Abstract
Wolfram syndrome (WS) is a rare genetic disease characterized by diabetes, optic atrophy and deafness. Patients die at 35 years of age, mainly from respiratory failure or dysphagia. Unfortunately, there is no treatment to block the progression of symptoms and there is an urgent need for adequate research models. Here, we report on the phenotypical characterization of two loss-of-function zebrafish mutant lines: wfs1aC825X and wfs1bW493X. We observed that wfs1a deficiency altered the size of the ear and the retina of the fish. We also documented a decrease in the expression level of unfolded protein response (UPR) genes in basal condition and in stress condition, i.e. after tunicamycin treatment. Interestingly, both mutants lead to a decrease in their visual function measured behaviorally. These deficits were associated with a decrease in the expression level of UPR genes in basal and stress conditions. Interestingly, basal, ATP-linked and maximal mitochondrial respirations were transiently decreased in the wfs1b mutant. Taken together, these zebrafish lines highlight the critical role of wfs1a and wfs1b in UPR, mitochondrial function and visual physiology. These models will be useful tools to better understand the cellular function of Wfs1 and to develop novel therapeutic approaches for WS.
Introduction
Wolfram syndrome (WS) is an autosomal recessive genetic disorder characterized by diabetes mellitus, diabetes insipidus, optic nerve atrophy, hearing loss and neurodegeneration (1). This disease is progressive and severe, leading to the premature death of affected individuals around 35 years of age, with severe neurological disabilities, including bulbar dysfunction and organic brain syndrome (2). More than 200 variants in the WFS1 gene, which encodes the endoplasmic reticulum (ER)-resident glycoprotein Wolframin (WFS1) (3,4), have been associated so far with the disease (5).
The unfolded protein response (UPR) is an adaptive response to ER stress, with activation of RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK), activating transcription factor 6 (ATF6) and/or inositol-requiring kinase 1 (IRE1), preventing the accumulation of stress-induced unfolded proteins and improving ER homeostasis (6). Under ER stress, ATF6 dissociates from WFS1 and is cleaved and released from the ER to regulate stress signaling targets in the nucleus. As ER homeostasis is restored, WFS1 expression is induced by ER stress, which results in the degradation of ATF6 (7). In WS, WFS1 is not functional. ATF6 is no longer degraded via WFS1 and is therefore overactivated, regardless of emergency stress conditions, leading to the death of pancreatic β cells (8).
WFS1 also plays a critical role in calcium homeostasis regulation. Under physiological conditions, WFS1 interacts with the Ca2+ sensor NCS1 (9) and prevents its degradation, both proteins forming a complex with the inositol-1,4,5 triphosphate receptor (IP3R) to activate ER-Ca2+ transfer to mitochondria. When the tripartite WFS1/NCS1/IP3R complex and VDAC1 are functional, Ca2+ can properly transfer from the ER to the mitochondria and activate the tricarboxylic acid cycle and mitochondrial oxidative respiration. We have previously shown that under WS pathophysiological conditions, the WFS1/NCS1/IP3R complex loses its effectiveness, leading to the degradation of NCS1 and a decrease in ER-Ca2+ transfer to mitochondria, triggering in fine cell death (9,10).
Currently, no treatment for WS is available and patients have only access to medical devices that would improve their quality of life. The urgency to find a cure is obvious. Cellular (11–13) and murine models (14–17) have been developed to reproduce various deficits observable in humans and, therefore, to understand the pathological mechanisms and alleviate the alterations with active molecules.
In recent years, the zebrafish (Danio rerio) has become an alternative research model. It offers a unique combination of different characteristics, including short generation time, transparency, small size and very efficient breeding methods, and its genome is very close to that of humans. These properties make it ideal for studying genes associated with human diseases (18–20), particularly neurodegenerative diseases and neurological disorders (21–24). They also confer great advantages to the zebrafish model compared with other models for finding new treatments, most particularly for testing a very large number of potential therapeutic molecules quickly and at lower cost as well as directly testing on a whole living organism with all its complexity (25,26).
We here characterized two zebrafish lines that carry stop codon mutations in orthologs of the WFS1 gene, which are responsible for WS. Their precise phenotyping was necessary to better understand the physiopathology and to realize their usefulness as a screening model for the validation of novel therapeutic strategies, such as repurposed drugs, ER stress chaperones or potential mitoprotectants. As the zebrafish genome is duplicated, the impact of wfs1 mutations in both copies of the gene, wfs1a and wfs1b, could be addressed using two mutant lines: wfs1aC825X and wfs1bW493X. We investigated, at the behavioral level, their roles in sensory functions such as vision or hearing and locomotion and, at the cellular level, their impact on metabolic markers like ER stress factors, glycemia and mitochondrial function.
Results
Morphology and grossly observable phenotypes of wfs1a and wfs1b mutant zebrafish lines
Various mutant lines for wfs1a and wfs1b were generated through ENU mutagenesis (27). For each gene, the line carries a premature stop codon, leading, potentially, to a non-functional protein. We selected the wfs1aC825X (sa11465) and wfs1bW493X (sa16422) lines (Fig. 1A) in which point mutations lead to a premature stop codon and potentially to a non-functional protein. These mutations abolished restriction sites for MwoI and BsrI, respectively, allowing each genotype to be validated by PCR followed by enzymatic digestion (Supplementary Material, Fig. S1A). Sanger sequencing confirmed the non-sense mutations (Supplementary Material, Fig. S1B).
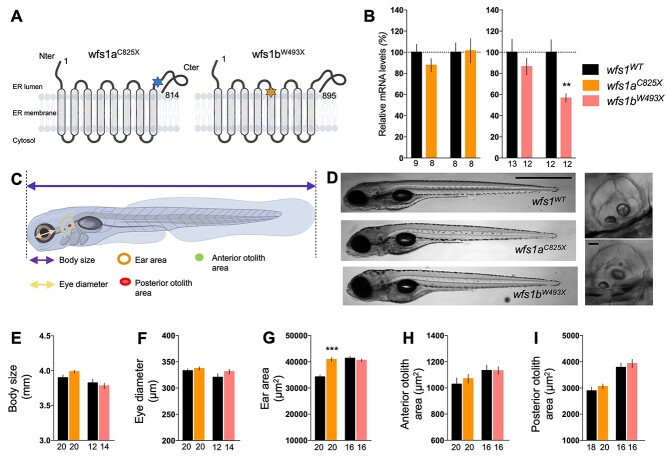
Figure 1.
Characterization of wfs1 mutant zebrafish lines. (A) Schematic molecular structure of Wfs1a and Wfs1b proteins with mutation. The stars represent the punctual mutations, blue for Wfs1a mutation and orange for Wfs1b. (B) Relative wfs1a and wfs1b mRNA levels assessed by qPCR in wfs1aC825X and wfs1bW493X zebrafish larvae, normalized against zef1α reference gene. (C) Schematic representation of the different measurements of the larva. (D) Representative images of wild-type and homozygous mutant wfs1 larvae. Images to the right show the ear in the wild-type (upper image) and wfs1aC825X mutant (the lower image). (E) Measurement of body size, (F) eye diameter, (G) ear area in lateral view, (H) anterior and (I) posterior otoliths area. Scale bars, 200 μm in (D), 10 μm in the insets. Error bars represent mean ± SEM and the number of fish is indicated below the columns. **P < 0.01, ***P < 0.0001; unpaired t-test.
In order to determine if the mutations induced mRNA decay, relative wfs1a and wfs1b mRNA levels were measured by quantitative real-time PCR (qPCR) in both mutant lines (Fig. 1B). Surprisingly, neither wfs1a nor wfs1b gene expression was altered in wfs1aC825X mutants, whereas both genes were downregulated in wfs1bW493X fish, suggesting a genetic interaction of wfs1b on the wfs1a gene. The protein expression level of Wfs1 could not be assessed as no effective antibodies specifically labeling the zebrafish protein are presently available. However, owing to the premature stop codons, we predicted that both proteins, when expressed, are not functional.
In order to determine if mutations in wfs1 have an impact on zebrafish larval morphology, different parameters, detailed in Figure 1C, were measured at 5 dpf using a binocular magnifier (Fig. 1D) and were analyzed using Fiji software (Fig. 1E–I). Overall, the mutations did not impact the observable gross morphology of the larva, with the exception of the enlarged ear area for wfs1aC825X compared with controls (Fig. 1D and G).
wfs1a and wfs1b are expressed in distinct but overlapping domains during zebrafish development
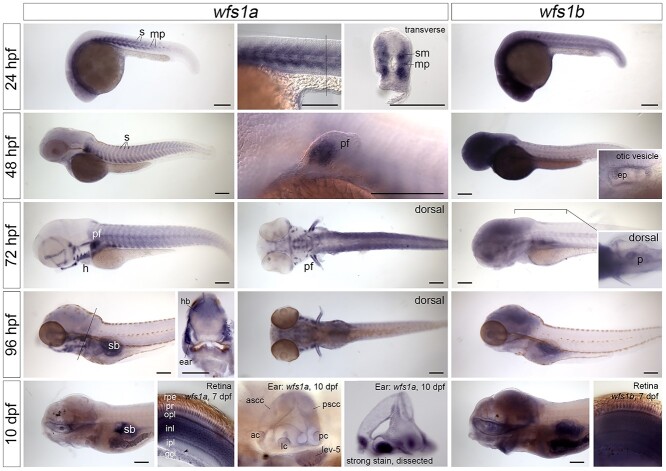
To characterize the mRNA expression patterns of wfs1a and wfs1b during development, two antisense RNA probes for each gene were generated and were used for in situ hybridization on zebrafish embryos, larvae and juveniles at different developmental stages (Fig. 2). At early stages [24 h post-fertilization (hpf)], expression patterns of the two paralogs were quite distinct, with specific expression of wfs1a in developing skeletal muscle, whereas expression of wfs1b was more ubiquitous. Transverse sections through the trunk at 24 hpf showed wfs1a expression in the developing somites, most strongly in the slow-twitch muscle fibers and muscle pioneer cells. Strong expression in muscles of the pectoral fin was evident from 48 hpf. Expression in the craniofacial skeletal muscles, including those articulating the eye and the jaw, was present from 72 hpf. By 96 hpf, the expression of wfs1a in trunk somitic muscle was reduced, but a strong expression in craniofacial skeletal muscle was maintained. Weak expression of wfs1a was also present in the heart (most likely myocardium) from 72 hpf, and in the mouth, swimbladder, gut and brain from 96 hpf. Expression of wfs1b was ubiquitous at early stages, with strong expression in the central nervous system (brain and retina), persisting in the brain up to 10 days post-fertilization (10 dpf). From 48 hpf, expression of wfs1b was also present in the exocrine pancreas, which was visible in a dorsal view as a characteristic comma-shaped organ on the right-hand side of the body (see Fig. 2, inset). At later stages, there were several regions of overlap in the expression patterns of wfs1a and wfs1b. Both genes were expressed in the retina at 7 dpf, with wfs1a showing different levels of expression in the different retinal cell layers. Both genes were also weakly expressed in otic epithelium, including in the epithelial projections that lead to the formation of the semicircular canals at 48 hpf (see inset). Using an adapted staining protocol (see Materials and Methods), we detected expression of both zebrafish genes throughout the brain, but not the spinal cord, at 7 and 10 dpf. In the brain, expression marked the cell bodies but not the areas of neuropil (Supplementary Material, Fig. S2). Other sites of co-expression included the somites, pectoral fin and lens of the eye at 24–48 hpf and the brain, swimbladder, gut and gills at 7–10 dpf (Fig. 2, Supplementary Material, Fig. S2).
Figure 3.
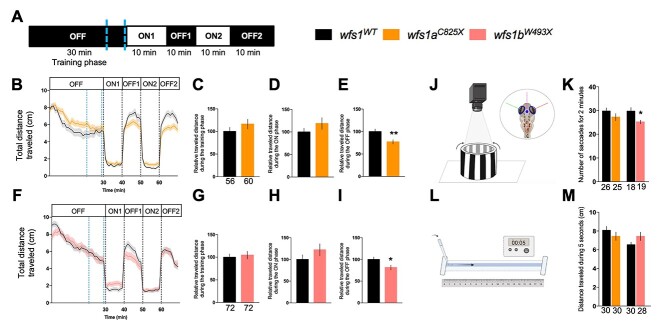
Behavioral analyses of 5 dpf wfs1 mutant larvae. (A) Light protocol: the activity is measured for 70 min, with 30 min of training in the dark (OFF), then two cycles of light/dark (ON/OFF) of 10 min each. Analysis of the distance traveled by (B) the wfs1aC825X and (F) wfs1bW493X larvae at 5 dpf during the light/dark cycle in the VMR test. Relative distance traveled was measured during some experiment phases as (C, G) the training phase [blue-dotted lines in (B, F) between 21 and 29 min]; (D, H) the ON phase (the averaged ON1 and ON2 phases); (E, I) the OFF phase (the averaged OFF1 and OFF2 phases). (J) Illustration of the OKR test. Four larvae are immobilized in petri dish and placed in an arena with moving black and white gratings. (K) The number of saccades performed in 2 min. (L) Illustration of the touch escape test. The larva’s tail is touched with a tip and (M) the traveled distance of the larva in the rail is measured for 5 s, repeated for three times per larva and averaged. Relative distances were expressed as % of associated controls. Error bars represent ± SEM calculated from three replicas. The number of animals is indicated below the columns. *P < 0.05; **P < 0.01; unpaired t-test.
Figure 2.
Expression of wfs1a and wfs1b mRNA in the zebrafish embryo, larva and juvenile. In situ hybridizations showing the expression patterns of wfs1a (left-hand and middle columns) and wfs1b (right-hand column) between 24 hpf and 10 dpf. All images are lateral views with anterior to the left, except for the inset showing expression of wfs1b in the pancreas (dorsal view, anterior to the left) and the sections through the retina. The position of the transverse hand-cut sections through the trunk at 24 and 96 hpf (dorsal to the top) is marked by a thin line in the preceding image. The eyes have been removed from the 10 dpf embryos, to show expression in the mouth and brain. The hand-cut thick sections through the retina, and the second image of the ear at 10 dpf, were taken from samples stained using an alternative protocol that resulted in stronger staining intensity (see Materials and Methods). Expression of wfs1a in the retina at 7 dpf is strong in the photoreceptors, outer plexiform and inner nuclear layers but is weaker or missing from the inner plexiform and ganglion cell layers (GCLs) and is very weak or absent from displaced amacrine cells (arrowhead). Expression of wfs1b in the retina at 7 dpf appears ubiquitous. Expression in the inner ear appears ubiquitous for both genes and may include some trapping. All images are bright-field images apart from the images of the trunk at 24 hpf, pectoral fin at 48 hpf and retinae at 7 dpf, which were taken with DIC optics. Abbreviations: ac, anterior crista; ascc, anterior semicircular canal; ep, epithelial projections in the otic vesicle; gcl, ganglion cell layer; h, heart; hb, hindbrain; inl, inner nuclear layer; ipl, inner plexiform layer; lc, lateral crista; lev-5, levator arcus branchialis 5; mp, muscle pioneer cells; opl, outer plexiform layer; p, pancreas; pc, posterior crista; pf, pectoral fin; pr, photoreceptors; pscc, posterior semicircular canal; rpe, retinal pigmented epithelium; s, somites; sb, swimbladder; sm, slow-twitch muscle fibers. Scale bars, 200 μm in all images.
The wfs1 mutations impacted visual and locomotor functions of zebrafish larvae
The locomotor response of larvae after visual stimulation was analyzed using the visual motor response (VMR) assay. At 5 dpf, larvae were placed individually in the wells of a 96-well plate and their individual activity was measured according to the described protocol. A training phase of 30 min in the dark (OFF, 0% light) allowed the larva to acclimate to its new environment, followed by two sequences of light (ON, 100% light, 10 min) and dark periods (OFF, 0% light, 10 min) (Fig. 3A). For both lines, the traveled distance profiles are shown in Figure 3B and F; and the quantification is shown in Figure 3C and G for the training phases; in Figure 3D and H, for the ON periods; and in Figure 3E and I, for the OFF periods. In the wfs1aC825X (Fig. 3B–E) and wfs1bW493X mutant lines (Fig. 3F–I), the relative distance traveled during the training (Fig. 3C and G) and ON (Fig. 3D and H) phases were not altered. However, the distance traveled during the OFF period (Fig. 3E and I) phase was significantly decreased in both wfs1a and wfs1b mutant larvae.
Figure 5.
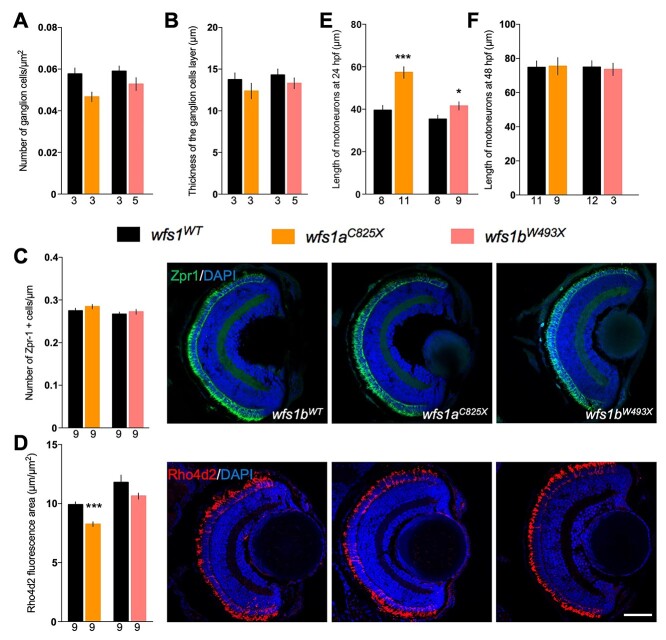
Molecular characterization of the retina and measure of the motor neuron length for both wfs1 mutant zebrafish lines. Retina measurements in wfs1 mutants with (A) quantification of the number of ganglion cells, (B) the thickness of the associated layer and (C) photoreceptors cells as red and green cones labeled with Zpr-1 antibody and (D) rods with anti-Rho4d2 antibody. Quantification of the tail motor neuron length of 24 hpf (E) and 48 hpf (F) zebrafish. For each fish, five to six neurons were measured, and the average length was calculated. Scale bar, 50 μm. The number of animals is indicated below the columns. *P < 0.05; ***P < 0.001; unpaired t-test.
The visual acuity of the two wfs1 mutant zebrafish lines was measured using the optokinetic response (OKR) assay. The eye movement of the larva was induced by the rotation of black-and-white stripes retroprojected on a fixed cylinder (Fig. 3J). The number of saccades in 2 min reflected the visual acuity of the immobilized larva. The wfs1a mutant larvae did not show alteration in the number of saccades, but wfs1b mutants showed a significant decrease (Fig. 3K), suggesting a specific visual deficit in this line.
A reflex motor response was also measured using the touch-evoked escape behavior (Fig. 3L). After manual stimulation, none of the mutant line exhibited an alteration of the distance traveled, suggesting an absence of motor alteration (Fig. 3M).
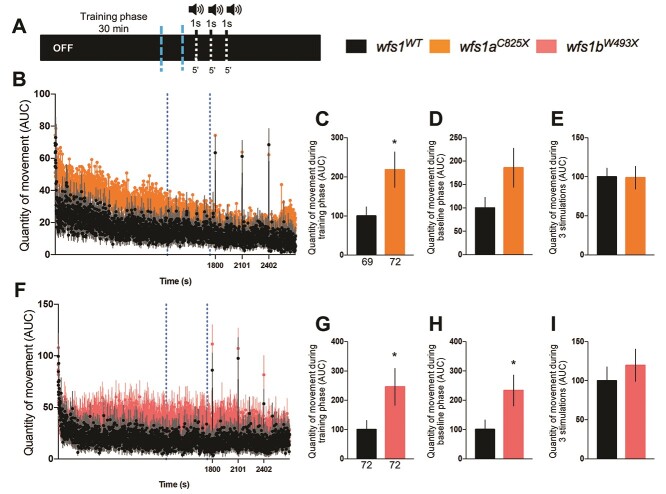
The acoustic startle response (ASR) of the larvae, induced by a repetitive 1 s duration noise (Fig. 4A), was assessed by measuring the quantity of movement in both wfs1 mutant larvae (Fig. 4B and F). The quantity of movement during the training and baseline phases was increased in both wfs1 mutant lines compared with controls (Fig. 4C, D, G and H). The wfs1 mutations had, however, no impact on the quantity of movement after stimulation in either wfs1 mutant line (Fig. 4E and I).
Figure 6.
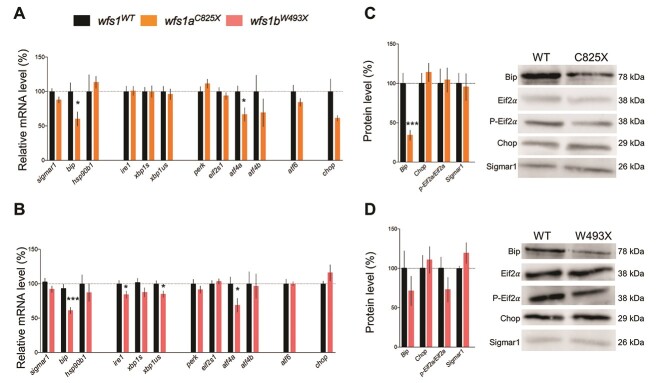
Relative mRNA and protein expression levels of larvae zebrafish ER stress factors in physiological condition at 5 dpf. mRNA levels were analyzed by qPCR and protein contents by western blot in (A, C) wfs1aC825X and (B, D) wfs1bW493X. zeif2α and Stain-free™ were used as controls in qPCR and western blot analysis, respectively. The expression levels of mRNA and protein are represented as the fold change from the control group. Data are expressed in mean ± SEM, n = 5–9 in each group. *P < 0.05, ***P < 0.001; unpaired t-test.
Figure 4.
Analysis of the quantity of movement of 5 dpf wfs1 mutant larvae during the noise cycle in the ASR test. (A) Sound protocol: the activity is measured for 45 min in a total dark (OFF) condition, with a training phase for 30 min silently, then three cycles of white sounds (90 dB) of 1 s each and 5 min interspersed. Quantity of movement per second for (B) the wfs1aC825X and (F) wfs1bW493X larvae according to the sound protocol. Relative quantity of movement during (C, G) the training phase [blue-dotted lines in (B, F) between 21 and 29 min]; (D, H) the baseline phase, period of 2 min before each sound (the averaged three baseline phases); (E, I) the three stimulations (the averaged sounds phases). Activity was expressed as % of associated controls. Error bars represent ± SEM calculated from three replicas. The number of animals is indicated below the columns. *P < 0.05; unpaired t-test.
The wfs1aC825X mutation disrupted eye development of zebrafish larvae
Immunohistochemical analysis of the developing eye was performed for both wfs1a and wfs1b lines (Fig. 5, Supplementary Material, Fig. S3). At 5 dpf, we observed a trend to a decrease in the number of retinal ganglion cells in wfs1aC825X compared with wfs1aWT controls (Fig. 5A, Supplementary Material, Fig. S3) without alteration of the thickness of the cell layer (Fig. 5B, Supplementary Material, Fig. S3). The number of red and green cones, labeled with Zpr-1 antibody, remained the same between mutant and control larvae (Fig. 5C). In contrast, the number of rods, labeled with Rho4d2 antibody, was significantly decreased (Fig. 5D).
Figure 7.
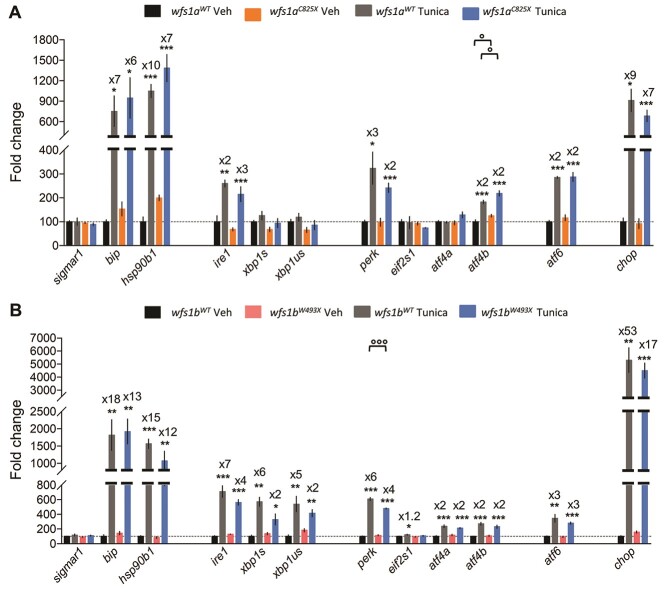
Relative gene expression levels of ER stress factors in zebrafish larvae at 5 dpf exposed to tunicamycin during 24 h. Expression analysis of the selected genes using cDNA prepared from wfs1aC825X (A) and wfs1bW493X (B) zebrafish larvae. The relative expression levels of mRNA are represented as the fold change from the control vehicle group. Data are expressed in mean ± SEM, n = 5 in each group. *P < 0.05, **P < 0.01, ***P < 0.001 versus DMSO condition, °P < 0.05, °°°P < 0.001 versus WT condition; two-way ANOVA, unpaired t-test.
The wfs1 mutations disrupted motor neuron length of 24 hpf zebrafish larvae
At 24 and 48 hpf, the immunohistochemical analysis of the developing motor neurons was performed for both wfs1a and wfs1b lines using SV2 antibodies (Fig. 5E and F). At 24 hpf, we observed a statistically significant increase in the length of wfs1aC825X and wfs1bW493X larvae motor neurons compared with their respective controls (Fig. 5E). This difference is no longer present at 48 hpf (Fig. 5F).
The wfs1 mutations affected the expression of ER stress genes
The expression levels of genes involved in the regulation cascade of ER stress were analyzed by qPCR in both mutant zebrafish lines (Fig. 6A and B). Expression levels of the UPR pathway inducer, hsp90b1, were not modified in mutant larvae in all zebrafish lines, whereas bip mRNA levels were decreased in wfs1aC825X and wfs1bW493X compared with the controls (Fig. 6A and B). Sigmar1 mRNA level was unchanged in either mutant line. Among the primary effectors of the UPR pathway, the expression of ire1 was decreased only in wfs1bW493X compared with WT controls, while perk, atf6 and chop were unchanged in both mutant lines. Among the secondary effectors, xbp1us was decreased in wfs1bW493X mutants compared with the controls (Fig. 6B) and atf4α was decreased in both wfs1aC825X and wfs1bW493X, while xbp1s, eif2s1 and atf4β were unchanged in both lines.
Western blot analyses showed that the Bip protein level was significantly decreased in wfs1aC825X line and non-significantly altered in wfs1bW493X compared with controls, while there was no impact of wfs1 mutations on expression levels of p-Eif2α/Eif2α and Sigmar1 (Fig. 6C and D).
The wfs1 mutations impact the tunicamycin-induced ER stress response
Larvae were treated with 2 μg/ml of tunicamycin for 24 h to analyze the response of wfs1 mutant lines to ER stress (Fig. 7). The mRNA levels of bip, hsp90b1, ire1, perk, atf4b, atf6 and chop (Fig. 7A and B) were drastically increased in both mutant and control lines after the tunicamycin treatment, with the exception of eif2s1 and sigmar1 (Fig. 7A and B), which were unchanged with the treatment regardless of the genotype.
Interestingly, the genotype has an impact on the ER stress response to tunicamycin treatment. In the wfs1a mutant line, primary sensors ire1, perk and atf6 were increased but not the secondary effectors (Fig. 7A). In the wfs1b mutant line, perk was less induced by the tunicamycin treatment, but secondary effectors of both the ire1 and perk pathways were significantly induced by tunicamycin (Fig. 7B). Taking into account the percentages of induction provoked by tunicamycin for each effector in the mutant and control lines, specified in Figure 7A and B, we summarized the observations in synthetic schemes presented for the wfs1aC825X and wfs1bW493X mutant lines. It appeared that UPR is marginally affected in the wfs1aC825X mutant line (Fig. 8A), while both ire1- and perk-dependent pathways are altered in the wfs1bW493X mutant line (Fig. 8B). These observations suggested that wfs1 participated in ER stress regulation and that the wfs1b mutant zebrafish line is an adequate model to analyze the protein involvement in UPR (Fig. 8).
Figure 8.

Schematic summary of ER stress pathways alterations observed in basal condition or after tunicamycin treatment in wfs1aC825X (A) and wfs1bW493X (B) zebrafish line.
Only wfs1b mutants showed a transient mitochondrial defect
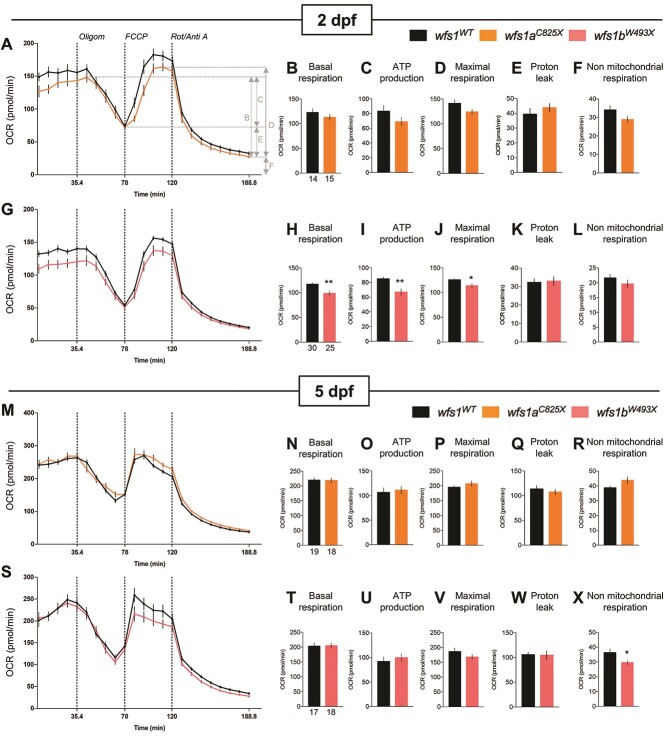
Mitochondrial oxidative respiration rates were measured in zebrafish larvae to determine the consequences of wfs1 mutations on mitochondrial function at two developmental ages (2 and 5 dpf) (Fig. 9). The oxygen consumption rate (OCR) was measured before and after the addition of inhibitors (the ATP synthase inhibitor oligomycin; the uncoupling agent FCCP; antimycin A and rotenone) (Fig. 9A, G, M and S) to derive several parameters of mitochondrial respiration (as shown in Fig. 9A). At 2 dpf, wfs1a mutant larvae showed no alteration of basal respiration (Fig. 9B), ATP production (Fig. 9C), maximal respiration (Fig. 9D), proton leak (Fig. 9E) or non-mitochondrial respiration (Fig. 9F). wfs1b mutant larvae showed significant decreases in basal respiration (Fig. 9H), ATP production (Fig. 9I) and maximal respiration (Fig. 9J) but showed no change in proton leak (Fig. 9K) and non-mitochondrial respiration (Fig. 9L). This suggested that wfs1b mutant larvae showed a marked alteration of mitochondrial respiration.
Figure 9.
Analysis of mitochondrial respiration in zebrafish larvae at 2 and 5 dpf with Seahorse XF mito stress test. OCR profiles of (A–L) wfs1aC825X at 2 dpf (A–F) and 5 dpf (G–L); and OCR profiles of (M–X) wfs1bW493X at 2 dpf (M–R) and 5 dpf (S–X) during the assay, with (B, H) basal respiration, (C, I) ATP production, (D, J) maximal respiration, (E, K) proton leak and (F, L) non-mitochondrial respiration. Error bars represent ± SEM calculated from three replicas. The number of animals is indicated below the columns. *P < 0.05, **P < 0.01; unpaired t-test.
At 5 dpf, both wfs1a and wfs1b mutant larvae showed no alteration of basal respiration (Fig. 9N and T), ATP production (Fig. 9O and U), maximal respiration (Fig. 9P and V), proton leak (Fig. 9Q and W) and non-mitochondrial respiration (Fig. 9R), with the exception of a decrease in the non-mitochondrial respiration in wfs1bW493X larvae (Fig. 9X). This suggested that, at 5 dpf, both mutant lines retained a similar metabolic capacity as WT zebrafish.
Discussion
The aim of the study was to characterize novel zebrafish models of WS. The zebrafish genome contains two wfs1 genes, wfs1a and wfs1b. The wfs1a gene contains 10 exons and encodes a protein of 1061 amino acids, whereas the wfs1b gene contains 8 exons and encodes a protein of 895 amino acids. The human WFS1 gene contains 8 exons and encodes a protein of 890 amino acids that has a similarity to zebrafish wfs1a of 52.47% and to wfs1b of 52.80%. This suggested that wfs1b gene structure is closer to human WFS1 than wfs1a. We selected the wfs1aC825X and wfs1bW493X mutant lines to analyze the effect of a loss of function of each protein on the development of zebrafish larvae, their visual, auditory and locomotor functions and their impact on mitochondrial activity and the ER stress response. During our work, a complementary study to ours was reported by Cairns and colleagues (28), with the characterization of the same wfs1bW493X fish but with a different wfs1a mutant line: wfs1aW692X (sa10021). While the time points as well as the different wfs1a model studied make the direct comparison difficult, their results and ours provide an overall picture of wfs1 roles in zebrafish during development and adulthood.
Our study shows that the expression patterns of wfs1a and wfs1b genes are quite distinct and dynamic during early development, which may reflect different functions for the two zebrafish paralogs. Particularly striking is the muscle-specific expression of wfs1a, whereas the expression of wfs1b is more ubiquitous (Fig. 2, Supplementary Material, Fig. S2). This difference lasts in time as, by 4 months old, wfs1a is still highly expressed in muscle compared with wfs1b (28). Expression of Wfs1 in the mouse is also seen in a multitude of different tissues, including skeletal and cardiac muscle (29,30) (see also http://www.informatics.jax.org/gxd/marker/MGI:1328355). Wfs1-deficient mice show alterations in protein expression (31) and energy metabolism (32) in both muscle tissue types. In vitro assays have also demonstrated that the mammalian Wfs1 protein functions in the nuclear envelope to reposition muscle-specific genes during myogenesis (33). The respiratory failure or dysphagia in WS patients, however, is thought to involve a neurodegenerative rather than muscular pathology, as reviewed in Pallotta et al. (34). Dominant missense mutations in human WFS1 gene are also associated with a distinct Wolfram-like syndrome that includes hypotonia (35). Although there are no overt morphological muscle phenotypes in the single gene mutant zebrafish analyzed here, a metabolic defect in muscle could contribute to the locomotor deficits observed. It is also possible that the mild zebrafish phenotypes reflected transcriptional adaptation triggered by mutant mRNA decay (36), although we did not find evidence for any compensatory up-regulation of paralogous transcript levels by qPCR in the mutants.
Expression of the wfs1b gene in the zebrafish pancreas and of wfs1a and wfs1b genes in the eye and inner ear also appeared to be conserved with that of Wfs1 in the mammal. Expression of zebrafish wfs1b gene was present in the exocrine pancreas from 48 hpf to, at least, 10 dpf. More detailed sections and colocalization studies would be needed to determine expression in the endocrine pancreas and β cells. Apart from the early ubiquitous expression of wfs1b, neither gene showed strong expression in the ear or eye at early larval stages (3–4 dpf). However, both genes were expressed in the retina at 7 and 10 dpf (the last stage analyzed) and throughout the otic vesicle. In the retina, the expression of wfs1b appeared ubiquitous, whereas levels of wfs1a expression differed between the different cell layers. In adulthood, wfs1b expression is higher in the eye than wfs1a expression. However, these results of reverse transcriptase-PCR do not provide details of the specific sub-localization of each gene expression (28). In the mouse, expression of Wfs1 mRNA and protein has been reported in all neuronal cell types of the retina (37) and in many cell types of the inner ear (38), including the cochlea and organ of Corti, which have no direct counterpart in the zebrafish ear. Both genes were expressed in the brain at the stages we analyzed (24 hpf–10 dpf), with expression most evident for wfs1b at the earlier stages. Cairns et al. have reported that expression of wfs1b in the eye and brain is stronger than that of wfs1a in 4-month-old fish using qPCR (28).
WS is an emblematic disease characterized by optic atrophy, diabetes and neurosensory hearing loss (1). The wfs1a loss of function had no effect on the gross morphology of the zebrafish larva except for the size of the ear area that was increased compared with control. No difference in auditory function was observed using ASR, suggesting that the structural anomaly was not correlated to a functional one. Interestingly, a decrease in the size of the rod outer segments was observed in the retina, suggesting that rods degenerated or that the outer segment was shortened. Although these findings are outside the scope of most cases of WS, they are still pertinent to the human pathology. WFS1 expression was faintly detected in human photoreceptors as well as in other species, such as the Cynomolgus monkey (39) and mouse (37,40). In addition, some electroretinogram abnormalities have been reported in human, suggesting some deficit at the photoreceptor level (41). In wfs1a larvae, the morphological alteration correlated with a visual deficit, estimated by VMR.
Similar to what we observed with wfs1a larvae, wfs1b loss of function did not induce any gross morphological alteration. These results are in contrast with those reported by Cairns et al., who described shorter larvae for both lines in the early stages of the larvae (30, 50 and 80 hpf). While some alterations, like the head-trunk angle, were fully recovered by 80 hpf, it is not stated whether this difference is the consequence of a developmental delay or a permanent characteristic of the fish (28). Our study would suggest that the developmental defect is transient and limited to the early stages as, by 5 dpf (120 hpf), no morphological differences were noted.
Like wfs1a mutant larvae, wfs1b mutants have no auditory deficits when analyzed by ASR compared with control. No difference in the number of rods, cones or Retinal Ganglion Cells (RGCs) was observed. Nevertheless, their visual function was altered when measured by VMR and OKR. Indeed, wfs1aC825X and wfs1bW493X zebrafish mutants showed a decrease in their distance swum in the dark phase and, for wfs1bW493X larvae only, in their number of saccades, suggesting a deficit in their visual acuity. In addition, the absence of difference between the mutant lines during the touch escape assay suggested that this alteration is not linked to pure locomotor activity but rather to a pure retinal and/or to a retinal and retinotectal neural network impairment. Indeed, the retinotectal neural network connects RGCs to the optic tectum, a zebrafish brain structure that creates a visual map retinotopically (42).
Cairns and colleagues reported that both wfs1a and wfs1b mutant fish, at 4 and 12 months old, exhibited a decrease in the RGC density, which was associated with thinner RGC layers and ganglion cell layer (GCL). In addition, in the case of wfs1b fish, these morphological observations were correlated to a significantly decreased visual function, recorded by OKR (28). However, we cannot exclude that the absence of wfs1a function will not have an impact on the visual function as the size of the rod outer segments was decreased at 5 dpf as well as the RGC density was decreased at 4 and 12 months old. Taken together, these data suggest that the wfs1b line might undergo a more severe degenerative process leading to morphological and functional alteration earlier than wfs1a fish.
The absence of any gross morphological defect in our fish is also in favor of a retinal and/or retinotectal impairment rather than a muscular or locomotor anomaly. Like Cairns et al. (28), we observed a transient developmental defect of the length of the motor neurons of the tail in the 24 hpf larvae, which was completely restored as early as 48 hpf. However, contrary to their study where they reported a decrease in the motor neuron length of the wfs1b line (28), we observed an increase in the length of both mutant zebrafish larvae tail motor neurons. This discrepancy between our two studies might be explained by a difference in husbandry conditions (number of larvae/tank and feeding) or the region of the trunk that was analyzed in our study compared with Cairns. For wfs1aC825X, another possible explanation would be that since the mutation is not the same, it may induce a different phenotype. Nevertheless, since this defect is transitory as evidenced by the normal length of the motor neurons at 48 hpf, more work needs to be done in order to decipher the exact mechanism leading to this early and transient alteration.
Deafness is another hallmark of WS. In humans, hearing loss appears in 46% of the patients (43) in the second decade of life. In addition, in a recent study on a rat model of WS, null for WFS1, hearing loss was observed starting at 6.5 months old, in the higher frequencies, and worsened with age (44). Despite being a prominent feature of WS, none of the characterization studies of the several mouse models of the pathology report any hearing deficit, suggesting either a mild or non-existent hearing loss, or a late onset of the phenotype. In the case of the zebrafish models, it is difficult to determine if the absence of hearing loss is linked to the age of the larvae (5 dpf) compared with humans or if the structure/function of the zebrafish inner ear is too different to that of humans. A longitudinal study, with not only broadband but also frequency-specific stimuli, would help characterize more precisely the impact of the absence of each wfs1 gene copy on the auditory pathway.
WFS1 protein is involved in the regulation of the UPR. Indeed, it has been shown that WFS1 regulates the ubiquitin ligase HRD1 in order to control the protein level of ATF6. When WFS1 is deficient, HRD1 is less able to address ATF6 to the proteasome (11), leading to an increased activation of ATF6 and to an uncontrolled UPR. In our study, we observed that in basal condition, wfs1a gene loss of function impacted to some extent the expression level of bip, atf4α and chop, suggesting that the PERK pathway was altered. In contrast, in the same basal condition, wfs1b loss of function impacted the relative expression level of bip, atf4α, ire1 and xbp1us, suggesting that both the IRE1 and PERK pathways were altered. In stress condition, wfs1a gene loss of function marginally impacted the induction of only perk and atf4β, leading to a preserved UPR. The impact of ER stress was more marked in wfs1b mutants since the relative expression level of bip, ire1, xbp1s, xbp1us, perk, atf4α and chop was significantly or tended to be altered, confirming that both PERK and IRE1 pathways were affected.
In addition to its role in ER stress, WFS1 is involved in the regulation of the mitochondrial activity in human fibroblasts (9). Therefore, we took advantage of the capacity of the seahorse XFe24 to measure the OCR in the living zebrafish (45) and to decipher the impact of the loss of function of wfs1a and wfs1b genes on the different mitochondrial parameters. At 2 dpf, wfs1bW493X mutant larvae showed a decrease in basal, ATP-linked and maximal respiration. wfs1aC825X did not exhibit any respiration alteration compared with controls. Intriguingly, at 5 dpf, the only observable difference was a decrease in the non-mitochondrial respiration in wfs1bW493X mutants. These data were in agreement with the decrease in complex I and complex II activities previously reported in patients’ fibroblasts (9) but only in very early developmental stages. In addition, based on our in situ hybridization experiments, the expression of wfs1a seems to be more tissue-specific than that of wfs1b, suggesting a broader impact on mitochondrial physiology for the latter. We can hypothesize that the mitochondrial alteration in tissue(s) expressing wfs1a is diluted out in the whole organism experiment.
In conclusion, it appeared that wfs1a and wfs1b genes recapitulated most of the mammalian WFS1 roles. However, wfs1b may likely be closer to mammal WFS1 than the wfs1a ortholog in terms of cellular expression, function and physiopathology. Taken collectively, our data highlighted the potential of wfs1b mutant line as a relevant and promising model to identify a novel therapeutic solution.
Materials and Methods
Zebrafish lines and husbandry
The present study followed the recommendations of the ARRIVE guidelines (46) and the European Union Directive 2010/63. Both zebrafish (D. rerio) wfs1aC825X (sa11465) and wfs1bW493X (sa16422) lines were generated by N-ethyl-N-nitrosourea (ENU)-induced mutagenesis (Sanger Institute Zebrafish Mutation Project) (27) and were acquired from Zebrafish International Resource Center (University of Oregon, Eugene, OR, USA). A cysteine codon at amino acid position 825 has been replaced with a stop codon for the wfs1a line and tryptophan codon has been replaced with a stop codon at position 493 for the wfs1b line. Numbering is based on the following accession numbers (XM_690160.9; XP_695252.5) and (XM_679418.7; XP_684510.2) for wfs1a and wfs1b, respectively. Adult zebrafish were bred and maintained under standard conditions in an automated fish tank system (ZebTEC, Tecniplast) at 28°C, pH 7, conductivity around 500 mS and with a 14 h:10 h light:dark cycle. Eggs were obtained by natural spawning and were maintained in E3 medium (5 mm NaCl, 0.17 mm KCl, 0.33 mm CaCl2, 0.33 mm MgSO4, 0.05% methylene blue) at 28°C. Each experimental procedure was carried out in triplicate using larvae from three different crosses.
Genotyping
For isolated homozygous wfs1a and wfs1b mutant larvae, genomic DNA was extracted from tail fins at 2 months of age and the tissue was lysed in 50 mm NaOH at 95°C for 1 h. Fragments of genomic DNA for the wfs1a and wfs1b genes were amplified by PCR using GoTaq Green Master Mix (Promega, Madison, WI, USA) and the following primer sequences: wfs1a, 5′-TGCGACTCTGGACTCGATTTAG-3′ (forward); 5′-CGGATAAACCTCACCGTAGAGA-3′ (reverse); wfs1b, 5′-GGCTATGGTTGCAGTCTTCTTC-3′ (forward); 5′-GCTGAACCAGCATTACAGCTGC-3′ (reverse). These primers amplify a 392 bp region encompassing the wfs1a mutation site and a 455 bp region encompassing the wfs1b mutation site. These fragments were digested with the restriction enzymes MwoI for wfs1a DNA (ER1732, Thermo Fisher Scientific) and BsrI for wfs1b DNA (ER0881, Thermo Fisher Scientific). The DNA fragments were separated by electrophoresis on a 3% agarose gel and the genotypes were analyzed. The mutations were confirmed by Sanger sequencing.
In situ hybridization
In situ hybridization was performed on AB (ZSB-GENO-960809-7) or mifta−/− (nacrew2, lacking melanophores) (47) fish. For RNA probe preparation, two fragments from each gene (wfs1a, wfs1b) were amplified in a nested RT-PCR that incorporated a T7 polymerase site into the antisense primer and T3 polymerase site into the sense primer. Primers were designed that avoided repetitive sequences, using the reference sequences XM_690160.9 and XM_679418.7 for wfs1a and wfs1b, respectively. Sequences used were as follows: wfs1a 5′ fragment outer primers, 944F 5′-TGTCTTCAGAGAGTAAGTTTGAGCA-3′, 1919R 5′-CTTAATGCTGTGAAGAAAATGGCCA-3′; wfs1a 5′ fragment inner primers, 975FT3 5′-AATTAACCCTCACTAAAGGAGGAGAGCAGCCATGACCATGTAC-3′, 1919 T7 5′-TAATACGACTCACTATAGCTTAATGCTGTGAAGAAAATGGCCA-3′; wfs1a 3′ fragment outer primers, 1585F 5′-ATCATCTCCAATCTCACCATCGATT-3′, 3327R 5′-GGTCTTCTTTACACGTTGATCCTAGCA-3′; wfs1a 3′ fragment inner primers, 1895FT3 5′-AATTAACCCTCACTAAAGTGGCCATTTTCTTCACAGCATTAAG-3′, 3327RT7 5′-TAATACGACTCACTATAGGGTCTTCTTTACACGTTGATCCTAGCA-3′; wfs1b 5′ fragment outer primers, 39F 5′-GAGGGTTTATTTAGTCAGCTGCATC-3′, 2164R 5′-GGTGGAGTTGTATACTTTCATCCCT-3′; wfs1b 5′ fragment inner primers, 178FT3 5′-AATTAACCCTCACTAAAGCTTAAAAATGGACACGTCTCTGCTT-3′, 1980RT7 5′-TAATACGACTCACTATAGGGCACACTGTTACGATCATTTTGGTCA-3′; wfs1b 3′ fragment outer primers, 1810F 5′-TACGCTGCTGCAAGAGTCCACTGTA-3′, 4248R 5′-TTAGAAATTTTGGCATCGGTTCACA-3′; wfs1b 3′ fragment inner primers 1849FT3 5′-AATTAACCCTCACTAAAGCTCAGTTGTGGGCTACCTACTTTTT-3′, 3123RT7 5′-TAATACGACTCACTATAGAGCACATTAGGAAAGAGCTCCATTA-3′. Antisense probes generated with T7 polymerase were tested for specific expression patterns, and pairs of probes (5′ and 3′ fragments) for each gene were pooled. In situ hybridization was performed as previously described (48,49).
For the stronger staining shown in the retina at 7 dpf and ear at 10 dpf in Figure 2, and in the brain in Supplementary Material, Figure S2, an adapted protocol for larvae post-5 dpf (50) was used. Embryos were mounted in glycerol and were imaged on an Olympus BX51 compound microscope using either bright-field or differential interference contrast (DIC) optics using a Micropublisher 6 camera and Ocular software. Thick sections were cut by hand with a scalpel blade.
VMR assay
The locomotor activity of zebrafish larvae was quantified with the VMR assay using an infrared (IR) tracking system (Zebralab®, Viewpoint, Lissieu, France) as described previously (51). In brief, at 5 dpf, larvae were transferred in a 96-well plate (Whatman, #7701-1651) with 300 μl E3 medium and the locomotor behavior was monitored with an automated videotracking device (Zebrabox®, ViewPoint). The response to light changes was recorded by an IR camera under IR light illumination. The light protocol was as follows: 30 min of acclimatization in the dark (0% light intensity), then two cycles of 10 min duration light ON (100% light intensity) or light OFF (0%) periods. The total duration of the experiment was 70 min. Activity during the experiment was measured in mm/s. The values obtained during OFF periods were subtracted for each larva from their values registered during ON periods to remove inter- and intra-group variability in basic locomotion.
ASR assay
The locomotor activity of zebrafish larvae was quantified with the ASR assay in the ZebraBox®. Experimental conditions were similar to those used in the VMR assay. The experiment consisted, first, in acclimating larvae during 30 min with no sound (35 dB ambient), followed by a 1 s stimulation with a white sound at 90 dB, repeated for three times with an inter-trial time interval of 5 min. The quantity of movements during the entire experiment was measured for each larva. Baseline activity levels were subtracted from the activity levels during the sound stimulations (2 min before each stimulation) in order to normalize the values.
OKR assay
At 5 dpf, zebrafish larvae were immersed in groups of four in a Petri dish (35 mm diameter) containing 2.5% methylcellulose (#9004-65-3, Sigma Aldrich, St Louis, MI, USA). Larvae were placed dorsal side up, forming an X to avoid touching and interfering with each other. All measurements were done in the afternoon between 2:00 p.m. and 6:00 p.m. The room temperature was 28°C and the light was OFF. The visual system performance of larval zebrafish was assessed using a videotracking device (VisioBox®, ViewPoint). Forty 6 mm wide black and white strips were projected at 2 rpm for 1 min clockwise and then for 1 min anti-clockwise. Larvae were illuminated with IR from below and responses were tracked using a FL3-U3-32S2M 1/2.8-inch Monochrome camera (FLEA3, Flir) at 25 frames/s. The number of saccades was manually recorded (PHIVisualize software) and the average number of saccades per 2 min quantified.
Touch response
To measure the touch escape, 5 dpf larvae were transferred to a rail developed with a 3D printer (18 × 0.4 cm) with 200 μl of E3 medium and placed to the extremity of the rail. The tail of larvae was touched with a tip and the distance traveled was measured during 5 s. The same procedure was repeated three times per larvae, each test separated by 1 min to reduce stress. Each larva was assessed individually and the three values per larva were averaged. The light was ON and the temperature was 28°C.
Immunohistochemistry
Whole larvae were fixed in paraformaldehyde at 4°C for 48 h, cryoprotected in 30% sucrose and mounted in O.C.T.™ medium (Sakura, Tissue-Tek, Alphen aan den Rijn, The Netherlands). Larvae were transversely sectioned in 10 μm thick slices using a cryostat (Leica, Wetzlar, Germany) at −20°C and were mounted on glass slides. Cryosections were blocked with a solution containing 0.1% Triton X-100 in phosphate-buffered saline (PBS) with 5% Horse serum for 30 min at room temperature. They were subsequently incubated at 4°C overnight with the following primary antibodies: mouse anti-Rho4d2 (1:7000; ab98887, Abcam, Cambridge, UK) and mouse anti-Zpr-1 (1:500; ab174435, Abcam). After several washes, sections were incubated with specific secondary antibodies: Cy3-conjugated anti-mouse IgG (1:800; 715-165-150, Jackson ImmunoResearch, West Grove, PA, USA), Cy3-conjugated anti-rabbit IgG (1:1000; 711-166-152, Jackson ImmunoResearch) or Alexa Fluor-488 conjugated anti-mouse IgG (1:1000; 715–545-150, Jackson ImmunoResearch). Nuclei were counterstained with 40,6-diamidino-2 phenylendole (DAPI; 1:5000; Sigma Aldrich). The emitted fluorescence was measured using a confocal microscope (LSM880 Fast Airyscan, Carl Zeiss, Jena, Germany).
Whole-mount immunohistochemistry
Whole larvae at 24 and 48 hpf were dechorionated and fixed in paraformaldehyde at 4°C for 24 h, washed with PBS, permeabilized for 2 h in 1% Triton X-100 and then blocked with a solution containing 0.1% Triton X-100 in PBS with 10% Horse serum and 1% BSA for 1 h at room temperature. They were subsequently incubated at 4°C overnight with the mouse anti-SV2 (AB_2315387, 1:50; Developmental Studies Hybridoma Bank, Iowa City, IA, USA). After several washes, larvae were incubated with specific secondary antibodies: Cy3-conjugated anti-mouse IgG (1:500; 715-165-150, Jackson ImmunoResearch), and nuclei were counterstained with DAPI (1:5000; Sigma Aldrich). Z-stack images were acquired using a confocal microscope (LSM880 Fast Airyscan, Carl Zeiss).
Cell counts
Cone cells, immunolabeled with Zpr-1 antibody, were quantified individually and the total area of rod outer segments, immunolabeled with Rho4d2 antibody, was evaluated and both measures were normalized to the length of the associated retina. Ganglion cells, highlighted by DAPI counterstaining, were counted in three specific regions of the retina, consistent from one sample to another (Supplementary Material, Fig. S2). The total number of ganglion cells from all regions was averaged per larva. The thickness of the GCL was also quantified in the same specific regions of the retina.
Seahorse XF Mito stress test
The OCR of 5 dpf larvae was measured with the Seahorse XFe24 Extracellular Flux Analyzer (Agilent). Larvae were placed singly in wells of a Seahorse XFe24 spheroid microplate containing 500 μl of E3 medium. A grid was placed manually on larvae to keep them at the bottom of the wells throughout the experiment. The blanks were two empty wells of the plate. Four basal cycle readings were recorded, then five recording cycles following oligomycin (25 μm) injection, five recording cycles after carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (8 μm) injection and nine recording cycles after rotenone + antimycin A (1.5 μm) injection. Calculations for specific parameters, such as non-mitochondrial respiration, basal respiration, maximal respiration, proton leak and ATP production, were made. The room temperature was controlled at 28°C. Measurements of total zebrafish OCR were started immediately and performed according to the manufacturer’s instructions.
Chemical treatment
To induce ER stress, larvae were incubated, at 4 dpf, for 24 h with 2 μg/ml of tunicamycin (sc-3606, Santa Cruz Biotechnology, Dallas, TX, USA) diluted in 0.1% dimethylsulfoxide (DMSO) directly in the E3 medium. Control larvae were treated with 0.1% DMSO diluted in E3 medium.
RT-PCR and qPCR
At 5 dpf, total RNA from 20 whole wfs1aC825X, wfs1bW493X homozygous mutant fish and its associated phenotypically wild-type controls were extracted using a Nucleospin® RNA Kit (Macherey-Nagel, Hoerdt, France) according to the manufacturer’s instructions. RNA concentration and purity were evaluated using the Agilent RNA 6000 Nano® Kit (Agilent Technologies, Santa Clara, CA, USA). RNA samples (1 μg/μl) were denatured 5 min at 70°C and reverse transcribed into cDNA for 1 h at 37°C using M-MLV reverse transcriptase (Promega). Primer sequences are indicated in Supplementary Material, Table S1. Control reactions were conducted with sterile water to determine the signal background and DNA contamination. The standard curve of each gene was confirmed to be in a linear range, while zef1α gene was selected as a reference.
Western blot
To measure protein expression, 20 whole larvae per condition, at 5 dpf, were homogenized on ice for 15 s in 100 μl of lysis buffer [62.5 mm Tris–HCl, 25% glycerol, 2% sodium dodecyl sulfate, 0.04% cOmplete™ (Roche, Basel, Switzerland), 0.1% PhosSTOP™ (Roche) pH 6.8]. Total proteins were separated on a 1.5 mm 12% running gel and 4% acrylamide stacking gel at 100 V. Proteins were transferred onto a nitrocellulose membrane at 100 V for 1 h in transfer buffer and were blocked in 5% non-fat milk solution for 1 h. Immunoblotting was performed with primary antibodies as follows: rabbit anti-Sigmar1 antibody (1:500, 15168-1-AP; Proteintech, Rosemont, IL, USA), rabbit anti-GADD153 antibody (1:1000, G6916, Sigma Aldrich), rabbit anti-Bip antibody (1:700, SPC-180, Biosciences), rabbit anti-Eif2α antibody (D9G8) (1:500, #3398, Cell Signaling), rabbit anti-p-Eif2α antibody (D9G8) (1:500, #9722, Cell Signaling), in buffer (0.1% TBS/Triton X-100) pH 7.4, overnight at 4°C. After several washes, membranes were incubated with horseradish peroxidase (HRP) conjugated goat anti-rabbit secondary antibody (1:2000; ab6721, Abcam) or goat anti-mouse (1:2000; ab6789, Abcam) secondary antibody for 1 h at room temperature, and the proteins were detected with the indicated HRP detection reagent (10776189, Merck, Germany) and the Bio-Rad imaging system (Bio-Rad, CA, USA). Relative intensities of each band were quantified using Image lab v6.1 software (Bio-Rad) and were normalized to the total protein quantity (Stain-Free™, Bio-Rad) (Supplementary Material, Fig. S4).
Statistical analyses
Data were expressed as mean ± SEM. Statistical significance between groups was determined by unpaired Student’s t-test or two-way ANOVA. The levels of statistical significance considered were *P < 0.05, **P < 0.01 and ***P < 0.001. Statistical analyses were performed using Prism v7.0 software (GraphPad, San Diego, CA, USA).
Supplementary Material
Acknowledgements
We thank Emily Noël and Ryan MacDonald for help in interpreting gene expression patterns and the Sheffield Aquarium Team for expert care of the fish. We acknowledge the imaging facility MRI, member of the national infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04, ‘Investments for the future’). We thank the ZebraSens zebrafish phenotyping platform at MMDN (Montpellier, France) and the qPHd quantitative PCR platform at University of Montpellier (Montpellier, France).
We thank Cyril Beuvry for setting up and optimizing the touch response protocol, Yoan Arribat for providing us with the SV2A antibodies and Méghane Bataillard for helping with motor neuron length measurements.
Conflict of Interest statement. None declared.
Contributor Information
Lucie Crouzier, MMDN, Université Montpellier, EPHE, INSERM, Montpellier, France.
Elodie M Richard, MMDN, Université Montpellier, EPHE, INSERM, Montpellier, France.
Camille Diez, MMDN, Université Montpellier, EPHE, INSERM, Montpellier, France.
Hala Alzaeem, MMDN, Université Montpellier, EPHE, INSERM, Montpellier, France.
Morgane Denus, MMDN, Université Montpellier, EPHE, INSERM, Montpellier, France.
Nicolas Cubedo, MMDN, Université Montpellier, EPHE, INSERM, Montpellier, France.
Thomas Delaunay, IES, Université Montpellier, CNRS, Montpellier, France.
Emily Glendenning, Development, Regeneration and Neurophysiology, School of Biosciences, University of Sheffield, Sheffield S10 2TN, UK.
Sarah Baxendale, Development, Regeneration and Neurophysiology, School of Biosciences, University of Sheffield, Sheffield S10 2TN, UK.
Jean-Charles Liévens, MMDN, Université Montpellier, EPHE, INSERM, Montpellier, France.
Tanya T Whitfield, Development, Regeneration and Neurophysiology, School of Biosciences, University of Sheffield, Sheffield S10 2TN, UK.
Tangui Maurice, MMDN, Université Montpellier, EPHE, INSERM, Montpellier, France.
Benjamin Delprat, MMDN, Université Montpellier, EPHE, INSERM, Montpellier, France.
Funding
University of Montpellier (Fondation Pour l’Audition FPA RD-2019-13, Retina France Association and Foundation); Association Syndrome de Wolfram and the Région Occitanie (to L.C.); the Snow Foundation and Eye Hope Foundation (E.M.R.); BBSRC Project Grant (BB/S007008/1 to T.T.W. and S.B.).
Ethics statement
All animal work at the University of Sheffield was undertaken under license from the UK Home Office and animals were maintained according to the recommended husbandry guidelines (52).
Authors’ contributions
B.D. and T.M. conceived the project. B.D., T.M, S.B. and T.T.W. designed the experiments and analyzed the data. L.C. performed most of the in vivo experiments and analyses. L.C, C.D., M.D. and H.A. performed visual motor response experiments. E.G. and S.B. performed in situ hybridization experiments. E.G. prepared Figure 2. N.C. took care of the zebrafish. J.-C.L. helped with the novel touch escape protocol. T.D. helped with a novel ASR device. L.C., E.M.R., B.D., T.M and T.T.W. prepared the figures, wrote and edited the manuscript. E.M.R. implemented the manuscript. All authors contributed to the preparation of the figures and manuscript writing.
References
- 1. Barrett, T.G., Bundey, S.E. and Macleod, A.F. (1995) Neurodegeneration and diabetes: UK nationwide study of Wolfram (DIDMOAD) syndrome. Lancet Lond. Engl., 346, 1458–1463. [DOI] [PubMed] [Google Scholar]
- 2. Kinsley, B.T., Swift, M., Dumont, R.H. and Swift, R.G. (1995) Morbidity and mortality in the Wolfram syndrome. Diabetes Care, 18, 1566–1570. [DOI] [PubMed] [Google Scholar]
- 3. Inoue, H., Tanizawa, Y., Wasson, J., Behn, P., Kalidas, K., Bernal-Mizrachi, E., Mueckler, M., Marshall, H., Donis-Keller, H., Crock, P.et al. (1998) A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat. Genet., 20, 143–148. [DOI] [PubMed] [Google Scholar]
- 4. Strom, T.M., Hörtnagel, K., Hofmann, S., Gekeler, F., Scharfe, C., Rabl, W., Gerbitz, K.D. and Meitinger, T. (1998) Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum. Mol. Genet., 7, 2021–2028. [DOI] [PubMed] [Google Scholar]
- 5. Urano, F. (2016) Wolfram syndrome: diagnosis, management, and treatment. Curr. Diab. Rep., 16, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schröder, M. and Kaufman, R.J. (2005) ER stress and the unfolded protein response. Mutat. Res., 569, 29–63. [DOI] [PubMed] [Google Scholar]
- 7. Oslowski, C.M. and Urano, F. (2011) The binary switch that controls the life and death decisions of ER stressed beta cells. Curr. Opin. Cell Biol., 23, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fonseca, S.G., Fukuma, M., Lipson, K.L., Nguyen, L.X., Allen, J.R., Oka, Y. and Urano, F. (2005) WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic β-cells. J. Biol. Chem., 280, 39609–39615. [DOI] [PubMed] [Google Scholar]
- 9. Angebault, C., Fauconnier, J., Patergnani, S., Rieusset, J., Danese, A., Affortit, C.A., Jagodzinska, J., Mégy, C., Quiles, M., Cazevieille, C.et al. (2018) ER-mitochondria cross-talk is regulated by the Ca2+ sensor NCS1 and is impaired in Wolfram syndrome. Sci. Signal., 11, eaaq1380. [DOI] [PubMed] [Google Scholar]
- 10. Delprat, B., Rieusset, J. and Delettre, C. (2019) Defective endoplasmic reticulum–mitochondria connection is a hallmark of Wolfram syndrome. Contact, 2, 2515256419847407. [Google Scholar]
- 11. Fonseca, S.G., Ishigaki, S., Oslowski, C.M., Lu, S., Lipson, K.L., Ghosh, R., Hayashi, E., Ishihara, H., Oka, Y., Permutt, M.A.et al. (2010) Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J. Clin. Invest., 120, 744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fonseca, S.G., Urano, F., Weir, G.C., Gromada, J. and Burcin, M. (2012) Wolfram syndrome 1 and adenylyl cyclase 8 interact at the plasma membrane to regulate insulin production and secretion. Nat. Cell Biol., 14, 1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13. Shang, L., Hua, H., Foo, K., Martinez, H., Watanabe, K., Zimmer, M., Kahler, D.J., Freeby, M., Chung, W., LeDuc, C.et al. (2014) β-Cell dysfunction due to increased ER stress in a stem cell model of Wolfram syndrome. Diabetes, 63, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonnet Wersinger, D., Benkafadar, N., Jagodzinska, J., Hamel, C., Tanizawa, Y., Lenaers, G. and Delettre, C. (2014) Impairment of visual function and retinal ER stress activation in Wfs1-deficient mice. PLoS One, 9, e97222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishihara, H., Takeda, S., Tamura, A., Takahashi, R., Yamaguchi, S., Takei, D., Yamada, T., Inoue, H., Soga, H., Katagiri, H.et al. (2004) Disruption of the WFS1 gene in mice causes progressive β-cell loss and impaired stimulus–secretion coupling in insulin secretion. Hum. Mol. Genet., 13, 1159–1170. [DOI] [PubMed] [Google Scholar]
- 16. Ivask, M., Hugill, A. and Kõks, S. (2016) RNA-sequencing of WFS1-deficient pancreatic islets. Physiol. Rep., 4, e12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luuk, H., Koks, S., Plaas, M., Hannibal, J., Rehfeld, J.F. and Vasar, E. (2008) Distribution of Wfs1 protein in the central nervous system of the mouse and its relation to clinical symptoms of the Wolfram syndrome. J. Comp. Neurol., 509, 642–660. [DOI] [PubMed] [Google Scholar]
- 18. Lieschke, G.J. and Currie, P.D. (2007) Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet., 8, 353–367. [DOI] [PubMed] [Google Scholar]
- 19. Phillips, J.B. and Westerfield, M. (2014) Zebrafish models in translational research: tipping the scales toward advancements in human health. Dis. Model. Mech., 7, 739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. White, R.M. (2015) Cross-species oncogenomics using zebrafish models of cancer. Curr. Opin. Genet. Dev., 30, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bai, Q. and Burton, E.A. (2011) Zebrafish models of Tauopathy. Biochim. Biophys. Acta, 1812, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bandmann, O. and Burton, E.A. (2010) Genetic zebrafish models of neurodegenerative diseases. Neurobiol. Dis., 40, 58–65. [DOI] [PubMed] [Google Scholar]
- 23. Das, S. and Rajanikant, G.K. (2014) Huntington disease: Can a zebrafish trail leave more than a ripple? Neurosci. Biobehav. Rev., 45, 258–261. [DOI] [PubMed] [Google Scholar]
- 24. Laird, A.S., Mackovski, N., Rinkwitz, S., Becker, T.S. and Giacomotto, J. (2016) Tissue-specific models of spinal muscular atrophy confirm a critical role of SMN in motor neurons from embryonic to adult stages. Hum. Mol. Genet., 25, 1728–1738. [DOI] [PubMed] [Google Scholar]
- 25. Lam, P.-Y. and Peterson, R.T. (2019) Developing zebrafish disease models for in vivo small molecule screens. Curr. Opin. Chem. Biol., 50, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patton, E.E., Zon, L.I. and Langenau, D.M. (2021) Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials. Nat. Rev. Drug Discov., 20, 611–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kettleborough, R.N.W., Busch-Nentwich, E.M., Harvey, S.A., Dooley, C.M., deBruijn, E., vanEeden, F., Sealy, I., White, R.J., Herd, C., Nijman, I.J.et al. (2013) A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature, 496, 494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cairns, G., Burté, F., Price, R., O’Connor, E., Toms, M., Mishra, R., Moosajee, M., Pyle, A., Sayer, J.A. and Yu-Wai-Man, P. (2021) A mutant wfs1 zebrafish model of Wolfram syndrome manifesting visual dysfunction and developmental delay. Sci. Rep., 11, 20491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Diez-Roux, G., Banfi, S., Sultan, M., Geffers, L., Anand, S., Rozado, D., Magen, A., Canidio, E., Pagani, M., Peluso, I.et al. (2011) A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol., 9, e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamada, T., Ishihara, H., Tamura, A., Takahashi, R., Yamaguchi, S., Takei, D., Tokita, A., Satake, C., Tashiro, F., Katagiri, H.et al. (2006) WFS1-deficiency increases endoplasmic reticulum stress, impairs cell cycle progression and triggers the apoptotic pathway specifically in pancreatic beta-cells. Hum. Mol. Genet., 15, 1600–1609. [DOI] [PubMed] [Google Scholar]
- 31. Eimre, M., Kasvandik, S., Ivask, M. and Kõks, S. (2018) Proteomic dataset of wolframin-deficient mouse heart and skeletal muscles. Data Brief, 21, 616–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tepp, K., Puurand, M., Timohhina, N., Aid-Vanakova, J., Reile, I., Shevchuk, I., Chekulayev, V., Eimre, M., Peet, N., Kadaja, L.et al. (2020) Adaptation of striated muscles to Wolframin deficiency in mice: alterations in cellular bioenergetics. Biochim. Biophys. Acta, 1864, 129523. [DOI] [PubMed] [Google Scholar]
- 33. Robson, M.I., deLas Heras, J.I., Czapiewski, R., Lê Thành, P., Booth, D.G., Kelly, D.A., Webb, S., Kerr, A.R.W. and Schirmer, E.C. (2016) Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Mol. Cell, 62, 834–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pallotta, M.T., Tascini, G., Crispoldi, R., Orabona, C., Mondanelli, G., Grohmann, U. and Esposito, S. (2019) Wolfram syndrome, a rare neurodegenerative disease: from pathogenesis to future treatment perspectives. J. Transl. Med., 17, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Franco, E., Flanagan, S.E., Yagi, T., Abreu, D., Mahadevan, J., Johnson, M.B., Jones, G., Acosta, F., Mulaudzi, M., Lek, N.et al. (2017) Dominant ER stress-inducing WFS1 mutations underlie a genetic syndrome of neonatal/infancy-onset diabetes, congenital sensorineural deafness, and congenital cataracts. Diabetes, 66, 2044–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. El-Brolosy, M.A., Kontarakis, Z., Rossi, A., Kuenne, C., Günther, S., Fukuda, N., Kikhi, K., Boezio, G.L.M., Takacs, C.M., Lai, S.-L.et al. (2019) Genetic compensation triggered by mutant mRNA degradation. Nature, 568, 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kawano, J., Tanizawa, Y. and Shinoda, K. (2008) Wolfram syndrome 1 (Wfs1) gene expression in the normal mouse visual system. J. Comp. Neurol., 510, 1–23. [DOI] [PubMed] [Google Scholar]
- 38. Cryns, K., Thys, S., Van Laer, L., Oka, Y., Pfister, M., Van Nassauw, L., Smith, R.J.H., Timmermans, J.-P. and Van Camp, G. (2003) The WFS1 gene, responsible for low frequency sensorineural hearing loss and Wolfram syndrome, is expressed in a variety of inner ear cells. Histochem. Cell Biol., 119, 247–256. [DOI] [PubMed] [Google Scholar]
- 39. Yamamoto, H., Hofmann, S., Hamasaki, D.I., Yamamoto, H., Kreczmanski, P., Schmitz, C., Parel, J.-M. and Schmidt-Kastner, R. (2006) Wolfram syndrome 1 (WFS1) protein expression in retinal ganglion cells and optic nerve glia of the Cynomolgus monkey. Exp. Eye Res., 83, 1303–1306. [DOI] [PubMed] [Google Scholar]
- 40. Schmidt-Kastner, R., Kreczmanski, P., Preising, M., Diederen, R., Schmitz, C., Reis, D., Blanks, J. and Dorey, C.K. (2009) Expression of the diabetes risk gene wolframin (WFS1) in the human retina. Exp. Eye Res., 89, 568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scaramuzzi, M., Kumar, P., Peachey, N., Nucci, P. and Traboulsi, E.I. (2019) Evidence of retinal degeneration in Wolfram syndrome. Ophthalmic Genet., 40, 34–38. [DOI] [PubMed] [Google Scholar]
- 42. Förster, D., Helmbrecht, T.O., Mearns, D.S., Jordan, L., Mokayes, N. and Baier, H. (2020) Retinotectal circuitry of larval zebrafish is adapted to detection and pursuit of prey. elife, 9, e58596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chaussenot, A., Bannwarth, S., Rouzier, C., Vialettes, B., Mkadem, S.A.E., Chabrol, B., Cano, A., Labauge, P. and Paquis-Flucklinger, V. (2011) Neurologic features and genotype-phenotype correlation in Wolfram syndrome. Ann. Neurol., 69, 501–508. [DOI] [PubMed] [Google Scholar]
- 44. Jagomäe, T., Seppa, K., Reimets, R., Pastak, M., Plaas, M., Hickey, M.A., Kukker, K.G., Moons, L., De Groef, L., Vasar, E.et al. (2021) Early intervention and lifelong treatment with GLP1 receptor agonist liraglutide in a Wolfram syndrome rat model with an emphasis on visual neurodegeneration, sensorineural hearing loss and diabetic phenotype. Cell, 10, 3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bond, S.T., McEwen, K.A., Yoganantharajah, P. and Gibert, Y. (2018) Live metabolic profile analysis of zebrafish embryos using a seahorse XF 24 extracellular flux analyzer. Methods Mol. Biol. Clifton NJ, 1797, 393–401. [DOI] [PubMed] [Google Scholar]
- 46. Kilkenny, C., Browne, W.J., Cuthill, I.C., Emerson, M. and Altman, D.G. (2010) Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. J. Pharmacol. Pharmacother., 1, 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lister, J.A., Robertson, C.P., Lepage, T., Johnson, S.L. and Raible, D.W. (1999) Nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development, 126, 3757–3767. [DOI] [PubMed] [Google Scholar]
- 48. Oxtoby, E. and Jowett, T. (1993) Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res., 21, 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thisse, C. and Thisse, B. (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc., 3, 59–69. [DOI] [PubMed] [Google Scholar]
- 50. Elizondo, M.R., Arduini, B.L., Paulsen, J., MacDonald, E.L., Sabel, J.L., Henion, P.D., Cornell, R.A. and Parichy, D.M. (2005) Defective skeletogenesis with kidney stone formation in dwarf zebrafish mutant for trpm7. Curr. Biol., 15, 667–671. [DOI] [PubMed] [Google Scholar]
- 51. Crouzier, L., Diez, C., Richard, E.M., Cubedo, N., Barbereau, C., Rossel, M., Delaunay, T., Maurice, T. and Delprat, B. (2021) Loss of Pde6a induces rod outer segment shrinkage and visual alterations in pde6aQ70X mutant zebrafish, a relevant model of retinal dystrophy. Front. Cell Dev. Biol., 9, 675517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aleström, P., D’Angelo, L., Midtlyng, P.J., Schorderet, D.F., Schulte-Merker, S., Sohm, F. and Warner, S. (2020) Zebrafish: housing and husbandry recommendations. Lab. Anim., 54, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.