Abstract
F and R100-1 are closely related, derepressed, conjugative plasmids from the IncFI and IncFII incompatibility groups, respectively. Heteroduplex mapping and genetic analyses have revealed that the transfer regions are extremely similar between the two plasmids. Plasmid specificity can occur at the level of relaxosome formation, regulation, and surface exclusion between the two transfer systems. There are also differences in pilus serology, pilus-specific phage sensitivity, and requirements for OmpA and lipopolysaccharide components in the recipient cell. These phenotypic differences were exploited in this study to yield new information about the mechanism of pilus synthesis, mating pair stabilization, and surface and/or entry exclusion, which are collectively involved in mating pair formation (Mpf). The sequence of the remainder of the transfer region of R100-1 (trbA to traS) has been completed, and the complete sequence is compared to that of F. The differences between the two transfer regions include insertions and deletions, gene duplications, and mosaicism within genes, although the genes essential for Mpf are conserved in both plasmids. F+ cells carrying defined mutations in each of the Mpf genes were complemented with the homologous genes from R100-1. Our results indicate that the specificity in recipient cell recognition and entry exclusion are mediated by TraN and TraG, respectively, and not by the pilus.
The F plasmid is a naturally derepressed conjugative plasmid of 100 kb that was used in the early 1970s to investigate the genetics of bacterial conjugation. With the complete sequence of the F transfer region becoming available in 1994 (28), the wealth of data on the five discernible functions in the F transfer region: DNA transport, F-pilus assembly and retraction, mating pair stabilization, surface and entry exclusion, and regulation could be reconciled with information gleaned from computer analyses and data bank searches for homologues of known function. While homologies between different transfer systems were often noted, the functions of only a few gene products were discerned by using this approach.
Studies on the F pilus revealed that it is composed of a major subunit, pilin, of 70 amino acids. Pilin is processed from a 121-amino-acid precursor, propilin, encoded by the traA gene. Propilin is inserted in the inner membrane through the action of TraQ, an F-pilin-specific chaperone in a process which requires ATP and an active proton motive force (43). Propilin is cleaved to pilin by the host leader peptidase and is acetylated at its N terminus by TraX (54). Mature pilin is stored as a large pool in the inner membrane, where it is assembled into a functional pilus filament by the assembly proteins TraL, -E, -K, -B, -V, -C, -F, -W, -U, and -H, TrbC, and the N-terminal portion of TraG. The F pilus is also capable of retraction or disassembly presumably into the host cell; however, it has been difficult to discern which proteins are responsible for this process, with only TrbI being named as a candidate (49). TraN and the C-terminal portion of TraG are important for mating pair stabilization (Mps), a process that allows F+ cells to mate more efficiently in liquid media and to resist disaggregation by shear forces or the addition of chaotropic agents such as sodium dodecyl sulfate (50). Two proteins are involved in preventing redundant plasmid exchange between F+ cells: TraT, an outer membrane protein, blocks mating pair stabilization (surface exclusion) between donor cells, while TraS, an inner membrane protein, blocks the signal for DNA transfer between donor cells (entry exclusion) (2).
While substantial progress has been made in understanding the protein-DNA interactions at the oriT (origin of transfer) region prior to transfer, as well as the regulation of this process, little is known about the functions of the proteins involved in pilus assembly, mating pair stabilization, or exclusion. In most cases, mutations in the genes for these functions resulted in a loss of pili and/or DNA transfer leading to the conclusion that the F pilus is intimately involved in DNA transfer. Computer analysis of the F transfer region sequence has revealed that orf169 encodes a transglycolase related to the lysozyme family (8); TraB is a homologue and/or analogue of VirB10, suggesting that it might be part of a periplasmic complex similar to that for the VirB proteins (13). However, little else is known about the function of the majority of mating pair formation (Mpf) gene products.
The R100 (100-kb) plasmid, also known as NR1, R222 (84), which carries multiple drug resistance, was able to repress F transfer through the process of fertility inhibition (58). The pili of the derepressed mutant, R100-1 (73), are serologically distinct from F pili (27). Cells carrying R100-1 show reduced sensitivity to F-specific phages. Unlike F, R100-1 pili do not attach the isometric RNA phages such as R17 laterally and, although the filamentous phages such as f1 or M13 bind to the pilus tip, they do not infect the cells efficiently (83). Unlike F+ cells, R100-1+ cells do not require the outer membrane protein, OmpA, or the pyrophosphorylethanolamine (PPEA) group in the inner core of the lipopolysaccharide (LPS) of the recipient cell for efficient conjugation (5). Both plasmids specify different surface (TraT) and entry (TraS) exclusion systems (83). The genes for DNA transport (TraI, -M, and -Y but not TraD) and regulation of conjugation (TraJ, -M, and -Y and the antisense RNA, FinP, but not FinO) are specific for their cognate plasmid (reviewed in reference 39).
In a study on the allelic specificity of related F-like plasmids, Willetts and Maule (83) concluded that, unlike the genes that conferred plasmid specificity, the genes for Mpf were interchangeable. However, that analysis used the complete R100-1 plasmid to complement mutations in F, where subtle differences between the two plasmids could have been overlooked. Studies on the effect of complementing a traA (pilin) mutation in F with cloned pilin genes from R100-1 and ColB2, another F-like plasmid, revealed that the sequence differences in pilin do not determine the requirement for OmpA and PPEA in the recipient cell, surface exclusion specificity or sensitivity to f1 phage infection (5). These results suggested that there was another protein either at the pilus tip or in the outer membrane of the donor cell responsible for identifying these moieties on the recipient cell surface. Consequently, we undertook to complete the sequence of the R100-1 transfer region, which included the remaining genes for Mpf. The sequences for traA (27), traL to traP (4), and orfJ to traF (39) were combined with the sequence of the 6.4-kb region (trbA to traS) reported here to generate a complete view of the transfer region of R100-1. Complementation data revealed that the 15 genes known to be involved in pilus synthesis and assembly are interchangeable between the two plasmids and that none of these proteins imparts specificity to the mating system. TraN was found to be involved in the requirement for OmpA and PPEA in the recipient cell (40). TraG is apparently involved in recognizing TraS during the process of entry exclusion, a phenomenon which provides interesting clues to the mechanism of signal transmission during the initiation of the transfer process.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The Escherichia coli strains used in this study include ED2149 [F− lacΔU124 Δ(nadA aroG gal att λ bio)], ED24 (F− Lac− Spcr), XK1200 (ED2149 gyrA), JE2571 (leu thr str fla pil), MC4100 (F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flb3501 deoC1 ptsF25 rbsR), and DH5α (hsdR17 ΔlacU169 φ80 lacZΔM15 recA1 supE44). A list of F and R100-1 plasmids and chimeric plasmids derived from F and R100-1 are given in Tables 1 and 2, respectively. Bacterial cultures were grown in Luria-Bertani (LB) medium with appropriate antibiotics at the following concentrations: ampicillin (50 μg/ml), kanamycin (30 μg/ml), streptomycin (200 μg/ml), and naladixic acid (40 μg/ml).
TABLE 1.
Derivatives of the F plasmid carrying mutations in tra and trb genes involved in pilus synthesis
| Gene | Flac mutanta | kanb or CATc insertion mutant | Reference |
|---|---|---|---|
| traA | pOX38A::CATc | 5 | |
| Flac traA1 | 82 | ||
| traL | Flac traL311d | 81 | |
| traE | Flac traE18 | 82 | |
| traK | Flac traK105d | 82 | |
| traB | Flac traB2 | 82 | |
| traV | Flac traV569 | 19 | |
| traC | Flac traC5 | 82 | |
| Flac traC1044 | 70 | ||
| trbI | pOX38-trbI463b | 49 | |
| traW | Flac traW546 | 53 | |
| traU | Flac traU526 | pOX38-traU347b | 55 |
| trbC | pOX38-trbC460b | 48 | |
| traN | pOX38N::CATc | 40 | |
| traF | Flac traF13 | 82 | |
| traQ | pOX38-traQ238b | 37 | |
| traH | Flac traH80 | 81 | |
| traG | Flac traG101d | 25 | |
| FlactraG106 | |||
| traX | pOX38-traX482b | 46 |
Except where noted, all Flac tra mutants carry an amber mutation. These mutants were tested in strain ED2149.
kan (kanamycin resistance) insertion mutants were tested in strain XK1200.
CAT insertion mutants were tested in strain XK1200.
traK105 and traG106 are frameshift mutations, and traL311 is a UGA nonsense mutation (54).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pBK8 | 3.5-kb NsiI fragment from R100-1 (traN-traF) in pBS/SK+; Apr | 40 |
| pKAF2 | 3.0-kb fragment from F (traK-traP) in BamHI-EcoRI sites of pUC118; Apr | This work |
| pKAF3 | 0.88-kb fragment from F (traX) in SmaI-HindIII sites of pT7-3; Apr | This work |
| pKAR1 | 11.6-kb EcoRI fragment from R100-1 (traK-trbC) in pT7.4; Apr | This work |
| pKAR2 | 3.0-kb fragment from R100-1 (traK-traP) in BamHI-EcoRI sites of pUC118; Apr | This work |
| pKAR3 | 2.4-kb BamHI-PstI fragment from R100-1 (traY-traK) in pACYC177; Kmr | This work |
| pKAR4 | 2.1-kb SmaI-SacI fragment from R100-1 (trbE-trbA) in SmaI site of pACYC177; Apr | This work |
| pKAR5 | 5.1-kb SacI-EcoRI fragment from R100-1 (traQ-traH) in SmaI-HindIII sites of pACYC177; Apr | This work |
| pKAR6 | 3.0-kb fragment from R100-1 (traG) in BamHI-HindIII sites of pT7.3; Apr | This work |
| pKAR7 | 0.82-kb fragment from R100-1 (traX) in PstI-HindIII sites of pT7-3; Apr | This work |
| pKI164 | 2.6-kb HincII fragment from F (traP-traR) in pACYC177; Apr | 57 |
| pKI175 | 6.2-kb AvaI fragment from F (traW-traQ) in pACYC177; Apr | 85 |
| pKI375 | 3.0-kb Asp7001 fragment carrying F (trbC-traF) genes; pBS/SK+; Apr | 47 |
| pLF164 | 1.4-kb PstI fragment of F (traA-traE) in pUC8, Apr | This work |
| pLF107 | 1.5-kb SmaI fragment of R100-1 (traA-traE) in pUC8, Apr | This work |
| pRS29 | EcoRI f1 and f15 fragments from F (traV-traH) in pSC101; Tcr | 72 |
| pRS27 | EcoRI f6 and f15 fragments from F (traM-traR) in pSC101; Tcr | 72 |
| pRS31 | EcoRI f2, f17, and f19 fragments from F (traS-finO::IS3) in pSC101; Tcr | 72 |
| pRS1670 | F traG in pGEX-1 vector; Apr | 25 |
| pBluescript SK(+) | ColE1 ori; Apr | Stratagene |
| pACYC177 | p15A ori; Apr Kmr | 11 |
| pT7-3, pT7-4 | T7 promoter; ColE1 ori; Apr | 74 |
| pSC101 | Tcr | 15 |
| pOX38-Km | pOX38-Km Tra+ RepFIA+ Kmr f1 HindIII fragment of F plus HindIII fragment of Tn5 | 10 |
| R100-1 | R100-1 Tra+ Cmr Far Smr Spr Sur Tcr | 4 |
DNA preparation.
The 100-kb R100-1 plasmid was purified by the method of Humphries et al. (34). A 2-ml overnight culture of E. coli JE2571 harboring the R100-1 plasmid was inoculated into 1 liter of LB broth containing streptomycin (20 μg/ml), and the culture was grown overnight. Cells were harvested by centrifugation at 2,000 × g for 15 min at 4°C. The cell pellet was resuspended in 20 ml of 25% sucrose–50 mM Tris-HCl (pH 8.0) and incubated on ice for 5 min. Three milliliters of lysozyme solution (20 mg of lysozyme per ml in 0.25 M EDTA [pH 8.0]) was added and mixed by inversion, and the mixture was further incubated on ice for 5 min. Then, 20 ml of 0.25 M EDTA (pH 8.0) was added, mixed by inversion, and incubated on ice for 30 min. After the addition of 30 ml of Triton lysis solution (4 ml of Triton X-100; 50 ml of 0.25 EDTA, pH 8.0; 10 ml of 1 M Tris-HCl, pH 8.0; and 2 ml of a 5-mg/ml mixture of RNase A per 200 ml of solution), the mixture was further incubated on ice for 60 min with periodic mixing by gentle inversion. The supernatant was collected after centrifugation at 12,000 × g for 2 h at 4°C. After the addition of 12 ml of 50% polyethylene glycol 8000 and 4 ml of 5 M NaCl, the DNA was precipitated at 4°C overnight. The precipitate was collected following centrifugation at 3,000 × g for 15 min at 4°C. The precipitate was dissolved in 23 ml of CsCl in TE buffer (10 mM Tris-HCl–1 mM EDTA, pH 7.5; density, 1.397 g/cm3) and was recentrifuged at 12,000 × g for 15 min at 4°C. The liquid portion was transferred to a solid-wall ultracentrifuge tube, to which 200 μl of ethidium bromide (10 mg/ml) was added. The gradient was formed in a 50.2 Ti fixed angle rotor in a Beckman ultracentrifuge at 38,000 rpm for 48 h at 18°C. The appropriate band was drawn off with a syringe and was dialyzed extensively in TES buffer (40 ml of 1 M Tris-HCl [pH 8.0]–16 ml of 0.25 M EDTA [pH 8.0]–8 ml of 0.5 M NaCl in 4 liters of water). After phenol extraction, the DNA was recovered by ethanol precipitation and stored in TE buffer at −20°C. Small preparations of DNA were prepared by the method of Birnboim and Doly (9) or by using a Qiagen kit (40).
Recombinant DNA techniques, DNA sequencing, and PCR.
Techniques for cloning restriction fragments or PCR fragments were as described previously (68). Restriction endonuclease digestion, generation of blunt ends, and ligation of DNA molecules were performed as described by Ausubel et al. (7). All enzymes were supplied by Boehringer Mannheim except Vent polymerase (New England Biolabs). The method for generating 3′ deletions in pKAR-5, as well as sequencing, either by the dideoxy sequencing method or automated sequencing, has been described previously (39). All sequences that diverged from the F sequence were sequenced in both directions, while sequences that were nearly completely homologous to F were sequenced more than once in the same direction. All restriction sites used in cloning were sequenced over by using other, overlapping clones or PCR products to ensure that the sequences were complete. The sequences of primers used in this project are available upon request.
Immunogold electron microscopy.
The procedure for immunogold electron microscopy was as described in Rondot et al. (66). The monoclonal antibody (MAb) JEL92 (29) was used at a 1:500 dilution and gold-conjugated anti-mouse goat antibodies (EY Labs) were used at a 1:100 dilution.
Phage sensitivity, mating, and surface exclusion assays.
Mating assays, as well as surface exclusion assays, have been described by Anthony et al. (5) and Klimke and Frost (40). The surface exclusion index is the ratio of transconjugants formed in the absence or the presence of an exclusion system. Flac donors were selected on MacConkey lactose medium (68) containing either ampicillin or kanamycin, while transconjugants were selected on M9 minimal medium (7) supplemented with 0.2% lactose and appropriate antibiotics.
Phage sensitivity assays for f1 and R17 phages were determined by using a modified procedure described by Frost and Paranchych (30). Approximately 107 cells of the bacteria to be tested were mixed with 3 ml of 0.7% LB top agar (7) and used as an overlay lawn on LB agar plates containing appropriate antibiotics. Both f1 and R17 phage suspensions containing approximately 1011 PFU/ml were serially diluted in phosphate-buffered saline, pH 7.3 (PBS [7]), and 10 μl of the various dilutions (ranging from 109 to 101 PFU) were spotted onto the lawn. After incubation at 37°C overnight, lysis at various dilutions were observed, and the efficiency of plating was scored as follows: S, fully sensitive, lysis observed with 102 PFU; S (1%), moderately sensitive, lysis observed with 104 PFU; (R), weakly sensitive, lysis observed with 108 PFU; and R, resistant, no lysis.
Sensitivity to M13K07 (79) was determined by incubating 107 cells (typically 200 μl in a 1.5-ml tube) with M13K07 at a multiplicity of infection of 10 for 10 min on ice to allow attachment. The cells were then incubated at 37°C for 10 min to allow infection. The cells were pelleted at low speed in an Eppendorf microcentrifuge and washed once with fresh LB broth. They were incubated for a further 30 min at 37°C, serially diluted, and spotted on plates containing kanamycin. The sensitivity to M13K07 was reported as the number of kanamycin-resistant colonies per 107 cells.
RESULTS
Updating the sequences of the R100-1 and F transfer regions.
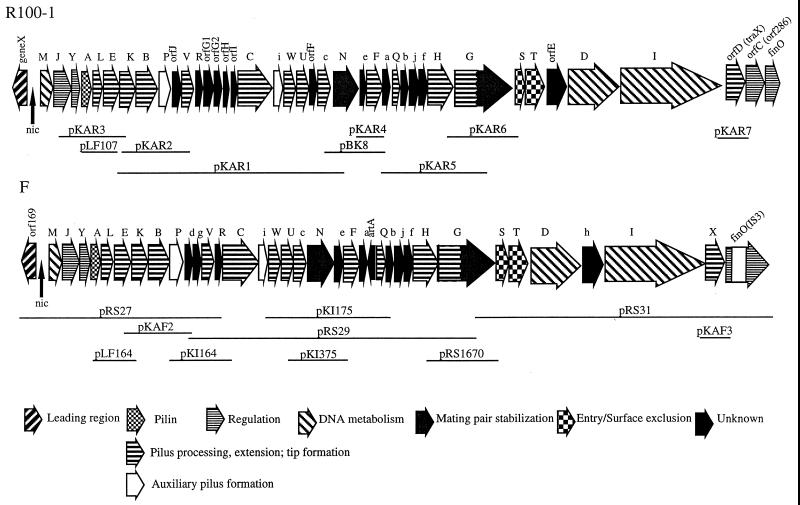
Portions of the sequence of the R100 or R100-1 transfer regions have been presented previously. These include geneX (22), the oriT region (51), traM (21), traJ and traY (23, 35), traA (27), traL-traP (4), orfJ-traF (39), traT (60), traT-traI (86), and finO-orfA (52, 86). The entire sequence including trbA-traS which is determined in this study is included under the accession number AF112468. A comparison of the entire R100-1 and F transfer regions is presented in Fig. 1. Four sequencing errors were identified in the F traD sequence first presented by Jalajakumari and Manning (36) and reprinted in Frost et al. (28). These errors result in frameshift mutations over short regions of the gene. When corrected (sequence correction incorporated into U01159) the homology between TraD of R100-1 and F increases, with the two proteins being virtually identical over the N-terminal region. Two important mutations in traD were also sequenced. The traD8 mutation, which is associated with loss of DNA transfer and increased piliation (6), is due to a G-to-A transition which changes a tryptophan codon to an amber at position 103. The temperature-sensitive mutation traD39 characterized by decreased DNA transfer at the nonpermissive temperature (63) is due to a C-to-T transition which results in a proline to leucine substitution at position 396.
FIG. 1.
Comparison of the F and R100-1 transfer operons with the chimeric plasmids used in this study depicted below. Detailed information on the clones is given in Table 2.
Differences between F and R100-1 transfer regions.
A compilation of the gene products encoded by F and R100-1 plasmids, the percent similarity and identity between each protein, and a brief description of their function or important characteristics is given in Table 3. In the new sequence presented here (trbA-traS), major differences in the sequences of the inessential genes trbA and trbF were identified between F and R100-1. In addition, artA, a gene which maps between trbA and traQ in F and is oriented opposite to the direction of transcription of the transfer region, is missing in R100-1. We have continued to use the orf designations initiated by Yoshioka et al. (86), where the gene order is orfC, finO, orfB, and orfA at the distal end of the operon. orfC is designated orf286 in R6-5 and is missing in F (76). The fertility inhibition gene finO is interrupted by an IS3 element in F (12) and contains a frameshift mutation in R100-1 (87) which is responsible for the derepressed phenotype of both plasmids. The orfD gene in R100-1 is the homologue of F traX (54), whose gene product is required for N-terminal acetylation of pilin. The orfE gene lies between traT and traD in R100-1 and has no known function. orfF, orfG1, orfG2, orfH, orfI, and orfJ have been discussed in Klimke et al. (39) and are summarized in Table 3.
TABLE 3.
Comparison of the gene products of the R100-1/F transfer regions
| R100-1/F gene product | No. of aa residues | % Similarity/identity | Function, comments, and location | Referencea |
|---|---|---|---|---|
| GeneX/Orf169 | 169/169 | 96.5/94.1 | Transglycosylase; IMb | 8, 22, 42 |
| TraM | 127/127 | 95.3/89.0 | Relaxosome formation; interacts with TraD | 1, 17, 18, 21, 51, 64 |
| TraJ | 223/229 | 47.5/22.6 | Positive regulator of tra operon | 35, 75 |
| TraY | 75/131 | 58.5/38.6c | Positive regulator; relaxosome formation; homologous to N and C domains of F TraY | 35, 59, 75 |
| TraA | 119/121 | 92.4/89.1 | Pilin subunit; IM | 27, 44, 66, 71 |
| TraL | 91/91 | 100/98.9 | Pilus assembly; IM; pilus locator | 4 |
| TraE | 188/188 | 95.2/93.6 | Pilus assembly; IM | 4 |
| TraK | 242/242 | 99.2/98.8 | Pilus assembly; periplasm | 4 |
| TraB | 486/475 | 97.9/97.3 | Pilus assembly, IM | 4 |
| TraP | 195/196 | 88.7/82.1 | Inessential; IM | 4 |
| OrfJ/TrbD-TrbGd | 106/65+83 | 83.1/78.5 (D) | F TrbD and TrbG are fused in R100-1 | 39 |
| 49.4/24.7 (G) | No known function | |||
| TraV | 171/171 | 97.7/97.1 | Pilus assembly; OM lipoprotein | 19, 39 |
| TraR | 79/73 | 97.3/97.3 | C4-type zinc finger; no known function | 19, 39 |
| OrfG1 | 138/− | No F equivalent; no known function | 39 | |
| OrfG2 | 149/− | OrfG2 = OrfG1 (71.5/61.3% similarity/identity) | 39 | |
| OrfH | 73/− | No F equivalent; no known function | 39 | |
| OrfI | 115/− | No F equivalent; no known function | 39 | |
| TraC | 876/875 | 99.3/98.8 | Pilus assembly/outgrowth; IM in presence of F transfer proteins; ATP/GTP binding motif | 39 |
| TrbI | 128/128 | 93.8/93.8 | Pilus retraction; IM | 39 |
| TraW | 210/210 | 100/99.5 | Pilus assembly; periplasm | 39 |
| TraU | 330/330 | 99.1/98.5 | DNA transfer; periplasm | 39 |
| OrfF | 103/− | No F equivalent; no known function | 39 | |
| TrbC | 212/212 | 93.4/91.5 | Pilus assembly; periplasm | 39 |
| TraN | 617/602 | 82.3/73.6 | Mating pair stabilization; 20 conserved cys; OM | 39, 40 |
| TrbE | 63/55 | 95.3/93.0 | No known function | 39 |
| TraF | 247/247 | 99.2/98.4 | Pilus assembly; periplasm | 39 |
| TrbA | 115/115 | 74.8/53.0 | No known function | This work |
| ArtA | −/104 | No R100-1 equivalent; no known function | This work | |
| TraQ | 94/94 | 97.9/97.9 | Pilin chaperone; IM | This work |
| TrbB | 181/179 | 93.9/89.9 | No known function | This work |
| TrbJ | 95/93 | 97.9/95.7 | No known function | This work |
| TrbF | 130/126 | 64.3/46.0 | No known function | This work |
| TraH | 457/458 | 94.1/87.7 | Pilus assembly; periplasm | This work |
| TraG | 940/938 | 95.7/93.0 | Pilus assembly; Mps; IM | This work |
| TraG* | 489/487 | 93.4/89.7 | Mps; C-terminal domain of TraG; periplasm | This work |
| TraS | 163/149 | 40.7/13.1 | Entry exclusion; IM | This work and reference 60 |
| TraT | 243/244 | 99.6/99.2 | Surface exclusion; lipoprotein, OM | 60 |
| OrfE | 245/− | No F equivalent; no known function | 86 | |
| TraD | 738/717 | 97.6/96.6 | DNA transport; RNA (I) phageSe; two ATP/GTP binding motifs; docking protein | 18, 86 |
| TrbH | −/239 | No R100-1 equivalent; no known function | 86 | |
| TraI | 1756/1756 | 97.5/96.0 | oriT-specific relaxase; helicase; two nucleoside triphosphate binding motifs | 33, 59, 86 |
| TraI* | 802/802 | 97.6/96.8 | C-terminal domain of TraI; no known function | |
| OrfD/TraX | 248/248 | 93.9/90.3 | Acetylates N terminus of pilin; IM | 46, 86 |
| OrfC | 286/ | No F equivalent; increases FinO levels in cis | 76, 86 | |
| FinO | 186/186 | 96.8/96.2 | Represses TraJ via FinP antisense RNA; F finO interrupted by IS3 | 16, 52, 77, 86, 87 |
A more complete description of the F gene products is given in Frost et al. (28) and Firth et al. (24). The sources for the sequences for F not mentioned in Frost et al. (28) and for R100-1 are given. Recent references discussing function are also given.
IM, inner membrane; OM, outer membrane.
F TraY has homologous N- and C-terminal domains resulting from a gene duplication event. The percent similarity/identity values are an average for R100-1 TraY (aa 1 to 75) compared to the N-terminal domain of F TraY (aa 1 to 75), as well as the C-terminal domain (aa 56 to 131).
The OrfJ protein resembles a fusion of F TrbD and TrbG. The percent similarity/identity values are given for F TrbD (D) versus OrfJ (aa 1 to 65) and F TrbG (G) versus OrfJ (aa 24 to 83).
In terms of genes whose sequences are presented here and are involved in conjugation, only three varied in sequence. TraH varied in a small domain between amino acids (aa) 393 and 420; TraG varied between aa 620 and 673 and near the C terminus with both regions in TraG*, the putative periplasmic cleavage product of TraG (25). TraS, the entry exclusion protein found in the inner membrane, was nonhomologous between F and R100-1, suggesting that TraS may be responsible for the specificity of exclusion noted among F-like plasmids. The sequence of the C-terminal portion of traS reported here matched the partial sequence of traS presented in Ogata et al. (60).
The R100-1 transfer region is considerably longer than that of F, with three extra individual genes (orfCEF) and one cluster of four extra genes (orfG1G2HI), with orfG1 being homologous to orfG2 (39) while only artA and trbH are present in F but not in R100-1. Another interesting aspect of the R100-1 sequence is the fusion of trbD and trbG in F to form orfJ in R100-1 (39). These two cases are reminiscent of the gene duplication in traY of F in which the first and second halves of the TraY proteins are homologous to each other and to R100-1 TraY (35). The precision with which these extra genes are inserted or deleted between other genes is remarkable and argues for an evolutionary process whereby additions or deletions which affect transcription or translation of other genes are strongly selected against.
The changes in sequence between F and R100-1 transfer genes could be divided into three different types. In the first type, sequence changes are related to plasmid specificity. These include the genes for DNA metabolism (traMYI but not traD); regulation (traJYM and finP but not finO), and entry exclusion (traS). The evidence for specificity shown by these proteins has been discussed by Everett and Willetts (20) and Willetts and Maule (83). Other changes in sequence alter the phenotype of the transfer system but are not plasmid specific. For instance, TraT, which has a 1-aa difference between the F and R100-1 variants, can exclude its cognate plasmid more efficiently by preventing mating pair stabilization between donor cells (32). Similarly, small changes between R100-1 and F pilin (traA) affect pilus serology and phage infectivity but do not affect DNA transfer (5, 27, 30). The major change in sequence in the central region of F and R100-1 TraN can be correlated with the decreased mating efficiency of F with recipient cells carrying mutations in ompA and the inner core of the LPS (40). Here we show that the differences between F and R100-1 TraG, a protein involved in mating pair stabilization, define the plasmid specificity exhibited during entry exclusion. The third class of sequence changes leads to no discernible changes in the phenotype. These include traBP (4) and traD, where a QQP motif in F is repeated sequentially eight times in R100-1 (86).
Characterization of F tra mutants for sensitivity to the phagemid M13K07.
A transducing phage assay was originally used to demonstrate that f1 phage could infect F− cells at low efficiency (67). F-specific filamentous phages do not absolutely require the pilus for penetration into the cell but instead use the pilus to access a secondary receptor, TolA, located in the periplasm, more efficiently (80). Two domains in the attachment protein, pIII, are responsible for F-pilus tip recognition and TolA binding, as suggested by this two-step process (14, 65). A temperature-sensitive mutation in traC (traC1044) does not form F-pilus filaments but is fully sensitive to transducing phage such as M13K07 at the nonpermissive temperature (70). This mutation appears to permit pilus tip formation but prevents pilus extension beyond the cell surface. Thus, only the pilus tip exposed at the cell surface is theoretically required for successful f1 phage infection. The M13K07 assay provides a simple means to assess formation of the pilus tip prior to protrusion of the pilus filament and is more sensitive than assays based on plaque formation.
As a prelude to the complementation assays with either cloned F or R100-1 genes, the sensitivity of F tra mutants to M13K07 (kanamycin resistant) was tested to determine whether these mutations affected pilus tip formation or pilus extension (Table 4). All of the essential genes for pilus synthesis were tested except for trbC (pOX38-trbC460) and traU (pOX38-traU526), which are kanamycin resistant. The results suggested that some mutations block pilus tip assembly and give resistance to M13K07, while other mutations allow tip assembly and M13K07 sensitivity but not pilus elongation, as found for the traC1044 mutation. Thus, mutations traL311, traE18, traK105, traC5 and traG106 affect pilus tip formation, whereas traB2, traC1044, traW546, traH80, and traF13 affect pilus outgrowth or extension rather than tip formation. All of these mutations blocked pilus expression and severely decreased mating efficiency. The traV569 mutation did not block mating completely and was weakly sensitive to M13K07, suggesting that this mutation is leaky. Thus, it is difficult to classify whether the traV569 mutation affects pilus tip formation or outgrowth. Mutations in traU affect transfer efficiency to various degrees and phage sensitivity to some extent, as measured by the phage spot test, but have a minimal effect on pilus expression or M13K07 sensitivity. Thus, TraU might be involved primarily in DNA transfer as previously suggested (55).
TABLE 4.
Transfer proficiency and phage sensitivity of derivatives of F tra and trb mutants expressing the corresponding genes from F and R100-1 plasmids
| tra mutant with or without complementing plasmid | Pili (%) | Mating efficiencya (no. of transconjugants/100 donors) | Phage sensitivityb (%)
|
||
|---|---|---|---|---|---|
| f1 | R17 | M13K07c | |||
| Flac traA1 | No | 0.0 | R | R | 0 |
| pOX38A::CAT | No | 0.0 | R | R | 0 |
| pOX38A::CAT + pLF164 | 1.0 | S | S | ||
| pOX38A::CAT + pKAR3 | 0.9 | S | R (10) | ||
| Flac traL311 | No | 0.01 | R | R | 0 |
| Flac traL311 + pLF164 | 83.3 | S | S | ||
| Flac traL311 + pKAR3 | 67.8 | S | S | ||
| Flac traE18 | No | 0.0 | R | R | 0 |
| Flac traE18 + pRS27 | 3.1 | S | S | ||
| Flac traE18 + pKAR3d | 0.1 | ND | ND | ||
| Flac traK105 | No | 0.0 | R | R | 0 |
| Flac traK105 + pRS27d | 0.005 | ND | ND | ||
| Flac traK105 + pKAR1d | 0.3 | ND | ND | ||
| Flac traB2 | No | 0.0 | R | R | 104 (1) |
| Flac traB2 + pRS27 | 4.5 | S | S | ||
| Flac traB2 + pKAR1 | 2.4 | S | S | ||
| pOX38-traP474e | 4.4 | S | S | ND | |
| pOX38-traP474 + pRS27 | 33.0 | S | S | ||
| pOX38-traP474 + pKAR2 | 29.0 | S | S | ||
| Flac traV569 | No | 0.02 | R | R | 104 (1) |
| Flac traV569 + pKI164 | 16.7 | S | S | ||
| Flac traV569 + pKAR1 | 0.9 | S | S | ||
| Flac traC1044f | No | 0.0 | (R) | R | 105 (10) |
| Flac traC5 | No | 0.008 | R | R | 0 |
| Flac traC5 + pRS29 | 0.8 | S | S | ||
| Flac traC5 + pKAR1 | 16.7 | S | S | ||
| Flac traW546 | No | 0.02 | R | R | 105 (10) |
| Flac traW546 + pRS29 | 40.0 | S | S | ||
| Flac traW546 + pKAR1 | 20.0 | S | S | ||
| Flac traU526 | Yes (50) | 0.5 | (R) | (R) | 106 (100) |
| pOX38-traU347 | Yes (20) | 0.0 | R (1) | R (1) | ND |
| pOX38-traU347 + pRS29 | 12.5 | S | S | ||
| pOX38-traU347 + pKAR1 | 38.5 | S | S | ||
| pOX38-trbC460 | No | 0.0 | R | R | ND |
| pOX38-trbC460 + pRS29 | 0.7 | S | S | ||
| pOX38-trbC460 + pKAR1 | 21.4 | S | S | ||
| pOX38N::CAT | Yes | 0.0 | S | S | ND |
| pOX38N::CAT + pKI375 | 160 (ompA; 0.04)g | ||||
| pOX38N::CAT + pBK8 | 56 (ompA; 99.0) | ||||
| Flac traF13 | No | 0.00 | R | R | 106 (100) |
| Flac traF13 + pKI175 | 100.0 | S | S | ||
| Flac traF13 + pKAR4 | 100.0 | S | S | ||
| pOX38-traQ238 | No | 0.003 | R | R | ND |
| pOX38-traQ238 + pRS29 | 6.4 | S | S | ||
| pOX38-traQ238 + pKAR5 | 46.2 | S | S | ||
| Flac traH80 | No | 0.0 | R | R | 104 (1) |
| Flac traH80 + pRS29 | 0.1 | S | S | ||
| Flac traH80 + pKAR5 | 13.7 | S | S | ||
| Flac traG101 | Yes | 0.0 | S | S | ND |
| Flac traG106 | No | 0.0 | R | R | 0 |
| Flac traG106 + pRS1670 | 36.1 | S | S | ||
| Flac traG106 + pKAR6 | 25.9 | S | S | ||
| pOX38-traX482 | Yes | 67.0 | S | S | ND |
| pOX38-traX482 + pKAF3 | 100.0 | S | S | ||
| pOX38-traX482 + pKAR7 | 100.0 | S | S | ||
| pOX38-Km/F | Yes | 67.3 | S | S | 106 (100) |
| R100-1 | Yes | 97.2 | R (1) | R (10) | 105 (10) |
All Flac mutants were in Escherichia coli ED2149 and all pOX38 derivatives were in E. coli XK1200, while R100-1 was in E. coli JE2571. The recipient for matings was ED24, while for R100-1 ED24 carrying the pT7-3 vector was the recipient. A 100% mating efficiency was assigned to the highest value among the strains.
Phage sensitivity was determined by qualitative spot tests. “S” represents 70 to 100% of the sensitivity exhibited by pOX38-Km. Intermediate sensitivities are given as percentages in parentheses. R, resistant; (R), partially resistant when the level of resistance is undefined; ND, not determined.
M13K07 is a phagemid described by Vieira and Messing (79) which yields kanamycin-resistant colonies upon infection. The number of kanamycin-resistant colonies resulting from infection of 107 cells with 107 phage is given, as is the percentage of infection of wild-type pOX38-Km (in parentheses).
Phage sensitivity profiles for Flac traE18 + pKAR3, Flac traK105 + pKAR1, and Flac traK105 + pRS27 could not be discerned from spot tests due to low levels of complementation. Pili from these cells, when examined by electron microscopy, attached both R17 and f1 phages.
The data on traP is taken from Anthony et al. (4). traP mutants have fragile pili that appear to fall off the cell easily during preparation for electron microscopy (unpublished observation).
Flac traC1044 assembles a pilus tip but does not elongate the pilus (70).
The mating efficiency with recipient strains carrying an ompA mutation is given in parentheses (40).
Mutations in genes essential for pilin expression, such as traA (pilin) or traQ (pilin chaperone), were either tested with M13K07 and were found to confer resistance or were not tested. Similarly, mutations which do not affect F piliation or phage sensitivity were not tested (i.e., traG101, traN::CAT, traP474, and traX482 [pilin acetylation]). Interestingly, R100-1 had a moderate sensitivity to M13K07 similar to Flac traC1044 and Flac traW546 (10%), whereas its sensitivity to f1 phage by the traditional spot test was 1% of F as previously reported (83).
Complementation of F tra mutants with R100-1 variants.
In this study we concentrated on the genes for Mpf, which includes the genes for pilus synthesis, mating pair stabilization, and entry and/or surface exclusion. The plasmid specificities of genes involved in DNA metabolism and regulation of the transfer region have been discussed previously (83).
A series of chimeric plasmids were constructed containing different portions of the R100-1 transfer region as described in Table 2. Constructs containing either F or R100-1 DNA were carried on similar vectors of low to medium copy number. Where possible, conserved or approximately equivalent restriction sites were used to construct these plasmids. When no convenient restriction sites were present, PCR fragments derived from segments of the F or R100-1 transfer regions were cloned and used in complementation experiments.
Each of the tra mutations in JCFL0 or pOX38 (Table 1) were complemented with chimeric plasmids carrying either the F or R100-1 variant of the gene (Table 2). Although complementation was usually not complete, there was a clear increase in mating efficiency, as well as phage sensitivity (Table 4). In almost all cases, the R100-1 and F clones complemented each mutation equally well. Complementations by pRS27 or pRS29 (F) of the traK105, traC5, trbC460, traQ238, and traH80 mutations were lower than with the equivalent R100-1 clones and were interpreted as complete complementation by both constructs. The traE18 and traV569 mutations were complemented significantly better by chimeric plasmids derived from the F transfer region compared to those derived from R100-1. The discrepancy in mating efficiency assays for traE18 could be ascribed to the differences in sequence between F and R100-1 TraE (10 nonconservative and 2 conservative amino acid replacements) (Table 1), since both expressed equivalent numbers of pili which bound phage equally well (Table 4). However, the low level of complementation of the traE18 mutation does not allow a determination of plasmid specificity to be conferred on the TraE protein at this time. The TraV proteins from F and R100-1 are almost completely homologous (97.1% identity), suggesting that the complementation results are due to differing levels of expression between pKI164 and pKAR1.
Clear differences between F and R100-1 genes included traA (pilin) as previously noted (5, 26) and traN with its differing requirement for OmpA and LPS in the recipient cell (40). The difference in sequence between F and R100-1 pilin accounts for the reduced ability of R100-1 pili to attach and be infected by R17 phage (30). However, f1 phage, which attaches to the tip of F and R100-1 pili equally well, infects R100-1 cells poorly. In a complementation assay, both the F and R100-1 pilins conferred complete sensitivity to pOX38A::CAT, suggesting that pilin is not the true primary receptor for the f1 phage (5). No other transfer protein appeared to be involved in f1 infection, i.e., gave complete complementation, as measured by mating efficiency assays, but was f1 resistant. Interestingly, the expression of either F or R100-1 TraF from a multicopy plasmid gave complete f1 resistance but did not affect mating efficiency or R17 sensitivity (data not shown). This suggests that overexpression of the periplasmic TraF might interfere with the f1 infection process, possibly by blocking access to the secondary receptor, TolA (80).
Surface exclusion assays were also carried out for the F mutants complemented with genes derived from F or R100-1. In almost every case, the surface exclusion indices were comparable for both the F and R100-1 clones (Table 5 and data not shown). One exception was the surface exclusion assay for Flac traG106 (compare to data for pOX38A::CAT) (Table 5), suggesting that traG has a role in entry, surface exclusion, or both. In this assay, the recipient cell contained a clone expressing both F TraS and T. These cells could exclude Flac traG106 in the presence of F traG (pRS1670) but not R100-1 traG (pKAR6). The 600-fold difference in the surface exclusion indices is characteristic of TraS, the entry exclusion determinant, since it excludes its cognate plasmid 10-50 times more efficiently than TraT (2, 3).
TABLE 5.
Surface exclusion properties of F mutants expressing the corresponding genes from F and R100-1 plasmids
| Donor strain | Mating efficiencya (no. of transconjugants/ 100 donors)
|
Surface exclusion indexb | |
|---|---|---|---|
| pSC101 | pRS31 | ||
| pOX38-Km/F | 51.7 | 4.8 × 10−2 | 1,069.8 |
| R100-1 | 24.6 | 2.1 | 11.6 |
| pOX38A::CAT + pLF164 | 4.9 | 6.7 × 10−3 | 731.3 |
| pOX38A::CAT + pLF107 | 7.9 | 8.3 × 10−3 | 951.8 |
| Flac traG106 + pRS1670 | 27.6 | 4.8 × 10−3 | 5,787.8 |
| Flac traG106 + pKAR6 | 10.0 | 1.1 | 9.4 |
The mating efficiencies of F tra mutants to recipients harboring either pSC101 vector control or to recipients expressing TraT and TraS from pRS31 are indicated.
The surface exclusion index was calculated by dividing the mating efficiency to recipients carrying pSC101 by the mating efficiency to recipients carrying pRS31.
Characteristics of pili synthesized by an F traX mutant.
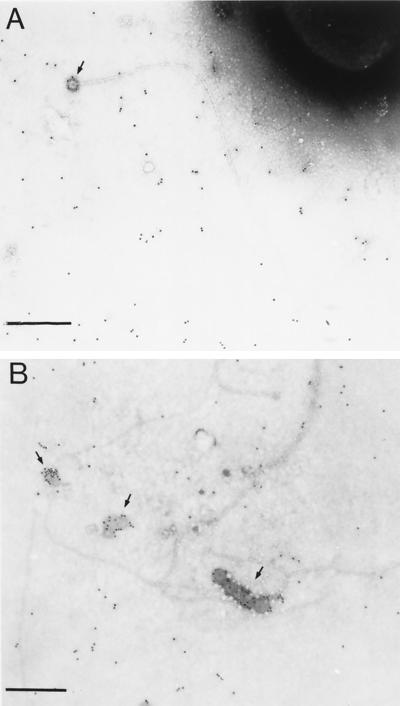
Pili expressed by pOX38-traX482 contain unacetylated pilin subunits, but these pili are proficient in conjugation and attach both f1 and R17 phages well (54). However, overexpression of pili in the absence of traX results in a subtle change in the conformation of the pilin subunits within the filament such that R17 no longer attaches and the N-terminal domain is exposed on the sides of the pilus, as monitored by the use of JEL92 MAbs (31). Since F and R100-1 TraX proteins diverge in sequence near their N termini, their ability to acetylate F and R100-1 pilin might vary, leading to slight conformational changes of pilin within the pilus filament. Acetylation of F pilin expressed by pOX38-traX482 was restored by supplying either F or R100-1 traX in trans, suggesting that the F and R100-1 enzymes are equally active (data not shown). Electron micrographs of pili expressed by pOX38-traX482 revealed that they were more kinked than wild-type pili and contained more vesicle-like structures (Fig. 2). Like wild-type F pili, pOX38-traX482 pili did not bind the MAb JEL92 on their sides (Fig. 2), a finding which is in contrast to the data presented by Grossman et al. (31). Thus, overexpression of pili in the absence of traX alters the configuration of the pilin subunits within the pilus.
FIG. 2.
The effect of pilin acetylation on structural characteristics of pili. Immunogold electron microscopy was carried out as described in Materials and Methods. The binding of MAb JEL92 to pili elaborated by MC4100 cells harboring either pOX38-Km (A) or pOX38-traX482 (B) is shown. Arrows indicate the terminal knobs or vesicular materials coated with gold particles. Magnification is ×14,500, and the size bar represents 1 μm.
DISCUSSION
The analysis of the sequence of the R100-1 transfer region has revealed a number of instances where genes have been inserted or deleted when compared to F. In addition, there are instances of gene fusions (orfJ), serial repeats of genes (R100-1 orfG1 and orfG2) and duplications within genes (F traY). Other sequences showed mosaicism with clearly defined stretches of sequence being nonhomologous (traB, traP, traN, trbA, and traG). Several proteins known to have a role in conjugation (TraJ, -Y, -M, and -S) vary considerably in sequence, which defines the specificity of those proteins for their cognate plasmids. Others vary in sequence with little to no effect on the conjugative process (TraB and -P). The differences in TraN between F and R100-1 are striking and have been related to the requirement for OmpA and LPS in the recipient cell. The differences in TraG, the only other protein besides TraN involved in mating pair stabilization, are localized to the C-terminal half of the protein which corresponds to TraG* and is responsible for this step in the conjugation process.
A major goal of this study was to identify a transfer protein that could confer resistance to f1 phage without affecting functional pilus production, as seen with the R100-1 plasmid. Complementation studies revealed that neither pilin nor any other pilus synthesis genes from R100-1 gave the desired phenotype. Thus, the precise nature of the pilus tip remains unknown. Proposed models for the nature of the pilus tip include a unique configuration of pilin molecules which is not dependent on pilin sequence or a host-encoded moiety at the tip. Alternatively, more than one transfer protein may be required to generate the precise tip structure which gives each transfer system its characteristic pattern of filamentous phage sensitivity.
One interesting aspect of these results is the characterization of the F tra mutants with M13K07. Our results suggest that mutations in traB, traF, traH, traW and possibly traV, as well as traC1044 (70), allow pilus tip formation but block pilus outgrowth beyond the cell surface. Similarly, mutations in traL, traE, traK, traC, and traG106 appear to affect formation of the tip structure. None of these mutations affect the level of pilin subunits in the inner membrane pools (56). TraL is an interesting protein which has never been detected in in vivo expression experiments. An mRNA processing site lies within the coding sequence for TraL, and its initiating methionine abuts a complex secondary structure thought to stabilize the highly translated traA message (41). One function for such a rarely expressed protein could be to limit the number of sites for pilus assembly, since there are usually fewer than three pili/cell under derepressed conditions. Mutations in trbI, an inessential transfer gene, give longer-than-usual pili, suggesting a defect in pilus retraction or length determination (49). Mutations in traP yield pili which function almost normally but have a tendency to be easily dislodged from the cell during preparation of specimens for electron microscopy (reference 4 and unpublished observation).
While much remains to be discovered about the precise function of the genes involved in Mpf, the various genes are beginning to fall into functional subgroups within the five major classes of transfer genes. For the Mpf genes, there are subgroups involved in pilin maturation (TraQ and -X), pilus tip formation (TraL, -E, -K, -C, and -G) pilus outgrowth (TraB, -F, -H, and -W and possibly TraV), retraction (TrbI), pilus stabilization (TraP), and possibly DNA transfer (TraU). There are also surface exclusion (TraT) and entry exclusion (TraS) proteins that block transfer between donor cells at different stages.
It appears to be important to differentiate between the initiation of DNA synthesis and DNA transport, which are separate events controlled by different mechanisms (3, 38, 69). These reports suggested that the pilus was required for generating a signal that initiated DNA synthesis (62). Mutations in the mating pair stabilization genes (traN548 and traG101) allowed this signal to occur after loose cell-to-cell contacts had formed but blocked DNA transport to the recipient cell (38). The surface exclusion and entry exclusion proteins prevented stabilization and signalling from occurring, respectively. TraT in the recipient cell blocks mating pair stabilization but not the signal for DNA synthesis in the donor cell (2, 3); TraS in the recipient cell blocks a plasmid-specific signal for DNA synthesis and subsequent transport (3, 61). Nonpiliated traG mutants such as traG106, when complemented by R100-1 traG, are unaffected by F surface exclusion genes (most probably TraS) in the recipient cell. Thus, the inner membrane protein, TraG, and/or the periplasmic derivative TraG* (25), in the donor cell can differentiate between F and R100-1 TraS, an inner membrane protein, in the recipient cell. This suggests that TraG might be translocated into the recipient cell or that TraG and TraS communicate with each other perhaps via the pilus.
The reason for the acetylation of F pilin has been puzzling since traX mutants have no obvious phenotype (46). Our results suggest that acetylation by TraX might ensure correct assembly of the pilus. The deformities in pilus structure appear to be proportional to the level of tra gene expression in the absence of traX with the traX mutation in single-copy pOX38 leading to kinked pili and overexpression of traA-traG in pTG801 (31), giving F pili with a very different structure. Thus, TraX might act as a governor on the speed and accuracy of pilus assembly. In agreement with this, the pili expressed by pOX38-tra715, where the entire transfer region including traX is overexpressed from a T7 promoter (45), appear to be normal (unpublished observations), suggesting that the levels of pilin and TraX are balanced in this system and are capable of correctly assembling the F pilus.
The nature of the pilus tip has eluded identification for many years. One clue has been a decrease in mating pair formation by the addition of 1 mM Zn2+, which is thought to bind to the pilus tip (62). Cross-linking experiments with f1 or M13 pIII protein have been inconclusive (unpublished observations), and the complementation analyses presented here have not yielded any clues. It is interesting that f1 phage attach equally well to F+ and R100-1-containing cells but that R100-1 is resistant (1%) to these phages. Perhaps f1 attached to the F pilus tip mimics a recipient cell and triggers pilus retraction, while the R100-1 pilus remains immune. These results suggest that a signal is transmitted down the pilus, perhaps by a conformational change in the pilin subunits triggered at the tip and passing through the length of the pilus. Since pilin can assume more than one configuration in a functional pilus, as seen in the experiments of Grossman et al. (31), this remains an attractive possibility. Another question that has continually eluded researchers is the nature of the conjugative pore. While there is probably a role for the pilus in this process, an analysis of previous results plus the complementation data presented here suggests that the mating pair stabilization genes might form this elusive channel. If the pilus is involved in signalling that DNA synthesis should begin in the donor cell, the mechanism by which it transmits this signal to TraI, bound at oriT, to begin unwinding the DNA as a prelude to conjugative DNA replication and/or transfer remains the central question in this process. Our results suggest that TraG might have an important role in this signalling mechanism.
ACKNOWLEDGMENTS
We thank Kirsten Heilesen for resequencing F traD.
This research was supported by NSERC and the MRC of Canada. W.A.K. is supported by an NSERC Studentship.
REFERENCES
- 1.Abo T, Ohtsubo E. Repression of the traM gene of plasmid R100 by its own product and integration host factor at one of the two promoters. J Bacteriol. 1993;175:4466–4474. doi: 10.1128/jb.175.14.4466-4474.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman M, Kennedy N, Skurray R. Cell-cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc Natl Acad Sci USA. 1977;74:5104–5108. doi: 10.1073/pnas.74.11.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achtman M, Skurray R. A redefinition of the mating phenomenon in bacteria. In: Reissig J L, editor. Microbial interactions: receptors and recognition. Vol. 3. London, England: Chapman and Hall; 1977. pp. 233–279. [Google Scholar]
- 4.Anthony K G, Kathir P, Moore D, Ippen-Ihler K, Frost L S. Analysis of the traLEKBP sequence and the TraP protein from three F-like plasmids: F, R100-1, and ColB2. J Bacteriol. 1996;178:3194–3200. doi: 10.1128/jb.178.11.3194-3200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anthony K G, Sherburne C, Sherburne R, Frost L S. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994;13:939–953. doi: 10.1111/j.1365-2958.1994.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong G D, Frost L S, Sastry P A, Paranchych W. Comparative biochemical studies on F and EDP208 conjugative pili. J Bacteriol. 1980;141:333–341. doi: 10.1128/jb.141.1.333-341.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 8.Bayer M, Eferl R, Zellnig G, Teferle K, Dijkstra A, Koraimann G, Hogenauer G. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage infection. J Bacteriol. 1995;177:4279–4288. doi: 10.1128/jb.177.15.4279-4288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1517. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler M, Galas D. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J Mol Biol. 1983;170:61–91. doi: 10.1016/s0022-2836(83)80227-7. [DOI] [PubMed] [Google Scholar]
- 11.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheah K-C, Skurray R A. The F plasmid carries an IS3 insertion within finO. J Gen Microbiol. 1986;132:3269–3275. doi: 10.1099/00221287-132-12-3269. [DOI] [PubMed] [Google Scholar]
- 13.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Click E M, Webster R E. Filamentous phage infection: required interactions with the TolA protein. J Bacteriol. 1997;179:6464–6471. doi: 10.1128/jb.179.20.6464-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen S N, Chang A C Y. Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol. 1977;132:734–737. doi: 10.1128/jb.132.2.734-737.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cram D S, Loh S M, Cheah K-C, Skurray R A. Sequence and conservation of genes at the distal end of the transfer region on plasmids F and R6-5. Gene. 1991;104:85–90. doi: 10.1016/0378-1119(91)90469-r. [DOI] [PubMed] [Google Scholar]
- 17.Dempsey W B. Regulation of R100 conjugation requires traM in cis to traJ. Mol Microbiol. 1994;13:987–1000. doi: 10.1111/j.1365-2958.1994.tb00490.x. [DOI] [PubMed] [Google Scholar]
- 18.Disqué-Kochem C, Dreiseikelmann B. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J Bacteriol. 1997;179:6133–6137. doi: 10.1128/jb.179.19.6133-6137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doran T, Loh S, Firth N, Skurray R. Molecular analysis of the F plasmid traVR region: traV encodes a lipoprotein. J Bacteriol. 1994;176:4182–4186. doi: 10.1128/jb.176.13.4182-4186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R, Willetts N. Characterisation of an in vivo system for nicking at the origin of conjugal DNA transfer of the sex factor F. J Mol Biol. 1980;136:129–150. doi: 10.1016/0022-2836(80)90309-5. [DOI] [PubMed] [Google Scholar]
- 21.Fee B E, Dempsey W B. Cloning, mapping, and sequencing of plasmid R100 traM and finP genes. J Bacteriol. 1986;167:336–345. doi: 10.1128/jb.167.1.336-345.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fee B E, Dempsey W B. Nucleotide sequence of geneX of antibiotic resistance plasmid R100. Nucleic Acids Res. 1988;16:4726. doi: 10.1093/nar/16.10.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finlay B B, Frost L S, Paranchych W. Origin of transfer of IncF plasmids and nucleotide sequences of the type II oriT, traM, and traY alleles from ColB4-K98 and the type IV traY allele from R100-1. J Bacteriol. 1986;168:132–139. doi: 10.1128/jb.168.1.132-139.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Firth N, Ippen-Ihler K, Skurray R A. Structure and function of the F factor and mechanism of conjugation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaecter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2377–2401. [Google Scholar]
- 25.Firth N, Skurray R. Characterization of the F plasmid bifunctional gene, traG. Mol Gen Genet. 1992;232:145–153. doi: 10.1007/BF00299147. [DOI] [PubMed] [Google Scholar]
- 26.Frost L S. Conjugative pili and pilus-specific phages. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 189–221. [Google Scholar]
- 27.Frost L S, Finlay B B, Opgenorth A, Paranchych W, Lee J S. Characterization and sequence analysis of pilin from F-like plasmids. J Bacteriol. 1985;164:1238–1247. doi: 10.1128/jb.164.3.1238-1247.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frost L S, Ippen-Ihler K, Skurray R S. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frost L S, Lee J, Scraba D, Paranchych W. Two monoclonal antibodies specific for different epitopes within the amino terminal region of F pilin. J Bacteriol. 1986;168:192–198. doi: 10.1128/jb.168.1.192-198.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frost L S, Paranchych W. DNA sequence analysis of point mutations in traA, the F pilin gene, reveal two domains involved in F-specific bacteriophage attachment. Mol Gen Genet. 1988;213:134–139. doi: 10.1007/BF00333409. [DOI] [PubMed] [Google Scholar]
- 31.Grossman T H, Frost L S, Silverman P M. Structure and function of conjugative pili: monoclonal antibodies as probes for structural variants of F pili. J Bacteriol. 1990;172:1174–1179. doi: 10.1128/jb.172.3.1174-1179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison J L, Taylor I M, Platt K, O’Connor C D. Surface exclusion specificity of the TraT lipoprotein is determined by single alterations in a five amino acid region of the protein. Mol Microbiol. 1992;6:2825–2832. doi: 10.1111/j.1365-2958.1992.tb01462.x. [DOI] [PubMed] [Google Scholar]
- 33.Howard M T, Nelson W C, Matson S W. Stepwise assembly of a relaxosome at the F plasmid origin of transfer. J Biol Chem. 1995;270:28381–28386. [PubMed] [Google Scholar]
- 34.Humphries G, Willshaw G A, Anderson E S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochem Biophys Acta. 1975;383:457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- 35.Inamoto S, Yoshioka Y, Ohtsubo E. Identification and characterization of the products from the traJ and traY genes of plasmid R100. J Bacteriol. 1988;170:2749–2757. doi: 10.1128/jb.170.6.2749-2757.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jalajakumari M B, Manning P A. Nucleotide sequence of the traD region in the E. coli F sex factor. Gene. 1989;81:195–202. doi: 10.1016/0378-1119(89)90179-0. [DOI] [PubMed] [Google Scholar]
- 37.Kathir P, Ippen-Ihler K. Construction and characterization of derivatives carrying insertion mutations in F plasmid transfer region genes, trbA, artA, traQ, and trbB. Plasmid. 1991;36:40–54. doi: 10.1016/0147-619x(91)90035-u. [DOI] [PubMed] [Google Scholar]
- 38.Kingsman A, Willetts N. The requirements for conjugal DNA synthesis in the donor strain during Flac transfer. J Mol Biol. 1978;122:287–300. doi: 10.1016/0022-2836(78)90191-2. [DOI] [PubMed] [Google Scholar]
- 39.Klimke W A, Anthony K G, Fekete R, Manchak J, Frost L S. Plasmid specificity and interaction: the similarities and differences between the transfer regions of two compatible plasmids, F and R100-1. In: Syvanen M, Kado C, editors. Horizontal gene transfer. New York, N.Y: Chapman and Hall; 1998. pp. 25–39. [Google Scholar]
- 40.Klimke W A, Frost L S. Genetic analysis of the role of the transfer gene traN of the F plasmid in mating pair stabilization during conjugation. J Bacteriol. 1998;180:4036–4043. doi: 10.1128/jb.180.16.4036-4043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koraimann G, Högenauer G. A stable core region of the tra operon mRNA of plasmid R1-19. Nucleic Acids Res. 1989;17:1283–1298. doi: 10.1093/nar/17.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loh S, Cram D, Skurray R. Nucleotide sequence of the leading region adjacent to the origin of transfer on plasmid F and its conservation among conjugative plasmids. Mol Gen Genet. 1989;219:177–186. doi: 10.1007/BF00261174. [DOI] [PubMed] [Google Scholar]
- 43.Majdalani N, Ippen-Ihler K. Membrane insertion of the F-pilin subunit is Sec independent but requires leader peptidase B and the proton motive force. J Bacteriol. 1996;178:3742–3747. doi: 10.1128/jb.178.13.3742-3747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malmborg A-C, Söderlind E, Frost L S, Borrebaeck C A K. Selective phage infection mediated by epitope expression on F pilus. J Mol Biol. 1997;273:544–551. doi: 10.1006/jmbi.1997.1332. [DOI] [PubMed] [Google Scholar]
- 45.Maneewannakul K, Maneewannakul S, Ippen-Ihler K. Sequence alterations affecting F plasmid transfer gene expression: a conjugation system dependent on transcription by the RNA polymerase of phage T7. Mol Microbiol. 1992;6:2961–2973. doi: 10.1111/j.1365-2958.1992.tb01755.x. [DOI] [PubMed] [Google Scholar]
- 46.Maneewannakul K, Maneewannakul S, Ippen-Ihler K. Characterization of traX, the F plasmid locus required for acetylation of F pilin subunits. J Bacteriol. 1995;177:2957–2964. doi: 10.1128/jb.177.11.2957-2964.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maneewannakul S, Kathir P, Ippen-Ihler K. Characterization of the F plasmid mating aggregation gene traN and of the new F transfer region locus trbE. J Mol Biol. 1992;225:299–311. doi: 10.1016/0022-2836(92)90923-8. [DOI] [PubMed] [Google Scholar]
- 48.Maneewannakul S, Maneewannakul K, Ippen-Ihler K. Characterization of trbC, a new F plasmid tra operon gene that is essential to conjugative transfer. J Bacteriol. 1991;173:3872–3878. doi: 10.1128/jb.173.12.3872-3878.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maneewannakul S, Maneewannakul K, Ippen-Ihler K. Characterization, localization and sequence of F transfer region products: the pilus assembly gene, TraW, and a new product, TrbI. J Bacteriol. 1992;174:5567–5574. doi: 10.1128/jb.174.17.5567-5574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manning P A, Morelli G, Achtman M. traG protein of the F sex factor of Escherichia coli K-12 and its role in conjugation. Proc Natl Acad Sci USA. 1981;78:7487–7491. doi: 10.1073/pnas.78.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McIntire S A, Dempsey W B. oriT sequence of the antibiotic plasmid R100. J Bacteriol. 1987;169:3829–3832. doi: 10.1128/jb.169.8.3829-3832.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McIntire S A, Dempsey W B. Fertility inhibition gene of plasmid R100. Nucleic Acids Res. 1987;15:2029–2042. doi: 10.1093/nar/15.5.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miki T, Horiuchi T, Willetts N. Identification and characterization of four new tra cistrons on the E. coli K-12 sex factor. Plasmid. 1978;1:316–323. doi: 10.1016/0147-619x(78)90048-3. [DOI] [PubMed] [Google Scholar]
- 54.Moore D, Hamilton C H, Maneewannakul K, Mintz Y, Frost L S, Ippen-Ihler K. The Escherichia coli K-12 F plasmid gene traX is required for acetylation of F-pilin. J Bacteriol. 1993;175:1375–1383. doi: 10.1128/jb.175.5.1375-1383.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore D, Maneewannakul K, Maneewannakul S, Wu J H, Ippen-Ihler K, Bradley D E. Characterization of the F-plasmid conjugative transfer gene traU. J Bacteriol. 1990;172:4263–4270. doi: 10.1128/jb.172.8.4263-4270.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore D, Sowa B A, Ippen-Ihler K. The effect of tra mutations on the synthesis of F pilin membrane polypeptide. Mol Gen Genet. 1981;184:260–264. doi: 10.1007/BF00272914. [DOI] [PubMed] [Google Scholar]
- 57.Moore D, Wu J, Kathir P, Hamilton C M, Ippen-Ihler K. Analysis of transfer genes and gene products within the traB-traC region of the Escherichia coli fertility factor, F. J Bacteriol. 1987;169:3994–4002. doi: 10.1128/jb.169.9.3994-4002.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakaya R, Nakamura A, Murata Y. Resistance transfer agents in Shigella. Biochem Biophys Res Commun. 1960;3:654–659. doi: 10.1016/0006-291x(60)90081-4. [DOI] [PubMed] [Google Scholar]
- 59.Nelson W C, Howard M T, Sherman J A, Matson S W. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J Biol Chem. 1995;270:28374–28380. [PubMed] [Google Scholar]
- 60.Ogata R T, Winters C, Levine R P. Nucleotide sequence analysis of the complement resistance gene from plasmid R100. J Bacteriol. 1982;151:819–827. doi: 10.1128/jb.151.2.819-827.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ou J T. Mating signal and DNA penetration deficiency in conjugation between male Escherichia coli and minicells. Proc Natl Acad Sci USA. 1975;72:3721–3725. doi: 10.1073/pnas.72.9.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ou J T, Anderson T F. Effect of Zn2+ on bacterial conjugation: inhibition of mating pair formation. J Bacteriol. 1972;111:177–185. doi: 10.1128/jb.111.1.177-185.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panicker M M, Minkley E G., Jr DNA transfer occurs during a cell surface contact stage of F sex factor-mediated bacterial conjugation. J Bacteriol. 1985;162:584–590. doi: 10.1128/jb.162.2.584-590.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pölzleitner E, Zechner E, Renner W, Fratte R, Jauk B, Högenauer G, Koraimann G. TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol Microbiol. 1997;25:495–507. doi: 10.1046/j.1365-2958.1997.4831853.x. [DOI] [PubMed] [Google Scholar]
- 65.Riechmann L, Holliger P. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell. 1997;90:351–360. doi: 10.1016/s0092-8674(00)80342-6. [DOI] [PubMed] [Google Scholar]
- 66.Rondot S, Anthony K G, Dubel S, Ida N, Beyreuther K, Frost L S, Little M, Breitling F. Epitopes fused to F-pilin are incorporated into functional recombinant pili. J Mol Biol. 1998;279:589–603. doi: 10.1006/jmbi.1998.1773. [DOI] [PubMed] [Google Scholar]
- 67.Russel M, Whirlow H, Sun T-P, Webster R E. Low frequency infection of F− bacteria by transducing particles of filamentous bacteriophages. J Bacteriol. 1988;170:5312–5316. doi: 10.1128/jb.170.11.5312-5316.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sambrook J, Fritsch J E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 69.Sarathy P V, Siddiqi O. DNA synthesis during bacterial conjugation. III. Is DNA replication in the Hfr obligatory for chromosome transfer? J Mol Biol. 1973;78:443–451. doi: 10.1016/0022-2836(73)90467-1. [DOI] [PubMed] [Google Scholar]
- 70.Schandel K A, Maneewannakul S, Ippen-Ihler K, Webster R E. A traC mutant that retains sensitivity to f1 bacteriophage but lacks F pili. J Bacteriol. 1987;169:3151–3159. doi: 10.1128/jb.169.7.3151-3159.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Silverman P M. Towards a structural biology of bacterial conjugation. Mol Microbiol. 1997;23:423–429. doi: 10.1046/j.1365-2958.1997.2411604.x. [DOI] [PubMed] [Google Scholar]
- 72.Skurray R A, Nagaishi H, Clark A J. Molecular cloning of DNA form F sex factor of Escherichia coli K-12. Proc Natl Acad Sci USA. 1976;73:64–68. doi: 10.1073/pnas.73.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugino Y, Hirota Y. Conjugal fertility associated with resistance factor R in Escherichia coli. J Bacteriol. 1962;84:902–910. doi: 10.1128/jb.84.5.902-910.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taki K, Abo T, Ohtsubo E. Regulatory mechanisms in expression of the traY-I operon of sex factor plasmid R100: involvement of traJ and traY gene products. Genes Cells. 1998;3:331–345. doi: 10.1046/j.1365-2443.1998.00194.x. [DOI] [PubMed] [Google Scholar]
- 76.van Biesen T, Frost L S. Differential levels of fertility inhibition among F-like plasmids are related to the cellular concentration of finO mRNA. Mol Microbiol. 1992;6:771–780. doi: 10.1111/j.1365-2958.1992.tb01527.x. [DOI] [PubMed] [Google Scholar]
- 77.van Biesen T, Frost L S. The FinO protein of IncF plasmids binds FinP antisense RNA and its target, traJ mRNA, and promotes duplex formation. Mol Microbiol. 1994;14:427–436. doi: 10.1111/j.1365-2958.1994.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 78.van Duin J. Single-stranded RNA bacteriophages. In: Calendar R, editor. The bacteriophages. I. New York, N.Y: Plenum Press, Inc.; 1988. pp. 117–167. [Google Scholar]
- 79.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 80.Webster R E. The tol gene products and the import of macromolecules into Escherichia coli. Mol Microbiol. 1991;5:1005–1011. doi: 10.1111/j.1365-2958.1991.tb01873.x. [DOI] [PubMed] [Google Scholar]
- 81.Willetts N. Characterization of the F transfer cistron traL. Genet Res. 1973;21:205–213. doi: 10.1017/s0016672300013379. [DOI] [PubMed] [Google Scholar]
- 82.Willetts N, Achtman M. Genetic analysis of transfer by the Escherichia coli sex factor F, using P1 transductional complementation. J Bacteriol. 1972;110:843–851. doi: 10.1128/jb.110.3.843-851.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Willetts N, Maule J. Specificities of IncF plasmid conjugation genes. Genet Res. 1986;47:1–11. doi: 10.1017/s0016672300024447. [DOI] [PubMed] [Google Scholar]
- 84.Womble D D, Rownd R H. Genetic and physical map of the IncFII antibiotic resistance plasmids. Microbiol Rev. 1988;52:433–451. doi: 10.1128/mr.52.4.433-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu J H, Moore D, Lee T, Ippen-Ihler K. Analysis of Escherichia coli K-12 F factor transfer genes: traQ, trbA and trbB. Plasmid. 1987;18:54–69. doi: 10.1016/0147-619x(87)90078-3. [DOI] [PubMed] [Google Scholar]
- 86.Yoshioka Y, Fujita Y, Ohtsubo E. Nucleotide sequence of the promoter-distal region of the tra operon of plasmid R100, including traI (DNA helicase I) and traD genes. J Mol Biol. 1990;214:39–53. doi: 10.1016/0022-2836(90)90145-C. [DOI] [PubMed] [Google Scholar]
- 87.Yoshioka Y, Ohtsubo H, Ohtsubo E. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J Bacteriol. 1987;169:619–623. doi: 10.1128/jb.169.2.619-623.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]