Abstract
A combined genetic and physical map of the Agrobacterium tumefaciens A348 (derivative of C58) genome was constructed to address the discrepancy between initial single-chromosome genetic maps and more recent physical mapping data supporting the presence of two nonhomologous chromosomes. The combined map confirms the two-chromosome genomic structure and the correspondence of the initial genetic maps to the circular chromosome. The linear chromosome is almost devoid of auxotrophic markers, which probably explains why it was missed by genetic mapping studies.
Most of the work on Agrobacterium tumefaciens, since its identification as the causal agent in crown gall disease of dicotyledonous plants at the turn of the century, has rightfully focused on the mechanism of tumor induction (52; for recent reviews of all aspects of the disease, see references 2, 10, 23, 39, and 58). The virulence mechanism turns out to be unique among interactions between prokaryotic pathogens and eukaryotic hosts. Since most of the virulence genes lie on the Ti plasmid, the chromosomal complement of A. tumefaciens has been relatively understudied.
Initial chromosomal maps for A. tumefaciens, based on chromosome mobilization and recombination of genetic markers, suggested a single circular chromosome (11, 29, 44, 47, 48). However, recent physical mapping data strongly suggests that A. tumefaciens has two chromosomes, one circular chromosome of ∼3 Mbp and one linear chromosome of ∼2.1 Mbp (1, 31). This chromosome organization appears to be a conserved trait throughout the genus (32). While multiple chromosomes have been found in some other eubacteria (7–9, 43, 54, 57), we were interested in the discrepancy between the initial genetic maps and the more recent physical mapping data. We hypothesized that the original genetic mapping techniques somehow missed the linear chromosome. To test this hypothesis, we constructed a combined genetic and physical map of the A. tumefaciens genome by collecting a large number of transposon-mediated auxotrophic mutations, using a transposon carrying rare restriction sites, and then physically mapping the transposon insertions by using pulsed-field gel electrophoresis (PFGE). Our results confirm the two-chromosome genome organization, and we found that almost all the auxotrophic markers lie on the circular chromosome. We put forward an explanation for the discrepancy between the initial genetic maps and the physical mapping data and suggest some hypotheses for the gene organization in this bacterial species.
(Partial results of this work were presented at the 19th Annual Crown Gall Meeting [24] and at the Microbial Genomes III Conference [25].)
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The A. tumefaciens strain and plasmids used in this study are described in Table 1. A. tumefaciens cultures were grown in a modified Luria-Bertani (LB) medium (only 5 g of NaCl/liter) at 30°C. Screens for A. tumefaciens auxotrophic mutants were carried out in M9 minimal medium with sucrose as a carbon source (45). The antibiotics carbenicillin, kanamycin, rifampin, and tetracycline were used as needed at 50, 50, 20, and 10 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Pertinent characteristics | Reference(s) |
|---|---|---|
| A. tumefaciens | ||
| A348 | Prototrophic biovar 1 strain; C58 derivative cured of pTiC58, with pTiA6 introduced by mating; Rfr | 22 |
| Plasmids | ||
| pUT::Tn5(pfm) | λpir-dependent replicon carrying a mini-Tn5 derivative with rare restriction sites and a separate transposase gene; plasmid confers Apr, and Tn5(pfm) confers Kanr and Cmr; maintained in E. coli S17-1 | 15, 60 |
| R68.45 | Broad-host-range conjugable plasmid; confers Cbr, Tcr, and Kanr | 27 |
| pG644 | Cosmid clone of A. tumefaciens chromosomal sequences containing the att gene cluster in pVK102; confers Kanr | 36, 41 |
| pCP13.101 | Cosmid clone of A. tumefaciens chromosomal sequences containing the cel gene cluster in pCP13; confers Tcr | 21, 40 |
| pRK290 | Broad-host-range mobilizable RK2-based plasmid; parent plasmid to pCP13 and pVK102 cosmid vectors; confers Tcr | 17 |
| pRK2013 | Helper plasmid for mobilization of RK2-based plasmids carrying the rlx locus; ColE1 replicon; confers Kanr | 19 |
Matings.
Donor (either Escherichia coli or A. tumefaciens carrying a plasmid) and recipient (A. tumefaciens) strains were mixed by streaking on a modified LB agar plate and incubated at 30°C for 48 h. When necessary (when the donor plasmid was a mobilizable pRK290 derivative), a third strain, E. coli carrying pRK2013, was included in the mating. The cell mixture was scraped off, resuspended in M9 minimal medium without a carbon source, diluted, and plated on the appropriate selective medium.
Mutant isolation and characterization.
A. tumefaciens A348 was mated with E. coli S17-1/pUT::Tn5(pfm). A. tumefaciens carrying Tn5(pfm) insertions were selected on modified LB medium containing kanamycin [selective for the presence of Tn5(pfm)] and rifampin (selective for A. tumefaciens). Single colonies were screened for auxotrophy by plating on M9 and modified LB medium. Potential Tn5(pfm)-induced auxotrophs were tested on M9 plates with various nutrient pools and later supplemented with specific pathway intermediates (13).
Confirmation of the linkage between Tn5(pfm) insertion and auxotrophic mutation.
A. tumefaciens auxotrophic mutant strains were grown overnight at 30°C in 2-ml cultures. Total genomic DNA was isolated from each strain and resuspended at ∼0.4 to 0.5 μg/ml (16). Approximately 4 to 5 μg (10 μl) of each sample was electroporated into competent wild-type A. tumefaciens A348 cells, and the transformed cells were plated on modified LB medium containing kanamycin (5). Three days later, the few resulting colonies were picked from each transformation and streaked onto modified LB plates containing kanamycin, M9 plates containing kanamycin, and M9 plates containing kanamycin and the specific nutrient required by the original A. tumefaciens auxotrophic mutant.
PFGE of intact and digested DNAs.
Wild-type and mutant A. tumefaciens strains used in physical mapping were grown for 48 h at 30°C in 2-ml cultures. Cells were pelleted, suspended in 2% agarose plugs, digested overnight with pronase E (2 mg/ml) at 50°C and washed (53). Restriction enzyme digest of genomic DNA in the agarose plugs by PacI and SwaI (New England Biolabs) were carried out at 25°C for 24 h. PFGE was carried out in a contour-clamped homogeneous electric field apparatus (Bio-Rad), in 0.5× TBE buffer (45 mM Tris, 45 mM borate, 1.25 mM EDTA [pH 8.3]). Restriction enzyme digests of genomic DNA were electrophoresed through 1% agarose gels with a ramp of 40 to 90 s for 22 h at 180 V. Lambda ladder (Bio-Rad) served as size markers.
Plasmid-mediated mobilization of chromosomal markers.
R68.45, a conjugable plasmid used in several of the original genetic mapping experiments, was mobilized into several A. tumefaciens Tn5(pfm)-induced auxotrophic mutants in independent overnight matings with E. coli harboring R68.45. The presence of R68.45 in the A. tumefaciens strains was selected by growth on modified LB medium containing kanamycin [selective for the presence of Tn5(pfm)], carbenicillin (selective for the presence of R68.45), and rifampin (selective for A. tumefaciens). A. tumefaciens strains carrying R68.45 were used as donors in overnight matings on modified LB medium with recipient A. tumefaciens strains harboring different auxotrophic mutations. Transconjugants in which the auxotrophic marker of the recipient strain had been replaced by the wild-type counterpart from the donor chromosome were selected by plating dilutions of a mating mixture onto M9-sucrose medium.
RESULTS
Construction of a combined genetic and physical map for A. tumefaciens A348.
To build on the results of Allerdet-Servent et al. (1), we first repeated their experiments with A. tumefaciens A348. We obtained identical results for strain A348 (data not shown), which differs from strain C58 only by having a chromosomal rifampin resistance mutation and a different Ti plasmid (22). Next, we devised a strategy for physical localization of genetic markers with digestions by PacI and SwaI, the same enzymes used in the initial physical mapping experiments (1). Tn5(pfm), a minitransposon carrying selectable markers and several rare restriction sites, was introduced by mating into A. tumefaciens A348 (60). From 30 independent matings, approximately 11,000 kanamycin-resistant colonies were replicated onto LB and M9-sucrose minimal media. Of these, 103 were identified as Tn5(pfm)-induced auxotrophs. A total of 56 independently isolated auxotrophs were chosen for further analysis, with 45 eventually being characterized down to the affected biosynthetic pathway and the remainder having unknown requirements (Tables 2 and 3).
TABLE 2.
Biochemical complementation and physical mapping of Tn5(pfm)-induced auxotrophic mutations
| Mutationb | Biochemical complementation | Restriction fragment abolished by Tn5(pfm) insertiona and new fragments created (kb)
|
|
|---|---|---|---|
| PacI | SwaI | ||
| ade-101::Tn5C1 | Adenine only | Hc | B; 855, 35 |
| ade-102::Tn5C2 | Adenine or hypoxanthine | A; 760, 660 | B; 790, 105 |
| ade-103::Tn5L1 | Adenine or hypoxanthine | B; 565, 400 | C; 540, 230 |
| ade-104::Tn5C3 | Adenine or hypoxanthine | Hc | B; 790, 105 |
| aah-101::Tn5C4, C5 | Adenine + histidine | C; 500, 290 | A; 825, 300 |
| aah-102::Tn5C6 | Adenine + histidine | A; 1090, 330 | A; 840, 285 |
| aat-101::Tn5C7 | Adenine + thiamine | A; 970, 450 | B; 780, 115 |
| aat-102::Tn5C8 | Adenine + thiamine | H; 230, 20 | B; 820, 75 |
| cys-101::Tn5C9 | Cysteine, thiosulfate, or sulfite | A; 1265, 155 | B; 470, 425 |
| cys-102::Tn5C10 | Cysteine or thiosulfate | A; 1330, 90 | A; 725, 400 |
| glt-101::Tn5C11, C12, C13 | Glutamate or glutamine | C; 410, 380 | A; 975, 150 |
| gln-101::Tn5C14 | Glutamine | A; 1225, 195 | B; 485, 410 |
| gln-102::Tn5C15 | Glutamine | A; 1050, 370 | A; 845, 280 |
| his-101::Tn5C16 | Histidine | A; 1090, 330 | A; 840, 285 |
| ilv-101::Tn5C17, C18 | Isoleucine + valine | E; 295, 90 | Hc |
| leu-101::Tn5C19, C20, C21 | Leucine or α-ketoisocaproate | C; 400, 390 | A; 970, 155 |
| met-101::Tn5C22 | Methionine or cystathione | A; 1220, 200 | A; 710, 415 |
| met-102::Tn5C23, C24 | Methionine or cystathione | C; 395, 395 | A; 1035, 90 |
| pan-101::Tn5C25 | Pantothenate or β-alanine | C; 470, 320 | A; 1070, 55 |
| pan-102::Tn5L2 | Pantothenate or β-alanine | B; 765, 200 | D; 460, 180 |
| prx-101::Tn5C26 | Pyridoxine | A; 890, 530 | —d |
| ser-101::Tn5L3, L4 | Serine, glycine, or threonine | B; 510, 455 | C; 680, 90 |
| ser-102::Tn5C27 | Serine or glycine | Ec | Hc |
| ser-103::Tn5C28 | Serine only | A; 1290, 130 | B; 565, 330 |
| thi-101::Tn5C29, C30, C31 | Thiamine | C; 530, 260 | — |
| thi-102::Tn5C32 | Thiamine | C; 490, 300 | A; 840, 285 |
| thr-101::Tn5L5 | Threonine | B; 490, 475 | C; 660, 110 |
| trp-101::Tn5C33, C34, C35 | Tryptophan or anthranilate | Ec | G; 255, 70 |
| trp-102::Tn5C36, C37 | Tryptophan only | C; 400, 390 | A; 790, 335 |
| trp-103::Tn5C38 | Tryptophan only | H; 235, 15 | — |
| trp-104::Tn5C39 | Tryptophan or shikimate | E; 355, 20 | — |
| ura-101::Tn5C40 | Uracil or orotic acid | A; 1185, 235 | A; 655, 470 |
PacI and SwaI restriction fragments are designated by the letters used in the nomenclative system of Allerdet-Servant et al. (1).
We used the standard genetic nomenclature except for two novel auxotrophic phenotypes, for which we derived new genetic abbreviations (aah and aat). The C or L designation for each insertion indicates its location on the circular or linear chromosome, respectively.
We were unable to detect and measure the new restriction fragments due to their small size.
—, there was no detectable difference from the wild-type restriction pattern, most probably due to the Tn5(pfm) insertion being too close to the end of a restriction fragment.
TABLE 3.
Physical mapping of Tn5(pfm)-induced mutations with unknown auxotrophic requirements
| Mutationb | Restriction fragments abolished by Tn5(pfm) insertiona and new fragments created (kb)
|
|
|---|---|---|
| PacI | SwaI | |
| aux-101::Tn5C41 | A; 950, 470 | A; 1040, 85 |
| aux-102::Tn5L6 | F; 210, 140 | D; 460, 180 |
| aux-103::Tn5C42 | C; 430, 360 | A; 1025, 100 |
| aux-104::Tn5C43 | C; 470, 320 | A; 1070, 55 |
| aux-105::Tn5C44 | C; 730, 60 | A; 655, 470 |
| aux-106::Tn5C45 | A; 1290, 130 | B; 495, 400 |
| aux-107::Tn5C46 | A; 1250, 170 | B; 460, 435 |
| aux-108::Tn5L7 | B; 750, 215 | C; 440, 330 |
| aux-109::Tn5C47 | C; 540, 250 | —d |
| aux-110::Tn5C48 | A; 940, 480 | B; 820, 75 |
| aux-111::Tn5C49 | Ac | B; 640, 255 |
PacI and SwaI restriction fragments are designated by the letters used in the nomenclative system of Allerdet-Servant et al. (1).
The C or L designation for each insertion indicates its location on the circular or linear chromosome, respectively.
We were unable to detect and measure the new restriction fragments due to their small size.
There was no detectable difference from the wild-type restriction pattern, most probably due to the Tn5(pfm) insertion being too close to the end of a restriction fragment.
To confirm that the auxotrophy was due to the Tn5(pfm) insertion, genomic DNA was individually isolated from a random subset of the auxotrophic strains (aah-102::Tn5C6, gln-102::Tn5C15, met-102::Tn5C23, ser-101::Tn5L4, trp-101::Tn5C34, and aux-102::Tn5L6). Each genomic DNA sample was electroporated into wild-type A. tumefaciens A348, and the transformants were plated on LB medium containing kanamycin to select for cells in which the transposase-less Tn5(pfm) insertion had been recombined into the recipient genome (5). Kanamycin-resistant colonies were then screened for coinheritance of the proper auxotrophic marker. In all cases, the kanamycin-resistant colonies from a given electroporation carried the auxotrophic marker corresponding to the auxotrophic strain whose genomic DNA had been used in that electroporation.
Tn5(pfm) insertions were mapped by PFGE of PacI- and SwaI-digested genomic DNA. Since Tn5(pfm) carries PacI and SwaI restriction sites, the insertion of the transposon leads to an altered restriction pattern compared to the wild type. A total of 56 Tn5(pfm)-induced auxotrophic mutations and 28 prototrophic Tn5(pfm) insertions were mapped (Tables 2 to 4). The PacI and SwaI digestion patterns were consistent with single Tn5(pfm) insertions in each mutant and served to identify the fragments harboring the transposon (Fig. 1).
TABLE 4.
Physical mapping of Tn5(pfm) prototrophic insertions
| Insertionb | Restriction fragments abolished by Tn5(pfm) insertiona and new fragments created (kb)
|
|
|---|---|---|
| PacI | SwaI | |
| Tn5C50 | A; 1270, 120 | —c |
| Tn5C51 | A; 1150, 270 | —c |
| Tn5C52 | Ac | B; 585, 310 |
| Tn5C53 | C; 770, 20 | Gc |
| Tn5C54 | A; 1230, 190 | Bc |
| Tn5C55 | Ac | Bc |
| Tn5C56 | Ec | Hc |
| Tn5C57 | —c | Ac |
| Tn5C58 | Cc | —c |
| Tn5C59 | —c | Ac |
| Tn5C60 | —c | Bc |
| Tn5C61 | —c | Bc |
| Tn5C62 | —c | Bc |
| Tn5L8 | Fc | D; 420, 220 |
| Tn5L9 | B; 690, 275 | D; 560, 80 |
| Tn5L10 | B; 530, 435 | C; 555, 215 |
| Tn5L11 | B; 510, 455 | C; 530, 240 |
| Tn5L12 | D; 475, 235 | E; 430, 85 |
| Tn5L13 | Fc | D; 430, 210 |
| Tn5L14 | Bc | D; 410, 230 |
| Tn5L15 | F; 210, 140 | D; 195, 455 |
| Tn5L16 | Bc | C; 450, 320 |
| Tn5L17 | Dc | Ec |
| Tn5L18 | Bc | Dc |
| Tn5L19 | Dc | Ec |
| Tn5L20 | Bc | Cc |
| Tn5L21 | —c | Ec |
| Tn5L22 | —c | Cc |
PacI and SwaI restriction fragments are designated by the letters used in the nomenclature system of Allerdet-Servant et al. (1).
The C or L designation for each insertion indicates its location on the circular or linear chromosome, respectively.
We did not characterize the insertion further.
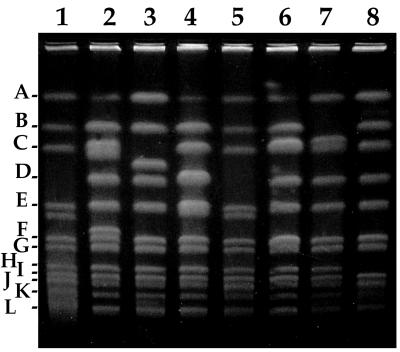
FIG. 1.
Example of PFGE of SwaI-digested genomic DNA from A. tumefaciens A348 strains carrying Tn5(pfm)-induced auxotrophic mutations: aux-102::Tn5L6 (lane 1), met-101::Tn5C22 (lane 2), thr-101::Tn5L5 (lane 3), aux-105::Tn5C44 (lane 4), pan-102::Tn5L2 (lane 5), ura-101::Tn5C40 (lane 6), ade-104::Tn5C3 (lane 7), and ilv-101::Tn5C18 (lane 8). The positions of the wild-type SwaI restriction fragments are shown on the left, in the nomenclature of Allerdet-Servant et al. (1). Only one wild-type restriction fragment is missing in each mutant strain.
Essential features of the map.
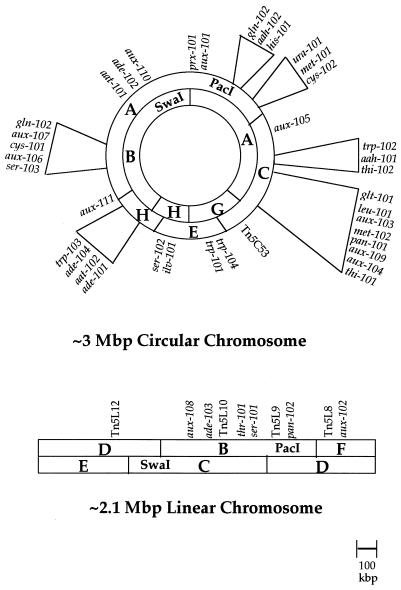
The results of the mapping were entirely consistent with the findings of earlier studies indicating two independent chromosomes, a 3.0-Mb circle and a 2.1-Mb linear structure (1). Furthermore, the restriction enzyme digestions of the genomic DNA of the various Tn5(pfm) mutants allowed us to order the PacI and SwaI fragments on each of the chromosomes and to localize a large number of the transposon insertions (Fig. 2). Insertions were found on both chromosomes. None of the transposon insertions localized to the small SwaI fragments J, K (doublet), L, M, and N. All but one of these small fragments had been assigned to chromosomes earlier by Southern hybridization (1), but we were unable to assign specific map positions for them.
FIG. 2.
Best-fit combined genetic and physical maps of the two A. tumefaciens A348 chromosomes. The PacI and SwaI restriction fragments are designated by the letters used in the nomenclature system of Allerdet-Servant et al. (1). The locations of all auxotrophic and some prototrophic Tn5(pfm) insertions are indicated by their appropriate abbreviations along the outer edge of each map. The small SwaI restriction fragments J, K (doublet), L, and M, which were previously localized by hybridization (1) to the circular (J, K1, and L) and linear (K2 and M) chromosomes, are not shown on these maps, since we were unable to obtain a Tn5(pfm) insertion in any of them.
Auxotrophic markers were found for almost all biosynthetic pathways, with the major exceptions being the arginine, lysine, and proline pathways. Auxotrophic markers are present on both chromosomes but predominantly (conservative estimate of 37 of 43 loci) on the circle. The essential genes on the circular chromosome are widely scattered, with little evidence of pathway-specific gene clusters. In contrast, we were unable to find auxotrophic markers on over one-third of the linear chromosome. This was not due to the lack of Tn5(pfm) insertions in the linear chromosome, since prototrophic Tn5(pfm) insertions were found on the two chromosomes at comparable frequencies, with 13 hits on the circular chromosome and 15 hits on the linear chromosome (Table 4).
Correspondence of the physical-genetic map to previous genetic maps.
It seemed reasonable to suggest that the circular chromosome recognized by physical mapping and further characterized in this study is the same as the circular chromosome from earlier genetic mapping studies. To further anchor our physical and genetic map in comparison to previous genetic maps, we tested whether a methionine biosynthetic gene used in previous genetic mapping studies was the same as any of the methionine biosynthetic genes identified by Tn5(pfm) mutagenesis. The chemically induced auxotrophic mutation met6 had previously been shown to map very close to both the att gene cluster, required for initial binding of A. tumefaciens to plant cells, and the cel gene cluster, encoding a cellulose biosynthesis pathway (48). Both the att and cel gene clusters have been separately isolated from genomic cosmid libraries (40, 41). To determine if the cosmid clones carrying the att and cel gene clusters also harbored the wild-type met6 gene and whether that gene would complement any of our Tn5(pfm)-induced methionine auxotrophs, the att gene cluster cosmid pG644, the cel gene cluster cosmid pCP13.101, and pRK290, the parent plasmid on which the cosmids are based (17, 21, 36), were independently mobilized into A. tumefaciens A348 strains carrying the auxotrophic mutations met-101::Tn5C22, met-102::Tn5C23, and met-102::Tn5C24. The parent plasmid pRK290 and the att gene cluster cosmid pG644 failed to complement any of the mutations. However, the cel gene cluster cosmid pCP13.101 complemented the met-102::Tn5C23 and met-102::Tn5C24 mutations, which have transposon insertions at the same location.
Ability of the circular and linear chromosomes to be mobilized.
One possible explanation for the failure of the original genetic mapping experiments to detect both chromosomes may be a reduced ability of the linear chromosome to be mobilized. We used R68.45, the same conjugable plasmid used in many of the original genetic mapping experiments, in experiments to determine if the circular and linear chromosomes each could be mobilized (27). The basic strategy was to mate donor and recipient strains carrying different Tn5(pfm) insertions. The donor strain also harbored R68.45. Chromosome mobilization was determined by selection for recipients in which the Tn5(pfm) insertion site of the recipient strain had been replaced by its wild-type counterpart from the donor strain (Table 5). In a control experiment where the donor (cys-101::Tn5C9) and recipient (cys-101::Tn5C9) strains carried the exact same transposon insertion, no wild-type transconjugants were found, as expected. This also showed that Tn5(pfm) insertions do not revert at a measurable frequency, which makes sense since this minitransposon lacks a transposase gene (60). In experiments where the donor and recipient strains carried different Tn5(pfm) insertions, the circular and linear chromosomes were mobilized at comparable frequencies. The only exceptions were cases in which the auxotrophic markers in the donor and recipient strains were near each other on the same chromosome (trp-101::Tn5C33 × cys-101::Tn5C9, cys-101::Tn5C9 × trp-101::Tn5C33, and aux-108::Tn5L7 × thr-101::Tn5L5).
TABLE 5.
Frequency of R68.45-mediated-mobilization and recombination of markers on the circular and linear chromosomes
| Mutation in donor strain carrying R68.45a | Mutation in recipient straina | Frequency of wild-type transconjugants per recipient cell |
|---|---|---|
| cys-101::Tn5C9 | cys-101::Tn5C9 | <2.3 × 10−9b |
| gln-102::Tn5C15 | cys-101::Tn5C9 | 6.0 × 10−6 |
| trp-101::Tn5C33 | cys-101::Tn5C9 | 1.5 × 10−8 |
| aux-108::Tn5L7 | cys-101::Tn5C9 | 4.2 × 10−6 |
| cys-101::Tn5C9 | thr-101::Tn5L5 | 1.3 × 10−6 |
| gln-102::Tn5C15 | thr-101::Tn5L5 | 4.6 × 10−6 |
| trp-101::Tn5C33 | thr-101::Tn5L5 | 1.3 × 10−6 |
| aux-108::Tn5L7 | thr-101::Tn5L5 | 8.3 × 10−8 |
| cys-101::Tn5C9 | aux-108::Tn5L7 | 4.6 × 10−7 |
| cys-101::Tn5C9 | gln-102::Tn5C15 | 7.7 × 10−6 |
| cys-101::Tn5C9 | trp-101::Tn5C33 | <4.1 × 10−9b |
The C or L designation for each insertion indicates its location on the circular or linear chromosome, respectively.
No transconjugants were recovered, and so the number given is an estimate of the upper limit possible.
DISCUSSION
We were able to confirm the two-chromosome genome organization in A. tumefaciens by constructing a combined genetic and physical map of the circular and linear chromosomes (Fig. 2). A strong case can be made that the circular chromosome is the chromosome on which previous genetic maps are based. The previous genetic maps are congruent with one another and are consistent with a circular chromosome (11, 29, 44, 47, 48). Furthermore, the chvAB genes, encoding enzymes involved in extracellular β-glucan production, had been placed on one of the genetic maps and were later shown in the initial physical mapping to hybridize to PacI fragment A and SwaI fragment A of the circular chromosome (1, 11, 18). Finally, we were able to prove that the met6 gene, located on one of the genetic maps, is the same as one of the methionine biosynthetic genes we physically localized to the circular chromosome (48).
We can rule out some explanations of why the genetic mapping approaches missed the linear chromosome. First, the chromosome mobilization experiment shows that genetic markers on the linear chromosome can be mobilized by R68.45 at frequencies comparable to those for markers on the circular chromosomes (Table 5). This shows that the linear chromosome is not recalcitrant to conjugation-based mobilization due to its topology. Second, we found prototrophic Tn5(pfm) insertions at similar frequencies for both chromosomes. This discounts the possibility of transpositional bias between the chromosomes. Therefore, we are left with a simple but intriguing hypothesis. Since the genetic mapping approaches mainly used auxotrophic markers and we found that virtually all such markers (∼86%) lie on the circular chromosome, it is possible that the small collections of auxotrophic strains used for the genetic maps do not contain any examples of mutations on the linear chromosome. We believe that this hypothesis is robust based on the large number of independent auxotrophic markers, both characterized and uncharacterized, that we physically mapped. The fact that some biosynthetic pathways (arginine, lysine, phenylalanine, proline, and tyrosine) are not represented in our collection is not worrisome, since it is possible to find such auxotrophic markers for these pathways in A. tumefaciens and since several of these markers were mapped to the circular chromosome by purely genetic approaches (11, 29, 44, 47, 48).
The paucity of auxotrophic markers on the linear chromosome brings up the question of the origin of the two-chromosome state in this genus (32). If the current chromosomes resulted from a splitting of a single ancestral chromosome, one might expect those two chromosomes to have similar densities of auxotrophic markers. To see if this is true for single chromosome genomes, we looked at the distribution of putative auxotrophic markers (i.e., genes involved in amino acid, cofactor or vitamin, and nucleotide biosynthesis that, when mutated, would lead to auxotrophy) in the published genomic sequences of several members of the Eubacteria and Archaea (3, 4, 14, 20, 33, 35, 37, 51, 56). In the genomes analyzed, auxotrophic markers are rarely separated by more than 100 kbp, with the largest gap being less than 300 kbp. An even better comparison is the recent low-resolution sequencing of approximately one-third of the ∼0.9-Mb chromosome II of Rhodobacter sphaeroides 2.4.1T (8, 9, 54). Putative auxotrophic markers were found at a density slightly lower than but comparable to that for the single-chromosome genomes. In contrast, we found only six auxotrophic markers on the 2.1-Mbp linear chromosome of A. tumefaciens, with approximately one-third of the linear chromosome being devoid of such markers. As detailed genetic maps or complete genomic sequences become available for other species with multiple non-homologous chromosomes, such as Brucella melitensis (31, 43), Burkolderia (formerly Pseudomonas) cepacia (7), and Vibrio cholera (57), it will be interesting to see if they show asymmetry in the distribution of auxotrophic markers between their chromosomes.
One hypothesis to explain the lack of auxotrophic markers on one-third of the linear chromosome is a bias against Tn5(pfm) jumping into this region due to a different base composition. The only data we obtained that can address this idea is the distribution of randomly chosen prototrophic Tn5(pfm) insertions. Of 15 such insertions on the linear chromosome, 4 mapped to the region lacking auxotrophic markers (Table 4). This number closely matches that expected for random insertion of the transposon. While this small data set cannot disprove the hypothesis, it is highly suggestive that transpositional bias is not the cause of the asymmetrical distribution of auxotrophic markers on the linear chromosome.
An alternative hypothesis is the acquisition or evolution of a large cluster of nonessential genes either on the ancestral chromosome before the split into two chromosomes or on the linear chromosome after the split. For example, the linear chromosome may contain a large gene cluster specifically involved in the interaction of A. tumefaciens with plant tissue. This is seen in several animal pathogens, where many virulence genes are clustered into “pathogenicity islands” (28, 38). Of the known A. tumefaciens chromosomal virulence genes, only the chvAB operon, the att gene cluster, the cel gene cluster, and the ros gene have been mapped, and all are located on the circular chromosome (1, 11, 48). To further test this hypothesis, the other known chromosomal virulence genes and nonvirulence genes implicated in the plant-microbe interaction need to be localized on the physical map (6, 26, 30, 34, 42, 46, 49, 50, 55).
Ultimately, a fuller understanding of the genetic structure and role of the two chromosomes in the ecology of A. tumefaciens will require genomic sequencing. We have initiated such an effort for the ∼710-kbp PacI fragment D of the linear chromosome. We hope to verify the real size of the auxotrophic marker gap on the linear chromosome. Other benefits will include the identification of (i) additional genes that can be used for structure-function, evolutionary, and comparative genomic studies, (ii) a bacterial telomere, and (iii) genes involved in the interaction of A. tumefaciens with plant tissues.
ACKNOWLEDGMENTS
Research support was provided by a University of Richmond Faculty Research Grant to B.W.G.; University of Richmond Undergraduate Research Grants to M.C.F., B.P.M., J.L.R., and L.M.H.; and University of Richmond Summer Undergraduate Research Fellowships to B.P.M. and B.A.S.
We are grateful to Jeff Elhai, Todd Steck, Ann Matthysse, Kwong Kwok Wong, and Michael McClelland for the gifts of strains and plasmids and for many helpful suggestions. We also thank the students of the BIOL315 course at University of Richmond for help in the initial auxotrophic mutant screens, Bill Shanabruch for helpful instruction on PFGE, two anonymous reviewers for their comments, and Dahlia Doughty and Charlaine Scott for help with the experiments needed to address the reviewers’ comments.
REFERENCES
- 1.Allardet-Servent A, Michaux-Charachon S, Jumas-Bilak E, Karayan L, Ramuz M. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J Bacteriol. 1993;175:7869–7874. doi: 10.1128/jb.175.24.7869-7874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binns A N, Thomashow M F. Cell biology of Agrobacterium infection and transformation of plants. Annu Rev Microbiol. 1988;42:575–606. [Google Scholar]
- 3.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 5.Charles T C, Doty S L, Nester E W. Construction of Agrobacterium strains by electroporation of genomic DNA and its utility in analysis of chromosomal virulence mutations. Appl Environ Microbiol. 1994;60:4192–4194. doi: 10.1128/aem.60.11.4192-4194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charles T C, Nester E W. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1993;175:6614–6625. doi: 10.1128/jb.175.20.6614-6625.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng H-P, Lessie T G. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J Bacteriol. 1994;176:4034–4042. doi: 10.1128/jb.176.13.4034-4042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary M, Mackenzie C, Nereng K S, Sodergren E, Weinstock G M, Kaplan S. Multiple chromosomes in bacteria: structure and function of chromosome II of Rhodobacter sphaeroides 2.4.1T. J Bacteriol. 1994;176:7694–7702. doi: 10.1128/jb.176.24.7694-7702.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhary M, Mackenzie C, Nereng K, Sodergren E, Weinstock G M, Kaplan S. Low-resolution sequencing of Rhodobacter sphaeroides 2.4.1T: chromosome II is a true chromosome. Microbiology. 1997;143:3085–3099. doi: 10.1099/00221287-143-10-3085. [DOI] [PubMed] [Google Scholar]
- 10.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooley M B, Kado C I. Mapping of the ros virulence regulatory gene of A. tumefaciens. Mol Gen Genet. 1991;230:24–27. doi: 10.1007/BF00290645. [DOI] [PubMed] [Google Scholar]
- 12.Cooley M B, Souza M R D, Kado C I. The virC and virD operons of the Agrobacterium Ti plasmid are regulated by the ros chromosome gene: analysis of the cloned ros gene. J Bacteriol. 1991;173:2608–2616. doi: 10.1128/jb.173.8.2608-2616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis R W, Botstein D, Roth J R. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1980. pp. 206–208. [Google Scholar]
- 14.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 15.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiRita V J, Gelvin S B. Deletion analysis of the mannopine synthase gene promoter in sunflower crown gall tumors and Agrobacterium tumefaciens. Mol Gen Genet. 1987;207:233–241. doi: 10.1007/BF00331583. [DOI] [PubMed] [Google Scholar]
- 17.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglas C J, Staneloni R J, Rubin R A, Nester E W. Identification and genetic analysis of an Agrobacterium tumefaciens chromosomal virulence region. J Bacteriol. 1985;161:850–860. doi: 10.1128/jb.161.3.850-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Suadek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 21.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 22.Garfinkel D J, Simpson R B, Ream L W, White F F, Gordon M P, Nester E W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981;27:143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 23.Gelvin S B. Crown gall disease and hairy root disease: a sledgehammer and a tackhammer. Plant Physiol. 1990;92:281–285. doi: 10.1104/pp.92.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodner B, Markelz B, Racette J, Flanagan C, Crowell C, Mellors S, Schilling B, Halfon L. Proceedings of the 19th Annual Crown Gall Meeting. 1998. Chromosome organization in Agrobacterium; p. 1. [Google Scholar]
- 25.Goodner B W, Markelz B P, Flanagan M C, Crowell C B, Racette J L, Schilling B A, Mellors J S, Lappas C M. Microbial Genomes III: Sequencing, Functional Characterization, and Comparative Genomics. February 1999. 1999. An Agrobacterium tumefaciens genome project at a primarily undergraduate institution—current progress and future goals; p. C24. [Google Scholar]
- 26.Gray J, Wang J, Gelvin S B. Mutation of the miaA gene of Agrobacterium tumefaciens results in reduced vir gene expression. J Bacteriol. 1992;174:1086–1098. doi: 10.1128/jb.174.4.1086-1098.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas D, Holloway B W. Chromosome mobilization by the R plasmid R68.45: a tool in Pseudomonas genetics. Mol Gen Genet. 1978;158:229–237. doi: 10.1007/BF00267194. [DOI] [PubMed] [Google Scholar]
- 28.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function, and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 29.Hooykaas P J J, Peerbolte R, Regensburg-Tuink A J G, de Vries P, Schilperoort R A. A chromosomal linkage map of Agrobacterium tumefaciens and a comparison with the maps of Rhizobium spp. Mol Gen Genet. 1982;188:12–17. [Google Scholar]
- 30.Huang M W, Cangelosi G A, Halperin W, Nester E W. A chromosomal Agrobacterium tumefaciens gene required for effective plant signal transduction. J Bacteriol. 1990;172:1814–1822. doi: 10.1128/jb.172.4.1814-1822.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jumas-Bilak E, Maugard C, Michaux-Charachon S, Allardet-Servent A, Perrin A, O’Callaghan D, Ramuz M. Study of the organization of the genomes of Escherichia coli, Brucella melitensis and Agrobacterium tumefaciens by insertion of a unique restriction site. Microbiology. 1995;141:2425–2432. doi: 10.1099/13500872-141-10-2425. [DOI] [PubMed] [Google Scholar]
- 32.Jumas-Bilak E, Michaux-Charachon S, Bourg G, Ramuz M, Allerdet-Servent A. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J Bacteriol. 1998;180:2749–2755. doi: 10.1128/jb.180.10.2749-2755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 34.Kemner J M, Liang X, Nester E W. The Agrobacterium tumefaciens virulence gene chvE is part of a putative ABC-type sugar transport operon. J Bacteriol. 1997;179:2452–2458. doi: 10.1128/jb.179.7.2452-2458.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 36.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 37.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S-K, Codani J-J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S-Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo M-F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S-M, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Ouderga B, Park S-H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A M, Prescecan E, Pujic P, Purnelle B, Rapaport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror S J, Serror P, Shin B-S, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognoni A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa H-F, Zumstein E, Yoshikawa H, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 38.Lee C A. Pathogenicity islands and the evolution of bacterial pathogens. Infect Agents Dis. 1996;5:1–7. [PubMed] [Google Scholar]
- 39.Matthysse A G. Initial interactions of Agrobacterium tumefaciens with plant host cells. Crit Rev Microbiol. 1986;13:281–307. doi: 10.3109/10408418609108740. [DOI] [PubMed] [Google Scholar]
- 40.Matthysse A G, White S, Lightfoot R. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matthysse A G, Yarnall H A, Young N. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J Bacteriol. 1996;178:5302–5308. doi: 10.1128/jb.178.17.5302-5308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metts J, West J, Doares S H, Matthysse A G. Characterization of three Agrobacterium tumefaciens avirulent mutants with chromosomal mutations that affect induction of vir genes. J Bacteriol. 1991;173:1080–1087. doi: 10.1128/jb.173.3.1080-1087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michaux S, Paillisoon J, Carles-Nurit M-J, Bourg G, Allardet-Servent A, Ramuz M. Presence of two independent chromosomes in the Brucella melitensis 16M genome. J Bacteriol. 1993;175:701–705. doi: 10.1128/jb.175.3.701-705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller I S, Fox D, Saeed N, Borland P A, Miles C A, Sastry G R K. Enlarged map of Agrobacterium tumefaciens C58 and the location of the chromosomal regions which affect tumorigenicity. Mol Gen Genet. 1986;205:153–159. [Google Scholar]
- 45.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 437–439. [Google Scholar]
- 46.Parke D. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J Bacteriol. 1995;177:3808–3817. doi: 10.1128/jb.177.13.3808-3817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pischl D L, Farrand S K. Characterization of transposon Tn5-facilitated donor strains and development of a chromosomal linkage map for Agrobacterium tumefaciens. J Bacteriol. 1984;159:1–8. doi: 10.1128/jb.159.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson J L, Holliday T, Matthysse A G. Mapping of Agrobacterium tumefaciens chromosomal genes affecting cellulose synthesis and bacterial attachment to host cells. J Bacteriol. 1988;170:1408–1411. doi: 10.1128/jb.170.3.1408-1411.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rong L, Carpita N C, Mort A, Gelvin S B. Soluble cell wall compounds from carrot roots induce the picA and pgl loci of Agrobacterium tumefaciens. Mol Plant-Microbe Interact. 1994;7:6–14. [Google Scholar]
- 50.Rong L, Karcher S J, Gelvin S B. Genetic and molecular analyses of picA, a plant-inducible locus on the Agrobacterium tumefaciens chromosome. J Bacteriol. 1991;173:5110–5120. doi: 10.1128/jb.173.16.5110-5120.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G, Daniels C, Mao J, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith E F, Townsend C O. A plant tumor of bacterial origin. Science. 1907;25:671–673. doi: 10.1126/science.25.643.671. [DOI] [PubMed] [Google Scholar]
- 53.Sobral B W S, Atherly A G. A rapid and cost-effective method for preparing genomic DNA from gram-negative bacteria in agarose plugs for pulsed-field gel electrophoresis. BioTechniques. 1989;7:938. [PubMed] [Google Scholar]
- 54.Suwanto A, Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: presence of two unique circular chromosomes. J Bacteriol. 1989;171:5850–5859. doi: 10.1128/jb.171.11.5850-5859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomashow M F, Karlinsey J, Marks J R, Hurlbert R E. Identification of a new virulence locus in Agrobacterium tumefaciens that affects polysaccharide composition and plant cell attachment. J Bacteriol. 1987;169:3209–3216. doi: 10.1128/jb.169.7.3209-3216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 57.Trucksis M, Mickalski J, Deng Y K, Kaper J B. The Vibrio cholerae genome contains two unique circular chromosomes. Proc Natl Acad Sci USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winans S C. Two-way chemical signalling in Agrobacterium-plant interactions. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winans S C, Kerstetter R A, Nester E W. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J Bacteriol. 1988;170:4047–4054. doi: 10.1128/jb.170.9.4047-4054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong K K, McClelland M. Dissection of the Salmonella typhimurium genome by use of a Tn5 derivative carrying rare restriction sites. J Bacteriol. 1992;174:3807–3811. doi: 10.1128/jb.174.11.3807-3811.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]