Abstract
The type-specific capsular polysaccharide (CP) of a group B streptococcus, Streptococcus agalactiae type Ia, is a high-molecular-weight polymer consisting of the pentasaccharide repeating unit 4)-[α-d-NeupNAc-(2→3)-β-d-Galp-(1→4)-β-d-GlcpNAc-(1→3)]-β-d-Galp-(1→4)-β-d-Glcp-(1. Here, cloning, sequencing, and transcription of the type Ia-specific capsular polysaccharide synthesis (cps) genes and functional analysis of these gene products are described. A 26-kb DNA fragment containing 18 complete open reading frames (ORFs) was cloned. These ORFs were designated cpsIaA to cpsIaL, neu (neuraminic acid synthesis gene) A to D, orf1 and ung (uracil DNA glycosylase). The cps gene products of S. agalactiae type Ia were homologous to proteins involved in CP synthesis of S. agalactiae type III and S. pneumoniae serotype 14. Unlike the cps gene cluster of S. pneumoniae serotype 14, transcription of this operon may start from cpsIaA, cpsIaE, and orf1 because putative promoter sequences were found in front of these genes. Northern hybridization, reverse transcription-PCR, and primer extension analyses supported this hypothesis. DNA sequence analysis showed that there were two transcriptional terminators in the 3′ end of this operon (downstream of orf1 and ung). The functions of CpsIaE, CpsIaG, CpsIaI, and CpsIaJ were examined by glycosyltransferase assay by using the gene products expressed in Escherichia coli JM109 harboring plasmids containing various S. agalactiae type Ia cps gene fragments. Enzyme assays suggested that the gene products of cpsIaE, cpsIaG, cpsIaI, and cpsIaJ are putative glucosyltransferase, β-1,4-galactosyltransferase, β-1,3-N-acetylglucosaminyltransferase, and β-1,4-galactosyltransferase, respectively.
Encapsulated bacteria are frequently associated with serious diseases in both humans and animals. The capsular polysaccharides (CPs) of pathogenic bacteria confer resistance to complement-mediated opsonophagocytosis (35). In addition, some bacteria have CPs that mimic host molecules to avoid the specific immune system of the host (10). Bacterial CPs are generally composed of repeating oligosaccharides consisting of two to ten monosaccharides and are sometimes complemented with other components.
Group B streptococci, Streptococcus agalactiae, are human pathogens causing invasive diseases such as sepsis, meningitis, and pneumonia in infants (8). These gram-positive bacteria have two distinct polysaccharide antigens. One of these, group antigen (C substance), composed of a number of rhamnose units, is common to all strains. The others are type-specific CPs that separate S. agalactiae into eight serotypes. The chemical structures of these polysaccharides have been determined (11, 18–20, 24, 45, 48, 49).
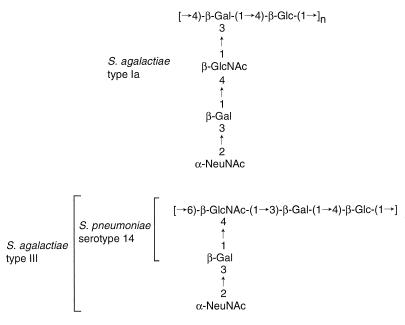
We are particularly interested in the type-specific CP of S. agalactiae type Ia, whose polysaccharide has a linear backbone of a 4)-β-d-Glcp-(1-4)-β-d-Galp-(1 repeating unit with trisaccharide side chains of α-NeupNAc-(2-3)-β-d-Galp-(1-4)-β-d-GlcpNAc- (1 linked to C3 of each β-d-galactose residue of the backbone (19). Although Streptococcus pneumoniae strains have divergent CPs, the unit structure of the S. agalactiae type Ia CP is similar to those of CPs of S. pneumoniae serotype 14 and S. agalactiae type III (Fig. 1). However, the polysaccharide is distinct from that of S. pneumoniae serotype 14, since the latter does not contain sialic acid. In S. agalactiae type III, the same sugar units polymerize through a linkage different from that of type Ia, which results in a different CP structure (Fig. 1).
FIG. 1.
Subunit structures of CPs from S. agalactiae type Ia, type III and S. pneumoniae serotype 14. Glc, glucose; Gal, galactose; GlcNAc, N-acetylglucosamine; NeuNAc, N-acetylneuraminic acid.
Recently, the genes involved in CP synthesis (cps) and the mechanisms of biosynthesis have been reported for many bacteria (2, 6, 7, 14, 15). In fact, cps gene clusters have been analyzed in many S. pneumoniae strains (3, 16, 25–27, 33, 34, 36) but only partially identified in S. agalactiae type III (38). The biosynthesis of CPs is a complex enzymatic pathway starting with the uptake or synthesis of the monosaccharides and their activation to nucleotide derivatives. Membrane-bound transferase complexes then catalyze the successive coupling of the monosaccharides to a membrane-bound lipid carrier, followed by polymerization of the sugar subunits and subsequent export and attachment of the complete CP to the cell surface (6, 7).
In this study the structure and the transcription of the cps gene cluster required for synthesis of S. agalactiae type Ia CP were studied and compared with those of S. pneumoniae serotype 14. The functions of several gene products were also determined by measuring enzyme activities.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
S. agalactiae type Ia strain OI1 was isolated from a vaginal swab from a patient with no symptoms of infection. This strain was confirmed to express the type Ia capsule by using type Ia-specific antiserum (Denka Seiken Co.), which was prepared with CP of a type strain from the WHO collaborate center, The Czech Republic National Collection of Type Cultures at the Institute of Hygiene. S. agalactiae type Ia strain OI1 was cultured in Todd-Hewitt broth (Becton Dickinson) supplemented with 2% glucose and 1.5% Na2HPO4 at 37°C (46). Escherichia coli DH5α (39) was used as the host for pBluescript II KS (+) or SK (+) (Stratagene). E. coli JM109 (39) was used as the host for the expression plasmids. All E. coli clones were routinely grown in Luria-Bertani broth (39) containing appropriate antibiotics.
DNA manipulations.
Most DNA manipulations were performed according to standard procedures (39). Chromosomal DNA was isolated as reported previously (4). 32P-labeled probes were prepared with a BcaBEST labeling kit (Takara). PCR was performed with Takara Long and Accurate Taq according to the manufacturer’s instructions.
DNA sequencing.
The DNA sequences of both strands were determined by using an ABI 373S automated DNA sequencer (Perkin-Elmer, Applied Biosystems Division). The sequencing data were compared with those in the DDBJ, EMBL, and GenBank databases by using the BLAST network service at the National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md.
Construction of expression plasmids.
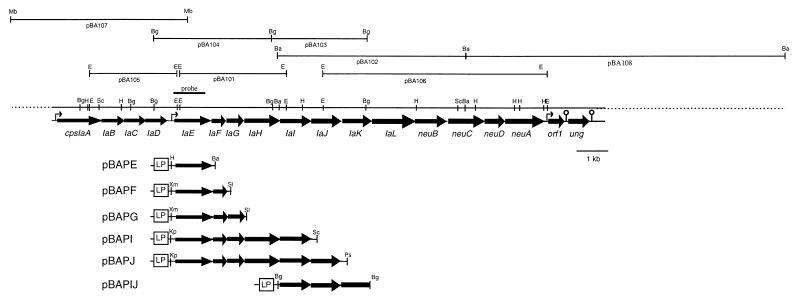
For construction of expression plasmids, various cps gene DNA fragments were directly amplified with chromosomal DNA from S. agalactiae type Ia by using PE-FWD1 (5′-CCCAAGCTTGTGGCTATCTTGAAGAGT-3′ [the HindIII site is underlined]), PE-FWD2 (5′-TCCCCGGGTGGCTATCTTGAAGAGT-3′ [the XmaI site is underlined]), or PE-FWD3 (5′-GGGGTACCGTGGCTATCTTGAAGAGT-3′ [the KpnI site is underlined]) as specific primers for the 5′ end of cpsIaE. PE-REV (5′-CGGGATCCTCCTTTCAAACCTTACCT-3′ [the BamHI site is underlined]), PF-REV (5′-ACGCGTCGACTGACAATTCTGAGGTTC-3′ [the SalI site is underlined]), PG-REV (5′-ACGCGTCGACAACGAGTTAAAAGCTGC-3′ [the SalI site is underlined]), PI-REV (5′-GGGGGGAGCTCATAGGTACAATTACACTT-3′ [the SacI site is underlined]), and PJ-REV (5′-AGGCTGCAGAACAATTCGTGGTCACTC-3′ [the PstI site is underlined]) were used as specific primers for the 3′ ends of the respective cps genes. The PCR products were digested with an appropriate restriction endonuclease to cleave within each primer sequence and ligated to pBluescript II SK (+) or KS (+). The expression plasmids containing cpsIaE alone and cpsIaE to IaF, to IaG, to IaI, and to IaJ were designated pBAPE, pBAPF, pBAPG, pBAPI, and pBAPJ, respectively (Fig. 2). These plasmids were sequenced to check whether any mutations were introduced. The plasmid containing cpsIaI and IaJ was constructed by ligating the DNA fragment of pBA103 into pBluescript II SK (+) in the correct orientation with respect to the lac promoter and designated pBAPIJ. E. coli JM109 was transformed with these plasmids. The cps genes in these plasmids were under the control of the lac promoter of pBluescript. Membranes of the recombinant E. coli cells were isolated 2 h after induction with 1 mM IPTG for analysis of sugar intermediates.
FIG. 2.
Restriction map of the cps locus of S. agalactiae type Ia. The locations of ORFs and the direction of transcription are shown by the arrows. Gene designations are indicated below the arrows. The sites of putative promoters ( ) and terminators ( ) are marked. The DNA probe used for colony hybridization is shown above the restriction map. An overview of the expression plasmids used for the glycosyltransferase assays is indicated below the restriction map. LP is the lac promoter of pBluescript. Abbreviations for restriction sites are as follows: Ba, BamHI; Bg, BglII; E, EcoRI; H, HindIII; Kp, KpnI; Mb, MboI; Ps, PstI; Sc, SacI; Sl, SalI; Xm, XmaI.
Glycosyltransferase assays with 14C-labeled UDP-monosaccharides.
Sugar intermediates formed by the recombinant E. coli were analyzed to assess the enzyme activities of several cps gene products. Preparation of the E. coli membrane fractions and glycosyltransferase activity assays were performed essentially as described by Kolkman et al. (26, 27). E. coli containing the plasmid pBAPIJ was used as a negative control. For competition analyses, unlabeled UDP-glucose, UDP-galactose, and UDP-N-acetylglucosamine were added at a final concentration of 500 μM.
Isolation of RNA, Northern hybridization, reverse transcription (RT)-PCR, and primer extension.
Total cellular RNA was prepared from 50-ml cultures of exponentially-growing S. agalactiae type Ia cells by using an RNeasy Midi kit (QIAGEN). All RNA isolation steps were performed according to the manufacturer’s instructions, except that 50 U of mutanolysin (Sigma)/ml was used to degrade the cell walls in addition to lysozyme and incubation time was extended to 1 h. The isolated RNA was treated with RNase-free DNase I (Sigma) at 25°C for 2 h.
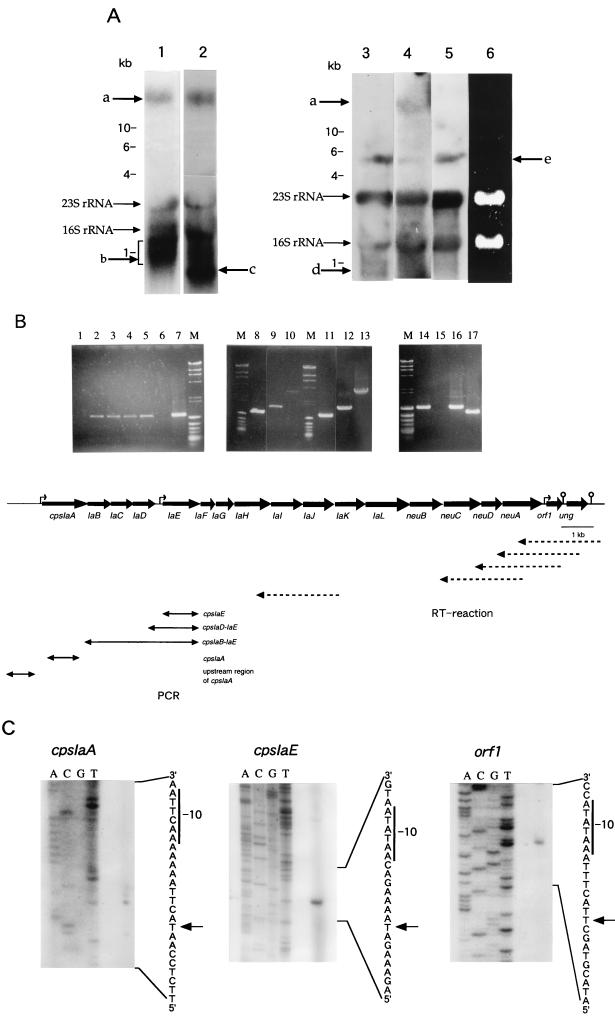
For RT-PCR, reverse transcription was performed with primers derived from the downstream flanking region of cpsIaJ (5′-CTACAAGCTCCATCACTTCTTCA-3′), the internal region of neuA (5′-TTTTTCCCTAATGGCATAATCG-3′), the 3′-end region of orf1 (5′-GAGCCAAATCAGATAAGGACACTG-3′), the internal region of ung (5′-TGACAGCATCAGTAAAAGGTTCCC-3′ and the downstream region of ung (5′-CGCTGGGGTTTTGCTAGGATT-3′) by using ReverTra Ace (Toyobo) (Fig. 3B). PE-FWD1 and PE-REV primers were used for amplification of cpsIaE, PD-FWD (5′-TGATGGTCGTTCCTT-3′) and PE-REV primers were used for the 3′ region of cpsIaD and cpsIaE, and PB-FWD (5′-TCTAGCTTATCTAATGCAAAAT-3′) and PE-REV primers were used for cpsIaB to IaE. For cpsIaA, PA-FWD (5′-GGCATTTAGACACCTGAACG-3′) and PA-REV (5′-GTTTGAACGGATGTTTGGAGCTGTG-3′) were used. For the upstream region of cpsIaA, primers designed according to the partially sequenced upstream region (PUA-FWD, 5′-ACAATCTCAGGACTGTTTA-3′; PUA-REV, 5′-TGGTAGCATGAATGAAGCCGC-3′) were used (Fig. 3B). As controls, each locus was amplified with the same primers by using chromosomal DNA of S. agalactiae type Ia as the template. As negative controls, reaction product without reverse transcriptase was used as the template.
FIG. 3.
Analysis of transcription of the S. agalactiae type Ia cps gene cluster. (A) Northern blotting analysis of total RNA of S. agalactiae type Ia strain OI1. RNA was hybridized with cpsIaA (lane 1), orf1 (lane 2), cpsIaA upstream (lane 3), ung (lane 4), and ung downstream (lane 5) probes. In lane 6, total RNA was electrophoresed on a 1% agarose gel containing 2.2 M formaldehyde and detected by ethidium bromide staining. With cpsIaA, orf1 and ung probes, a long transcript which seemed to correspond to the cpsIa gene cluster was observed (band a). Such a band was not observed with cpsIaA upstream and ung downstream probes. The smeared band in lane 1 (band b) may be degradation products. Band c in lane 2 seemed to correspond to a transcript from the orf1 gene. Band d in lane 3 seemed to be produced from the upstream region of cpsIaA. Unknown bands (e) were observed with several different probes. The sizes of RNA standards (Takara) are indicated. (B) RT-PCR of cps genes. Amplification of the cpsIaE locus was performed with RT reaction products produced by using the ung 3′-downstream primer (lane 1), the ung internal primer (lane 2), the orf1 3′-end primer (lane 3), the neuA internal primer (lane 4), and the cpsIaJ 3′-downstream primer (lane 5). Amplification was also performed with a sample that was not reverse transcribed as a negative control (lane 6). cpsIaE (lane 8), cpsIaD to IaE (lane 9), cpsIaB to IaE (lane 10), cpsIaA (lane 14), and the upstream region of cpsIaA (lane 15) were amplified from the RT reaction product produced with the cpsIaJ 3′-downstream primer. These loci were also amplified by using chromosomal DNA (1 μg) of S. agalactiae type Ia as the template (lanes 7 and 11, cpsIaE; lane 12 cpsIaD to IaE; lane 13, cpsIaB to IaE; lane 16, cpsIaA; lane 17, upstream region of cpsIaA) as controls. The direction and start sites of the RT reaction are indicated by broken arrows. PCR-amplified regions are shown by solid arrows. (C) Primer extension analyses of the transcriptional initiation sites in the sequences upstream of the cpsIaA, cpsIaE, and orf1 genes. The arrows indicate the transcriptional start sites, and the −10 consensus sequences are also shown.
Northern blotting analyses were carried out by using formaldehyde-1% agarose gels as reported previously (39). DNA probes were amplified by using PA-FWD and PA-REV primers for cpsIaA, PORF1-FWD (5′-CCGGGGATCCTCAGAGAAACCAGAAACA-3′) and PORF1-REV (5′-GCGGCTCGAGTTAATCTTCGTCCTTAAG-3′) for orf1, PU-FWD (5′-CCAGTGTCCTTATCTGATTTGGCTC-3′) and PU-REV (5′-TGACAGCATCAGTAAAAGGTTCCC-3′) for ung, and PUA-FWD and PUA-REV for upstream of cpsIaA. Primers PDU-FWD (5′-GTTTAAGCCAAACGGAACCAA-3′) and PDU-REV (5′-AGTGGTAATACTGGCACCA-3′) were synthesized for amplification of the downstream region of ung. These probes were labeled with [α-32P]dCTP (>3,000 Ci/mmol) (Amersham).
For primer extension, the oligodeoxyribonucleotide primers (5′-TCACCCGTAGAGGTGTATG-3′, 5′-GGAAAGTCGTGTCGTTG-3′, and 5′-AATTTCATCACGAAACAAGG-3′) were designed based on the sequences downstream of the cpsIaA, cpsIaE, and orf1 initiation codons, respectively. These oligonucleotide primers (50 pmol) were 5′-end labeled with [γ-32P]ATP (>3,000 Ci/mmol) (Amersham) by using a Megalabel kit (Takara). Then, 14 μl of total RNA (20 μg) was mixed with 4 μl of labeled oligonucleotide primers (2 × 104 cpm) and 4 μl of 5× reaction buffer for reverse transcriptase (ReverTra Ace; Toyobo). The mixture was denatured for 2 min at 80°C, and then 10 μl of deoxynucleoside triphosphate mixture (2.5 mM each), 3 μl of 5× reaction buffer, and 1.5 μl of reverse transcriptase (150 U) were added, and the reaction mixture was incubated for 1 h at 42°C. The final products were denatured for 3 min at 95°C and loaded onto a 6% polyacrylamide–8 M urea sequencing gel. Sequencing was performed with the same oligonucleotide primer by using a BcaBEST dideoxy sequencing kit (Takara) and [α-32P]dCTP (>3,000 Ci/mmol) (Amersham). In the sequencing reaction, pBA104, pBA107, and pBA108 were used as templates for the promoter regions upstream of cpsIaE, cpsIaA, and orf1, respectively.
Nucleotide sequence accession number.
The sequence reported here was submitted to the GenBank database through DDBJ with accession no. AB028896.
RESULTS AND DISCUSSION
Cloning of the cps locus from S. agalactiae type Ia.
The DNA sequence of a part of the cps locus (cpsA to -D) of S. agalactiae type III has been reported previously (38). To obtain DNA for use as a probe for cps genes of S. agalactiae type Ia strain OI1, PCR was performed with chromosomal DNA of the strain by using primers cpsDIII-FWD (5′-GGGGGATCCAATGGTATTGAAATACAG-3′) and cpsDIII-REV (5′-AATCTGCAGACTTAGCTCCTGTCCCGAGT-3′), which were designed according to the previously reported DNA sequence of the cpsD gene of S. agalactiae type III. To clone cps genes, an EcoRI genomic DNA library of S. agalactiae type Ia was constructed with pBluescript II SK (+) as a vector, and this library was screened by colony hybridization with the PCR product. The location of this probe is shown in Fig. 2. One positive clone containing a 3.5-kb EcoRI fragment was selected and designated pBA101. The gene corresponding to cpsD of S. agalactiae type III in this clone was designated cpsIaE to avoid confusion, because the overall structure of the cps gene cluster has been studied in detail in S. pneumoniae and the corresponding genes of pneumococcal bacteria are named cpsE (e.g., cps14E of S. pneumoniae serotype 14 and cps19fE of S. pneumoniae serotype 19F). Sequence analysis of the plasmid pBA101 revealed that the 3.5-kb EcoRI fragment contained almost all of cpsIaE and three complete open reading frames (ORFs) immediately downstream of cpsIaE, designated cpsIaF to cpsIaH, but did not contain the entire type Ia cps locus. Therefore, BamHI, BglII, and MboI genomic DNA libraries of S. agalactiae type Ia were constructed with pBluescript II SK (+) to clone both upstream and downstream regions of the 3.5-kb EcoRI fragment. From these libraries, 6.1-kb BamHI, 3.1-kb BglII, and 3.9-kb BglII fragments were cloned in pBluescript, with the DNA insert of pBA101 used as a probe. This yielded plasmids pBA102, pBA103, and pBA104, respectively (Fig. 2). EcoRI DNA fragments of 2.8 and 7.3 kb were also cloned by using pBA104 and pBA103 as probes and designated pBA105 and pBA106, respectively (Fig. 2). Furthermore, the 5.7-kb MboI fragment designated pBA107 and the 10.9-kb BamHI fragment designated pBA108 were cloned by using pBA105 and pBA106 as probes, respectively. By gene walking experiments, a DNA region of 26 kb containing the cps locus was obtained.
DNA sequence analysis.
The DNA sequence of 17,826 nucleotides was completely determined on both strands with overlapping clones covering the cps gene locus. Sequence analysis showed 18 complete ORFs, designated cpsIaA to cpsIaL, neuB, neuC, neuD, and neuA, orf1, and ung. All ORFs were in the same orientation and were spaced one behind the other at short distances, except for the region of 227 bp between cpsIaD and cpsIaE (Fig. 2). Possible −35 (TTGTTT) and −10 (TATATT) sequences were identified in this gap region. Two other potential −35 (ATGATA and TTGCGA) and −10 (TAAGTT and TATATT) sequences were identified upstream of cpsIaA and orf1, respectively. Two putative Rho-independent transcription terminator sequences were found downstream of orf1 (ΔG = −24.5 kcal/mol) and ung (ΔG = −32.5 kcal/mol) (Fig. 2). These observations suggested that at least 17 ORFs from cpsIaA to orf1 may constitute one polycistronic operon and that transcription may start from cpsIaA, cpsIaE, and orf1.
A possible Shine-Dalgarno sequence was identified just upstream of the potential initiation codon of each ORF. All ORFs were preceded by ATG codons. The usage of terminator codons was in agreement with the usual E. coli preference. TAA was used 13 times, TAG was used three times, and TGA was used twice. The average G+C content of the sequenced area was 31.7%. The percent G+C content of the cps cluster agreed well with that of chromosomal DNA of S. agalactiae (34.0%) (37).
The amino acid sequence of each ORF was deduced, and an overview of all cpsIa genes, with their properties and translation products, is shown in Table 1. The cpsIaA gene product was identical to CpsX of S. agalactiae type III (28) and also showed a high degree of similarity to Cps14A (50.2% identity) and Cps19fA (50.6% identity) of S. pneumoniae serotype 14 (27) and 19F (16). Furthermore, CpsIaA exhibited similarity to LytR (32.9% identity) of Bacillus subtilis (29), as was the case with Cps14A. The pneumococcal CpsA proteins are thought to be involved in the regulation of CP expression, and the LytR protein is a transcription attenuator for the autolysin (lytABC) operon (29). Therefore, the CpsIaA protein may also have a specific role in the transcriptional regulation of cpsIa genes of S. agalactiae. The hydropathy profile of CpsIaA demonstrated the presence of three N-terminal hydrophobic segments as reported with the corresponding gene of S. agalactiae type III (28). This suggested that the putative regulatory protein may bind to the cell membrane, similarly to LytR. However, the mechanism by which these membrane proteins regulate transcription is still unclear.
TABLE 1.
Properties of the ORFs in the cps locus of S. agalactiae type Ia and homologies with gene products of other bacteria
| ORF | Nucleotide position in sequence | No. of amino acids | Proposed function of gene product | Similar gene product(s) (% identity) | Reference |
|---|---|---|---|---|---|
| cpsIaA | 151–1605 | 485 | Regulationb | Streptococcus agalactiae type III CpsXd (100%) | 28 |
| Streptococcus pneumoniae serotype 14 Cps14Ad (50.2%) | 27 | ||||
| Streptococcus pneumoniae serotype 19 Cps19fAd (50.6%) | 16 | ||||
| Bacillus subtilisLytRd (32.9%) | 29 | ||||
| cpsIaB | 1614–2342 | 243 | Unknown | Streptococcus agalactiae type III CpsAd (99.2%) | 38 |
| Streptococcus pneumoniae serotype 14 Cps14Bd (62.6%) | 26 | ||||
| Streptococcus pneumoniae serotype 19 Cps19fBd (64.2%) | 16 | ||||
| cpsIaC | 2354–3043 | 230 | Chain length regulatorb | Streptococcus agalactiae type III CpsBd (99.1%) | 38 |
| Streptococcus pneumoniae serotype 14 Cps14Cd (44.2%) | 26 | ||||
| Streptococcus pneumoniae serotype 19 Cps19fCd (45.6%) | 16 | ||||
| Rhizobium melilotiExoPde (22.5%) | 15 | ||||
| cpsIaD | 3057–3743 | 229 | Chain length regulator or exportb | Streptococcus agalactiaetype III CpsCd (99.1%) | 38 |
| Streptococcus pneumoniae serotype 14 Cps14Dd (55.9%) | 26 | ||||
| Streptococcus pneumoniae serotype 19 Cps19fDd (56.8%) | 16 | ||||
| Rhizobium melilotiExoPdf (29.6%) | 15 | ||||
| cpsIaE | 3973–5145 | 391 | Glucosyltransferasea | Streptococcus agalactiae type III CpsDd (98.9%) | 38 |
| Streptococcus pneumoniae serotype 14 Cps14Ec (49.1%) | 25 | ||||
| Streptococcus pneumoniae serotype 19 Cps19fEd (49.1%) | 16 | ||||
| Salmonella entericaRfbPd (32.1%) | 47 | ||||
| cpsIaF | 5172–5618 | 149 | Unknown | Streptococcus pneumoniae serotype 14 Cps14F (83.9%) | 26 |
| Sphingomonas S88 SpsKde (33.1%) | 50 | ||||
| cpsIaG | 5621–6160 | 180 | β-1,4-galactosyltransferasea | Streptococcus pneumoniae serotype 14 Cps14Gc (53.2%) | 26 |
| Sphingomonas S88 SpsKdf (23.6%) | 50 | ||||
| cpsIaH | 6202–7338 | 379 | Putative CP polymeraseb | Streptococcus pneumoniaeserotype 14 Cps14Hd (23.6%) | 26 |
| Salmonella typhimuriumRfcd (19.5%) | 9 | ||||
| Shigella flexneriRfcd (16.1%) | 32 | ||||
| cpsIaI | 7341–8339 | 333 | β-1,3-N-acetylglucosaminyltransferasea | Streptococcus thermophilus Sfi6 EpsId (34.2%) | 42 |
| Streptococcus pneumoniae serotype 14 Cps14Ic (26.4%) | 27 | ||||
| Rhizobium melilotiExoWd (25.5%) | 14 | ||||
| Rhizobium melilotiExoUd (23.7%) | 14 | ||||
| Neisseria meningitidisLgtAd (20.7%) | 21 | ||||
| cpsIaJ | 8347–9291 | 315 | β-1,4-galactosyltransferasea | Streptococcus pneumoniae serotype 14 Cps14Jc (37.0%) | 27 |
| Streptococcus thermophilus Sfi6 Eps Id (30.3%) | 42 | ||||
| Neisseria meningitidisLgtAd (26.5%) | 21 | ||||
| Rhizobium melilotiExoOd (26.5%) | 14 | ||||
| Rhizobium melilotiExoUd (26.7%) | 14 | ||||
| cpsIaK | 9378–10331 | 318 | Unknown | Haemophilus influenzae type b Orf4f (20.8%) | 44 |
| cpsIaL | 10331–11728 | 466 | Repeat unit transporterb | Shigella dysenteriaeRfbXd (19.8%) | 23 |
| Staphylococcus aureusCapFd (22.2%) | 30 | ||||
| Streptococcus pneumoniae serotype 14 Cps14Ld (16.4%) | 27 | ||||
| neuB | 11731–12753 | 341 | Sialic acid synthesisb | Escherichia coliNeuBd (56.0%) | 1 |
| Neisseria meningitidis group B NeuBd (35.7%) | 12 | ||||
| neuC | 12833–13984 | 384 | Sialic acid synthesisb | Escherichia coliNeuCd (43.6%) | 51 |
| Neisseria meningitidis group B NeuCd (28.5%) | 12 | ||||
| neuD | 13984–14610 | 209 | Acetyltransferaseb | Escherichia coliNeuDd (33.0%) | 1 |
| neuA | 14624–15862 | 413 | CMP-sialic acid synthetaseb | Escherichia coliNeuAc (33.0%) | 52 |
| Streptococcus agalactiae type III CpsFc (99.2%) | 17 | ||||
| orf1 | 15979–16461 | 161 | Unknown | Escherichia coliEvgSde (28.2%) | 43 |
| ung | 16563–17213 | 217 | Uracil DNA glycosylaseb | Streptococcus pneumoniaeUngd (75.8%) | 31 |
| Bacillus subtilisUngd (52.6%) | 13 |
Experimentally determined in this study.
Predicted by sequence similarity.
Enzymatic activities have been determined.
Enzymatic activity was proposed.
Homology refers to the N-terminal part of the gene product.
Homology refers to the C-terminal part of the gene product.
The cpsIaB to cpsIaD gene products were almost identical to CpsA to CpsC of S. agalactiae type III, respectively. These products also showed high degrees of similarity to the corresponding gene products of S. pneumoniae serotypes 14 and 19F (Table 1). Among these previously reported gene products, those showing homology to CpsIaC and CpsIaD have been suggested to play important roles in the determination of chain length and the export of CPs. The ExoP protein of Rhizobium meliloti is composed of two domains, and CpsC and CpsD proteins of streptococcal bacteria were similar to its N-terminal and C-terminal domains, respectively. The N-terminal domain of ExoP is expected to have a role in determining chain length, based on its homology to other bacterial proteins (5), and the C-terminal domain is supposed to have a regulatory function (5). As CpsIaC and CpsIaD also showed similarity to the respective domains of ExoP protein, as suggested with Cps14C and Cps14D, these proteins may function in determining chain length and in exporting type Ia CP.
The cpsIaE gene product showed significant identity (98.9%) to type III CpsD (38). Furthermore, the gene product showed close similarity to Cps14E (49.1% identity) of S. pneumoniae serotype 14 (25). CpsD of S. agalactiae type III was suggested to be a galactosyltransferase based on the result of mutational analysis (38), and Cps14E of S. pneumoniae serotype 14 appeared to be a glucosyltransferase (25). Polypeptides that show homology to CpsIaE of S. agalactiae type Ia are known to catalyze linkage of the first sugar to the lipid carrier (Table 1), suggesting that the gene product may transfer glucose and/or galactose to the lipid carrier.
The gene products encoded by cpsIaF and cpsIaG were homologous to the proteins Cps14F (83.9% identity) and Cps14G (53.2% identity) of S. pneumoniae serotype 14, respectively (Table 1). Since Cps14G protein is a β-1,4-galactosyltransferase (26), CpsIaG of S. agalactiae type Ia was expected to have the same enzyme activity. CpsIaF of S. agalactiae type Ia contained a hydrophobic region in the center of the molecule, as was also reported for Cps14F of S. pneumoniae serotype 14 (26). This hydrophobic region of Cps14F is thought to be anchored in the membrane as reported with SpsK of Sphingomonas strain S88 (26).
S. agalactiae type Ia CpsIaH showed a degree of homology (23.6% identity) to Cps14H of S. pneumoniae serotype 14. Although these two proteins did not show close homology over the entire region, the 12 membrane-spanning domains observed in Cps14H were also detected in S. agalactiae type Ia CpsIaH (data not shown). In S. pneumoniae serotype 14, Cps14H is supposed to be a CP polymerase, based on its homology to O antigen polymerase (Rfc) of Shigella flexneri (32) and Salmonella typhimurium (9). Since CpsIaH of S. agalactiae type Ia showed homology to the Rfc proteins, CpsIaH may also be a CP polymerase.
The gene products encoded by cpsIaI and cpsIaJ showed similarity to several putative glycosyltransferases involved in the biosynthesis of CPs, lipopolysaccharides, and exopolysaccharides in numerous bacterial species (Table 1). Especially, CpsIaI and CpsIaJ of S. agalactiae type Ia showed moderate similarity to Cps14I and Cps14J of S. pneumoniae serotype 14, respectively. In S. pneumoniae serotype 14, Cps14I and Cps14J have β-1,3-N-acetylglucosaminyltransferase and β-1,4-galactosyltransferase activity, respectively (27). This suggested that cpsIaI and cpsIaJ may encode an N-acetylglucosaminyltransferase and a galactosyltransferase required for addition of the third and the fourth sugar residues in the oligosaccharide side chain of type Ia CP, respectively.
DXD, DXS, and ED sequences have been found in many glycosyltransferases involved in biosynthesis of CPs, lipopolysaccharides, and exopolysaccharides (22, 27). It was previously reported that α-glycosyltransferases contain two sets of DXD which are essential for enzyme activity (40, 41). On the other hand, β-glycosyltransferases contain one set of DXD, DXS and, sometimes, ED sequences aligned at suitable distances (22, 27). All these sequences were found in CpsIaI and CpsIaJ, but only DXD and DXS were found in CpsIaE. The lack of ED in CpsIaE was in accordance with the previous observation by Keenleyside and Whitfield that glycosyltransferases to lipid carriers often do not contain this sequence (22). Neither of these conserved sequences was found in the CpsIaG galactosyltransferase of S. agalactiae type Ia. CpsIaG may belong to a different glycosyltransferase family than CpsIaE, CpsIaI, and CpsIaJ.
The cpsIaL gene of S. agalactiae type Ia encoded a protein which showed similarities to RfbX of Shigella dysenteriae and CapF of Staphylococcus aureus (Table 1). Furthermore, the hydropathy profile of CpsIaL was similar to those of RfbX-related proteins (data not shown). The RfbX of S. dysenteriae is thought to be involved in one of the later CP synthesis steps, such as transfer of the repeating unit to the cell surface (23). Thus, CpsIaL may be involved in a later stage of CP synthesis.
It is noteworthy that the levels of homology between CpsIaA to CpsIaG of S. agalactiae type Ia and corresponding gene products of S. pneumoniae serotype 14 were high (44 to 85%). However, their downstream gene products (CpsIaH to CpsIaL) showed less homology (15 to 40%) to those of S. pneumoniae serotype 14.
In addition to these ORFs, six ORFs were identified downstream of cpsIaL (Fig. 2). Among these, the first four ORFs were found to be related to sialic acid synthesis. These ORFs were designated neuB, neuC, neuD, and neuA, based on their similarity to gene products required for polysialic acid synthesis in E. coli K1. In E. coli K1, neuB and neuC gene products are involved in sialic acid synthesis (1, 51), and it is likely that the corresponding gene products of S. agalactiae type Ia have the same function. NeuD of S. agalactiae type Ia showed similarity (33.0% identity) to NeuD of E. coli K1. The NeuD protein of E. coli showed homology to several bacterial acetyltransferases, but its target is unknown. The neuA gene product of S. agalactiae type Ia was almost identical to previously reported CpsF (99.2%) of S. agalactiae type III (17) and showed homology (33.0%) to NeuA of E. coli K1 (52). These well-studied proteins have been confirmed to be CMP-sialic acid synthetases.
Additional ORFs were identified downstream of neuA and designated orf1 and ung. ORF1 showed similarity (28.2% identity) to the N-terminal cytoplasmic domain of EvgS, which seems to be a sensor protein of a two-component regulatory system responding to environmental stimuli (43). However, the function and involvement of the ORF1 protein in CP synthesis are not yet clear. An ORF showing high similarity to ung gene products of S. pneumoniae (31) and Bacillus subtilis (13) was found downstream of the orf1 gene. The Ung proteins function in DNA mismatch repair. The role of the gene in the cpsIa cluster is also still obscure.
Functional analysis of streptococcal glycosyltransferase expressed in E. coli.
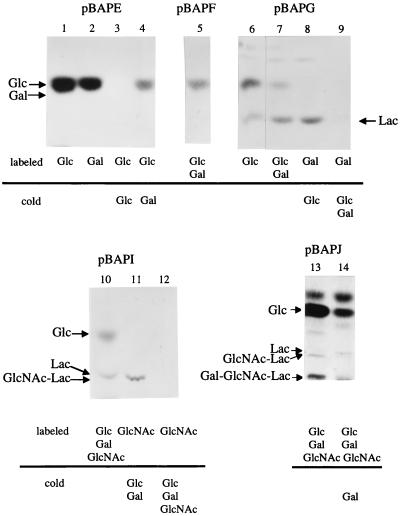
Repeating units of bacterial CPs are known to be synthesized on lipid carriers on the cell surface (6, 7). Since CpsIaE, CpsIaG, CpsIaI, and CpsIaJ showed homology to several glycosyltransferases of S. pneumoniae serotype 14, the functions of these molecules were examined by analysis of the intermediates in synthesis of the oligosaccharide subunit formed by membrane fractions of E. coli harboring expression plasmids of these cps genes. Membrane fractions were used as sources of enzymes and acceptors, and oligosaccharide intermediates added to lipid carriers were extracted in lipid fractions as described by Kolkman et al. After the release of lipid carriers by treatment with trifluoroacetic acid, these intermediates were analyzed by thin-layer chromatography (TLC) (26).
Membranes of the E. coli clone carrying the plasmid pBAPE showed incorporation of [14C]glucose and [14C]galactose (Table 2). However, [14C]glucose was the only labeled sugar detected by TLC analysis (Fig. 4, lanes 1 and 2). Furthermore, the incorporation of [14C]glucose or [14C]galactose into the lipid carrier was almost completely inhibited by excess cold glucose, but galactose showed only partial inhibition (Fig. 4, lanes 3 and 4, and Table 2). These results suggested that CpsIaE had glucosyltransferase activity but not galactosyltransferase activity. Added UDP-[14C]galactose was probably converted to UDP-[14C]glucose by an intrinsic UDP-galactose-4-epimerase of E. coli as suggested with S. pneumoniae serotype 14 (26).
TABLE 2.
Incorporation of [14C]glucose and [14C]galactose into the glycolipid fraction of membranes of E. coli clones expressing streptococcal cps genes
| E. coli JM109 + clone | Amt of incorporated
radioactivity (cpm/μg of protein)
|
|||||
|---|---|---|---|---|---|---|
| [14C]Glc | [14C]Gal | [14C]Glc + [14C]Gal | Glc + [14C]Gal | [14C]Glc + Glc | [14C]Glc + Gal | |
| pBAPE | 105 | 100 | 135 | 1 | 10 | 45 |
| pBAPG | 30 | 29 | 47 | 21 | ||
| pBAPIJ | 8 | 5 | 7 | 1 | ||
FIG. 4.
Thin-layer chromatogram of 14C-labeled sugar intermediates of CP synthesis with isolated membranes of various E. coli strains expressing type Ia cps genes (pBAPE, pBAPF, pBAPG, pBAPI, and pBAPJ). TLC plates were developed twice for the clone pBAPJ. Added UDP-monosaccharides are shown below the chromatograms. Lac, lactose.
Membranes of the E. coli clone carrying the plasmid pBAPG showed incorporation of radioactivity upon incubation with UDP-[14C]glucose and/or UDP-[14C]galactose (Table 2). TLC analyses indicated synthesis of lactose and glucose intermediates (Fig. 4, lanes 6 and 7). Only lactose was detected when UDP-[14C]galactose and cold glucose were added (Fig. 4, lane 8). Furthermore, cold galactose inhibited lactose intermediate formation (Fig. 4, lane 9). On the other hand, lactose was not detected with pBAPF (Fig. 4, lane 5). Taken together, these results showed that cpsIaG encoded a galactosyltransferase which catalyzed the transfer of galactose as the second monosaccharide. Lactose synthesis was detected when only UDP-[14C]glucose was added. This seemed to be due to the conversion of UDP-glucose to UDP-galactose by the epimerase mentioned above. The E. coli clone carrying pBAPIJ, which lacked cpsIaE to cpsIaH, did not show incorporation of radioactivity (Table 2).
The E. coli clone carrying pBAPI showed incorporation of radioactivity into the lipid carriers by a reaction with 14C-labeled UDP-N-acetylglucosamine, UDP-glucose, and UDP-galactose (data not shown). Following incubation with UDP-N-[14C]acetylglucosamine, cold UDP-glucose, and UDP-galactose, GlcpNAc-Lac trisaccharide was detected by TLC (Fig. 4, lane 11). A band with a similar Rf was detected in addition to glucose when all three 14C-labeled UDP-monosaccharides were added (Fig. 4, lane 10). This band may have been a mixture of lactose and GlcpNAc-Lac, since these di- and trisaccharides were very close on TLC under these conditions. Excess cold N-acetylglucosamine inhibited the trisaccharide intermediate formation (Fig. 4, lane 12). These results indicated that CpsIaI had N-acetylglucosaminyltransferase activity.
The membrane fraction of the E. coli clone carrying pBAPJ included an activity, which produced the tetrasaccharide Galp-GlcpNAc-Lac (Fig. 4, lane 13). Since the clone carrying pBAPI did not show tetrasaccharide-forming activity, and galactose inhibited tetrasaccharide formation (Fig. 4, lane 14), cpsIaJ seemed to encode a galactosyltransferase, which catalyzed the transfer of galactose as the fourth monosaccharide.
Because the intermediate products were present at low levels, their precise chemical structures could not be determined by these glycosyltransferase assays. However, the precise structure of type Ia CP has been determined directly (19), and the unit structure is identical to that of CP of S. pneumoniae serotype 14, except for the terminal sialic acid. As judged from these observations and the results of homology analysis and glycosyltransferase assays, it is likely that CpsIaE, CpsIaG, CpsIaI, and CpsIaJ are glucosyltransferase, β-1,4-galactosyltransferase, β-1,3-N-acetylglucosaminyltransferase, and β-1,4-galactosyltransferase, respectively.
Transcription of cps genes.
Analysis of the DNA sequence suggested that the cps genes, cpsIaA to orf1 or to ung may be organized as a single operon. To confirm this, RNA was isolated from S. agalactiae type Ia and hybridized with DNA probes from various regions of the cps gene cluster. As shown in Fig. 3A, long transcripts (about 15 kb) were observed with cpsIaA (lane 1), orf1 (lane 2), and ung (lane 4) probes but not with cpsIaA upstream or ung downstream probes (lanes 3 and 5), which showed that 18 genes (cpsIaA to ung) constitute a single transcription unit. The orf1 probe also hybridized to a short transcript (0.5 kb) (Fig. 3A, lane 2), indicating that the promoter upstream of orf1 may be functional. The size of this short transcript was consistent with that of the orf1 gene (0.5 kb), suggesting that the terminator signal downstream of orf1 might be functional. Then, RT-PCR was performed to characterize the transcripts expressed from the cps gene cluster in detail. For RT reaction, five primers for the cpsIaJ, neuA, orf1, and ung loci and the downstream region of ung were used (Fig. 3B). The cpsIaE gene (1.2 kb) was almost equally amplified from all RT reaction products except for that downstream of ung (Fig. 3B, lanes 1 to 5), suggesting that termination of transcription occurred downstream of ung. Taken together, these results suggest that 18 genes (cpsIaA to ung) constitute a single transcription unit.
To confirm the 5′ end of the cps locus, the following two RT-PCR experiments were performed. From the chromosomal DNA of S. agalactiae type Ia, cpsIaE (lanes 7 and 11), cpsIaD to cpsIaE (1.7 kb, lane 12), and cpsIaB to cpsIaE (3.6 kb, lane 13) loci were amplified as shown in Fig. 3B. These were used as controls for the size effect of band density on gels. The same loci were then amplified from the RT product obtained by using the primer designed from 3′-flanking region of cpsIaJ. The extent of the amplification of cpsIaD to cpsIaE and that of cpsIaB to cpsIaE was less than that of cpsIaE (Fig. 3B, lanes 8 to 10), indicating that RT partially stopped between the 3′ end of cpsIaD and the 5′ end of cpsIaE. This result suggested that transcription of the cps genes started from two sites, upstream of cpsIaA and upstream of cpsIaE, consistent with the presence of possible promoter sequences upstream of cpsIaA and of cpsIaE. The cpsIaA gene and the upstream region of cpsIaA were also amplified from the RT product prepared by using the 3′-flanking region of cpsIaJ. As shown in Fig. 3B, cpsIaA was amplified, but its upstream region was not (lanes 14 and 15). The cpsIaA gene was also amplified from the RT product prepared with the 3′ end of ung (data not shown). These results indicated that the 5′ end of the transcript is upstream of cpsIaA. No potential termination site was found in the gap region between cpsIaD and cpsIaE of S. agalactiae type Ia. This suggested that a transcript corresponding to cpsIaA to cpsIaD may not be formed and that cpsIaA to IaD may be expressed as a long transcript covering the entire cps locus.
Based on the results of Northern hybridization and RT-PCR, we determined the transcriptional start sites located just upstream of cpsIaA, cpsIaE, and orf1 by primer extension. The results shown in Fig. 3C indicated that the start sites of transcription resided 14, 56, and 46 nucleotides upstream of the cpsIaA, cpsIaE, and orf1 start codons, respectively (Fig. 3C). Typical −10 sequences (TAAGTT, TATATT, and TATATT) were found eight or ten nucleotides upstream of the three transcriptional start sites (Fig. 3C), which corresponded to the putative promoters identified by DNA sequence analysis.
The entire cps gene cluster of S. pneumoniae serotype 14 (cps14A to 14L) was suggested to be expressed as one transcriptional unit, since a putative promoter is located just upstream of cps14A and a potential terminator is located just downstream of cps14L (27). However, in the cps cluster of S. agalactiae type Ia, alternative transcription may occur from at least three promoters. As the internal promoter sequences and the long gap region detected in the cpsIa cluster were not found in the cps14 cluster, this is specific for the cps loci of S. agalactiae type Ia. Furthermore, the genes required for the synthesis of sialic acid were included in the cps polycistronic operon, and the cps cluster of S. agalactiae type Ia was much larger than the S. pneumoniae serotype 14 cps gene cluster. In these respects, the operon structure of cpsIa genes differs from that of cps14 genes.
In this study, the sialyltransferase required for the addition of the fifth monosaccharide was not identified. None of the ORFs reported here showed significant similarity to previously reported sialyltransferases. Furthermore, no ORF encoding a product with sialyltransferase activity was detected within the sequenced DNA region. Screening for a sialyltransferase gene is currently in progress in our laboratory.
ACKNOWLEDGMENTS
We thank Manami Goto for preparing the manuscript and figures.
This work was supported in part by a grant-in-aid from the Ministry of Education, Science and Sports of Japan (10134218).
REFERENCES
- 1.Annunziato P W, Wright L F, Vann W F, Silver R P. Nucleotide sequence and genetic analysis of the neuD and neuB genes in region 2 of the polysialic acid gene cluster of Escherichia coliK1. J Bacteriol. 1995;177:312–319. doi: 10.1128/jb.177.2.312-319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. Genomic organization of the Klebsiella pneumoniae cpsregion responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol. 1995;177:1788–1796. doi: 10.1128/jb.177.7.1788-1796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrecubieta C, García E, López R. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniaetype 3. Gene. 1995;167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 5.Becker A, Niehaus K, Pühler A. Low-molecular-weight succinoglycan is predominantly produced by Rhizobium melilotistrains carrying a mutated ExoP protein characterized by a periplasmic N-terminal domain and a missing C-terminal domain. Mol Microbiol. 1995;16:191–203. doi: 10.1111/j.1365-2958.1995.tb02292.x. [DOI] [PubMed] [Google Scholar]
- 6.Boulnois G J, Jann K. Bacterial polysaccharide capsule synthesis, export and evolution of structural diversity. Mol Microbiol. 1989;3:1819–1823. doi: 10.1111/j.1365-2958.1989.tb00168.x. [DOI] [PubMed] [Google Scholar]
- 7.Boulnois G J, Roberts I S. Genetics of capsular polysaccharide production in bacteria. Curr Top Microbiol Immunol. 1990;150:1–18. doi: 10.1007/978-3-642-74694-9_1. [DOI] [PubMed] [Google Scholar]
- 8.Boyer K M, Gadzala C A, Burd L I, Fisher D E, Paton J B, Gotoff S P. Selective intrapartum chemoprophylaxis of neonatal group B streptococcal early-onset disease. I. Epidemiologic rationale. J Infect Dis. 1983;148:795–801. doi: 10.1093/infdis/148.5.795. [DOI] [PubMed] [Google Scholar]
- 9.Collins L V, Hackett J. Molecular cloning, characterization, and nucleotide sequence of the rfc gene, which encodes an O-antigen polymerase of Salmonella typhimurium. J Bacteriol. 1991;173:2521–2529. doi: 10.1128/jb.173.8.2521-2529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross A S. The biologic significance of bacterial encapsulation. Curr Top Microbiol Immunol. 1990;150:87–95. doi: 10.1007/978-3-642-74694-9_5. [DOI] [PubMed] [Google Scholar]
- 11.Di Fabio J L, Michon F, Brisson J-R, Jennings H J. Structure of the capsular polysaccharide antigen of type IV group B Streptococcus. Can J Chem. 1989;67:877–882. [Google Scholar]
- 12.Ganguli S, Zapata G, Wallis T, Reid C, Boulnois G, Vann W F, Roberts I S. Molecular cloning and analysis of genes for sialic acid synthesis in Neisseria meningitidisgroup B and purification of the meningococcal CMP-NeuNAc synthetase enzyme. J Bacteriol. 1994;176:4583–4589. doi: 10.1128/jb.176.15.4583-4589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaser P, Kunst F, Arnaud M, Coudart M P, Gonzales W, Hullo M F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, Presencan E, Santana M, Schneider E, Schweizer J, Vertes A, Rapoport G, Danchin A. Bacillus subtilisgenome project: cloning and sequencing of the 97 kb region from 325 degrees to 333 degrees. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 14.Glucksmann M A, Reuber T L, Walker G C. Family of glycosyl transferases needed for the synthesis of succinoglycan by Rhizobium meliloti. J Bacteriol. 1993;175:7033–7044. doi: 10.1128/jb.175.21.7033-7044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glucksmann M A, Reuber T L, Walker G C. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J Bacteriol. 1993;175:7045–7055. doi: 10.1128/jb.175.21.7045-7055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidolin A, Morona J K, Morona R, Hansman D, Paton J C. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniaetype 19F. Infect Immun. 1994;62:5384–5396. doi: 10.1128/iai.62.12.5384-5396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haft R F, Wessels M R, Mebane M F, Conaty N, Rubens C E. Characterization of cpsF and its product CMP-N-acetylneuraminic acid synthetase, a group B streptococcal enzyme that can function in K1 capsular polysaccharide biosynthesis in Escherichia coli. Mol Microbiol. 1996;19:555–563. doi: 10.1046/j.1365-2958.1996.395931.x. [DOI] [PubMed] [Google Scholar]
- 18.Jennings H J. Capsular polysaccharides as vaccine candidates. Curr Top Microbiol Immunol. 1990;150:97–127. doi: 10.1007/978-3-642-74694-9_6. [DOI] [PubMed] [Google Scholar]
- 19.Jennings H J, Katzenellenbogen E, Lugowski C, Kasper D L. Structure of native polysaccharide antigens of type Ia and type Ib group B Streptococcus. Biochemistry. 1983;22:1258–1264. doi: 10.1021/bi00274a042. [DOI] [PubMed] [Google Scholar]
- 20.Jennings H J, Lugowski C, Kasper D L. Conformational aspects critical to the immunospecificity of the type III group B streptococcal polysaccharide. Biochemistry. 1981;20:4511–4518. doi: 10.1021/bi00519a001. [DOI] [PubMed] [Google Scholar]
- 21.Jennings M P, Hood D W, Peak I R A, Virji M, Moxon E R. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 22.Keenleyside W J, Whitfield C. A novel pathway for O-polysaccharide biosynthesis in Salmonella entericaserovar borreze. J Biol Chem. 1996;271:28581–28592. doi: 10.1074/jbc.271.45.28581. [DOI] [PubMed] [Google Scholar]
- 23.Klena J D, Schnaitman C A. Function of the rfb gene cluster and the rfe gene in the synthesis of O antigen by Shigella dysenteriae1. Mol Microbiol. 1993;9:393–402. doi: 10.1111/j.1365-2958.1993.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 24.Kogan G, Brisson J-R, Kasper D L, von Hunolstein C, Orefici G, Jennings H J. Structural elucidation of the novel type VII group B Streptococcuscapsular polysaccharide by high resolution NMR spectroscopy. Carbohydr Res. 1995;277:1–9. doi: 10.1016/0008-6215(95)00195-y. [DOI] [PubMed] [Google Scholar]
- 25.Kolkman M A B, Morrison D A, van der Zeijst B A M, Nuijten P J M. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolkman M A B, van der Zeijst B A M, Nuijten P J M. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniaeserotype 14. J Biol Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 27.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cpslocus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 28.Koskiniemi S, Sellin M, Norgren M. Identification of two genes, cpsX and cpsY, with putative regulatory function on capsule expression in group B streptococci. FEMS Immunol Med Microbiol. 1998;21:159–168. doi: 10.1111/j.1574-695X.1998.tb01162.x. [DOI] [PubMed] [Google Scholar]
- 29.Lazarevic V, Margot P, Soldo B, Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J Gen Microbiol. 1992;138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- 30.Lin W S, Cunneen T, Lee C Y. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Méjean V, Rives I, Claverys J-P. Nucleotide sequence of the Streptococcus pneumoniae unggene encoding uracil-DNA glycosylase. Nucleic Acids Res. 1990;18:6693. doi: 10.1093/nar/18.22.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniaetype 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 34.Morona J K, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniaetype 19B. J Bacteriol. 1997;179:4953–4958. doi: 10.1128/jb.179.15.4953-4958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moxon E R, Kroll J S. The role of bacterial polysaccharide capsules as virulence factors. Curr Top Microbiol Immunol. 1990;150:65–85. doi: 10.1007/978-3-642-74694-9_4. [DOI] [PubMed] [Google Scholar]
- 36.Ramirez M, Tomasz A. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J Bacteriol. 1998;180:5273–5278. doi: 10.1128/jb.180.19.5273-5278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rotta J. Pyrogenic hemolytic streptococci. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. p. 1051. [Google Scholar]
- 38.Rubens C E, Heggen L M, Haft R F, Wessels M R. Identification of cpsD, a gene essential for type III capsule expression in group B streptococci. Mol Microbiol. 1993;8:843–855. doi: 10.1111/j.1365-2958.1993.tb01631.x. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Saxena I M, Brown R M, Jr, Fevre M, Geremia R A, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibayama K, Ohsuka S, Tanaka T, Arakawa Y, Ohta M. Conserved structural regions involved in the catalytic mechanism of Escherichia coliK-12 WaaO (RfaI) J Bacteriol. 1998;180:5313–5318. doi: 10.1128/jb.180.20.5313-5318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stingele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilussfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Utsumi R, Katayama S, Taniguchi M, Horie T, Ikeda M, Igaki S, Nakagawa H, Miwa A, Tanabe H, Noda M. Newly identified genes involved in the signal transduction of Escherichia coliK-12. Gene. 1994;140:73–77. doi: 10.1016/0378-1119(94)90733-1. [DOI] [PubMed] [Google Scholar]
- 44.Van Eldere J, Brophy L, Loynds B, Celis P, Hancock I, Carman S, Kroll J S, Moxon E R. Region II of the Haemophilus influenzaetype b capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol Microbiol. 1995;15:107–118. doi: 10.1111/j.1365-2958.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 45.von Hunolstein C, Ascenzi S D, Wagner B, Jelínková J, Alfarone G, Recchia S, Waner M, Orefici G. Immunochemistry of capsular type polysaccharide and virulence properties of type VI Streptococcus agalactiae (group B Streptococcus) Infect Immun. 1993;61:1272–1280. doi: 10.1128/iai.61.4.1272-1280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Hunolstein C, Nicolini L, D’Ascenzi S, Volpe C, Alfarone G, Orefici G. Sialic acid and biomass production by Streptococcus agalactiaeunder different growth conditions. Appl Microbiol Biotechnol. 1993;38:458–462. [Google Scholar]
- 47.Wang L, Liu D, Reeves P R. C-terminal half of Salmonella enterica wbaP(rfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J Bacteriol. 1996;178:2598–2604. doi: 10.1128/jb.178.9.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wessels M R, DiFabio J L, Benedi V-J, Kasper D L, Michon F, Brisson J-R, Jelíková J, Jennings H J. Structural determination and immunochemical characterization of the type V group B Streptococcuscapsular polysaccharide. J Biol Chem. 1991;266:6714–6719. [PubMed] [Google Scholar]
- 49.Wessels M R, Pozsgay V, Kasper D L, Jennings H J. Structure and immunochemistry of an oligosaccharide repeating unit of the capsular polysaccharide of type III group B Streptococcus. J Biol Chem. 1987;262:8262–8267. [PubMed] [Google Scholar]
- 50.Yamazaki M, Thorne L, Mikolajczak M, Armentrout R W, Pollock T J. Linkage of genes essential for synthesis of a polysaccharide capsule in Sphingomonasstrain S88. J Bacteriol. 1996;178:2676–2687. doi: 10.1128/jb.178.9.2676-2687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zapata G, Crowley J M, Vann W F. Sequence and expression of the Escherichia coli K1 neuCgene product. J Bacteriol. 1992;174:315–319. doi: 10.1128/jb.174.1.315-319.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zapata G, Vann W F, Aaronson W, Lewis M S, Moos M. Sequence of the cloned Escherichia coli K1 CMP-N-acetylneuraminic acid synthetase gene. J Biol Chem. 1989;264:14769–14774. [PubMed] [Google Scholar]