Abstract
The timing of the appearance of the FtsZ ring at the future site of division in Escherichia coli was determined by in situ immunofluorescence microscopy for two strains grown under steady-state conditions. The strains, B/rA and K-12 MC4100, differ largely in the duration of the D period, the time between termination of DNA replication and cell division. In both strains and under various growth conditions, the assembly of the FtsZ ring was initiated approximately simultaneously with the start of the D period. This is well before nucleoid separation or initiation of constriction as determined by fluorescence and phase-contrast microscopy. The durations of the Z-ring period, the D period, and the period with a visible constriction seem to be correlated under all investigated growth conditions in these strains. These results suggest that (near) termination of DNA replication could provide a signal that initiates the process of cell division.
The cell division cycle of Escherichia coli includes more than a dozen genes which are essential for the cell division process (for reviews, see references 26 and 32). The functions of the majority of the proteins involved in cell division are not yet known. However, the biochemical and structural characterization and the in situ visualization of these proteins are rapidly advancing. FtsZ has thus far been identified as the earliest gene product involved in cell division. It was shown to assemble into a ring at midcell by immunogold labeling (4), by in situ immunofluorescence (22), and by production of a fusion protein of FtsZ and green fluorescence protein (27). FtsZ has a tubulin-like structure (24), and, as for tubulin, its polymerization is GTP dependent (28). The cytoplasmic actin-like (36) protein FtsA (27, 47) and the cytoplasmic membrane-bound protein ZipA (16, 17) were reported to colocalize with and be dependent on FtsZ assembly. The integral membrane proteins FtsK (54) and FtsW (46) also depend on FtsZ for their localization.
PBP 3 (or FtsI) is a periplasmic, membrane-anchored peptidoglycan transpeptidase that is specifically involved in cell division (1) and has been shown to localize at midcell at least during the period in which the constriction is visible (46, 48, 49). Three other periplasmic membrane-anchored proteins, FtsQ (7, 8), FtsN (2), and FtsL (14), which are essential for cell division but have otherwise completely unknown functions, also localize at midcell in constricting cells. In rapidly growing cells at 30°C, 45% of the cells showed an FtsN ring at midcell (2), compared to about 80% of FtsZ ring-containing cells. FtsQ could be detected only at midcell in cells with a visible constriction (7). Therefore, these proteins seem to be required at a later stage in the cell division process.
Because FtsZ is one of the proteins that appear very early in the cell cycle and all other cell division proteins thus far depend on its localization at midcell for their own localization, FtsZ seems to be a suitable indicator of cell division initiation. Analysis of the localization of FtsZ by immunofluorescence microscopy as a function of cell length can therefore be used to determine the timing of the cell division process in the cell cycle.
In the so-called nucleoid occlusion model (30, 53), it has been postulated that the timing and the position of cell division are dependent on the influence of the nucleoids on peptidoglycan synthesis. According to this model, repression of peptidoglycan synthesis in the nucleoid region of the cell results in the inhibition of constriction initiation. Constriction can occur only at a site where the repressive effect of the nucleoids has diminished sufficiently as a result of nucleoid segregation. Accordingly, constriction is initiated between segregated nucleoids by a division signal which is supposedly produced upon termination of DNA replication. This model implies that termination of DNA replication occurs before the appearance of an FtsZ ring at midcell.
To validate this model, the correlation between the timing of the appearance of the FtsZ ring and termination of DNA replication should be established. According to the model of Helmstetter and Cooper (19), DNA replication (or the C period) of E. coli cells growing with doubling times of between 20 and 60 min takes about 40 min. After termination of DNA replication, the cell needs another 20 min (the D period) to divide into two daughter cells. Bacteria that grow with a generation time of 20 min will therefore have multiple-fork DNA replication and up to three overlapping cell cycles. Consequently, to determine the average age at which the cell initiates the process of cell division and to correlate this to other cyclic events like the DNA replication cycle, it is essential to grow the cells at long generation times to ensure the absence of overlapping cell cycles.
In this study, the localization of FtsZ and FtsA of E. coli as a function of cell age and with respect to other cell cycle parameters has been investigated by in situ immunofluorescence microscopy. For this purpose, two E. coli strains for which cell cycle parameters have been determined accurately under steady-state slow-growth conditions were chosen. E. coli B/rA was chosen because its DNA replication cycle has been extensively studied (5, 13, 19, 21). The second strain, E. coli K-12 MC4100 lysA, was chosen because it is the parental strain of several isogenic fts mutants and because its peptidoglycan synthesis cycle has been analyzed (42). In addition, its DNA replication cycle and length distribution (20) and constriction period at a growth rate of 85 min (41) have been determined. Based on the analysis of the appearance of the FtsZ ring at various growth rates, we show that the FtsZ ring appears approximately simultaneously with the termination of DNA replication and with the increase of peptidoglycan synthesis at midcell. Implications for the cell division process and the nucleoid occlusion model are discussed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli K-12 MC4100 [F− araD139 Δ(argF-lac)U169 deoC1 flbB5301 lysA ptsF25 rbsR relA1 rpsL150] and its isogenic derivative MC4100 pbpB2158(Ts), a temperature-sensitive mutant (41), were grown to steady state in glucose minimal medium containing 6.33 g of K2HPO4 · 3H2O, 2.95 g of KH2PO4, 1.05 g of (NH4)2SO4, 0.10 g of MgSO4 · 7H2O, 0.28 mg of FeSO4 · 7H2O, 7.1 mg of Ca(NO3)2 · 4H2O, 4 mg of thiamine, 4 g of glucose, and 50 mg of lysine per liter at pH 7.0 and 28°C. E. coli B/rA (ATCC 12407) was grown to steady state in the same minimal medium without thiamine and lysine and with the osmolarity adjusted to 300 mosM with NaCl. The generation time was dependent on the carbon source used (0.4% [wt/vol] glucose or glycerol, 0.08% l-alanine, or a mixture of 0.04% l-alanine and 0.04% l-proline) and the temperature of growth (28, 30, or 37°C). Absorbance was measured at 450 nm with a 300-T-1 microsample spectrophotometer (Gilford Instrument Laboratories Inc., Oberlin, Ohio), and cell numbers were measured with an electronic particle counter (orifice diameter, 30 μm). Cultures were considered to be in steady-state growth if the average cell mass remained constant over time.
Fixation and permeabilization.
Cells were fixed in 2.8% formaldehyde–0.04% glutaraldehyde for 15 min at room temperature. For the permeabilization, the cells were collected at 7,000 rpm for 5 min, washed twice in phosphate-buffered saline (PBS) (pH 7.2), and subsequently incubated in 0.1% Triton X-100 in PBS for 45 min at room temperature. The cells were washed three times in PBS and incubated in PBS containing 100 μg of lysozyme per ml and 5 mM EDTA for 45 min at room temperature. Finally, the cells were washed three times in PBS.
In situ immunofluorescence labeling.
Nonspecific binding sites were blocked by incubating the cells in 0.5% (wt/vol) blocking reagents (Boehringer, Mannheim, Germany) in PBS for 30 min at 37°C. Incubation with primary antibodies, i.e., either a monoclonal antibody (MAb) against FtsZ (45) or a polyclonal antibody against FtsA (a gift from M. Vicente, Consejo Superior de Investigaciones Científicas, Madrid, Spain), diluted in blocking buffer was carried out for 60 min at 37°C. The cells were washed three times with PBS containing 0.05% (vol/vol) Tween 20. Incubation with secondary antibodies, i.e., donkey antimouse antibody conjugated with Cy3 (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) or goat antimouse or goat antirabbit antibody conjugated with Alexa 546 (Molecular Probes, Eugene, Oreg.), diluted in blocking buffer was carried out for 30 min at 37°C. The cells were washed three times in PBS–0.05% Tween 20. The nucleoids were stained with DAPI (4′,6-diamino-2-phenylindole) at a final concentration of 0.5 μg/ml in H2O. The cells were washed once in H2O and resuspended in PBS.
Microscopy and image analysis.
Cells were immobilized on agarose slides as described by Van Helvoort and Woldringh (44) and photographed with a cooled charge-coupled device camera (Princeton Instruments, SARL, Utrecht, The Netherlands) mounted on an Olympus BX-60 fluorescence microscope. Images were taken by using the program IPlab 3.1a (Signal Analytics, Vienna, Va.). In all experiments the cells were photographed first in the phase-contrast mode, then with a DAPI fluorescence filter (U-MWU; excitation at 330 to 385 nm), and finally with an Alexa filter (U-MNG; excitation at 530 to 550 nm). The three photographs were stacked, and the length of each cell was measured with the phase-contrast image, the nucleoid separation was determined with the DAPI image, and the presence of rings or foci was determined with the fluorescence image. Interactive measurements were performed as “structured point collection” on a Macintosh 7200 computer by using the public domain program Object-Image1.62 by Norbert Vischer (University of Amsterdam) (44a), which is based on NIH Image by Wayne Rasband.
RESULTS
Effect of the immunolabeling procedure on the length distribution of E. coli cells.
To be able to correlate cell division protein (divisome) assembly with cellular processes like DNA replication, nucleoid segregation, peptidoglycan synthesis, and cell constriction, care should be taken to study these processes under comparable conditions. For cell cycle parameters this requires steady-state growth (with defined growth media, osmolarities, temperatures, and growth rates). Cells grown in steady state will have a constant age distribution, and the relative frequency of cells in a certain age class will also remain constant, despite the fact that the absolute cell number in the population increases. Thus, for a fraction of cells (constricting cells and Z-ring-containing cells) at the end of the cell cycle with ages (ax) of between Td − tx and Td, tx can be calculated by using
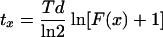 |
1 |
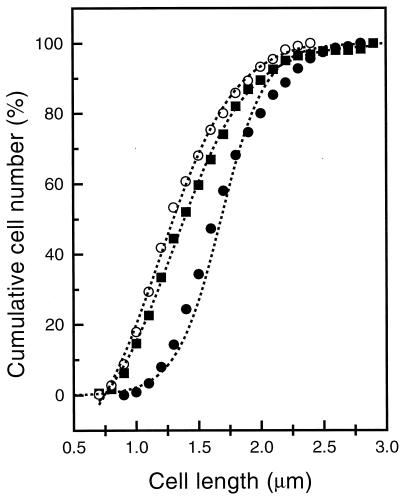
where Td is the generation time and F(x) is the fraction of cells with a certain characteristic which appears at age ax and which lasts until the cells have separated into two daughter cells (34). This approach is valid only if the experimental procedure for the immunolabeling of the cells has no influence on the length distribution of these cells. E. coli B/rA was used to correlate the assembly of the FtsZ ring with the DNA replication cycle, because its DNA replication cycle has been investigated most thoroughly (5, 19, 21). To assess whether the immunolabeling procedure had any effect on the cell length distribution, E. coli B/rA was grown to steady state in minimal medium with a generation time of 130 to 135 min. The cells were harvested at an optical density at 450 nm of 0.1 and divided into three parts. One portion of the cells was fixed with glutaraldehyde and formaldehyde, the second part was fixed and permeabilized, and the third part was fixed, permeabilized, and in addition labeled with MAb F168-12 against FtsZ (45) and a secondary antibody conjugated with Alexa. To assay whether the three treatments had different effects on the shape of the bacteria, the length distribution of at least 500 cells from each treatment was determined. The length distribution of the fixed cells completely overlapped with the distribution described by Koppes et al. (21). Hardly any difference in length distribution between the fixed cells and cells which were fixed and permeabilized could be observed. However, the distribution of cells which had been labeled with antibodies appeared to deviate (Fig. 1). The extra wash and centrifugation steps for labeling of the permeabilized cells increased the length and the width of the cells somewhat. The cumulative cell number plotted against the cell length in Fig. 1 shows that the labeled cells are 0.3 μm longer than the permeabilized cells, irrespective of cell length class. Small cells therefore increase in size relatively more than larger cells. Another explanation for the relative increase in cell length could be that some of the small cells were lost due to the centrifugation steps. This would result in an overestimation of the number of larger cells and an underestimation of the number of smaller cells. If this had occurred, it would be expected that the number of constricting cells would also be higher in the permeabilized and labeled culture than in the permeabilized culture. However, the same percentage of constricting cells (12%) (data not shown) was found for all three treatments. Also, the percentage of cells which had separated nucleoids, based on fluorescence microscopy analysis of cells with DAPI-stained DNA, was 13% for all three treatments (data not shown). Therefore, the various cell length classes are influenced with respect to cell size by the immunolabeling procedure other than by the loss of small cells. It can be concluded that the percentage of cells showing a morphologically detectable cell cycle parameter (i.e., constriction or a Z ring) gives a valid representation of the fraction of cells in the steady-state population with this characteristic. Similar results were found for E. coli K-12 grown to steady state in minimal medium with a generation time of 85 min (results not shown).
FIG. 1.
Cumulative length distribution of E. coli B/rA grown to steady state with a generation time of 130 min. The cells were either fixed (○), fixed and permeabilized (■), or fixed, permeabilized, labeled with MAb F168-12 against FtsZ, and fluorescence stained with a Cy3 secondary antibody (●). Data are for 514, 505, and 723 cells, respectively.
Determination of the timing of FtsZ ring and FtsA ring assembly by in situ immunofluorescence microscopy.
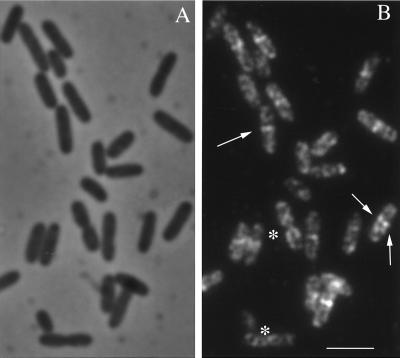
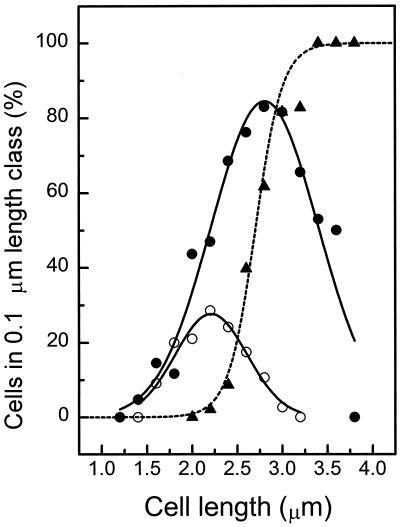
E. coli B/rA was grown at a range of generation times. Apart from the cell length, the number of cells showing a constriction (Fig. 2A [phase-contrast images]), separated nucleoids, or an open FtsZ ring or a closed FtsZ ring (Fig. 2B [Alexa images]) was determined. An open FtsZ ring is defined as two Z dots opposite each other near the membrane at midcell (Fig. 2B). The relative position of the FtsZ ring in the cell was very precise, at 0.50 ± 0.02 of the cell length. The cells were arranged according to length classes of 0.1-μm width, and the number of cells per length class with a certain characteristic was determined. For example, at a generation time of 130 min, the average length of the cells with a closed Z ring was 14% larger than that of cells with an open ring (Fig. 3), indicating that the open ring precedes the closed ring. The decrease in the number of cells with an FtsZ ring among the longest cells suggests that the FtsZ ring does not persist up to the separation into two daughter cells (Fig. 2B and 3).
FIG. 2.
E. coli B/rA grown to steady state with a generation time of 75 min. (A) Phase-contrast microscopy image of the cells; (B) corresponding Alexa-labeled FtsZ fluorescence. Bar, 2.5 μm. An example of a cell showing an open ring with two distinct dots at midcell is indicated by two opposite arrows; a cell showing a closed FtsZ ring is indicated by one arrow. Two cells (∗) with visible constrictions do not show detectable FtsZ at midcell. The cytosolic nonpolymerized FtsZ is somewhat more apparent in B/rA than in K-12 because of the weaker intensity of the FtsZ ring in the former strain.
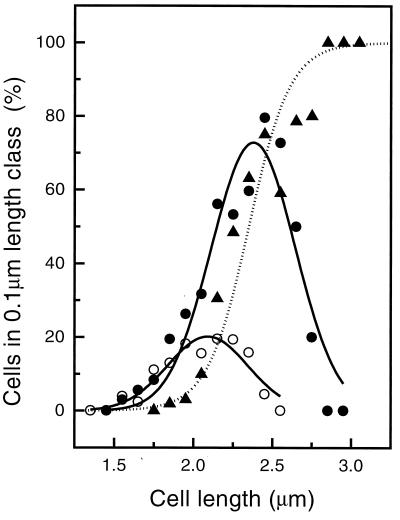
FIG. 3.
Appearance of the FtsZ ring as a function of cell length in E. coli B/rA grown to steady state at a generation time of 130 min. For 1,422 cells, the number of cells with an open FtsZ ring (○), a closed FtsZ ring (●), or a constriction (▴) in a particular length class was determined. The dotted line shows a sigmoidal fit through the data points for the constricting cells, and the solid lines show Gaussian fits through the FtsZ data points.
For each growth rate, the fractions of cells with an open FtsZ ring (two dots), a closed FtsZ ring, a visible constriction, and a visible separation of the nucleoids were determined for at least 500 cells. Since the open ring precedes the closed ring, the age at which the FtsZ ring appears is represented by the sum of both fractions. In the example shown in Fig. 3, this results in a fraction of closed FtsZ rings of 0.198 and a total fraction of closed and open FtsZ rings of 0.273. Because the FtsZ ring is not detectable during the last stage of the division process, the age at which the FtsZ ring appears will be underestimated by using equation 1. However, the cells without FtsZ dots, FtsZ rings, or a constriction represent the fraction of cells that do not yet have an assembled FtsZ ring. For this fraction of cells at the beginning of the cell cycle with ages (ax) of between 0 and tx, the time tx after birth at which the FtsZ ring assembles can be calculated by using
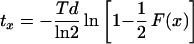 |
2 |
where the parameters are defined as for equation 1. Using equation 2, the average cell age ax at which the FtsZ ring appears in cells growing with a Td of 130 min is 85 min, and the age at which the ring can no longer be detected is 128 min. The latter age is calculated by addition of the fraction of cells that do not have an FtsZ ring or a constriction and the fraction of cells containing an FtsZ ring.
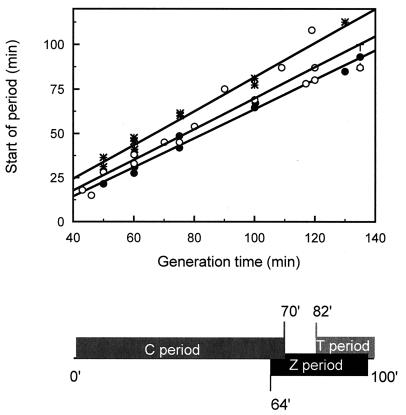
The initiation of the appearance of the FtsZ ring, or the Z period, was likewise determined for the various generation times with which the bacteria had been grown and was plotted as a function of the generation time (Fig. 4). Least-squares analysis of the data reveals a linear relation between the start of the Z period and the generation time of ax = 0.82x − 18.5, with r = 0.993. The initiation of the Z period occurs at the relative cell age of 0.45 for a Td of 50 min up to 0.68 for a Td of 135 min. Similarly, the age at which the nucleoids were visibly separated and a visible constriction appears (the T period) was calculated and plotted as function of generation time (Fig. 4). Least-squares analysis of the data shows linear relations between (i) the separation of the nucleoids and the generation time and (ii) the start of constriction and the generation time of ax = 0.88x − 8.8, with r = 0.991, and ax = 0.96x − 13.9, with r = 0.991, respectively. The cells start to constrict at a relative cell age of 0.68 for a Td of 50 min up to 0.85 for a Td of 135 min. Nucleoid separation precedes constriction at all generation times (except Td = 50 min) with a fraction of the relative cell age. It can be concluded that for all generation times the FtsZ ring is initiated well before the nucleoids are separated and the constriction is visible by phase-contrast microscopy.
FIG. 4.
Correlation between the appearance of the FtsZ ring, the termination of DNA replication (i.e., the start of the D period), and the start of a visible constriction. The start of the Z period (●), the start of the D period (or the end of the C period) (○), and the start of the constriction or T period (∗) are plotted as a function of the generation time. The lines are the linear least-squares fits through the data points for the FtsZ ring (ax = 0.82x − 18.5, with r = 0.993), the D period (ax = 0.86x − 16.9, with r = 0.982), and the T period (ax = 0.96x − 13.9, with r = 0.991). The nucleoid separation more or less coincides with the start of constriction (ax = 0.88x − 8.8, with r = 0.991) (not shown). The lower panel shows a schematic overview of the C, Z, and T periods in an individual cell growing with a generation time of 100 min derived from the linear fits in the upper panel. The values for the D period were obtained from reference 18. The value for the length of the C period (i.e., time between the initiation and the termination of DNA replication) was obtained from reference 5.
The appearance of the FtsA ring in E. coli B/rA was determined over the same range of growth rates as that for appearance of the FtsZ ring. No significant difference in the timing of the appearance of the FtsA ring was observed, although the open-ring period seemed to be on average somewhat longer for FtsA than for FtsZ (results not shown). The FtsA ring was localized with a polyclonal antibody, whereas FtsZ was located with a MAb. Since no difference in the timings of the FtsA and FtsZ rings was observed, the binding of the MAb to a single epitope does not seem to restrict the resolution of FtsZ detection.
Correlation between the DNA replication cycle and the appearance of the FtsZ ring.
What determines the timing of the assembly and the position of the FtsZ ring at the constriction site? Mulder and Woldringh (29) have suggested that septal peptidoglycan synthesis is repressed in the vicinity of the nucleoids and that termination of DNA replication could release a positive cell division initiation signal (the nucleoid occlusion model). According to this model, the cell is not able to determine the site of division unless DNA replication has terminated and the nucleoids have segregated. To assess whether there is indeed a correlation between termination of DNA replication and initiation of cell division or the assembly of the FtsZ ring, the average cell ages at which these events occur were compared.
The start of the D period (i.e., the period between termination of DNA replication and cell division) for B/rA at a wide range of generation times was obtained from reference 18 and plotted as a function of the generation time (Fig. 4). A least-squares analysis of the data gives a linear correlation between the termination of DNA replication and the generation time of ax = 0.86x − 16.9, with r = 0.982. The average cell has a replicated genome at a relative cell age of 0.53 for a Td of 50 min up to 0.74 for a Td of 135 min.
At all generation times the open FtsZ ring appears somewhat before DNA replication has terminated, and a closed FtsZ ring is found just after the termination. Therefore, it can be concluded that termination of DNA replication more or less coincides with the assembly of the FtsZ ring at midcell.
Correlation between the peptidoglycan synthesis cycle and FtsZ ring assembly in E. coli K-12.
Peptidoglycan synthesis during the cell cycle has been described for E. coli K-12 MC4100 lysA growing at a generation time of 85 min. Peptidoglycan synthesis starts to increase at midcell approximately 36 min after cell birth, reaches a maximum rate in the constricting cells, and maintains this rate throughout the constriction period (42, 50). The constriction becomes visible by electron microscopy 31 min before the cells separate into two daughter cells (41).
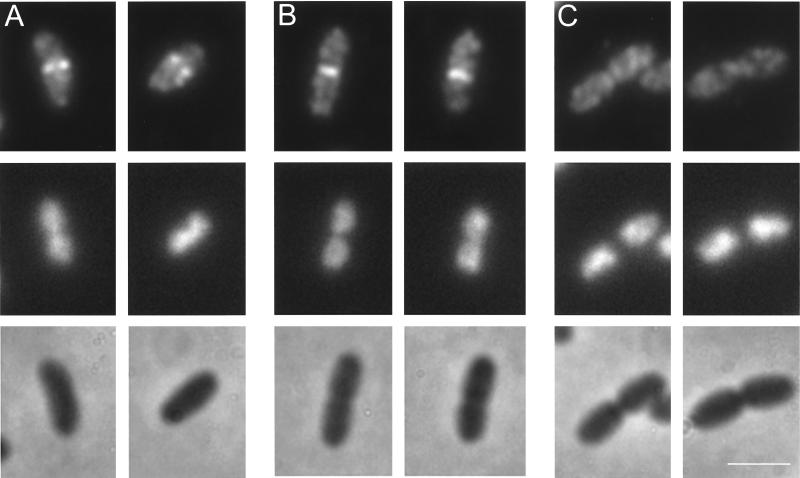
To determine the correlation between the appearance of the FtsZ ring and the onset of the increase in peptidoglycan synthesis at midcell, the same strain was grown to steady state with a generation time of 85 min and labeled with MAb F168-12 against FtsZ (Fig. 5). The percentage of E. coli K-12 cells with an FtsZ ring in the middle was 53% ± 3% (n = 4; ≥500 cells). Discrimination between open and closed FtsZ rings revealed that the fractions of the cells with open and closed rings are 0.135 and 0.395, respectively. The average length of the cells with an open ring is 18% less than that of those with a closed ring (Fig. 6). By using equations 1 and 2, it follows that the assembly of the FtsZ ring commences in E. coli K-12 at 32.8 ± 2.3 min of the generation time or at the relative cell age of 0.39 (Table 1; Fig. 7). Since the onset of the increased peptidoglycan synthesis occurs at the relative cell age of 0.42, it can be concluded that the FtsZ ring assembly coincides approximately with the increase in peptidoglycan synthesis.
FIG. 5.
E. coli K-12 MC4100 grown to steady state with a generation time of 85 min at 28°C. The upper panels show Alexa-labeled FtsZ fluorescence, the middle panels show the corresponding DAPI fluorescence microscopy images, and the lower panels show the corresponding phase-contrast microscopy images of the cells. Bar, 2.0 μm. (A) Examples of cells showing an open ring with two distinct dots at midcell; (B) cells showing a closed FtsZ ring; (C) cells with visible constrictions and segregated nucleoids without detectable FtsZ at midcell.
FIG. 6.
Appearance of the FtsZ ring as a function of cell length in E. coli K-12 grown to steady state at a generation time of 85 min. For 516 cells, the number of cells with an open FtsZ ring (○), a closed FtsZ ring (●), or a constriction (▴) in a particular length class was determined. The dotted line shows a sigmoidal fit through the data points for the constricting cells, and the solid lines show Gaussian fits through the FtsZ data points.
TABLE 1.
Correlation of cell cycle parameters and events with the relative cell ages of E. coli K-12 and B/rA at a generation time of 85 min
FIG. 7.
Schematic representation of the cell cycles of E. coli B/rA and K-12 growing with a generation time of 85 min. The C period (5) shows the duration of the DNA replication cycle. The D period is the time required between the termination of DNA replication and the separation into two daughter cells. The Z period shows the duration of the period in which FtsZ can be detected at midcell. The P period (42) shows the period of septal peptidoglycan synthesis, and the T period is the duration of the constriction.
Similar durations of the D period and the Z period.
Although E. coli K-12 and B/rA have been reported to have similar DNA replication periods, or C periods (5), the FtsZ ring period is 1.6 times longer in the K-12 strain than in the B/rA strain (Table 1). If a relationship between termination of DNA replication and assembly of the FtsZ ring, exists, then the two strains should exhibit a large difference in the durations of the D period.
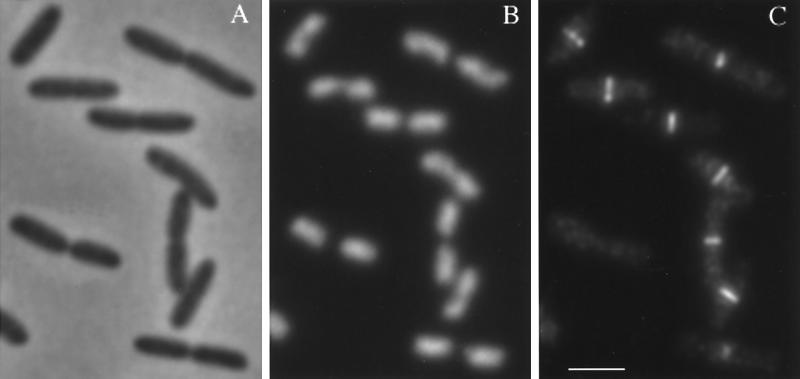
Huls et al. (20) showed recently that at a generation time of 84 min at 28°C, E. coli MC4100 has a C period of 70 min and a D period of about 47 min, which are 1.6 times longer than those in E. coli B/rA. Therefore, the initiation of the assembly of the FtsZ ring at midcell coincides more or less with the termination of DNA replication, like in B/rA (Table 1; Fig. 7). In the same article (20), the DNA replication cycle of the temperature-sensitive mutant MC4100 pbpB2158(Ts) was analyzed. Due to a modest delay in DNA segregation, this strain has an increased average genome content per cell and a very long D period of about 90 min, which exceeds its doubling time of 78 min at 28°C. We determined the percentage of cells with an FtsZ ring in this strain under exactly the same growth conditions by immunofluorescence microscopy (Fig. 8). Only 8.5% of the cells did not show an FtsZ ring at midcell, and about 2.5% of the cells showed FtsZ rings at one-quarter and three-quarters of the cell length. This indicates that the FtsZ ring assembly is initiated around cell birth, or again more or less simultaneously with the termination of DNA replication. After termination of DNA replication, the MC4100 and MC4100 pbpB2158(Ts) cells need about 18 and 40 min, respectively, before the nucleoids are completely separated (20). Therefore, it can be concluded that complete nucleoid separation seems not to be a prerequisite for FtsZ ring assembly.
FIG. 8.
E. coli K-12 MC4100 pbpB2158(Ts) grown to steady state with a generation time of 78 min at 28°C. (A) Phase-contrast microscopy image of the cells; (B) the corresponding DAPI-stained nucleoids; (C) the corresponding Alexa-labeled FtsZ fluorescence. Bar, 2.5 μm.
DISCUSSION
FtsZ is one of the proteins that can be detected very early in the cell cycle as a ring at the future division site. Thus far, all detected cell division proteins depend on the localization of FtsZ for their own localization at midcell (2, 8, 16, 17, 27, 35, 46, 48, 49, 54). It is likely that FtsZ ring formation and division-specific peptidoglycan synthesis go hand in hand (4, 31). Thus, for a number of reasons, FtsZ seems to be a good indicator for the initiation of cell division.
Little is known about the timing of cell division and its correlation with other cyclic events like DNA replication (5) and the increase in peptidoglycan synthesis at midcell (42, 50). In fast-growing cells, multiple cell cycles are overlapping, because the replication of the E. coli genome requires at least about 40 min (C period) and there is another 20 min (D period) before the cells are able to divide (13, 19). A comparison between the timing of the appearance of the FtsZ ring and the timing of other cyclic events is therefore experimentally feasible only for cells growing with a generation time of more than 60 min under defined growth conditions.
Here we have cultivated E. coli populations under steady-state conditions in minimal medium with various carbon sources and temperatures to allow an evaluation of the cell age at which the FtsZ ring appears. In addition, we have assessed whether the timing of FtsA appearance at midcell is simultaneous with that of FtsZ in slowly growing cells, as has been reported for fast-growing cells (27). The presence of FtsZ and FtsA was detected by immunofluorescence microscopy and image analysis. Using steady-state growth, the cell age at which the ring appears was calculated by using equation 2 from the fraction of cells showing no ring and no constriction (see Results).
Three morphologically different stages in the appearance of the ring can be discriminated.
At all growth rates, three different FtsZ ring stages could be discriminated. In the first stage the ring appeared as two dots located at opposite sites at the future site of division (Fig. 2B). This stage precedes the second stage, in which the ring is closed. Depending on the generation time, the first stage takes about 2 to 8 min. The assembly of the FtsZ ring has been described as starting from a central point and polymerizing bidirectionaly until the ring is closed (25). This process has been reported to take place within 1 min (3) even at the very low growth rate of 120 min per generation (39). To encircle E. coli B/rA, which has a circumference of about 1.88 μm at this growth rate (43), within 1 min, FtsZ should polymerize at a rate of about 2 μm/min. This rate is similar to the growth rate of 1.2 μm/min for freely growing microtubules in vitro (12). Since the duration of the first stage is much longer than 1 min, it seems unlikely that it represents the polymerization of FtsZ into a ring. What could cause the ring to be visualized as two opposite dots? The fluorescence images are two-dimensional representations of three-dimensional fluorescence intensities that cause the bacterial borders of the bacterium to be overexposed compared with the central surface. Sun and Margolin (39) have presented similar images of living cells with central FtsZ-green fluorescence protein fluorescence gaps as optical artifacts. Our fixation procedure, using a combination of glutaraldehyde and formaldehyde, cross-links proteins and might reorient the FtsZ molecules of the nascent ring to opposite membrane locations. However, the observation that this stage occurs in all experiments at a clearly shorter average cell length than the second stage, with a closed FtsZ ring, suggests that these stages are morphologically and functionally different. Possibly, the first stage represents a difference in the stability of the FtsZ ring because not all divisome proteins have yet assembled.
The last stage of the FtsZ ring is its depolymerization at the end of the constriction period. Depending on the growth rate, during the last 2 to 4 min of the constriction period no central FtsZ ring or spot could be observed (Fig. 2B). Either the amount of FtsZ is not discernible from the background fluorescence of the soluble cytosolic FtsZ molecules or the protein is not required for the separation into two daughter cells. Similar morphological stages for the FtsA ring could be discriminated for all growth rates studied.
No difference in the timing of the appearance or of the morphology of the FtsA ring could be observed compared to FtsZ. ZipA and FtsA are the only cell division proteins that have been shown to interact directly with FtsZ (16, 47) and that are recruited simultaneously with FtsZ to the division site (17). The period between the appearance of the FtsZ or -A ring and the assembly of other division proteins might correspond to the stage in which the ring appears as two opposite dots at midcell. The function of the ring during this stage could be dual. First, the ring clearly marks the position and the circumference of the future division site. Normal cell division requires a particular ratio of FtsA to FtsZ protein (9, 11). Colocalization of FtsZ and FtsA in the ring (47) could recruit other cell division proteins to the divisome assembly site and possibly determine the number of divisome subassemblies required for the synthesis of two new cell poles. Second, this position also determines the location of the two compartments into which the nucleoids should be segregated. Proteins involved in nucleoid segregation could use the FtsZ or -A ring as an anchoring point. For instance, the SMC-like protein MukB, which is involved in chromosome segregation or condensation, binds FtsZ in vitro with a very high affinity (23), and MukB mutants do not survive in the FtsZ84(Ts) mutant at a permissive temperature (40).
Timing of the FtsZ assembly and cell shape.
The timing of the assembly of the FtsZ ring is quite different in E. coli K-12 than in B/rA (Table 1; Fig. 7). Although both are considered to be wild-type E. coli strains, K-12 cells are somewhat longer and 1.5 times wider than B/rA cells (41, 52). Therefore, K-12 has to synthesize a much larger new polar surface than B/rA at the same growth rate. It is tempting to speculate that this is why K-12 has a constriction period 1.76 times longer than that of B/rA. Strikingly, the duration of the Z period is also 1.6 times longer in E. coli K-12 than in B/rA. The relative cell age at which the ring appears in E. coli B/rA increases with longer generation times (Fig. 4). Accordingly, the duration of the constriction period (Fig. 4) and the width of the cells (52) decrease at longer generation times. It seems likely, therefore, that the timing of the appearance of the FtsZ ring depends on the morphology of the bacterium or the amount of polar surface that has to be synthesized. A similar correlation has been found between the amount of polar surface to be synthesized or the growth rate and the concentration of penicillin needed to reduce the rate of cell division by 50% without inhibiting growth (15).
FtsZ ring assembly and peptidoglycan synthesis.
The ring assembly more or less coincides with the increase in peptidoglycan synthesis at the future site of the constriction (Table 1, Fig. 7). Admittedly, the age at which the peptidoglycan synthesis increases at midcell has not been determined with great precision due to the low cell number analyzed by autoradiography of radioactive pulse-labeled sacculi (42, 50). The recently published method of De Pedro et al. (10), in which sacculi are labeled with biotinylated d-Cys, possibly facilitates the analysis of a much larger number of cells. This would enable a more accurate determination of the timing of the onset of increased peptidoglycan synthesis at midcell at different growth rates. Nevertheless, the notion that for cell division to occur an interplay between FtsZ ring formation and division-specific peptidoglycan synthesis is needed (31) appears to be reinforced. However, the expected functional link between the peptidoglycan and cytoplasmic compartments remains elusive. FtsW might be a possible candidate, not only because of its membrane topology but also because, like FtsZ, it localizes to the midcell in an early stage of the division cycle (46).
Timing of FtsZ ring assembly and DNA replication.
In three strains with very different cell cycles, the initiation of the FtsZ ring assembly at midcell coincides more or less with termination of DNA replication and occurs well before the cells show a visible constriction and separated nucleoids. This indicates that an event near termination of DNA replication could provide a positive signal needed for the onset of division, as suggested in the nucleoid occlusion model (30). For instance, the disassembly of the DNA replicon located at midcell (33) could create the space and possibly allow a local increase in the FtsZ concentration. The increase in FtsZ concentration might induce the polymerization of FtsZ at the potential division site. Our results also show that complete segregation of the nucleoids is not a requirement for the assembly of the FtsZ ring as proposed in the nucleoid occlusion model (30). Since DNA replication and nucleoid segregation go hand in hand (51), it can be surmised that a large part of the nucleoids segregated. This suggests but does not prove that a certain separation of the replicating DNA masses (6) is sufficient to allow midcell localization of FtsZ. Resolution of the two circular chromosomes into monomers is blocked if cell division is inhibited with cephalexin (38), by FtsZ(Ts) mutations (38), and in the pbpB2158(Ts) mutant (20). Recently it was shown that FtsK is involved in the resolution of chromosome dimers (37). Overall, it seems more likely that the assembly of the divisome and the process of cell constriction assist in the last stage of segregation of the nucleoids than vice versa.
In this study we have shown that termination of DNA replication could provide a signal for the initiation of cell division or FtsZ ring polymerization as suggested in the nucleoid occlusion model. Complete segregation of the nucleoids seems not to be a requirement for the initiation of FtsZ ring assembly. Rather, it is more likely that the divisional structure assists in the segregation of the nucleoids.
ACKNOWLEDGMENTS
We thank Miguel Vicente for the gift of the polyclonal serum against FtsA and Conrad L. Woldringh for critically reading the manuscript and helpful discussions.
This work was supported by the Life Sciences Foundation (SLW) (grant 805-33-221P), which is subsidized by the Netherlands Organization for Scientific Research (NWO) and by the European Community (EEG) (Cell Factory contract no. CT96-0122).
REFERENCES
- 1.Adam M, Damblon C, Jamin M, Zorzi W, Dusart V, Galleni M, El Kharroubi A, Piras G, Spratt B G, Keck W, Coyette J, Ghuysen J M, Nguyen-Distèche M, Frère J M. Acyltransferase activities of the high-molecular-mass essential penicillin-binding proteins. Biochem J. 1991;279:601–604. doi: 10.1042/bj2790601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinal S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 3.Addinal S G, Cao C, Lutkenhaus J. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J Bacteriol. 1997;179:4277–4284. doi: 10.1128/jb.179.13.4277-4284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 5.Bipatnath M, Dennis P P, Bremer H. Initiation and velocity of chromosome replication in Escherichia coli B/r and K-12. J Bacteriol. 1998;180:265–273. doi: 10.1128/jb.180.2.265-273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouché J-P, Pichoff S. On the birth and fate of bacterial division sites. Mol Microbiol. 1998;29:19–26. doi: 10.1046/j.1365-2958.1998.00874.x. [DOI] [PubMed] [Google Scholar]
- 7.Buddelmeijer N, Aarsman M E G, Kolk A H J, Vicente M, Nanninga N. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J Bacteriol. 1998;180:6107–6116. doi: 10.1128/jb.180.23.6107-6116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J C, Weiss D S, Ghigo J-M, Beckwith J. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J Bacteriol. 1999;181:521–530. doi: 10.1128/jb.181.2.521-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai K, Lutkenhaus J. The proper ratio of FtsZ and FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Pedro M A, Quintela J C, Höltje J-V, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewar S J, Begg K J, Donachie W D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dogterom M, Yurke B. Measurement of the force-velocity relation for growing micotubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- 13.Donachie W D. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 14.Ghigo J-M, Weiss D S, Chen J C, Yanow J G, Beckwith J. Localization of FtsL to the Escherichia coli septal ring. Mol Microbiol. 1999;31:725–738. doi: 10.1046/j.1365-2958.1999.01213.x. [DOI] [PubMed] [Google Scholar]
- 15.Hadas H, Einav M, Fishov I, Zaritsky A. Division-inhibition capacity of penicillin in Escherichia coli is growth-rate dependent. Microbiology. 1995;141:1081–1083. doi: 10.1099/13500872-141-5-1081. [DOI] [PubMed] [Google Scholar]
- 16.Hale C A, de Boer P A J. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 17.Hale C A, De Boer P A J. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmstetter C E. Timing of synthetic activities in the cell cycle. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1627–1639. [Google Scholar]
- 19.Helmstetter C E, Cooper S. DNA synthesis during the division cycle of rapidly growing Escherichia coli B/r. J Mol Biol. 1968;31:507–518. doi: 10.1016/0022-2836(68)90424-5. [DOI] [PubMed] [Google Scholar]
- 20.Huls, P. G., N. O. E. Vischer, and C. L. Woldringh. Delayed nucleoid segregation in Escherichia coli. Mol. Microbiol., in press. [DOI] [PubMed]
- 21.Koppes L J H, Woldringh C L, Nanninga N. Size variations and correlation of different cell cycle events in slow-growing Escherichia coli. J Bacteriol. 1978;134:423–433. doi: 10.1128/jb.134.2.423-433.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin A P, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtiliss. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 23.Lockhardt A, Kendrick-Jones J. Interaction of the N-terminal domain of MukB with the bacterial tubulin homologue FtsZ. FEBS Lett. 1998;430:278–282. doi: 10.1016/s0014-5793(98)00677-2. [DOI] [PubMed] [Google Scholar]
- 24.Löwe J, Amos L A. crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 25.Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol Microbiol. 1993;9:403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 26.Lutkenhaus J, Mukherjee A. Cell division. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1615–1626. [Google Scholar]
- 27.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulder E, Woldringh C L. Actively replicating nucleoids influence the positioning of division sites in DNA-less cell forming filaments of Escherichia coli. J Bacteriol. 1989;171:4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulder E, Woldringh C L. Autoradiographic analysis of diaminopimelic acid incorporation in filamentous cells of Escherichia coli: repression of peptidoglycan synthesis around the nucleoid. J Bacteriol. 1991;173:4751–4756. doi: 10.1128/jb.173.15.4751-4756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanninga N. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol. 1991;5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 32.Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niki H, Hiraga S. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 1998;12:1036–1045. doi: 10.1101/gad.12.7.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powell E O. Growth rate and generation time of bacteria, with special reference to continuous culture. J Gen Microbiol. 1956;15:492–511. doi: 10.1099/00221287-15-3-492. [DOI] [PubMed] [Google Scholar]
- 35.Raskin D M, de Boer A J. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–964. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez M, Valencia A, Ferrandiz M J, Sander C, Vicente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steiner W W, Liu G, Donachie W D, Kuempel P L. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- 38.Steiner W W, Kuempel P L. Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli. Mol Microbiol. 1998;27:257–268. doi: 10.1046/j.1365-2958.1998.00651.x. [DOI] [PubMed] [Google Scholar]
- 39.Sun Q, Margolin W. FtsZ dynamics during the division cycle of live Escherichia coli cells. J Bacteriol. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Q, Yu X-C, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 41.Taschner P E M, Huls P G, Pas E, Woldringh C L. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J Bacteriol. 1988;170:1533–1540. doi: 10.1128/jb.170.4.1533-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taschner P E M, Ypenburg N, Spratt B G, Woldringh C L. An amino acid substitution in penicillin-binding protein 3 creates pointed polar caps in Escherichia coli. J Bacteriol. 1988;170:4828–4837. doi: 10.1128/jb.170.10.4828-4837.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trueba F J, Woldringh C L. Changes in cell diameter during the division cycle of Escherichia coli. J Bacteriol. 1980;142:869–878. doi: 10.1128/jb.142.3.869-878.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Helvoort J M L M, Woldringh C L. Nucleoid partitioning in Escherichia coli during steady-state growth and upon recovery from chloramphenicol treatment. Mol Microbiol. 1994;13:577–583. doi: 10.1111/j.1365-2958.1994.tb00452.x. [DOI] [PubMed] [Google Scholar]
- 44a.Vischer N. [Online.] Object-Image1.62. Amsterdam, The Netherlands: University of Amsterdam; 25 March 1997. http://simon.bio.uva.nl/object-image.html . [18 June 1999, last date accessed.] [Google Scholar]
- 45.Voskuil J L A, Westerbeek C A M, Wu C, Kolk A H J, Nanninga N. Epitope mapping of Escherichia coli cell division protein FtsZ with monoclonal antibodies. J Bacteriol. 1994;176:1886–1893. doi: 10.1128/jb.176.7.1886-1893.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Khattar M K, Donachie W D, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss D S, Chen J C, Ghigo J-M, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss D S, Pogliano K, Carson M, Guzmán L-M, Fraipont C, Nguyen-Distèche M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 50.Wientjes F B, Nanninga N. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J Bacteriol. 1989;171:3412–3419. doi: 10.1128/jb.171.6.3412-3419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woldringh C L. Morphological analysis of nuclear separation and cell division during the life cycle of Escherichia coli. J Bacteriol. 1976;125:248–257. doi: 10.1128/jb.125.1.248-257.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woldringh C L, De Jong M A, Van der Berg W, Koppes L. Morphological analysis of the division cycle of two Escherichia coli substrains during slow growth. J Bacteriol. 1977;131:270–279. doi: 10.1128/jb.131.1.270-279.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woldringh C L, Mulder E, Huls P G, Vischer N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol. 1991;142:309–320. doi: 10.1016/0923-2508(91)90046-d. [DOI] [PubMed] [Google Scholar]
- 54.Yu X, Tran A H, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]