Abstract
RhaS, an AraC family protein, activates rhaBAD transcription by binding to rhaI, a site consisting of two 17-bp inverted repeat half-sites. In this work, amino acids in RhaS that make base-specific contacts with rhaI were identified. Sequence similarity with AraC suggested that the first contacting motif of RhaS was a helix-turn-helix. Assays of rhaB-lacZ activation by alanine mutants within this potential motif indicated that residues 201, 202, 205, and 206 might contact rhaI. The second motif was identified based on the hypothesis that a region of especially high amino acid similarity between RhaS and RhaR (another AraC family member) might contact the nearly identical DNA sequences in one major groove of their half-sites. We first made targeted, random mutations and then made alanine substitutions within this region of RhaS. Our analysis identified residues 247, 248, 250, 252, 253, and 254 as potentially important for DNA binding. A genetic loss-of-contact approach was used to identify whether any of the RhaS amino acids in the first or second contacting motif make base-specific DNA contacts. In motif 1, we found that Arg202 and Arg206 both make specific contacts with bp −65 and −67 in rhaI1, and that Arg202 contacts −46 and Arg206 contacts −48 in rhaI2. In motif 2, we found that Asp250 and Asn252 both contact the bp −79 in rhaI1. Alignment with the recently crystallized MarA protein suggest that both RhaS motifs are likely helix-turn-helix DNA-binding motifs.
The AraC family of transcription activators (sometimes also called the AraC/XylS family) was found to consist of more than 100 members in the last published homology search (performed in March 1997) (13). Activators in this family are found in a wide variety of bacterial species. Many members of this family regulate expression of genes involved in carbon metabolism, stress responses, or pathogenesis. Examples of proteins that regulate carbon metabolism (and the carbon source and species they are found in) are AraC (l-arabinose, Escherichia coli), RhaS and RhaR (l-rhamnose, E. coli), XylS (benzoates, Pseudomonas putida), and MelR (melibiose, E. coli) (2, 14, 33, 36). Some of the proteins that regulate stress response genes are SoxS (oxidative stress, E. coli), MarA (multiple antibiotic resistance, E. coli), Ada (alkylating agents, E. coli), and Rob (originally identified as binding to oriC but cross-activates SoxS- and MarA-regulated genes, E. coli) (6, 18–21, 30, 39). Finally, some AraC family proteins that regulate virulence factors in pathogens include CfaD (fimbriae, enterotoxigenic E. coli) and VirF (invasion proteins, Shigella dysenteriae) (11, 17, 31, 32).
The RhaS protein, one of the AraC family members, activates expression of the E. coli rhaBAD and rhaT operons (10, 38). rhaT encodes the l-rhamnose transport protein (35). rhaBAD encodes the enzymes required for catabolism of the sugar l-rhamnose (25, 28); hence RhaS binding and activation of rhaBAD occurs only in the presence of l-rhamnose (9, 10). In the absence of l-rhamnose, there is apparently insufficient active RhaS to saturate the rhaBAD promoter, resulting in a lag in rhaBAD mRNA accumulation after l-rhamnose addition (10). RhaR, another AraC family protein (12, 13, 29, 36), activates rhaSR expression (37). Although RhaS and RhaR are encoded in the same operon, there apparently is sufficient RhaR to saturate the rhaSR promoter in the absence of l-rhamnose, as there is no detectable lag in rhaSR mRNA accumulation after l-rhamnose induction (10). As a result, l-rhamnose induction of rhaBAD expression first requires induction of rhaSR expression, which leads to accumulation of RhaS and finally activation of rhaBAD expression.
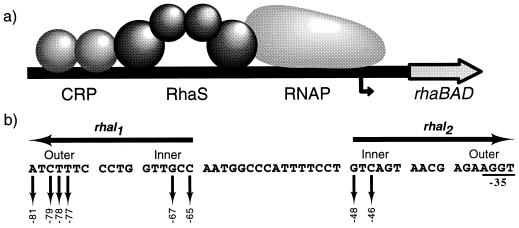
Egan and Schleif (9) used DNase I footprinting and point mutations to locate the DNA-binding site for RhaS (rhaI) to a region spanning −32 to −81 relative to the rhaBAD transcription start site (Fig. 1). The symmetry of important single-base mutations in rhaI led to the prediction that the RhaS binding site is an inverted repeat of two 17-bp half-sites (rhaI1 and rhaI2) separated by 16 bp of uncontacted DNA. Based on the fact that AraC binds to DNA as a dimer (16) and the size of the RhaS binding site, it is predicted that RhaS binds to rhaI as a dimer, with each RhaS monomer contacting one half-site. When viewed from one face of the DNA helix, each rhaI half-site consists of two major grooves, separated by a minor groove. All of the base pairs found important for RhaS binding lie in the major grooves of rhaI (9), suggesting that RhaS makes all of its sequence-specific DNA contacts within the major grooves (Fig. 1). This is in agreement with the previous findings that AraC and RhaR also interact with their DNA binding sites within two major grooves of each half-site (3, 37). We expected that in order to contact two major grooves, each RhaS monomer would possess two DNA-contacting motifs.
FIG. 1.
(a) Model for transcription activation at rhaBAD, showing the two activator proteins, cyclic AMP receptor protein (CRP) and RhaS, and RNA polymerase (RNAP) shown. The bent arrow represents the rhaBAD transcription start site. (b) DNA sequence of the RhaS binding site (rhaI). The two half-sites, rhaI1 and rhaI2, are inverted repeats, as indicated by the arrows above the sequence. Sequences within the half-sites are divided into two major grooves separated by a minor groove, with the outer and inner major grooves indicated. Down arrows indicate bases previously identified as important for interaction with RhaS (9); their positions relative to the rhaBAD transcription start site are indicated below. Bases overlapping the −35 region of the promoter were not analyzed in this previous study since their effects on RhaS binding and RNA polymerase binding could not easily be distinguished by the analysis used. Notice that all of the bases important in rhaI1 are identical in rhaI2.
One DNA-binding motif in AraC has been predicted to be a helix-turn-helix (H-T-H) motif consisting of residues 196 to 215 (3). Ser208 and His212 of this motif have been shown to make base-specific contacts with DNA (3, 27). Asp256 of AraC has also been shown to be important for DNA sequence recognition and was predicted to be a part of a second H-T-H motif (H-T-H 2) (27). Recently Rhee et al. (30) determined the crystal structure of the first AraC family member, MarA. MarA is one of relatively few AraC family members that consists of a DNA-binding domain without any dimerization domain, and it activates transcription as a monomer (6, 23). The MarA structure includes two H-T-H DNA-binding motifs; the first (H-T-H 1) aligns with AraC Ser208 and His212, and the second (H-T-H 2) aligns with AraC Asp256.
In this study, we used a genetic analysis to identify RhaS amino acids that make specific contacts with rhaI. Our results provide further evidence that AraC family transcription activators utilize two H-T-H motifs to contact DNA. Further, this work is the most extensive functional analysis of the second AraC family H-T-H motif to date. Our data suggest that Arg202 and Arg206 of RhaS, which align with H-T-H 1 of MarA and AraC, make base-specific contacts with rhaI. Both of these amino acids contact the inner major grooves of the inverted repeat RhaS binding site (Fig. 1). Our results further show that residues within the 247 to 254 region of RhaS are crucial for contact with rhaI. We found that Asp250 and Asn252, which lie in this region, make specific contacts with the outer major grooves of rhaI and are part of H-T-H 2 of RhaS.
MATERIALS AND METHODS
Culture media.
Cultures for β-galactosidase assay were grown in 1× MOPS [3-(N-morpholino)propanesulfonic acid]-buffered medium (26) (40 mM MOPS, 4 mM Tricine, 0.01 mM FeSO4, 9.5 mM NH4Cl, 0.276 mM K2SO4, 0.5 μM CaCl2, 0.528 mM MgCl2, 50 mM NaCl, 3 × 10−9 M Na2Mo4, 4 × 10−7 M H3BO3, 3 × 10−8 M CoCl2, 10−8 M CuSO4, 8 × 10−8 M MnCl2, 10−8 M ZnSO4, 1.32 mM K2HPO4, 10 mM NaHCO3, 0.2% Casamino Acids, 0.002% thiamine). Ampicillin and tetracycline were used at 125 and 12.5 μg/ml, respectively, when necessary. For other experiments (cloning, strain construction, Ter test, etc.), cells were grown in tryptone-yeast extract medium (22) with or without antibiotic or in TB-maltose (0.8% Bacto-Tryptone, 0.5% NaCl, 0.2% maltose).
DNA sequencing.
IRD41 dye-labeled primers for DNA sequencing (Table 1) were custom made by LI-COR, Inc. (Lincoln, Neb.). DNA sequences were verified by automated dideoxy sequencing on a LI-COR 4000L sequencer. Sequencing reactions were performed with a Thermo Sequenase fluorescent-labelled primer cycle sequencing kit from Amersham Life Science.
TABLE 1.
Oligonucleotides used in this study
| Designation | Sequence, 5′-3′a | Use |
|---|---|---|
| 898 | TGAGTAAAGCTTTTATTGCAGAAAGCCATCCCG | Amplify rhaS, downstream |
| 1170 | CCGGAATTCTTGTGGTGATGTGATGCTCAC | Amplify rhaS, upstream |
| 2064 | TCTCTTTCACTGGCTACGCTACATCG | Arg202Ala |
| 2065 | CGTACGCTACATGCGCAGCTTAAGCAG | Arg206Ala |
| 2066 | CAATTTTCTCTTTCAGCGCGTACGCTACAT | Leu201Ala |
| 2067 | CCTATCGCTGTGGATTCAGCGACAGTAACCACTTTTCGACGCTTTb | H-T-H 2 random mutations |
| 2068 | ATGACCGTATTACATAGTGTGGATc | rhaS sequencing |
| 2069 | TTATTGCAGAAAGCCATCCCGTCCc | rhaS sequencing |
| 2073 | GGATTCAGCGCCAGTAACCAC | Asp250Ala |
| 2074 | TGGTTGCACAGATGGAACAGCc | rhaS sequencing |
| 2075 | GTTGAGACGTGATGCGCTGTTc | rhaS sequencing |
| 2087 | TTCAGCGACAGTGCCCACTTT | Asn252Ala |
| 2088 | TGTGGAGCCAGCGACAGTAAC | Phe248Ala |
| 2089 | AGTAACGCCTTTTCGACGCTTTTT | His253Ala |
| 2090 | AACCACGCTTCGACGCTTTTT | Phe254Ala |
| 2091 | AGTAACCACTTTGCGACGCTTT | Ser255Ala |
| 2093 | TATCGCGCTGGATTCAGC | Cys246Ala |
| 2120 | CGTACGCTAGCTCGGCAGCTTA | His205Ala |
| 2125 | CCACTTTTCGGCGCTTTTTCG | Thr256Ala |
For oligonucleotides used for PCR amplification, sequences complementary to rhaS are underlined; for those used for site-directed mutagenesis, positions of mutations are underlined.
During synthesis, the wild-type nucleotide as well as a low concentration of each of the other three nucleotides was added at each of the underlined positions, resulting in a pool of oligonucleotides with an average of approximately 1.5 mutations each.
IRD41 dye labeled for use in a LI-COR automated sequencer.
Plasmids, phage, and strains.
Wild-type rhaS was amplified by PCR using primers 1170 and 898 and plasmid pSE101 as the template. The PCR product was digested at the EcoRI site in 1170 and the HindIII site in 898 and cloned between the EcoRI and HindIII sites of Tetr pALTER-1, resulting in pSE160. An Apr version of the clone (pSE159) was made by using Promega Altered Sites II in vitro mutagenesis system. The DNA sequence of the cloned rhaS gene in both pSE159 and pSE160 was verified by DNA sequencing on both strands.
All strains used were derivatives of ECL116 (1) (Table 2). Translational rhaB-lacZ fusions with point mutations within rhaI (9) were initially present on derivatives of plasmid pSE105 (Table 2). These fusions were transferred to phage λRS45 (λimm21) by in vivo recombination (34) to generate recombinant λ phages (Table 2). The final strains were constructed by transducing SME1082 with the corresponding recombinant λ phage. Lysogens were selected by spreading the transduction mixture on a plate carrying a lawn of λgt30 (λimm21). Lysogens were differentiated from λ phage-resistant cells by their sensitivity to a heteroimmune phage upon cross-streaking. Single lysogens were identified by the Ter test (15). In the case of (rhaB-lacZ), fusions with single point mutations at −65 and −78, a lysogen was first made by transducing ECL116 with the corresponding recombinant λ phage. Single lysogens were identified by β-galactosidase assay and confirmed by the Ter test (15). The single lysogen was then introduced into SME1082 by phage P1-mediated generalized transduction (24).
TABLE 2.
Strains, phage, and plasmids used in this study
| Strain, phage, or plasmid | Genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| ECL116 | F− ΔlacU169 endA hsdR thi | 1 |
| SME1082 | ECL116 ΔrhaSa | Laboratory collection |
| SME1212 | SME1082 λSME103 C(−46)Gb | This study |
| SME1213 | SME1082 λSME103 T(−78)Gb | This study |
| SME1214 | SME1082 λSME103 C(−65)Tb | This study |
| SME1221 | SME1082 λSME103 T(−77)Gb | This study |
| SME1222 | SME1082 λSME103 wtc | This study |
| SME1724 | SME1082 λSME103 G(−48)Tb | This study |
| SME1725 | SME1082 λSME103 A(−81)Cb | This study |
| SME1726 | SME1082 λSME103 G(−67)Ab | This study |
| SME1733 | SME1082 λSME103 C(−79)Ab | This study |
| Phage | ||
| λRS45 | bla′-lacZscatt+ int+ imm21 | 34 |
| λSME103 | λRS45 Φ(rhaB-lacZ)Δ110 | 10 |
| Plasmids | ||
| pRS414 | Apr, ′lacZ lacY+ lacA+ | 34 |
| pALTER-1 | Aps TetrlacZ f1 ori | Promega Corp. |
| pSE101 | AprrhaBAD+ rhaSR+ | 10 |
| pSE105 | pRS414 Φ(rhaB-lacZ) (wt) | 10 |
| pSE159 | Apr pALTER-1 rhaS (wt) | This study |
| pSE160 | Tetr pALTER-1 rhaS (wt) | This study |
| pSE165 | pSE160 (Leu201Ala) | This study |
| pSE166 | pSE159 (Arg202Ala) | This study |
| pSE167 | pSE159 (His205Ala) | This study |
| pSE168 | pSE160 (Arg206Ala) | This study |
| pSE169 | pSE159 (Cys246Ala) | This study |
| pSE170 | pSE160 (Gly247Ala) | This study |
| pSE171 | pSE159 (Phe248Ala) | This study |
| pSE172 | pSE160 (Asp250Ala) | This study |
| pSE173 | pSE159 (Asn252Ala) | This study |
| pSE174 | pSE159 (His253Ala) | This study |
| pSE175 | pSE159 (Phe254Ala) | This study |
| pSE176 | pSE159 (Ser255Ala) | This study |
| pSE177 | pSE159 (Thr256Ala) | This study |
| pSE178 | pSE159 (Gly247Arg) | This study |
| pSE179 | pSE160 (Phe248Val) | This study |
| pSE180 | pSE160 (Ser249Gly) | This study |
| pSE181 | pSE160 (Ser251Gly) | This study |
| pSE182 | pSE159 (Asn252His) | This study |
| pSE183 | pSE159 (Asn252Ser) | This study |
| pSE184 | pSE160 (Asn252Ile) | This study |
| pSE185 | pSE160 (His253Gln) | This study |
β-Galactosidase assay.
Starter cultures of strains to be assayed were grown in tryptone-yeast extract broth containing appropriate antibiotic for approximately 7 h at 37°C; 40 μl of starter culture was then inoculated into 2.5 ml of overnight medium and grown for approximately 17 h. Overnight medium consisted of 1× MOPS medium containing 0.04% glycerol as the carbon source and appropriate antibiotic. A 100-μl volume of the overnight culture was inoculated into 10 ml of growth medium. Growth medium consisted of 1× MOPS medium containing 0.4% glycerol as the carbon source with 0.2% l-rhamnose added as inducer in all cases and appropriate antibiotic. Cultures were grown at 37°C with vigorous shaking in baffled, 125-ml Erhlenmeyer flasks to an A600 of approximately 0.4. Growth medium cultures were then centrifuged, and the cells were resuspended in Z buffer (24). β-Galactosidase activity was determined as described by Miller (24) except that incubation with substrate o-nitrophenyl-β-d-galactopyranoside was at room temperature. Specific activities were averaged from at least three independent assays, with two replicates in each assay.
Mutagenesis of rhaS.
Oligonucleotide primers used for random and site-directed mutagenesis (Table 1) were synthesized by Oligos Etc. Inc. A 45-nucleotide oligonucleotide with degenerate sequence (5) over the region to be mutagenized was used to generate random mutations in the rhaS region encoding amino acids 246 to 255 (Table 1). The oligonucleotide was designed to introduce an average of 1.5 mutations within the 30-bp region and was used in a site-directed mutagenesis procedure (Promega Altered Sites II in vitro mutagenesis system) to create random mutations.
Alanine substitutions within the first and second possible DNA-binding motifs of RhaS were created by site-directed mutagenesis (Promega Altered Sites II and GeneEditor in vitro mutagenesis systems) (Table 1). Double-stranded DNA template was used to construct all amino acid substitutions except Arg202Ala and Thr256Ala, for which a single-stranded DNA template was used. Both random and alanine substitutions were entirely sequenced on both strands to confirm each mutation and to ensure that there were no additional amino acid changes. One random mutant (Phe248Val) had two silent mutations in addition to the substitution at residue 248, but all other mutants had DNA sequences identical to that of the wild-type rhaS clone.
RESULTS
First RhaS DNA-contacting motif.
AraC amino acids Ser208 and His212, which correspond to amino acids 2 and 6 of the DNA recognition helix of an H-T-H motif, have been shown to make specific contacts with base pairs within the AraC binding site (araI) (3, 27). These AraC amino acids align with RhaS Arg202 and Arg206. Hence, we created alanine substitutions at Arg202 and Arg206 of RhaS. Amino acid 1 of the recognition helix often is involved in making specific DNA contacts, and so we also made an alanine substitution at Leu201. Further, it has been recently shown that Gln45 of MarA makes specific contact with DNA (30). Since MarA Gln45 aligns with RhaS His205, we also created an alanine substitution at His205 of RhaS.
We measured the ability of each of the above RhaS mutants to activate transcription from a wild-type Φ(rhaB-lacZ) fusion to determine whether the mutant residues might be important for RhaS to contact DNA (Table 3). All four of the mutations tested (Leu201Ala, Arg202Ala, His205Ala, and Arg206Ala) were at least twofold defective in transcription activation at the wild-type Φ(rhaB-lacZ) fusion (Table 3). Arg202Ala and Arg206Ala had especially large defects. These data are consistent with the hypothesis that Leu201, Arg202, His205, and Arg206 are part of a RhaS H-T-H DNA-binding motif involved in contacting rhaI, although other explanations are possible on the basis of this result alone.
TABLE 3.
Alanine substitutions in RhaS H-T-H 1 analyzed at wild-type rhaB-lacZ
| RhaS derivative | % of wild-type activitya (sp act [U]) |
|---|---|
| Leu201Ala | 48 (238) |
| Arg202Ala | 4 (14) |
| His205Ala | 28 (97) |
| Arg206Ala | 0.1 (0.7) |
β-Galactosidase specific activity was measured from a single-copy rhaB-lacZ fusion in a rhaS deletion strain transformed with plasmids encoding either a wild-type or mutant form of RhaS. Cultures were grown in MOPS growth medium containing glycerol, l-rhamnose, and appropriate antibiotic. The activity of RhaS mutants is represented as the percentage of the activity of wild-type RhaS. Standard errors were less than 10% of the average units.
Random mutagenesis of the second possible RhaS DNA-contacting motif.
As described above, RhaS is expected to have two DNA-binding motifs. It seemed likely that the RhaS H-T-H involving amino acids 201 to 206 would contact the inner major grooves of rhaI (Fig. 2a), based on the expectation that the promoter proximal RhaS and AraC monomers would bind to DNA in the same relative orientation (and possibly make conserved contacts with RNA polymerase). Egan and Schleif (9) previously observed that the outer major grooves of rhaI are nearly identical to the corresponding major grooves of the site for RhaR binding (Fig. 2a). This suggests that RhaS and RhaR may share a conserved DNA-binding motif. We found a 10-residue region which was highly conserved in RhaS (residues 246 to 255) and RhaR (residues 282 to 291) (Fig. 2b). Eight of the ten amino acids in this region are identical whereas the other two are similar—a degree of sequence similarity that is unusually high, even for the very similar RhaS and RhaR proteins. Therefore, we hypothesized that residues 246 to 255 might be part of the second DNA-binding motif of RhaS.
FIG. 2.
(a) Comparison of the DNA-binding sites (bs) recognized by RhaS, RhaR, and AraC. Horizontal arrows indicate the half-sites and their relative orientations. Notice that the downstream half-sites are all in the same relative orientation whereas the upstream half-sites are not. Bracketed bases are conserved in the RhaS and RhaR DNA-binding sites. The outer and inner major grooves of the RhaS binding site are indicated. The −35 regions of the promoters are shown. Vertical arrows indicate amino acid-base pair contacts between AraC H-T-H 1 and its DNA-binding site. (b) Sequence of amino acids 246 to 255 of RhaS, showing high similarity with the aligned region of RhaR (amino acids 282 to 291). Solid lines indicate identical amino acids; broken lines represent similar amino acids.
We used an oligonucleotide with degenerate sequence (2067 [Table 1]) in a site-directed mutagenesis procedure to create random mutations in this conserved region of RhaS. All of the mutations generated were analyzed, without any functional selection. We isolated mutations at 7 of the 10 amino acid residues in the region (Table 4). As we had already constructed an alanine substitution at amino acid 250 of RhaS (see below), we did not assay the random substitution that we isolated at position 250 (Asp250Glu). To test if RhaS residues from positions 246 to 255 might contact rhaI, we first analyzed the abilities of the random mutants to activate transcription from a wild-type Φ(rhaB-lacZ) fusion (Table 4). With the exception of Ser249Gly and Ser251Gly, all of the random mutants were considerably defective in activating transcription from wild-type Φ(rhaB-lacZ). Defects greater than 10-fold were found with mutations at amino acids Gly247, Phe248, and Asn252. This suggests that these residues and possibly His253, which had a threefold defect, might be involved in DNA binding by RhaS. These residues are candidates for the second DNA-binding motif of RhaS.
TABLE 4.
Random mutations in RhaS H-T-H 2 analyzed at wild-type rhaB-lacZ
| RhaS derivative | % of wild-type activitya (sp act [U]) |
|---|---|
| Gly247Arg | 0.9 (3.8) |
| Phe248Val | 3.3 (17) |
| Ser249Gly | 93 (536) |
| Ser251Gly | 87 (505) |
| Asn252His | 0.4 (2.0) |
| Asn252Ser | 37 (184) |
| Asn252Ile | 0.3 (2.0) |
| His253Gln | 32 (185) |
β-Galactosidase specific activity was measured from a single-copy rhaB-lacZ fusion in a rhaS deletion strain transformed with plasmids encoding either a wild-type or mutant form of RhaS. Cultures were grown in MOPS growth medium containing glycerol, l-rhamnose, and appropriate antibiotic. The activity of RhaS mutants is represented as the percentage of the activity of wild-type RhaS. Standard errors were less than 21% of the average units.
Alanine substitutions in the second RhaS H-T-H motif.
We made individual alanine substitutions at most of the amino acid positions in the 246–255 region. Alanine substitutions were not constructed at Ser249 or Ser251 since random Gly substitutions at these positions had very little effect on RhaS function. We did not construct an alanine substitution at Gly247 since an alanine substitution at this position was isolated in the random mutagenesis described above. While we were in the process of creating and analyzing the alanine substitutions, publication of the MarA crystal structure showed that Arg96 makes specific contacts with DNA (30). Since, MarA Arg96 aligns with RhaS Thr256 (13), we also made an alanine substitution at this position.
We tested these RhaS alanine mutants for the ability to activate transcription at wild-type Φ(rhaB-lacZ). Alanine mutants at Gly247, Phe248, Asp250, Asn252, His253, and Phe254 were all significantly defective for transcription activation from the wild-type fusion (Table 5). This result supports the hypothesis that these amino acids might be part of a second DNA-binding motif in RhaS, which we predict to be an H-T-H motif based on alignment with MarA (30).
TABLE 5.
Alanine substitutions in RhaS H-T-H 2 analyzed at wild-type rhaB-lacZ
| RhaS derivative | % of wild-type activitya (sp act [U]) |
|---|---|
| Cys246Ala | 66 (174) |
| Gly247Ala | 6.2 (33) |
| Phe248Ala | 0.9 (2.3) |
| Asp250Ala | 8.3 (44) |
| Asn252Ala | 31 (80) |
| His253Ala | 48 (153) |
| Phe254Ala | 1.2 (3.2) |
| Ser255Ala | 80 (210) |
| Thr256Ala | 61 (208) |
β-Galactosidase specific activity was measured from a single-copy rhaB-lacZ fusion in a rhaS deletion strain transformed with plasmids encoding either a wild-type or mutant form of RhaS. Cultures were grown in MOPS growth medium containing glycerol, l-rhamnose, and appropriate antibiotic. The activity of RhaS mutants is represented as the percentage of the activity of wild-type RhaS. Standard errors were less than 8% of the average units.
Base-specific contacts by RhaS.
To test whether base-specific contacts are made by any of the amino acids thought to be important in DNA binding, we used a genetic loss-of-contact approach (7, 8). The rationale behind the genetic loss-of-contact approach is that a protein with a single amino acid substitution will lose the ability to discriminate between the wild-type and mutant base pairs only at the position or positions in the DNA that are normally contacted by the substituted amino acid. As a result, any DNA positions normally contacted by the substituted amino acid can be mutated with relatively little effect, while mutations at other DNA positions will have large effects. This analysis will determine whether some of the amino acids that we have identified are directly involved in DNA binding and also identify the base pair positions contacted by such base-specific amino acids.
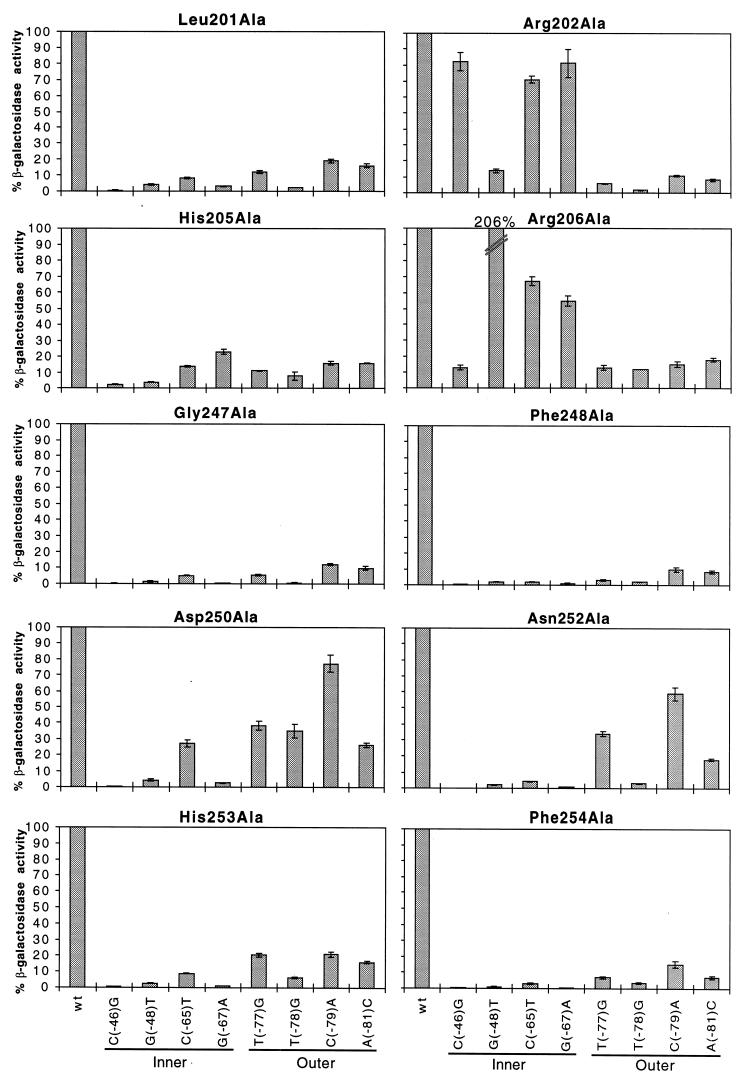
Each of the alanine substitutions found to be at least twofold defective at wild-type Φ(rhaB-lacZ) was tested at eight derivatives of Φ(rhaB-lacZ) (Fig. 3). The derivatives contained single point mutations at the important base positions in rhaI1 and in the upstream major groove of rhaI2 (Fig. 1). Within RhaS H-T-H 1, we found that Leu201Ala and His205Ala gave considerably lower activity at all of the mutant Φ(rhaB-lacZ) fusions compared with wild-type Φ(rhaB-lacZ) (Fig. 3). This indicates that Leu201 and His205 do not make any specific contacts with rhaI. In contrast, point mutations at −65 and −67 within rhaI1 had relatively little effect on activation by both Arg202Ala and Arg206Ala (Fig. 3). In rhaI2, the point mutation at −46 had relatively little effect on activation by Arg202Ala and the point mutation at −48 had relatively little effect on activation by Arg206Ala. These data suggest that Arg202 contacts the base pairs at −46, −65, and −67, and Arg206 contacts the base pairs at −48, −65, and −67. This result strongly suggests that Arg202 and Arg206 are both directly involved in DNA binding and do not simply have indirect effects on DNA binding by RhaS.
FIG. 3.
Alanine substitutions in RhaS H-T-H 1 and 2 analyzed at mutant rhaB-lacZ fusions. The x axis (labeled at the bottom) represents either the wild-type rhaBAD promoter or the position of point mutations in rhaI found to be important for RhaS binding (Fig. 1). Locations of the point mutations in either the inner or outer major grooves of rhaI are indicated. The y axis (labeled on the left) represents the percent β-galactosidase specific activity for each RhaS alanine mutant at mutant rhaB-lacZ compared with the same mutant protein at the wild-type rhaB-lacZ fusion. The first bar in each graph represents the RhaS alanine mutant protein assayed at the wild-type rhaB-lacZ promoter and is set to 100%. Error bars are shown.
We also tested the defective mutants in the second H-T-H motif of RhaS at mutant Φ(rhaB-lacZ) fusions to determine if some or all of them make specific contacts with rhaI (Fig. 3). Gly247Ala, Phe248Ala, His253Ala, and Phe254Ala all gave significantly lower β-galactosidase specific activities from all mutant Φ(rhaB-lacZ) fusions compared with the wild-type fusion. This result suggests that these amino acids do not make any specific contacts with rhaI. With Asp250Ala and Asn252Ala, on the other hand, a point mutation at −79 had relatively little effect on activation. We conclude that both Asp250 and Asn252 are directly involved in DNA binding and make specific contacts with the base pair at position −79 within rhaI. In addition, Asn252Ala gave a significantly higher level of activity with the point mutation at −77 than the remaining base changes in rhaI, suggesting that Asn252 may make a second contact with the base pair at position −77.
DISCUSSION
In this work, we set out to determine the amino acids in the RhaS protein that contact rhaI, as well as the specific bases in rhaI that are contacted by those amino acids. Each of the rhaI half-sites is 17 bp long (Fig. 1), and therefore the face of the DNA helix likely to be contacted by RhaS will consist of two major grooves of DNA (9). This suggests that the RhaS protein contains two distinct DNA-contacting motifs in each monomer.
H-T-H 1 of RhaS is a DNA-binding motif.
We constructed alanine substitutions at four amino acid positions within the recognition helix of a RhaS H-T-H DNA-binding motif proposed based on alignment with AraC (3). We found that mutations in the first (Leu201), second (Arg202), fifth (His205), and sixth (Arg206) amino acids of the recognition helix resulted in defects in RhaS activation from the wild-type rhaBAD promoter (Table 3). This result is consistent with the hypothesis that these amino acids may be involved in sequence-specific DNA contacts by RhaS. As discussed below, our results indicate that this motif contacts the inner major grooves of the rhaI inverted repeat.
H-T-H 2 of RhaS is also a DNA-binding motif.
At the time we began these experiments, the second DNA-contacting motif had not been identified in any AraC family proteins. We hypothesized that the RhaS and RhaR proteins might use a conserved DNA-contacting motif to recognize the conserved outer major grooves of their binding sites (Fig. 2). We identified amino acids 246 to 255 of RhaS as a region having unusually high sequence similarity with RhaR and proposed that this region might be a part of the second DNA-contacting motif. We found that random substitutions at positions Gly247, Phe248, Asn252, and His253 resulted in a defect in the ability of RhaS to activate expression from the wild-type rhaBAD promoter (Table 4). Ala mutants at all of the amino acids from residues 247 to 254, except for 249 and 251 (which were tested as random substitutions only), were found to be at least twofold defective for activation from the rhaBAD promoter, and most were more than 10-fold defective (Table 5). We propose that this region is part of a second DNA-binding motif of RhaS that, based on alignment with the crystal structure of MarA (30), is likely an H-T-H motif.
Identification of DNA contacts by RhaS.
Further analysis of the mutants that we constructed in the two H-T-H motifs was performed by using a genetic loss-of-contact approach (outlined in Results) (7, 8). A genetic approach was chosen for initial characterization of these mutants since it is more informative than comparison of binding affinities of the mutants to wild-type and much easier than in vitro missing - contact probing (3, 4). In addition, a genetic approach was particularly useful in this case since the RhaS protein is severely insoluble (9).
Using the genetic loss-of-contact approach, we have determined that 4 of the 10 amino acids identified as potentially important for DNA binding by RhaS make specific contacts with base pairs in rhaI. In H-T-H 1, amino acids Arg202 and Arg206 were each found to contact the base pairs at positions −65 and −67 in rhaI1. The contacts in rhaI2 were not exactly symmetric, with Arg202 contacting only −46 and Arg206 contacting only −48 (Fig. 1 and 4). Since the contacted bases are identical in both half-sites, the asymmetry in the contacts is likely to be due to either the DNA context of the contacted bases or the influence of proteins that may contact RhaS (cyclic AMP receptor protein and RNA polymerase). Thus, RhaS H-T-H 1 contacts the inner major grooves of the rhaI inverted repeat site (Fig. 4), consistent with the expectation that the promoter proximal monomers of RhaS and AraC bind in the same relative orientation.
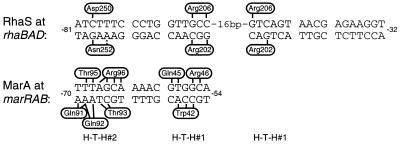
FIG. 4.
Summary of specific amino acid-base pair contacts by RhaS and comparison with MarA DNA contacts. DNA half-sites are divided into two major grooves and an intervening minor groove. Vertical and angled black lines represent amino acid-base pair contacts. The broken bar indicates a less conclusive DNA contact by RhaS. The side of the DNA sequence on which each amino acid is represented is arbitrary and is not meant to represent the base within the pair that is contacted. For MarA, the base(s) within each pair that is contacted is known (30); however, this information is not represented for simplicity and because the corresponding information is not known for RhaS. Relative positions of H-T-H 1 and 2 are shown below the two DNA sequences.
In RhaS H-T-H 2, Asp250 and Asn252 are both predicted to make base-specific contact with the base pair at position −79 of rhaI1. In addition, our results somewhat weakly suggest that Asn252 may also interact with the base pair at position −77. These base pairs are within the outer major groove of the inverted repeat rhaI site, consistent with the finding that H-T-H 1 contacts the inner major grooves. The important bases for RhaS binding in the outer major groove of rhaI2 have not been identified due to their overlap with the −35 region of the promoter. Since the important bases in rhaI1 are identical in rhaI2, we initially predicted that the contacts would be symmetric; however, the finding of asymmetry in the inner major grooves suggests that further investigation is necessary to identify the contacts in this region.
Comparison with MarA structure.
Both of the amino acids in H-T-H 1 of RhaS that make sequence-specific DNA contacts align with MarA residues also predicted to make specific DNA contacts. Arg202 of RhaS aligns with MarA Trp42. One of the base pairs contacted by each of these amino acids is a G-C base pair in the analogous position in the two binding sites (−56 at marRAB and −67 at rhaBAD), while each of these residues also makes additional, different contacts (Fig. 4). Arg206 of RhaS aligns with Arg46 of MarA. In this case, the identical amino acid contacts the identical base pair in the analogous position in the two binding sites (G-C at −56 in marRAB and −67 in rhaBAD), and each amino acid also contacts a C-G base pair (−55 at marRAB and −65 at rhaBAD). MarA has a third amino acid in H-T-H 1 that contacts DNA (Gln45). MarA Gln45 aligns with RhaS His205, which was found to be important for DNA binding by RhaS but not specific for any base positions. It seems likely that only Arg202 and Arg206 of RhaS H-T-H 1 make DNA contacts since two amino acid partners have been identified for each of the two important base pairs in the inner major grooves.
Asn252 of RhaS aligns with Gln92 of MarA, which also makes specific DNA contacts (Fig. 4). Each of these amino acids contacts one base pair at analogous positions in the two binding sites (−68 at marRAB and −79 at rhaBAD), as well as a possible second contact by Asn252 (at −77) and one additional contact by Gln92 (at −69). Asp250 of RhaS is the only DNA-contacting amino acid that does not align with a contacting amino acid in MarA. In fact, the alignment of RhaS and MarA (13) predicts that Asp250 lies within the turn prior to the recognition helix of H-T-H 2. There are no DNA contacts (neither specific nor phosphate backbone) by amino acids in this turn of MarA. However, Asp250 of RhaS aligns with Asp256 of AraC, which has also been found important in contacting DNA (27). Interestingly, more than one-fourth of the proteins aligned by Gallegos et al. (13) have an aspartic acid in this position, suggesting that other family members may use this amino acid to contact DNA as well.
There are four amino acids in MarA H-T-H 2 which make base-specific DNA contacts and align with RhaS amino acids that do not make contacts (Fig. 4). These amino acids align with RhaS amino acids judged not to be important for DNA binding (Ser251, Ser255, and Thr256) or found to be important for DNA binding but not base specific (His253). It is not surprising that MarA, which binds to DNA as a monomer (23), has more amino acid-DNA contacts per monomer than RhaS, which is predicted to bind as a dimer. Our current analysis shows an equal number of amino acids in the MarA monomer and the RhaS dimer that make DNA contacts; however, the total number of base-specific contacts made by the MarA monomer (20 contacts) is greater than the number (12 to 14) made by the RhaS dimer. There is reason to believe that the binding strength of these two proteins may be more similar than the number of contacts suggests. First, we do not think that we have identified all of the DNA contacts made by RhaS with the outer major groove of rhaI (see below). Second, MarA would be expected to need more than twice the binding strength of each RhaS monomer to make up for the cooperativity gained by RhaS binding as a dimer.
It is not surprising to find that (i) three of the five base pairs for which we conclusively identified contacts were contacted by two different amino acids and (ii) two of the amino acids in RhaS contact two base pairs separated by an uncontacted base pair. The DNA contacts made by MarA are in accord with both of these findings. There are five different examples in MarA of base pairs contacted by two amino acids, and even one example of a base pair contacted by three amino acids (30) (Fig. 4). Further, two of the amino acids in MarA (Trp42 and Arg96) contact three and four adjacent base pairs, respectively, although with Arg96 there is a water molecule involved in one of the contacts (30).
Have all of the base-specific contacts been identified?
Curiously, at least two (or three if −77 is included) important base pairs in the outer major grooves of rhaI do not yet have identified amino acid partners. We have tested all of the amino acids ranging from the last amino acid of the first helix, through the turn, up to the sixth amino acid of the recognition helix, without finding any contacts with these base pairs. It is possible that additional amino acids of RhaS make DNA contacts; however, these amino acids are not located in the most common positions for H-T-H contacts. Alternatively, it is possible that amino acids that we tested make DNA contacts that were not detected by our analysis for some reason.
Nonspecific amino acid contacts.
Our analysis identified six amino acids in RhaS that were defective for transcription activation at the rhaBAD promoter but that did not show evidence of base-specific DNA contacts. It is possible, although certainly not probable, that some of these amino acids are involved in transcription activation rather than DNA binding by RhaS. It is also possible that some of these amino acids make contacts with the phosphate backbone of the DNA, and therefore are directly involved in DNA binding, but do not make base-specific contacts. Eleven different amino acids in MarA make only phosphate backbone contacts with the DNA (30). Finally, it is possible that some of these amino acid substitutions alter the conformation of RhaS and therefore indirectly affect DNA binding. Inspection on the MarA structure of the amino acids that aligns with each of these nonspecific RhaS amino acids (by using RasMol version 2.6) indicates that all of them except Phe254 are significantly surface exposed and near the DNA backbone. Substitutions at Phe254 most likely alter the conformation of RhaS and thereby indirectly reduce DNA binding. Each of the other nonspecific amino acids could possibly make phosphate backbone contacts.
ACKNOWLEDGMENTS
We thank the members of our laboratory for critical discussions and comments on the manuscript, Susan Bear for constructing pSE159, pSE160, and pSE172, and our reviewers for thoughtful comments. We thank Bonnie Liscek and other members of the University of Kansas Biochemical Research Service Laboratory for help with automated DNA sequencing.
This work was supported by Public Health Service grant GM55099 from the National Institute of General Medical Sciences, the National Science Foundation under grant EPS-9550487 and matching support from the State of Kansas, a General Research Fund award from the University of Kansas, and the Franklin Murphy Molecular Biology Endowment, all to S.M.E.
REFERENCES
- 1.Backman K, Chen Y-M, Magasanik B. Physical and genetic characterization of the gln A-glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1981;78:3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourgerie S J, Michan C M, Thomas M S, Busby S J W, Hyde E I. DNA binding and DNA bending by the MelR transcription activator protein from Escherichia coli. Nucleic Acids Res. 1997;25:1685–1693. doi: 10.1093/nar/25.9.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunelle A, Schleif R. Determining residue-base interactions between AraC protein and araI DNA. J Mol Biol. 1989;209:607–622. doi: 10.1016/0022-2836(89)90598-6. [DOI] [PubMed] [Google Scholar]
- 4.Brunelle A, Schleif R. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci USA. 1987;84:6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang L W, Kovari I, Howe M M. Mutagenic oligonucleotide-directed PCR amplification (Mod-PCR): an efficient method for generating random base substitution mutations in a DNA sequence element. PCR Methods Appl. 1993;2:210–217. doi: 10.1101/gr.2.3.210. [DOI] [PubMed] [Google Scholar]
- 6.Cohen S P, Hachler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebright R H. Identification of amino acid-base pair contacts by genetic methods. Methods Enzymol. 1991;208:620–640. doi: 10.1016/0076-6879(91)08032-d. [DOI] [PubMed] [Google Scholar]
- 8.Ebright R H. Use of “loss-of-contact” substitutions to identify residues involved in an amino acid-base pair contact: effect of substitution of Gln18 of Lac repressor by Gly, Ser, and Leu. J Biomol Struct Dyn. 1985;3:281–297. doi: 10.1080/07391102.1985.10508417. [DOI] [PubMed] [Google Scholar]
- 9.Egan S M, Schleif R F. DNA-dependent renaturation of an insoluble DNA binding protein. Identification of the RhaS binding site at rhaBAD. J Mol Biol. 1994;243:821–829. doi: 10.1006/jmbi.1994.1684. [DOI] [PubMed] [Google Scholar]
- 10.Egan S M, Schleif R F. A regulatory cascade in the induction of rhaBAD. J Mol Biol. 1993;234:87–98. doi: 10.1006/jmbi.1993.1565. [DOI] [PubMed] [Google Scholar]
- 11.Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi C O. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallegos M-T, Michán C, Ramos J L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Pérez M M, Ramos J L, Gallegos M T, Marqués S. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J Biol Chem. 1999;274:2286–2290. doi: 10.1074/jbc.274.4.2286. [DOI] [PubMed] [Google Scholar]
- 15.Gottesman M E, Yarmolinsky M B. The integration and excision of the bacteriophage lambda genome. Cold Spring Harbor Symp Quant Biol. 1968;33:735–747. doi: 10.1101/sqb.1968.033.01.084. [DOI] [PubMed] [Google Scholar]
- 16.Hendrickson W, Schleif R. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc Natl Acad Sci USA. 1985;82:3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordi B J A M, van der Zeijst B A M, Gaastra W. Regions of the CFA/I promoter involved in the activation by the transcriptional activator CfaD and repression by the histone-like protein H-NS. Biochimie. 1994;76:1052–1054. doi: 10.1016/0300-9084(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 18.Landini P, Busby S J. The Escherichia coli Ada protein can interact with two distinct determinants in the ς70 subunit of RNA polymerase according to promoter architecture: identification of the target of Ada activation at the alkA promoter. J Bacteriol. 1999;181:1524–1529. doi: 10.1128/jb.181.5.1524-1529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemotte P K, Walker G C. Induction and autoregulation of ada, a positively acting element regulating the response of Escherichia coli K-12 to methylating agents. J Bacteriol. 1985;161:888–895. doi: 10.1128/jb.161.3.888-895.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Demple B. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol Microbiol. 1996;20:937–945. doi: 10.1111/j.1365-2958.1996.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Demple B. SoxS, an activator of superoxide stress genes in Escherichia coli: purification and interaction with DNA. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 22.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 23.Martin R G, Jair K-W, Wolf R E, Jr, Rosner J L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 25.Moralejo P, Egan S M, Hidalgo E, Aguilar J. Sequencing and characterization of a gene cluster encoding the enzymes for l-rhamnose metabolism in Escherichia coli. J Bacteriol. 1993;175:5585–5594. doi: 10.1128/jb.175.17.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niland P, Huhne R, Muller-Hill B. How AraC interacts specifically with its target DNAs. J Mol Biol. 1996;254:667–674. doi: 10.1006/jmbi.1996.0668. [DOI] [PubMed] [Google Scholar]
- 28.Power J. The l-rhamnose genetic system in Escherichia coli K-12. Genetics. 1967;55:557–568. doi: 10.1093/genetics/55.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramos J L, Rojo F, Zhou L, Timmis K N. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coli AraC activators. Nucleic Acids Res. 1990;18:2149–2152. doi: 10.1093/nar/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee S, Martin R G, Rosner J L, Davies D R. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai T, Sasakawa C, Yoshikawa M. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton virF protein. Mol Microbiol. 1988;2:589–597. doi: 10.1111/j.1365-2958.1988.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 32.Savelkoul P H M, Willshaw G A, McConnell M M, Smith H R, Hamers A M, van der Zeijst B A M, Gaastra W. Expression of CFA/I fimbriae is positively regulated. Microb Pathog. 1990;8:91–99. doi: 10.1016/0882-4010(90)90073-y. [DOI] [PubMed] [Google Scholar]
- 33.Schleif R. Two positively regulated systems, ara and mal. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1300–1309. [Google Scholar]
- 34.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 35.Tate C G, Muiry J A R, Henderson P J F. Mapping, cloning, expression, and sequencing of the rhaT gene which encodes a novel l-rhamnose-H+ transport protein in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1992;287:6923–6932. [PubMed] [Google Scholar]
- 36.Tobin J F, Schleif R F. Positive regulation of the Escherichia colil-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J Mol Biol. 1987;196:789–799. doi: 10.1016/0022-2836(87)90405-0. [DOI] [PubMed] [Google Scholar]
- 37.Tobin J F, Schleif R F. Transcription from the rha operon psr promoter. J Mol Biol. 1990;211:1–4. doi: 10.1016/0022-2836(90)90003-5. [DOI] [PubMed] [Google Scholar]
- 38.Via P, Badia J, Baldoma L, Obradors N, Aguilar J. Transcriptional regulation of the Escherichia coli rhaT gene. Microbiology. 1996;142:1833–1840. doi: 10.1099/13500872-142-7-1833. [DOI] [PubMed] [Google Scholar]
- 39.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]