Abstract
The competence and sporulation factor (CSF) of Bacillus subtilis is an extracellular pentapeptide produced from the product of phrC. CSF has at least three activities: (i) at low concentrations, it stimulates expression of genes activated by the transcription factor ComA; at higher concentrations, it (ii) inhibits expression of those same genes and (iii) stimulates sporulation. Because the activities of CSF are concentration dependent, we measured the amount of extracellular CSF produced by cells. We found that by mid-exponential phase, CSF accumulated to concentrations (1 to 5 nM) that stimulate ComA-dependent gene expression. Upon entry into stationary phase, CSF reached 50 to 100 nM, concentrations that stimulate sporulation and inhibit ComA-dependent gene expression. Transcription of phrC was found to be controlled by two promoters: P1, which precedes rapC, the gene upstream of phrC; and P2, which directs transcription of phrC only. Both RapC and CSF were found to be part of autoregulatory loops that affect transcription from P1, which we show is activated by ComA∼P. RapC negatively regulates its own expression, presumably due to its ability to inhibit accumulation of ComA∼P. CSF positively regulates its own expression, presumably due to its ability to inhibit RapC activity. Transcription from P2, which is controlled by the alternate sigma factor ςH, increased as cells entered stationary phase, contributing to the increase in extracellular CSF at this time. In addition to controlling transcription of phrC, ςH appears to control expression of at least one other gene required for production of CSF.
Response to high cell density (quorum sensing) is widespread in bacteria, regulating many diverse processes. In Bacillus subtilis, sporulation and the development of genetic competence (the natural ability to import exogenous DNA) are stimulated as cells grow to high cell density. As with other gram-positive bacteria, cell density control in B. subtilis is mediated by extracellular peptides (4, 13, 26, 31).
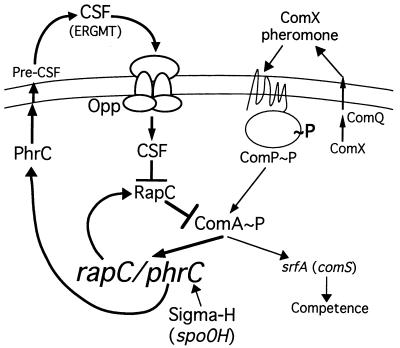
The development of genetic competence is stimulated by two peptide factors, ComX pheromone, a modified 10-amino-acid peptide (15), and the competence and sporulation factor (CSF), an unmodified five-amino-acid peptide (32) (Fig. 1). ComX pheromone and CSF stimulate transcription of the srfA operon (15, 32). The srfA operon encodes a small protein ComS, which is required for competence development (3, 7), and the surfactin biosynthetic enzymes (18). Transcription from the srfA promoter is activated by the phosphorylated form of the response regulator ComA (5, 19–21, 28, 38), and activity of ComA is controlled by a kinase, ComP, and a phosphatase, RapC (Fig. 1). ComP, a membrane-bound histidine protein kinase (39), is required for activation of ComA. RapC, a putative aspartyl-phosphate phosphatase (24), inhibits activation of ComA (32). ComX pheromone stimulates gene expression, apparently by stimulating the kinase ComP (15, 33), whereas CSF stimulates gene expression, apparently by inhibiting the phosphatase RapC (32) (Fig. 1). ComX pheromone most likely works at the cell surface, and CSF is actively transported into the cell by the oligopeptide permease Opp (also known as Spo0K), where it interacts with intracellular receptors (14).
FIG. 1.
Model of the regulation of synthesis of and response to RapC and CSF. CSF is synthesized as a precursor protein PhrC with a signal sequence and putative peptidase cleavage sites. Pre-CSF is cleaved to produce the active CSF pentapeptide (ERGMT). The transcription factor ComA is activated (phosphorylated) by the membrane-bound histidine protein kinase ComP and by ComX pheromone, which probably activates ComP. ComQ is needed for production of the active ComX pheromone. CSF is transported into the cell by the oligopeptide permease (also known as Spo0K), where it stimulates expression of genes activated by ComA∼P, probably by inhibiting activity of the phosphatase RapC, which is a negative regulator of ComA∼P. Results presented in this report indicate that ComA∼P activates transcription of both rapC and phrC. Furthermore, we show that RapC negatively regulates it own synthesis, presumably by dephosphorylating ComA∼P. CSF, in contrast, was shown to positively regulate transcription of itself and rapC, presumably by inhibiting the phosphatase activity of RapC. ςH, the spo0H gene product, activates transcription of phrC.
CSF has a remarkable constellation of activities for a five-amino-acid peptide (14, 32). At relatively low extracellular concentrations (1 to 5 nM), CSF stimulates expression of srfA by inhibiting the phosphatase RapC. However, high concentrations of CSF (>20 nM) inhibit expression of srfA, perhaps by inhibiting activity of the kinase ComP. In addition to its effects on expression of ComA∼P-controlled genes, high concentrations of CSF (>20 nM) can stimulate sporulation under some conditions (14, 32). CSF appears to stimulate sporulation, at least in part, by inhibiting the activity of the phosphatase RapB (23), an aspartyl-phosphate phosphatase that dephosphorylates Spo0F∼P (25), an essential component of the phosphorelay that controls sporulation (1, 4, 10).
Because CSF has these different activities at different extracellular concentrations, we were interested in characterizing the regulation and production of CSF. Production of mature CSF involves several steps, starting with transcription and translation of phrC, the gene encoding the precursor of CSF (32). The 40-amino-acid primary product of phrC has a signal sequence and putative peptidase cleavage sites, indicating that a 11- to 25-amino-acid peptide is exported via a Sec-dependent pathway (24). Steps after initial processing that result in release of the mature pentapeptide are not yet known.
We have measured the extracellular concentration of CSF produced during growth and compared this to the transcriptional control of phrC. phrC is in an operon with rapC (24). We found that transcription of the rapC phrC operon is activated by high cell density through ComA∼P and that rapC and phrC regulate their own expression (Fig. 1). RapC, by negatively regulating ComA∼P, is part of a homeostatic autoregulatory loop. PhrC (CSF) stimulates ComA activity and positively regulates its own expression. Furthermore, we show that by mid-exponential phase, CSF is at concentrations that stimulate competence gene expression and that as cells enter stationary phase, the extracellular concentrations of CSF reach levels approaching 100 nM, concentrations that are known to stimulate sporulation and inhibit early competence gene expression.
MATERIALS AND METHODS
Strains construction.
B. subtilis strains used are listed in Table 1; unless otherwise indicated, all are derived from the parent strain JH642 and contain trpC and pheA mutations. Strains were constructed by transformation with chromosomal DNA by standard protocols (8). ΔcodY was transferred by linkage to unkU::spc (30). The presence of the ΔcodY mutation was confirmed by PCR analysis of chromosomal DNA.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype |
|---|---|
| AG174 (JH642) | trpC2 pheA1 |
| IRN216 | amyE::(rapC-lacZΩ1 neo) (P1-lacZ) |
| IRN217 | amyE::(P1-lacZ) rapC::pJS49 (cat) |
| IRN218 | amyE::(P1-lacZ) ΔphrC::erm |
| IRN219 | amyE::(P1-lacZ) comA::cat |
| IRN220 | amyE::(P1-lacZ) comP::spc |
| IRN222 | amyE::(P1-lacZ) comQ::spc |
| IRN224 | amyE::(P1-lacZ) Δopp358::erm |
| IRN289 | amyE::(P1-lacZ) unkU::spc |
| IRN299 | amyE::(P1-lacZ) unkU::spc ΔcodY |
| IRN238 | amyE::(phrC-lacZΩ2 neo) (P2-lacZ) |
| IRN243 | amyE::(P2-lacZ) spo0H::cat::spc |
| IRN249 | amyE::(P2-lacZ) spo0A(D56N)-cat::spc |
| IRN250 | amyE::(P2-lacZ) comA::cat |
| IRN252 | amyE::(P2-lacZ) spo0A(D56N)-cat::spc abrB:: Tn917 (MLSR) |
| IRN235 | (ΔphrC::erm)::pGK50 (P1, P2-lacZ) |
| IRN246 | (P1, P2-lacZ) spo0A(D56N)-cat::spc |
| IRN273 | (P1, P2-lacZ) spo0H::cat::spc |
| IRN277 | (P1, P2-lacZ) ΔcomA::spc |
| BALB6 | (P1, P2-lacZ) spo0AΔ204 abrB703 |
| JRL192 | comA::cat |
| AG665 | spo0H::cat::spc |
| BAL125 | amyE::(srfA-lacZΩ374 neo) ΔphrC::erm |
Construction of lacZ fusions.
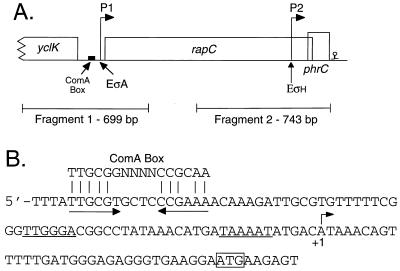
Three lacZ transcriptional fusions were constructed to analyze regulation of the rapC phrC operon. To analyze expression from the P1 promoter, a 699-bp fragment (fragment 1 [Fig. 2A]), from 446 bp upstream to 253 bp downstream of the start codon of rapC, was cloned upstream of lacZ and integrated into the chromosome at amyE. The P1 fragment was amplified by PCR using Vent polymerase (New England Biolabs) and primers rapC3 (5′-AGAAGCTTACGGTGACATTTGGCTG-3′) and rapC4 (5′-GGGATCCAATTCAGACAGGCTTGGC-3′) with restriction sites (underlined) added at the ends. This PCR fragment was subcloned between the HindIII and BamHI sites of pKS2 (15) making plasmid pGK47. pGK47 was linearized and transformed into wild-type B. subtilis, selecting for Neor transformants. The transformants were screened for an amylase-deficient phenotype to confirm that the plasmid had integrated into the chromosome at the amyE locus.
FIG. 2.
(A) The rapC phrC operon. The location of each of the two promoters is indicated by the arrows. The location of P1 was determined by primer extension analysis and is inferred, based on sequence, to depend on the major sigma factor ςA (see panel B). The ςH-dependent promoter P2 was mapped previously (2). A putative Rho-independent terminator is located downstream of phrC, and a putative ComA-binding site (ComA Box) is upstream of rapC. Fragment 1 and 2 indicate the regions used for making promoter fusions to lacZ. Fragment 2 includes the entire phrC gene. (B) Sequence of the P1 promoter region. Primer extension analysis was used to map the 5′ end of the rapC mRNA (see Materials and Methods). The 5′ end is indicated by the arrow and +1. The putative −10 and −35 regions are underlined, and the ATG start codon for rapC is boxed. The consensus ComA box (17) is aligned with the putative ComA box in the rapC promoter region.
To analyze expression from the P2 promoter, a 743-bp fragment (fragment 2 [Fig. 2A]), from 619 bp upstream to 124 bp downstream of the start codon of phrC, was cloned upstream of lacZ and integrated into the chromosome at amyE. This fragment extends two base pairs beyond the stop codon of phrC, resulting in the introduction of an additional copy of phrC. lacZ fusions with downstream junctions internal to phrC had β-galactosidase activity similar to the fusion with the additional copy of phrC (data not shown). The P2 fragment was amplified by PCR using Vent polymerase and primers rapC5 (5′-GGAATTCTACGTGGAGCAGGAAAC-3′) and phrC4 (5′-GGGATCCTCTTACGTCATTCCTCTT-3′), with restriction sites (underlined) added at the ends. This PCR fragment was cloned between the EcoRI and BamHI sites of pKS2, making plasmid pGK52. pGK52 was introduced into wild-type B. subtilis as described above for pGK47.
Expression of phrC under the control of the P1 and P2 promoters was analyzed by using a lacZ fusion integrated into the B. subtilis chromosome by single crossover immediately downstream of phrC. To construct this fusion, the same 743-bp P2 fragment as described above was subcloned into the EcoRI and BamHI sites of pJM783 (22) to create plasmid pGK50. A ΔphrC::erm mutant JMS751 (14) was transformed with pGK50, selecting for Cmr transformants. The resulting transformant has lacZ fused to the entire operon, including a wild-type copy of phrC.
Media.
Cells were grown at 37°C in defined S7 minimal medium with glucose (1%), glutamate (0.1%), and required amino acids (Trp and Phe), and MOPS (morpholinepropanesulfonic acid) buffer was used at 50 rather than 100 mM (12, 37). Casamino Acids were added to 0.1% when indicated.
Primer extension analysis.
The transcription start site for the rapC P1 promoter was determined by primer extension analysis. Total RNA was prepared from strain JMS682 (a wild-type strain with amy::srfA-lacZ), which was grown in defined minimal medium until 2 h after the end of exponential growth. The RNA was purified using an RNeasy Maxi kit (Qiagen) according to the manufacturer’s instructions; 40 μg of this RNA was analyzed by primer extension as described previously (34), with the following exceptions. The primer, RM2 (5′-ACTTCGGCATCCGGCACGCTGAATG-3′), which corresponds to +126 to +102 relative to the start of transcription, was radiolabeled with polynucleotide kinase and [γ-32P]ATP, and the unincorporated nucleotides were separated from the primer on a MicroSpin G-50 column (Pharmacia). Primer extension reactions and sequencing reactions with the RM2 primer were run on an 8% polyacrylamide gel, and radioactivity was detected by autoradiography.
β-Galactosidase assays.
β-Galactosidase specific activity was measured essentially as described previously (12, 15, 16) and is presented as (ΔA420 per minute per milliliter of culture per unit of optical density at 600 nm [OD600]) × 1,000.
Extracellular factor assays.
The activity of extracellular factors was assayed essentially as described previously (15, 32). Briefly, cells containing a lacZ fusion were grown to low cell density (OD600 of ∼0.1), and 0.25 ml of culture was mixed with an equal volume of a test sample, either conditioned medium, partly fractionated signaling peptides, or synthetic CSF. Samples were incubated at 37°C for 70 min (15), and β-galactosidase specific activity was measured. Conditioned medium (culture supernatant) was prepared by growing cells in defined minimal medium, then removing the cells by centrifugation, and subjecting the supernatant to filtration through a 0.2-μm-pore-size filter. Synthetic CSF was resuspended in defined minimal medium containing 50 μg of bovine serum albumin per ml prior to mixing with cells.
Quantitation of extracellular CSF.
Cells were grown in defined minimal medium, samples (7 ml) were taken at different times during growth, and cells were removed as described above. To separate CSF from ComX pheromone in the culture supernatant, the pH was adjusted to 2 with trifluoroacetic acid (TFA), and each sample was run over a 1-ml C18 reverse-phase column (Sep-Pak Plus; Millipore) that had been equilibrated with aqueous 0.1% TFA. After being washed with 3 column volumes of 0.1% TFA, CSF was eluted with 3 column volumes of buffer containing 0.1% TFA and 11% acetonitrile. To assay the column eluate for CSF activity, samples were dried in a Speed-Vac to remove TFA and acetonitrile and then resuspended in minimal medium containing 50 μg of bovine serum albumin ml. The samples were then diluted, and CSF activity was measured (as described above), using strain BAL125 (ΔphrC::erm srfA-lacZ). The activity of the column eluates was compared to the activity of known concentrations of synthetic CSF. CSF activity was linear in this assay from 0.05 to 1.5 nM CSF.
CSF was not lost to a significant extent (<5%) during fractionation and sample preparation. This was determined by adding CSF to 100 nM (the highest concentrations of CSF present in culture supernatants) to conditioned medium prepared from a strain unable to make CSF (ΔphrC). This sample was fractionated and prepared as described above and then tested for CSF activity. Greater than 95% of the CSF activity added to the conditioned medium was present in the column fraction (data not shown). Conditioned medium from the ΔphrC strain had no activity in this assay in the absence of added CSF (data not shown and reference 32).
RESULTS
lacZ fusions to the rapC phrC regulatory regions.
The rapC phrC operon is predicted to have at least two promoters. Immediately upstream of rapC is a promoter, P1, with the recognition sequence for the vegetative sigma factor, ςA (Fig. 2A). The location of the start of transcription for the P1 promoter was determined by primer extension analysis, using a primer internal to rapC (Fig. 2B). Just upstream from this promoter is a potential binding site for the transcription factor ComA. This sequence matches the consensus binding site for ComA (17) in 10 of 12 positions (Fig. 2B). In addition to the ςA-dependent promoter, there is a ςH-dependent promoter, P2, upstream of phrC (2), internal to rapC (Fig. 2A). Expression of phrC should be controlled by both P1 and P2.
To measure transcription of rapC and phrC, we constructed three different fusions to lacZ (Materials and Methods). One fusion contains P1 (Fig. 2A, fragment 1) and was integrated into the chromosome at a heterologous site (amyE). The second fusion contains P2 (Fig. 2A, fragment 2) and was integrated into the chromosome, also at amyE. The third fusion places lacZ under control of the entire operon and was integrated into the chromosome just downstream of phrC at the normal chromosomal location. The fusions are referred to as P1-lacZ, P2-lacZ, and (P1, P2)-lacZ, respectively.
Regulation of rapC by cell density and ComA.
Transcription from P1 increased as cells grew to high cell density in defined minimal glucose medium, as judged by accumulation of β-galactosidase specific activity from the P1-lacZ fusion (Fig. 3A). Indeed, expression of P1-lacZ in cells at low density increased in response to the addition of conditioned medium from cells at high density (data not shown). This pattern of expression is similar to that of the promoter for the srfA operon (15, 32, 33), which is activated by the phosphorylated form of the transcription factor ComA (reference 28 and references therein). This pattern of expression and the presence of a putative ComA binding site suggest that P1 is activated by ComA∼P.
FIG. 3.
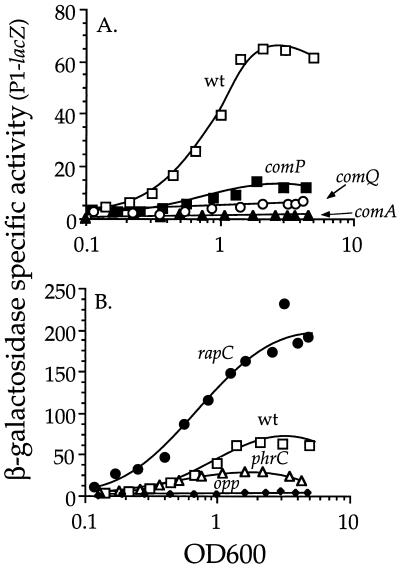
Regulation of rapC P1 by ComA∼P. Strains containing a P1-lacZ fusion were grown in defined minimal medium, and β-galactosidase specific activity is plotted as a function of OD600. Shown are data only from the exponential phase of growth. The data are from one of at least three independent experiments. (A) wt, wild type (IRN216); comP, IRN220; comQ, IRN222; comA, IRN219. (B) rapC, IRN217; phrC, IRN218; opp, IRN224; wt, wild type (IRN216) (same data as in panel A).
Transcription from P1 was altered in mutants affected in the ComA regulatory pathway. The most severe effect was in a comA null mutant; accumulation of β-galactosidase from P1-lacZ was reduced to ∼1% of that in wild-type cells (Fig. 3A). comP encodes a histidine protein kinase (39) that is required for activation (phosphorylation) of ComA and full transcription from ComA-dependent promoters (Fig. 1) (19, 39). As expected, a null mutation in comP also reduced expression from P1-lacZ (Fig. 3A). ComP activity is stimulated by an extracellular signaling peptide, the ComX pheromone (Fig. 1) (15, 33). Expression of P1-lacZ was reduced in a mutant (comQ) unable to make the ComX pheromone (Fig. 3A). Interestingly, expression from P1 was lower in the comQ (pheromone) mutant than the comP (kinase) mutant, indicating that ComP could have an inhibitory role, perhaps as a phosphatase, in addition to its stimulatory role. Alternatively, ComQ could have a role in stimulating ComA-dependent gene expression that is independent of ComP. The mutations have a similar effect on expression of srfA (33).
In addition to ComP and ComX pheromone, expression of genes activated by ComA∼P is also affected by CSF, the oligopeptide permease (Opp) that transports CSF into the cell, and the putative phosphatase RapC, which is a negative regulator of ComA∼P (Fig. 1) (14, 15, 32, 33). We found that transcription from P1 was affected by all of these components. A null mutation in rapC caused increased transcription from P1 such that β-galactosidase specific activity was approximately threefold greater throughout growth than that in wild-type cells (Fig. 3B). This result indicates a negative autoregulatory loop; that is, RapC negatively regulates (indirectly) its own expression, presumably by inhibiting accumulation of ComA∼P (Fig. 1) (32). In contrast, a null mutation in phrC, which eliminates production of CSF (32), caused decreased transcription from P1 (Fig. 3B), indicating a positive autoregulatory loop. That is, CSF (phrC) stimulates its own expression, most likely by inhibiting activity of the phosphatase RapC, leading to increased accumulation of ComA∼P and increased transcription of the rapC phrC operon from P1 (Fig. 1).
The ability of cells to respond to CSF depends on the oligopeptide permease, Opp (14, 33). An opp null mutation decreased expression of P1-lacZ to ∼6% of that in wild-type cells (Fig. 3B), similar to effects on expression of srfA (32). This effect is much greater than that of the phrC null mutation and indicates that the loss of the oligopeptide permease has pleiotropic effects, perhaps causing other phosphatases of the Rap family to be hyperactive and dephosphorylate ComA∼P (32).
CSF and the control of rapC transcription.
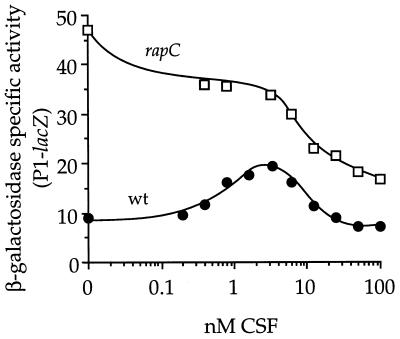
CSF is known to have two effects on expression of the ComA-controlled gene srfA. Low concentrations of CSF stimulate, and high concentrations inhibit, transcription of srfA (14, 32). We found that CSF also had these two effects on transcription from P1 (Fig. 4), indicating that this regulation was not specific for srfA. When added to cells at low cell density, relatively low concentrations of CSF (2 to 5 nM) stimulated transcription from P1 ∼2.5-fold in our standard assay (Fig. 4). The maximum potential induction caused by CSF may be partly masked by the inhibitory activity of CSF.
FIG. 4.
Regulation of rapC P1 by CSF. The β-galactosidase specific activity from P1-lacZ in cells at low density was measured 70 min after the addition of synthetic CSF (see Materials and Methods). Shown are data from one of at least three independent experiments. wt, wild type (IRN216); rapC, IRN217.
The inhibitory activity of CSF was tested in a rapC mutant background because the level of P1 expression was higher and the competing ability of CSF to stimulate rapC expression was removed (Fig. 4). In the rapC mutant background, addition of greater than 10 nM CSF inhibited expression from P1 (Fig. 4). High concentrations of CSF also inhibited expression of another ComA-controlled gene, rapA (data not shown). This indicates that the inhibitory activity of CSF affects the activity of a protein that regulates transcription of rapC, srfA, and rapA. A known negative regulator of these genes, CodY (29) (see below), is not required for the inhibitory activity of CSF (data not shown). We suspect that high concentrations of CSF might be inhibiting activity of the kinase ComP.
Regulation of rapC by CodY.
Under some growth conditions, the transcription factor CodY represses expression of several genes known to be activated by ComA∼P. In growth medium containing complex mixtures of amino acids, expression of srfA and rapA is inhibited during exponential growth and induced upon entry into stationary phase. The inhibition during exponential growth is mediated by CodY (29).
We tested the effects of amino acids and a codY null mutation on expression from rapCP1. The presence of casamino acids in the growth medium greatly reduced expression of P1-lacZ during exponential growth, and this effect was relieved in a codY null mutant (Fig. 5). The codY mutation had no noticeable effect on expression from P1 in the growth medium without Casamino Acids (data not shown). Despite the ability of CodY to repress rapC when Casamino Acids were added to the medium, codY did not affect expression of rapC during growth in nutrient sporulation medium (data not shown), similar to previous findings on rapA expression (30). CodY has been shown to bind to the srfA promoter region in vitro (29), and although a binding sequence has not been defined, it seems plausible that CodY also binds to the rapC promoter region.
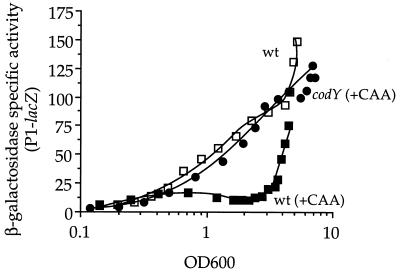
FIG. 5.
Control of rapC P1 by CodY. Strains containing a P1-lacZ fusion were grown in minimal medium with the indicated additions, and β-galactosidase specific activity is plotted as a function of OD600. Shown are data from one of at least three independent experiments. wt, wild type (IRN289) grown without and with Casamino Acids (+CAA); ΔcodY (+CAA), IRN299 grown with Casamino Acids. unkU::spc, which was used as a selectable maker in the transformation of the ΔcodY marker, had no effect on expression of P1-lacZ (data not shown). codY had no effect in the absence of Casamino Acids (data not shown). Apparent differences in P1-lacZ expression between Fig. 3 and 5 at high ODs are due to extra data points after the end of cell growth in Fig. 5 and daily variation.
Regulation of P2, the ςH-dependent promoter upstream of phrC.
The ςH-dependent promoter upstream from phrC was identified previously in a search for promoters transcribed in vitro by RNA polymerase containing the alternate sigma factor ςH, the spo0H gene product (2). We found that in defined minimal medium, expression of P2-lacZ was relatively constant during exponential growth and increased as cells approached stationary phase (Fig. 6). Expression was virtually undetectable in a spo0H null mutant (Fig. 6A), consistent with previous findings (2). Increased expression from P2 as cells approach stationary phase is most likely due to an increase in levels of ςH protein (9, 40). Expression from P2 did not appear to be regulated by cell density, as a comA null mutation had little or no effect on expression of P2-lacZ (Fig. 6A), nor did addition of conditioned medium to cells at low density (data not shown).
FIG. 6.
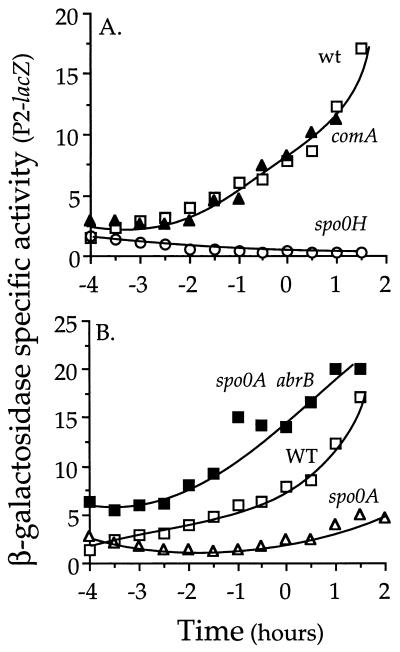
Expression of phrC P2. Strains containing a P2-lacZ fusion were grown in defined minimal medium, and β-galactosidase specific activity is plotted as a function of time relative to entry into stationary phase. The data are not plotted as a function of cell density because upon entry into stationary phase there is little increase in culture density but a significant increase in specific activity. Shown are data from one of at least three independent experiments. Time zero is defined as the end of exponential growth. (A) wt, wild type (IRN238); comA, IRN250; spo0H, IRN243. (B) WT, wild type (IRN238); spo0A, IRN249; spo0A abrB, IRN252.
A spo0A null mutation caused a decrease in expression of P2-lacZ, and this effect was relieved by a null mutation in abrB (Fig. 6B), consistent with the effect of these gene products on transcription of spo0H. Transcription of spo0H is controlled by gene products that regulate the initiation of sporulation. spo0H is repressed by AbrB (40), which, in turn, is repressed by the activated (phosphorylated) form of the transcription factor Spo0A (27). The activity of Spo0A is controlled by a phosphorelay (1) that integrates many signals that stimulate sporulation, including starvation (4, 11).
Regulation of phrC transcription.
Transcription of phrC is control by both the ComA-regulated promoter P1, and the ςH-dependent promoter P2 (Fig. 2A). To understand how these two promoters affect the regulation of phrC, we constructed a lacZ fusion [(P1, P2)-lacZ] that is controlled by both promoters. This was done by integrating a plasmid containing a phrC-lacZ fusion into the chromosome at the phrC locus.
A comA null mutation reduced expression of the (P1,P2)-lacZ fusion early in growth to less than 10% of the level of β-galactosidase of an otherwise wild-type strain (Fig. 7A). Expression of the (P1,P2)-lacZ fusion in a comA null mutant increased after entry into stationary phase, presumably due to the activation of ςH and increased transcription from P2.
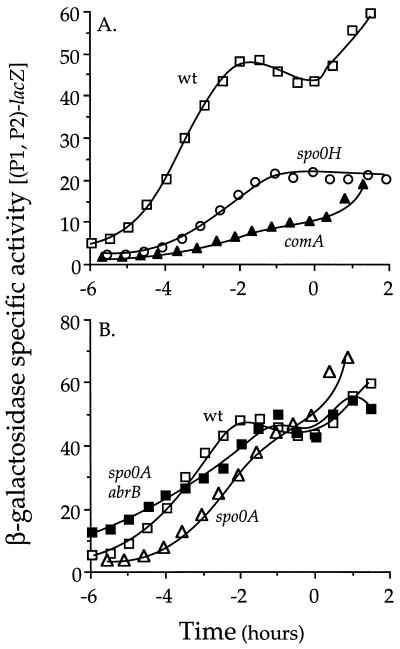
FIG. 7.
Expression of (P1, P2)-lacZ. Strains containing the (P1, P2)-lacZ fusion were grown in defined minimal medium, and β-galactosidase specific activity is plotted as a function of time. Shown are data from one of at least three independent experiments. Time zero is defined as the end of exponential growth. (A) wt, wild type (IRN235); comA, IRN277; spo0H, IRN273. (B) wt, wild type (IRN235); spo0A, IRN246; spo0A abrB, (BAL6).
A spo0H mutant also affected the expression of (P1,P2)-lacZ (Fig. 7A). While expression of phrC was reduced in a spo0H mutant during growth to about 35% of the level in an otherwise wild-type strain, expression still increased during exponential growth, peaking about 1 h before the onset of stationary phase, presumably due to the activation of ComA and increased transcription from P1. The effects of spo0H on phrC(P1,P2)-lacZ are largely due to its requirement for transcription of the P2 promoter. However, spo0H also indirectly regulates P1 through CSF effects on ComA∼P.
The transcription factors Spo0A and AbrB affect expression from the P2 promoter (see above); however, our results indicate that the major effect of spo0A and abrB on CSF production is postranscriptional. Previously, it was shown that accumulation of extracellular CSF in a spo0A mutant is reduced to ∼3 to 4% of that in wild-type cells, and this defect is suppressed by null mutations in abrB (33). Surprisingly, the effect of the spo0A null mutation on expression of phrC was quite modest. Expression of the (P1, P2)-lacZ fusion in the spo0A mutant was approximately 60% of that in wild-type cells (Fig. 7B). This defect was relieved in the spo0A abrB double mutant (Fig. 7B). These results indicate that Spo0A and AbrB likely affect transcription of at least one gene other than phrC that is involved in CSF production. This gene could affect export and/or processing of CSF.
Production of extracellular CSF.
The analysis of physiological responses to CSF involved addition of different concentrations of chemically synthesized peptide to cells (14, 32). To determine if these concentrations are physiologically relevant, we measured the CSF activity present in culture supernatants during exponential growth and upon entry into stationary phase. Culture supernatant was fractionated to separate CSF from ComX pheromone. The activity of CSF found in partly fractionated culture supernatant was an accurate reflection of the concentration of CSF. Reconstruction experiments were done to estimate recovery of CSF from culture supernatant. A known amount of CSF was added to culture supernatant from a ΔphrC mutant (unable to make CSF). At least 95% of the expected CSF activity was recovered following fractionation (see Materials and Methods). Activity was compared to a standard curve determined with known concentrations of synthetic CSF (Materials and Methods).
In a culture of wild-type cells, the concentration of extracellular CSF increased during exponential growth, reached 1 to 5 nM approximately two to three generations before entry into stationary phase (Fig. 8A), and increased to ∼90 nM shortly after entry into stationary phase (Fig. 8A). These concentrations are well within the range previously shown to elicit regulatory responses (14, 32).
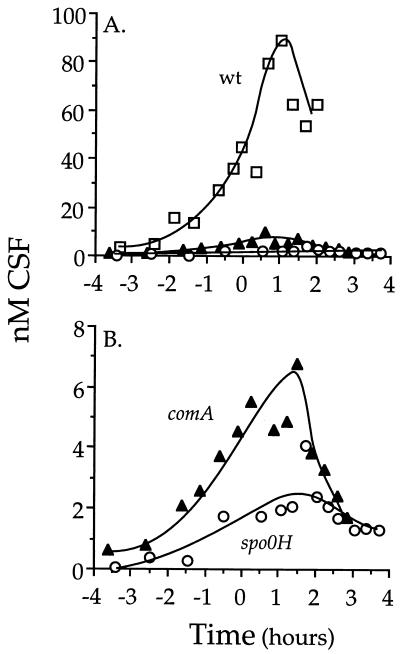
FIG. 8.
Accumulation of extracellular CSF. Conditioned medium was collected at different times during growth and assayed for CSF (Materials and Methods). Shown are data from one of three independent experiments. The levels of CSF measured for a particular strain during exponential growth varied by ∼10% from experiment to experiment. During stationary phase, this variability increased to ∼25%. Time zero is defined as the end of exponential growth. (A) open squares, wild type (wt; AG174); filled triangles, comA (JRL192); open circles, spo0H (AG665). (B) Data from panel A replotted to compare comA and spo0H mutants.
ComA and ςH affect the accumulation of extracellular CSF, but the effect of ςH on accumulation is greater than its effect on the transcription of phrC. The accumulation of extracellular CSF was severely reduced in a comA mutant strain to ∼7% of the level of a wild-type strain (Fig. 8). This is similar to the effect of a comA mutation on the expression of phrC [the (P1, P2)-lacZ fusion] (Fig. 7A). spo0H mutants also had severely reduced levels of extracellular CSF (Fig. 8). A spo0H mutant strain had less than 2% of the normal amount of CSF. However, this effect of a spo0H mutation on extracellular CSF production is more severe than the effect on transcription of phrC (Fig. 7A), indicating that spo0H probably is required for expression of genes, in addition to phrC, that are involved in CSF production. We cannot rule out that ComA also has an affect on the expression of genes required for CSF production other than phrC, but this effect would have to be small compared to ComA’s effect on transcription of phrC.
DISCUSSION
CSF of B. subtilis was initially identified as an extracellular peptide factor that stimulates transcription from the srfA promoter when added to cells at low cell density (32, 33). CSF is a pentapeptide produced from the C-terminal 5 amino acids of the 40-amino-acid product of phrC (32). CSF has at least three activities: (i) at low concentrations, it stimulates expression of genes activated by ComA∼P; at higher concentrations, it (ii) inhibits expression of those same genes and (iii) under some conditions stimulates sporulation (14, 32). Genetic evidence indicates that CSF stimulates expression of genes activated by ComA∼P by inhibiting the activity of the phosphatase RapC (32). CSF stimulates sporulation (14, 32), in part, by inhibiting activity of the phosphatase RapB, a negative regulator of the phosphorelay required for the initiation of sporulation (23). It is not known how CSF inhibits ComA-controlled gene expression, but we suspect that it might inhibit activity of the histidine protein kinase ComP. Measuring production of CSF during cell growth, we found that CSF accumulates in culture medium, reaching concentrations approaching 100 nM. These concentrations are in the range of those previously shown to cause the three different regulatory responses (14, 32).
RapC and CSF are part of an autoregulatory loop.
We found that both rapC and phrC affect their own expression by affecting transcription from the upstream operon promoter P1 that is activated by ComA∼P. RapC and PhrC (CSF) appear to accomplish this by affecting the level of ComA∼P (Fig. 1). A rapC null mutation causes increased expression from P1 (and other ComA∼P-activated promoters [32]), indicating that RapC negatively regulates its own transcription, creating a homeostatic loop. At low cell densities, CSF is secreted and diluted into the culture medium, causing the extracellular CSF concentration to be low. When CSF levels are too low to inhibit RapC activity, RapC should inhibit accumulation of ComA∼P, thus decreasing transcription from the ComA∼P-controlled rapC promoter. As the concentration of RapC falls, transcription from the rapC promoter would increase. This homeostatic loop should cause the level of RapC to remain relatively constant while cells are at low densities and extracellular CSF levels are below those that inhibit RapC activity.
A null mutation in phrC causes decreased expression from the upstream operon promoter P1 (and other promoters activated by ComA∼P [32]), indicating that CSF positively regulates its own synthesis. As cells grow and become more dense, the extracellular concentration of CSF increases. This is partly because the number of cells making CSF increases and partly because ComX pheromone is accumulating in the culture medium, activating ComA∼P and, in turn, activating transcription of phrC. The increased concentrations of CSF should inhibit the phosphatase activity of RapC, increasing the level of ComA∼P and thus transcription of the rapC phrC operon. It may be that once a level of CSF capable of partially inhibiting RapC is achieved, more CSF is synthesized to further inactivate RapC and commit cells to full activation of ComA∼P-controlled genes.
When added exogenously to cells at low cell density, concentrations of CSF between 2 and 5 nM cause maximal stimulation of ComA∼P-controlled gene expression (14, 32). In growing cultures, this concentration range is reached during mid-exponential growth, several generations before the onset of stationary phase. These levels of CSF, in combination with ComX pheromone, contribute to the activation of ComA∼P-controlled gene expression during exponential growth. One of the operons activated by ComA∼P, srfA, then contributes to activation of the competence transcription factor ComK and competence development (6, 35, 36).
As cells enter stationary phase, the concentration of CSF reaches a level that inhibits expression of ComA∼P-controlled genes and stimulates sporulation. This inhibition is likely a reflection of an activity of the CSF pentapeptide and not due to an impurity. The inhibitory activity is found in the CSF pentapeptide synthesized chemically and in CSF purified from culture supernatants (32). Furthermore, single alanine substitutions at each position of CSF results in some peptides that retain and others that lose inhibitory activity (14). In addition, expression of one of the alanine mutant peptides in vivo from a synthetic gene encoding a six-amino-acid peptide that is predicted to have the N-terminal formylmethionine removed, results in production of an intracellular peptide that inhibits expression of ComA-controlled genes (14).
The significance of this inhibitory activity of CSF for ComA∼P-controlled genes is unclear. phrC mutants appear to have decreased expression of ComA∼P-controlled genes at all phases of growth, based on decreased expression of promoter fusions to lacZ (Fig. 3B) (14, 32). Isolation of a mutant defective in inhibiting ComA∼P-controlled genes in response to CSF will help dissect the role of this activity of CSF in the regulation of ComA∼P-controlled genes.
Transcriptional and posttranscriptional control of CSF production by ςH.
Transcription of phrC is controlled, in part, by RNA polymerase containing the alternate sigma factor ςH (Fig. 2A). ςH is present at low levels during exponential growth, and its concentration increases upon entry into stationary phase (9, 40). The increase in active ςH during the transition to stationary phase is most likely what causes increased transcription from the phrC promoter P2. The extracellular concentrations of CSF shortly after entry into stationary phase (when ςH is most active) were as high as 50 to 100 nM, concentrations at which CSF stimulates sporulation.
In addition to ςH’s role in transcription of phrC, our results indicate that it has at least one other role in production of CSF. The levels of CSF produced in a spo0H mutant were reduced much more severely than can be explained by the effect of ςH on transcription of phrC. This indicates that ςH affects transcription of at least one other gene required for production (translation, processing, and secretion) of the CSF peptide. PhrC has a putative signal sequence and peptidase cleavage sites, indicating that a secreted pre-CSF peptide of between 11 and 25 amino acids is secreted by the Sec-dependent pathway (24). Pre-CSF is then processed to the mature five-amino-acid form. It seems likely that there is a specific peptidase that recognizes and cleaves pre-CSF. Such a peptidase would be a good candidate for a ςH-controlled gene product required for CSF production. The small amount of CSF made in a spo0H mutant could be due to processing of pre-CSF by a nonspecific or alternative peptidase or to processing by lower amounts of the normal peptidase. The additional gene(s) involved in production of mature CSF is probably also controlled by spo0A and abrB, as these genes also have much greater effects on accumulation of extracellular CSF than they do on transcription of phrC. It will be interesting to determine if genes involved in production of the CSF pentapeptide are also involved in production of other peptide signaling factors.
ACKNOWLEDGMENTS
We thank members of the laboratory for useful discussions, and especially K. P. Lemon, T. Palmer, and K. Bacon, for comments on the manuscript.
B.A.L. was supported, in part, by fellowship DRG1384 from the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation, and this work was supported in part by Public Health Service grant GM50895 from the NIH.
REFERENCES
- 1.Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 2.Carter H L, III, Tatti K M, Moran C P., Jr Cloning of a promoter used by sigma-H RNA polymerase in Bacillus subtilis. Gene. 1991;96:101–105. doi: 10.1016/0378-1119(90)90347-t. [DOI] [PubMed] [Google Scholar]
- 3.D’Souza C, Nakano M M, Zuber P. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:9397–9401. doi: 10.1073/pnas.91.20.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grossman A D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 5.Hahn J, Dubnau D. Growth stage signal transduction and the requirements for srfA induction in development of competence. J Bacteriol. 1991;173:7275–7282. doi: 10.1128/jb.173.22.7275-7282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn J, Kong L, Dubnau D. The regulation of competence transcription factor synthesis constitutes a critical control point in the regulation of competence in Bacillus subtilis. J Bacteriol. 1994;176:5753–5761. doi: 10.1128/jb.176.18.5753-5761.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamoen L W, Eshuis H, Jongbloed J, Venema G, van Sinderen D. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol Microbiol. 1995;15:55–63. doi: 10.1111/j.1365-2958.1995.tb02220.x. [DOI] [PubMed] [Google Scholar]
- 8.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd.; 1990. [Google Scholar]
- 9.Healy J, Weir J, Smith I, Losick R. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor sH in Bacillus subtilis. Mol Microbiol. 1991;5:477–487. doi: 10.1111/j.1365-2958.1991.tb02131.x. [DOI] [PubMed] [Google Scholar]
- 10.Hoch J A. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 11.Ireton K, Rudner D Z, Siranosian K J, Grossman A D. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 12.Jaacks K J, Healy J, Losick R, Grossman A D. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazazzera B A, Grossman A D. The ins and outs of peptide signaling. Trends Microbiol. 1998;6:288–294. doi: 10.1016/s0966-842x(98)01313-4. [DOI] [PubMed] [Google Scholar]
- 14.Lazazzera B A, Solomon J M, Grossman A D. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 15.Magnuson R, Solomon J, Grossman A D. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 16.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 17.Mueller J P, Bukusoglu G, Sonenshein A L. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J Bacteriol. 1992;174:4361–4373. doi: 10.1128/jb.174.13.4361-4373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano M M, Magnuson R, Myers A, Curry J, Grossman A D, Zuber P. srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J Bacteriol. 1991;173:1770–1778. doi: 10.1128/jb.173.5.1770-1778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano M M, Xia L, Zuber P. Transcription initiation region of the srfA operon, which is controlled by the ComP-ComA signal transduction system in Bacillus subtilis. J Bacteriol. 1991;173:5487–5493. doi: 10.1128/jb.173.17.5487-5493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano M M, Zuber P. Cloning and characterization of srfB, a regulatory gene involved in surfactin production and competence in Bacillus subtilis. J Bacteriol. 1989;171:5347–5353. doi: 10.1128/jb.171.10.5347-5353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano M M, Zuber P. The primary role of ComA in establishment of the competent state in Bacillus subtilis is to activate expression of srfA. J Bacteriol. 1991;173:7269–7274. doi: 10.1128/jb.173.22.7269-7274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perego M. Integration vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive organisms. Washington, D.C: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 23.Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci USA. 1997;94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perego M, Glaser P, Hoch J A. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system of Bacillus subtilis. Mol Microbiol. 1996;19:1151–1157. doi: 10.1111/j.1365-2958.1996.tb02460.x. [DOI] [PubMed] [Google Scholar]
- 25.Perego M, Hanstein C, Welsh K M, Djavakhishvill T, Glaser P, Hoch J A. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 26.Perego M, Hoch J A. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perego M, Spiegelman G B, Hoch J A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 28.Roggiani M, Dubnau D. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J Bacteriol. 1993;175:3182–3187. doi: 10.1128/jb.175.10.3182-3187.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serror P, Sonenshein A L. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J Bacteriol. 1996;178:5910–5915. doi: 10.1128/jb.178.20.5910-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slack F J, Serror P, Joyce E, Sonenshein A L. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol. 1995;15:689–702. doi: 10.1111/j.1365-2958.1995.tb02378.x. [DOI] [PubMed] [Google Scholar]
- 31.Solomon J M, Grossman A D. Who’s competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 1996;12:150–155. doi: 10.1016/0168-9525(96)10014-7. [DOI] [PubMed] [Google Scholar]
- 32.Solomon J M, Lazazzera B A, Grossman A D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 33.Solomon J M, Magnuson R, Srivastava A, Grossman A D. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 1995;9:547–558. doi: 10.1101/gad.9.5.547. [DOI] [PubMed] [Google Scholar]
- 34.Triezenberg S J. Primer extension. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1992. pp. 4.8.1–4.8.5. [Google Scholar]
- 35.van Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, Hamoen L. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol Microbiol. 1995;15:455–462. doi: 10.1111/j.1365-2958.1995.tb02259.x. [DOI] [PubMed] [Google Scholar]
- 36.van Sinderen D, Venema G. comK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J Bacteriol. 1994;176:5762–5770. doi: 10.1128/jb.176.18.5762-5770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasantha N, Freese E. Enzymes changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J Bacteriol. 1980;144:1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinrauch Y, Guillen N, Dubnau D. Sequence and transcription mapping of Bacillus subtilis competence genes comB and comA, one of which is related to a family of bacterial regulatory determinants. J Bacteriol. 1989;171:5362–5375. doi: 10.1128/jb.171.10.5362-5375.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinrauch Y, Penchev R, Dubnau E, Smith I, Dubnau D. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 1990;4:860–872. doi: 10.1101/gad.4.5.860. [DOI] [PubMed] [Google Scholar]
- 40.Weir J, Predich M, Dubnau E, Nair G, Smith I. Regulation of spo0H, a gene coding for the Bacillus subtilis ςH factor. J Bacteriol. 1991;173:521–529. doi: 10.1128/jb.173.2.521-529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]