is not the transcriptional regulation of gene transcription.
Fig. 4. AFF3 protein directly binds to the switch region.
(A) CD43− B cells were stained with CFSE and cultured. The cells were analyzed by flow cytometry. (B) Transcriptome analysis of Aff3-deficient B cells. CD43− B cells were cultured with LPS and IL-4. After 3 days of culture, total RNA was prepared and analyzed on a cDNA microarray. (C) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) and Western blotting analyses of Aicda (n = 3 in each group for RT-PCR). CD43− B cells were cultured with LPS (10 μg/ml) and IL-4 (10 ng/ml). Western blotting was performed with anti-AID antibody, and the membrane was reprobed with anti–extracellular signal–regulated kinase 1/2 (ERK1/2) antibody to assess the loading amounts. nt, nucleotide. (D) AFF3 binds to switch regions. CD43− B cells were cultured with LPS (10 μg/ml) and IL-4 (10 ng/ml) for 3 days. Empty (purple) or 3xFLAG-tagged cDNA of the C-terminal half of Aff3 (red) was transduced. The efficiency of transduction was estimated on the basis of the expression of enhanced GFP (EGFP), which was coexpressed with AFF3 by IRES (internal ribosomal entry site) insertion. Then, the cells were analyzed by ChIP with an anti-FLAG antibody. The amount of DNA precipitated was measured by real-time PCR (n = 3 in each group), with primers specific to the switch regions or the Acta (encoding α-actin) gene serving as a negative control. Sμ-SR indicates the switch region of IgM (table S4) (70). Sγ1-c and Sγ1-d indicate the switch region of IgG1 (fig. S7 and table S4). In (C and D), the data are shown as means ± SEM. ****P < 0.001. The P values were calculated using unpaired t tests with Welch’s correction. The data shown in this figure are representative of at least two experiments except for those in (B). The primers used in the experiments are listed in table S4.
As no substantial changes in B cell proliferation or activation were induced by Aff3 deficiency, we next assessed whether AFF3 is directly involved in CSR. Therefore, we investigated whether AFF3 could directly bind to switch regions. We added a 3xFLAG tag to the beginning of the C-terminal half of the AFF3 protein, which includes the DNA/RNA binding region (36–38), and we overexpressed the resulting construct in B cells. Binding to switch regions was assessed in a chromatin immunoprecipitation–quantitative polymerase chain reaction (ChIP-qPCR) assay with an anti-FLAG antibody. We detected significant increases in the signals near the switch regions of IgM and IgG1, indicating that AFF3 can bind to these regions (Fig. 4D).
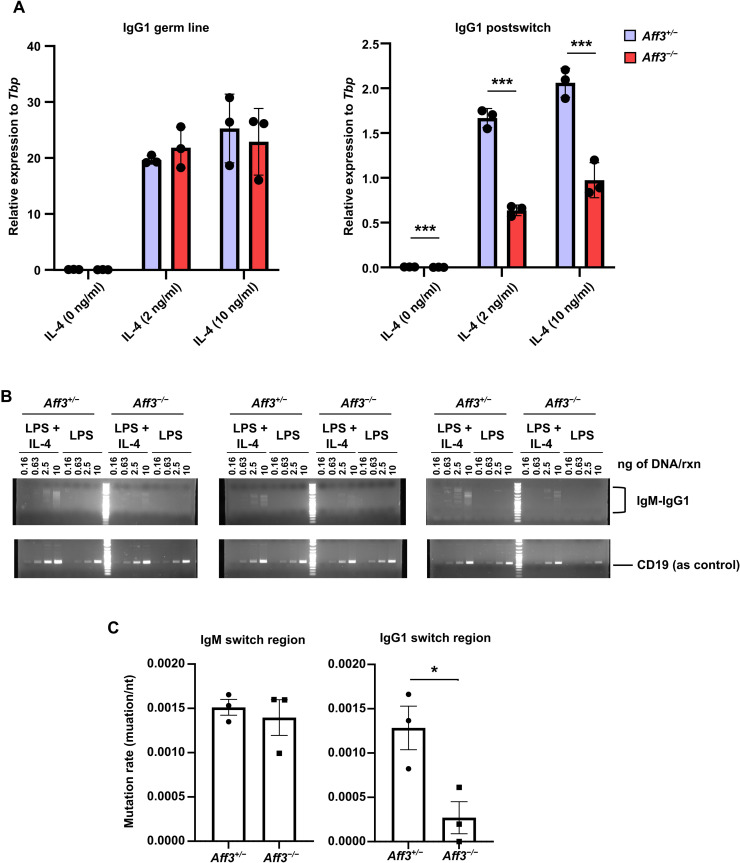
Aff3 deficiency reduces mutation rates in the switch region of IgG1
To examine which CSR step was regulated by AFF3, we first compared the germline and postswitch transcript levels of the IgG1 switch region in B cells stimulated with LPS and IL-4 as determined by quantitative reverse transcription PCR (qRT-PCR). No differences in the expression levels of germline transcripts were observed, whereas significant decreases in the expression levels of postswitch transcripts were observed in the Aff3-deficient cells (Fig. 5A). Several papers have suggested that RNAPII temporarily stalls in switch regions and forms an R loop, which promotes mutation and recombination in these regions (39, 40). Thus, we analyzed whether RNAPII binding to switch regions was decreased by Aff3 deficiency. However, the amount of RNAPII binding to switch regions in Aff3-deficient B cells was equivalent to that in control cells, as determined by ChIP-qPCR (fig. S7).
Fig. 5. Aff3 deficiency affects CSR.
(A) qRT-PCR was used to analyze the germline and postswitch transcript levels of IgG1 (n = 3 in each group). Aff3+/− (purple) or Aff3−/− (red) CD43− B cells were cultured with LPS (10 μg/ml) and the indicated amount of IL-4. After 3 days of culture, total RNA was extracted from the cells and analyzed. (B) Semiquantitative PCR was performed to analyze CSR. CD43 B cells were cultured as described in (A), and genomic DNA was extracted. PCR of the IgM and IgG1 switch regions in combination was performed with the indicated primers. The genomic DNA was serially diluted and used as the template for PCR. Primers for CD19 were used as an internal control. (C) Genomic DNA was prepared as described in (B). Fragments of IgM-IgG1 chimeric switch regions were amplified by nested PCR. The fragments were sequenced, and the mutation rates per nucleotide were plotted (n = 3 in each group). The detailed data are shown in table S1. In (A and C), the data are shown as means ± SEM. *P < 0.05 and ***P < 0.005. The P values were calculated using unpaired t tests with Welch’s correction. The data in (A and B) are representative of at least two experiments. The primers used are listed in table S4.
Regarding the other CSR step, we examined whether DNA recombination was affected by Aff3 deficiency. We prepared genomic DNA from LPS ± IL-4–stimulated B cells and performed a semiquantitative PCR analysis of the IgM-G1 chimeric switch region. We expected to detect smear bands corresponding to DNA recombination because the positions related to recombination were not fixed within the switch regions; chimeric switch regions of various lengths should therefore have appeared (41). The formation of smear bands derived from the chimera was decreased by Aff3 deficiency, indicating a reduction in DNA recombination in Aff3-deficient B cells (Fig. 5B). We also examined whether the efficiency of AID-induced mutation in switch regions was affected by Aff3 deficiency. The recombined IgM-G1 switch junction was amplified by PCR, and the obtained DNA fragments were cloned and sequenced. The mutation rate in the IgM switch region was not significantly affected by Aff3 deficiency, but that in the IgG1 switch region was significantly decreased in Aff3-deficient B cells (Fig. 5C and table S1). These results indicate that AFF3 regulates mutagenesis in switch regions in an isotype-dependent manner.
Aff3 deficiency reduces the recruitment of AID to switch regions
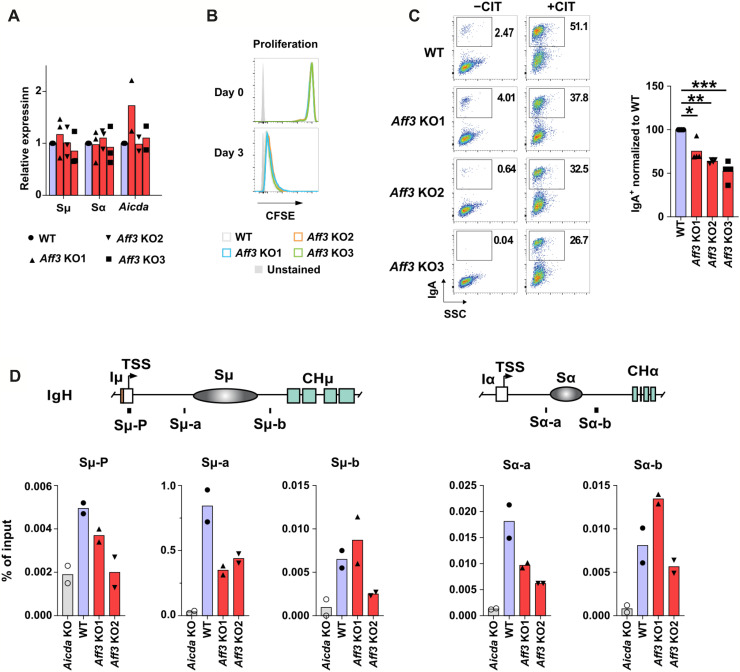
AID is an essential molecule for CSR (24), and its deletion leads to the almost complete disappearance of CSR among all isotypes. Notably, it has been reported that when functional AID levels are reduced by half, IgG2c levels are markedly decreased in an influenza infection model (42) similar to the case in Aff3-deficient mice immunized with CFA/OVA or CFA/4-hydroxy-3-nitrophenyl acetyl–chicken gamma globulin (CFA/NP-CGG) (Fig. 1D and fig. S5A). As the mRNA and protein expression of Aicda (AID) was not affected by Aff3 deficiency (Fig. 4C), we sought to assess the involvement of AFF3 in the recruitment of AID to switch regions. As the ChIP assay of AID was not successful in primary B cells, even when FLAG-tagged AID was used, we used the CH12 mouse B cell line, in which IgM is switched to IgA by stimulation with anti-CD40, IL-4, and transforming growth factor–β (TGF-β). The expression of germline transcripts and AID and the proliferation of cells were not altered in Aff3-deficient CH12 cells (Fig. 6, A and B, and fig. S8). On the other hand, we observed slightly but significantly reduced class switching to IgA in three independently generated Aff3−/− clones (Fig. 6C). Although we did not observe a statistically significant reduction in IgA in Aff3-deficient mice, the average IgA level tended to be decreased (Figs. 1C and 3A and figs. S5A and S6B). As IgA levels are affected by many factors in vivo, the observed variations might have affected the statistical significance of the results. In CH12 cells, the coloniality and relatively stable culture conditions of these cells allowed a moderate difference in IgA to be detected between the control and Aff3−/− cells. We subsequently performed ChIP-qPCR analysis of AID and found that the binding of AID to the IgM and IgA switch regions was reduced in Aff3-deficient cells (Fig. 6D), indicating that AFF3 is required for the maximal AID occupancy of switch regions.
Fig. 6. Aff3 deficiency decreased AID recruitment in the CH12 B cell line.
(A) Effects of Aff3 deficiency on mRNA expression in CH12 B cells (n = 3 in each group). The Aff3 gene was disrupted by CRISPR-Cas9 (fig. S8), and the transcripts of the switch regions of IgM, IgA, and AID of wild-type (WT) (purple) or knockout (KO) clones (red) were evaluated by qRT-PCR. (B) Effect of Aff3 deficiency on the proliferation of CH12 cells. The cells were stained with CFSE and analyzed after being cultured for 3 days. WT clones are shown in gray, and KO clones are indicated with blue, orange, and green lines. (C) Effect of Aff3 deficiency on the CSR to IgA in CH12 B cells (n = 3 in each group). WT (purple) or KO clones (red) were stimulated with CIT (anti-CD40 antibody, IL-4, and TGF-β) for 3 days and then analyzed by flow cytometry. (D) The binding of AID to switch regions was assessed by ChIP-qPCR with an anti-AID antibody (n = 2 in each group). Aicda-KO, WT, and KO clones are represented by the gray, purple, and red columns, respectively. In (C), *P < 0.05, **P < 0.01, and ***P < 0.005. The P values were calculated using one-way ANOVA with the Holm-Sidak multiple comparison test. The primers used in the experiments are listed in table S4.
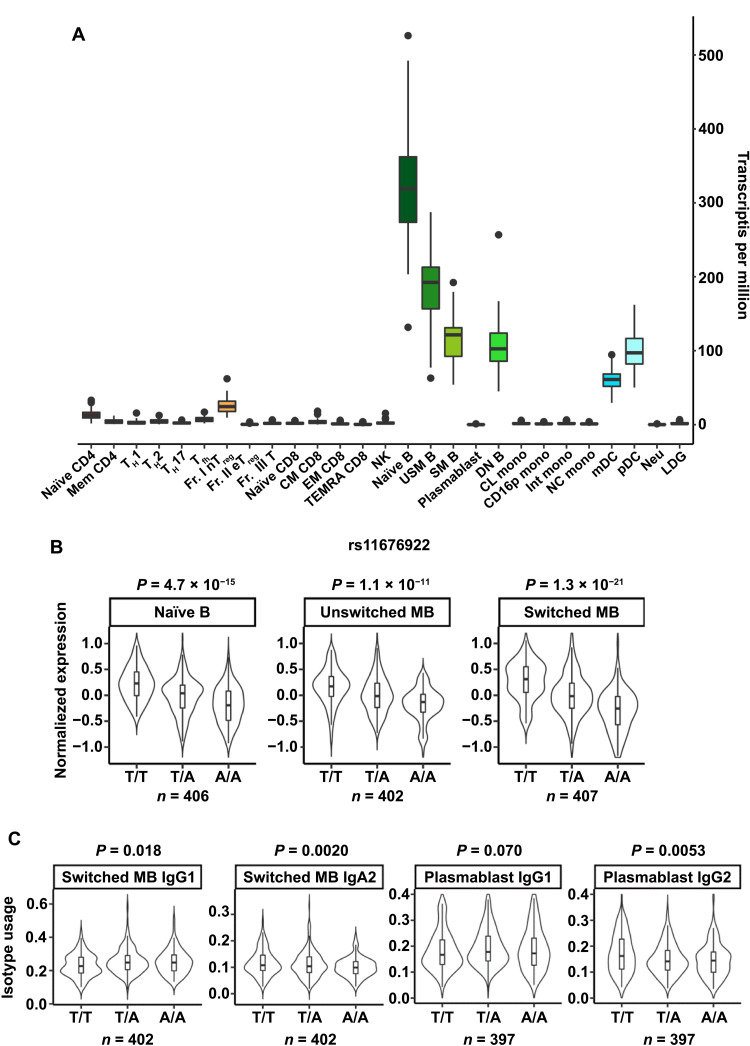
A risk allele for rheumatoid arthritis is associated with the mRNA expression of AFF3, IGHG2, and IGHA2 in B cells
Last, we assessed the association of a risk allele for rheumatoid arthritis with AFF3 mRNA expression and isotype usage in humans. We used a dataset including gene expression information from immune cell subsets and SNPs obtained from healthy donors and patients with immune-mediated diseases (ImmuNexUT; www.immunexut.org/) (43). First, we assessed the expression of AFF3 mRNA in this human database and found that AFF3 was particularly highly expressed in B cells and was expressed at very low levels in T cells (Fig. 7A). Next, we analyzed the expression quantitative trait loci (eQTLs) of the AFF3 gene. We found that the “T” allele of SNP rs11676922, associated with susceptibility to rheumatoid arthritis, was significantly associated with high AFF3 expression (Fig. 7B). We then analyzed the relationship of this allele with isotype usage, which revealed that the T risk allele was associated with highly frequent use of IgG1 and IgA2 (Fig. 7C). We next investigated whether there could be a causal relationship between AFF3 levels and IgG2 levels by overexpressing AFF3 or the empty viral control in Aff3+/− cells. We found that AFF3 overexpression was sufficient to increase switching to IgG1 (fig. S9). These results are consistent with the hypothesis that AFF3 regulates CSR in B cells and is involved in the pathogeneses of rheumatoid arthritis and other antibody-associated immune diseases in humans.
Fig. 7. AFF3 gene expression and its disease association in humans.
(A) AFF3 mRNA expression in human immune cells as determined from the ImmuNexUT (www.immunexut.org/) database (43). The full names of the cells are listed in table S5. (B) AFF3 mRNA expression in human B cell subpopulations stratified by genotype. The indicated genotypes (T/T, T/A, and AA) are for the rs11676922 SNP. T is the risk allele for rheumatoid arthritis. n indicates each sample size. (C) Isotype usage in B cell subpopulations stratified by genotype. The indicated genotypes (T/T, T/A, and AA) are the same as those in (B).
DISCUSSION
Studies on immune-mediated diseases performed using GWAS data have provided many candidate loci associated with each investigated disease (44, 45). The contributions of these loci to the pathogenesis of immune-mediated diseases are, however, largely unclear because of their small effects on gene expression and functions. Here, we screened genes among candidates that were associated with at least two autoimmune diseases based on the following criteria: expression restricted to immune cells and the lack of a known function in immune cells. Among nine genes meeting these criteria, we focused on the AFF3 gene because its mRNA expression is higher in lymphocytes than in other types of cells (12, 13), suggesting that AFF3 directly regulates acquired immunity. We found that AFF3 is required for optimal CSR through the regulation of AID recruitment or retention in switch regions. AFF3 is thought to be involved in transcription elongation because its paralogs AFF1/4 regulate this process (14, 46) and form a complex with P-TEFb, which is an essential regulator of transcription elongation (14). In our studies, neither Aff1 nor Aff4 complemented Aff3 deficiency, demonstrating that these genes are not redundant. Although Aff3 deficiency did not completely abolish CSR, it significantly affected susceptibility to infection by a malaria parasite. Furthermore, a risk allele for rheumatoid arthritis is correlated with high AFF3, IGHG2, and IGHA2 expression in human B cells. These data indicate that AFF3 is a substantial facilitator of CSR with an isotype preference and suggest AFF3 as a target molecule for the treatment of autoantibody-mediated diseases.
The regulation of CSR can be divided into several steps: (i) the regulation of AID expression, (ii) the transcription of switch regions to open their DNA structure, (iii) the recruitment of AID to the switch regions, (iv) the introduction of C-to-U mutations in the switch regions by AID, (v) the processing of the mutations by the DNA repair machinery to generate DNA breaks, and (vi) recombination joining of two breaks in different switch regions (24). In Aff3-deficient B cells, steps (i) and (ii) were unaffected, while the occurrence of step (iii) and the subsequent steps seemed to be decreased to some extent. While the mechanism by which AID is recruited to switch regions is still unclear, the following mechanisms have been proposed: (i) enhanced formation of the R loop and stalling of RNAPII, resulting in easy access by AID (25, 39); (ii) formation of G4-DNA in switch regions with high affinity to AID (42, 47, 48); (iii) binding of switch region–derived G4-RNA to AID and switch regions (40, 42); (iv) recruitment of AID by the RNA exosome coupled with RNAPII (49, 50); (v) binding of transcription elongation factors to AID and switch regions (51–53); and (vi) binding of cohesin and its regulators to AID and switch regions (54). Concerning mechanism (i), we did not observe a change in the ChIP signal corresponding to RNAPII in the switch regions, suggesting that R loop formation induced by the stalling of RNAPII is not affected by Aff3 deficiency. Regarding mechanisms (ii) and (iii), AFF3 was shown to preferentially bind to G4-DNA/RNA (55) and may therefore support the binding of AID to these nucleic acid structures, although our data do not provide evidence that either confirms or rules out this possibility. In mechanism (iv), the transcription elongation factors SPT5 (suppressor of Ty 5) and SPT6 and the polymerase associated factor (PAF) complex are required for the recruitment of AID (51–53, 56–58). In addition to these factors, Methot and colleagues (52) performed a BioID assay and detected AID and CDK9 in their vicinity. Since the proximity of CDK9 was reduced in AID mutants that failed to occupy switch regions but were normally recruited to the transcription start site, P-TEFb might be involved in linking AID to transcription elongation. AFF3 might regulate the recruitment of AID in collaboration with these factors. With respect to mechanism (v), one previous study has reported that cohesin directly interacts with CDK9 and other SEC components (59). Concerning mechanism (vi), as AID binds to cohesin and its related factors and as the knockdown of these factors reduces the recruitment of AID to switch regions, which is followed by the impairment of CSR (54), AFF3 may regulate CSR by facilitating the interaction of AID with cohesin factors. Further investigation remains to be carried out to clarify which factors are involved in AFF3-facilitated AID recruitment.
We identified an isotype- and context-dependent requirement of AFF3 for CSR. For instance, a reduction in IgG2c was observed under all conditions, while a reduction in IgG1 was observed in the serum of unimmunized Aff3-deficient mice but not in that of OVA-immunized mice. These results might be explained by previous reports showing that a reduction in AID abundance differentially affects the CSR of each isotype in a condition-dependent manner. In AicdaGV/+ mice, which exhibit half the level of functional AID found in Aicda+/+ mice, the abundance of each isotype was shown to be reduced to a different extent, and the size of the reduction depended on the conditions of the experiments (42). Specifically, the authors showed that among IgG1, IgG2b, and IgG2c, only IgG2c exhibited significantly reduced abundance in an influenza virus infection model. They also showed that no isotypes were affected in unimmunized mice but that the levels of IgG1, IgG3, and IgA were reduced to different degrees in ex vivo culture. Furthermore, knockdown of Spt5 in CH12 cells reduces (but does not completely abolish) AID recruitment to IgM and IgA switch regions (51), whereas the numbers of DNA breaks are decreased in the switch region of IgA but not in that of IgM (57). These results are consistent with our results, showing that Aff3 deficiency decreased the mutation rate in the switch region of IgG1 but not in that of IgM. The reason why the reduction in AID abundance differentially affects each isotype in a condition-dependent manner remains unclear. The concentrations and/or modifications of AID and CSR regulators differ among different target sites and conditions. Concerning SHM, Aff3 deficiency did not affect the affinity maturation of immunoglobulins in NP-CGG–immunized mice. As AicdaGV/+ mice show no significant reduction in SHM in either unimmunized or immunized conditions (42), SHM might also be less sensitive to AID reduction than to CSR to IgG2c.
We found that the risk allele for rheumatoid arthritis was correlated with high expression levels of AFF3 and frequent utilization of IgG2 and IgA2 in B cells. In patients with rheumatoid arthritis, rheumatoid factor IgG and anticyclic citrullinated peptide IgG antibodies are detected even before the onset of the disease and are related to disease severity, suggesting that specific classes of autoantibodies are related to the pathogenesis (60–63). Moreover, islet-specific autoantibodies (anti-insulin, glutamate decarboxylase, islet antigen 2, and islet-specific zinc transporter antibodies) have been detected in patients with T1D (64–66) and are considered to be the best available markers for predicting the disease. Thus, the regulation of CSR to IgG by AFF3 might be related to these potentially pathogenic antibodies. Although the autoantibodies detected in very early phase or before/without onset of the diseases suggest that the autoantibodies alone are not enough to cause the diseases (21, 65), these autoantibodies might be cooperatively involved in onset and progression of the diseases. Therefore, it is important to further investigate the class-specific effects of autoantibodies and the role of AFF3-related CSR in the pathogeneses of autoimmune diseases. Although B cell depletion therapy with an anti-CD20 antibody is effective for some types of autoimmune diseases (20, 21, 65), targeting a specific molecule that is more relevant to disease pathogenesis is desirable, considering that the immunodeficiency induced by B cell depletion may have detrimental effects, such as an increased risk of infection. In this context, the development of drugs targeting AFF3 may serve as an additional treatment strategy for autoantibody-related diseases that reduces adverse reactions (67).
MATERIALS AND METHODS
Mice
Aff3-floxed and null mice were generated at the Laboratory Animal Resource Center of the University of Tsukuba (fig. S2). A loxP gene fragment was inserted into the mouse Aff3 locus via the CRISPR-Cas9 method. The loxP-containing donor DNA fragments, guide RNA (the target sequences are listed in table S4), and Cas9 were introduced into mouse prenuclear-stage fertilized eggs by microinjection. Genetically modified mice (C57BL/6J background) were obtained by implanting fertilized eggs into the uteri of pseudo-parent mice. The modified Aff3 loci were evaluated by PCR and DNA sequencing. Mice carrying flox insertions and mice harboring deletions of exons 2 to 4 were obtained. The mice were housed under specific pathogen–free conditions and used for experiments at 8 to 12 weeks of age. All animal experiments were performed in accordance with the guidelines established by the animal research committee of Tokushima University. Aicda−/− mice, Cd4-cre transgenic mice, Cd19-cre knock-in mice, and R26LSL-tdTomato mice were gifts from T. Honjo (Kyoto University), M. Kubo (RIKEN), S. Chiba (The University of Tokyo), and M. Murakami (Hokkaido University), respectively.
Flow cytometry
Single-cell suspensions were prepared from mouse lymph node, spleen, or thymus tissue. Splenic cells were resuspended in lysis buffer (150 mM NH4Cl, 14 mM NaHCO3, and 0.1 mM EDTA) to lyse red blood cells for 1 min at room temperature and then washed with RPMI medium. The cells were stained with fluorescence-activated cell sorting (FACS) buffer [phosphate-buffered saline (PBS) with 1% fetal bovine serum and 0.1% NaN3] for 20 to 30 min at 4°C, washed and resuspended in FACS buffer, and analyzed by flow cytometry (FACSCanto II, BD). The data were analyzed using FACSDiva (BD) or Weasel software (version 3.6.2; https://frankbattye.com.au/Weasel/). The antibodies used in the experiments are listed in table S2.
Immunization
OVA (100 μg per mouse; Sigma-Aldrich, A5503) or NP-conjugated chicken gamma-globulin ratio 49 (100 μg per mouse; LGC Biosearch Technologies, N5055E-5) was emulsified in CFA (Sigma-Aldrich, F5881). Thereafter, the mice were immunized intraperitoneally.
Enzyme-linked immunosorbent assay
ELISAs were performed in flat-bottom 96-well plates (Falcon, 353072), and the antibodies used are listed in table S3. The plates were coated with anti-immunoglobulin capture antibodies or antigens at 4°C overnight. The plates were then washed five times with 0.05% PBST (PBS containing 0.05% Tween 20) and blocked with PBS containing 1% bovine serum albumin (BSA) for 2 hours at room temperature. The plates were next washed five times, loaded with diluted serum samples or standards, and incubated for 2 hours at room temperature. The plates were subsequently washed five times, loaded with secondary antibodies, and incubated for 1 hour at room temperature. Then, the plates were washed five times and developed with 3,3′,5,5′-tetramethylbenzidine substrate solution (Invitrogen, 00-4201-56); the reaction was stopped by the addition of 0.25 M H2SO4 (WAKO, 192-04755). The plates were lastly read at 450 nm on a SpectraMax ABS plate reader (Molecular Devices). For the NP-specific assay, plates were coated with NP(2)-BSA (LGC Biosearch Technologies, N-5050XL-10) or NP(25)-BSA (LGC Biosearch Technologies, N-5050H-10) resuspended in PBS.
Mouse malaria model
C57BL/6 mice were used as infected hosts, and P. yoelii 17X was used as the malaria parasite. At the time of P. yoelii inoculation, 10,000 sporozoites were intravenously administered to the mice. The course of infection was monitored by microscopic examination of Giemsa-stained tail blood smears. Mouse serum samples were collected at 50 days after inoculation for ELISA.
Mouse influenza infection
Mice were anesthetized by the inhalation of sevoflurane and then administered the A/Aichi/2/1968 (H3N2) influenza virus (1 × 104 plaque-forming units) via their nasal cavity using a pipette. The mice were weighed daily after infection. After euthanasia by deep anesthesia with sevoflurane or cervical dislocation 14 days after infection, blood was collected and used for the measurement of virus antigen-specific immunoglobulins (IgM, IgG, and IgA) after serum isolation.
B cell culture
For primary B cell culture, single-cell suspensions of mouse splenocytes were prepared by filtering through a 70-μm cell strainer (Falcon, 352350). The cells were centrifuged and resuspended in 1 ml of red blood cell lysis buffer for 1 min at room temperature and then washed two times with RPMI complete medium. Naïve B cells were purified by negative selection using a biotin-conjugated anti-CD43 antibody (Miltenyi Biotec, 130-101-954) and anti-biotin magnetic beads (Miltenyi Biotec, 130-090-485) according to the manufacturer’s protocol. B cells were plated at a density of 5 × 104 to 20 × 104 cells per well in a 24-well plate and cultured in RPMI complete medium. B cells were stimulated with LPS (Sigma-Aldrich, L2654) with or without IL-4 (Miltenyi Biotec, 130-097-757) or IFN-γ (BioLegend, 575304).
Western blotting
Cell pellets were lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer (Nacalai Tesque, 16488-34) with protease inhibitor (Roche, 11836170001), and the lysates were boiled in SDS loading dye. Twenty micrograms of the sample per lane was resolved by SDS–polyacrylamide gel electrophoresis, and the blots were incubated with an anti-AID antibody (clone 1AID-2E11; Invitrogen, MA3-081). They were then incubated with a horseradish peroxidase (HRP)–conjugated goat anti-mouse IgG antibody (31430, Pierce Biotechnology). After detecting AID, the antibodies were removed from the membrane with WB stripping solution (Nacalai Tesque, 05364-55), and the membrane was reprobed with an anti–extracellular signal–regulated kinase 1/2 antibody (clone 137F5; Cell Signaling Technology, #4695) and an HRP-conjugated goat anti-rabbit IgG antibody (Bio-Rad, 170-6515) as a loading control. The bands were detected with ECL Prime Western blotting detection reagent (Sigma-Aldrich, GERPN2236) and with the Odyssey XF (LI-COR). The bands were quantified with ImageJ (68).
Plasmid construction
Mouse Aff1, Aff3, and Aff4 cDNA was amplified from mouse splenic RNA using ReverTra Ace qPCR RT master mix with genomic DNA remover (TOYOBO, FSQ-301) and DNA polymerase KOD Plus Neo (TOYOBO, KOD-401) according to the manufacturer’s instructions. The fragments were cloned into the pGEM-T-easy vector (Promega, A1360) and then subcloned into the pMXs-IRES-GFP retroviral vector (Cell Biolabs, RTV-013). The primers used in the experiments are listed in table S4.
Gene transduction into B cells
Retroviruses were prepared via the transfection of pMXs-IRES-GFP harboring various cDNAs into Plat-E (69) packaging cells with GeneJuice Transfection Reagent (Millipore, 70967-6) according to the manufacturer’s instructions. After changing the medium at 24 hours, the supernatant was collected at 48 and 72 hours after transfection. The debris in the supernatant was removed by using 0.45-μm filters (Sartorius, S7598). Polybrene transfection reagent (Millipore, TR-1003-G) was added to a final concentration of 10 μg/ml. B cells were cultured in 24-well plates for 24 and 48 hours, and the retroviral supernatant was added to each well. The plates were centrifuged at 2600 rpm for 90 min at 30°C. After centrifugation, half of the supernatant was removed, and fresh RPMI complete medium supplemented with LPS and IL-4 was added.
cDNA microarray analysis
RNA was extracted, and genomic DNA was degraded using the RNeasy Plus Mini Kit (QIAGEN, 74134). The quality of the RNA was evaluated with an Agilent 2100 Bioanalyzer. Probe preparation and microarray analyses were performed with a 4x44K whole-genome mRNA microarray (Agilent Technologies). The resulting data were normalized using GeneSpring (Agilent Technologies) software and deposited into the Gene Expression Omnibus database (GSE193735).
Quantitative reverse transcription polymerase chain reaction
RNA was purified from cells using the RNeasy Plus Mini Kit (QIAGEN, 74134) according to the manufacturer’s instructions. cDNA was synthesized using SuperScript IV VILO master mix with ezDNase (Invitrogen, 11766050) according to the manufacturer’s instructions. qPCR experiments were performed with a StepOne (Applied Biosystems) or QuantStudio 5 (Applied Biosystems) system. Two microliters of cDNA and FAST SYBR Green Master Mix (ABI, 4385612) were used in these assays. The data were normalized to Tbp mRNA expression. The primers used in the experiments are listed in table S4.
Semiquantitative PCR
CD43− B cells from mouse splenocytes were cultured at a density of 6.8 × 104 cells per well in 24-well plates with RPMI complete medium containing LPS (10 μg/ml) (Sigma-Aldrich, L2654) and IL-4 (±10 ng/ml; Miltenyi Biotec, 130-097-757) for 4 days. After culture, the cells were harvested and disrupted with sodium chloride tris-EDTA buffer (10 mM tris-HCl, 10 mM EDTA, and 150 mM NaCl) containing proteinase K (0.2 mg/ml). The cells were incubated at 55°C for 2 hours, after which genomic DNA was extracted and purified with phenol/chloroform and isopropanol. Then, the DNA was treated with ribonuclease and purified. IgM-IgG1 chimeric switch regions were amplified from serially diluted genomic DNA with KOD Plus Neo (TOYOBO, KOD-401) according to the manufacturer’s instructions. The samples were analyzed by 1.2% agarose gel electrophoresis. The primers used in the experiments are listed in table S4.
Chromatin immunoprecipitation–quantitative polymerase chain reaction
ChIP was performed with a ChIP-IT High Sensitivity kit (Active Motif, 53040) according to the manufacturer’s instructions. Briefly, the cultured cells were cross-linked with formaldehyde (Thermo Scientific, 28906) at room temperature for 15 min. The reaction was stopped by adding stop solution and shaking for 5 min. The cells were washed with wash buffer and then lysed in chromatin prep buffer. The cells were centrifuged, resuspended in ChIP buffer in an AFA microtube (Covaris, 520045) and sonicated with an S220 instrument (Covaris) with the following parameters: duty cycle, 2%; peak incident power (PIP), 105 W; cycles per burst (CBP), 200; processing time, 12 min; and temperature, 4°C. After removing debris by centrifugation, the supernatant was diluted with ChIP buffer. The indicated antibody (anti-FLAG clone M2 antibody, Sigma-Aldrich, F1804: anti-RNAPII clone D8L4Y, Cell Signaling Technology, 14958) was added to the supernatant, which was then incubated at 4°C overnight with rotation. Then, protein G agarose beads were added, and incubation was performed for 2 hours at 4°C. The beads were washed with AM1 buffer, and the DNA/protein complexes were then eluted with AM4 buffer. Proteinase K was added to the eluted samples, which were then incubated at 55°C for 30 min and at 80°C for 2 hours. The DNA was purified with QIAGEN PB (QIAGEN, 19066) buffer and a gel extraction kit (QIAGEN, 28706). qPCR was performed as described for the qRT-PCR protocol. The primers used in the experiments are listed in table S4.
B cell proliferation
Purified CD43− B cells were suspended in PBS at a concentration of 107 cells/ml, and CellTrace CFSE (Invitrogen, C34570) was added to a final concentration of 5 μM. The cells were incubated for 8 min at 37°C in the dark. Then, RPMI complete medium was added to the cells, which were subsequently incubated for 3 min to stop the reaction. The cells were washed three times with RPMI complete medium and cultured at a density of 2.5 × 104 cells per well in a 48-well plate in the presence of LPS (10 μg/ml) and IL-4 (10 ng/ml). After 3 to 5 days of culture, the dilution of CFSE was assessed by flow cytometry.
Estimation of mutation rates in switch regions
Genomic DNA from class-switching B cells was prepared as described for semiquantitative PCR. IgM-IgG1 chimeric switch regions were amplified from the DNA with KOD Plus Neo (TOYOBO, KOD-401) beginning with the primers IgM-IgG1 SR1 forward and reverse (table S4) for the first 12 cycles followed by the primers IgM-IgG1 SR2 forward and reverse (table S4) for an additional 20 cycles. The amplified DNA fragments were cloned into pCR4 using a Zero Blunt TOPO PCR cloning kit (Thermo Fisher Scientific, 450245). The samples were then sequenced with the T7 primer.
CRISPR and CSR assays in CH12F3-2 cells
CH12F3-2 cells (a gift from T. Honjo, Kyoto University, Japan) were grown at 37°C under 5% (v/v) CO2 in RPMI 1640 (Wisent) supplemented with 10% fetal bovine serum (Wisent), 100 nM β-mercaptoethanol (Bioshop), and 1% penicillin-streptomycin (Wisent). To produce Aff3-knockout clones, 2 million CH12F3-2 cells were transiently transfected using the Amaxa Nucleofector Kit V (Lonza) with 1 μg each of two pSpCas9(BB)-2A-GFP plasmids (pX458; Addgene, #48138) containing two guide RNAs targeting mouse Aff3 exons 2 and 4. The gRNAs were cloned into pX458 with Bbs I digestion. Green fluorescent protein–positive (GFP+) cells were then subcloned by sorting into a 96-well plate. The clones were genotyped for a 1635–base pair (bp) deletion in the Aff3 gene by PCR and verified by qRT-PCR of the Aff3 transcript. The CSR assay was performed in these cells by stimulating them with CIT [rat anti-CD40 (1 μg/ml; clone 1C10, eBioscience), IL-4 (10 ng/ml), and TGF-β1 (1 ng/ml; R&D Systems)] and measuring the proportion of IgA+ cells 3 days later by flow cytometry using anti-IgA conjugated with R-phycoerythrin (Southern Biotech). For the proliferation assay, cells were stained with 5 μM CFSE (CellTrace CFSE Cell Proliferation Kit; Invitrogen). The initial staining and CFSE dilution were measured after 3 days via flow cytometry. The gRNA sequences and primers used for qRT-PCR and genotyping are listed in table S4.
ChIP-qPCR in CH12F3-2 cells
For AID ChIP, 4 million WT and Aff3−/− CH12F3-2 cells were stimulated in 10 ml of medium with CIT and cross-linked with 1% formaldehyde for 8 min at room temperature at 16 hours after stimulation. The reaction was stopped by the addition of glycine to a 125 mM final concentration. The cells were washed twice with cold PBS, harvested, resuspended in RIPA buffer [150 mM NaCl, 1% (v/v) IGEPAL CA-630, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 50 mM tris-HCl (pH 8), 5 mM EDTA, and 1× CPI (complete protease inhibitor)] and sonicated with a Covaris E220 instrument with the following parameters to generate DNA with 200- to 600-bp fragments: time, 10 min; duty factor, 10%; PIP, 140 W; and CBP, 200. The samples were then clarified by the addition of a 1/10 volume of 10% Triton X-100 followed by centrifugation at 20,000 rpm (4°C) for 15 min. An aliquot (5%) was taken to assess the DNA concentration and sonication pattern. For immunoprecipitation, samples were precleared with 20 μl of Dynabeads Protein G (Thermo Fisher Scientific, 10004D) for 1 hour (4°C). For each sample, 5 μg of anti-AID antibody (clone 328.8b; Active Motif, 39885) was coupled to 25 μl of Dynabeads Protein G for 30 min and blocked for 1 hour (4°C) with BT buffer [0.5% Triton X-100, 10 mM tris (pH 8), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, BSA (15 μg/ml), transfer RNA (3 μg/ml), and 1× CPI]. Immunoprecipitation was performed by overnight incubation of 20 μg of precleared chromatin with blocked antibody-bead complexes. The beads were then washed twice (15 min at 4°C) with RIPA buffer, three times with ChIP wash buffer [100 mM tris-HCl (pH 8.5), 0.5 M LiCl, 1% (v/v) IGEPAL CA-630, and 1% (w/v) sodium deoxycholate], and once with 1× tris-EDTA buffer (TE). The immunocomplexes were eluted by incubation for 10 min at 65°C with 100 μl of elution buffer [1% (w/v) SDS, 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 5 μg of proteinase K] followed by incubation overnight at 65°C. DNA was purified using a QIAquick PCR purification kit (QIAGEN) and resuspended in 50 μl of tris-HCl (pH 8) to use as a template in qPCR. Briefly, 10 μl of PCR mixtures containing 1× SYBR Green Mix (Thermo Fisher Scientific, A25741), a 1/10 fraction of ChIP-enriched DNA, and 100 nM primers were set up in 384-well plates. The primers used in the experiments are listed in table S4.
Human eQTL and isotype usage analysis
Gene expression data of immune cells in the ImmuNexUT database (61) were used for the analysis. AFF3 gene expression was compared among immune cells using healthy control samples. eQTL analysis was performed as previously reported (61). The isotype usage of each B cell sample was quantified using MiXCR (version 3.0.13) software. We treated each unique CDR3 sequence as a single event irrespective of the read depth. We defined isotype usage as the ratio of sequences with each isotype among all sequences in an individual. Samples with more than 500 unique clonotypes were used for the analysis. The association between isotype usage and genotype was assessed with a linear regression model adjusted for the clinical diagnosis, technical batch effects, and age.
Statistics
Statistical analyses were performed, and all graphs were constructed with GraphPad Prism 9 (GraphPad Software Inc.). The samples are presented as the mean with or without error bars indicating the SEM. The statistical method used to analyze each data point is shown in the figure legends.
Acknowledgments
We thank C. Kinouchi and M. Toyozaki for technical and editorial assistance.
Funding: This work was supported by the Japan Science and Technology Agency (Moonshot R&D, JPMJMS2025) (to K.Y.), JSPS KAKENHI (JP18K08388) (to S.-i.T.), and the Canadian Institute of Health Research (PJ-155944) (to J.M.D.N.). N.S. and P.G.S. are doctoral fellows, and J.M.D.N. is a distinguished scholar from the Fonds de Recherche du Québec-Santé (FRQ-S).
Author contributions: S.-i.T. and K.Y. conceived and designed the studies. P.G.S., N.S., and J.M.D.N. performed and analyzed the CH12 experiments. M.T., Y.Ma., and C.I. performed and analyzed the Plasmodium infection experiments. Y.Mo. and Y.I. performed and analyzed influenza virus infection experiments. M.O. and K.F. performed the human genetic analyses. S.-i.T., P.G.S., N.S., J.M.D.N., and K.Y. wrote the paper, and all authors reviewed the paper. K.Y. supervised all experiments.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Requests for the CH12-F3 cells could be fulfilled by J.M.D.N. following written consent from the original provider, T. Honjo, Center for Cancer Immunotherapy and Immunobiology, Kyoto University. A material transfer agreement might be required by the original provider.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S5
References
REFERENCES AND NOTES
- 1.Reay W. R., Cairns M. J., Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet. 22, 658–671 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Claussnitzer M., Cho J. H., Collins R., Cox N. J., Dermitzakis E. T., Hurles M. E., Kathiresan S., Kenny E. E., Lindgren C. M., MacArthur D. G., North K. N., Plon S. E., Rehm H. L., Risch N., Rotimi C. N., Shendure J., Soranzo N., McCarthy M. I., A brief history of human disease genetics. Nature 577, 179–189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai X., Kim J., Yang Z., Jurynec M. J., Akie T. E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A., Zhou Y., Grunwald D. J., Lin S., Cantor A. B., Orkin S. H., Zon L. I., TIF1γ controls erythroid cell fate by regulating transcription elongation. Cell 142, 133–143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalleri V., Bettini L. R., Barboni C., Cereda A., Mariani M., Spinelli M., Gervasini C., Russo S., Biondi A., Jankovic M., Selicorni A., Thrombocytopenia and Cornelia de Lange syndrome: Still an enigma? Am. J. Med. Genet. A. 170, 130–134 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Al Ismail A., Husain A., Kobayashi M., Honjo T., Begum N. A., Depletion of recombination-specific cofactors by the C-terminal mutant of the activation-induced cytidine deaminase causes the dominant negative effect on class switch recombination. Int. Immunol. 29, 525–537 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Casellas R., Basu U., Yewdell W. T., Chaudhuri J., Robbiani D. F., Di Noia J. M., Mutations, kataegis and translocations in B cells: Understanding AID promiscuous activity. Nat. Rev. Immunol. 16, 164–176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P. B., Chen H. V., Acharya D., Rando O. J., Fazzio T. G., R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 22, 999–1007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assing K., Nielsen C., Kirchhoff M., Madsen H. O., Ryder L. P., Fisker N., CD4+ CD31+ recent thymic emigrants in CHD7 haploinsufficiency (CHARGE syndrome): A case. Hum. Immunol. 74, 1047–1050 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Bottardi S., Mavoungou L., Milot E., IKAROS: A multifunctional regulator of the polymerase II transcription cycle. Trends Genet. 31, 500–508 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Astle W. J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A. L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M. A., Lambourne J. J., Sivapalaratnam S., Downes K., Kundu K., Bomba L., Berentsen K., Bradley J. R., Daugherty L. C., Delaneau O., Freson K., Garner S. F., Grassi L., Guerrero J., Haimel M., Janssen-Megens E. M., Kaan A., Kamat M., Kim B., Mandoli A., Marchini J., Martens J. H. A., Meacham S., Megy K., O’Connell J., Petersen R., Sharifi N., Sheard S. M., Staley J. R., Tuna S., van der Ent M., Walter K., Wang S.-Y., Wheeler E., Wilder S. P., Iotchkova V., Moore C., Sambrook J., Stunnenberg H. G., Di Angelantonio E., Kaptoge S., Kuijpers T. W., Carrillo-de-Santa-Pau E., Juan D., Rico D., Valencia A., Chen L., Ge B., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yang Y., Guigo R., Beck S., Paul D. S., Pastinen T., Bujold D., Bourque G., Frontini M., Danesh J., Roberts D. J., Ouwehand W. H., Butterworth A. S., Soranzo N., The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167, 1415–1429.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polychronakos C., Li Q., Understanding type 1 diabetes through genetics: Advances and prospects. Nat. Rev. Genet. 12, 781–792 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Ma C., Staudt L. M., LAF-4 encodes a lymphoid nuclear protein with transactivation potential that is homologous to AF-4, the gene fused to MLL in t(4;11) leukemias. Blood 87, 734–745 (1996). [PubMed] [Google Scholar]
- 13.Hiwatari M., Taki T., Taketani T., Taniwaki M., Sugita K., Okuya M., Eguchi M., Ida K., Hayashi Y., Fusion of an AF4-related gene, LAF4, to MLL in childhood acute lymphoblastic leukemia with t(2;11)(q11;q23). Oncogene 22, 2851–2855 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Luo Z., Lin C., Guest E., Garrett A. S., Mohaghegh N., Swanson S., Marshall S., Florens L., Washburn M. P., Shilatifard A., The super elongation complex family of RNA polymerase II elongation factors: Gene target specificity and transcriptional output. Mol. Cell. Biol. 32, 2608–2617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He N., Liu M., Hsu J., Xue Y., Chou S., Burlingame A., Krogan N. J., Alber T., Zhou Q., HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 38, 428–438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F. X., Smith E. R., Shilatifard A., Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 19, 464–478 (2018). [DOI] [PubMed] [Google Scholar]
- 17.ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Z., Lin C., Woodfin A. R., Bartom E. T., Gao X., Smith E. R., Shilatifard A., Regulation of the imprinted Dlk1-Dio3 locus by allele-specific enhancer activity. Genes Dev. 30, 92–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Wang C., Liu X., Yang Q., Ji H., Yang M., Xu M., Zhou Y., Xie W., Luo Z., Lin C., AFF3-DNA methylation interplay in maintaining the mono-allelic expression pattern of XIST in terminally differentiated cells. J. Mol. Cell Biol. 761–769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee D. S. W., Rojas O. L., Gommerman J. L., B cell depletion therapies in autoimmune disease: Advances and mechanistic insights. Nat. Rev. Drug Discov. 20, 179–199 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin S. J. S., Bloom M. S., Robinson W. H., B cell checkpoints in autoimmune rheumatic diseases. Nat. Rev. Rheumatol. 15, 303–315 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Saha T., Sundaravinayagam D., Di Virgilio M., Charting a DNA repair roadmap for immunoglobulin class switch recombination. Trends Biochem. Sci. 46, 184–199 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Chen Z., Wang J. H., Signaling control of antibody isotype switching. Adv. Immunol. 141, 105–164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y., Seija N., Di Noia J. M., Martin A., AID in antibody diversification: There and back again. Trends Immunol. 41, 586–600 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu K., Lieber M. R., Current insights into the mechanism of mammalian immunoglobulin class switch recombination. Crit. Rev. Biochem. Mol. Biol. 54, 333–351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenter A. L., Kumar S., Wuerffel R., Grigera F., AID hits the jackpot when missing the target. Curr. Opin. Immunol. 39, 96–102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortune M. D., Guo H., Burren O., Schofield E., Walker N. M., Ban M., Sawcer S. J., Bowes J., Worthington J., Barton A., Eyre S., Todd J. A., Wallace C., Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat. Genet. 47, 839–846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis S. N., Nsoesie E., Weeks C., Qiao D., Zhang L., Prediction of disease and phenotype associations from genome-wide association studies. PLOS ONE 6, e27175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hertweck A., Evans C. M., Eskandarpour M., Lau J. C. H., Oleinika K., Jackson I., Kelly A., Ambrose J., Adamson P., Cousins D. J., Lavender P., Calder V. L., Lord G. M., Jenner R. G., T-bet activates Th1 genes through mediator and the super elongation complex. Cell Rep. 15, 2756–2770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen R., Bélanger S., Frederick M. A., Li B., Johnston R. J., Xiao N., Liu Y.-C., Sharma S., Peters B., Rao A., Crotty S., Pipkin M. E., In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity 41, 325–338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demarta-Gatsi C., Smith L., Thiberge S., Peronet R., Commere P. H., Matondo M., Apetoh L., Bruhns P., Ménard R., Mécheri S., Protection against malaria in mice is induced by blood stage- arresting histamine-releasing factor (HRF)-deficient parasites. J. Exp. Med. 213, 1419–1428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White W. I., Evans C. B., Taylor D. W., Antimalarial antibodies of the immunoglobulin G2a isotype modulate parasitemias in mice infected with Plasmodium yoelii. Infect. Immun. 59, 3547–3554 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waki S., Uehara S., Kanbe K., Nariuch H., Suzuki M., Interferon-gamma and the induction of protective IgG2a antibodies in non-lethal Plasmodium berghei infections of mice. Parasite Immunol. 17, 503–508 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Cavinato R. A., Bastos K. R. B., Sardinha L. R., Elias R. M., Alvarez J. M., D’Império Lima M. R., Susceptibility of the different developmental stages of the asexual (schizogonic) erythrocyte cycle of Plasmodium chabaudi chabaudi to hyperimmune serum, immunoglobulin (Ig)G1, IgG2a and F(ab′)2 fragments. Parasite Immunol. 23, 587–597 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Wang Q., Kieffer-Kwon K. R., Oliveira T. Y., Mayer C. T., Yao K., Pai J., Cao Z., Dose M., Casellas R., Jankovic M., Nussenzweig M. C., Robbiani D. F., The cell cycle restricts activation-induced cytidine deaminase activity to early G1. J. Exp. Med. 214, 49–58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou S., Upton H., Bao K., Schulze-Gahmen U., Samelson A. J., He N., Nowak A., Lu H., Krogan N. J., Zhou Q., Alber T., HIV-1 Tat recruits transcription elongation factors dispersed along a flexible AFF4 scaffold. Proc. Natl. Acad. Sci. U.S.A. 110, E123–E131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi S., Li Z., Schulze-Gahmen U., Stjepanovic G., Zhou Q., Hurley J. H., Structural basis for ELL2 and AFF4 activation of HIV-1 proviral transcription. Nat. Commun. 8, 14076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He N., Chan C. K., Sobhian B., Chou S., Xue Y., Liu M., Alber T., Benkirane M., Zhou Q., Human polymerase-associated factor complex (PAFc) connects the super elongation complex (SEC) to RNA polymerase II on chromatin. Proc. Natl. Acad. Sci. U.S.A. 108, E636–E645 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavri R., R loops in the regulation of antibody gene diversification. Genes 8, 154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ribeiro de Almeida C., Dhir S., Dhir A., Moghaddam A. E., Sattentau Q., Meinhart A., Proudfoot N. J., RNA helicase DDX1 converts RNA G-quadruplex structures into R-loops to promote IgH class switch recombination. Mol. Cell 70, 650–662.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Begum N. A., Izumi N., Nishikori M., Nagaoka H., Shinkura R., Honjo T., Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J. Biol. Chem. 282, 731–742 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Yewdell W. T., Kim Y., Chowdhury P., Lau C. M., Smolkin R. M., Belcheva K. T., Fernandez K. C., Cols M., Yen W. F., Vaidyanathan B., Angeletti D., McDermott A. B., Yewdell J. W., Sun J. C., Chaudhuri J., A hyper-IgM syndrome mutation in activation-induced cytidine deaminase disrupts G-quadruplex binding and genome-wide chromatin localization. Immunity 53, 952–970.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ota M., Nagafuchi Y., Hatano H., Ishigaki K., Terao C., Takeshima Y., Yanaoka H., Kobayashi S., Okubo M., Shirai H., Sugimori Y., Maeda J., Nakano M., Yamada S., Yoshida R., Tsuchiya H., Tsuchida Y., Akizuki S., Yoshifuji H., Ohmura K., Mimori T., Yoshida K., Kurosaka D., Okada M., Setoguchi K., Kaneko H., Ban N., Yabuki N., Matsuki K., Mutoh H., Oyama S., Okazaki M., Tsunoda H., Iwasaki Y., Sumitomo S., Shoda H., Kochi Y., Okada Y., Yamamoto K., Okamura T., Fujio K., Dynamic landscape of immune cell-specific gene regulation in immune-mediated diseases. Cell 184, 3006–3021.e17 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Buniello A., Macarthur J. A. L., Cerezo M., Harris L. W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., Suveges D., Vrousgou O., Whetzel P. L., Amode R., Guillen J. A., Riat H. S., Trevanion S. J., Hall P., Junkins H., Flicek P., Burdett T., Hindorff L. A., Cunningham F., Parkinson H., The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orrù V., Steri M., Sidore C., Marongiu M., Serra V., Olla S., Sole G., Lai S., Dei M., Mulas A., Virdis F., Piras M. G., Lobina M., Marongiu M., Pitzalis M., Deidda F., Loizedda A., Onano S., Zoledziewska M., Sawcer S., Devoto M., Gorospe M., Abecasis G. R., Floris M., Pala M., Schlessinger D., Fiorillo E., Cucca F., Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat. Genet. 52, 1036–1045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin C., Smith E. R., Takahashi H., Lai K. C., Martin-Brown S., Florens L., Washburn M. P., Conaway J. W., Conaway R. C., Shilatifard A., AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 37, 429–437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pucella J. N., Chaudhuri J., AID invited to the G4 summit. Mol. Cell 67, 355–357 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Qiao Q., Wang L., Meng F.-L., Hwang J. K., Alt F. W., Wu H., AID recognizes structured DNA for class switch recombination. Mol. Cell 67, 361–373.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pefanis E., Wang J., Rothschild G., Lim J., Chao J., Rabadan R., Economides A. N., Basu U., Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514, 389–393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basu U., Meng F. L., Keim C., Grinstein V., Pefanis E., Eccleston J., Zhang T., Myers D., Wasserman C. R., Wesemann D. R., Januszyk K., Gregory R. I., Deng H., Lima C. D., Alt F. W., The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144, 353–363 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., San-Martin B. R., Barreto V., Nieland T. J., Root D. E., Casellas R., Nussenzweig M. C., Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143, 122–133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Methot S. P., Litzler L. C., Subramani P. G., Eranki A. K., Fifield H., Patenaude A.-M., Gilmore J. C., Santiago G. E., Bagci H., Côté J.-F., Larijani M., Verdun R. E., Di Noia J. M., A licensing step links AID to transcription elongation for mutagenesis in B cells. Nat. Commun. 9, 1248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willmann K. L., Milosevic S., Pauklin S., Schmitz K. M., Rangam G., Simon M. T., Maslen S., Skehel M., Robert I., Heyer V., Schiavo E., Reina-San-Martin B., Petersen-Mahrt S. K., A role for the RNA pol II-associated PAF complex in AID-induced immune diversification. J. Exp. Med. 209, 2099–2111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas-Claudepierre A.-S., Schiavo E., Heyer V., Fournier M., Page A., Robert I., Reina-San-Martin B., The cohesin complex regulates immunoglobulin class switch recombination. J. Exp. Med. 210, 2495–2502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melko M., Douguet D., Bensaid M., Zongaro S., Verheggen C., Gecz J., Bardoni B., Functional characterization of the AFF (AF4/FMR2) family of RNA binding proteins: Insights into the molecular pathology of FRAXE intellectual disability. Hum. Mol. Genet. 20, 1873–1885 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Begum N. A., Stanlie A., Nakata M., Akiyama H., Honjo T., The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation. J. Biol. Chem. 287, 32415–32429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stanlie A., Begum N. A., Akiyama H., Honjo T., The DSIF subunits Spt4 and Spt5 have distinct roles at various phases of immunoglobulin class switch recombination. PLOS Genet. 8, e1002675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okazaki I. M., Okawa K., Kobayashi M., Yoshikawa K., Kawamoto S., Nagaoka H., Shinkura R., Kitawaki Y., Taniguchi H., Natsume T., Iemura S. I., Honjo T., Histone chaperone Spt6 is required for class switch recombination but not somatic hypermutation. Proc. Natl. Acad. Sci. U.S.A. 108, 7920–7925 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Izumi K., Nakato R., Zhang Z., Edmondson A. C., Noon S., Dulik M. C., Rajagopalan R., Venditti C. P., Gripp K., Samanich J., Zackai E. H., Deardorff M. A., Clark D., Allen J. L., Dorsett D., Misulovin Z., Komata M., Bando M., Kaur M., Katou Y., Shirahige K., Krantz I. D., Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat. Genet. 47, 338–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim A., Han L., Yu K., Immunoglobulin class switch recombination is initiated by rare cytosine deamination events at switch regions. Mol. Cell. Biol. 40, e00125-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wigton E. J., Mikami Y., McMonigle R. J., Castellanos C. A., Wade-Vallance A. K., Zhou S. K., Kageyama R., Litterman A., Roy S., Kitamura D., Dykhuizen E. C., Allen C. D. C., Hu H., O’Shea J. J., Ansel K. M., MicroRNA-directed pathway discovery elucidates an miR-221/222–mediated regulatory circuit in class switch recombination. J. Exp. Med. 218, e20201422 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mandal R., Becker S., Strebhardt K., Targeting CDK9 for anti-cancer therapeutics. Cancers 13, 2181 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Almeida L. N., Clauder A., Meng L., Ehlers M., Arce S., Manz R. A., MHC haplotype and B cell autoimmunity: Correlation with pathogenic IgG autoantibody subclasses and Fc glycosylation patterns. Eur. J. Immunol. 52, 197–203 (2022). [DOI] [PubMed] [Google Scholar]
- 64.Vandamme C., Kinnunen T., B cell helper T cells and type 1 diabetes. Scand. J. Immunol. 92, e12943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dayan C. M., Besser R. E. J., Oram R. A., Hagopian W., Vatish M., Bendor-Samuel O., Snape M. D., Todd J. A., Preventing type 1 diabetes in childhood. Science 373, 506–510 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Bluestone J. A., Buckner J. H., Herold K. C., Immunotherapy: Building a bridge to a cure for type 1 diabetes. Science 373, 510–516 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Liang K., Smith E. R., Aoi Y., Stoltz K. L., Katagi H., Woodfin A. R., Rendleman E. J., Marshall S. A., Murray D. C., Wang L., Ozark P. A., Mishra R. K., Hashizume R., Schiltz G. E., Shilatifard A., Targeting processive transcription elongation via SEC disruption for MYC-induced cancer therapy. Cell 175, 766–779.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morita S., Kojima T., Kitamura T., Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Xu J., Husain A., Hu W., Honjo T., Kobayashi M., Ape1 is dispensable for S-region cleavage but required for its repair in class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 111, 17242–17247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin L., Gerth A. J., Peng S. L., CpG DNA redirects class-switching towards “Th1-like” Ig isotype production via TLR9 and MyD88. Eur. J. Immunol. 34, 1483–1487 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Wang N. S., McHeyzer-Williams L. J., Okitsu S. L., Burris T. P., Reiner S. L., McHeyzer-Williams M. G., Divergent transcriptional programming of class-specific B cell memory by T-bet and RORα. Nat. Immunol. 13, 604–611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R. F., Nussenzweig A., Nussenzweig M. C., H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197, 1767–1778 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dinkelmann M., Spehalski E., Stoneham T., Buis J., Wu Y., Sekiguchi J. M., Ferguson D. O., Multiple functions of MRN in end-joining pathways during isotype class switching. Nat. Struct. Mol. Biol. 16, 808–813 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rickert R. C., Roes J., Rajewsky K., B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25, 1317–1318 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cortizas E. M., Zahn A., Hajjar M. E., Patenaude A.-M., Di Noia J. M., Verdun R. E., Alternative end-joining and classical nonhomologous end-joining pathways repair different types of double-strand breaks during class-switch recombination. J. Immunol. 191, 5751–5763 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S5
References