Abstract
Background
Influenza-associated pulmonary aspergillosis (IAPA) and COVID-19-associated pulmonary aspergillosis (CAPA) affect about 15% of critically ill patients with influenza or COVID-19, respectively. These viral–fungal coinfections are difficult to diagnose and are associated with increased mortality, but data on their pathophysiology are scarce. We aimed to explore the role of lung epithelial and myeloid innate immunity in patients with IAPA or CAPA.
Methods
In this observational study, we retrospectively recruited patients who had been admitted to the intensive care unit (ICU) of University Hospitals Leuven, Belgium, requiring non-invasive or invasive ventilation because of severe influenza or COVID-19, with or without aspergillosis, between Jan 1, 2011, and March 31, 2021, whose bronchoalveolar lavage samples were available at the hospital biobank. Additionally, biobanked in vivo tracheobronchial biopsy samples from patients with IAPA or CAPA and invasive Aspergillus tracheobronchitis admitted to ICUs requiring invasive ventilation between the same dates were collected from University Hospitals Leuven, Hospital Network Antwerp (Belgium), and Amiens-Picardie University Hospital (France). We did nCounter gene expression analysis of 755 genes linked to myeloid innate immunity and protein analysis of 47 cytokines, chemokines, and growth factors on the bronchoalveolar lavage samples. Gene expression data were used to infer cell fractions by use of CIBERSORTx, to perform hypergeometric enrichment pathway analysis and gene set enrichment analysis, and to calculate pathway module scores for the IL-1β, TNF-α, type I IFN, and type II IFN (IFNγ) pathways. We did RNAScope targeting influenza virus or SARS-CoV-2 RNA and GeoMx spatial transcriptomics on the tracheobronchial biopsy samples.
Findings
Biobanked bronchoalveolar lavage samples were retrieved from 166 eligible patients, of whom 40 had IAPA, 52 had influenza without aspergillosis, 33 had CAPA, and 41 had COVID-19 without aspergillosis. We did nCounter gene expression analysis on bronchoalveolar lavage samples from 134 patients, protein analysis on samples from 162 patients, and both types of analysis on samples from 130 patients. We performed RNAScope and spatial transcriptomics on the tracheobronchial biopsy samples from two patients with IAPA plus invasive Aspergillus tracheobronchitis and two patients with CAPA plus invasive Aspergillus tracheobronchitis. We observed a downregulation of genes associated with antifungal effector functions in patients with IAPA and, to a lesser extent, in patients with CAPA. We found a downregulated expression of several genes encoding proteins with functions in the opsonisation, recognition, and killing of conidia in patients with IAPA versus influenza only and in patients with CAPA versus COVID-19 only. Several genes related to LC3-associated phagocytosis, autophagy, or both were differentially expressed. Patients with CAPA had significantly lower neutrophil cell fractions than did patients with COVID-19 only. Patients with IAPA or CAPA had downregulated IFNγ signalling compared with patients with influenza only or COVID-19 only, respectively. The concentrations of several fibrosis-related growth factors were significantly elevated in the bronchoalveolar lavage fluid from patients with IAPA versus influenza only and from patients with CAPA versus COVID-19 only. In one patient with CAPA, we visualised an active or very recent SARS-CoV-2 infection disrupting the epithelial barrier, facilitating tissue-invasive aspergillosis.
Interpretation
Our results reveal a three-level breach in antifungal immunity in IAPA and CAPA, affecting the integrity of the epithelial barrier, the capacity to phagocytise and kill Aspergillus spores, and the ability to destroy Aspergillus hyphae, which is mainly mediated by neutrophils. The potential of adjuvant IFNγ in the treatment of IAPA and CAPA should be investigated.
Funding
Research Foundation Flanders, Coronafonds, the Max Planck Society, the Fundação para a Ciência e a Tecnologia, the European Regional Development Fund, “la Caixa” Foundation, and Horizon 2020.
Research in context.
Evidence before this study
We searched PubMed without language restrictions for articles published between database inception and June 8, 2022, using the search terms “influenza” OR “COVID-19” AND “aspergillosis”, which resulted in 556 hits. Numerous observational studies have reported on the incidence of aspergillosis in critically ill patients with influenza or COVID-19. Two influenza-associated pulmonary aspergillosis (IAPA) mouse models have been created. Despite the abundance of reviews hypothesising on IAPA and COVID-19-associated pulmonary aspergillosis (CAPA) pathophysiology, no studies actually investigating IAPA and CAPA pathophysiology using human lower respiratory tract samples have been published.
Added value of this study
To our knowledge, this study is the first to use human lower respiratory tract samples to investigate epithelial and myeloid innate immunology in patients with IAPA or CAPA. Using state-of-the-art techniques to analyse bronchoalveolar lavage and in-vivo tracheobronchial biopsy samples, we discovered evidence of a multilevel breach in antifungal immunity in patients with IAPA and, to a lesser extent, in patients with CAPA. We observed a downregulation of numerous genes with antifungal effector functions, implicating impairments in macrophages, monocytes, and neutrophils. Although only minor differences were observed in the concentrations of major cytokines in the bronchoalveolar lavage samples, patients with IAPA or CAPA had reduced IFNγ signalling compared with patients with influenza only or COVID-19 only, respectively. The concentrations of several profibrotic growth factors were significantly increased in the bronchoalveolar lavage fluid from patients with IAPA versus influenza only and from patients with CAPA versus COVID-19 only, providing an indirect, additional cause for the higher mortality in patients with coinfections versus monoinfections. We also show that SARS-CoV-2-induced epithelial barrier disruption might facilitate tissue-invasive aspergillosis.
Implications of all the available evidence
Our results represent a first step towards understanding the pathophysiology of human IAPA and CAPA. Differences in immunological responses by aspergillosis status have diagnostic and therapeutic implications. Our dataset lays the foundations for the ensuing discovery and development of novel diagnostic and prognostic biomarkers. Moreover, our results point towards the potential of using recombinant IFNγ as an adjuvant to antifungal treatment. In future, high-resolution multiomics and functional studies will be necessary to further unravel the pathophysiology of IAPA and CAPA.
Introduction
Severe influenza infections cause around 400 000 deaths per year, and, according to WHO's COVID-19 Dashboard, as of June, 2022, COVID-19 has resulted in more than 6 300 000 deaths worldwide.1 Invasive pulmonary aspergillosis, a fungal infection that typically affects people who are severely immunocompromised, has been described in a subset of critically ill patients with influenza or COVID-19. In studies of patients with influenza or COVID-19 with high rates of adequate mycological work-up, the incidence of influenza-associated pulmonary aspergillosis (IAPA) is around 20% and the incidence of COVID-19-associated pulmonary aspergillosis (CAPA) is around 15%.2, 3, 4, 5, 6 Diagnosing IAPA or CAPA is difficult and mainly relies on invasive techniques such as bronchoalveolar lavage. Treatment options are limited to antifungals, which have substantial side-effects and multiple drug–drug interactions.7, 8 Because mortality is around 50% in patients with IAPA or CAPA versus 25–35% in patients with severe influenza or COVID-19 without aspergillosis, coinfection might have contributed to numerous deaths.6
The question remains as to why a subset of patients with severe influenza or COVID-19, including those without a pre-existing immunocompromised status, develop this fungal superinfection. Answering this question could lead to the identification and development of new biomarkers and novel (immunomodulatory) therapeutic targets or prompt the reallocation of existing immunomodulatory drugs. We therefore aimed to unravel the pathogenesis of IAPA and CAPA by performing gene expression and cytokine profiling, RNAScope, and spatial transcriptomics on lower respiratory tract samples from patients with severe influenza or COVID-19, with or without aspergillosis, focusing on the epithelium and the main driver of antifungal immunity: the myeloid innate immune system.
Methods
Study design and participants
In this observational study, we retrospectively and non-consecutively recruited patients who had been admitted to the intensive care unit (ICU) of University Hospitals Leuven, Belgium, requiring non-invasive or invasive ventilation because of influenza or COVID-19, with or without aspergillosis, between Jan 1, 2011, and March 31, 2021, whose bronchoalveolar lavage samples were available at the hospital biobank (appendix 1 pp 1–2). Diagnosis of probable or proven IAPA was based on the expert consensus criteria by Verweij and colleagues7 and diagnosis of probable or proven CAPA was based on criteria from the European Confederation of Medical Mycology and the International Society for Human and Animal Mycology.8 Additionally, biobanked in vivo tracheobronchial biopsy samples from patients with IAPA or CAPA and invasive Aspergillus tracheobronchitis admitted to ICUs requiring invasive ventilation between Jan 1, 2011, and March 31, 2021, were collected from University Hospitals Leuven, Hospital Network Antwerp (Belgium), and Amiens-Picardie University Hospital (France), with approval from their respective ethical committees. The study protocol was approved and the need for informed consent waived by the ethical committee of University Hospitals Leuven, Belgium (S65588).
Procedures
Bronchoalveolar lavage samples and biopsies were obtained by bronchoscopy as part of clinical routine. For patients with only influenza or COVID-19, we used the first bronchoalveolar lavage sample obtained during ICU admission. For patients with IAPA or CAPA, we used the first available bronchoalveolar lavage sample showing mycological evidence of aspergillosis (defined by a positive culture or a galactomannan optical index of ≥1·0) or the concomitant tracheobronchial biopsy with visualisation of invading Aspergillus hyphae (appendix 1 p 2). Demographic, clinical, treatment, and outcome data were derived from patient electronic medical records.
We did nCounter (NanoString; Seattle, WA, USA) gene expression analysis of 755 genes linked to myeloid innate immunity and protein analysis using ELISA or multiplex bead-based assays of 47 cytokines, chemokines, and growth factors on the bronchoalveolar lavage samples (see appendix 2 for a list of genes). Gene expression data were used to infer cell fractions by use of CIBERSORTx,9 to perform hypergeometric enrichment pathway analysis by use of the ClueGO plug-in (with Gene Ontology biological process as the pathway database) in Cytoscape10 and gene set enrichment analysis (with Reactome canonical pathways as the pathway database),11 and to calculate pathway module scores for the IL-1β, TNF-α, type I IFN, and type II IFN (IFNγ) pathways. Correlations between the different bronchoalveolar lavage results were calculated.
We did RNAScope (ACD; Newark, NJ, USA) ultrasensitive single molecule RNA in-situ hybridisation targeting influenza virus or SARS-CoV-2 RNA on formalin-fixed, paraffin-embedded sections of in vivo tracheobronchial biopsy samples from patients with IAPA or CAPA and invasive Aspergillus tracheobronchitis. We performed RNAScope to investigate whether Aspergillus hyphae colocalise with the presence of influenza virus or SARS-CoV-2 transcripts. We also did GeoMx (NanoString; Seattle, WA, USA) spatial transcriptomics (aligning regions of interest in epithelial zones and zones with inflammatory infiltrate) with a whole human transcriptome atlas plus a SARS-CoV-2 gene panel spike-in on these tracheobronchial biopsy samples, later performing gene set enrichment analysis (with Reactome canonical pathways as the pathway database). For more detail on participants, study design, and procedures, please see appendix 1 (pp 1–5).
Statistical analysis
There were no formal sample size calculations. We interrogated correlations between continuous variables using Spearman's correlation. Unless stated otherwise in the supplementary methods (appendix 1 pp 1–5), statistical testing was corrected for multiple testing by use of the more stringent Benjamini–Hochberg false discovery rate method for our primary analysis of gene expression (nCounter and GeoMx) and pathways (hypergeometric enrichment pathway analysis and gene set enrichment analysis) and by use of the less conservative Benjamini–Krieger–Yekutieli two-stage step-up method for our secondary analyses of CIBERSORTx cell fractions, pathway module scores, and protein concentrations. We made four comparisons in our main analysis: IAPA versus influenza only, CAPA versus COVID-19 only, IAPA versus CAPA, and influenza only versus COVID-19 only. We compared the differentially expressed genes from the IAPA versus influenza only and CAPA versus COVID-19 only comparisons with an existing dataset of the transcriptional response of peripheral blood mononuclear cells 24 h after stimulation with Aspergillus fumigatus conidia.12
In post-hoc sensitivity analyses of gene expression, we corrected for the use of corticosteroids in the comparisons of IAPA versus CAPA and influenza only versus COVID-19 only, and for the timepoint of sampling in relation to the start of mechanical ventilation in the comparisons of IAPA versus influenza only and CAPA versus COVID-19 only.
Adjusted p values are reported as q values. A two-sided alternative hypothesis at the 5% significance level was used for all statistical analyses, except for gene set enrichment analysis, for which the recommended 25% significance level was used.11 Unless stated otherwise in the supplementary methods (appendix 1 pp 1–5), statistical analyses were done by use of GraphPad Prism software (version 9.2.0) and R (version 4.1.0).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Biobanked bronchoalveolar lavage samples were retrieved from 166 eligible patients, of whom 40 had IAPA (30 probable and ten proven), 52 had influenza without aspergillosis (influenza only), 33 had CAPA (30 probable and three proven), and 41 had COVID-19 without aspergillosis (COVID-19 only). We did nCounter gene expression analysis on bronchoalveolar lavage samples from 134 patients, protein analysis on bronchoalveolar lavage samples from 162 patients, and both types of analysis on bronchoalveolar lavage samples from 130 patients (appendix 1 pp 13, 17). Additionally, we performed RNAScope and spatial transcriptomics on in-vivo tracheobronchial biopsy samples from two patients with IAPA plus invasive Aspergillus tracheobronchitis and from two patients with CAPA plus invasive Aspergillus tracheobronchitis during their ICU stay (appendix 1 pp 13, 17). More patients with influenza than with COVID-19 in the bronchoalveolar lavage samples analysis had European Organisation for Research and Treatment of Cancer risk factors for aspergillosis and chronic obstructive pulmonary disease (appendix 1 pp 6, 8), whereas more patients with COVID-19 than with influenza had received corticosteroids in ICU before bronchoalveolar lavage sampling (appendix 1 pp 7, 9). Timing of bronchoalveolar lavage sampling tended to be later after hospital and ICU admission and initiation of mechanical ventilation for patients with COVID-19 compared with patients with influenza (appendix 1 pp 7, 9, 13, 18). The timing of sampling was more heterogeneous in the aspergillosis groups than in the non-aspergillosis groups (appendix 1 pp 13, 18). In appendix 1, we report the clinical course of each included patient (pp 13, 18) and summary data (pp 6–10).
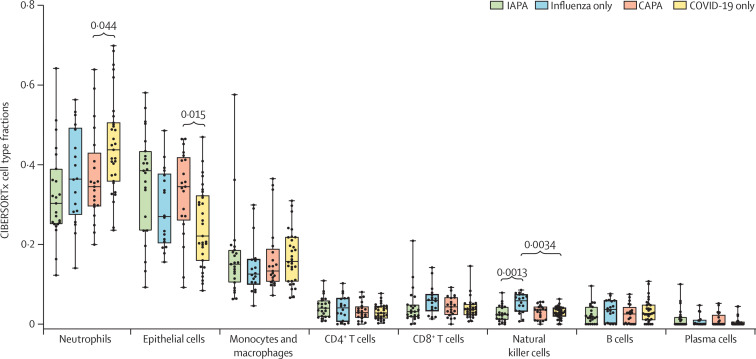
We used CIBERSORTx to estimate the proportions of immune and epithelial cell types in the bronchoalveolar lavage samples on the basis of gene expression signatures.9 Fractions of the eight major cell subtypes are shown in figure 1 . Patients with IAPA had numerically lower neutrophil cell fractions (Benjamini–Krieger–Yekutieli q=0·21) and numerically higher epithelial cell fractions (q=0·11) than patients with influenza only (figure 1). Similarly, patients with CAPA had significantly lower neutrophil cell fractions (Benjamini–Krieger–Yekutieli q=0·044) and significantly higher epithelial cell fractions (q=0·015) than patients with COVID-19 only (figure 1). Furthermore, we found significantly lower natural killer cell fractions in the samples from patients with IAPA compared with those from patients with influenza only (figure 1). The fraction of neutrophils was negatively correlated with the fractions of epithelial cells, macrophages and monocytes, and plasma cells (appendix 1 pp 13, 19).
Figure 1.
Cell type fractions in bronchoalveolar lavage fluid samples from patients with IAPA, influenza only, CAPA, or COVID-19 only
Box plots represent median and IQR, with whiskers set from minimum to maximum. Individual points represent each sample. q values were generated by Kruskall–Wallis tests, corrected by use of the Benjamini–Krieger–Yekutieli method. Only significant q values are shown. CAPA=COVID-19-associated pulmonary aspergillosis. IAPA=influenza-associated pulmonary aspergillosis.
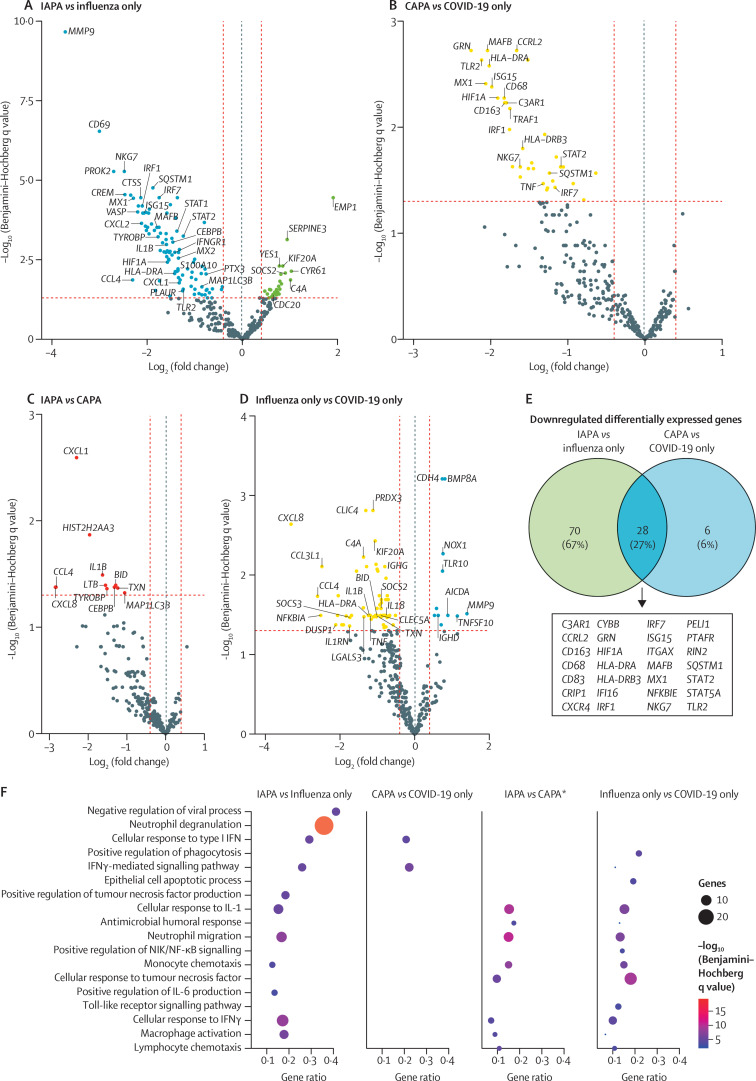
Differentially expressed genes were identified at a Benjamini–Hochberg q value of less than 0·05 for four comparisons: IAPA versus influenza only, CAPA versus COVID-19 only, IAPA versus CAPA, and influenza only versus COVID-19 only (figure 2 ; appendix 1 pp 13, 20–21). In sensitivity analyses in which we corrected for the use of corticosteroids in the IAPA versus CAPA and influenza only versus COVID-19 only comparisons and for the timepoint of sampling in relation to the start of mechanical ventilation in the IAPA versus influenza only and CAPA versus COVID-19 only comparisons, the number of differentially expressed genes stayed largely the same (appendix 1 pp 11–14, 22–23).
Figure 2.
Differentially expressed genes in bronchoalveolar lavage fluid samples from patients with IAPA, influenza only, CAPA, or COVID-19 only
Volcano plots showing differentially expressed genes for the comparisons of IAPA versus influenza only (A), CAPA versus COVID-19 only (B), IAPA versus CAPA (C), and influenza only versus COVID-19 only (D). A dotted blue line marks a log2(fold change) value of zero. A dotted red line crossing the y-axis marks a Benjamini–Hochberg q value of 0·05. The two dotted red lines crossing the x-axis marks a log2(fold change) of 0·4 and –0·4, respectively. We selected genes to present if they were linked to interesting pathways related to antiviral and antifungal immunity. (E) Venn diagram showing the downregulated differentially expressed genes shared in the comparisons of IAPA versus influenza only and CAPA versus COVID-19 only. (F) Dot plot showing ClueGO hypergeometric enrichment pathway analysis based on the downregulated differentially expressed genes of the four disease comparisons, with Gene Ontology biological process as the pathway database. *For the comparison of IAPA versus CAPA, differentially expressed genes with q values (Benjamini–Hochberg) of less than 0·20 were included to generate the analyses. Differentially expressed genes with q values (Benjamini–Hochberg) of less than 0·05 were used for all other comparisons. CAPA=COVID-19-associated pulmonary aspergillosis. IAPA=influenza-associated pulmonary aspergillosis. NIK=NF-κB-inducing kinase.
We discovered 98 significantly downregulated and 26 significantly upregulated differentially expressed genes for the IAPA versus influenza only comparison (figure 2A). Genes related to downstream type I IFN (eg, STAT1, STAT2, ISG15, and MX1) and type II IFN signalling (eg, IFNGR1, CEBPB, and HLA-DRA) were downregulated in patients with IAPA compared with patients with influenza only but the transcripts encoding IFNβ and IFNγ were not. Several genes related to LC3-associated phagocytosis, autophagy, or both were differentially expressed. MAP1LC3B, which encodes LC3 (also known as microtubule-associated proteins 1A/1B light chain 3B), and SQSTM1, which encodes an autophagy substrate that interacts with LC3 and targets bacteria during LC3-associated phagocytosis, were downregulated and CDC20, which encodes a protein that targets LC3 for ubiquitination and degradation, was upregulated.13, 14, 15 LC3-associated phagocytosis is a non-canonical autophagy pathway important for the killing of phagocytised Aspergillus conidia.16 We also found that TLR2, which is involved in the innate immune detection of Aspergillus and macrophage activation,17 and genes encoding proteins related to complement functions (eg, C5AR1, C3AR1, PLAUR, PLAU, and ITGAX) and PTX3-mediated phagocytosis of conidia (PTX3 and FCGR2A) were downregulated in patients with IAPA (figure 2A; appendix pp 13, 20).
Furthermore, VASP, which encodes an actin polymerase involved in phagocytosis and the lateral transfer of Aspergillus conidia between macrophages,18 and HIF1A, whose gene product is necessary for the prevention of neutrophil apoptosis at the site of aspergillosis,19 were downregulated in patients with IAPA. Other downregulated differentially expressed genes encoded proteins serving neutrophil effector functions: matrix metalloproteinases (MMP9), cathepsins (CTSA, CTSD, and CTSS), and alarmins (S100A4, S100A10, and S100A11; figure 2A; appendix pp 13, 20). We also found the downregulation of IL1B, which produces the proinflammatory cytokine IL-1β.
ClueGO Gene Ontology pathway analysis of the downregulated differentially expressed genes confirmed the downregulation of type I IFN, type II IFN (IFNγ), and IL-1 responses in patients with IAPA versus influenza only (figure 2F; appendix 1 pp 14, 24). For cell-specific processes, the Gene Ontology terms neutrophil degranulation and, to a lesser extent, neutrophil migration and macrophage activation were downregulated in IAPA versus influenza only in ClueGO hypergeometric enrichment pathway analysis (figure 2F; appendix 1 pp 14, 24). Gene set enrichment analysis of all genes using Reactome corroborated the downregulation of genes linked to IFN signalling and neutrophil degranulation in patients with IAPA (appendix 1 pp 14, 25).
For the comparison of patients with CAPA versus patients with COVID-19 only, we identified 34 significantly downregulated differentially expressed genes and no significantly upregulated genes (figure 2B). 28 of these differentially expressed genes overlapped with the downregulated differentially expressed genes of the IAPA versus influenza only comparison and were mainly genes related to downstream type I and II IFN signalling (eg, MX1 and HLA-DRA), macrophage activation (eg, CD163), and antigen presentation (eg, HLA-DRB3; figure 2E; appendix 1 pp 13, 21). TLR2 and HIF1A were also downregulated in both comparisons (figure 2E). Among the six other significantly downregulated differentially expressed genes were TNF (encoding TNF-α) as well as TRAF1 and CDKN1A (both upregulated upon TNF-α signalling). ClueGO hypergeometric enrichment pathway analysis of the downregulated differentially expressed genes showed significant downregulation of responses to type I and II IFN in patients with CAPA versus patients with COVID-19 only (figure 2F; appendix 1 pp 14, 24). Gene set enrichment analysis corroborated this finding and also showed the downregulation of neutrophil degranulation (appendix 1 pp 14, 26).
We identified 11 significantly downregulated differentially expressed genes in the comparison of IAPA versus CAPA, including IL1B and MAP1LC3B and the chemokine-encoding genes, CXCL1, CXCL8, and CCL4 (figure 2C). Due to the low number of differentially expressed genes with Benjamini–Hochberg q values of less than 0·05, we performed ClueGO pathway analysis with downregulated differentially expressed genes with q values of less than 0·20. ClueGO pathway analysis showed the downregulation of neutrophil migration, monocyte chemotaxis, macrophage activation, and cellular responses to IL-1 and IFNγ in patients with IAPA versus patients with CAPA (figure 2F; appendix 1 pp 14, 24). Gene set enrichment analysis showed the downregulation of pathways linked to interleukin and interferon signalling and neutrophil degranulation (appendix 1 pp 14, 27–28).
The comparison of influenza only versus COVID-19 only revealed 11 significantly upregulated and 63 significantly downregulated differentially expressed genes (figure 2D). As expected from earlier studies comparing the immune profiles of bronchoalveolar lavage fluid in these patient groups,20 there was a downregulation of the proinflammatory genes TNF and IL1B and the chemokine gene CXCL8 in patients with influenza only compared with patients with COVID-19 only. Other significantly downregulated differentially expressed genes that encode chemokines were CCL2, CCL3L1, CCL4, and CCL20. There was also a significant downregulation of genes that are upregulated in type II IFN signalling (eg, HLA-DRA). ClueGO pathway analysis of the downregulated differentially expressed genes confirmed the downregulation of cellular responses to TNF-α, IL-1, and IFNγ and the downregulation of neutrophil migration and macrophage activation in patients with influenza only versus COVID-19 only (figure 2F; appendix 1 pp 14, 24). However, in gene set enrichment analysis, only pathways related to the extracellular matrix were significantly downregulated in patients with influenza only compared with patients with COVID-19 only (appendix 1 pp 14, 29).
We compared differentially expressed genes from the IAPA versus influenza only and CAPA versus COVID-19 only comparisons with an existing dataset of the transcriptional response of peripheral blood mononuclear cells 24 h after stimulation with Aspergillus fumigatus conidia.12 We found a substantial overlap between downregulated differentially expressed genes in the IAPA versus influenza only and CAPA versus COVID-19 only comparisons and the genes that are upregulated in peripheral blood mononuclear cells in response to conidia—eg, CCRL2, GRN, and SQSTM1 for both comparisons and MMP9, CXCL2, and PLAU for IAPA versus influenza only specifically (appendix 1 pp 14, 30). This corroborates that the downregulation of these genes seen in the IAPA versus influenza only and CAPA versus COVID-19 only comparisons contributes to the development of aspergillosis, as they are transcriptional components of a normal antifungal host response.
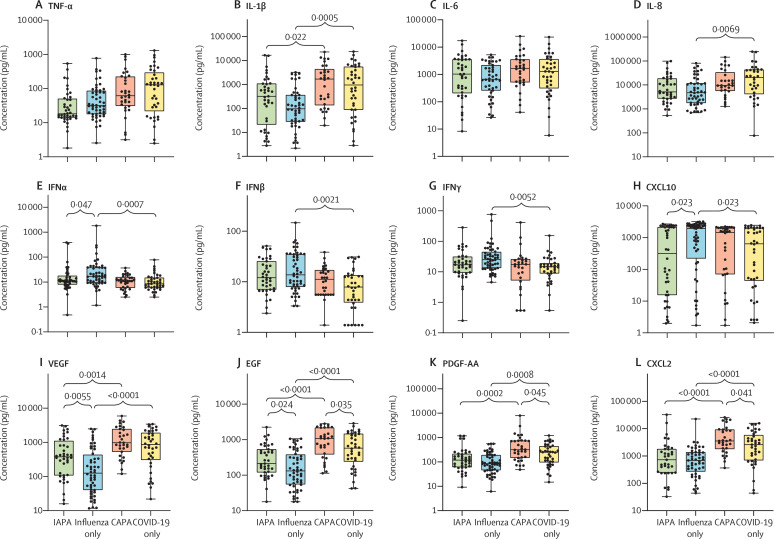
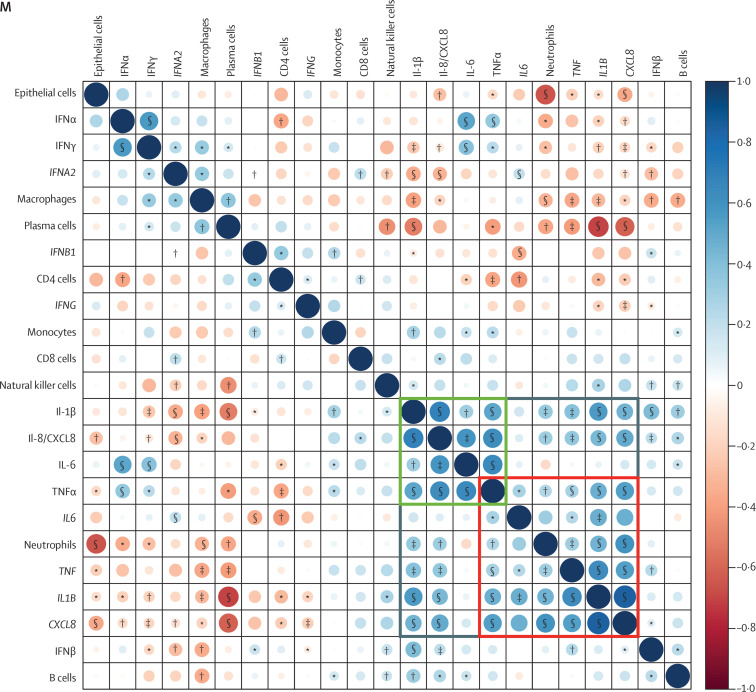
We assessed the concentrations and the correlations of multiple cytokines, chemokines, and growth factors in the bronchoalveolar lavage samples (figure 3A–L ; appendix 1 pp 15, 31). Generally, samples from patients with COVID-19 with or without aspergillosis had numerically higher concentrations of multiple proinflammatory cytokines and chemokines (eg, TNF-α, IL-1β, and IL-8) than did samples from patients with influenza with or without aspergillosis (figure 3A–D). Samples from patients with COVID-19 only had significantly lower concentrations of IFNα, IFNβ, and IFNγ than did those from patients with influenza only, a difference that was not seen when comparing patients with CAPA versus IAPA (figure 3E–G). When comparing patients with IAPA versus influenza only, we found significantly lower concentrations of IFNα and the interferon-induced cytokine CXCL10, but no differences in IFNβ and IFNγ concentrations (figure 3E–H). Concentrations of epidermal growth factor (EGF) and vascular endothelial growth factor (VEGF) were significantly higher in IAPA samples than in influenza only samples and concentrations of EGF and platelet-derived growth factor AA (PDGF-AA) were significantly higher in CAPA samples than in COVID-19 only samples (figure 3I–K). The concentrations of all other proteins, including IL-6, did not differ between CAPA samples and COVID-19 only samples, except for the higher concentration of CXCL2 in CAPA samples versus COVID-19 only samples (figure 3L; appendix 1 pp 15, 31). The concentrations of pentraxin-related protein PTX3 were very high in all groups, but did not significantly differ between groups, meaning that PTX3 could not discriminate between viral pneumonia with or without aspergillosis (appendix 1 pp 15, 31). Despite the differences we found in neutrophil fractions (figure 1), we did not find differences in the concentrations of most chemokines between the IAPA versus influenza only samples and the CAPA versus COVID-19 only samples (figure 3; appendix 1 pp 15, 31). We found significant correlations between the bronchoalveolar lavage fluid concentrations and the respective gene expressions of IL-1α, IL-1β, TNF-α, IFNβ, and several chemokines (eg, CCL4, CCL20, and CXCL2; figure 3M; appendix 1 pp 15, 32–36), reflecting substantial local production of these factors. Significant correlations between the bronchoalveolar lavage fluid concentrations and the respective gene expressions of IL-6, IFNγ, and most growth factors in our panel were not observed, meaning that these protein concentrations must be determined by extraluminal production rather than local intraluminal production (figure 3M; appendix 1 pp 15, 32–36).
Figure 3.
Concentrations and correlations of cytokines, chemokines, and growth factors in bronchoalveolar lavage samples
(A–L) Concentrations of major cytokines, chemokines, and growth factors. Box plots represent median and IQR, with whiskers set from minimum to maximum. Individual points represent each sample. q values were generated by Kruskall–Wallis tests, corrected by use of the Benjamini–Krieger–Yekutieli method. Only significant q values are shown. The scale between points is a log scale. (M) Correlogram showing the correlations between the CIBERSORTx-derived cell fractions, major cytokine concentrations, and gene expressions of the corresponding genes in bronchoalveolar lavage samples with all three types of results available. Components are hierarchically clustered. The grey box represents a major proinflammatory cluster. The green and red boxes represent two pro-inflammatory subclusters, with the green box representing proinflammatory cytokines and the red box representing the deconvoluted neutrophil fraction and the gene expression of proinflammatory cytokines. CAPA=COVID-19-associated pulmonary aspergillosis. EGF=epidermal growth factor. IAPA=influenza-associated pulmonary aspergillosis. PDGF-AA=platelet-derived growth factor AA. VEGF=vascular endothelial growth factor. *p<0·05. †p<0·01. ‡p<0·001. §p<0·0001.
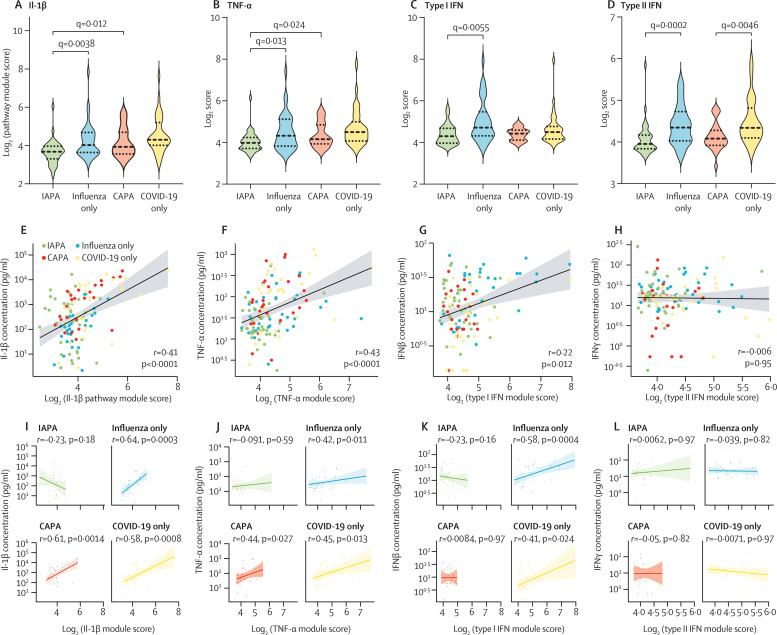
To quantify the downstream signalling pathways of four major cytokines with important roles in antifungal and antiviral immunity (IL-1β, TNF-α, type I IFN, and type II IFN), we calculated pathway module scores for each patient (figure 4 ; appendix 1 pp 15–16, 37). We found these pathway module scores to largely reflect the results of the pathway analyses done on the differentially expressed genes, thereby validating the robustness of our approach. IAPA samples showed significantly lower scores for all pathways compared with influenza only samples, but CAPA samples showed significantly lower scores compared with COVID-19 only samples for type II IFN only (figure 4; appendix 1 pp 15–16, 37). In general, the inferred neutrophil fractions correlated well with pathway module scores for IL-1β and TNF-α (appendix 1 pp 15–16, 37). Pathway module scores for IL-1β and TNF-α also significantly correlated with concentrations of the respective inducing cytokine for influenza only, CAPA, and COVID-19 only samples, but not for IAPA samples, potentially reflecting a blunted cellular response to both proinflammatory cytokines (figure 4I–J). Moreover, we observed a significant correlation between IFNβ concentration (not significant for IFNα) and the corresponding pathway module score for patients with influenza only or COVID-19 only, which was absent in patients with IAPA or CAPA (figure 4K; appendix 1 pp 15–16, 37). By contrast, type II IFN pathway module scores did not correlate with IFNγ concentrations in any patient group (figure 4L).
Figure 4.
Pathway module scores and their correlation with concentration for IL-1β, TNF-α, type I IFN, and type II IFN
Pathway module scores for IL-1β (A), TNF-α (B), type I IFN (C), and type II IFN (D). q values were generated by Kruskall–Wallis tests, corrected by use of the Benjamini–Krieger–Yekutieli method. Only significant q values are shown. Spearman rank correlation between the major cytokine concentration and the corresponding module score for all patients for IL-1β (E), TNF-α (F), type I IFN (G), and type II IFN (H). Spearman rank correlation between the major cytokine concentration and the corresponding module score per disease group for IL-1β (I), TNF-α (J), type I IFN (K), and type II IFN (L). The line represents the regression line and the shading represents the 95% CI. CAPA=COVID-19-associated pulmonary aspergillosis. IAPA=influenza-associated pulmonary aspergillosis.
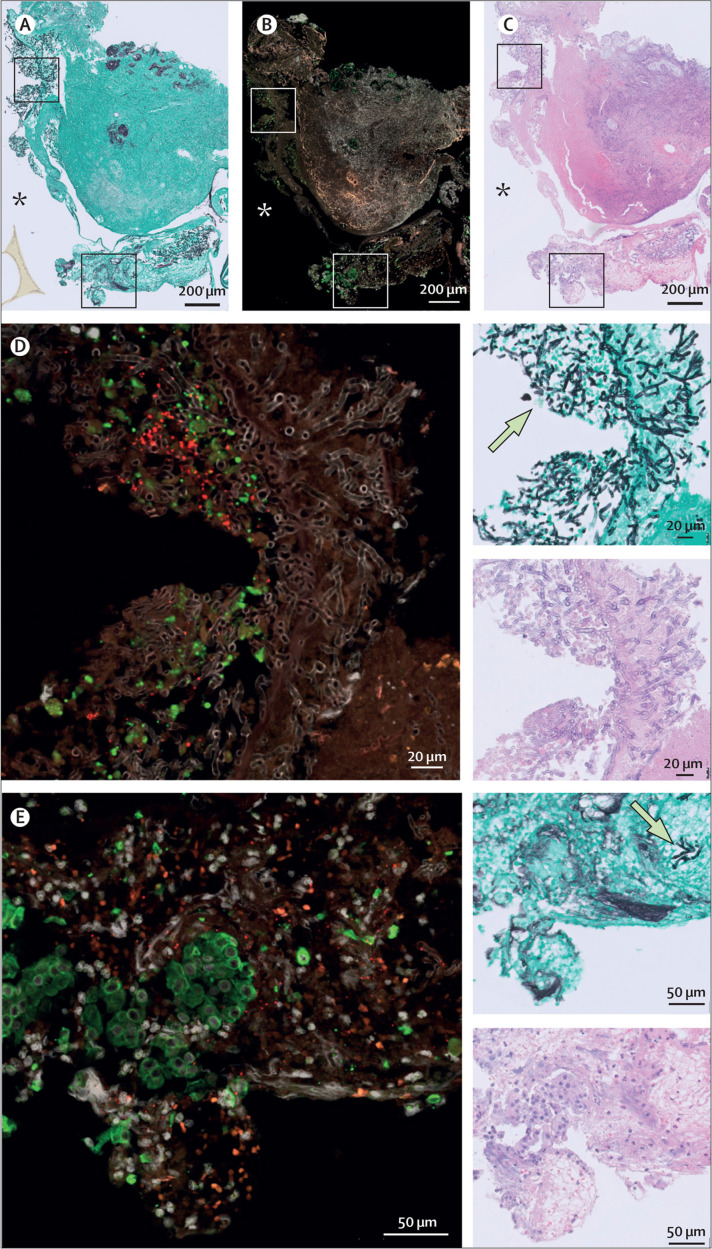
To further examine differences between IAPA and CAPA and the role of the epithelial barrier, we analysed in vivo tracheobronchial biopsies from two patients with IAPA and invasive Aspergillus tracheobronchitis and two patients with CAPA and invasive Aspergillus tracheobronchitis. Invasive Aspergillus tracheobronchitis is a particularly lethal form of IAPA and CAPA, with Aspergillus lesions apparent in the larger airways.21, 22 In formalin-fixed, paraffin-embedded sections of these biopsies, we performed RNAScope to investigate whether Aspergillus hyphae colocalise with the presence of influenza virus or SARS-CoV-2 transcripts. In one patient with CAPA, we visualised an active or very recent SARS-CoV-2 infection damaging the epithelium (figure 5 ). SARS-CoV-2 nucleocapsid RNA puncta colocalised with Aspergillus hyphae invading the tissue in this biopsy. As the biopsies from patients with IAPA no longer contained any epithelial tissue at the site of Aspergillus invasion, we did not identify influenza virus RNA in these zones.
Figure 5.
In vivo tracheobronchial biopsy from patient with CAPA and invasive Aspergillus tracheobronchitis
(A) Slide stained with Grocott–Gomori's methenamine silver, which makes fungi appear black and the background tissue green. (B) RNAScope image, in which the red puncta reflect SARS-CoV-2 nucleocapsid RNA. Epithelial cells are labelled green by anti-KRT8 antibody staining. DAPI was used as a nuclear stain (grey). (C) Slide stained with haematoxylin and eosin, which is used to look at damaged areas. (D–E) Two magnified areas of the CAPA biopsy, with respective haematoxylin and eosin-stained, Grocott-Gomori's methenamine silver-stained, and RNAScope images. Examples of fungi are indicated on the Grocott-Gomori's methenamine silver-stained images by arrows. The asterisks localise the lumen of the respiratory tract. CAPA=COVID-19-associated pulmonary aspergillosis.
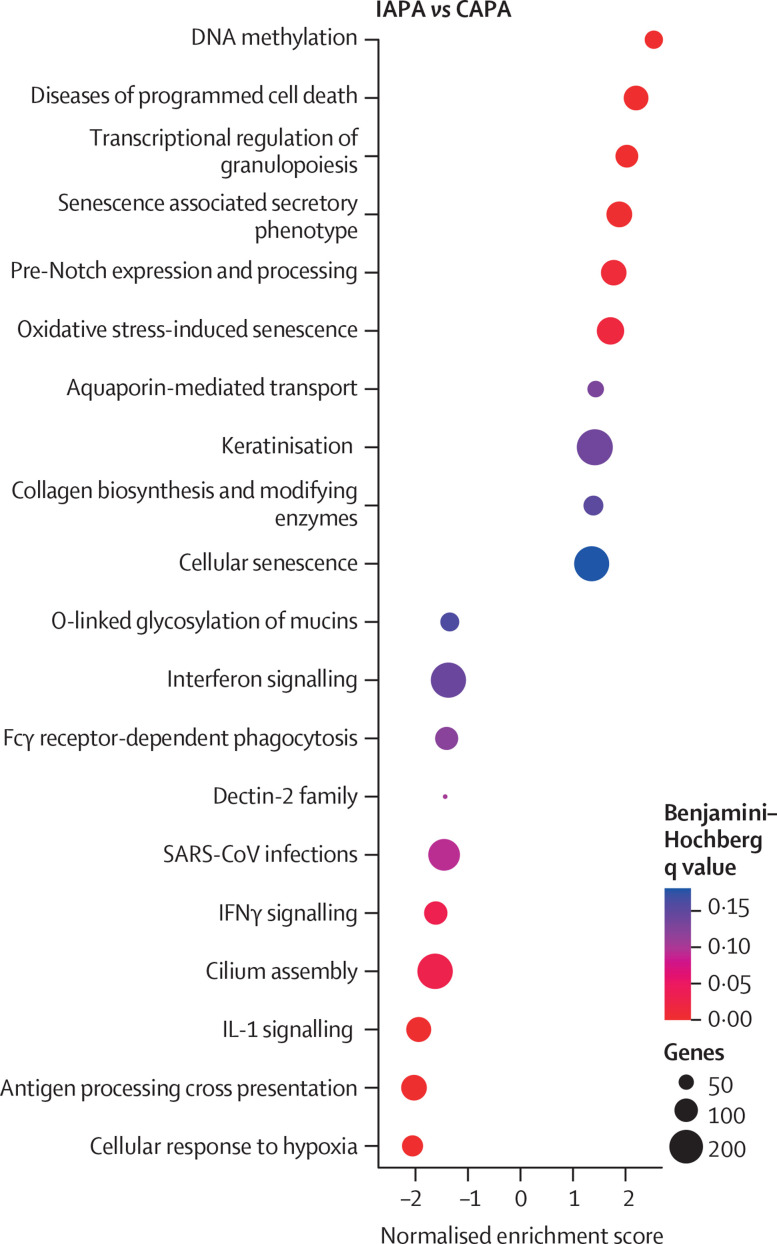
We additionally conducted spatial transcriptomics on these biopsies from patients with IAPA or CAPA and invasive Aspergillus tracheobronchitis (figure 6 ; appendix 1 pp 16, 38–43). By performing gene set enrichment analysis (with Reactome as library), we discovered a clear difference in the epithelial transcriptional response, with increased expression of keratinisation and programmed cell death pathways and decreased expression of IL-1 signalling, IFNγ signalling, and cilium assembly in IAPA samples versus CAPA samples (figure 6). However, we did not identify major differences in transcriptional responses for the inflammatory infiltrate in IAPA versus CAPA samples (appendix 1 pp 16, 41).
Figure 6.
Dot plot of gene set enrichment analysis based on gene expression of epithelial regions of interest in IAPA versus CAPA samples
Reactome canonical pathways was the pathway database. CAPA=COVID-19-associated pulmonary aspergillosis. IAPA=influenza-associated pulmonary aspergillosis.
Discussion
In this observational study, we conducted an explorative immune phenotyping of patients with IAPA or CAPA, with a focus on innate myeloid immunity, using transcriptomic and proteomic methods on lower respiratory tract samples. By analysing bronchoalveolar lavage fluid samples from a cohort of critically ill patients with influenza or COVID-19 with or without aspergillosis and several tracheobronchial biopsy samples from patients with IAPA or CAPA and invasive Aspergillus tracheobronchitis, we identified key patterns in the lower airway immune profiles of these patients. Our results uncover a three-level breach in innate antifungal immunity as a predisposing factor to invasive aspergillosis in patients with severe influenza or COVID-19.
A first line of immune defence against invasive aspergillosis is represented by the epithelium. An individual inhales hundreds of Aspergillus conidia each day, and these spores are constantly cleared by the epithelium lining the respiratory tract.23 The epithelial cells function as a pure physical barrier against aspergillosis and also contribute to the clearance of Aspergillus conidia by means of phagocytosis and mucociliary clearance.24 Epithelial destruction has therefore been proposed as one of the main mechanisms by which severe viral pneumonia predisposes to the development of aspergillosis.25 Using RNAScope technology, we obtained direct evidence that Aspergillus hyphae invade the mucosa through damaged epithelium that is concurrently or has recently been infected by SARS-CoV-2.
Because patients with respiratory failure caused by influenza or COVID-19 show a higher risk of developing aspergillosis than do patients with non-viral acute respiratory distress syndrome,2 epithelial disruption might not be the only viral factor predisposing to IAPA or CAPA. We have uncovered evidence that the second line of the antifungal host response, namely phagocytosis and the subsequent killing of Aspergillus conidia by phagocytes, could also be affected in patients with IAPA or CAPA. We observed a downregulated expression of several genes encoding proteins with functions in the opsonisation, recognition, and killing of conidia in patients with IAPA versus influenza only and in patients with CAPA versus COVID-19 only.
Once Aspergillus conidia have developed into hyphae, the fungus can no longer be destroyed by phagocytosis. The third line of the antifungal host response relies on extracellular mechanisms, mainly mediated by neutrophils, to kill the fungus.23 Cell fractions derived from the gene expression data revealed lower neutrophil fractions in patients with coinfections versus monoinfections (significant for CAPA vs COVID-19 only and non-significant for IAPA vs influenza only) and pathway analysis revealed that several neutrophil effector pathways (eg, degranulation and migration) were downregulated in patients with IAPA compared with patients with influenza only. In general, the inferred neutrophil fractions correlated well with pathway module scores for IL-1β and TNF-α, suggesting that these pathways are to a large extent activated by neutrophils.
Both IAPA and CAPA are associated with increased mortality compared with influenza or COVID-19 alone, but data on the effect of antifungal treatment on outcomes for affected patients are less clear.6 Analysing bronchoalveolar lavage samples, we found that patients with IAPA had significantly higher concentrations of EGF and VEGF than did patients with influenza only and that patients with CAPA had significantly higher concentrations of EGF and PDGF-AA than did patients with COVID-19 only. Therefore, we hypothesise that patients with these coinfections might be more severely affected by late-stage lung fibrosis than patients with monoinfections, which could in part explain the increased mortality.
We observed an aberrant transcriptional response to several key cytokines in bronchoalveolar lavage samples from patients with IAPA or CAPA. Severe influenza and COVID-19 involve hyperinflammatory states characterised by high bronchoalveolar lavage concentrations of IL-1β, IL-6, and TNF-α. This state has been proposed as a potential fertile ground for aspergillosis.25, 26 Nevertheless, we found blunted transcriptional responses to IL-1β, TNF-α, type I IFN, and type II IFN (IFNγ) in patients with IAPA versus influenza only. Of interest, patients with CAPA also had downregulated type II IFN signalling compared with patients not affected by aspergillosis. The role of IFNγ in COVID-19 remains unclear, with findings ranging from a beneficial to a detrimental role in disease progression.27, 28 IFNγ is generally seen as an important component of the antifungal host response, for instance via inducing LC3-associated phagocytosis.29 As such, adjuvant treatment with IFNγ is currently being investigated in candidaemia (NCT04979052) and has been propounded as a potential adjuvant in the treatment of invasive aspergillosis.26 Therefore, the role of diminished IFNγ signalling in the development of IAPA or CAPA warrants further investigation in animal models to assess its clinical relevance. Moreover, the findings of lower neutrophil fractions and reduced transcription of LC3-associated phagocytosis components in patients with IAPA versus influenza only or CAPA versus COVID-19 only calls for the investigation of neutrophil extracellular trap formation and LC3-associated phagocytosis in vitro, in animal models, and in patients. Future research is urgently needed to further unravel the pathophysiology of IAPA and CAPA and to translate this knowledge into clinical practice.
Our study has several limitations. First, the use of bulk multiplex gene expression yields a low-resolution view of transcription dynamics in the lower respiratory tract and does not easily allow for cell–cell interaction analyses. The cell fractions estimated by cellular deconvolution should therefore be interpreted cautiously. Second, we used retrospectively collected samples that had been obtained at different timepoints after the initiation of mechanical ventilation. This limitation could be addressed in future research by the use of prospectively collected, timepoint-matched samples. Third, we only focused on epithelial and myeloid innate immunity as these are the main components of antifungal immunity. However, adaptive lymphoid immunity might also be important in IAPA and CAPA. Finally, the use of human samples means we could not capture the very first stage of coinfection; we are only able to detect the fungus in bronchoalveolar lavage fluid after it is abundantly present in its growing form, hyphal form, or both. To describe the early infection dynamics of IAPA and CAPA (particularly epithelial or phagocytic uptake and escape of Aspergillus conidia), in vitro and animal model work will be necessary.
In conclusion, we obtained insight into the pathophysiology of both IAPA and CAPA using patient samples. Reduced type II IFN signalling and increased concentrations of fibrosis-associated growth factors in the lower respiratory tract are common traits for IAPA and CAPA. We provide evidence of a three-level breach in innate antifungal immunity, affecting epithelium, macrophages, and neutrophils, as a factor predisposing the development of IAPA and CAPA. Our observations could lead to the identification of novel host immune risk factors and eventually to the discovery of new diagnostic and prognostic biomarkers for IAPA and CAPA. Moreover, our results could be translated into in-depth research of the use of immunomodulatory targets, such as recombinant IFNγ, as adjuvant antifungal treatments. In future, high-resolution multiomics and functional studies will be necessary to further unravel the pathophysiology of IAPA and CAPA.
Data sharing
All data can be shared upon reasonable request to the corresponding author. nCounter gene expression data (raw and normalised counts) are available in appendix 2. GeoMx gene expression data (raw and third quartile normalised counts) have been deposited in Gene Expression Omnibus with accession number GSE198096.
Declaration of interests
SF received travel grants from Pfizer. KL received consultancy fees from MRM Health, MSD, and Gilead; speaker fees from FUJIFILM WAKO, Pfizer, and Gilead; and a service fee from Thermo Fisher Scientific. LV received travel grants from Pfizer and Gilead. JW received investigator-initiated grants, speaker's fees, and travel fees from Pfizer, Gilead, and MSD and declares participation in advisory boards for Pfizer and Gilead and the receipt of study drugs from MSD. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
This research project was funded by Research Foundation Flanders (FWO) under grant G053121N to SH-B and JW and grant G0A0621N to JVW; the Max Planck Society to PMo; the Coronafonds of KU Leuven and University Hospitals Leuven to JW; the Fundação para a Ciência e a Tecnologia (FCT; UIDB/50026/2020 and UIDP/50026/2020) to AC; the Northern Portugal Regional Operational Programme (NORTE 2020) under the Portugal 2020 Partnership Agreement through the European Regional Development Fund (NORTE-01–0145-FEDER-000039) to AC; the “la Caixa” Foundation (ID 100010434) and the FCT under the agreement LCF/PR/HR17/52190003 to AC; and the EU's Horizon 2020 research and innovation programme (grant agreement number 847507 [Host-Directed Medicine in Invasive Fungal Infections]) to FLVDV, AC, and JW. SF is funded by an FWO PhD fellowship (grant number 11M6922N). SMG is supported by an FCT PhD fellowship (SFRH/BD/136814/2018). CC is supported by the FCT (PTDC/SAU-SER/29635/2017 and CEECIND/04058/2018) and the Gilead Research Scholars Program—Anti-Fungals. GH is funded by an FWO Fundamental Clinical Research fellowship (1805116N). LS is supported by an FWO PhD fellowship (grant number 1186121N). KT acknowledges receipt of a mandate of the Industrial Research Fund, KU Leuven (IOFm/05/022). CV and JW received a research grant of the Klinische Onderzoeksraad UZ Leuven. LV is supported by an FWO PhD fellowship (grant number 11E9819N). FLVDV is supported by a Vidi grant of the Netherlands Association for Scientific Research. We thank Dide Jaubin (KU Leuven, Leuven, Belgium) for aid with the nCounter data analysis; Lauren Van Der Sloten (University Hospitals Leuven, Leuven, Belgium) for aid with the construction of the database with clinical information; Bo Corbeels (KU Leuven, Leuven, Belgium) and Kathleen Van Den Eynde (KU Leuven, Leuven, Belgium) for aid with the preparation of the tissue slides; Anna P Pavenko (NanoString; Seattle, WA, USA) for GeoMx benchwork; Marty Ross (Nanostring; Seattle, WA, USA) for GeoMx data analysis; and Wei Yang (Nanostring; Seattle, WA, USA), Yan Liang (Nanostring; Seattle, WA, USA), and Tony Zucca (Nanostring; Seattle, WA, USA) for scientific support with the GeoMx project.
Contributors
SF, DL, PMo, AC, JVW, and JW contributed to study conceptualisation and methodology. SF, SMG, MK, SC, BB, DC, CC, YD, GH, MH, SH-B, CJ, KL, LM, JM, PMe, RN, MRS, CV, LV, NVR, AV, SV, AW, DL, and GDH contributed to the investigation. SF, SMG, MK, SC, CC, BB, SH-B, LS, KT, GVV, and SV contributed to the formal analysis. SF, AC, and JVW contributed to data curation. SF wrote the original draft of the manuscript. SF, FLVDV, PMo, AC, JVW, and JW reviewed and edited the manuscript. SF, MK, SC, CJ, and LS made the figures. AC, JVW, and JW supervised. SH-B, PMo, FLVDV, AC, JVW, and JW acquired funding. LV and JVW directly accessed and verified the underlying data. All authors critically revised and approved the final manuscript. SF, AC, JVW, and JW were responsible for the decision to submit the manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Supplementary Material
References
- 1.Paget J, Spreeuwenberg P, Charu V, et al. Global mortality associated with seasonal influenza epidemics: new burden estimates and predictors from the GLaMOR Project. J Glob Health. 2019;9 doi: 10.7189/jogh.09.020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schauwvlieghe AFAD, Rijnders BJA, Philips N, et al. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respir Med. 2018;6:782–792. doi: 10.1016/S2213-2600(18)30274-1. [DOI] [PubMed] [Google Scholar]
- 3.Gangneux J-P, Dannaoui E, Fekkar A, et al. Fungal infections in mechanically ventilated patients with COVID-19 during the first wave: the French multicentre MYCOVID study. Lancet Respir Med. 2022;10:180–190. doi: 10.1016/S2213-2600(21)00442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prattes J, Wauters J, Giacobbe DR, et al. Risk factors and outcome of pulmonary aspergillosis in critically ill coronavirus disease 2019 patients—a multinational observational study by the European Confederation of Medical Mycology. Clin Microbiol Infect. 2022;28:580–587. doi: 10.1016/j.cmi.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janssen NAF, Nyga R, Vanderbeke L, et al. Multinational observational cohort study of COVID-19-associated pulmonary aspergillosis. Emerg Infect Dis. 2021;27:2892–2898. doi: 10.3201/eid2711.211174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feys S, Almyroudi MP, Braspenning R, et al. A visual and comprehensive review on COVID-19-associated pulmonary aspergillosis (CAPA) J Fungi (Basel) 2021;7 doi: 10.3390/jof7121067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verweij PE, Rijnders BJA, Brüggemann RJM, et al. Review of influenza-associated pulmonary aspergillosis in ICU patients and proposal for a case definition: an expert opinion. Intensive Care Med. 2020;46:1524–1535. doi: 10.1007/s00134-020-06091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koehler P, Bassetti M, Chakrabarti A, et al. Defining and managing COVID-19-associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect Dis. 2021;21:e149–e162. doi: 10.1016/S1473-3099(20)30847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruno M, Horst R, Pekmezovic M, et al. Data of common and species-specific transcriptional host responses to pathogenic fungi. Data Brief. 2021;35 doi: 10.1016/j.dib.2021.106928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonilla DL, Bhattacharya A, Sha Y, et al. Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity. 2013;39:537–547. doi: 10.1016/j.immuni.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prajsnar TK, Serba JJ, Dekker BM, et al. The autophagic response to Staphylococcus aureus provides an intracellular niche in neutrophils. Autophagy. 2021;17:888–902. doi: 10.1080/15548627.2020.1739443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Y-P, Lai S, Lin Q-Y, et al. CDC20 regulates cardiac hypertrophy via targeting LC3-dependent autophagy. Theranostics. 2018;8:5995–6007. doi: 10.7150/thno.27706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyrmizi I, Gresnigt MS, Akoumianaki T, et al. Corticosteroids block autophagy protein recruitment in Aspergillus fumigatus phagosomes via targeting dectin-1/Syk kinase signaling. J Immunol. 2013;191:1287–1299. doi: 10.4049/jimmunol.1300132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier A, Kirschning CJ, Nikolaus T, Wagner H, Heesemann J, Ebel F. Toll-like receptor (TLR) 2 and TLR4 are essential for Aspergillus-induced activation of murine macrophages. Cell Microbiol. 2003;5:561–570. doi: 10.1046/j.1462-5822.2003.00301.x. [DOI] [PubMed] [Google Scholar]
- 18.Shah A, Kannambath S, Herbst S, et al. Calcineurin orchestrates lateral transfer of Aspergillus fumigatus during macrophage cell death. Am J Respir Crit Care Med. 2016;194:1127–1139. doi: 10.1164/rccm.201601-0070OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepardson KM, Jhingran A, Caffrey A, et al. Myeloid derived hypoxia inducible factor 1-alpha is required for protection against pulmonary Aspergillus fumigatus infection. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cambier S, Metzemaekers M, de Carvalho AC, et al. Atypical response to bacterial coinfection and persistent neutrophilic bronchoalveolar inflammation distinguish critical COVID-19 from influenza. JCI Insight. 2022;7 doi: 10.1172/jci.insight.155055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyga R, Maizel J, Nseir S, et al. Invasive tracheobronchial Aspergillosis in critically ill patients with severe influenza. A clinical trial. Am J Respir Crit Care Med. 2020;202:708–716. doi: 10.1164/rccm.201910-1931OC. [DOI] [PubMed] [Google Scholar]
- 22.van de Veerdonk FL, Brüggemann RJM, Vos S, et al. COVID-19-associated Aspergillus tracheobronchitis: the interplay between viral tropism, host defence, and fungal invasion. Lancet Respir Med. 2021;9:795–802. doi: 10.1016/S2213-2600(21)00138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latgé J-P, Chamilos G. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev. 2019;33:ee00140. doi: 10.1128/CMR.00140-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croft CA, Culibrk L, Moore MM, Tebbutt SJ. Interactions of Aspergillus fumigatus conidia with airway epithelial cells: a critical review. Front Microbiol. 2016;7:472. doi: 10.3389/fmicb.2016.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dewi IM, Janssen NA, Rosati D, et al. Invasive pulmonary aspergillosis associated with viral pneumonitis. Curr Opin Microbiol. 2021;62:21–27. doi: 10.1016/j.mib.2021.04.006. [DOI] [PubMed] [Google Scholar]
- 26.van de Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latgé JP. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol. 2017;15:661–674. doi: 10.1038/nrmicro.2017.90. [DOI] [PubMed] [Google Scholar]
- 27.Sadanandam A, Bopp T, Dixit S, et al. A blood transcriptome-based analysis of disease progression, immune regulation, and symptoms in coronavirus-infected patients. Cell Death Discov. 2020;6:141. doi: 10.1038/s41420-020-00376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamond MS, Kanneganti T-D. Innate immunity: the first line of defense against SARS-CoV-2. Nat Immunol. 2022;23:165–176. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oikonomou V, Moretti S, Renga G, et al. Noncanonical fungal autophagy inhibits inflammation in response to IFN-γ via DAPK1. Cell Host Microbe. 2016;20:744–757. doi: 10.1016/j.chom.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data can be shared upon reasonable request to the corresponding author. nCounter gene expression data (raw and normalised counts) are available in appendix 2. GeoMx gene expression data (raw and third quartile normalised counts) have been deposited in Gene Expression Omnibus with accession number GSE198096.