SUMMARY
Adoptive T cell therapies (ACT) have been curative for a limited number of cancer patients. The sensitization of cancer cells to T cell killing may expand the benefit of these therapies for more patients. To this end, we use a three-step approach to identify cancer genes that disfavor T cell immunity. First, we profile gene transcripts upregulated by cancer under selection pressure from T cell killing. Second, we identify potential tumor gene targets and pathways that disfavor T cell killing using signaling pathway activation libraries and genome-wide loss-of-function CRISPR-Cas9 screens. Finally, we implement pharmacological perturbation screens to validate these targets and identify BIRC2, ITGAV, DNPEP, BCL2, and ERRα as potential ACT-drug combination candidates. Here, we establish that BIRC2 limits antigen presentation and T cell recognition of tumor cells by suppressing IRF1 activity and provide evidence that BIRC2 inhibition in combination with ACT is an effective strategy to increase efficacy.
Graphical Abstract
In brief
Kishton et al. use systems approaches to identify tumor genes and signaling pathways that protect cancer from T cell-mediated killing. Targeting these tumor defenses with combination approaches increases T cell elimination of tumors and may be a strategy to improve the clinical efficacy of immunotherapy.
INTRODUCTION
Adoptive T cell immunotherapies have driven improved clinical outcomes in patients with certain advanced cancers, including melanoma and lymphoid malignancies (Consortium, 2013; Maude et al., 2014; Rosenberg et al., 2011; Topalian et al., 2015). Despite these successes, many patients still receive limited benefit from existing immunotherapy approaches (Park et al., 2018; Yamamoto et al., 2019). Consequently, strategies to improve and broaden treatment efficacy are critical.
Multiple factors determine the efficacy of T cell-based cancer therapies, including features of antitumor T cells (Kishton et al., 2017; Vodnala et al., 2019) and the presence of inhibitory factors in the tumor microenvironment (Facciabene et al., 2012) or excess potassium (Eil et al., 2016), along with tumor-intrinsic mechanisms, including loss of function in tumor antigen presentation and inflammatory response pathways (Restifo et al., 1996). While loss of function in key tumor genes can mediate resistance to immunotherapy, it has also recently become apparent that tumor expression of genes can limit the efficacy of immunotherapy. Tumor expression of PTNP2 (Manguso et al., 2017), TRAF2 (Vredevoogd et al., 2019), and PBRM1 (Pan et al., 2018) each play an active role in limiting tumor susceptibility to immunotherapy. Tumor expression of these genes disfavors effective antitumor immune responses, and we hypothesized that a number of genes may have a similar impact in regulating immunotherapy. In addition, in the context of targeted inhibitors, tumors can drive treatment resistance through the induced expression of genes that promote cell survival, mediating resistance to treatment (Singleton et al., 2017). We hypothesized that adoptive T cell therapies (ACT), in which antitumor T cells engage and eliminate tumor cells, could similarly drive the transcription of tumor-intrinsic genes disfavoring immune-mediated cytolysis of tumor cells. These inducible tumor-expressed genes that disfavor T cell-mediated cytolysis (iD-CTL [cytotoxic T cell] genes) would therefore act as a dynamically regulated defense against T cell-mediated lysis and would represent compelling potential targets for combination therapy strategies to increase T cell antitumor efficacy.
Here, we present an integrated systems approach to mine iD-CTL genes. To determine whether iD-CTL mechanisms may exist in human cancers, we analyzed human melanoma datasets from The Cancer Genome Atlas (TCGA) and found a tumor gene set negatively associated with T cell cytolytic activity in patients with high T cell infiltrates. Using RNA sequencing (RNA-seq) analysis, we profiled genes and pathways that are dynamically induced in cancer cells upon exposure to antigen-specific T cells. Functional screens using gain-of-function constructs driving constitutive activity in key growth and survival-linked cellular signaling pathways in tandem with genome-wide loss-of-function CRISPR-Cas9 screens enabled us to uncover iD-CTL genes. We found that targeting iD-CTL genes, including BIRC2 (baculoviral inhibitor of apoptosis domain [IAP] repeat containing 2), ITGAV (integrin subunit alpha V), DNPEP, BCL2 (B cell lymphoma 2), and ESRRA (estrogen-related receptor alpha), through genetic or pharmacological inhibition markedly increased tumor cell susceptibility to T cell-mediated killing. Finally, we describe a mechanism by which one of the iD-CTL genes, BIRC2, acts to promote tumor survival during T cell engagement by limiting interferon (IFN) regulatory factor 1 (IRF1)-mediated upregulation of antigen presentation pathway genes and reducing T cell recognition of tumor target antigens.
RESULTS
Identification of tumor genes that disfavor T cell cytolytic activity in “immune hot” human cancers
When tumor-specific T cells recognize cognate tumor antigens, they can mediate cytolytic activity (CYT) against tumor, resulting in tumor cell death. However, interactions between T cells and tumor exist in an equilibrium, where CYT can be favored or disfavored depending on numerous factors. When CYT is disfavored, cancer immune escape can occur, resulting in a lack of disease control. To better understand how T cell CYT is influenced by tumor cells, we analyzed TCGA along with published datasets of human immunotherapy patients (Figure 1A). Analysis of melanoma patients treated with α-CTLA-4 blockade (Van Allen et al., 2015) revealed that T cell abundance, as measured by the intratumoral expression of CD3E, is not significantly associated with patient outcome, whereas CYT (defined as the geometric mean expression of PRF1 and GZMA) has a significant positive clinical association (Figure 1B). This suggested that mechanisms beyond T cell infiltration alone regulate tumor immune susceptibility. To further understand tumor genetic mechanisms that limit cytolytic activity of T cells at the tumor site, we mined the TCGA Cutaneous Melanoma dataset for tumor genes whose expression disfavored CYT even in heavily immune-infiltrated tumors (Figure 1C). We selected melanomas with above-median levels of T cell infiltration as measured by CD3E expression (Figure S1A) and performed transcriptome-wide correlation analysis to identify genes negatively correlated with CYT in the context of these immune “hot” tumors. The expression of a number of genes was found to be negatively correlated with CYT in this context (Figure 1D; Table S1), including the oncogene MYC, the most commonly amplified gene across all TCGA cancer datasets (Figure S1B; Table S2). Pathway analysis of negatively correlated genes revealed enrichment for anabolic growth and MYC signaling pathways, along with genes inhibited by the aminopeptidase inhibitor tosedostat (CHR2797) (Figure 1E). Importantly, the high expression of genes negatively correlated with CYT in T cell “hot” tumors was associated with reduced overall survival in melanoma patients (Figure 1F). In contrast, genes with the highest levels of copy-number loss across the TCGA dataset were associated with immune cell activity (Figure S1C). These human patient data suggest that tumor-expressed genes can limit T cells recognition and killing of tumor cells even in the context of highly T cell-infiltrated tumors.
Figure 1. Subset of tumor-expressed genes disfavor CYT in highly immune-infiltrated human melanomas.
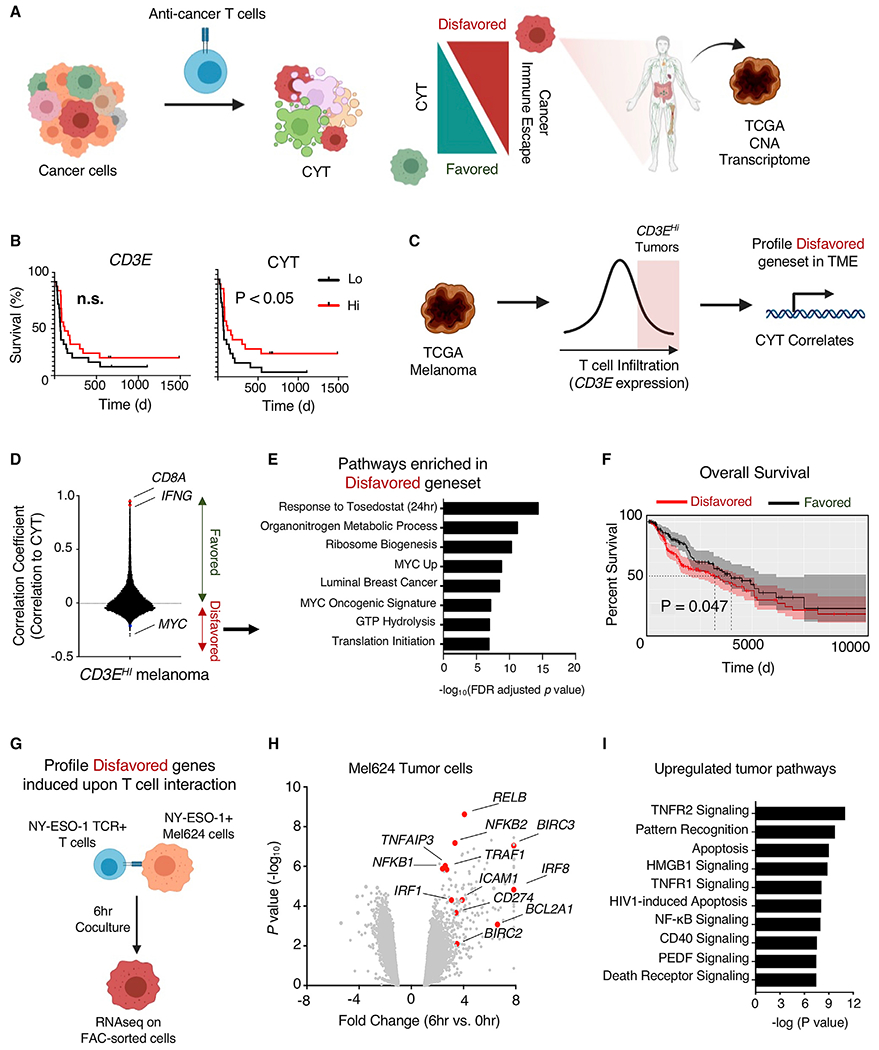
(A) To uncover tumor-expressed genes disfavoring T cell CYT against tumor cells, the transcriptional profile of TCGA melanoma samples, copy-number alteration (CNA) of TCGA tumors, and the transcriptome of tumor cells engaged with T cells were analyzed.
(B) Published data of melanoma patients treated with ipilimumab (Van Allen et al., 2015) were analyzed by stratifying patient tumors based on high and low expression of CD3E and CYT (geometric mean of GZMA and PRF1, with high and low representing above or below median values, respectively, with n = 21 patients in each group) and examining survival. Statistical analysis was performed using log rank (Mantel-Cox) test. n.s., not significant.
(C) TCGA melanoma patient samples were analyzed based on expression of CD3E and stratified based on above median expression. Stratified samples were profiled for genes whose expression was significantly correlated with CYT.
(D) Correlation coefficients (Pearson correlation) were calculated for all genes in tumors with above median CD3E expression. Genes with positive correlations favor T cell CYT, while genes with negative correlations disfavor CYT.
(E) Pathway enrichment analysis was performed on genes with significant negative correlations, and top pathways were plotted.
(F) TCGA melanoma patient survival was calculated based on normalized expression of genes favoring CYT comparing groups with below and above median gene expression. Statistical analysis was performed using log rank (Mantel-Cox) test.
(G) Transcriptional analysis of tumor-expressed genes disfavoring CYT that were induced by T cell interaction was performed by co-culturing NY-ESO-1 TCR-engineered T cells with NY-ESO-1 antigen-positive Mel624 cells at a 1:3 E:T ratio for 6 h. Tumor and T cells were purified by FACS sorting, and each population was subjected to RNA sequencing (RNA-seq).
(H) Volcano plot representing differentially expressed genes analyzed by RNA-seq of Mel624 cells that were co-cultured with ESO T cells for 0 or 6 h. Abundance is represented as relative fold change (x axis) versus significance (y axis).
(I) Pathway enrichment analysis (Ingenuity Pathway Analysis) of Mel624 genes significantly upregulated by T cell interaction.
While correlational analysis of gene expression in human cancer patients revealed genes negatively associated with cytolytic activity, the dynamic regulation of tumor genes in the context of T cell interaction remained unclear. To analyze the kinetics of tumor gene expression during T cell engagement, we used dynamic transcriptional analysis (DTA) of a two-cell type (2CT) system in which primary human T cells transduced with an HLA-A*02:01-restricted T cell receptor (TCR) specific for the NY-ESO-1 antigen (NY-ESO-1:157-165 epitope, ESO T cells hereafter) were co-cultured with NY-ESO-1+ Mel624 human melanoma tumor cells (Patel et al., 2017) (Figure 1G). We co-cultured ESO T cells with Mel624 cells for 0 or 6 h and sorted each population by fluorescence-activated cell sorting (FACS) (Figure S1D). mRNA was extracted and sequenced from the isolated cell populations. Consistent with ESO T cell recognition of tumors, we found increased T cell expression of inflammatory cytokines, including IL2 and IFNG, cytolytic molecules such as GZMB, and markers of T cell activation CD69 and TNFRSF9 (encoding 4-1BB), with the downregulation of genes associated with T cell quiescence such as TCF7, LEF1, and KLF2 (Figure S1E). Pathway analysis of transcriptional alterations in tumor-engaged T cells revealed the upregulation of anabolic growth pathways (Figure S1F). After co-culture with ESO T cells, Mel624 tumor cells increased the expression of genes associated with canonical inflammatory response pathways, including tumor necrosis factor receptor (TNFR) and nuclear factor κB (NF-κB) pathway signaling, along with the notable upregulation of genes such as BIRC2, BIRC3, and BCL2A1 that have been associated with resistance to cell death (Figures 1H and 1I). Tumor cells repressed the expression of Notch pathway genes after T cell engagement (Figure S1G).
Identification of iD-CTL through functional genomics
We next used functional genomics tools to interrogate the role of key tumor cell signaling pathways and genes in potentially disfavoring T cell-mediated tumor cytolysis. First, we conducted a systematic gain-of-function screen in A375 melanoma cells, an additional human melanoma line expressing HLA-A*02:01 and NY-ESO-1 antigen, by activating pro-growth and survival tumor cellular signaling pathways through lentiviral transduction of constitutive pathway activating constructs (Martz et al., 2014) and studying the impact on T cell killing of tumors (Figure 2A). We determined that activating several tumor cell signaling pathways could alter susceptibility to T cell killing. Constitutive expression of anti-apoptotic genes BCL2 or BCL-XL resulted in notable tumor cell resistance to T cell-mediated destruction, as did the activation of transforming growth factor-β receptor (TGFβR) signaling, while constitutive NOTCH1 signaling activity sensitized tumor cells to cellular immunotherapy (Figures 2B and 2C). Interestingly, the constitutive activation of NOTCH3 signaling promoted tumor cell resistance to T cell killing, suggesting that NOTCH1 and NOTCH3 oncogenic signaling may have opposing roles in tumor susceptibility to immunotherapy. We further assessed the role of tumor oncogenic signaling in regulating sensitivity to T cell recognition and destruction of tumor cells by profiling how the activation of each tumor signaling pathway affected the expression of β-2-microglobulin (β2M) and programmed cell death ligand 1 (PD-L1). Inhibition of caspase death pathway signaling through the mutation of caspase 3 resulted in a significant increase in β2M expression, while additional pathways drove trends toward increased expression (Figure S2A). Constitutive activation of NOTCH1 signaling drove a marked decrease in PD-L1 expression, while the expression of androgen receptor splice variant 7 (AR-V7), and oncogenic HRAS resulted in increased PD-L1 (Figure S2B). Examining the ratio of β2M to PD-L1 expression driven by each oncogenic signaling pathway revealed that constitutive NOTCH1 signaling increased this ratio, favoring T cell CYT, while AR-V7 and mutant HRAS reduced it (Figure S2C). To test whether significant hits from the ESO T cell killing screen generalized to an unrelated TCR antigen MART1, we expressed activating constructions in MART1+ Mel624 melanoma cells and co-cultured them with MART1 TCR-transduced human T cells (Johnson et al., 2006). We determined that the overexpression of BCL-xL and BCL-2 disfavored T cell CYT against tumor cells in this additional setting, while constitutively active NOTCH1 increased tumor susceptibility to CYT (Figure 2D).
Figure 2. Activation of tumor oncogenic pro-growth and survival pathways alters tumor cell susceptibility to T cell CYT.
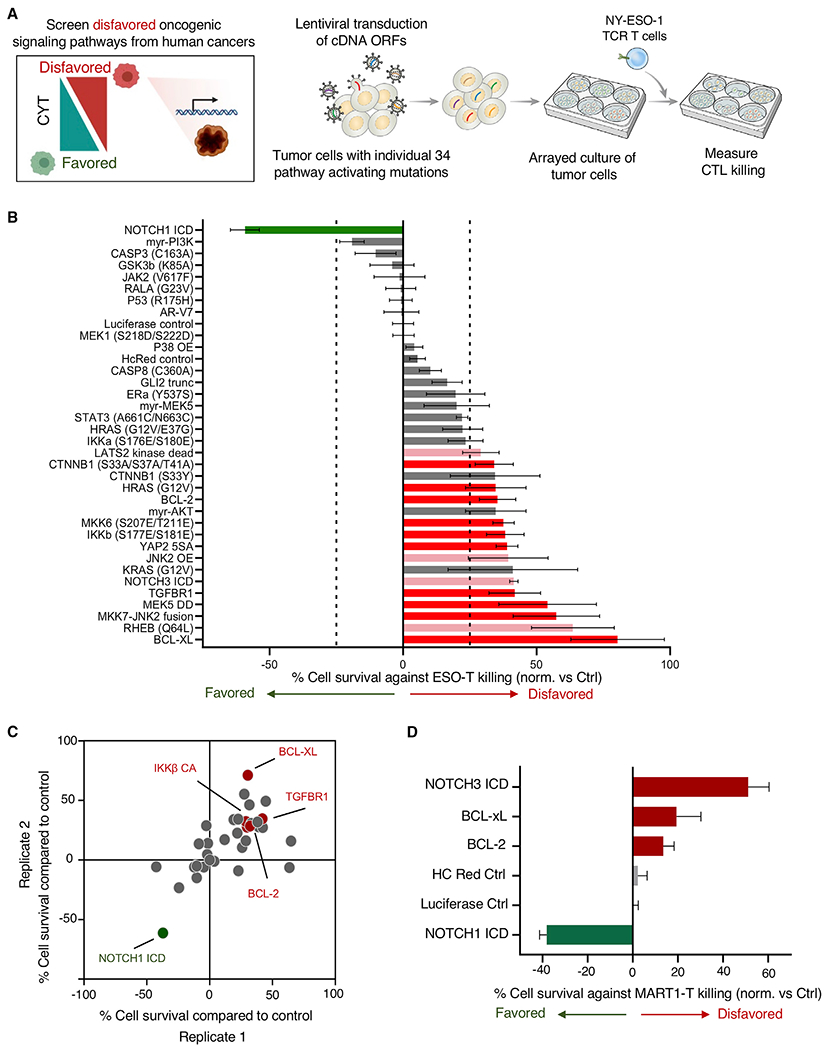
(A) Systematic screening of tumor signaling pathways for potential to alter tumor vulnerability to T cell CYT was performed by transducing A375 melanoma cells with lentiviral cDNA open reading frames (ORFs) encoding proteins driving constitutive activity across pro-growth and survival signaling pathways. Transduced cells were co-cultured with NY-ESO-1 TCR-engineered T cells in an arrayed format, and cytotoxic T cell (CTL) killing of tumor cells was measured.
(B) The impact of each alteration on tumor death after co-culture was determined. Modifications to tumor cells disfavoring T cell killing are shown in red and pink (red depicts modifications found to be statistically significant across 2 independent screens; pink indicates modifications driving statistically significant resistance to killing in 1 screen), and modifications increasing tumor cell killing by T cells are indicated in green. Statistical analysis was performed by 2-tailed Student’s t test, with comparison made to luciferase control. Significant hits were called with p < 0.05. Error bars depict standard deviation.
(C) Results of biological replicate screens are depicted.
(D) Mel624 melanoma cells were transduced with selected pathway activating constructs and susceptibility to MART-1 TCR-engineered T cells was measured. Error bars depict standard deviation.
We next used a 2CT genome-scale loss-of-function CRISPR-Cas9 screen to identify tumor genes that contribute to survival in the context of T cell engagement. In our previous study, we identified genes whose expression by tumors is essential for immunotherapy by using a strong T cell-selective pressure that eliminated approximately 75% of tumor cells. Analysis of single-guide RNAs (sgRNAs) that were enriched in surviving tumor cells relative to control uncovered genes whose loss promoted tumor cell survival (Patel et al., 2017). Here, we sought to identify genes whose loss promoted tumor cell death in the context of T cell engagement. We optimized the 2CT assay for this purpose by reducing T cell selective pressure by shortening co-culture duration to 6 h, resulting in the elimination of approximately 25% of tumor cells at a 1:3 effector T cell/tumor cell (E:T) ratio (Figure S3A). We transduced Mel624 cells with the Genome-scale CRISPR Knockout (GeCKOv2) library (Figure 3A) and exposed the transduced tumor cells to ESO T cells under these lower selective pressure conditions and compared sgRNA library representation in tumor cells before and after T cell co-culture. Consistent with our previous findings demonstrating that low T cell selective pressure generated only a modest impact on overall representation of sgRNAs in tumor cells (Patel et al., 2017), this 6-h co-culture did not markedly alter the overall representation of guides (Figures S3B and S3C). We reasoned that individual sgRNAs targeting genes whose expression disfavored T cell killing of tumor cells would have reduced abundance in surviving tumor cells (Figure 3B) and we ranked all genes by two different scoring metrics: second-most depleted sgRNA (Figure 3C) and the RNAi Gene Enrichment Ranking (RIGER) metric (Figure S3D). Top hits identified from the screen included genes such as TRAF2, BIRC2, ALG11, and mir1299. Gene Ontology analysis of significantly depleted genes indicated enrichments in death receptor and eukaryotic initiation factor 2 (EIF2) signaling, along with endoplasmic reticulum stress pathways and the unfolded protein response (Figure S3E). We compared significantly depleted genes from a replicate screen (Table S3) and found that the top genes identified in the 2CT screen, including mir1299, FAM32A, EIF3I, BIRC2, and BCL2, were also depleted in a replicate screen (Figure S3F).
Figure 3. Genome-scale 2CT assay uncovers tumor genes disfavoring T cell CYT.
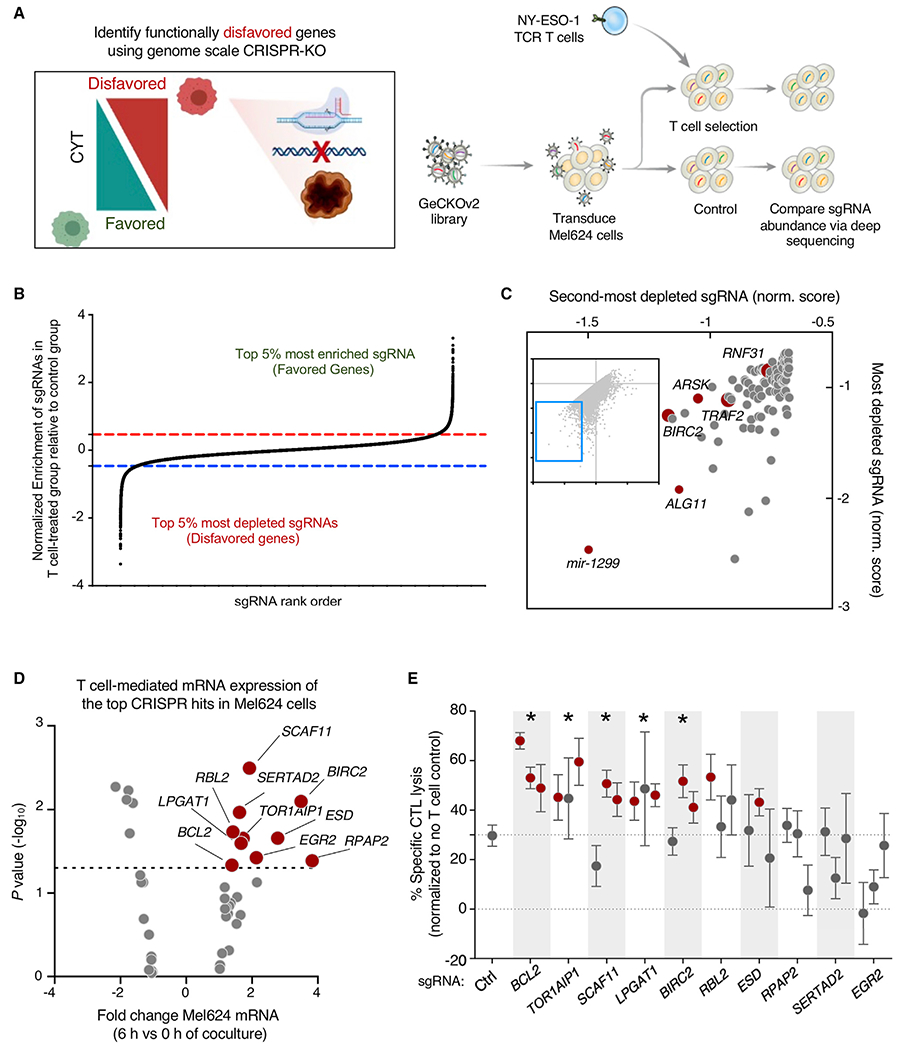
(A) Comprehensive functional genomics analysis of the potential for individual tumor genes to disfavor tumor killing by T cells. Mel624 melanoma cells were transduced with the Genome-scale CRISPR Knockout (GeCKOv2) library and cultured with ESO T cells for 6 h. sgRNA representation was measured in control and ESO T cell-treated tumor cells. Enrichment and depletion of sgRNA representation in ESO T cell-treated tumor cells was compared with control treated cells.
(B) Representation of sgRNA abundance in T cell-treated tumor cells relative to control tumor cells. The top 5% most depleted sgRNAs (targeting tumor genes whose expression disfavors T cell killing of tumor) and top 5% most enriched sgRNAs (targeting tumor genes whose expression favors T cell killing of tumor) are indicated.
(C) Scatterplot of the normalized enrichment of the most-depleted sgRNA versus the second-most depleted sgRNA for all genes after ESO T cell treatment (top 100 most depleted genes depicted in enlarged region.
(D) Volcano plot representing differentially expressed tumor genes (identified in Figure 1H) that were identified as being significantly depleted from duplicated CRISPR 2CT screens by RIGER analysis (n = 42 genes). Expression of genes was analyzed by RNA-seq, with abundance represented as relative fold change (x axis) versus significance (y axis). Ten genes that met CRISPR screen depletion criteria were significantly induced by T cell engagement (red dots; p < 0.05, fold change >1.3.
(E) The effect of gene knockout on tumor susceptibility to killing by T cells was measured by flow cytometry. Tumor genes identified in (D) were targeted with 3 independent sgRNAs. A375 melanoma cells were transduced with each sgRNA and Cas9, co-cultured with ESO T cells, and tumor killing assayed. Statistically significant sgRNAs for individual genes are indicated by red dots, as they resulted in a significant increase in tumor killing by T cells relative to non-targeting sgRNA (p < 0.05, Student’s 2-tailed t test). A gene was considered validated (indicated with *) if ≥2 sgRNAs targeting that gene resulted in statistically significant increases in T cell killing. Error bars depict standard deviation.
Data are representative of 2 independent experiments, with 4 technical replicates per experiment.
iD-CTL genes that functionally disfavor T cell CYT against tumor cells survival represent an acute mechanism to promote immunotherapy resistance. To identify such genes, we assessed the T cell-induced transcriptional changes in tumor genes whose targeting sgRNAs were significantly depleted in biological replicate CRISPR screens. Ten genes had statistically significant increases in mRNA expression after T cell engagement (Table S4), including SCAF11, BIRC2, RPAP2, and BCL2 (Figure 3D). We validated the functional role of these genes in promoting tumor resistance to T cell-mediated killing by transducing A375 cells with Cas9 and three newly designed sgRNA constructs targeting each candidate gene. We conducted an arrayed co-culture screen with ESO T cells and assessed the impact of gene knockout on tumor cell survival. Knockout of five of the candidate genes, BCL2, BIRC2, LPGAT1, SCAF11, and TOR1AIP1, resulted in significantly increased tumor cell death upon T cell co-culture (Figure 3E). Analysis of editing efficiency for validated sgRNAs revealed generally high levels of indel formation, confirming efficient editing of targeted genes (Figure S3G).
Targeting iD-CTL genes identify combination immunotherapy targets
We assessed the potential for targeting active tumor defense mechanisms in combination with immunotherapy by developing a pipeline for identifying potential targets for small molecule or biologic inhibition based on these datasets. As a proof of principle demonstration that this strategy can successfully identify targets for combination therapy, we considered the five validated iD-CTL genes (Figure 3E) along with the top 250 genes from duplicate CRISPR screens as ranked by rank-sum analysis of second-most depleted guide. We mapped genes to proteins (Table S5A) and queried proteins for druggability using the Drug Gene Interaction Database (DGIDB) (Griffith et al., 2013) (Table S5B), followed by manual curation. We identified 15 targets for which inhibitors were available, including BIRC2, BCL2, ITGAV, and DNPEP (Table S5C). When possible, we obtained two independent inhibitors per target to mitigate drug-specific effects. We assessed the impact of inhibitors on T cell elimination of tumor cells by performing a high-throughput assay in which ESO T cells were co-cultured with A375 melanoma cells in the presence of vehicle or one of two concentrations of each inhibitor for 16 h (Figure 4A). We determined that several inhibitors increased ESO T cell elimination of tumors (Figures 4B and S4A). We concluded that inhibitors of interest must improve tumor cell killing as a combination with ESO T cells relative to ESO T cell killing alone and should improve tumor cell killing as a combination with ESO T cells relative to inhibitor alone. Several inhibitors met these criteria, including those targeting BIRC2 (birinapant and LCL161), ITGAV (cilengitide and α-ITGAV monoclonal antibody), DNPEP (CHR2797), BCL2 (Abt199), and ERRα (C29) (Figures 4C and S4B).
Figure 4. Targeting tumor-expressed genes disfavoring T cell killing augments CYT.
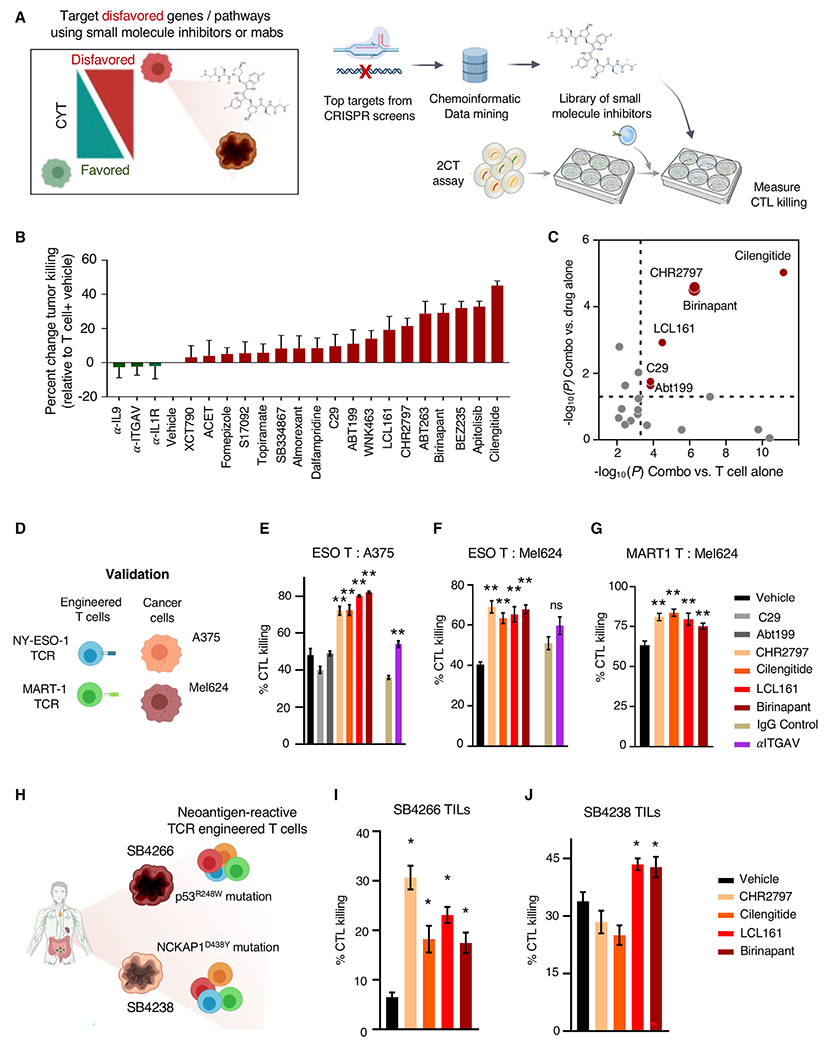
(A) Workflow used to identify and screen inhibitors for capacity to increase T cell-mediated destruction of tumor cells. Top 250 hits from CRISPR screen were filtered to identify targets with readily available inhibitors. Tumor cells were co-cultured with antigen-specific T cells, and tumor killing was measured.
(B) A375 melanoma cells were co-cultured with ESO T cells at a 1:2 E:T ratio for 16 h in the presence or absence of inhibitors. Co-cultures were washed and tumor cell viability was measured with WST1 viability reagent. The impact of inhibitors on ESO T cells of A375 cells was measured at 500-nM inhibitor concentrations (except for α-ITGAV at 250 ng/mL and fomepizole and dalfampridine at 500 μM.
(C) Plot of p values (significance threshold p < 0.05, dotted lines) between tumor elimination by the combination of inhibitor and ESO T cell treatment versus ESO T cells alone (x axis) and tumor elimination by the combination treatment versus inhibitor alone (y axis).
(B and C) p values calculated by 2-tailed Student’s t test. Data are representative of 3 independent experiments, with 4 technical replicates per sample.
(D) Depiction of experimental design for validation experiments. Tumor cells were co-cultured with T cells at an E:T ratio of 1:2 for 16 h in the presence or absence of inhibitors, and tumor cell viability was measured by flow cytometry.
(E) A375 melanoma cells were co-cultured with ESO T cells along with inhibitors at 5 μM, except for α-ITGAV at 2.5 μg/mL, and tumor cell elimination was measured by flow cytometry.
(F) Mel624 melanoma cells were co-cultured with ESO T cells along with inhibitors at 5 μM, except for α-ITGAV at 2.5 μg/mL, and tumor cell elimination was measured by flow cytometry.
(G) Mel624 melanoma cells were co-cultured with MART-1 T cells along with inhibitors at 5 μM, except for α-ITGAV at 2.5 μg/mL, and tumor cell elimination was measured by flow cytometry.
(H) Primary colon cancer cell lines SB4266 and SB4238 (expressing indicated mutations driving generation of neoantigens) were co-cultured with T cells engineered to express neoantigen-reactive TCRs in the presence or absence of inhibitors, and tumor killing was measured by flow cytometry.
(I) Colon cancer cell line SB4266 was co-cultured with T cells transduced with a TCR reactive against p53R248W mutation along with inhibitors at 5 μM. Tumor cell death was measured by flow cytometry.
(J) Colon cancer cell line SB4238 was co-cultured with T cells transduced with a TCR reactive against NCKAP1D438Y along with inhibitors at 5 μM. Tumor cell death was measured by flow cytometry.
Data are representative of 4 (E and F), 3 (G), or 2 (I and J) independent experiments, with 4 technical replicates per group. Error bars depict standard deviation.
Statistical significance calculated by 1-way ANOVA with multiple comparisons corrected with Dunnett’s adjustment. *p < 0.05; **p < 0.01.
We validated the impact of combination therapy targeting iD-CTL genes with these inhibitors in 2CT assays across several tumor antigens and cancer types (Figure 4D). To allow for single-cell analysis of tumor killing, we measured tumor cell viability after 2CT co-culture using FACS. Consistent with our initial findings, inhibition of BIRC2 (birinapant and LCL161), ITGAV (cilengitide and α-ITGAV), and DNPEP (CHR2797) increased ESO T cell killing of A375 melanoma cells across four independent human T cell donors (representative data from one donor depicted in Figure 4E, all donors with variability in killing between each healthy donor T cell product shown in Figure S4C). We determined that increased tumor cell death from combination therapy was dependent on the presence of T cells for each validated hit (Figure S4D). Importantly, we also found that except for α-ITGAV, each inhibitor increased ESO T cell killing of HLA-A*0201 and NY-ESO-1 antigen expressing Mel624 melanoma cells (Figure 4F). Demonstrating that the impact of the inhibitors as combination therapy was not tumor antigen-specific, combination treatment with T cells transduced with a TCR conferring recognition of melanoma antigen recognized by T cells 1 (MART-1) also increased the elimination of MART-1 expressing Mel624 (Figure 4G).
To determine whether combination treatment could increase neo-antigen-reactive T cell elimination of epithelial tumor cells, we used two independent patient-derived colon cancer cell lines, SB4238 and SB4266. These lines were co-cultured with T cells engineered to express TCRs reactive against mutated proteins expressed by each tumor line (Figure 4H). In SB4238, a non-synonymous mutation in NCKAP1 (G1312T, Figure S4E) resulted in an D438Y amino acid substitution that was recognized by a TCR identified in tumor-infiltrating lymphocytes (TILs) (Figures S4F and S4G). A TCR reactive against SB4266 through specific recognition of p53 R248W was previously described (Malekzadeh et al., 2019). TCR-transduced T cells alone eliminated a modest percentage of SB4266 cells, and this increased in the presence of inhibitors of BIRC2 (birinapant and LCL161), DNPEP (CHR2797), and ITGAV (cilengitide) (Figure 4I). However, only BIRC2 inhibitors birinapant and LCL161 increased neo-antigen-reactive T cell elimination of SB4238 tumor cells (Figure 4J). Secondary genetic validation of the impact of targeting BIRC2 and ITGAV through CRISPR-Cas9-mediated deletion demonstrated that the loss of either target sensitized tumor cells to T cell-mediated killing (Figures S4H and S4I). To better understand the specificity of the BIRC2 inhibitors, which can also inhibit other IAP family proteins at higher concentrations, we profiled the expression of BIRC2 and BIRC3 in A375 and Mel624 melanoma. RNA-seq analysis demonstrated that in each cell line, BIRC2 was predominantly expressed (Figure S4J). We also studied the impact of LCL161 and birinapant on BIRC2, BIRC3, and X-linked inhibitor of apoptosis protein (XIAP) protein expression, finding that treatment with either drug resulted in selective reduction in BIRC2 expression without altering BIRC3 or XIAP protein levels (Figure S4K).
Tumor expression of BIRC2 disfavors T cell CYT through suppression of IRF1-mediated antigen presentation pathway activity
As BIRC2 was identified and validated as iD-CTL gene and a potent target for combinatorial inhibition with T cell treatment, we next assessed the mechanism by which BIRC2 regulates T cell CYT. We observed that protein expression of BIRC2 was induced by the treatment of tumor cells with cytokines produced by T cells, including IFN-γ or TNF-α (Figure 5A), suggesting that T cell recognition and response to tumors can mediate the upregulation of BIRC2 expression. While several studies have previously demonstrated that BIRC2 can alter tumor cell susceptibility to inflammatory cytokines (Vredevoogd et al., 2019) and suppress chemokines that attract immune cells (Samanta et al., 2020), we determined that BIRC2 inhibition resulted in directly increased T cell recognition of tumor cells as determined by the production of IFN-γ (Figure 5B) or TNF-α (Figure S5A) after a 6-h co-culture with A375 melanoma. Importantly, this increased cytokine production was not a result of inhibitor action on T cells, as stimulation of ESO T cells with phorbol myristate acetate (PMA)/ionomycin in the presence of LCL161 did not result in increased IFN-γ (Figure S5B) or TNF-α (Figure S5D). Similarly, the stimulation of ESO T cells with αCD3/CD28 in the presence of LCL161 did not result in increased IFN-γ or TNF-α (Figures S5C and S5E). Genetic targeting of BIRC2 expression through CRISPR-Cas9 editing of A375 melanoma also resulted in increased T cell recognition of tumors as measured by increased IFN-γ (Figure 5C) and TNF-α (Figure S5F) after co-culture. We found that BIRC2 inhibition increased the ability of neoantigen-reactive T cells to recognize colon cancer 4238 as measured by 4-1BB upregulation (Figure S5G), confirming that this effect is not limited to melanoma or the NY-ESO-1 antigen. Importantly, BIRC2 inhibition did not increase either 4-1BB upregulation or tumor killing by donor-matched T cells transduced with GFP control (Figures S4G and S4H).
Figure 5. Tumor BIRC2 expression inhibits T cell recognition of tumor through negative regulation of IRF1-mediated upregulation of antigen presentation pathway genes.
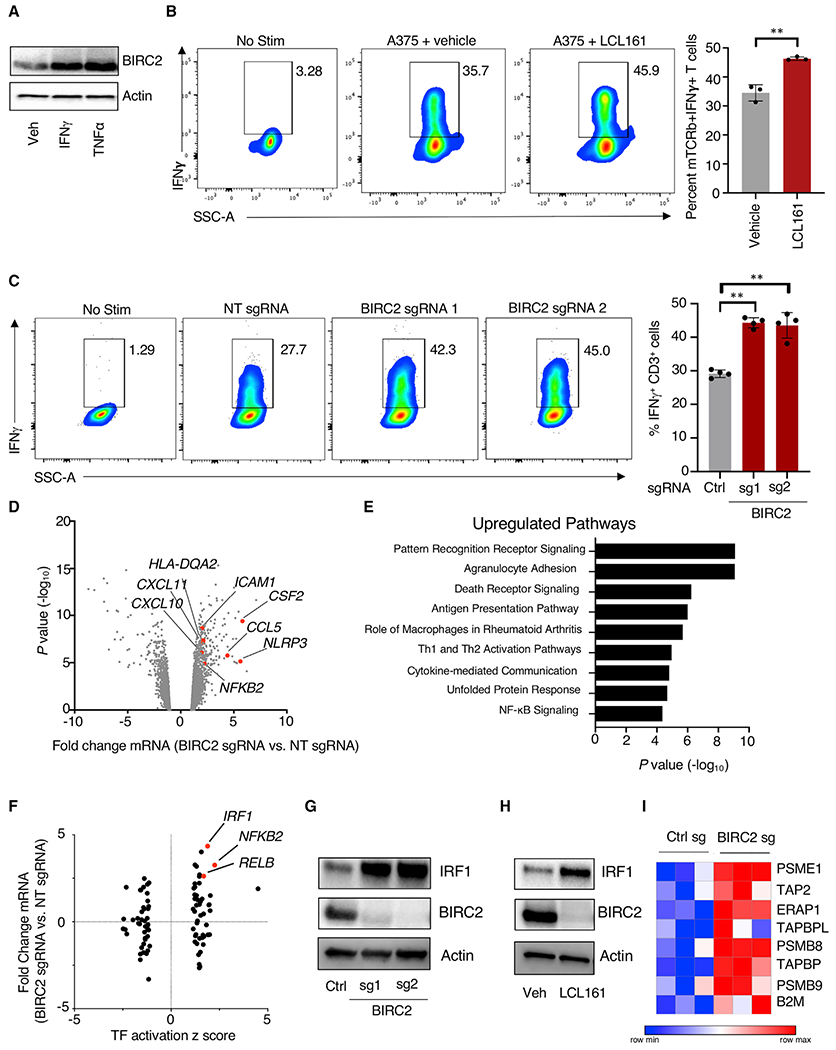
(A) A375 melanoma cells were treated with IFN-γ (100 ng/mL) or TNF-α (100 ng/mL) for 8 h, and western blot analysis was performed.
(B) ESO T cells were co-cultured with A375 melanoma cells in the presence or absence of 5 μM LCL161 for 5 h along with brefeldin A and monensin, and T cell intracellular production of IFN-γ was measured by flow cytometry.
(C) A375 melanoma cells transduced with Cas9 and non-targeting (NT) or BIRC2-targeting sgRNAs were co-cultured with ESO T cells for 5 h in the presence of brefeldin A and monensin, and T cell intracellular production of IFN-γ was measured by flow cytometry.
(D) Volcano plot representing differentially expressed genes in A375 cells transduced with Cas9 and BIRC2 targeting sgRNA versus NT sgRNA. Expression of genes was analyzed by RNA-seq and abundance represented as relative fold change (x axis) versus significance (y axis).
(E) Ingenuity Pathway Analysis of genes significantly upregulated in BIRC2-targeting sgRNA transduced A375 cells.
(F) Ingenuity transcription factor (TF) analysis of genes significantly upregulated in BIRC2-targeting sgRNA-transduced A375 cells. TF activation Z scores are depicted on the x axis and the fold change in mRNA for each TF is shown on the y axis.
(G) A375 cells were transduced with Cas9 and NT sgRNA or BIRC2-targeted sgRNAs, and western blot analysis was performed.
(H) A375 cells were treated with vehicle or 5 μM LCL161 for 16 h and western blot analysis was performed.
(I) Heatmap of mRNA expression of differentially expressed genes in IRF1-related antigen presentation pathway (Rettino and Clarke, 2013) in A375 cells transduced with Cas9 and NT sgRNA or BIRC2-targeted sgRNAs.
Data are representative of 3 independent experiments (A–C and G–H) or are pooled from a single experiment, with 3 biological replicates per group (D–F and I).
Error bars depict standard deviation. Statistical significance was evaluated by 2-tailed Student’s t test. **p < 0.01.
Having demonstrated that pharmacological and genetic perturbation of BIRC2 resulted in increased T cell recognition of tumors, we next sought to identify the mechanistic underpinnings of this phenotype. We conducted RNA-seq on A375 melanoma cells transduced with Cas9 and a non-targeting control or the BIRC2-targeted sgRNA construct. BIRC2-perturbed tumor cells exhibited increased expression of genes associated with inflammation (NLRP3) and inflammatory chemokines (CCL5 and CXCL10), along with additional immune regulators (Figure 5D). Pathway analysis of genes upregulated by BIRC2-perturbed revealed enrichment for antigen presentation pathway genes and other inflammatory signaling pathways (Figure 5E). Importantly, bioinformatic analysis of RNA-seq data demonstrated strong transcriptional upregulation of the transcription factor (TF) IRF1 and a marked increase in IRF1-related transcriptional activity (Figure 5F). We confirmed that CRISPR-mediated targeting of BIRC2 (Figure 5G), along with pharmacological inhibition (Figure 5H), each resulted in strong upregulation of IRF1 protein. Genetic depletion of BIRC2 resulted in the increased expression of previously annotated IRF1 target genes (Rettino and Clarke, 2013) related to the antigen presentation pathway (Figure 5I). Consistent with BIRC2 inhibition resulting in increased IRF1-mediated antigen presentation pathway activity, we observed that pharmacological inhibition of BIRC2 resulted in increased tumor cell surface expression of human leukocyte antigen (HLA) class I (Figure S6A) and β2M (Figure S6B). Genetic targeting of BIRC2 also resulted in significant increases in tumor cell surface expression of HLA class I (Figure S6C) and β2M (Figure S6D). Genetic (Figure S6E) or pharmacological (Figure S6F) targeting of BIRC2 also increased the total expression of HLA class I, along with increasing the expression of intercellular adhesion molecule 1 (ICAM1), a protein that can increase T cell binding and lysis of tumors upon antigen encounter (Lefor and Fabian, 1998). Proteomics analysis of BIRC2-perturbed A375 cells similarly demonstrated increased the protein-level expression of antigen presentation pathway-associated genes (Figures S6G and S6H). We found that pharmacological inhibition of BIRC2 rapidly increased the tumor expression of IRF1, with notable increases in protein expression as soon as 1 h after the addition of LCL161 (Figure S6I). Using a pulse-chase assay with cycloheximide to stop the synthesis of new proteins, we found that BIRC2 inhibition resulted in the increased protein stability of IRF1 (Figure S6J). Importantly, the expression of IRF1 is necessary for the increased tumor susceptibility to T cell lysis driven by BIRC2 inhibition, as CRISPR-mediated targeted on IRF1 abrogated the ability of LCL161 to increase the T cell killing of A375 cells (Figure S6K). In line with previous work (Samanta et al., 2020), we also observed that genetic perturbation of BIRC2 resulted in the increased tumor cell production of inflammatory chemokines CCL2, CCL5, and CXCL10 (Figure S6L) and resulted in increased T cell migration toward tumor targets (Figure S6M).
In vivo targeting of BIRC2 enhances ACT in murine model
Next, we studied the effects of BIRC2 inhibition in vivo. We established that BIRC2 inhibitors were capable of increasing murine tumor cell susceptibility to T cell-mediated killing. Co-culture of B16 melanoma cells with pmel TCR-transgenic T cells in the presence of BIRC2 inhibitors birinapant or LCL161 significantly increased tumor cell lysis (Figure S6N). We found that the in vivo inhibition of BIRC2 with either LCL161 or birinapant (Figure 6A) improved the ability of pmel-transgenic T cells to control B16 melanoma tumor growth (Figures 6B and S6O) and improved overall survival (Figure 6C), with a majority of mice treated with combination therapy, demonstrating complete tumor regressions (Figure 6D). We next studied the impact of genetic perturbation of tumor BIRC2 in vivo. We used CRISPR-Cas9 to deplete BIRC2 in B16 melanoma (Figure S6P) and inoculated mice with either control or BIRC2-depleted tumors (Figure S6Q). Genetic perturbation of BIRC2 in B16 melanoma resulted in increased tumor control by pmel T cells (Figure 6E) and an increase in mouse survival (Figure 6F). Collectively, our data support the conclusion that BIRC2-mediated restraint of IRF1-driven tumor antigen presentation pathway gene expression limits the efficacy of ACT, making BIRC2 a rational target for inhibition in conjunction with immunotherapy.
Figure 6. Targeting BIRC2 in combination with adoptive transfer of T cells increases antitumor efficacy.
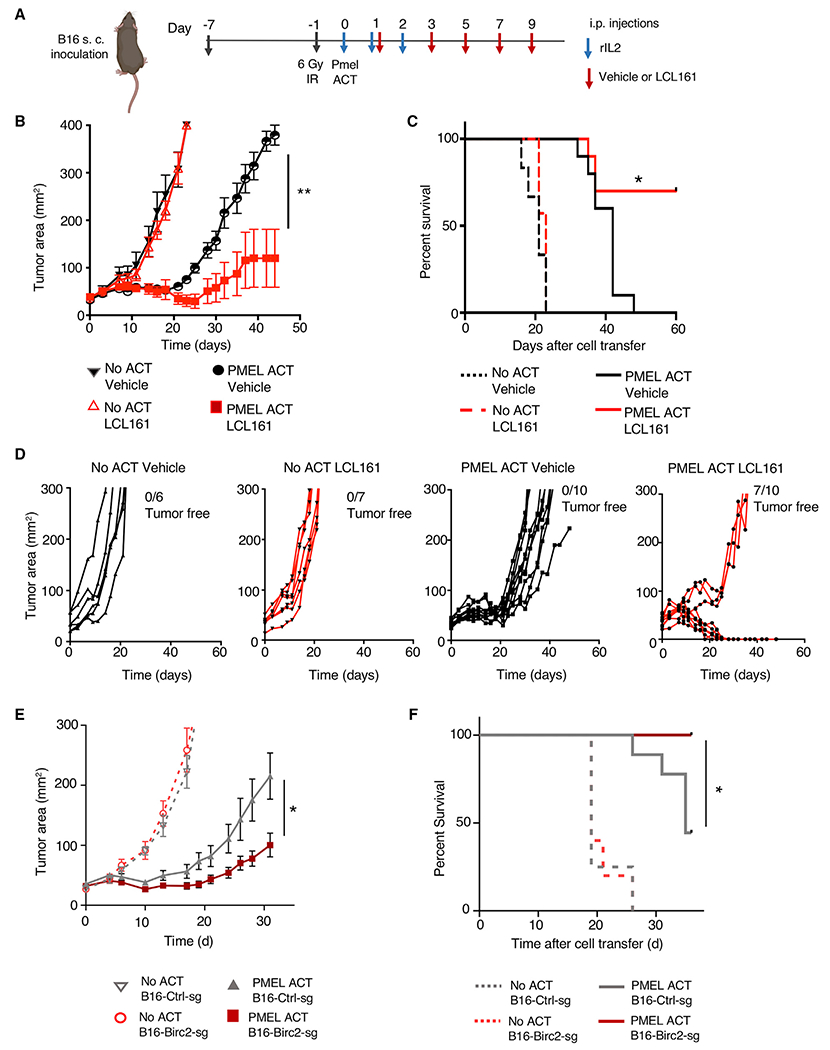
(A) Experimental design for small-molecule in vivo combination studies. Tumor growth of hybrid gp100-expressing B16 melanoma (Hanada et al., 2019) in mice receiving adoptive cell transfer of pmel1 T cells along with vehicle or LCL161 (30 mg/kg body weight). Mice were treated with LCL161 via intraperitoneal (i.p.) injection every 2 days beginning 24 h after pmel1 T cell infusion for a total of 5 doses. Mice also received 3 doses of rIL-2.
(B–D) Tumor area (B), (C) mouse survival, and (D) individual outcomes for each treated mouse are depicted.
(E and F) Tumor growth curve (E) and (F) survival of mice with subcutaneous B16 melanoma tumors modified with Cas9 and non-targeting sgRNA or BIRC2-targeting sgRNA treated with pmel1 T cells.
Data are representative of 3 (B–F) experiments, with at least 5 mice per group. Error bars depict SEM. Statistical significance was calculated using Wilcoxon rank-sum test (B and E) or log rank test (C and F). *p < 0.05; **p < 0.01.
DISCUSSION
Understanding the mechanisms tumors use to escape immunotherapy and developing strategies to combat these mechanisms can lead to optimized treatment strategies. Our goal was to identify tumor-expressed genes that disfavor the T cell cytolysis of tumors. We used bioinformatics-based screening of TCGA datasets to identify genes inversely correlated with T cell CYT in highly immune-infiltrated tumors. While these analyses suggest that intratumoral expression of genes can limit T cell cytolytic activity even in patient tumors highly infiltrated with T cells, the cellular origin of gene expression cannot be determined from this analysis and single-cell profiling of the tumor microenvironment may allow for better determination of mechanisms of tumor resistance to T cell CYT. Gain- and loss-of-function 2CT genomics screens and integration with dynamic transcriptional analysis of tumor responses to T cells allowed us to identify tumor genes disfavoring CYT that act as an active defense against immunotherapy. Alongside genes such as BCL2 and BIRC2 that have been implicated in tumor resistance to conventional therapies (Jung et al., 2015; Sartorius and Krammer, 2002), we found that genes with no previously described role in resistance acted to promote tumor survival in this context, including TOR1AIP1, SCAF11, and LPGAT1. Taken together, these genes represent components of an orchestrated tumor cell defense mechanism that limits the cytotoxic effect of T cells on tumors.
The observation that constitutive activity of various pro-growth and survival cell signaling pathways can promote tumor resistance to immunotherapy is also notable. Tumor cell expression of BCL2 and BCL-xL protected from T cell killing independently of the PD-L1 expression level, suggesting that pathways previously observed to protect tumor cells from chemotherapy and targeted inhibitors (Martz et al., 2014; Singleton et al., 2017) can function similarly in immunotherapy. The observation that constitutive NOTCH1-driven signaling in melanoma strongly sensitized tumor cells to T cell-mediated killing was unexpected and could reflect decreased tumor expression of PD-L1 (Figure S2B). However, given the key role of Notch signaling in regulating T cell differentiation and activity (Kelliher and Roderick, 2018), additional mechanisms may play a role in this observation.
We assessed the therapeutic potential of targeting tumor-expressed genes disfavoring T cell CYT to augment the efficacy of immunotherapy. Inhibiting several of these targets, including BIRC2, ITGAV, and DNPEP, increased T cell-mediated destruction of tumor cells in vitro. These inhibitors mediated increased T cell cytolytic activity across several tumor types and antigens, suggesting that combination therapy could sensitize tumors that respond poorly to immunotherapy. It is notable that gene targets inhibited by tosedostat (CHR2797) were associated with limited CYT in highly T cell-infiltrated tumors in human melanoma patients (Figure 1E), and we determined that the addition of CHR2797 to T cell/tumor cell co-cultures functionally improved tumor lysis by T cells (Figures 4E–4I). This orthogonal evidence of the activity of CHR2797 as a combination agent with antitumor T cells merits further investigation to evaluate the potential as a clinical strategy. Careful consideration of target selectivity and tumor specificity of cell therapies should be performed in the context of combination immunotherapies with inhibitors targeting BIRC2 or other tumor-resistance mechanisms. Mechanisms that lead to increased antitumor activity of T cells in the context of combination therapy may also conceivably increase the undesired recognition of normal tissues, particularly in the context of allogeneic cell products or targeting tumor antigens that are also expressed by normal tissues.
While previous functional genomics studies have identified BIRC2 as a negative regulator of susceptibility to immunotherapy (Manguso et al., 2017; Pan et al., 2018; Samanta et al., 2020; Vredevoogd et al., 2019), we show an additional role for BIRC2 in disfavoring the T cell killing of tumor cells. Tumor expression of BIRC2, potentiated by inflammatory cytokines produced by T cells, can act as an early suppressor of IRF1-mediated upregulation of antigen presentation pathway proteins that drive T cell recognition and killing of tumor cells. While we did not observe an impact of BIRC2 pharmacological inhibition on T cell activation or production of inflammatory cytokines in the absence of tumor target, previous work has suggested that BIRC2 may act as a T cell-intrinsic inhibitor of inflammatory activity (Dougan et al., 2010). These previous studies have observed T cell-intrinsic increases in cytokine production following prolonged stimulation in the presence of BIRC2 inhibitors, but our findings suggest that direct effects on T cell biology are not responsible for increased cytokine production upon short-term co-culture with tumors expressing cognate antigens.
Overall, our work here describes the integration of functional genomics with dynamic transcriptional analysis of the tumor response to T cells to identify tumor active defense mechanisms against cellular immunotherapy. Targeting these tumor defenses in combination with immunotherapy can increase the ability of T cells to destroy cancer. Recent work has demonstrated the possibility of conducting functional genomics studies in primary T cells (Shifrut et al., 2018), suggesting that similar approaches may be used to identify T cell-intrinsic genes that inhibit the ability to eliminate tumor cells and may also represent key targets for combination immunotherapy. Integrating functional genomics data from both tumor and T cell settings may help to optimize new immunotherapy strategies to promote increased therapeutic effects and improved patient outcomes.
Limitations of the study
While our work here establishes tumor mechanisms of resistance to cellular immunotherapy, our in vitro model systems are simplified compared with physiological conditions in human patients. Our studies used a binary interaction between T cells and tumor cells to allow for understanding the features of the tumor-T cell interaction, but in vivo T cell recognition and destruction of tumor cells takes place in a much more complex system, with numerous additional cell types potentially influencing the interaction. While we validate our findings using in vivo models, these may not fully reflect complex human biology. In addition, the incorporation of small-molecule inhibitors into cell therapy treatment regimens may affect patient morbidity, suggesting a careful analysis of risks and benefits should be performed before pursuing combination immunotherapy approaches in humans.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents may be directed to and will be fulfilled by the lead contact, Rigel J. Kishton (kishtonr@gmail.com).
Materials availability
Plasmids described in this manuscript will be shared upon request and completion of MTA.
Data and code availability
RNA sequencing data generated during this study are available at the NCBI-GEO database with the project accession ID GEO: GSE137824. Proteomics datasets generated during this study are available at MassIVE: MSV000089737.
All original code has been deposited at Github and is publicly available as of the date of publication (https://github.com/NIDAP-Community/Cancer-genes-disfavoring-T-cell-immunity).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal experiments were approved by the Institutional Animal Care and Use Committees of the NCI and were performed in accordance with NIH guidelines. C57BL/6NCR mice were obtained from Charles River Laboratories at NCI Frederick. B6.Cg-Thy1a/CyTg(TcraTcrb)8Rest/J (PMEL1) mice were purchased from Jackson Laboratory. All mice were maintained under specific pathogen-free conditions. Female mice aged 6–8 weeks were used for in vivo experiments.
Cell lines
Melanoma cell lines Mel624.38 were isolated from surgically resected metastases as previously described (Robbins et al., 2008) and were cultured in RPMI 1640 (Invitrogen) medium supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, UT), 2 mM L-glutamine and 1% penicillin-streptomycin. A375 melanoma cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured in RPMI 1640 medium supplemented with 10% FBS, 2 mM L-glutamine and 1% penicillin-streptomycin. 4238 and 4266 cells were generated from primary human tumor cultures and were cultured in RPMI 1640 medium supplemented with 10% FBS, 2 mM L-glutamine and 1% penicillin-streptomycin. Chimeric gp100 expressing B16 melanoma cells were generated as described in (Hanada et al., 2019) and were cultured in RPMI 1640 medium supplemented with 10% FBS, 2mM L-glutamine and 1% penicillin-streptomycin. Cell type identity of Mel624.38 and A375 melanoma was verified with STR analysis and all cell lines were confirmed as mycoplasma-free using a commercial testing kit.
Culture of primary human lymphocytes
Transduced T lymphocytes were cultured in RPMI 1640 supplemented with 10% FBS, 2 mM L-glutamine and 1% penicillin-streptomycin with 150 IU/mL interleukin 2.
Human specimens
Peripheral blood mononuclear cells (PBMCs) were isolated from healthy donors and tumor samples were isolated from patients with melanoma and GI cancers. All human specimens were collected with informed consent and procedures approved by the institutional review board (IRB) of the National Cancer Institute (NCI). Donor age and sex information for all human PBMC specimens was not collected due to anonymized collection procedures. Similarly, age and sex of patients providing tumor samples was not provided for this study in accordance with patient confidentiality.
Culture of Pmel T cells
Primary mouse T cells were cultured in RPMI supplemented with 10% FBS (Hyclone), 2mM L-glutamine, and 1% penicillin-streptomycin (Life Technologies), 1% MEM NEAA supplement (Life Technologies), 1% Sodium Pyruvate supplement (Life Technologies) and 0.1% 2-Mercaptoethanol (Life Technologies). Splenocytes from Pmel mice were stimulated with hgp10025-33 peptide (1μM) and 60 IU/mL recombinant human IL2 (Aldesleukin Proleukin; Novartis). CD8+ T cells were harvested on day 7 and used for further experiments.
METHOD DETAILS
The cancer genome Atlas bioinformatics analysis
Data analysis and visualization were performed in the NIH Integrated Analysis Portal (NIDAP) using R programs developed on Foundry (Palantir Technologies). Normalized RNA-Seq gene expression data for TCGA Skin Cutaneous Melanoma (SKCM) was downloaded from the FireBrowse portal (https://gdac.broadinstitute.org/) and subsequently log-normalized using limma-voom (Ritchie et al., 2015). Further downstream analyses were limited to samples having high (above median) CD3E gene expression. Correlation analysis was performed on all genes to the combined geometric mean of PRF1 and GZMA expression (CYT). Pathway enrichment of genes with significantly negative Pearson’s Correlation Coefficients (p<0.05) was performed using l2p (https://github.com/ccbr/l2p) on GO, KEGG, and various MSigDB pathway databases, including Hallmark (Liberzon et al., 2015), C2 and C7 collections. Survival analysis of samples with high vs. low expression of significantly negatively correlated genes was performed using survminer (Alboukadel Kassambara, Marcin Kosinski and Przemyslaw Biecek (2019). survminer: Drawing Survival Curves using ‘ggplot2’. R package version 0.4.6. https://CRAN.R-project.org/package=survminer). Code is available on Github (https://github.com/NIDAP-Community/Cancer-genes-disfavoring-T-cell-immunity).
Generation of TCR expression plasmids
Retroviral vectors for TCRs recognizing the HLA-A*02-restricted melanoma antigens NY-ESO-1 (NY-ESO-1:157-165 epitope) and MART-1 (MART-1:27-35 epitope, DMF5) were generated as previously described (Johnson et al., 2006; Robbins et al., 2008). Neoantigen-reactive TCRs were identified and expressed in primary human T cells as previously described (Lo et al., 2019; Malekzadeh et al., 2019).
Transduction of human T cells with TCRs
For transduction of human T cells, CD8+ T cells seeded at 2 × 106 cells per well in a 24-well plate were stimulated with anti-CD3 antibody OKT3 (5 ug/mL coated) and anti-CD28 antibody (5 ug/mL soluble) along with IL-2 (200 IU/mL) on day 0. Non-tissue culture treated 24-well plates were coated with 0.5 mL per well of 10 μg/mL RetroNectin (Takara) on day 1 and stored overnight at 4°C. Vector supernatant (1 mL per well) was added to plates on day 2 followed by centrifugation at 2000g for 2h at 32°C. 800 μL was aspirated and T cells were added at 1 × 106 cells/mL, centrifuged for 10 min at 1500 rpm and incubated overnight. A second transduction was performed the following day as described above. Cells were subsequently maintained in culture at 1 × 106 cells/mL and expanded until day 10, after which they were used or cryo-preserved for future use.
Lentiviral production and purification
Lentiviral particles were produced and purified as described previously (Patel et al., 2017). In brief, HEK293FT cells (Invitrogen) were cultured in DMEM supplemented with 10% FBS, 2 mM L-glutamine and 1% penicillin-streptomycin. One day prior to transfection, HEK293FT cells were seeded in T-225 flasks at 60% confluency. One hour prior to transfection, media was aspirated and replaced with 13 mL OptiMEM media (Invitrogen). Each flask of cells were transfected with 100 μL Lipofectamine 200 and 200 μL Plus reagent (Invitrogen) along with 20 μg of lentiCRISPRv2 plasmid or pooled plasmid human GeCKOv.2 (Genome-scale CRISPR knockout) library, 15 μg psPAX2 and 10 μg pMD2.G. 6–8 h after transfection, media was replaced with 20 mL of DMEM supplemented with 10% FBS and 1% BSA. Media containing viral particles was collected 48 h post-transfection and titer was assayed with Lenti-X GoStix (Clontech). Viral supernatant was centrifuged at 3,000 r.c.f. at 4°C for 10 min followed by filtration through 0.45 μm low-protein binding membrane. For pooled library plasmids, viral supernatants were concentrated by centrifugation at 4,000 r.c.f at 4°C for 35 min in Amicon Ultra-15 filters (Millipore Ultracel-100K). Concentrated viral supernatants were stored in aliquots at −80°C.
2CT T cell and tumor cell co-culture
Two days prior to co-culture, T cells were thawed in T cell media containing 3 U/mL DNase (Genentech Inc.) overnight. Tumor cells were seeded at desired density on this day in T cell media. After 24 h, T cells were cultured in T cell media supplemented with 300 IU/mL IL-2 for 24 h. T cells were co-cultured with tumor cells at various effector: target (E:T) ratios for specified durations. After co-culture, T cells were removed by washing tumor cells with PBS and tumor cells were detached using trypsin. Cells were stained with fixable Live/Dead dye (Invitrogen) followed by human anti-CD3 antibody (clone SK7, BD) in FACS staining buffer (PBS + 0.2% BSA). Cell counts were normalized with CountBright Absolute Counting Beads (Invitrogen) by FACS.
2CT GeCKOv.2 screens, genomic DNA extractions and screen analysis
2CT genome-wide CRISPR screens were performed as previously described (Patel et al., 2017), with some modifications. In brief, Mel624 cells were transduced independently with both A and B GeCKOv.2 libraries. For each screen, cells were split into two groups of 5 × 107 transduced Mel624 cells. One group was co-cultured with ESO T cells at an E:T ratio of 1:3 for each library. A second group of transduced Mel624 cells were cultured under the same density and conditions, but without ESO T cells. The co-culture phase was maintained for 6 h, after which the T cells were removed as described above. The recovery phase was maintained for another 48 h and surviving cells were frozen to evaluate sgRNA depletion. gDNA was extracted from frozen tumor cells using previously optimized (Chen et al., 2015) ammonium acetate and alcohol precipitation procedure to isolate gDNA with AL buffer (Qiagen) substituted for the initial cell lysis step. sgRNA abundance was detected as previously described (Patel et al., 2017).
Arrayed validation of targets with CRISPR/Cas9
Selected targets that were identified from the genome-scale CRISPR screen and DTA analysis were further studied using additional sgRNA targeting sequences. We utilized 3 newly designed sgRNA guide sequences for each gene as listed in Table S6. We cloned these sgRNAs into the lentiGuide-Puro vector as previously described (Sanjana et al., 2014). A375 cells were transduced with lentiCas9-Blast (Addgene) and selected with 5 μg/mL blasticidin for 10 days, after which cells were transduced with lentiGuide-Puro sgRNA constructs. Cells were then selected with 1 μg/mL puromycin for 7 days. In some experiments (Figures 5 and 6), sgRNA guide sequences were cloned into the lentiCRISPRv2 vector for mechanistic studies as previously described (Patel et al., 2017).
Negative control non-targeting sgRNA used in validation study was selected based on comparison of multiple non-targeting sgRNAs utilized in Patel et al. (2017). To control for off-target activity of sgRNAs and for intergenic double-strand breaks, we considered only those genes valid that have at least two validation sgRNAs showing sensitivity to T cell killing in 2CT assays.
RNA sequencing and analysis
mRNA-seq was performed on Mel624 melanoma cells and NY-ESO-1 TCR transduced primary human T cells that were co-cultured and separated into purified populations by FACS. Separate sequencing was performed on A375 melanoma cells that were transduced with Cas9 and non-targeting sgRNA or Cas9 and BIRC2-targeting sgRNA. Libraries were prepared using TruSeq RNA sample prep kit (FC-122-1001, Illumina). Approximately 40 million reads were sequenced and aligned to the human genome (hg19 for Mel624/NY-ESO-1 T cell experiment, hg38 for A375 experiments) with TopHat 2.0.11. Uniquely retained mapped reads were used to calculate differentially expressed genes using edgeR or Cuffdiff. Fisher’s exact test or t-tests were used to calculate significance with indicated p values and fold-change thresholds. To filter for RNA contamination derived from incomplete FACS-based purification of ESO T cells and Mel624 melanoma cells, we filtered out genes that had 100-fold enrichments in expression in the opposing cell type under basal conditions. RNA-sequencing raw data files are deposited at GEO-GSE137824.
Tumor cell chemokine and cytokine secretion
A375 cell production of chemokine proteins was quantified using a CBA Human Chemokine Kit (BD Biosciences Cat #552990) and GM-CSF production was measured using Human GM-CSF ELISA Kit (Abcam Cat #ab174448). In brief, A375 cells transduced with Cas9 and non-targeting sgRNA or BIRC2-targeting sgRNAs were plated at 4 × 105 in a 6-well dish and cultured for 24 h. Media supernatant was collected and spun at 1500rpm for 10 min to pellet cells and debris. Supernatant was collected for analysis. In some cases, media was diluted 1:5 using assay diluent to ensure detected values were within standard calibration curves.
Tumor pathway activation screen
Tumor pathway activation was performed as previously described (Martz et al., 2014), with minor modifications. In brief, lentiviral particles were generated for each pathway activation construct and A375 melanoma cells were transduced by spinfection at an MOI of 0.3. Transduced cells were incubated for 24 h, followed by puromycin selection. After selection, cells were plated and 2CT assay was conducted with ESO T cells at a 1:3 ratio for 16h.
Lentiviral transduction of tumor cells
Lentiviral transduction of tumor cells was performed as previously described (Patel et al., 2017). In brief, tumor cell culture media was replaced with RPMI 1640 media supplemented with 10% FBS and 8μg/mL polybrene. Lentiviral particles were added at an MOI of 0.3, followed by spinfection at 1500 rpm for 30 min. Following this, cells were incubated for 24 h, after which the culture media was aspirated and replaced with RPMI 1640 media supplemented with 10% FBS and selection antibiotic. For puromycin-resistant constructs, puromycin was added at 1μg/mL. For blasticidin-resistant constructs, blasticidin was added at 5μg/mL. Cells were maintained under antibiotic selection until non-transduced control cells were eliminated by drugs.
High throughput inhibitor screen
A375 melanoma cells were plated at 1 × 104 cells/well in 150μL of complete RPMI in a 96-well flat bottom plate and incubated for 24 h. Inhibitors were then added in 50μL of complete RPMI (concentrated to reach desired concentration after addition), followed by addition of ESO T cells at a 1:3 E:T ratio in complete RPMI to a total of 250μL. Cells were incubated for 16h, after which cells were washed twice with PBS, and WST1 reagent (Sigma Aldrich) was added in complete RPMI and cell viability was assessed.
Inhibitor sources
Inhibitors were purchased from commercial suppliers as listed below, or were provided (Compound 29, (Patch et al., 2011)) by the Donald McDonnell Laboratory (Duke University). ABT199 (Selleckchem #S8048), ABT263 (Selleckchem #S1001), LCL161 (Selleckchem #S7009), Birinapant (Selleckchem #S7015), Fomepizole (Selleckchem #S1717), CHR2797 (Tocris #3595), XCT790 (Sigma Aldrich #X4753), Topiramate (Selleckchem #S1438), ACET (Tocris #2728), Almorexant (Selleckchem #S2160), SB334867 (R&D Systems, #1960), anti-IL1R1 (R&D Systems, #AF269), anti-IL9 (R&D Systems, #AF209), anti-ITGAV (Abcam, #ab16821), Cilengitide (Selleckchem, #S7077), Guanidine HCl (Sigma Aldrich, #G3272), Dalfampridine (Sigma Aldrich, #275875), BEZ235 (Selleckchem, #S1009), Apitolisib (Selleckchem, #S2696), S17092 (Sigma Aldrich, #SML0181), WNK463 (Selleckchem, #S8358).
FACS-based 2CT inhibitor validations
Tumor cells were plated at 3 × 104 cells/well in 250μL of complete RPMI in a 48-well flat bottom plate and incubated for 24 h. Inhibitors were then added in 50μL of complete RPMI, followed by addition of TCR-transduced T cells at a 1:3 E:T ratio in complete RPMI to a total of 400μL. Cells were incubated for 16h, and analyzed as described for 2CT assay.
Murine ACT models
For in vivo immunotherapy experiments, we utilized previously described (Hanada et al., 2019) B16 melanoma cells expressing a chimeric mouse-human gp100 antigen (B16mhGP100). These engineered tumor target cells abrogate the necessity of co-administering a vaccine post-ACT and allow for lower therapeutic doses of Pmel T cells. We generated gene-deleted B16mhGP100 cells using lentiviruses encoding sgRNA (sequence TGAATGCCACCTCGTTCCAG) targeting Birc2 as described above. C57BL/6 mice were subcutaneously implanted with 5 × 105 B16mhGP100 cells and tumors were allowed to grow for 10 days. Mice were then irradiated (6 Gy) and injected intravenously with 5 × 106 Pmel CD8+ T cells. Mice received intraperitoneal injections of IL-2 in PBS (6 × 104 IU in 0.5mL) once daily for 3 consecutive days. In certain experiments, mice received intraperitoneal injections of LCL-161 or birinapant in Captisol (30% in H2O acidified to pH 4 with citric acid). Injections of inhibitors was performed every other day for 5 total injections. Tumor measurements were performed in a blinded fashion by an independent investigator approximately every two days after T cell transfer. Tumor area was calculated as length x width of the tumor. Mice with tumors in excess of 400 mm2 were euthanized.
Pathway enrichment analysis
For gene pathway analysis, enriched or depleted genes were examined for gene category over-representation using Ingenuity Pathway Analysis (QIAGEN). For analysis of CRISPR depleted genes, the top 250 significant genes by RIGER analysis were selected. For analysis of RNA-sequencing experiments, all differentially expressed genes were analyzed. Fisher’s exact test (p < 0.05) was used to compute significance for over-representation of genes in a pathway.
cBioportal analysis of human patient datasets
Analysis of human melanoma patient gene expression was performed using the Skin Cutaneous Melanoma geneset (TCGA, PanCancer Atlas). Samples were selected for which mRNA data was available and gene expression correlation analysis was performed. PanCancer analysis of CNV was performed using the PanCancer Atlas datasets.
Identification of druggable targets for screening
Validated tumor resistance genes (Figure 3D) along with the top 250 depleted genes from CRISPR screens as calculated by rank-sum of replicate screens was analyzed by PANTHER (Mi et al., 2019) to map gene list to protein coding genes. 237 identified protein coding genes were analyzed using DGIDB (Griffith et al., 2013) to identify genes that are druggable. Genes were analyzed as ‘druggable gene category results’ and filtered on categories ‘inhibitor, allosteric modulator, antagonist, blocker, channel blocker, desensitize the target, gating inhibitor, incorporation into and destabilization, inhibitor, competitive inhibitor, inhibitory allosteric modulator, inhibitory immune response, intercalation, inverse agonist, negative modulator, neutralizer, partial antagonist, reducer, suppressor’. Results were manually curated for commercial availability of inhibitors.
T cell migration
Tumor cells were plated at 1 × 104 cells in complete RPMI in a 96-well receiver plate (Corning #3382) and incubated for 24h. Following this, ESO T cells were added to a 96-well transwell plate (Corning #3387) at 1 × 105 cells/well in 100μL complete RPMI and the T cell plate was added to the tumor cell plate and incubated for 5 h. Media supernatant from tumor cell wells was collected, wells were washed with PBS which was also collected, then tumor cells were detached with trypsin and collected. Total contents of each well were stained with fixable viability dye and αCD3e, and migrated T cells were enumerated by FACS counting.
Flow cytometry
Tumor cells or T cells suspended in FACS staining buffer were stained with fluorochrome-conjugated antibodies against CD3e (SK7, BD) and murine TCRβ (12-5961-82, ThermoFisher). In some experiments, cells were stained for 4-1BB (309809, Biolegend), HLA Class I (11-9983-42, ThermoFisher), PD-L1 (12-5983-42, ThermoFisher) and β2M (316311, Biolegend). Cell viability was determined using propidium iodide exclusion or fixable Live/Dead kit (Invitrogen). Intracellular staining assay on ESO T cells was conducted after 5–6 h co-culture with A375 cells in the presence of monensin (BD, 512092KZ) and brefeldin A (BD, 512301KZ). Staining was performed using manufacturer’s instructions using antibodies against IFNγ (25723.11, BD) or TNFα (12-7349-41, ThermoFisher). Flow cytometric data were acquired using either a FACSCanto II or LSRII Fortessa cytometer (BD), and data were analyzed using FlowJo version 10.5.3 software (FlowJo LLC).
Proteomics analysis
TMT labeling-based proteomics analysis was performed as previously described (Shi et al., 2021). In brief, cell pellets were produced in duplicate from cell lines and lysed in 50 mM HEPES, pH 8.0 and 8M urea followed by sonication. Lysates were clarified by centrifugation and protein concentration was quantified using BCA method. One hundred micrograms of lysate were alkylated and digested by addition of trypsin at a ratio of 1:50 and incubating overnight at 37°C. Digestion was acidified by adding formic acid to a final concentration of 1% and desalted using peptide desalting columns (Thermo Fisher) according to protocol. Peptides were eluted from columns, dried in a speedvac and stored at −20°C. For TMT labeling, samples were reconstituted in 50 mM HEPES, ph 8.0 and TMTpro label (Thermo Fisher) in 100% ACN was added. After incubation, the reaction was terminated by adding hydroxylamine. Peptide samples were pooled and cleaned using peptide desalting columns. Peptides were fractionated using a Waters Acquity UPLC system coupled with a fluorescence detector. Samples were reconstituted in 0.1% TFA and subjected to nanoflow liquid chromatography coupled to an Orbitrap Exlipse mass spectrometer. Spectra were searched using the Real Time Search Node in the tune file using human Uniprot database using Comet search algorithm with TMR16 plex set as a static modification of lysine and the N-termini of the peptide. Acquired MS/MS spectra were searched against a human Uniprot protein database along with a contaminant protein database, using a SEQUEST and percolator validator algorithms in the Proteome Discoverer 2.4 software.
Immunoblotting
Immunoblotting was performed as described previously (Jacobs et al., 2008). Primary antibodies were followed by mouse- or rabbit-conjugated horseradish peroxidase (HRP) secondary antibodies. HRP-conjugated antibodies (anti-mouse or anti-rabbit IgG HRP conjugate, Promega) were detected by enhanced chemiluminescence detections (Thermofisher) using a Biorad ChemiDoc MP. This included the following antibodies: BIRC2 (108361, Abcam), BIRC3 (3130, Cell Signaling), HLA Class I (70328, Abcam), ICAM1 (4915, Cell Signaling), IRF1 (8478 Cell Signaling), XIAP (2042, Cell Signaling). Alternatively, primary antibodies were followed by fluorescently labeled anti-mouse or rabbit antibodies and imaged using a Biorad ChemiDoc MP. This included the following antibodies: β-actin (A5441, Sigma).
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample sizes were determined by prior experience and estimated power calculations. Statistical analysis was performed with Prism (GraphPad Software Inc.). Data were compared using either a two-tailed Student’s t test or one-way ANOVA with multiple comparisons corrected with Dunnett adjustment. p values <0.05 were considered significant. For adoptive transfer experiments, mice were randomized prior to cell transfer and treatments. Tumor treatment graphs were compared by using the Wilcoxon rank sum test and analysis of animal survival was analyzed by a log-rank test.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse IgG HRP conjugated secondary antibody | Promega | Cat# W4021; RRID: AB_430834 |
| Anti-rabbit IgG HRP conjugated secondary antibody | Promega | Cat# W4011; RRID: AB_430833 |
| Beta-Actin | Sigma Aldrich | Cat# A5441; RRID: AB_476744 |
| B2M | Biolegend | Cat# 316311; RRID: AB_10643412 |
| BIRC2 | Abcam | Cat# 108361; RRID: AB_10862855 |
| BIRC3 | Cell Signaling | Cat# 3130; RRID: AB_10693298 |
| HLA Class I (FACS-grade) | ThermoFisher | Cat# 11-9983-42; RRID: AB_1633402 |
| HLA Class I | Abcam | Cat# 70328; RRID: AB_1269092 |
| Human 4-1BB/CD137 | Biolegend | Cat# 309809; RRID: AB_830671 |
| Human CD3 clone SK7 | BD Biosciences | Cat# 345767; RRID: AB_2833003 |
| Human CD3 clone SK7 | BD Biosciences | Cat# 347347; RRID: AB_400287 |
| Human IFNg | BD Pharmingen | Cat# 340452; RRID: AB_400428 |
| Human TNFa | ThermoFisher | Cat# 12-7349-41; RRID: AB_10668834 |
| ICAM1 | Cell Signaling | Cat# 4915; RRID: AB_2280018 |
| IL1R1 | R&D Systems | Cat# AF269; RRID: AB_355286 |
| IL9 | R&D Systems | Cat# AF209; RRID: AB_2296123 |
| IRF1 | Cell Signaling | Cat# 8478; RRID: AB_10949108 |
| ITGAV | Abcam | Cat# ab16821; RRID: AB_443484 |
| PD-L1 | ThermoFisher | Cat# 12-5983-42; RRID: AB_11042286 |
| TCR beta monoclonal antibody (H57-597) | eBiosciences | Cat# 12-5961-82; RRID: AB_466066 |
| Anti-human CD3 Functional Grade Purified (clone OKT3) | eBiosciences | Cat# 16-0037; RRID: AB_468855 |
| XIAP1 | Cell Signaling | Cat# 2042; RRID: AB_2214870 |
| Biological samples | ||
| Human peripheral blood mononuclear cells (PBMCs) | NIH Blood bank | N/A |
| Primary tumor cell lines 4238 and 4266 | NCI Surgery Branch | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Abt199 | Selleckchem | Cat# S8048 |
| ACET | Tocris | Cat# 2728 |
| Almorexant | Selleckchem | Cat# S2160 |
| Apitolisib | Selleckchem | Cat# S2696 |
| BEZ235 | Selleckchem | Cat# S1009 |
| Birinapant | Selleckchem | Cat# S7015 |
| CHR2797 | Tocris | Cat# 3595 |
| Cilengitide | Selleckchem | Cat# S2160 |
| Compound 29 | Donald McDonnell Laboratory, Duke University | N/A |
| Dalfampridine | Sigma Aldrich | Cat # 275875 |
| Fomepizole | Selleckchem | Cat# S1717 |
| Golgi Stop | BD Biosciences | Cat# 512092KZ |
| Golgi Plug | BD Biosciences | Cat# 512301KZ |
| Guanidine HCL | Sigma Aldrich | Cat# G3272 |
| LCL161 | Selleckchem | Cat# S7009 |
| S17092 | Sigma Aldrich | Cat# SML0181 |
| SB334867 | R&D Systems | Cat# 1960 |
| Topiramate | Selleckchem | Cat# S1438 |
| WNK463 | Selleckchem | Cat# S8358 |
| XCT790 | Sigma Aldrich | Cat# S2160 |
| Proleukin (aldesleukin)- Human recombinant IL2 | Novartis | N/A |
| RetroNectin GMP grade, Recombinant Human Fibronectin Fragment CH-296 | Takara | Cat# T202 |
| Critical commercial assays | ||
| CBA Human Chemokine Kit | BD Biosciences | Cat# 552990 |
| Human GM-CSK ELISA Kit | Abcam | Cat# ab174448 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74134 |
| RNeasy Plus Micro kit | Qiagen | Cat# 74034 |
| WST-1 Reagent | Sigma Aldrich | Cat# 5015944001 |
| eBioscience Fixable Viability Dye eFluor 450 | Thermo Fisher Scientific | Cat# 65-0863-14 |
| Deposited data | ||
| Raw data files for RNA sequencing | This paper | GEO: GSE137824 |
| Raw proteomics data files | This paper | MassIVE: MSV000089737 |
| Experimental models: Cell lines | ||
| B16-mhgp100 melanoma | Eil et al., 2016 | N/A |
| A375 melanoma | ATCC | Cat# CRL-1619 |
| Mel624.38 melanoma | NCI Surgery Branch; Robbins et al., 2008 | N/A |
| 4238 primary tumor cell line | This paper; NCI Surgery Branch | N/A |
| 4260 primary tumor cell line | Malekzadeh et al., 2019; NCI Surgery Branch | N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6N | NCI/Charles River | N/A |
| Pmel-1 TCR-transgenic (Pmel, C57BL/6.Cg-/CyTg[TcraTcrb]8Rest/J | In house | N/A |
| Oligonucleotides | ||
| sgRNA sequences – see Table S6 | This paper | N/A |
| Recombinant DNA | ||
| MART-1 T cell receptor | Johnson et al., 2006 | N/A |
| NY-ESO-1 T cell receptor | Robbins et al., 2008 | N/A |
| Mutant p53 T cell receptor | Malekzadeh et al., 2019 | N/A |
| Mutant NCKAP1 T cell receptor | This paper | N/A |
| GeCKOv.2 Genome scale CRISPR knockout library | Sanjana et al., 2014 | N/A |
| lentiGuide-Puro | Sanjana et al., 2014 | N/A |
| lentiCas9-Blast | Sanjana et al., 2014 | Addgene Cat# 52962 |
| lentiGuide-Puro vectors with sgRNA inserts - sgRNAs identified in Table S6 | This paper | N/A |
| Pathway activating constructs | Kris Wood laboratory, Duke University; Martz et al., 2014 | N/A |
| Software and algorithms | ||
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/ |
| Ingenuity pathway analysis | Qiagen | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ |
| Prism 6 | GraphPad Software | http://www.graphpad.com |
| R-package DeSeq2 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Code for TCGA analysis | This paper | https://github.com/NIDAP-Community/Cancer-genes-disfavoring-T-cell-immunity |
Highlights.
Integrated systems approach profiles cancer-specific genes that disfavor T cell killing
Inhibiting BIRC2 sensitizes tumor cells to the cytotoxic activity of T cells
BIRC2 genetic or chemical ablation enhances adoptive T cell therapy in vivo efficacy
BIRC2 regulates antigen presentation via inhibition of IRF1 activity
ACKNOWLEDGMENTS
The authors of this article were supported by the Center for Cell-Based Therapy, NCI, NIH (Bethesda, MD, USA), the NIH Center for Regenerative Medicine, the Milstein Family Foundation, and the Intramural Research Program of the NCI (ZIA BC010763). The authors thank the NIH CCR Mass Spectrometry (Protein & Small Molecule) Dedicated Service for performing proteomics analysis of samples. We would like to thank Erina He for illustrations used in the manuscript and members of the Restifo, Rosenberg, and Yang labs for helpful discussions and suggestions. N.E.S. is supported by NYU and NYGC startup funds, NIH/NHGRI (R00HG008171 and DP2HG010099), NIH/NCI (R01CA218668), DARPA (D18AP00053), the Sidney Kimmel Foundation, the Melanoma Research Alliance, and the Brain and Behavior Foundation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111153.
DECLARATION OF INTERESTS
R.J.K., S.K.V., Y.P., and N.P.R. hold equity in and are currently employed by Lyell Immunopharma, South San Francisco, CA, USA. S.J.P. has previously held positions at Boehringer Ingelheim and NextCure Inc. and currently holds equity in NextCure Inc. N.E.S. is a scientific advisor for Vertex and Qiagen.
REFERENCES
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. (2015). Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, G.T (2013). The genotype-tissue expression (GTEx) project. Nat. Genet 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan M, Dougan S, Slisz J, Firestone B, Vanneman M, Draganov D, Goyal G, Li W, Neuberg D, Blumberg R, et al. (2010). IAP inhibitors enhance co-stimulation to promote tumor immunity. J. Exp. Med 207, 2195–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eil R, Vodnala SK, Clever D, Klebanoff CA, Sukumar M, Pan JH, Palmer DC, Gros A, Yamamoto TN, Patel SJ, et al. (2016). Ionic immune suppression within the tumour microenvironment limits T cell effect or function. Nature 537, 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facciabene A, Motz GT, and Coukos G (2012). T-regulatory cells: key players in tumor immune escape and angiogenesis. Cancer Res. 72, 2162–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M, Griffith OL, Coffman AC, Weible JV, McMichael JF, Spies NC, Koval J, Das I, Callaway MB, Eldred JM, et al. (2013). DGIdb: mining the druggable genome. Nat. Methods 10, 1209–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada KI, Yu Z, Chappell GR, Park AS, and Restifo NP (2019). An effective mouse model for adoptive cancer immunotherapy targeting neoantigens. JCI Insight 4, 124405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SR, Herman CE, Maciver NJ, Wofford JA, Wieman HL, Hammen JJ, and Rathmell JC (2008). Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol 180, 4476–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LA, Heemskerk B, Powell DJ Jr., Cohen CJ, Morgan RA, Dudley ME, Robbins PF, and Rosenberg SA (2006). Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J. Immunol 177, 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SA, Park YM, Hong SW, Moon JH, Shin JS, Lee HR, Ha SH, Lee DH, Kim JH, Kim SM, et al. (2015). Cellular inhibitor of apoptosis protein 1 (cIAP1) stability contributes to YM155 resistance in human gastric cancer cells. J. Biol. Chem 290, 9974–9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher MA, and Roderick JE (2018). NOTCH signaling in T-cell-mediated anti-tumor immunity and T-cell-based immunotherapies. Front. Immunol 9, 1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishton RJ, Sukumar M, and Restifo NP (2017). Metabolic regulation of T cell longevity and function in tumor immunotherapy. Cell Metab. 26, 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefor AT, and Fabian DF (1998). Enhanced cytolytic activity of tumor infiltrating lymphocytes (TILs) derived from an ICAM-1 transfected tumor in a murine model. J. Surg. Res 75, 49–53. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W, Parkhurst M, Robbins PF, Tran E, Lu YC, Jia L, Gartner JJ, Pasetto A, Deniger D, Malekzadeh P, et al. (2019). Immunologic recognition of a shared p53 mutated neoantigen in a patient with metastatic colorectal cancer. Cancer Immunol. Res 7, 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malekzadeh P, Pasetto A, Robbins PF, Parkhurst MR, Paria BC, Jia L, Gartner JJ, Hill V, Yu Z, Restifo NP, et al. (2019). Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J. Clin. Invest 129, 1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, Collins NB, Bi K, LaFleur MW, Juneja VR, et al. (2017). In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martz CA, Ottina KA, Singleton KR, Jasper JS, Wardell SE, Peraza-Penton A, Anderson GR, Winter PS, Wang T, Alley HM, et al. (2014). Systematic identification of signaling pathways with potential to confer anti-cancer drug resistance. Sci. Signal 7, ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, et al. (2014). Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med 371, 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, and Thomas PD (2019). Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc 14, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, Tsoucas D, Qiu X, Lim K, Rao P, et al. (2018). A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Rivière I, Gonen M, Wang X, Sénéchal B, Curran KJ, Sauter C, Wang Y, Santomasso B, Mead E, et al. (2018). Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med 378, 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patch RJ, Searle LL, Kim AJ, De D, Zhu X, Askari HB, O’Neill JC, Abad MC, Rentzeperis D, Liu J, et al. (2011). Identification of diaryl ether-based ligands for estrogen-related receptor alpha as potential antidiabetic agents. J. Med. Chem 54, 788–808. [DOI] [PubMed] [Google Scholar]
- Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, et al. (2017). Identification of essential genes for cancer immunotherapy. Nature 548, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restifo NP, Marincola FM, Kawakami Y, Taubenberger J, Yannelli JR, and Rosenberg SA (1996). Loss of functional beta 2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. J. Natl. Cancer Inst 88, 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettino A, and Clarke NM (2013). Genome-wide identification of IRF1 binding sites reveals extensive occupancy at cell death associated genes. J. Carcinog. Mutagen S6-009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PF, Li YF, El-Gamil M, Zhao Y, Wargo JA, Zheng Z, Xu H, Morgan RA, Feldman SA, Johnson LA, et al. (2008). Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J. Immunol 180, 6116–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, Citrin DE, Restifo NP, Robbins PF, Wunderlich JR, et al. (2011). Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res 17, 4550–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D, Huang TYT, Shah R, Yang Y, Pan F, and Semenza GL (2020). BIRC2 expression impairs anti-cancer immunity and immunotherapy efficacy. Cell Rep. 32, 108073. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorius UA, and Krammer PH (2002). Upregulation of Bcl-2 is involved in the mediation of chemotherapy resistance in human small cell lung cancer cell lines. Int. J. Cancer 97, 584–592. [DOI] [PubMed] [Google Scholar]
- Shi G, Chiramel AI, Majdoul S, Lai KK, Das S, Beare PA, Andresson T, Best SM, and Compton AA (2021). Rapalogs downmodulate intrinsic immunity and promote cell entry of SARS-CoV-2. Preprint at bioRxiv. 10.1101/2021.04.15.440067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifrut E, Carnevale J, Tobin V, Roth TL, Woo JM, Bui CT, Li PJ, Diolaiti ME, Ashworth A, and Marson A (2018). Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton KR, Crawford L, Tsui E, Manchester HE, Maertens O, Liu X, Liberti MV, Magpusao AN, Stein EM, Tingley JP, et al. (2017). Melanoma therapeutic strategies that select against resistance by exploiting MYC-driven evolutionary convergence. Cell Rep. 21, 2796–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Drake CG, and Pardoll DM (2015). Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, et al. (2015). Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350,207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodnala SK, Eil R, Kishton RJ, Sukumar M, Yamamoto TN, Ha NH, Lee PH, Shin M, Patel SJ, Yu Z, et al. (2019). T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363, eaau0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vredevoogd DW, Kuilman T, Ligtenberg MA, Boshuizen J, Stecker KE, de Bruijn B, Krijgsman O, Huang X, Kenski JCN, Lacroix R, et al. (2019). Augmenting immunotherapy impact by lowering tumor TNF cytotoxicity threshold. Cell 178, 585–599.e15. [DOI] [PubMed] [Google Scholar]
- Yamamoto TN, Kishton RJ, and Restifo NP (2019). Developing neoantigen-targeted T cell-based treatments for solid tumors. Nat. Med 25, 1488–1499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data generated during this study are available at the NCBI-GEO database with the project accession ID GEO: GSE137824. Proteomics datasets generated during this study are available at MassIVE: MSV000089737.
All original code has been deposited at Github and is publicly available as of the date of publication (https://github.com/NIDAP-Community/Cancer-genes-disfavoring-T-cell-immunity).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


